Abstract
Uniparental chromosome elimination occurs in several interspecific hybrids of plants. We studied the mechanism underlying selective elimination of the paternal chromosomes during the early development of Hordeum vulgare × Hordeum bulbosum embryos. The following conclusions regarding the role of the centromere-specific histone H3 variant (CENH3) in the process of chromosome elimination were drawn: (i) centromere inactivity of H. bulbosum chromosomes triggers the mitosis-dependent process of uniparental chromosome elimination in unstable H. vulgare × H. bulbosum hybrids; (ii) centromeric loss of CENH3 protein rather than uniparental silencing of CENH3 genes causes centromere inactivity; (iii) in stable species combinations, cross-species incorporation of CENH3 occurs despite centromere-sequence differences, and not all CENH3 variants get incorporated into centromeres if multiple CENH3s are present in species combinations; and (iv) diploid barley species encode two CENH3 variants, the proteins of which are intermingled within centromeres throughout mitosis and meiosis.
Keywords: kinetochore, interspecies hybridization, micronuclei, wide crosses
Chromosome elimination of one parental genome after fertilization of the egg by the sperm of another species is a fairly common phenomenon and results in the formation of haploid embryos. It has been exploited for barley (1) and other species (e.g., wheat, potato) to produce doubled haploids for breeding and mapping purposes (reviewed in 2). The advantage of doubled haploids for breeders is that homozygosity can be achieved in the first generation, whereas in breeding systems, such as pedigree or backcrossing, several selfed generations are needed to obtain high levels of homozygosity.
To produce haploids of barley, crosses are made with Hordeum bulbosum Linnaeus (bulbous barley grass), a close relative of cultivated barley in the secondary gene pool. Chromosomes of H. bulbosum are eliminated several days after pollination (1, 3–6) independent of the crossing direction (1), but hybrids combining both sets of parental chromosomes can be obtained (7). Chromosome elimination is known to depend on genetic factors (7) and temperature after fertilization (8).
Several explanations have been proposed to account for uniparental chromosome elimination [e.g., difference in timing of essential mitotic processes attributable to asynchronous cell cycling (9), asynchrony in nucleoprotein synthesis leading to a loss of the most retarded chromosomes (3, 10)]. Other hypotheses that have been put forward are the formation of multipolar spindles (5), spatial separation of genomes during interphase (11, 12), and genome elimination by nuclear extrusions (4, 13). In addition, degradation of alien chromosomes by host-specific nucleases (14), uniparental nondisjunction of anaphase chromosomes (15), and parent-specific inactivation of centromeres (11, 16–19) have been suggested. The actual cellular mechanism involved in the process of uniparental chromosome elimination remains poorly understood.
To test whether parent-specific inactivation of centromeres is involved in the mitosis-dependent process of chromosome elimination in interpecific hybrids, we analyzed the centromere-specific histone H3 variant (CENH3) [originally called CENP-A in humans (20) and HTR12 in Arabidopsis thaliana (21)] in chromosomally unstable and stable Hordeum vulgare × H. bulbosum combinations. CENH3 was selected for our study because in mammals (22), Caenorhabditis elegans (23), and Drosophila melanogaster (24), its loss results in the failure of centromere formation and chromosome segregation. A region in CENH3 defined as the centromere targeting domain (CATD) is critical for centromeric localization of CENH3 in various species (25–27). The CATD is composed of the loop1 linker and α2-helix of CENH3 (28, 29), and its substitution enabled the incorporation of an H3 chimera into centromeres (26). This domain mediates molecular recognition events before and after nucleosome assembly and is important for binding of CENH3 to centromeric DNA (27, 28, 30), to CENH3-specific chaperones (31–33), and to CENH3-stabilizing factors (34, 35).
Although centromeric DNA sequences are extremely diverse, all eukaryotic centromeres contain CENH3 (36). The chromosomal location of CENH3 is the assembly site for the kinetochore complex of active centromeres. Any error in histone gene transcription, translation, modification, or import could affect the ability to assemble intact CENH3 chromatin, which would result in the loss of CENH3 from centromeres and, hence, of centromere identity (reviewed in 37). In contrast to conventional histones, CENH3 is rapidly evolving and shows signatures of adaptive evolution in some species (38). ChIP data indicated that CENH3 interacts with H. vulgare with CEREBA, a centromeric retroelement-like element conserved among cereal centromeres, and with H. bulbosum- and H. vulgare-specific GC-rich centromeric satellite sequences (39).
The present work provides insights into the role of CENH3 in the process of selective elimination of paternal chromosomes during the development of H. vulgare × H. bulbosum hybrid embryos. In addition, we identified two functional CENH3s in both diploid barley species and assayed their incorporation into centromeres of alien chromosomes. We found that despite the presence of transcripts, not all parental CENH3 variants are incorporated into centromeres if multiple CENH3s coexist in species combinations. Thus, the lack of cross-species CENH3 incorporation might act as a barrier to species hybridization.
Results
Uniparental Elimination of Chromosomes in Unstable H. vulgare × H. bulbosum Hybrids Is Accompanied by Loss of CENH3 from Centromeres.
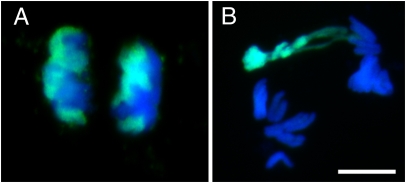
The fertilization of the H. vulgare egg by the H. bulbosum sperm is followed by complete elimination of the H. bulbosum genome. Depending on the genotype and on environmental conditions, most of the H. bulbosum chromatin is eliminated after pollination in almost all embryos within 1 wk (2). We first analyzed the mitotic behavior of H. bulbosum chromosomes in dividing cells of unstable hybrid embryos derived from H. vulgare × H. bulbosum (Cb2920/4). To promote chromosome elimination, a temperature above 18 °C was used for cultivation of pollinated H. vulgare plants. In addition to H. bulbosum chromosomes with normal mitotic movements (Fig. 1A), between 20% and 70% of anaphase cells showed abnormally segregating H. bulbosum chromosomes (Fig. 1B). Such chromosomes of H. bulbosum lagged behind H. vulgare chromosomes, and the sister chromatids segregated asymmetrically at anaphase. As previously described (4), the level of mitotic chromosome condensation partly differed between the parental genomes; chromosomes of H. bulbosum were often less condensed. These observations are consistent with a loss of paternal chromosomes during cell division via lagging chromosomes that later form micronuclei (3).
Fig. 1.
Anaphase chromosome segregation behavior of normally segregating (A) and lagging (B) H. bulbosum chromosomes in an unstable H. vulgare × H. bulbosum hybrid embryo. Chromosomes of H. bulbosum (green) were identified by genomic in situ hybridization using labeled genomic DNA of H. bulbosum. Chromosomes of H. vulgare are shown in blue. Note the lagging chromosomes of H. bulbosum in B. (Scale bar: 10 μm.)
