Abstract
The emergence of the unique H1N1 influenza A virus in 2009 resulted in a pandemic that has spread to over 200 countries. The constellation of molecular factors leading to the emergence of this strain is still unclear. Using a computational approach, we identified molecular determinants that may discriminate the hemagglutinin protein of the 2009 human pandemic H1N1 (pH1N1) strain from that of other H1N1 strains. As expected, positions discriminating the pH1N1 from seasonal human strains were located in or near known H1N1 antigenic sites, thus camouflaging the pH1N1 strain from immune recognition. For example, the alteration S145K (an antigenic position) was found as a characteristic of the pH1N1 strain. We also detected positions in the hemagglutinin protein differentiating classical swine viruses from pH1N1. These positions were mostly located in and around the receptor-binding pocket, possibly influencing binding affinity to the human cell. Such alterations may be liable in part for the virus’s efficient infection and adaptation to humans. For instance, 133A and 149 were identified as discriminative positions. Significantly, we showed that the substitutions R133AK and R149K, predicted to be pH1N1 characteristics, each altered virus binding to erythrocytes and conferred virulence to A/swine/NC/18161/02 in mice, reinforcing the computational findings. Our findings provide a structural explanation for the deficient immunity of humans to the pH1N1 strain. Moreover, our analysis points to unique molecular factors that may have facilitated the emergence of this swine variant in humans, in contrast to other swine variants that failed.
Keywords: machine learning, alternating decision trees
A pandemic H1N1 (pH1N1) human influenza virus was identified in April 2009 (1) and has since spread to over 200 countries and caused over 18,000 deaths (2). Evolutionary analysis of the pH1N1 strain indicates that its HA belongs to the classical swine lineage. HAs in this lineage are more similar to those of historical human strains such as the 1918 H1N1 strain than to those of circulating H1N1 strains from recent years (3). Prior to the emergence of the pH1N1 strain, only sporadic cases of human infection by swine influenza viruses had been reported (4–8). The molecular basis enabling this recent strain to efficiently infect and be transmitted between humans remains obscure (9–12).
Herein, we used a computational approach to uncover signature residues in the receptor-binding domain (RBD) of the pH1N1 HA protein. Using a machine-learning algorithm of alternating decision trees (ADTs) (13), we predicted positions that differentiated between HA protein sequences of the pH1N1 strain and HA sequences of other H1N1 strains. The algorithm combines moderately successful rules to produce highly accurate predictions (13). Here the rules represent correlations between the presence of specific amino acid type(s) in selected positions and the sequence annotation, e.g., pandemic versus seasonal strain. This is done by iteratively reweighting training examples (i.e., HA sequences), thus, concentrating on the sequences that are more “difficult” to classify. We subsequently ranked each predicted position according to its discriminative potency (see SI Text S5). First, we detected the alterations distinguishing the antigenicity of the pH1N1 strain from that of prior seasonal human strains. We used all human H1N1 sequences (seasonal H1N1 and pH1N1) available in the National Center for Biotechnology Information (NCBI) database (14) and trained the classification algorithm to detect residues in the HA sequence discriminating pH1N1 from those strains. Next, we carried out the same analysis to identify molecular features that distinguish this swine-origin virus from classical swine strains. The detected positions, which were situated mainly in and around the receptor-binding pocket, may reveal why the pandemic virus was able to emerge in the human population whereas other swine viruses were unsuccessful. Notably, we experimentally validated two of our predictions: We inserted either mutation R133AK or mutation R149K—predicted pH1N1 characteristics—into the HA of the swine strain A/swine/NC/18161/02. Each substitution altered virus binding to erythrocytes and resulted in a strain that was more virulent than the parental strain in DBA/2J mice. These results substantiate the computational method presented here.
Results
To identify signature sites we trained the ADT algorithm on the RBD of the HA sequence. We chose to focus our analysis on the RBD, because previous studies have shown that minor changes in this region can account for differences in virus transmissibility, which is a prerequisite for emergence (15, 16). Furthermore, the H1N1 HA includes four known antigenic sites (17) in the RBD region (Fig. 1), namely, Sa (residues 128, 129, 156–160, and 162–167), Sb (residues 187–198), Ca (residues 140–145, 169–173, 206–208, 224, 225, and 238–240), and Cb (residues 79–84) [numbering according to Protein Data Bank (PDB) ID code 3lzg].
