ABSTRACT
Highly pathogenic avian influenza A (HPAI) viruses of the H5N1 subtype have recently emerged from avian zoonotic reservoirs to cause fatal human disease. Adaptation of HPAI virus RNA-dependent RNA polymerase (PB1, PB2, and PA proteins) and nucleoprotein (NP) to interactions with mammalian host proteins is thought to contribute to the efficiency of viral RNA synthesis and to disease severity. While proteomics experiments have identified a number of human proteins that associate with H1N1 polymerases and/or viral ribonucleoprotein (vRNP), how these host interactions might regulate influenza virus polymerase functions and host adaptation has been largely unexplored. We took a functional genomics (RNA interference [RNAi]) approach to assess the roles of a network of human proteins interacting with influenza virus polymerase proteins in viral polymerase activity from prototype H1N1 and H5N1 viruses. A majority (18 of 31) of the cellular proteins tested, including RNA-binding (DDX17, DDX5, NPM1, and hnRNPM), stress (PARP1, DDB1, and Ku70/86), and intracellular transport proteins, were required for efficient activity of both H1N1 and H5N1 polymerases. NXP2 and NF90 antagonized both polymerases, and six more RNA-associated proteins exhibited strain-specific phenotypes. Remarkably, 12 proteins differentially regulated H5N1 polymerase according to PB2 genotype at mammalian-adaptive residue 627. Among these, DEAD box RNA helicase DDX17/p72 facilitated efficient human-adapted (627K) H5N1 virus mRNA and viral RNA (vRNA) synthesis in human cells. Likewise, the chicken DDX17 homologue was required for efficient avian (627E) H5N1 infection in chicken DF-1 fibroblasts, suggesting that this conserved virus-host interaction contributes to PB2-dependent host species specificity of influenza virus and ultimately to the outcome of human HPAI infections.
IMPORTANCE
Highly pathogenic avian influenza A (HPAI) viruses have recently emerged from wild and domestic birds to cause fatal human disease. In human patients, it is thought that adaptation of the viral polymerase, a complex of viral proteins responsible for viral gene expression and RNA genome replication, to interactions with mammalian rather than avian host proteins contributes to disease severity. In this study, we used computational analysis and RNA interference (RNAi) experiments to identify a biological network of human proteins that regulates an H5N1 HPAI virus polymerase, in comparison to a mammalian H1N1 virus. Of 31 proteins tested, 18 (58%) were required for polymerase function in both HPAI and H1N1 viruses. Remarkably, we also found proteins such as DDX17 that governed the HPAI virus polymerase’s adaptation to human cells. These virus-host interactions may thus control pathogenicity of HPAI virus in humans and are promising therapeutic targets for antiviral drugs in severe influenza infections.
Introduction
Influenza A viruses (Orthomyxoviridae) cause seasonal epidemics in humans and occasionally lethal global pandemics, such as those of 1918-1919 (H1N1), 1957 (H2N2), 1968 (H3N2), and 2009 (swine-origin H1N1). Currently, highly pathogenic avian influenza A (HPAI) virus strains of the H5N1 subtype circulating in wild and domestic birds are capable of crossing the species barrier into humans and causing significant pulmonary (1–3) and systemic (4) disease. Reverse-genetics studies have demonstrated that the viral polymerase (a heterotrimeric complex consisting of the PB1, PB2, and PA proteins) and nucleoprotein (NP) contribute to the pathogenicity of H5N1 HPAI viruses in humans and other mammals (5–7). Experimental evidence also suggests that adaptation of polymerase proteins and/or NP to interact with mammalian rather than avian host proteins contributes to efficient viral mRNA, cRNA, and/or viral RNA (vRNA) genome synthesis (8, 9). For avian viruses, adaptations in PB2 (e.g., E627K, D701N, and Q591K) allow efficient replication in mammalian systems (10, 11) and are correlated with disease severity in mammals (5–7, 12, 13). These mutations appear to be selected to facilitate H5N1 HPAI virus replication in humans (14).
In humans, the influenza A virus polymerase-host interactome consists of more than 50 host cell proteins identified primarily by mass spectrometry in protein complexes associated with H1N1 influenza A virus heterotrimeric polymerase, NP, or a viral ribonucleoprotein (vRNP) comprised of these proteins and a vRNA template (8, 15–21). Although the direct or indirect nature of many of these host protein interactions with viral polymerase subunits, NP, or viral RNAs requires experimental validation, the discovery of such a large number of interacting proteins suggests that the processes of influenza A virus transcription, replication, and vRNP import/export are potentially dependent on a wide spectrum of host factors over the course of the virus life cycle. In a few cases, notably those of RNA-binding proteins (16, 19, 21, 22) and molecular transporters (8, 15, 20, 23), the importance of these virus-host interactions to the activity of H1N1 polymerases has been characterized. However, the importance of the majority of newly identified factors for polymerase function is unknown, particularly with respect to H5N1 HPAI. Recent RNA interference (RNAi) analyses of a large number of human genes regulating the influenza A H1N1 virus life cycle considerably expanded the list of host factors that regulate virus replication (24–27). However, these high-throughput studies did not specifically examine host factors with respect to H1N1 or H5N1 viral polymerase RNA synthesis (viral gene expression and genome replication) functions.
The present study sought a deeper understanding of influenza virus polymerase-host interactions that regulate polymerase activity in human cells. We hypothesized that host factors interacting with H1N1 polymerase might regulate H1N1 and potentially H5N1 polymerase functions. Using a functional genomics approach, we describe a network of human proteins that modulate the polymerases of influenza A/WSN/1933 virus, a mammalian H1N1 strain, and A/Viet Nam/1203/2004 (A/VN/1203/04), an H5N1 HPAI virus isolated from a fatal human case (6). Given the importance of the polymerase genotype for HPAI infection in mammals, we also searched for a polymerase-interacting host factor(s) that governs adaptation of H5N1 polymerase to human cells. We identified host factors within the network that differentially regulated H5N1 HPAI viral polymerase in human cells depending on the PB2 amino acid 627 genotype, such as DEAD box RNA helicase 17 (DDX17).
RESULTS
H1N1 polymerase-interacting human proteins modulate H1N1 and H5N1 polymerase activity.
To discover how interacting host proteins regulate influenza virus polymerases, we first analyzed the H1N1 influenza A virus polymerase-host interactome (see Fig. S1 in the supplemental material). Using biological network analyses (27), we selected 31 of 54 human factors for RNAi phenotype studies (Fig. 1A). These 31 factors formed an interconnected network of proteins (Benjamini-Hochberg estimate of false discovery probability [PBH] < 0.011) encoding functions known to regulate influenza virus replication, including RNA processing, transcription and translation, molecular transport, cell cycle, and cell death (17, 26–28).
FIG 1 .
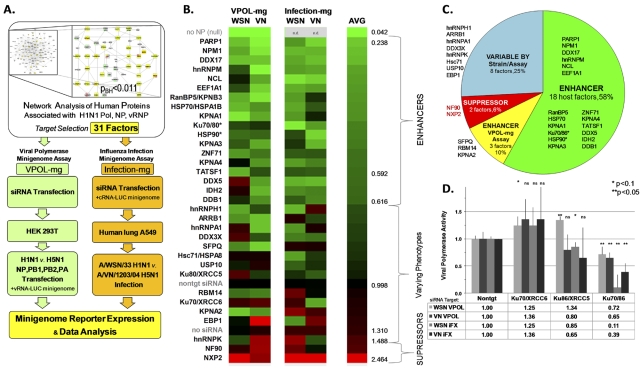
Host factors regulate H1N1 and H5N1 influenza virus polymerase activity in human cells. (A) Host proteins interacting with influenza A virus polymerase, NP, and/or vRNP were analyzed by network bioinformatics. A subset of 31 factors was selected to analyze their roles in H1N1 and H5N1 influenza virus polymerase activity. For assays for viral polymerase phenotypes, siRNAs targeting specific host factor transcripts were transfected into HEK 293T cells or human lung A549 cells. For the viral polymerase minigenome assay (VPOL-mg), after knockdown, HEK 293T cells were transfected with A/WSN/33 (H1N1) or A/VN/1203/04 (H5N1) nucleoprotein and polymerase expression plasmids in optimized ratios, a vRNA promoter minigenome luciferase reporter, and a constitutive Renilla internal control. For the influenza virus infection assay (infection-mg), A549 cells were cotransfected with siRNA, a cRNA promoter minigenome luciferase reporter, and a Renilla internal control and, after knockdown, infected with influenza A/WSN/33 (H1N1) or A/VN/1203/04 (H5N1) HALo virus (MOI = 0.5). Viral polymerase activity was assessed and normalized by dual-luciferase assay. (B) Heat map of average A/WSN/33 (H1N1) (WSN) or A/VN/1203/04 (H5N1) (VN) viral polymerase activity in a ratio to the nontarget siRNA value in VPOL-mg and infection-mg assays for the 31 RNAi targets, nontarget (nontgt) siRNA, and “no siRNA” controls. Targets were ordered and classified by averaging data from all four conditions (AVG). *, simultaneous targeting of both subunits of HSP90 or Ku70/86; n.d., not done. (C) Functional classification of phenotypes for the 31 host RNAi targets, reflecting composite data from all four experimental conditions. (D) Ku70/XRCC6 and Ku86/XRCC5 targeted individually or simultaneously (Ku70/86) by siRNA in VPOL-mg (VPOL) and infection-mg (IFX) assays for the A/WSN/33 (H1N1) (WSN) and A/VN/1203/04 (H5N1) (VN) strains, with significance indicated by an unpaired, 2-tailed t test.