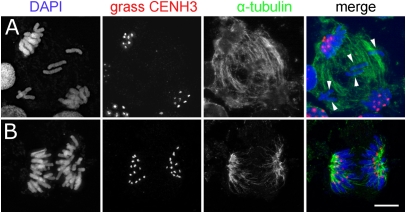
The centromere activity of hybrid cells undergoing uniparental chromosome elimination was determined via immunostaining with grass CENH3- and α-tubulin–specific antibodies. Before that, the cross-reactivity of the anti-grass CENH3 antibody used (40) with CENH3 of H. bulbosum was confirmed (Fig. S1 A and B). Fig. 2A shows typical results obtained in anaphase cells in 3- to 6-d-old unstable hybrid embryos. CENH3-positive active centromeres were found in segregating chromatids, whereas lagging chromosomes were depleted of CENH3 (Fig. 2A and Fig. S2 A–C). Neither a primary constriction nor a clear interaction between α-tubulin fibers and kinetochores of lagging H. bulbosum chromosomes was recognizable. As a control, all centromeres of stable H. vulgare × H. bulbosum (Cb3811/3) hybrids cultivated below 18 °C displayed CENH3 signals (Fig. 2B).
Fig. 2.
Anaphase chromosomes of an unstable (A) and stable (B) H. vulgare × H. bulbosum hybrid embryo after immunostaining with anti-grass CENH3 and anti–α-tubulin. The centromeres of lagging chromosomes (arrowheads) are CENH3-negative. (Scale bar: 10 μm.)
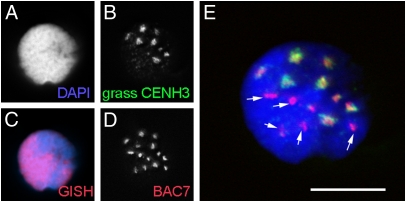
In addition, embryonic nuclei of unstable hybrids displayed two classes of CENH3 signals. Approximately seven discrete CENH3 signals of strong intensity, together with CENH3 signals of less intensity, per nucleus were found (Fig. 3 B and E). Subsequent FISH with differentially labeled genomic DNA of H. bulbosum (Fig. 3C) and with the Hordeum centromere-specific probe BAC 7 (41) (Fig. 3D) confirmed the hybrid nature of those nuclei. Only approximately 7 of the 14 more or less equally sized centromeric FISH signal clusters overlapped with the position of strong CENH3 signals (Fig. 3E), suggesting that interphase centromeres of H. bulbosum also carry less CENH3 protein. Thus, centromere inactivity of H. bulbosum chromosomes, by means of a loss of CENH3, triggers the mitosis-dependent process of uniparental chromosome elimination in unstable H. vulgare × H. bulbosum hybrids.
Fig. 3.
Interphase nucleus of an unstable H. vulgare × H. bulbosum hybrid embryo (A) after immunostaining with anti-grass CENH3 (B), genomic in situ hybridization with H. bulbosum DNA (C, red), and in situ hybridization with the Hordeum centromere-specific probe BAC7 (D). GISH, genomic in situ hybridization. (E) Only approximately 7 of the 14 more or less equally sized centromeric FISH signal clusters are overlapping with the position of strong CENH3 signals. Hence, interphase centromeres of H. bulbosum carry less CENH3 protein. BAC7-positive centromeres without CENH3-signals are shown (arrows). (Scale bar: 10 μm.)
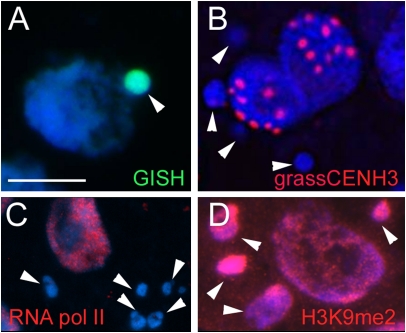
At the end of mitosis during reformation of nuclear membranes, lagging chromosomes form micronuclei (42, 43). To determine the centromeric and transcriptional activity of micronucleated H. bulbosum chromatin (Fig. 4A), the presence of CENH3 and of the RNA polymerase II was assayed. Most micronuclei were CENH3-negative (Fig. 4B), and only occasionally were weak CENH3 immunosignals seen. RNA polymerase II signals were distributed homogeneously throughout the nucleoplasm of normal nuclei (Fig. 4C), whereas micronuclei were weakly immunolabeled (Fig. 4C). Because heterochromatinization of eliminated chromatin is a well-known phenomenon in many nonplant organisms (44), we tested next whether micronuclei are enriched in the heterochromatin marker histone H3 dimethylated at lysine 9 (H3K9me2). Around 60% of 233 micronuclei analyzed revealed an enhanced level of H3K9me2 compared with normal nuclei (Fig. 4D). Hence, the transcriptional activity of micronucleated chromatin is strongly reduced. Because the centromeres of micronucleated chromosomes are inactive, no further segregation of H. bulbosum chromosomes is possible after micronucleus formation.
Fig. 4.
Characterization of micronuclei of unstable H. vulgare × H. bulbosum hybrid embryos. Micronuclei are H. bulbosum-positive after genomic in situ hybridization (A) CENH3-negative (B) and RNA polymerase II-negative (C) after immunostaining but enriched in H3K9me2-specific heterochromatin-specific markers (D). Arrowheads indicate micronuclei. (Scale bar: 10 μm.)
Both Hordeum Species Encode Two Active Variants of CENH3.
Because silencing or biased expression of homologous genes has been well documented in both natural and synthetic hybrids (45), we first isolated the corresponding genes with the aim of testing whether the CENH3 gene of H. bulbosum undergoes silencing in unstable hybrids.
Initial RT-PCR using a monocotyledon-specific CENH3 primer pair (46) (Table S1, primer pair 1/2) for isolating on Hordeum CENH3 amplified parts of the two CENH3 variants in H. vulgare called HvαCENH3 and HvβCENH3. BLAST analysis against the National Center for Biotechnology Information database resulted in the identification of the 5′ region of HvαCENH3 (GenBank accession no. BU996921). The coding sequence of HvβCENH3 (GenBank accession no. AK249602) was completed by 3′ RACE PCR using primer 3 (Table S1). Based on the H. vulgare CENH3 sequences, primers were designed to amplify both CENH3s of H. bulbosum, named HbαCENH3 and HbβCENH3 (Table S1, primer pairs 14/15 and 1/2 for HbαCENH3 and primer pair 6/7 and primer 3 for HbβCENH3 were used for RACE PCR). The deduced amino acid sequences of the identified CENH3s [GenBank accession nos. JF419328 (HvαCENH3), JF419329 (HvβCENH3), GU245882.1 (HbαCENH3), and JF419330 (HbβCENH3)] were compared with CENH3 of maize (47), rice (40), and sugar cane (48) to determine the conserved αN-helix, α1-helix, α2-helix, α3-helix, and CENH3-specific (26) loop1 regions (Fig. S3A). Despite the first five completely conserved amino acids, the N-terminal tails were highly variable. βCENH3 types have a shorter N-terminal region than αCENH3 types and a longer loop1 region.