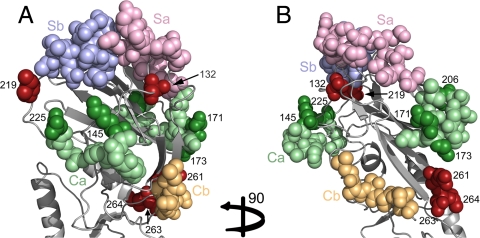
Fig. 1.
Detected positions, discriminating between prior human H1N1 circulating strains and the pH1N1 strain, correspond to known antigenic sites. The RBD of the HA protein from the human A/California/04/2009 H1N1 strain (PDB ID code 3lzg) is shown in cartoon representation. Front (A) and side (B) views of the RBD with the identified positions and antigenic sites presented as all-atom spheres. The Sa antigenic site is in pink, Sb site in blue, Ca site in green, and the Cb antigenic site in orange. The identified specificity determinants that overlap with H1N1 antigenic sites are shown using a dark variant of the site color (e.g., a position identified in Ca is colored in dark green, whereas the rest of the site is in pale green). Predictions that do not overlap with a known antigenic site are in dark red. All newly detected positions are located in and around the recognition hot spots of the immune system.
pH1N1 Versus Seasonal H1N1 Human Strains.
We compared pH1N1 isolates with isolates from human-seasonal H1N1. We selected the 10 most discriminative positions (see SI Text S5), whose combination provided near-perfect classification accuracy of 98%, on average. Mapping these positions on the HA structure, we discovered that all residues were situated in and around known H1N1 antigenic sites (17) (Fig. 1 and Table 1). Specifically, five residues were located in the known Ca antigenic site, one was in direct contact with the Sb site, and four were in the vicinity of these sites. These results are in agreement with a recent study showing that the majority of antigenic differences between pH1N1 and historical strains appear in the Ca antigenic site, whereas the Sa site has remained relatively conserved (18).
Table 1.
Highly ranked residues that discriminate the pH1N1 strain from circulating human H1N1 strains in the hemagglutinin RBD
| Residue number in structure 3lzg | Rank* | Circulating human strain characteristics† | pH1N1 characteristics‡ | Structural comments |
| 145 | 1 | S | K | In antigenic site Ca. A change from the (positively charged) K to (polar) S can change the stereochemistry of the antigenic site and affect receptor specificity. |
| 171 | 2 | N | D | In antigenic site Ca. May change the physicochemical characteristics of Ca. |
| 225 | 3 | — | — | In antigenic site Ca. Known specificity determinant for altering avian to human H1N1 sequences. |
| 219 | 4 | E | I | Close to antigenic site Sb. The E-to-I mutation may affect the antigenic site’s structure. |
| 261 | 5 | S | E | Residues 261 + 263 + 264 form a cluster in the structure. Residue 261 is very close to antigenic site Ca. From the other side, residues 263 and 264 are close to antigenic site Cb. These positions may be a continuous part of the antigenic sites. The alterations may affect the protein structure or the approximate antigenic sites. |
| 132 | 6 | I | P,S | In the vicinity of the Sa antigenic site. |
| 206 | 7 | — | — | In antigenic site Ca. |
| 173 | 8 | — | G | In antigenic site Ca. The mutation may alter the antibody binding. |
| 264 | 9 | F | A | See comments for position 261. |
| 263 | 10 | G | N |
Positions are numbered as in the A/California/04/2009 H1N1 strain (PDB ID code 3lzg) structure.
*Rank of contribution to discrimination given to the detected position (see Computational Methods).
†The amino acid suggested to characterize circulating human H1N1 strains by the algorithm.
‡The amino acid suggested to characterize the pH1N1 strain. The symbol “—” indicates that no characteristic amino acid was identified.
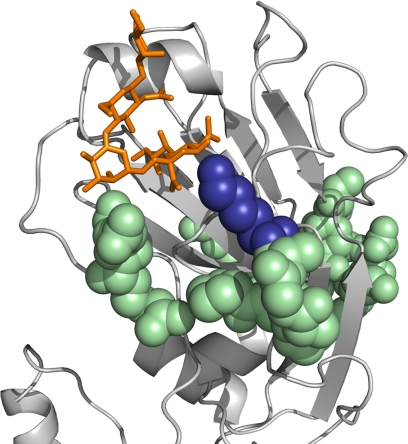
The physicochemical characteristics of many of the predicted alterations (Table 1) may influence the structure of the antigenic sites and consequently the antibody binding. For instance, our analysis suggests that in seasonal strains, serine appears in position 145, whereas in the pH1N1 strain lysine appears in this position. Position 145 is located in the Ca antigenic site (17) (Fig. 2), and the substitution of serine with lysine may alter the geometry and physicochemical characteristics of the site. Interestingly, this position is located in the site that binds the sialic acid of the host receptor (Fig. 1). As binding of the HA to the host’s negatively charged sialic acid mediates viral infection, the predicted substitution to the positively charged lysine may increase affinity to the host receptor in the pH1N1 strain.