We then measured the effects of short interfering RNA (siRNA) knockdown of each of the 31 host factors on A/WSN/33 (H1N1) and A/VN/1203/04 (H5N1) viral polymerase activity in human cells using minigenome reporter assays (Fig. 1A). We optimized minigenome assays to study host factor requirements for the polymerase by itself (VPOL-mg assay, including NP; see Fig. S2 in the supplemental material) and during influenza virus infection (infection-mg assay) (29). For each host factor targeted by siRNA (see Table S1), polymerase activity was expressed as a ratio to the nontarget (control) siRNA value (Fig. 1B), and phenotypic differences were tested for significance (see Table S2). Cell viability assays excluded only KPNA4 and IDH2 from subsequent analyses (see Fig. S3).
Our data rejected the null hypothesis that most host proteins identified in the interconnected network were bystanders, perhaps interacting but not affecting viral polymerase function. Instead, both A/WSN/33 (H1N1) and A/VN/1203/04 (H5N1) viral polymerases depended on a common set (18/31, or 58%) of human host factors (enhancers) for optimal function in both assays (Fig. 1C). Many enhancer factors were RNA binding or processing proteins (e.g., DDX17, NPM1, and heterogeneous nuclear ribonucleoprotein M [hnRNPM]), molecular transport or chaperone proteins (KPNA1, KPNA3, HSP90, HSP70, and RanBP5), or proteins responding to DNA damage or cellular stress (PARP1, the Ku70/86 complex, and DDB1). Only simultaneous knockdown of both the Ku70 and Ku86 subunits showed a significant phenotype, suggesting redundant functions for Ku proteins vis-à-vis the influenza virus polymerase (Fig. 1D). Viral polymerase activity significantly increased when NXP2 or NF90 was targeted by siRNA, suggesting these host proteins are antagonists, or suppressors, of both H1N1 and H5N1 polymerases. Eleven more factors displayed variability depending on the assay and/or strain (see Discussion). Hsc71/HSPA8 was required only for optimal polymerase function in infected cells, maybe due to this protein’s function in cellular stress (see Fig. S2 in the supplemental material).
A protein network that regulates H1N1 and H5N1 polymerases.
To provide a more coordinated picture of how H1N1 and H5N1 polymerases are regulated by host factors in human cells, we mapped RNAi viral polymerase phenotypes onto the interconnected network of host proteins extracted from the H1N1 influenza A virus polymerase-host interactome (see Fig. S1 in the supplemental material). This functionally enriched regulatory network was expanded to encompass additional proteins reported to affect influenza virus replication (Fig. 2A): DBT and RPS5 (25), hnRNPU (24), hnRNPF (30), KPNB1 (26), CRM1/XPO1 (31), the EIF4A complex (24, 32), host RNA polymerase II (28), and PABPC1 (32). Moreover, three proteins in the regulatory network (ARRB1, hnRNPK, and RBM14) that shared a high degree of connection (k > 3 edges) with H1N1 polymerase interactors also displayed significant polymerase phenotypes (Fig. 2A). This suggests that proximal or “hub-like” interactors may contribute to polymerase functions and in part could explain the relatively low overlap in gene hits among global influenza virus RNAi studies (33).
FIG 2 .
Regulatory network modulating H1N1 polymerase and H5N1 polymerase according to PB2 genotype at amino acid position 627. (A) Protein interaction network comprising influenza virus polymerase-associated cellular proteins and proximal nodes extracted from the human protein interactome by Ingenuity pathways analysis (IPA), with significance estimated by Benjamini-Hochberg test (PBH). H1N1 and H5N1 polymerase-host RNAi phenotype data from VPOL-mg (vPOL) and infection-mg (iFX) assays are indicated by color according to this study and published references (lit.; Refs. 16, 22, 24–28, 30–32). Edges between nodes are reported protein-protein interactions; Untested, phenotypic assay(s) not performed for interactor; Other, connecting protein; dashed links, indirect association reliant on proteins not shown. Inset table: enriched functions found by IPA and Gene Ontology (GO) analyses. (B) Viral polymerase minigenome reporter assays with 293T cells for two A/VN/1203/04 (H5N1) viral polymerase PB2 genotypes (627K, wild type/human isolate; 627E, avianized mutant). Fold induction, ratio to results of mock VPOL-mg assay. (C) Average VPOL-mg activity for PB2 627K and 627E genotypes, expressed as a ratio to the corresponding nontarget siRNA (Nontgt) value for 28 principal network host factors. Phenotypes are classified by significance estimated by an unpaired, 2-tailed t test (enhancer or suppressor, Pmax < 0.15; no significant difference from nontarget control, Pmax ≥ 0.15). Targets are ordered by magnitude/phenotype; y axis, log2 scale. The Gene Ontology functional cluster of RNA-binding/processing proteins (P value, group enrichment score) among indicated targets exhibiting differential phenotypes is shown.
Host factors that differentially regulate H5N1 polymerase according to PB2 genotype at mammalian-adaptive residue 627.
We then explored the hypothesis that host factors in the regulatory network could modulate not only functions of H1N1 and H5N1 influenza virus polymerases but also adaptation of HPAI viruses to human cells. We examined host factor RNAi phenotypes for two A/VN/1203/04 (H5N1) polymerases side-by-side: that of the wild-type (wt), human isolate, with a lysine (K) at position 627 in PB2, and a mutant with a change to glutamic acid (E), the consensus residue found in most avian H5N1 isolates (11, 34). Avianized (PB2|627E) polymerase displayed approximately 75-fold-lower activity than that of the wt/human isolate (PB2|627K) in human 293T cells but still provided sufficient dynamic range for VPOL-mg assays (Fig. 2B). In the regulatory network, 13/25 (52%) host factors conserved enhancer phenotypes for both wt/human isolate and avianized H5N1 polymerases (Fig. 2C). Surprisingly, 12/25 (48%) of factors that had been enhancers for the wt/human isolate showed no significant phenotype, or even antagonist phenotypes, for the avianized H5N1 polymerase. RNA-binding/processing proteins (P < 0.004, group enrichment score) and molecular transport/chaperone proteins had differential phenotypes.
Exogenous cDNA expression of four RNA-binding host proteins.
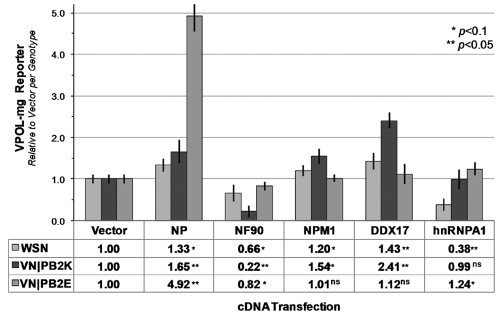
To complement siRNA experiments on RNA-binding proteins in the regulatory network, we overexpressed cDNA plasmids encoding human NPM1, DDX17, NF90, and hnRNPA1. As expected from VPOL-mg optimization experiments (see Fig. S2 in the supplemental material), increasing NP increased viral polymerase activity (Fig. 3). NF90 suppressed, and both DDX17 and NPM1 enhanced, H1N1 and wt/human isolate H5N1 polymerase activity in comparison to results with an empty DNA vector. Thus, for mammalian-adapted H1N1 and H5N1 polymerases, overexpression phenotypes for DDX17, NPM1, and NF90 were generally consistent with RNAi phenotypes (i.e., opposite to VPOL-mg phenotypes in Fig. 1B). The exception was hnRNPA1, which showed disparate phenotypes in cDNA assays. Along with results of siRNA experiments (Fig. 1B), these data suggest that hnRNPA1 may have pleiotropic functional interactions with influenza virus polymerase in human cells.