Phylogenetic analysis of the amino acids of Hordeum CENH3s with further plant CENH3s (Fig. S3B) showed that αCENH3s and βCENH3s of Hordeum species form two distinct subclusters. In addition, Hordeum αCENH3s cluster with the CENH3s of rice, maize, and sugar cane. Therefore, it is likely that α and β types of CENH3 diverged before the speciation of H. vulgare and H. bulbosum. Chromosome mapping of CENH3s confirmed this assumption (Fig. S4). αCENH3 is encoded by chromosome 1H in H. vulgare and in H. bulbosum; CENH3 of rice maps to chromosome 5, which is syntenic to barley 1H (49). Furthermore, βCENH3 is located on chromosome 6H in both Hordeum species.
CENH3s of Both Parental Genomes Are Transcribed in Chromosomally Stable and Unstable Hybrid Embryos.
To test whether H. bulbosum-specific CENH3 inactivation occurs, the transcriptional activity of parental CENH3s in unstable hybrids was determined. Stable and unstable hybrid embryos of different ages (5–7 d after pollination) were isolated, two to five embryos of each age and type were pooled, and RNA was isolated for cDNA synthesis. To detect transcripts, we used αCENH3 (primer pair 4/5) or βCENH3 (primer pair 6/7) type-specific primer pairs. H. vulgare- and H. bulbosum-derived CENH3 transcripts were discriminated via species-specific restriction sites. AlwI cleavage of the H. bulbosum-derived PCR products generates two fragments of 210 bp and 54 bp, leaving H. vulgare-derived αCENH3 fragments undigested (Fig. S5A). Similarly, BanII cleavage of H. bulbosum-derived βCENH3 amplicons results in fragments of 398 bp and 234 bp, whereas H. vulgare-derived transcripts are unaffected (Fig. S5B). BanII and AlwI cleavage of CENH3 transcripts amplified from stable or unstable H. vulgare × H. bulbosum hybrid revealed transcription of all four parental CENH3s (Fig. S5 A and B). The expression patterns were similar regardless of the embryo age. Thus, all CENH3 variants of both parental genomes undergo transcription in unstable hybrids, and uniparental silencing of HbCENH3 genes is not the cause of chromosome elimination in unstable hybrids.
Both CENH3 Variants Intermingle in Mitotic and Meiotic Centromeres of H. vulgare.
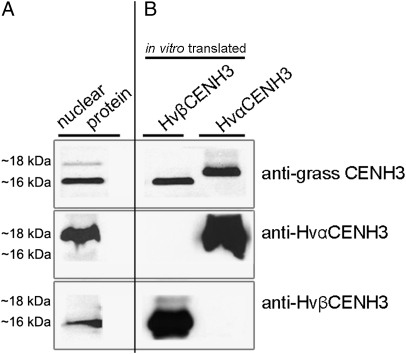
Duplication and maintenance of two or more copies of CENH3 seem to be rare events in both plant and animal species. Examples of diploid plants that possess two CENH3s are Arabidopsis halleri, Arabidopsis lyrata (50), and Luzula nivea (51). Both variants are transcribed, but it is not clear whether they are also functional. To characterize the chromosomal distribution of multiple CENH3s in a diploid organism, antibodies were generated for each variant of H. vulgare CENH3. Western blot analysis using in vitro translated HvαCENH3 and HvβCENH3 confirmed the specificity of both antibodies (Fig. 5) because each type of antibody recognized only the corresponding CENH3 variant, whereas anti-grass CENH3 reacted with both types of CENH3.
Fig. 5.
Western blot analysis demonstrating the specificity of anti-HvαCENH3-, anti-HvβCENH3-, and anti-grass CENH3-specific antibodies on nuclear (A) and in vitro translated HvαCENH3 and HvβCENH3 proteins (B).
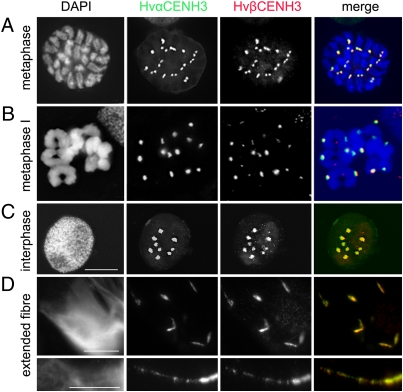
To determine the chromosomal distribution of αCENH3 and βCENH3 during mitosis and meiosis, immunofluorescence experiments were done. Double immunostaining revealed that αCENH3 and βCENH3 are present in the centromeres of all barley chromosomes at all mitotic stages analyzed (Fig. 6A and Fig. S6A). Signals of both CENH3 variants always overlapped with the position of the primary constriction. A similar distribution of both CENH3 variants was also detected at different stages of the first and second meiotic divisions (Fig. 6B and Fig. S6B). The amount of αCENH3 and βCENH3 proteins seems to be comparable, because the intensity of corresponding immunofluorescence signals was generally of similar intensity.
Fig. 6.
Distribution of αCENH3 and βCENH3 in mitotic (A) and meiotic (B) metaphase chromosomes and interphase nuclei of H. vulgare (C) as demonstrated by immunostaining. An overlap of αCENH3 and βCENH3 signals was found for centromeres as well as for (D) up to 12-fold artificially extended centromeres. (Scale bars: 10 μm.)
To decipher the organization of αCENH3- and βCENH3-containing centromeric chromatin at higher resolution, immunostaining experiments were done on extended chromatin fibers. For this purpose, chromatin fibers prepared from isolated nuclei of barley leaves were immunolabeled with both CENH3 antibodies. An overlap of αCENH3- and βCENH3-specific signals was observed on centromeres (Fig. 6C) but also on up to 12-fold artificially extended centromeres (Fig. 6D). We conclude that centromeres can incorporate αCENH3 and βCENH3 in all chromosomes at mitotic and meiotic stages. Both CENH3 variants are intermingled in centromeric chromatin and are apparently equally involved in the formation of barley centromeres.
Although Transcribed, Not All CENH3 Variants Are Deposited on Centromeres if Multiple CENH3s Coexist in Species Combinations.
To investigate whether both CENH3s of H. vulgare are incorporated into centromeres of alien chromosomes, we studied stable H. vulgare × H. bulbosum and Triticum aestivum × H. vulgare combinations. First, we confirmed the species specificity of the barley CENH3 antibodies. Both antibodies did not cross-react with H. bulbosum or T. aestivum CENH3, whereas anti-grass CENH3 recognized the corresponding protein in all species tested (Fig. S7). Next, double-immunostaining experiments were performed on root nuclei of a stable H. vulgare × H. bulbosum hybrid plant with anti-grass CENH3 (as a positive control) combined with either HvαCENH3- or HvβCENH3-specific antibodies. Up to14 HvαCENH3 and HvβCENH3 signals overlapped with the position of anti-grass CENH3 signals (Fig. S8A). Hence, both CENH3 variants of H. vulgare incorporate equally well into the centromeric nucleosomes of H. bulbosum.