Fig. 2.
Residue 145 (shown in blue spheres) has been detected as a discriminative position in our analysis. The HA receptor-binding domain of the H1 A/California/04/2009 H1N1 strain (PDB ID code 3lzg) is shown in gray cartoon representation, and the human receptor analogue is shown in orange. Interestingly, this amino acid is in the known antigenic Ca (in pale green spheres) and is located near the human receptor as well. The human receptor analogue was modeled into the RBD by superimposing the structures of A/California/04/2009 H1N1 and A/Brevig Mission/1/1918 H1N1 HAs (PDB ID codes 3lzg and 2wrg) with the α2-6 analogue bound.
Recent studies have shown that the 2009 pH1N1 strain lacks two glycosylation sites in the RBD that are conserved in human-seasonal H1N1 strains (18, 19). Specifically, in human-seasonal strains, threonine or serine at positions 131 and 165 introduce glycosylation sites at asparagine 129 and 163, respectively. The algorithm does not specifically account for changes that alter potential glycosylation patterns, and yet three of these four positions—129, 131, and 165—were detected as discriminative positions between pH1N1 and seasonal H1N1 human strains. However, these positions were assigned relatively low ranks (51, 42, and 50, respectively).
pH1N1 Versus Classical Swine Strains.
The molecular characteristics giving rise to a human-adapted swine virus are of major interest and concern. Therefore, we looked for positions in the HA sequence separating the pH1N1 strain from previous swine strains.
We selected the 13 most discriminative positions (see SI Text S5 and Table 2); their combination resulted in an average of approximately 90% classification accuracy. Almost all detected positions are located in the receptor-binding pocket, suggesting that they have some role in receptor-binding efficacy.
Table 2.
Highly ranked residues that discriminate between the pH1N1 and H1N1 swine strains
| Residue number in structure 3lzg | Rank* | Swine characteristics† | pH1N1 characteristics‡ | Structural comments |
| 149 | 1 | R | K | In close proximity to the Ca and Cb antigenic sites. |
| 171 | 2 | N | D | In antigenic site Ca. The alteration may interfere with antibody binding. |
| 225 | 3 | G, E | D | This position is in direct contact with the host cell receptor and known to affect avian to human receptor specificity. The alteration may alter receptor binding. Also in antigenic site Ca. |
| 132 | 4 | T | S | Residues 131 + 132 + 133A form a cluster in the structure, situated in the receptor-binding pocket and may affect binding. Also in vicinity to the Sa antigenic site and may affect this site. |
| 133A | 5 | R | — | |
| 188 | 6 | T | S | In contact with residue 189, which is in direct contact with the host receptor. An alteration in this position may indirectly affect receptor specificity and binding. Also in antigenic site Sb. |
| 226 | 7 | — | — | In direct contact with the sialic acid of the host cell receptor. |
| 206 | 8 | — | T | Residue 206 is in direct contact with residue 221 of the adjacent RBD, which is in the receptor-binding pocket. The alteration may indirectly affect the receptor binding. Additionally, this residue is located in antigenic site Ca; a mutation may affect antibody binding. |
| 208 | 9 | K | R | Binds position 206, which is in direct contact with the adjacent RBD, therefore, can affect binding. Located in antigenic site Ca and mutation here may alter antibody binding. |
| 200 | 10 | T | A | In direct contact with the Sb antigenic site; its alteration may affect antigenicity. |
| 131 | 11 | E | D | Residues 131 + 132 + 133A form a cluster in the structure (described above). |
| 189 | 12 | — | A | Binds the receptor; alterations may affect binding. Also in the Sb antigenic site. |
| 186 | 13 | P | S | Binds the receptor; a mutation may alter the binding. |
Positions are numbered as in the A/California/04/2009 H1N1 strain (PDB ID code 3lzg) structure sequence.
*Rank of contribution to discrimination given to the detected position (see Computational Methods).