FIG 3 .
Exogenous expression of host factor cDNA modulates influenza virus polymerase activity. Subconfluent 293T cells were transfected with a vRNA promoter luciferase minigenome, a constitutively active Renilla luciferase, cDNA expression plasmids encoding indicated host factors, or empty vector, and A/WSN/33 (WSN) or A/VN/1203/04 (H5N1) (VN) with either PB2 627K (PB2K, wt/human isolate) or 627E (PB2E, avianized mutant) polymerase plasmid. Viral polymerase activity was assessed as a ratio of normalized polymerase activities to the empty vector value, with significance estimated by an unpaired, 2-tailed t test. ns, not significant.
Impact of host factors in H5N1 virus replication.
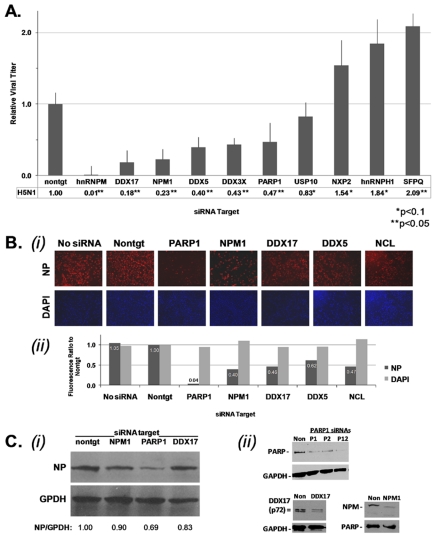
We validated the phenotypes of eight representative RNA-binding proteins from the regulatory network and two stress-related proteins, PARP1 and USP10, in H5N1 infection of human lung A549 cells. As with PARP1, knockdown of hnRNPM, DDX17, DDX5, DDX3X, and NPM1 each led to a significant reduction in the titer of A/VN/1203/04 (H5N1) HALo virus (i.e., virus modified by removal of the hemagglutinin [HA0] protein’s polybasic cleavage site) released from A549 cells (Fig. 4A), in concordance with phenotypes in both the VPOL-mg and infection-mg assays (Fig. 1B). Knockdown of NXP2 or hnRNPH1 allowed a mild increase in the virus titer, in agreement with the siRNA experiments, while targeting SFPQ, a protein involved in mRNA processing, resulted in a more significant increase in titers. For PARP1 and four RNA-binding proteins tested, reduced accumulation of NP in H5N1-infected A549 cells was observed, consistent with the hypothesis that these factors are required for optimal polymerase function (Fig. 4B). In A549 cells with substantial knockdown of PARP1, NPM1, or DDX17 at the protein level, viral nucleoprotein expression was still reduced 20 h after H5N1 infection (Fig. 4C). Since A/VN/1203/04 (H5N1) HALo virus undergoes only one life cycle in this system, these results suggest that the reduction in NP protein results from reduced viral gene expression and/or amplification of viral genomes.
FIG 4 .
Host factor RNAi silencing alters progression of the H5N1 virus life cycle. (A) Average relative titers of virus released 20 h p.i. from A549 cells targeted 36 h prior by siRNA transfection and infected by influenza A/VN/1203/04 (H5N1) HALo virus (MOI = 0.1). Values shown are averages of plaque and limiting dilution assay ratios for each host target siRNA relative to nontarget (nontgt) siRNA values, with significance estimated by an unpaired, 2-tailed t test. (B) Viral nucleoprotein (NP) expression in A549 cells mock-treated or transfected with nontarget (Nontgt) or siRNA-targeting selected host factors 24 h prior to infection with A/VN/1203/04 (H5N1) HALo virus (MOI = 2). Cultures were fixed 6 h p.i. for IFA with monoclonal anti-NP and Alexa-555-labeled secondary antibody and 4′,6-diamidino-2-phenylindole (DAPI) nuclear staining (i). Fluorescence intensity quantification of Alexa-555 in representative images as a ratio to the nontarget siRNA value (ii). (C) NP expression (20 h p.i.) in A549 cells infected with A/VN/1203/04 (H5N1) HALo (MOI = 0.5). Infected cell lysates were separated by SDS-PAGE and immunoblotted, and separated strips were probed with anti-NP or anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies. Densitometry depicting the ratio of NP to GAPDH for equal protein samples is indicated (i). A549 cells transfected with siRNA (Non, nontarget; P1 and P2 indicate two different siRNAs, while P12 indicates both pooled, targeting PARP1; DDX17, siRNA pool targeting DDX17; NPM1, siRNA pool targeting nucleophosmin) were lysed and run on an SDS-PAGE gel, and Western blots were probed with antibodies as indicated (ii).
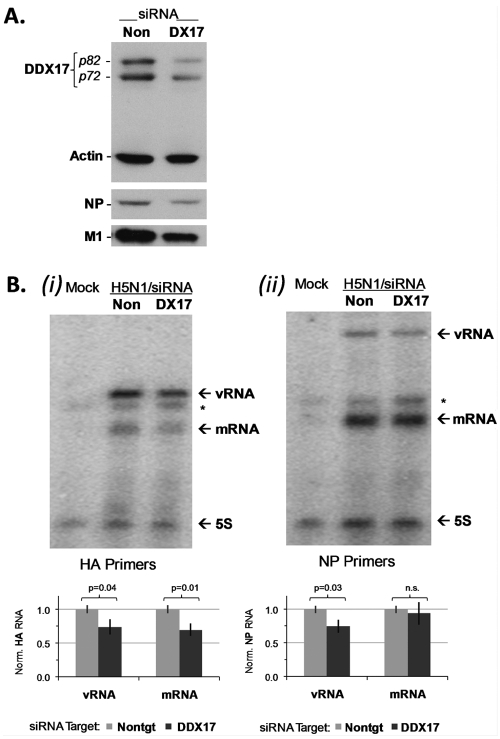
DDX17 is required for efficient H5N1 mRNA and vRNA synthesis during infection of human cells.
For DDX17 knockdown in the VPOL-mg assay, the divergence between H5N1 PB2|672E antagonist and PB2|627K enhancer phenotypes was approximately 6.4-fold (Fig. 2B). DDX17 thus exhibited characteristics of a species specificity factor for H5N1 HPAI polymerase, so we investigated mechanisms by which DDX17 allows optimal viral gene expression in human cells. Surprisingly, DDX17 interacted with avianized H5N1 polymerase complexes at least as well as, if not better than, the wt/human isolate in immunoprecipitation experiments (see Fig. S4 in the supplemental material). When DDX17 was targeted by siRNA in human 293T cells, expression of the NP and M1 proteins was reduced during A/VN/1203/04 (H5N1) HALo infection (Fig. 5A). Primer extension analysis showed an approximately 25% loss in synthesis of both hemagglutinin (HA) and NP vRNA segments and a 30% loss in HA mRNA synthesis, when DDX17 was targeted by siRNA (Fig. 5B). The decrease in NP mRNA was not significant at this time point (12 h p.i.). These data indicate that DDX17 is required for maximal viral mRNA and vRNA synthesis in human cells infected by H5N1 virus. However, it is also possible that DDX17 has an additional posttranscriptional role(s) facilitating viral gene expression.
FIG 5 .
DDX17 facilitates viral RNA synthesis in H5N1 infection of human cells. (A) 293T cells were targeted by Nontarget (Non) or DDX17 (DX17) siRNA. After 24 h of knockdown, cells were infected by A/VN/1203/04 (H5N1) HALo virus (MOI = 1). Cell protein lysates were analyzed 12 h p.i. for the DDX17, NP, M1, and actin proteins by Western blotting. (B) Primer extension analysis with specific primers for HA (i) or NP (ii) mRNA and vRNA synthesis, as indicated, for mock-infected control or 293T cells 12 h p.i. with A/VN/1203/04 (H5N1) HALo virus (MOI = 1). 5S, rRNA loading control; *, nonspecific background band. Graphs show RNA quantification by densitometry normalized to 5S rRNA and background; significance was estimated by an unpaired, 2-tailed t test from density histograms and standard deviations.