Next, a stable H. vulgare-H. bulbosum 7H substitution line was studied. Chromosome 7H of H. bulbosum does not encode any CENH3 gene. The genotype of the H. vulgare-H. bulbosum 7H substitution line was confirmed by in situ hybridization (Fig. S8C). After immunostaining, for each CENH3 antibody combination, up to 14 overlapping discrete signals per nucleus were detected (Fig. S8B). Hence, αCENH3 and βCENH3 of H. vulgare can functionally compensate for the missing CENH3 of H. bulbosum.
We then posed the question of whether barley centromeres still specifically incorporate HvCENH3 when barley chromosomes carrying CENH3 genes are added to the genome of T. aestivum, a species less closely related to H. vulgare than H. bulbosum. When hybrids are formed, the centromere-specific histones of each parental species may operate in the context of a dual set of centromeric sequences. We used a wheat-barley double-disomic 1H + 6H addition line. RT-PCR demonstrated (αCENH3: primer pair 8/9, βCENH3: primer pair 3/10) the presence of both H. vulgare CENH3 transcripts in a T. aestivum background (Fig. S8D). Finally, double immunostaining on nuclei of the characterized wheat-barley addition lines with grass CENH3 (as a positive control), and HvαCENH3- or HvβCENH3-specific antibodies was performed. Anti-HvαCENH3 and anti-grass CENH3 each detected up to 46 overlapping signals per nucleus (Fig. S8E), but no centromeric incorporation of HvβCENH3 was found in nuclei of wheat-barley 1H + 6H plants. Thus, no species-specific incorporation of CENH3 occurs if CENH3 of both parents coexists in stable hybrids. However, not all parental CENH3 variants necessarily undergo centromere incorporation if multiple CENH3s coexist.
Discussion
Diploid Hordeum Species Encode Two Functional CENH3 Variants.
This report describes two functional CENH3 variants in diploid grasses. Because two CENH3 variants exist in H. vulgare and H. bulbosum genes, duplication of αCENH3 must have occurred at least 7 million years ago, the time when H. vulgare and H. bulbosum diverged (52). An alternative explanation is that the second gene variant is the remainder of an earlier whole-genome duplication that occurred 20 million years before the divergence of Oryza, Brachypodium, and Hordeum from a common ancestor that existed ∼41–47 million years ago (53). Because Oryza sativa and Brachyopodium distachyon encode only one CENH3 variant each, it is possible that the second CENH3 variant in these grasses was lost. Loss of a second CENH3 gene copy also occurred in Zea mays (47). In maize, the allotetraploidization event probably occurred ∼11.4 million years ago (54). On the other hand, in other allopolyploid organisms, such as Arabidopsis suecica (21), wild rice (55), or Nicotiana tabacum (56), multiple CENH3 types were identified.
Despite considerable protein sequence differences in the CATD region, both CENH3 variants of H. vulgare are detectable in the centromeric nucleosomes of all mitotic, meiotic, and extended interphase chromosomes of barley. This observation suggests at least two possibilities regarding the centromeric localization of two CENH3 variants. Either both CENH3 variants are incorporated into the same nucleosomes forming heterodimers or barley centromeres are composed of alternate blocks of αCENH3- and βCENH3-containing nucleosomes. The centromeric composition of alternating blocks of CENH3- and H3-containing nucleosomes has previously been demonstrated for metazoan organisms (36, 57) and plants (58). However, because of the limited resolution of light microscopy, our immunostaining experiment on extended chromatin fibers does not exclude the possible formation of αCENH3- and βCENH3-containing heterodimers.
CENH3 Behavior in Stable Species Combinations.
Our analysis demonstrates that cross-species incorporation of CENH3s occurs in stable species combinations, because both αCENH3 and βCENH3 of H. vulgare were detected in all centromeres of stable interspecific H. vulgare × H. bulbosum plants. However, because antibodies specific for CENH3s of H. bulbosum were not available, it was not possible to determine whether CENH3s of H. bulbosum are incorporated into the centromeres of H. vulgare × H. bulbosum hybrids. A different situation was observed for αCENH3 and βCENH3 of H. vulgare in wheat-barley 1H + 6H addition lines. Despite transcription of both barley CENH3 variants, only HvαCENH3-specific immunosignals were detected. Because CENH3s of wheat are probably more similar in sequence to αCENH3 of H. vulgare, HvαCENH3 was preferentially incorporated into the centromeres of wheat and barley.
After sexual hybridization, the CENH3 of one parent can promote centromere functionality of the second parent despite centromere sequence differences. However, the CENH3 sequence dissimilarity must not be too severe. This finding is strongly supported by the availability of a large number of chromosome addition lines. Even remotely related species, such as oat and maize, can be sexually hybridized to produce fertile partial hybrids (59). In this species combination, CENH3 of oat compensates for the missing CENH3 of maize, because the maize CENH3 gene is silenced in the genetic background of oat in oat-maize chromosome addition lines (16). Similarly, the CENH3 protein from A. thaliana can be detected in the centromeres of all chromosomes of allotetraploid A. thaliana × Arabidopsis arenosa hybrid A. suecica (21). Our observation based on sexually generated hybrids supports previous cross-species CENH3 incorporation experiments in transgenic organisms (27, 51, 60–62), for which it was reported that CENH3s of closely related species can target centromeres in alien species.
It will be interesting to study how many different CENH3 variants are incorporated into centromeres of polyploid species. To what degree does the cross-capability between species depend on the ability of centromeres to incorporate different parental CENH3 variants? Does each CENH3 variant use its own set of assembly factors in hybrids? However, in a transgenic situation, CENH3 of a closely related species could get incorporated alone (62) or in combination with WT CENH3 (51, 60) with help from the loading machinery of the host organism. Transgenic complementation of an inactive version of CENH3 was only possible if the heterologous CENH3 originated from a closely related species (62). For instance GFP-CENH3 from the close relative A. arenosa complemented A. thaliana cenh3-1, which is consistent with the observation that A. thaliana CENH3 localizes to both A. thaliana and A. arenosa chromosomes in the allopolyploid species A. suecica (21). However, GFP-CENH3 from a closely related Brassica species was targeted to centromeres but did not complement cenh3-1, indicating that kinetochore localization and centromere function of alien CENH3 can be uncoupled (62).
Centromeric Loss of CENH3 Protein Is Involved in the Process of Uniparental Chromosome Elimination.
CENH3 immunostaining of H. vulgare × H. bulbosum embryos demonstrated that uniparental centromere inactivation is, as previously postulated (11, 16–19), a cause of mitosis-dependent chromosome elimination in wide hybrids. Active centromeres of H. vulgare and H. bulbosum were CENH3-positive, whereas inactive H. bulbosum centromeres were CENH3-negative or the amount of CENH3 was reduced in unstable hybrids.