†The amino acid suggested to characterize circulating swine H1N1 strains by the algorithm.
‡The amino acid suggested to characterize the pandemic H1N1 human strain. The symbol “—” indicates that no characteristic amino acid was identified.
Some of the positions were particularly interesting. Residues 131, 132, and 133A form a structural cluster (Fig. 3) located in the receptor-binding pocket. These positions may have mutated simultaneously, which in turn may have affected the structural conformation of the receptor-binding pocket, ultimately influencing the host cell binding. Furthermore, this cluster is near the Sb antigenic site, and the mutations might also be involved in antigenic variation.
Fig. 3.
Detected positions discriminating between swine H1N1 strains and the pH1N1 strain RBD of the HA protein from the human A/California/04/2009 H1N1 strain (PDB ID code 3lzg) is shown in cartoon representation. Front (A) and side (B) views of the RBD with the identified positions and antigenic sites presented as all-atom spheres (the orientations are similar to these in Fig. 1 A and B). The Sa antigenic site is in pink, Sb site in blue, Ca site in green, and the Cb antigenic site in orange. The identified specificity determinants that overlap with H1N1 antigenic sites are shown using a dark variant of the site color (e.g., a position identified in Ca is colored in dark green, whereas the rest of the site is in pale green). Predictions that do not overlap with a known antigenic site are in dark red.
Residues 206 and 208 (Fig. 3) are both located in antigenic site Ca and can affect antigenicity. More interestingly, these residues interact with positions 220, 221, and 229 of the receptor-binding pocket in the adjacent monomer (Fig. 4). Therefore, mutations in residues 206 and 208 may result in a structural change to the adjacent receptor-binding pocket or influence the interaction between the monomers.
Fig. 4.
Residues 206 and 208. Positions 206 and 208 were identified as discriminative between the swine and pH1N1 strains. Two adjacent monomers of the H1 A/California/04/2009 H1N1 (PDB ID code 3lzg) trimer are shown in cartoon representation (the third monomer is not shown for clarity). Each monomer is in a different color. The human receptor analogue is shown in an orange stick representation. Positions 206 and 208 are shown in red spheres, and positions 220, 221, and 229 from the adjacent monomer, which are in direct contact with the receptor, are shown in blue spheres. An alteration in residues 206 or 208 may affect the structural conformation of the receptor-binding pocket from the adjacent monomer.
Position 186 binds the receptor directly and is in direct contact with the Sb antigenic site (Fig. 3). Our analysis suggests that serine in this position is a characteristic of the pH1N1 strain. Indeed, a previous study has found serine in this position to be a human H1N1 characteristic, whereas proline in this position characterizes swine strains (20). Moreover, this position has been recently reported to affect growth in cell culture and eggs (21). Thus, the appearance of serine in pH1N1 may be one of the characteristics leading it to become a more “human-like” virus. Additionally, residues 188 and 189 are also located in the receptor-binding pocket (Fig. 3) and are both in antigenic site Sb. Mutations in these positions may affect the receptor binding and antigenicity as well.
Residues 225 and 226 both bind the host cell receptor (Fig. 3); therefore, alterations in these positions may induce a variation in the binding specificity to the receptor. Moreover, position 225 is already known to affect receptor-binding specificity of avian and human H1N1 viruses (22). In pH1N1, the amino acid in this position is characteristic of the human virus (23), which may account for its efficient adaption and transmission in humans.
We note that although a small number of the positions that were highly ranked by our computational approach have been reported to affect adaptation or transmissibility in humans, the majority of the detected positions in our analysis have not been previously described. However, the strategic locations of these predicted alterations, together with their characteristics, suggest a role for these features in the emergence of pH1N1 as an effective human pathogen.
We also conducted the same analyses (i.e., pH1N1 versus seasonal and pH1N1 versus swine) on the whole HA sequence (instead of the RBD alone). Reassuringly, most of the highly ranked positions in the former analyses appeared in the full HA analyses as well (see SI Text S1 and Tables S1 and S2).
In addition, we were interested to see whether positions identified in our analyses were located in or around T-cell and B-cell epitopes, which would provide additional support for the role of these positions in antigenicity. However, we found that 78% of the RBD sequence is covered by one or more epitopes reported in the Immune Epitope Database (www.immuneepitope.org) (see Fig. S1), rendering this analysis uninformative.
Experimental Validation.