Avian DDX17 is required for optimal H5N1 replication in avian cells.
While human DDX17 was required for optimal replication of the wt/human isolate H5N1 virus in human cells, we asked whether this host factor is required for optimal replication of avianized H5N1 virus in avian cells. The domestic chicken (Gallus gallus) has emerged as a host for H5N1 HPAI viruses (1). After identifying a DDX17 ortholog in the chicken genome by sequence comparison, we synthesized siRNAs specifically targeting the chkDDX17 transcript (see Table S1 in the supplemental material). Knockdown of the chkDDX17 protein in chicken DF-1 fibroblasts (Fig. 6A) resulted in a 3-fold-decreased viral titer (Fig. 6B) and significantly less NP expression (Fig. 6C) during A/Viet Nam/1203/04 (H5N1) HALo (PB2|627E) infection. The wild-type/human isolate (PB2|627K) virus showed a 2-fold decrease in viral titer (data not shown). Thus, a DDX17-like protein in chicken cells is required for optimal avian (PB2|627E) H5N1 virus replication.
FIG 6 .
DDX17 is required for optimal H5N1 infection in chicken fibroblasts. (A) Targeted knockdown of chicken DDX17. Nontarget siRNA (Ntgt) or specific siRNA targeting a Gallus gallus DDX17 ortholog (DX17) was synthesized, purified, pooled, and transfected into DF-1 cells. After 48 h of knockdown, cells were lysed and analyzed by Western blotting for chicken DDX17 and actin with cross-reactive, polyclonal antisera. (B) Decreased H5N1 virus replication with knockdown of chicken DDX17. Chicken DF-1 fibroblasts transfected by nontarget (Nontgt) or chicken DDX17 (chkDDX17) siRNA were infected after 48 h with A/VN/1203/04 (H5N1) HALo (PB2|627E) virus (MOI = 0.5). Viral titers (16 h p.i.) were estimated by limiting dilution relative to the nontarget siRNA control value, and significance was tested by an unpaired, 2-tailed t test. (C) DF-1 cells were infected with avianized A/VN/1203/04 (H5N1) HALo (PB2|627E) virus (MOI = 0.5) and fixed 20 h p.i. NP expression in infected DF-1 cells was analyzed by IFA with monoclonal anti-NP-labeled and Alexa-555-labeled anti-mouse secondary antibody and DAPI nuclear staining (i), and fluorescence intensity was quantified relative to the nontarget siRNA value, with significance tested by an unpaired, 2-tailed t test (ii).
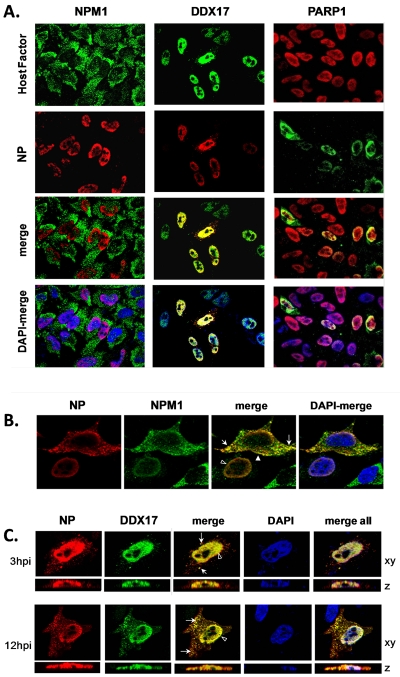
DDX17 and NPM1 colocalize with H5N1 NP in the cytoplasm late in infection.
Finally, we studied the subcellular interaction of DDX17 and NPM1 with viral NP during the H5N1 virus life cycle in human cells. According to the current understanding of viral ribonucleoprotein (vRNP) export and virion assembly (31, 35, 36), NP first accumulates in the nucleoplasm as a viral protein, and RNA syntheses progress. An unidentified signal induces NP to relocalize inside the nuclear periphery for subsequent export with viral RNA, mediated by NS2/NEP, to the cytoplasm for microtubule-mediated transport to the plasma membrane, where new virions bud. In uninfected, interphase HeLa cells, endogenous DDX17 exhibited a punctate, predominantly intranuclear distribution that was altered by mitosis; NPM1, an abundant RNA chaperone, was mostly cytoplasmic but also was present in the nucleoli and nucleoplasm (see Fig. S5 in the supplemental material). Early in the spatial context of H5N1 infection, NPM1 did not overlap with nuclear NP (Fig. 7A). However, by 12 h p.i., NPM1 closely associated with NP in the nuclear periphery and perinuclear region and in discrete punctae in the cytoplasm (Fig. 7B). DDX17 initially associated with NP in the nucleus and at micropunctae in the cytoplasm (Fig. 7A) and showed extensive colocalization with NP in the cytoplasm by 12 h p.i. (Fig. 7C). The relocalization of NPM1 and DDX17 in infected cells contrasted with the nuclear localization of another enhancer, PARP1 (Fig. 7A). These results suggested that both DDX17 and NPM1 associate with vRNP that have been exported to the cytoplasm late in the viral life cycle.
FIG 7 .
Colocalization of H5N1 NP with DDX17 and NPM1 during infection. (A) IFA of HeLa cells infected with influenza A/VN/1203/04 (H5N1) HALo virus (MOI = 0.5) were fixed 3 h p.i. and incubated with anti-NP monoclonal and anti-NPM1 or -DDX17 polyclonal antibodies or anti-PARP1 monoclonal and anti-NP polyclonal antibodies, Alexa-555-labeled (red) or Alexa-488-labeled (green) secondary antibodies, and DAPI nuclear stain (blue). Images were captured by structured illumination, providing defined submicron vertical (z-plane) optical sections imaged through the cell, and merged as indicated. (B) IFA of HeLa cells infected with A/VN/1203/04 (H5N1) HALo virus (MOI = 0.2) fixed 12 h p.i. Colocalization (yellow) of NP and NPM1 in the nuclear periphery and perinuclear region (open arrowhead), in discrete punctae in the cytoplasm (arrows), or in proximity to the plasma membrane (filled arrowhead) is indicated. (C) IFA of HeLa cells infected with A/VN/1203/04 (H5N1) HALo virus (MOI = 0.2) fixed at 3 h p.i. or 12 h p.i. Colocalization (yellow) of NP and DDX17 in the nucleus (open arrowhead) or in discrete punctae in the cytoplasm (arrows) is indicated. Structured illuminations of horizontal optical sections (xy plane) were stacked to reconstruct lateral (z-plane) images of infected cells.
DISCUSSION
Host protein regulation of influenza virus polymerases.
Virus-host interactions modulating the influenza A virus polymerase have the potential to govern the replication and species specificity of emerging HPAI virus strains. We found that a majority (58%) of the virus-host interactions explored herein positively regulated (enhanced) activities of both A/WSN/33 (H1N1) and A/VN/1203/04 (H5N1) polymerases (Fig. 1C). Significant phenotypes in the VPOL-mg assay (Fig. 1B; see also Table S2 in the supplemental material) suggested that many human host factors can directly regulate viral polymerase activity in the absence of other viral proteins or cellular pathways induced by infection. Though precise mechanisms remain to be resolved, one can hypothesize that by interacting with the enhancer proteins in the regulatory network (Fig. 2A), influenza virus polymerases interact with enriched cellular processes (e.g., RNA processing; see Table S3) to achieve a high level of intranuclear RNA synthesis. Conversely, antagonist proteins could activate antiviral or RNA stress pathways that inhibit polymerase functions, viral protein expression, or RNA traffic. Molecular transport and RNA-related cellular functions were also identified by functional genomics (RNAi) surveys (24–27, 33), suggesting the critical nature of these processes, at least for H1N1 virus infection.
RNA-associated cellular proteins have been found to positively or negatively affect viral RNA syntheses (16, 19, 21, 22). Indeed, we found that RNA-binding proteins, particularly heterogeneous nuclear ribonucleoproteins (hnRNPs) and DEAD box (DDX) RNA helicases, regulated both H1N1 and H5N1 polymerases (Fig. 2A and 3) and H5N1 infection in a human lung cell line (Fig. 4). NF90 (DRBP76/ILF3), an antiviral RNA-binding protein that restricts H1N1 and H5N1 NP from interacting with vRNA (22), antagonized polymerase (Fig. 1B and 3). Six RNAassociated proteins (EBP1 [16], hnRNPK [25], hnRNPH1, hnRNPA1, RBM14, and DDX3X) and ARRB1 (25), a signaling modulator, displayed significantly differential phenotypes between H1N1 and H5N1 viruses (Fig. 2A). The possibility that H1N1 and H5N1 viruses differ in functional adaptation to an RNA-related process (see Table S3 in the supplemental material) highlights the importance of exploring strain differences in antiviral drug development.