Elimination of H. bulbosum chromosomes is completed by 5–9 d after pollination (3–5). Because CENH3 is a stable protein (22), sperm-derived centromere-incorporated CENH3 proteins are likely to provide a residual kinetochore function of H. bulbosum until it falls below a level critical for correct chromosome segregation, resulting in chromosome elimination. That a limited fraction of parental H3 variants is transmitted to the progeny has been demonstrated for animal embryos (22). If preexisting CENH3 is partitioned equally between duplicated sister centromeres and no de novo incorporation of CENH3 into H. bulbosum centromeres occurs, its amount will be approximately halved with each cell division. In humans, even 10% of the endogenous CENH3 can aid in efficient kinetochore assembly (63). On the other hand, a reduced CENH3 amount can trigger centromere inactivity in neocentromeres of maize (64). Nevertheless, if a zygotic resetting of CENH3, as demonstrated for GFP-tagged CENH3 in A. thaliana (65, 66), also exists in grasses, active removal of both parental CENH3s before the first zygote division would occur and the reactivation of H. bulbosum centromeres in unstable hybrids would be diminished.
Despite transcription of all parental CENH3 genes in unstable H. vulgare × H. bulbosum hybrids and the existence of cross-species incorporation of CENH3s in stable hybrids, the centromeres of H. bulbosum in unstable hybrids had a reduced content of CENH3. Therefore, we assume that in unstable H. vulgare × H. bulbosum hybrids, no incorporation of CENH3 into the centromeres of H. bulbosum takes place. The regulation of CENH3 loading and assembly into centromeres is mediated by a number of proteins, and the erroneous function of any of these will result in a nonfunctional centromere (reviewed in 67). For example, inactivation of the metazoan CENH3 interacting partner CENP-H, CENP-I, CENP-K, or CENP-N results in reduced assembly of nascent CENH3 into centromeric chromatin and causes defects in kinetochore assembly and chromosome congression (34, 68). However, with the exception of Mis12 of A. thaliana (69), no other protein involved in the establishment and maintenance of CENH3 chromatin has been identified in plants.
In unstable hybrids, we noted a different degree of condensation between both parental chromosomes. The condensation process of H. bulbosum chromosomes was often delayed (4). In agreement with our observation, Bennett et al. (3) reported that H. bulbosum requires more time to complete the cell cycle than H. vulgare. Because the correct time of CENH3 deposition seems to be essential (25), cell cycle asynchrony (e.g., attributable to genotypic differences) might interfere with the loading of H. bulbosum nucleosomes with CENH3 in unstable hybrids. This, however, does not exclude the possibility that other factors may result in the failure to assemble active H. bulbosum centromeres in unstable hybrids. Notably, in interspecific somatic hybrids of different mammals, predominant loss of one parental genome occurs (reviewed in 70). However, it is not known whether a comparable process of chromosome elimination via loss of CENH3 is evolutionarily conserved and would also occur in animal hybrids.
The elimination process of chromosomes can be influenced by environmental conditions (8). A temperature above 18 °C during the early stages of hybrid embryo growth can promote chromosome elimination. How does temperature influence the process of chromosome elimination? Temperature-mediated changes in nucleosome composition via altered deposition of histone variants by chaperons have recently been demonstrated for plants (71). If temperature-mediated changes in centromeric nucleosome assembly occur, the temperature effect on the process of uniparental chromosomes could be explained. However, it is not known whether chaperones involved in CENH3 loading are temperature sensitive.
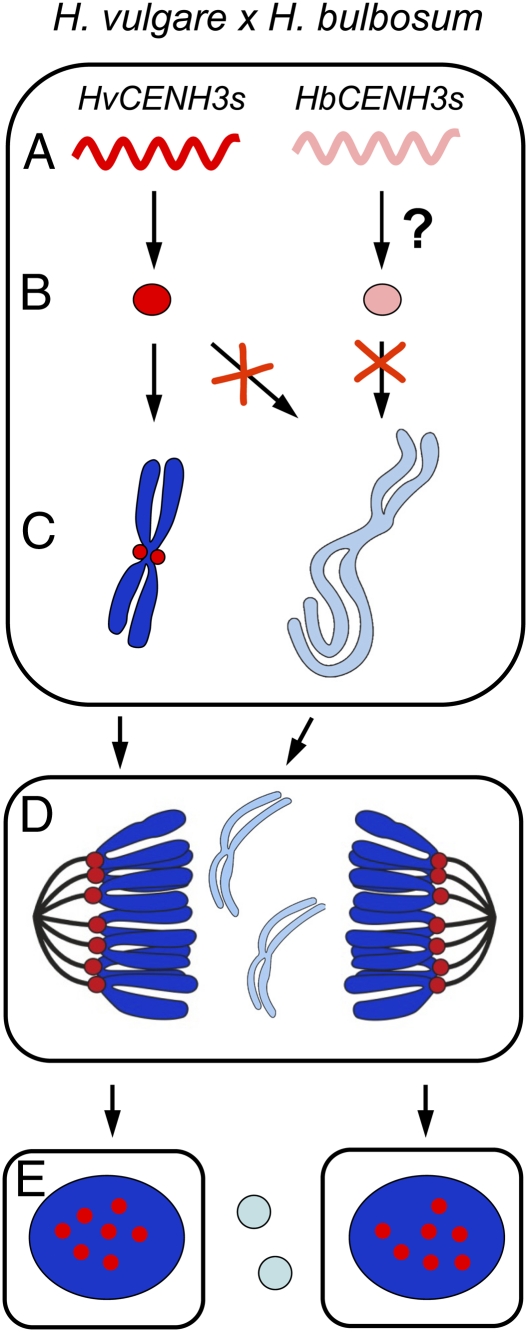
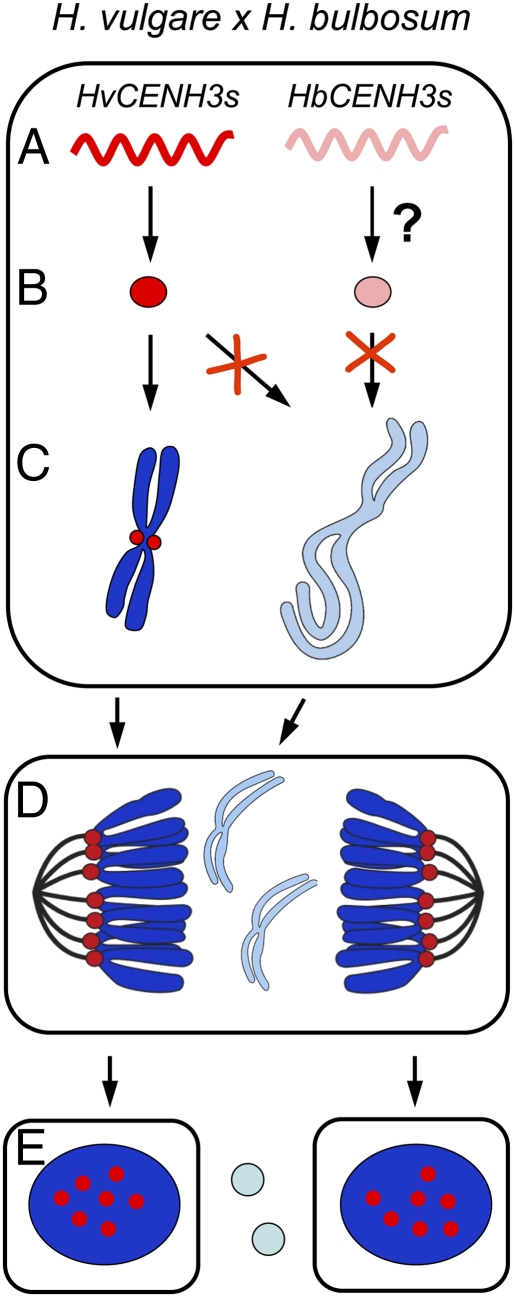
On the basis of the above-mentioned observations, we propose a possible model of how the mitosis-dependent process of uniparental chromosome elimination works in H. vulgare × H. bulbosum hybrid embryos (Fig. 7). After fertilization of the H. vulgare egg by the H. bulbosum sperm, all parental CENH3 genes are transcriptionally active. Translation of HvCENH3s occurs, but whether translation of HbCENH3s takes place is unknown. HvCENH3 is then loaded into the centromeres of H. vulgare but not of H. bulbosum. Because of cell cycle asynchrony of the two parental genomes, CENH3 incorporation probably only occurs in the centromeres of H. vulgare during G2. A low temperature during early embryogenesis supports centromere activity of both parental genomes. In unstable hybrids, H. bulbosum chromosomes lag because of centromere inactivity during anaphase, subsequently forming micronuclei. Finally, micronucleated H. bulbosum chromatin will degrade, and a haploid H. vulgare embryo will develop. Whether a comparable haploidization process takes place in A. thaliana cenh3-1 null mutants expressing altered CENH3 proteins that were crossed to WT plants remains to be studied (61).