In order to evaluate whether our computational findings had identified residues with phenotypic relevance, we altered an H1N1 virus with a classical swine-lineage HA, with the goal of rendering it more “pH1N1-like.” The experimental methods are described in SI Text S2. Residues 133A and 149 were detected in our analysis as discriminating between classical swine H1N1 and pH1N1 strains (Table 2). Accordingly, we generated two single amino acid HA mutants by inserting mutation R133AK or mutation R149K into the HA of A/swine/NC/18161/02, an endemic H1N1 swine virus. Both mutant viruses, rg-swine/NC/18161/02-HA133A and rg-swine/NC/18161/02-HA149, respectively, were successfully rescued. “Swine-like” pandemic reverse mutants rg-TN/560-1/09-HA133A and rg-TN/560-1/09-HA149 were also successfully generated.
We first compared the binding specificities of A/swine/NC/18161/02 with those of the two HA mutants. As a surrogate measure of receptor specificity, we carried out hemagglutination assays using erythrocytes from different species. Both swine HA mutant viruses showed decreased binding to chicken, goose, guinea pig, and human (type O) erythrocytes compared with the parental strain. Pandemic viruses containing the reverse mutations (i.e., mutation to the corresponding swine virus residue) were unaltered in their erythrocyte binding patterns as compared to the parental strain (Tables S3 and S4). The three viruses showed similar binding to turkey erythrocytes, and none of the viruses bound to horse erythrocytes (Table S3). Whereas horse erythrocytes contain α2-3-linked sialic acids (α2-3SAL, “avian-like” receptors) but no α2-6SAL (human-like receptors), chicken and goose erythrocytes contain more α2-3SAL than α2-6SAL, and human O, guinea pig, and turkey erythrocytes have more α2-6SAL than α2-3SAL (24, 25). It is important to note that these erythrocyte receptor generalizations are very much oversimplifications as highlighted by the lack of difference between the three viruses in binding to turkey erythrocytes. Therefore, these findings indicate that indeed the mutations presented here affect the binding to different erythrocytes; however, they do not point to a preference of a specific sialic acid receptor. Because HA residue 133A is situated in the receptor-binding pocket (Table 2 and Fig. 3), we anticipated that it would play a role in receptor binding; thus, the differences observed between the binding of the 133A mutant and that of the parental virus were in line with our expectations. Interestingly, mutation of residue 149 produced similar results in the hemagglutination assay, even though this residue is not located directly in the receptor-binding pocket.
We further wished to investigate the effect of these mutations on viral virulence. We used the DBA/2J mouse strain, as it has been shown to be highly susceptible to most influenza viruses (26). To measure the lethal dose of each virus, we infected mice with either 5 × 101, 5 × 102, 5 × 103, or 5 × 104 egg 50% infective dose (eID50) of A/swine/NC/18161/02, rg-A/swine/NC/18161/02-HA133A, or rg-A/swine/NC/18161/02-HA149. Both swine HA mutants were more virulent than their parental strain, with 50% mouse-lethal doses (MLD50s) of < 1.5, < 1.5, and 2.45 for rg-A/swine/NC/18161/02-HA133A, rg-A/swine/NC/18161/02-HA149, and rg-A/swine/NC/18161/02, respectively. DBA/2J mice were also infected with 101, 102, 103, and 104 eID50 of rg-TN/560-1/09-HA133A, rg-TN/560-1/09-HA149 (i.e., pandemic viruses with swine virus signature mutations), and rg-A/TN/560-1/09. These swine-like pandemic mutants were less pathogenic than their parental strain with MLD50s of < 3.4, 3.5, and 2.4 for rg-TN/560-1/09-HA133A, rg-TN/560-1/09-HA149, and rg-TN/560-1/09, respectively (Table S4). The critical biological roles of HA positions 133A and 149 were therefore confirmed in vivo.
Discussion
In this study we identified molecular features differentiating the HA protein of the pH1N1 from the HAs of other H1N1 strains. We detected alterations in amino acid positions in the receptor-binding pocket and antigenic sites of the pH1N1 HA that could be responsible in part for the successful transmission of the virus into humans. Moreover, we experimentally confirmed that two of the highly ranked positions indeed had a phenotypic effect in vivo.