Host factors regulate adaptation of an HPAI virus polymerase to human cells.
For an H5N1 HPAI virus, wild-type/human isolate (PB2|627K) and avianized (PB2|627E) polymerases differed considerably in their reliance on host factors in the regulatory network (Fig. 2B). Evidence has suggested that HPAI polymerases containing a mammal-adapted genotype at adaptive residues (e.g., PB2 591, 627, and 701) are able to interact with a host factor(s) to facilitate efficient RNA synthesis activity (11, 37). Host factors that exhibited a differential phenotype for avianized polymerase (Fig. 2B) included chaperone/transport proteins, consistent with models suggested in the literature (8, 38), and RNA-binding proteins, such as DDX17. This suggests that an RNA-related process may also contribute to the mechanistic basis of phenotypes related to the PB2 residue 627 genotype.
DDX17, a protein critical for H5N1 polymerase adaptation to human cells.
DDX17 (p72) and a related protein, DDX5 (p68), are abundant nuclear ATP-dependent DEAD-box RNA helicases that can heterodimerize (39) and catalyze RNA unwinding and rearrangement (40). DDX17 and DDX5 have been implicated in ribosomal biogenesis (41). Human DDX17 and DDX5 exhibited consistent, strong enhancer phenotypes in polymerase assays (Fig. 1B) that were confirmed in H5N1 infection, independently of minigenome reporters. DDX17 and DDX5 were required for A/VN/1203/04 (H5N1) wt/human isolate (PB2|627K) but not avianized mutant (PB2|627E) polymerase activity (Fig. 2B). When overexpressed in human cells, DDX17 also enhanced activity of wt/human isolate but not avianized H5N1 polymerase (Fig. 3). Mechanistically, human DDX17 facilitated both mRNA and vRNA synthesis during wt/human isolate H5N1 infection (Fig. 5B). DDX17 closely colocalized with NP in the cell nucleus early (3 h p.i.) in H5N1 infection (Fig. 7A), consistent with a role in viral RNA synthesis.
Proteomics and immunoprecipitation experiments have found that human DDX5 (17), DDX17, and NPM1 (18; data not shown) associate with WSN (H1N1) polymerase. Surprisingly, we found that DDX17 also associated with either wt/human isolate (PB2|627K) or avianized (PB2|627E) HPAI (H5N1) polymerase complexes and with either PB2 alone (see Fig. S5 in the supplemental material). However, the presence of another enhancer, NPM1, was greater in the wt/human isolate PB2 complexes. Thus, the failure of avian H5N1 polymerase to take advantage of DDX17’s ability to facilitate viral RNA synthesis is apparently not due simply to an inability of human DDX17 to interact with a heterotrimeric polymerase containing PB2|627E but perhaps involves association with another protein(s), such as NPM1 or DDX5. We also found that while the chicken DDX17 homologue was required for H5N1 protein expression (Fig. 6C) and viral replication (Fig. 6B), human DDX17 antagonized avianized polymerase in human cells (Fig. 2B). Thus, physical association of PB2|627E with avian DDX17 in avian cells might mechanistically enhance replication, while in contrast, interaction with human DDX17 could inhibit unadapted HPAI virus polymerase in human cells. This hypothesis might explain why avian influenza virus polymerases are blocked from efficient RNA synthesis in human cells by an antagonistic host factor(s) (42). Taken together, H5N1 virus requires a DDX17-like protein in both avian and human cells, suggesting conservation of a polymerase-host interaction critical for H5N1 survival in different host species.
Possible roles for RNA chaperones in vRNP export from the nucleus.
We found that two RNA-associated proteins (DDX17 and NPM1) may have additional functions during influenza virus vRNP export from the nucleus. Nuclear export of vRNP occurs in a CRM1-dependent manner (31). DDX17 and DDX5 function together in ribosomal pre-rRNA processing, and rRNA is also exported from the nucleus in a CRM1-dependent manner (41, 43). Meanwhile, the ribosomal protein RPL5, which associates with influenza virus vRNP (18), requires NPM1 for nuclear export (44). Late in H5N1 virus infection, DDX17 and NPM1 had redistributed in the cytoplasm in conjunction with NP (Fig. 7), likely signifying export of nascent vRNP from the nucleus (35). Thus, we are investigating the hypothesis that influenza viruses have evolved to hijack aspects of the CRM1-dependent export mechanism of another large ribonucleoprotein complex, the ribosome, in order to facilitate their own vRNP nuclear export.
Mapping functions in the influenza virus polymerase-host interactome.
Genomics experiments, from proteomics and genome-wide RNAi surveys to more targeted studies examining particular aspects of virus life cycles (e.g., influenza virus polymerase regulation), allow for the first time systematic analysis of the complexity of host-pathogen regulatory networks. While the present study identifies host factors important for influenza virus polymerase function and host species tropism, a comprehensive, dynamic model of how host interactions regulate influenza virus RNA synthesis, gene expression, and genome replication is far from complete. Adoption of common standards for RNAi data submission (45) would allow for more accurate meta-analysis and integration of multiple genomics data types to model complex virus-host interactions (27). We have demonstrated that functional genomics analysis of host proteins provides a novel means to study the phenotypes of influenza virus polymerase mutants and to map adaptation of emerging H5N1 HPAI polymerases to human cells. This approach has the potential to identify novel targets for therapeutic intervention in severe influenza virus infections.
MATERIALS AND METHODS
Viruses.
Attenuated influenza A/Viet Nam/1203/2004 (H5N1) HALo viruses (PB2|627K and PB2|627E) were generated by reverse genetics as described previously (13, 46), with removal of the hemagglutinin (HA0) protein’s polybasic cleavage site (GenBank accession no. CY077101); the virus undergoes only one round of replication in the absence of exogenous trypsin. All other wild-type viral gene segments were unmodified. Titers of H5N1 influenza viruses were determined by plaque assay on MDCK cells (46) or by limiting-dilution assays on A549 or chicken DF-1 cells. Titers of A/WSN/33 (H1N1) virus were determined as described previously (18, 26).
Bioinformatics analyses.
The H1N1 polymerase-host interactome was initially analyzed using Ingenuity pathways analysis (IPA7.6; Ingenuity Systems, Redwood, CA). The top-scoring, interconnected subnetwork was extracted, expanded by literature curation, and tested for significance by a Benjamini-Hochberg (PBH) estimate for false discovery probability, i.e., the probability that the linkages and functions assigned to the derived node-edge sets are due to chance alone. The DAVID software program (47) analyzed Gene Ontology (GO) term enrichment and functional group clustering.
Targeting host genes with siRNA.
Nontarget (scrambled) siRNA or siRNA targeting specific human or chicken transcripts (see Table S1 in the supplemental material) was transfected into 293T, A549, or DF-1 cells at 10 to 15 nM. For a subset of siRNA, efficacy of RNAi knockdown was evaluated by Western blotting, reverse transcription-PCR (RT-PCR), or siRNA targeting green fluorescent protein (GFP) (see Fig. S2). Cell viability (see Fig. S3) was measured by a Renilla reporter, CellTiter-Glo, and caspase-Glo 3/7 assays (Promega Corp., Madison, WI).
Minigenome reporter assays.
The VPOL-mg (vRNA-luciferase reporter) assay with HEK 293T cells (18) was optimized with plasmid ratios of 10:2:1:2 (NP/PB1/PB2/PA) to assay autonomous polymerase function (see Fig. S2 in the supplemental material). For cDNA experiments, ratios were 5:2:1:2. VPOL-mg and siRNA conditions were tested for two known polymerase interactors, PARP1 (18) and RanBP5 (15) (see Fig. S2). The infection-mg (cRNA-luciferase reporter) assay (29) was adopted to measure polymerase function in infected A549 cells. In minigenome reporter assays, reporter gene expression (“viral polymerase activity”) is dependent on viral polymerase-mediated transcription and replication of a model influenza virus RNA, measured by a Dual-Luciferase reporter assay (Promega Corporation, Madison, WI). Significance of differences relative to nontarget siRNA were estimated by an unpaired, 2-tailed t test to calculate P values, with a Pmax value of ≥0.15 regarded as no significant difference (see Table S2). Data for each RNAi target will be available in PubChem under Minimum Information About an RNAi Experiment (MIARE) data standards (45).