Fig. 7.
Proposed model of how the mitosis-dependent process of uniparental chromosome elimination operates in H. vulgare × H. bulbosum hybrid embryos. (A) After fertilization of the H. vulgare egg by the H. bulbosum sperm, all parental CENH3s are transcriptionally active. (B) Translation of HvCENH3s occurs; whether translation of HbCENH3s occurs is not known. (C) Loading of CENH3 (red) into the centromeres of H. vulgare but not of H. bulbosum occurs. As a result of cell cycle asynchrony of the two parental genomes, CENH3 incorporation probably only occurs in the centromeres of H. vulgare during G2. A reduced temperature during early embryogenesis promotes normal centromere activity of both parental genomes. (D) As a result of centromere inactivity in unstable hybrids, anaphase chromosomes of H. bulbosum lag and subsequently form micronuclei. (E) Micronucleated H. bulbosum chromatin finally degrades, and a haploid H. vulgare embryo will develop.
In summary, we report four major conclusions regarding the role of CENH3 in chromosomally stable and unstable interspecific combinations (1). Diploid barley species encode two CENH3 variants whose gene products are intermingled throughout mitotic and meiotic centromeres (2). In stable species combinations, cross-species incorporation of CENH3 occurs despite centromere-sequence differences. However, not all CENH3 variants get incorporated into centromeres if multiple CENH3s are present in species combinations (3). Centromere inactivity of H. bulbosum chromosomes triggers the mitosis-dependent process of uniparental chromosome elimination in unstable H. vulgare × H. bulbosum hybrids (4). Centromeric loss of CENH3 protein rather than uniparental silencing of CENH3 genes is causing centromere inactivity.
Materials and Methods
Plant Material, Crossing Procedure, and Preparation of Embryos.
A series of crosses were made between diploid H. vulgare “Emir” (female parent) and diploid H. bulbosum lines Cb2920/4 and Cb3811/3 (pollen donors) (72). For H. vulgare plants, two environments were used with different temperatures to control chromosome elimination after pollination. A temperature higher than 18 °C supports chromosome elimination, whereas a temperature lower than 18 °C promotes retention of the parental chromosomes after pollination with H. bulbosum (8). Barley spikes were emasculated 1–2 d before anthesis and pollinated 1 d later with freshly collected H. bulbosum pollen. Embryo development was stimulated by spraying the spikes with an aqueous solution of gibberellic acid at a rate of 75 ppm 1 d after pollination (details are provided in SI Materials and Methods). For subsequent in situ hybridization or indirect immunostaining, 6- to 13-d-old ovaries were isolated and fixed in ethanol/acetic acid (3:1) or 4% (wt/vol) paraformaldehyde in 1× microtubule-stabilizing buffer (MTSB), respectively. Embryos were isolated under a stereomicroscope using fine needles.
Different wheat-barley addition lines as well as barley-H. bulbosum substitution lines were used for mapping of CENH3 coding genes (details are provided in Fig. S4).
Isolation of Nuclei and Preparation of Stretched Chromatin Fibers.
Nuclei were isolated according to the method of Galbraith et al. (73). Extended chromatin fibers were prepared as described (39). The immunostaining procedure on the stretched chromatin fibers or on nuclei was the same as for squashed cells.
FISH and Indirect Immunostaining.
FISH and indirect immunostaining were carried out as described by Gernand et al. (4) and Ma et al. (74), respectively. FISH probes were labeled with Atto-590-dUTP or Atto-488-dUTP (Jena Bioscience) by nick translation. The following primary antibodies were used: rabbit anti-RNA polymerase II CDC phospho Ser5 (catalog no. 39233; diluted 1:100; ACTIVE MOTIF), rabbit anti-histone H3K9me2 (catalog no. 07-441, diluted 1:300; Upstate), mouse anti–α-tubulin (catalog no. T 9026, diluted 1:100; Sigma), and rabbit anti-grass CENH3 (40) (diluted 1:100), as well as HvαCENH3- and HvβCENH3-specific antibodies (diluted 1:100). Epifluorescence signals were recorded with a cooled CCD-camera (ORCA-ER; Hamamatsu). Imaging was performed using an Olympus BX61 microscope and an ORCA-ER CCD camera (Hamamatsu). Deconvolution microscopy was used for superior optical resolution of globular structures. All images were collected in gray scale and pseudocolored with Adobe Photoshop 6 (Adobe). Projections (maximum intensity) were done with the program AnalySIS (Soft Imaging System).
RNA Isolation, RT-PCR, and RACE PCR.
RNA was extracted by either the TRIzol method (75) or using the PicoPure RNA Isolation Kit (Arcturus). cDNA was prepared from DNase I-treated RNA using the Reverse Aid H Minus First Strand cDNA Synthesis Kit (Fermantas). PCR included the following steps: 94 °C for 3 min, 40 cycles at 94 °C for 1 min, annealing temperature for 1 min and 10 s, and 72 °C for 1 min and 30 s. Primers and annealing temperatures are given in Table S1. For RACE PCR, the SMART RACE cDNA Amplification Kit (Clontech Company) was used.
Analysis of PCR Fragments and Cleaved Amplified Polymorphic Sequence Analysis.
DNA fragments were ligated into the pGEM-T easy vector (Promega) and sequenced using the PGRC Sequencing Service (Leibnitz Institute of Plant Genetics and Crop Plant Research). For cleaved amplified polymorphic sequence analysis, purified PCR products were digested using AlwI or BanII, size-fractionated by gel electrophoresis, and recorded.
Generation of H. vulgare CENH3-Specific Antibodies.
Suitable CENH3 type-specific epitopes were identified [CQRRQETDGAGTSETPRRAGR and CAEGAPGEPTKRKPHRFR for generating HvαCENH3-specific antibodies (a double-peptide antibody) and CSKSEPQSQPKKKEKRAYR for generating HvβCENH3-specific antibodies]. The corresponding peptides were synthesized and used to immunize guinea pigs for generation of anti-HvαCENH3 antibodies and rabbits for generation of anti-HvβCENH3 antibodies to analyze both CENH3 types simultaneously. Peptide synthesis, immunization of animals, and peptide affinity purification of antisera were performed by Pineda (Antikörper-Service).
In Vitro Protein Synthesis and Western Blot Analysis.
For in vitro protein synthesis, the PURExpress cell-free transcription-translation system (New England Biolabs) was used. Quantified protein samples were separated on 10% (wt/vol) SDS/PAGE gels using Tricin-SDS/PAGE running buffers (76), blotted on nitrocellulose membranes, and then incubated first with primary antibodies (diluted 1:1,000) and then with secondary antibodies [anti-rabbit IgG: IRDye800 conjugated, diluted 1:5,000 (LI-COR); anti-guinea pig IgG: HRP-conjugated, diluted 1:5,000 (Dianova)].
Phylogenetic Analysis.
Evolutionary analyses was conducted using deduced amino acid sequences of CENH3 in Z. mays, Saccharum officinarum, O. sativa, A. thaliana, and α and β types of H. vulgare and H. bulbosum. Phylogenetic relationships were estimated by the neighbor joining method (Lasergene).
Supplementary Material
Acknowledgments
We thank members of the Cytogenetics Department of the Leibnitz Institute of Plant Genetics and Crop Plant Research for helpful comments on the manuscript. Further, we thank Lu Ma for performing the FISH experiments and A. K. M. R. Islam (Australia) and T. R. Endo (Japan) for providing the wheat-barley addition lines. This work was supported by the Deutsche Forschungsgemeinschaft (Grant HO 1779/9-1) and Marie Curie Actions (Grant 219313).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GeneBank data base (accession nos. JF419328, JF419329, GU245882.1, and JF419330).
See Commentary on page 13361.
1R.P. has been retired since 2010.
See Author Summary on page 13373.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103190108/-/DCSupplemental.
References
- 1.Kasha KJ, Kao KN. High frequency haploid production in barley (Hordeum vulgare L.) Nature. 1970;225:874–876. doi: 10.1038/225874a0. [DOI] [PubMed] [Google Scholar]
- 2.Devaux P, Pickering R. Haploids in the improvement of poaceae. In: Palmer CE, Keller WA, Kasha KJ, editors. Haploids in Crop Improvement II, Biotechnology in Agriculture and Forestry Series. Vol. 56. Berlin: Springer; 2005. pp. 215–242. [Google Scholar]
- 3.Bennett MD, Finch RA, Barclay IR. The time rate and mechanism of chromosome elimination in Hordeum hybrids. Chromosoma. 1976;54:175–200. [Google Scholar]
- 4.Gernand D, Rutten T, Pickering R, Houben A. Elimination of chromosomes in Hordeum vulgare x H. bulbosum crosses at mitosis and interphase involves micronucleus formation and progressive heterochromatinization. Cytogenet Genome Res. 2006;114:169–174. doi: 10.1159/000093334. [DOI] [PubMed] [Google Scholar]
- 5.Subrahmanyam NC, Kasha KJ. Selective chromosome elimination during haploid formation in barley following interspecific hybridization. Chromosoma. 1973;42:111–112. [Google Scholar]
- 6.Symko S. Haploid barley from crosses of Hordeum bulbosum (2x) by Hordeum vulgare (2x) Can J Genet Cytol. 1969;11:602–608. [Google Scholar]
- 7.Humphreys MW. Chromosome instability in Hordeum vulgare x H. bulbosum hybrids. Chromosoma. 1978;65:301–307. [Google Scholar]
- 8.Pickering RA. Partial control of chromosome elimination by temperature in immature embryos of Hordeum vulgare L. x H. bulbosum. Euphytica. 1985;14:869–874. [Google Scholar]
- 9.Gupta SB. Duration of mitotic cycle and regulation of DNA replication in Nicotiana plumbaginifolia and a hybrid derivative of N. tabacum showing chromosome instability. Can J Genet Cytol. 1969;11:133–142. [Google Scholar]
- 10.Laurie DA, Bennett MD. The timing of chromosome elimination in hexaploid wheat x maize crosses. Genome. 1989;32:953–961. [Google Scholar]
- 11.Finch RA. Tissue-specific elimination of alternative whole parental genomes in one barley hybrid. Chromosoma. 1983;88:386–393. [Google Scholar]
- 12.Linde-Laursen I, von Bothmer R. Orderly arrangement of the chromosomes within barley genomes of chromosome-eliminating Hordeum lechleri x barley hybrids. Genome. 1999;42:225–236. [Google Scholar]
- 13.Gernand D, et al. Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell. 2005;17:2431–2438. doi: 10.1105/tpc.105.034249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies DR. Chromosome elimination in inter-specific hybrids. Heredity. 1974;32:267–270. [Google Scholar]
- 15.Ishii T, Ueda T, Tanaka H, Tsujimoto H. Chromosome elimination by wide hybridization between Triticeae or oat plant and pearl millet: Pearl millet chromosome dynamics in hybrid embryo cells. Chromosome Res. 2010;18:821–831. doi: 10.1007/s10577-010-9158-3. [DOI] [PubMed] [Google Scholar]
- 16.Jin WW, et al. Maize centromeres: Organization and functional adaptation in the genetic background of oat. Plant Cell. 2004;16:571–581. doi: 10.1105/tpc.018937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim NS, Armstrong KC, Fedak G, Ho K, Park NI. A microsatellite sequence from the rice blast fungus (Magnaporthe grisea) distinguishes between the centromeres of Hordeum vulgare and H. bulbosum in hybrid plants. Genome. 2002;45:165–174. doi: 10.1139/g01-129. [DOI] [PubMed] [Google Scholar]
- 18.Mochida K, Tsujimoto H, Sasakuma T. Confocal analysis of chromosome behavior in wheat x maize zygotes. Genome. 2004;47:199–205. doi: 10.1139/g03-123. [DOI] [PubMed] [Google Scholar]
- 19.Schwarzacher Robinson T, Finch RA, Smith JB, Bennett MD. Genotypic control of centromere positions of parental genomes in Hordeum X Secale hybrid metaphases. J Cell Sci. 1987;87:291–304. [Google Scholar]
- 20.Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with Scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 21.Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell. 2002;14:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howman EV, et al. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blower MD, Daigle T, Kaufman T, Karpen GH. Drosophila CENP-A mutations cause a BubR1-dependent early mitotic delay without normal localization of kinetochore components. PLoS Genet. 2006;2:e110. doi: 10.1371/journal.pgen.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vermaak D, Hayden HS, Henikoff S. Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol. 2002;22:7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lermontova I, et al. Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell. 2006;18:2443–2451. doi: 10.1105/tpc.106.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 29.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henikoff S, Ahmad K, Malik HS. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 31.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 32.Foltz DR, et al. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagana A, et al. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat Cell Biol. 2010;12:1186–1193. doi: 10.1038/ncb2129. [DOI] [PubMed] [Google Scholar]