IFA and microscopy.
Infected A549 or DF-1 cells were fixed for immunofluorescence assay (IFA) and probed with specific primary antibodies (described in Text S1 [Materials and Methods] in the supplemental material) and Alexa Fluor-linked (488-nm anti-rabbit and 555-nm anti-mouse) IgG secondary antibodies (Molecular Probes, Eugene, OR). For subcellular localization studies, HeLa cells were infected with A/VN/1203/04 (H5N1) HALo virus (multiplicity of infection [MOI] of 0.5), and imaged by structured illumination on an Axioplan 2 microscope (Zeiss, Göttingen, Germany) providing optical, submicron vertical (z-plane) sections through the cell.
Molecular biology assays.
Primer extension assays were performed as described (23), using 32P-labeled DNA oligonucleotides to A/VN/1203/04 HA and NP, detecting vRNA segments and mRNA, and a 5S rRNA loading control. Immunoblot and immunoprecipitation assays were performed as described previously (18), with specific protocols described in Text S1 in the supplemental material (supplemental Materials and Methods).
Refer to Text S1 (supplemental Materials and Methods) for detailed descriptions of all methods and reagents.
SUPPLEMENTAL MATERIAL
Supplemental methods. Download Text S1, PDF file, 0.1 MB.
Interactome network comprising cellular proteins associated with H1N1 influenza A virus polymerase, NP, and/or vRNP. (A) H1N1 influenza virus polymerase-host interactome. Human interactome network comprising 54 proteins known to associate with influenza A virus nucleoprotein (NP), viral polymerase subunits (PB1, PB2, or PA), or viral ribonucleoprotein (vRNP) and immediate neighbors, synthesized from constituent human interactome subnetworks identified by Ingenuity pathways analysis and data curated from the literature. Significance of the hypothetical network was estimated by the Benjamini-Hochberg false discovery P value (PBH). Molecules targeted in this study are shaded yellow; untested identified viral protein interactors are shaded gray; other human proteins are white. Edges between nodes represent known protein-protein interactions; arrows, activation; dashed links, indirect interactions. (B) The table shows enrichment of functional processes among molecules in the H1N1 polymerase-host interactome that were discovered by Ingenuity pathways analysis (IPA) and Gene Ontology (GO) analyses. Significance of IPA categories or GO classifications was estimated by the Benjamini-Hochberg test (PBH) or Fisher’s exact test (P < 0.05), respectively. Download Figure S1, PDF file, 0.6 MB.
Optimization of viral polymerase assay and siRNA targeting host factors.(A) Schematic representation of influenza A virus polymerase-minigenome (VPOL-mg) assay with vRNA promoter-driven firefly luciferase reporter or infection-mg assay with cRNA-minigenome reporter. For optimization, viral polymerase activity was evaluated by dual-luciferase assay, normalized to a Pol II-driven Renilla internal control, in 24-well format. (B) Optimization of VPOL-mg assay in 293T cells. Dose-response of optimized pCAGGS–NP and polymerase (-PB1, -PB2, and -pa) plasmid transfection at a 10:2:1:2 ratio, determined for A/WSN/33 (WSN) and wild-type A/VN/1203/04 (VN1203) luciferase minigenome reporter (i). Comparison of various WSN NP and polymerase plasmids for magnitude of polymerase-mediated reporter gene expression relative to 100 ng plasmids at a 1:1:1:1 ratio (activity*, light bars) and for absolute amount of activity per 100 ng plasmids (Efficiency**, dark bars) (ii). For dependence on individual subunits, increasing amounts of pCAGGS-NP plasmid were transfected (in ng), with constant PB1, PB2, and PA in a [NP]:50 ng:50 ng:50 ng ratio (iii) or pCAGGS-PB2 plasmid in a 100 ng:100 ng:[PB2]:100 ng ratio for WSN and a 50 ng:50 ng:[PB2]:50 ng ratio for VN1203 (iv). Varying [PB1] or [PA] resulted in weak linear dependence (v). (C) Magnitudes of VPOL-mg assay data for polymerase complexes in 293T cells at optimized plasmid ratios. A/WSN/33 or A/VN/1203/04 PB2 genotype 627E (avianized mutant) or 627K (wild-type/human isolate) luciferase and constitutive Renilla values in a 24-well-format experiment. (D) VPOL-mg assay using (enhanced) GFP-minigenome reporter and constitutive red fluorescent protein (RFP) control for two A/VN/1203/04 (H5N1) viral polymerase PB2 genotypes (aa627K, wild type/human isolate; aa627E, avianized mutant). (E) Synthesis, transfection, and activity of siRNA. For lab-synthesized siRNA, in vitro synthesis, purification, and quantification of siRNA were performed as described in Text S1 (supplemental Materials and Methods). Examples of product siRNA analyzed by gel electrophoresis (2.5% agarose) stained with ethidium bromide show sensitivity of siRNA products to RNaseA/H treatment (i). A CY3-labeled nontarget siRNA transfected into HEK 293T cells indicates >95% transfection efficiency (ii). Lab-synthesized nontarget siRNA or siRNA targeting PARP1 was transfected into 293T cells: RT-PCR with primers detecting PARP1 (iii) and Western blotting to specifically detect PARP1 and GAPDH (iv) were carried out. (F) Influenza A virus polymerase minigenome assay in cells targeted by siRNA. Nontarget siRNA or siRNA targeting GFP, RanBP5/importin β3 (KPNB3), or PARP1 was cotransfected into subconfluent human embryonic kidney (HEK) 293T fibroblasts with plasmids expressing A/WSN/33 NP, PB1, PB2, and PA, a vRNA promoter minigenome encoding an (enhanced) GFP reporter, and a constitutive Pol II-driven DS-Red (pCAGGS-RFP) plasmid, and examined 18 h posttransfection (p.t.) by epifluorescence microscopy. (G) Increased A/WSN/33 viral polymerase activity in heat-shocked 293T cells is partly dependent on Hsc71/HSPA8. A VPOL-mg assay was carried out in 293T cells targeted with Nontgt, PARP1, or Hsc71 siRNA, either untreated at 37°C or heat shocked at 42°C for 30 min, 4 h prior to harvesting lysates for the dual-luciferase assay. Significance was estimated by an unpaired, 2-tailed t test between 37°C and 42°C (brackets) or in comparison to the nontarget control value for 37°C and 42°C, respectively (within the table). Download Figure S2, PDF file, 0.4 MB.
Cell viability and cytotoxicity analyses of siRNA transfection. (A) Cell viability was measured for individual siRNA or pools 48 h posttransfection with CellTiter-Glo, which measures intracellular energy output (ATP availability) via production of a luminescent reporter, and a Pol II-driven Renilla reporter, measuring cellular gene expression. Apoptosis was measured in parallel transfections by measuring total caspase 3 and 7 enzymatic activity via production of a luminescent product. Measurements were performed in duplicate on A549 cells at 95% confluence in a 96-well format and expressed as a ratio to nontarget (Nontgt) siRNA. The table shows siRNA possessing one or more assay results outside the normal range (nr) determined for nontarget siRNA under the given transfection conditions in A549 cells, with the type of cell compromise indicated. Staurosporine, treatment of A549 cells with 1 µM staurosporine for 12 h. (B) Average of normalized WSN and VN VPOL-mg results for each siRNA target plotted against a metric for increasing A549 cell compromise attributed to target siRNA transfection. Exponential regression analysis was done, with R2 values indicated; staurosporine (0.5 µM or 1 µM, 12 h) provided a positive control for induction of apoptosis. KPNA4 was subsequently excluded from network and functional enrichment analyses. Download Figure S3, PDF file, 0.2 MB.
Human DDX17 interacts with H5N1 PB2, polymerase, and endogenous NPM1. FLAG-tagged DDX17 immunoprecipitates A/VN/1203/04 (H5N1) polymerase complex, PB2 alone, and endogenous NPM1. Human DDX17-FLAG was transfected into 293T cells with NP or heterotrimeric polymerase (PB1, PB2, PA, and a vRNA minigenome) with either PB2 627K (wt/human isolate) or 627E (avianized mutant). Proteins immunoprecipitated by anti-FLAG monocolonal antibody matrix were analyzed by Western blotting with specific antibodies to PB2, NP, and NPM1. Download Figure S4, PDF file, 0.2 MB.