- 36.Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malik HS, Henikoff S. Conflict begets complexity: The evolution of centromeres. Curr Opin Genet Dev. 2002;12:711–718. doi: 10.1016/s0959-437x(02)00351-9. [DOI] [PubMed] [Google Scholar]
- 39.Houben A, et al. CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma. 2007;116:275–283. doi: 10.1007/s00412-007-0102-z. [DOI] [PubMed] [Google Scholar]
- 40.Nagaki K, et al. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- 41.Hudakova S, et al. Sequence organization of barley centromeres. Nucleic Acids Res. 2001;29:5029–5035. doi: 10.1093/nar/29.24.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heddle JA, Carrano AV. The DNA content of micronuclei induced in mouse bone marrow by gamma-irradiation: Evidence that micronuclei arise from acentric chromosomal fragments. Mutat Res. 1977;44:63–69. doi: 10.1016/0027-5107(77)90115-4. [DOI] [PubMed] [Google Scholar]
- 43.Schubert I, Oud JL. There is an upper limit of chromosome size for normal development of an organism. Cell. 1997;88:515–520. doi: 10.1016/s0092-8674(00)81891-7. [DOI] [PubMed] [Google Scholar]
- 44.Tobler H. The differentiation of germ and somatic cell lines in nematodes. In: Henning W, editor. Germ line-Soma Differentiation. Results and Problems in Cell Differentiation. Vol. 13. Berlin: Springer; 1986. pp. 1–69. [DOI] [PubMed] [Google Scholar]
- 45.Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagaki K, Kashihara K, Murata M. Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula nivea. Plant Cell. 2005;17:1886–1893. doi: 10.1105/tpc.105.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong CX, et al. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell. 2002;14:2825–2836. doi: 10.1105/tpc.006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagaki K, Murata M. Characterization of CENH3 and centromere-associated DNA sequences in sugarcane. Chromosome Res. 2005;13:195–203. doi: 10.1007/s10577-005-0847-2. [DOI] [PubMed] [Google Scholar]
- 49.Thiel T, et al. Evidence and evolutionary analysis of ancient whole-genome duplication in barley predating the divergence from rice. BMC Evol Biol. 2009;9:209. doi: 10.1186/1471-2148-9-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawabe A, Nasuda S, Charlesworth D. Duplication of centromeric histone H3 (HTR12) gene in Arabidopsis halleri and A. lyrata, plant species with multiple centromeric satellite sequences. Genetics. 2006;174:2021–2032. doi: 10.1534/genetics.106.063628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moraes IC, Lermontova I, Schubert I. Recognition of A. thaliana centromeres by heterologous CENH3 requires high similarity to the endogenous protein. Plant Mol Biol. 2011;75:253–261. doi: 10.1007/s11103-010-9723-3. [DOI] [PubMed] [Google Scholar]
- 52.Blattner FR. Phylogenetic analysis of Hordeum (Poaceae) as inferred by nuclear rDNA ITS sequences. Mol Phylogenet Evol. 2004;33:289–299. doi: 10.1016/j.ympev.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 53.Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hirsch CD, Wu YF, Yan HH, Jiang JM. Lineage-specific adaptive evolution of the centromeric protein CENH3 in diploid and allotetraploid Oryza species. Mol Biol Evol. 2009;26:2877–2885. doi: 10.1093/molbev/msp208. [DOI] [PubMed] [Google Scholar]
- 56.Nagaki K, Kashihara K, Murata M. A centromeric DNA sequence colocalized with a centromere-specific histone H3 in tobacco. Chromosoma. 2009;118:249–257. doi: 10.1007/s00412-008-0193-1. [DOI] [PubMed] [Google Scholar]
- 57.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan HH, et al. Intergenic locations of rice centromeric chromatin. PLoS Biol. 2008;6:2563–2575. doi: 10.1371/journal.pbio.0060286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kynast RG, et al. A complete set of maize individual chromosome additions to the oat genome. Plant Physiol. 2001;125:1216–1227. doi: 10.1104/pp.125.3.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagaki K, Terada K, Wakimoto M, Kashihara K, Murata M. Centromere targeting of alien CENH3s in Arabidopsis and tobacco cells. Chromosome Res. 2010;18:203–211. doi: 10.1007/s10577-009-9108-0. [DOI] [PubMed] [Google Scholar]
- 61.Ravi M, Chan SW. Haploid plants produced by centromere-mediated genome elimination. Nature. 2010;464:615–618. doi: 10.1038/nature08842. [DOI] [PubMed] [Google Scholar]
- 62.Ravi M, et al. The rapidly evolving centromere-specific histone has stringent functional requirements in Arabidopsis thaliana. Genetics. 2010;186:461–471. doi: 10.1534/genetics.110.120337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Topp CN, et al. Identification of a maize neocentromere in an oat-maize addition line. Cytogenet Genome Res. 2009;124:228–238. doi: 10.1159/000218128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aw SJ, Hamamura Y, Chen Z, Schnittger A, Berger F. Sperm entry is sufficient to trigger division of the central cell but the paternal genome is required for endosperm development in Arabidopsis. Development. 2010;137:2683–2690. doi: 10.1242/dev.052928. [DOI] [PubMed] [Google Scholar]
- 66.Ingouff M, et al. Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol. 2010;20:2137–2143. doi: 10.1016/j.cub.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 67.Mehta GD, Agarwal MP, Ghosh SK. Centromere identity: A challenge to be faced. Mol Genet Genomics. 2010;284:75–94. doi: 10.1007/s00438-010-0553-4. [DOI] [PubMed] [Google Scholar]
- 68.Okada M, et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 69.Sato H, Shibata F, Murata M. Characterization of a Mis12 homologue in Arabidopsis thaliana. Chromosome Res. 2005;13:827–834. doi: 10.1007/s10577-005-1016-3. [DOI] [PubMed] [Google Scholar]
- 70.Handmaker SD. Hybridization of eukaryotic cells. Annu Rev Microbiol. 1973;27:189–204. doi: 10.1146/annurev.mi.27.100173.001201. [DOI] [PubMed] [Google Scholar]
- 71.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 72.Sanei M, et al. Interspecific hybrids of Hordeum marinum ssp. marinum x H. bulbosum are mitotically stable and reveal no gross alterations in chromatin properties. Cytogenet Genome Res. 2010;129:110–116. doi: 10.1159/000313641. [DOI] [PubMed] [Google Scholar]
- 73.Galbraith DW, et al. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science. 1983;220:1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- 74.Ma L, et al. Synteny between Brachypodium distachyon and Hordeum vulgare as revealed by FISH. Chromosome Res. 2010;18:841–850. doi: 10.1007/s10577-010-9166-3. [DOI] [PubMed] [Google Scholar]
- 75.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 76.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]