Localization of human DDX17 and NPM1 in uninfected HeLa cells. (A) HeLa cells were fixed and incubated with anti-DDX17 polyclonal antisera, Alexa-488 (green)-labeled secondary antibodies, and DAPI stain (blue) to visualize nuclei. Images depicted were captured by structured illumination, providing defined vertical (z-plane) optical sectioning through the cell, and merged as indicated. DDX17 appears in discrete nuclear punctae in interphase cells (star), quite distinct from chromatin in postmitotic cell pairs (arrows). (B) HeLa cells were fixed and incubated with anti-NPM1 polyclonal antisera, Alexa-488 (green)-labeled secondary antibody, and DAPI (blue) to visualize nuclei; images depicted were captured by structured illumination. Download Figure S5, PDF file, 1.2 MB.
siRNA used in this study.
Viral polymerase activity assays in human cells targeted by siRNA.
RNA-related cellular functions enriched in the regulatory network (see Fig. 2A).
ACKNOWLEDGMENTS
We appreciate the generous gifts of the cRNA minigenome reporter from Megan Shaw and H. Heinrich Hoffmann (Mount Sinai) and DDX17 cDNA from Frances Fuller-Pace (University of Dundee, Dundee, United Kingdom). We thank Nigel Binns (University of Edinburgh, Edinburgh, United Kingdom) for assistance with MIARE data submission to PubChem. We also thank Mirco Schmolke, Masaki Mibayashi, Reed Shabman, and Svetlana Burmakina for reagents and technical advice, Richard Cadagan and Osman Lizardo for excellent technical support, PRiME (NIAID contract HHSN2662000500021C) for access to and Alyza Skaist for assistance with IPA, Rumana Huq of Mount Sinai Microscopy (5R24CA095823-04 and 1S10RR09145-01), and Xudong Li, Larry Leung, Alina Baum, Benjamin Hale, Rong Hai, Estanislao Nistal-Villán, and Peter Palese for helpful discussions.
This work was funded by CRIP, a NIAID-funded Center of Excellence for Influenza Research and Surveillance grant (HHSN266200700010C), and NIH (P01AI58113, U01AI074539, and U54AI057158, to A.G.-S.), BMBF FluResearchNet (to G.C. and M.S.), Northeast Biodefense Center (U54AI057158-L, to J.S.), and a NIAID postdoctoral fellowship, 5T32AI07647 (to E.B.).
Footnotes
Citation Bortz E, et al. 2011. Host- and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteins. mBio 2(4):e00151-11. doi:10.1128/mBio.00151-11.
REFERENCES
- 1. Neumann G, Chen H, Gao GF, Shu Y, Kawaoka Y. 2010. H5N1 influenza viruses: outbreaks and biological properties. Cell Res. 20:51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith GJ, et al. 2006. Emergence and predominance of an H5N1 influenza variant in China. Proc. Natl. Acad. Sci. U. S. A. 103:16936–16941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wan XF, et al. 2008. Evolution of highly pathogenic H5N1 avian influenza viruses in Vietnam between 2001 and 2007. PLoS One 3:e3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gao R, et al. 2010. A systematic molecular pathology study of a laboratory confirmed H5N1 human case. PLoS One 5:e13315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hatta M, Neumann G, Kawaoka Y. 2001. Reverse genetics approach towards understanding pathogenesis of H5N1 Hong Kong influenza A virus infection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:1841–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maines TR, et al. 2005. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J. Virol. 79:11788–11800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salomon R, et al. 2006. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 203:689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gabriel G, Herwig A, Klenk HD. 2008. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 4:e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rolling T, et al. 2009. Adaptive mutations resulting in enhanced polymerase activity contribute to high virulence of influenza A virus in mice. J. Virol. 83:6673–6680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li J, et al. 2009. Single mutation at the amino acid position 627 of PB2 that leads to increased virulence of an H5N1 avian influenza virus during adaptation in mice can be compensated by multiple mutations at other sites of PB2. Virus Res. 144:123–129 [DOI] [PubMed] [Google Scholar]
- 11. Yamada S, et al. 2010. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 6:e1001034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hatta M, Gao P, Halfmann P, Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840–1842 [DOI] [PubMed] [Google Scholar]
- 13. Steel J, Lowen AC, Mubareka S, Palese P. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 5:e1000252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Le QM, Sakai-Tagawa Y, Ozawa M, Ito M, Kawaoka Y. 2009. Selection of H5N1 influenza virus PB2 during replication in humans. J. Virol. 83:5278–5281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deng T, et al. 2006. Role of Ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex. J. Virol. 80:11911–11919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Honda A, Okamoto T, Ishihama A. 2007. Host factor Ebp1: selective inhibitor of influenza virus transcriptase. Genes Cells 12:133–142 [DOI] [PubMed] [Google Scholar]
- 17. Jorba N, et al. 2008. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics 8:2077–2088 [DOI] [PubMed] [Google Scholar]
- 18. Mayer D, et al. 2007. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 6:672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Momose F, et al. 2001. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J. Virol. 75:1899–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Momose F, et al. 2002. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 277:45306–45314 [DOI] [PubMed] [Google Scholar]
- 21. Naito T, et al. 2007. An influenza virus replicon system in yeast identified Tat-SF1 as a stimulatory host factor for viral RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 104:18235–18240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang P, et al. 2009. Nuclear factor 90 negatively regulates influenza virus replication by interacting with viral nucleoprotein. J. Virol. 83:7850–7861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chase G, et al. 2008. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology 377:431–439 [DOI] [PubMed] [Google Scholar]
- 24. Brass AL, et al. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karlas A, et al. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822 [DOI] [PubMed] [Google Scholar]
- 26. König R, et al. 2010. Human host factors required for influenza virus replication. Nature 463:813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shapira SD, et al. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amorim MJ, Read EK, Dalton RM, Medcalf L, Digard P. 2007. Nuclear export of influenza A virus mRNAs requires ongoing RNA polymerase II activity. Traffic 8:1–11 [DOI] [PubMed] [Google Scholar]
- 29. Hoffmann HH, Palese P, Shaw ML. 2008. Modulation of influenza virus replication by alteration of sodium ion transport and protein kinase C activity. Antiviral Res. 80:124–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee JH, et al. 2010. Direct interaction of cellular hnRNP-F and NS1 of influenza A virus accelerates viral replication by modulation of viral transcriptional activity and host gene expression. Virology 397:89–99 [DOI] [PubMed] [Google Scholar]
- 31. Elton D, et al. 2001. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 75:408–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burgui I, Aragón T, Ortín J, Nieto A. 2003. PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 84:3263–3274 [DOI] [PubMed] [Google Scholar]
- 33. Watanabe T, Watanabe S, Kawaoka Y. 2010. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7:427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Squires B, et al. 2008. BioHealthBase: informatics support in the elucidation of influenza virus host pathogen interactions and virulence. Nucleic Acids Res. 36:D497–D503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elton D, Amorim MJ, Medcalf L, Digard P. 2005. “Genome gating”; polarized intranuclear trafficking of influenza virus RNPs.Biol. Lett. 1:113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Momose F, et al. 2011. Apical transport of influenza A virus ribonucleoprotein requires Rab11-positive recycling endosome. PLoS One 6:e21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moncorge O, Mura M, Barclay WS. 2010. Evidence for avian and human host cell factors that affect the activity of influenza virus polymerase. J. Virol. 84:9978–9986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gabriel G, et al. 2011. Differential use of importin-alpha isoforms governs cell tropism and host adaptation of influenza virus. Nat. Commun. 2:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ogilvie VC, et al. 2003. The highly related DEAD box RNA helicases p68 and p72 exist as heterodimers in cells. Nucleic Acids Res. 31:1470–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rössler OG, Straka A, Stahl H. 2001. Rearrangement of structured RNA via branch migration structures catalysed by the highly related DEAD-box proteins p68 and p72. Nucleic Acids Res. 29:2088–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jalal C, Uhlmann-Schiffler H, Stahl H. 2007. Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res. 35:3590–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mehle A, Doudna JA. 2008. An inhibitory activity in human cells restricts the function of an avian-like influenza virus polymerase. Cell Host Microbe 4:111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murdoch K, Loop S, Rudt F, Pieler T. 2002. Nuclear export of 5S rRNA-containing ribonucleoprotein complexes requires CRM1 and the RanGTPase cycle. Eur. J. Cell Biol. 81:549–556 [DOI] [PubMed] [Google Scholar]
- 44. Yu Y, et al. 2006. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol. Cell. Biol. 26:3798–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haney SA. 2007. Increasing the robustness and validity of RNAi screens. Pharmacogenomics 8:1037–1049 [DOI] [PubMed] [Google Scholar]
- 46. Steel J, et al. 2009. Live attenuated influenza viruses containing NS1 truncations as vaccine candidates against H5N1 highly pathogenic avian influenza. J. Virol. 83:1742–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download Text S1, PDF file, 0.1 MB.
Interactome network comprising cellular proteins associated with H1N1 influenza A virus polymerase, NP, and/or vRNP. (A) H1N1 influenza virus polymerase-host interactome. Human interactome network comprising 54 proteins known to associate with influenza A virus nucleoprotein (NP), viral polymerase subunits (PB1, PB2, or PA), or viral ribonucleoprotein (vRNP) and immediate neighbors, synthesized from constituent human interactome subnetworks identified by Ingenuity pathways analysis and data curated from the literature. Significance of the hypothetical network was estimated by the Benjamini-Hochberg false discovery P value (PBH). Molecules targeted in this study are shaded yellow; untested identified viral protein interactors are shaded gray; other human proteins are white. Edges between nodes represent known protein-protein interactions; arrows, activation; dashed links, indirect interactions. (B) The table shows enrichment of functional processes among molecules in the H1N1 polymerase-host interactome that were discovered by Ingenuity pathways analysis (IPA) and Gene Ontology (GO) analyses. Significance of IPA categories or GO classifications was estimated by the Benjamini-Hochberg test (PBH) or Fisher’s exact test (P < 0.05), respectively. Download Figure S1, PDF file, 0.6 MB.
Optimization of viral polymerase assay and siRNA targeting host factors.(A) Schematic representation of influenza A virus polymerase-minigenome (VPOL-mg) assay with vRNA promoter-driven firefly luciferase reporter or infection-mg assay with cRNA-minigenome reporter. For optimization, viral polymerase activity was evaluated by dual-luciferase assay, normalized to a Pol II-driven Renilla internal control, in 24-well format. (B) Optimization of VPOL-mg assay in 293T cells. Dose-response of optimized pCAGGS–NP and polymerase (-PB1, -PB2, and -pa) plasmid transfection at a 10:2:1:2 ratio, determined for A/WSN/33 (WSN) and wild-type A/VN/1203/04 (VN1203) luciferase minigenome reporter (i). Comparison of various WSN NP and polymerase plasmids for magnitude of polymerase-mediated reporter gene expression relative to 100 ng plasmids at a 1:1:1:1 ratio (activity*, light bars) and for absolute amount of activity per 100 ng plasmids (Efficiency**, dark bars) (ii). For dependence on individual subunits, increasing amounts of pCAGGS-NP plasmid were transfected (in ng), with constant PB1, PB2, and PA in a [NP]:50 ng:50 ng:50 ng ratio (iii) or pCAGGS-PB2 plasmid in a 100 ng:100 ng:[PB2]:100 ng ratio for WSN and a 50 ng:50 ng:[PB2]:50 ng ratio for VN1203 (iv). Varying [PB1] or [PA] resulted in weak linear dependence (v). (C) Magnitudes of VPOL-mg assay data for polymerase complexes in 293T cells at optimized plasmid ratios. A/WSN/33 or A/VN/1203/04 PB2 genotype 627E (avianized mutant) or 627K (wild-type/human isolate) luciferase and constitutive Renilla values in a 24-well-format experiment. (D) VPOL-mg assay using (enhanced) GFP-minigenome reporter and constitutive red fluorescent protein (RFP) control for two A/VN/1203/04 (H5N1) viral polymerase PB2 genotypes (aa627K, wild type/human isolate; aa627E, avianized mutant). (E) Synthesis, transfection, and activity of siRNA. For lab-synthesized siRNA, in vitro synthesis, purification, and quantification of siRNA were performed as described in Text S1 (supplemental Materials and Methods). Examples of product siRNA analyzed by gel electrophoresis (2.5% agarose) stained with ethidium bromide show sensitivity of siRNA products to RNaseA/H treatment (i). A CY3-labeled nontarget siRNA transfected into HEK 293T cells indicates >95% transfection efficiency (ii). Lab-synthesized nontarget siRNA or siRNA targeting PARP1 was transfected into 293T cells: RT-PCR with primers detecting PARP1 (iii) and Western blotting to specifically detect PARP1 and GAPDH (iv) were carried out. (F) Influenza A virus polymerase minigenome assay in cells targeted by siRNA. Nontarget siRNA or siRNA targeting GFP, RanBP5/importin β3 (KPNB3), or PARP1 was cotransfected into subconfluent human embryonic kidney (HEK) 293T fibroblasts with plasmids expressing A/WSN/33 NP, PB1, PB2, and PA, a vRNA promoter minigenome encoding an (enhanced) GFP reporter, and a constitutive Pol II-driven DS-Red (pCAGGS-RFP) plasmid, and examined 18 h posttransfection (p.t.) by epifluorescence microscopy. (G) Increased A/WSN/33 viral polymerase activity in heat-shocked 293T cells is partly dependent on Hsc71/HSPA8. A VPOL-mg assay was carried out in 293T cells targeted with Nontgt, PARP1, or Hsc71 siRNA, either untreated at 37°C or heat shocked at 42°C for 30 min, 4 h prior to harvesting lysates for the dual-luciferase assay. Significance was estimated by an unpaired, 2-tailed t test between 37°C and 42°C (brackets) or in comparison to the nontarget control value for 37°C and 42°C, respectively (within the table). Download Figure S2, PDF file, 0.4 MB.
Cell viability and cytotoxicity analyses of siRNA transfection. (A) Cell viability was measured for individual siRNA or pools 48 h posttransfection with CellTiter-Glo, which measures intracellular energy output (ATP availability) via production of a luminescent reporter, and a Pol II-driven Renilla reporter, measuring cellular gene expression. Apoptosis was measured in parallel transfections by measuring total caspase 3 and 7 enzymatic activity via production of a luminescent product. Measurements were performed in duplicate on A549 cells at 95% confluence in a 96-well format and expressed as a ratio to nontarget (Nontgt) siRNA. The table shows siRNA possessing one or more assay results outside the normal range (nr) determined for nontarget siRNA under the given transfection conditions in A549 cells, with the type of cell compromise indicated. Staurosporine, treatment of A549 cells with 1 µM staurosporine for 12 h. (B) Average of normalized WSN and VN VPOL-mg results for each siRNA target plotted against a metric for increasing A549 cell compromise attributed to target siRNA transfection. Exponential regression analysis was done, with R2 values indicated; staurosporine (0.5 µM or 1 µM, 12 h) provided a positive control for induction of apoptosis. KPNA4 was subsequently excluded from network and functional enrichment analyses. Download Figure S3, PDF file, 0.2 MB.
Human DDX17 interacts with H5N1 PB2, polymerase, and endogenous NPM1. FLAG-tagged DDX17 immunoprecipitates A/VN/1203/04 (H5N1) polymerase complex, PB2 alone, and endogenous NPM1. Human DDX17-FLAG was transfected into 293T cells with NP or heterotrimeric polymerase (PB1, PB2, PA, and a vRNA minigenome) with either PB2 627K (wt/human isolate) or 627E (avianized mutant). Proteins immunoprecipitated by anti-FLAG monocolonal antibody matrix were analyzed by Western blotting with specific antibodies to PB2, NP, and NPM1. Download Figure S4, PDF file, 0.2 MB.
Localization of human DDX17 and NPM1 in uninfected HeLa cells. (A) HeLa cells were fixed and incubated with anti-DDX17 polyclonal antisera, Alexa-488 (green)-labeled secondary antibodies, and DAPI stain (blue) to visualize nuclei. Images depicted were captured by structured illumination, providing defined vertical (z-plane) optical sectioning through the cell, and merged as indicated. DDX17 appears in discrete nuclear punctae in interphase cells (star), quite distinct from chromatin in postmitotic cell pairs (arrows). (B) HeLa cells were fixed and incubated with anti-NPM1 polyclonal antisera, Alexa-488 (green)-labeled secondary antibody, and DAPI (blue) to visualize nuclei; images depicted were captured by structured illumination. Download Figure S5, PDF file, 1.2 MB.
siRNA used in this study.
Viral polymerase activity assays in human cells targeted by siRNA.
RNA-related cellular functions enriched in the regulatory network (see Fig. 2A).