Comparison of aligned sequences of viruses from various sources as a means to detect altered positions is a commonly used approach to guide experimental studies (e.g., refs. 10 and 23). A thorough investigation of all the thousands of available HA sequences would be desirable. As manual inspection of this volume of data is virtually impossible; it is necessary to use computational methods. Entropy (mutual-information)-based methods that use the amino acid frequency in each position are commonly used to this effect (27–29). These methods consider each position in the alignment separately, disregarding the possible relations between the sequence positions. Allen et al. (30) used a classification algorithm that did take such relations into account. They used it to detect host-specific alterations in the influenza A proteins, but none of the positions they detected were in the HA, the main protein responsible for the host cell binding. Our approach is similar to theirs in that it also takes into account the relations between the sequence positions. It selects a set of positions whose combination provides optimal discrimination between two groups (e.g., pH1N1 and swine-H1N1 sequences). For comparison we analyzed the same datasets using a mutual-information-based method (29). Reassuringly, the majority of the highly ranked positions presented here were also obtained in that analysis (SI Text S3 and Tables S5 and S6). The mutual-information method identified 49 positions distinguishing between the human-seasonal H1N1 and the pH1N1 dataset, and our methodology showed that 10 residues were sufficient to provide a very high classification accuracy of 98%. Eight of the residues we identified were also identified in the mutual-information analysis. For the swine H1N1 and pH1N1 dataset, the mutual-information analysis produced 14 discriminative amino acid positions, whereas our method produced 13, 6 of which were predicted by both methods. Intriguingly, position 133A, which our method highlighted, and whose subsequent substitution in an H1N1 virus led to a phenotypic effect (see Results), did not come up in the mutual-information analysis.
Recently, probabilistic approaches that explicitly take into account the phylogenetic relations between the taxa were used to identify host, clade, and drug-resistance signature residues in the various influenza proteins (31–33). This approach may solve problems caused due to uneven sampling in sequence space. However, because of the large quantity of available HA sequences and their high similarity, we do not believe that uneven sampling poses a major concern in our case. With these data we obtained an average accuracy of 90% for the discrimination of swine versus pH1N1 sequences (using only the 13 highly ranked positions), which is very high given the low diversification within the swine and pH1N1 groups and the relatively close evolutionary relationship between pH1N1 and other swine strains. The analysis of sequence datasets with more distant evolutionary relationships, such as human seasonal versus pH1N1 (see Results) and human seasonal versus swine, resulted in even better accuracy of 98% (SI Text S4).
An obvious concern regarding our approach would be that because antigenic sites evolve rapidly (5), they are prone to be detected by the algorithm. We therefore examined the correlation between the variability of selected positions and their discriminative rank (see Fig. S2). Reassuringly, we found a very weak negative correlation (r = -0.26; p = 0.04). Thus, the positions detected here were identified based on their discriminative power and not their variability.
Another caveat to our approach relates to the low diversity in the HA sequences of the pH1N1 strain, which results from the fact that the unique influenza strain has been circulating in the human population for a relatively short time, probably insufficient to mutate and substantially diverge. Presumably, some of the unique amino acid characteristics this virus acquired are not crucial for its functionality, but are merely founder effects, i.e., mutations that were randomly “fixed” during the transition to humans and have not mutated because they are not yet under antigenic pressure (32). Indeed, over time, some of these positions will diverge and may prove to be functionally insignificant, but others might provide a supportive genetic background for other functionally important mutations (31, 34). Bearing in mind these difficulties, it is encouraging that essentially all the positions identified in our analysis were located in functionally important regions.
We studied two different, albeit related, questions. First, we sought the distinctive properties of pH1N1 in comparison with prior human H1N1 strains. Vaccines for seasonal influenza strains induced little or no neutralizing antibody response to pH1N1 among children and adults under 64 y of age (35), thus indicating that the antigenic properties of pH1N1 differ from those of previous seasonal strains. Indeed, our analysis showed that all positions detected as discriminating between the pandemic and seasonal human strains are located in and around the known H1N1 antigenic sites. Considering that many of the characteristic substitutions of the pH1N1 viruses substantially affected the physicochemical nature of the amino acids in these positions (Table 1), these alterations could have resulted in variation of the stereochemistry of the antibody-binding sites. Thus, a host immune to either the swine or human virus lineages might not be able to recognize the pH1N1 virus, a phenomenon known as “antigenic drift” (36).
Second, we studied the molecular adaptations that enabled the virus to emerge in humans. The efficient adaptation and vast transmission of the swine-origin virus, pH1N1, into the human population has caused substantial concern. Prior to this pandemic, only isolated cases of infection by a swine virus had been seen in humans (4–8), and limited human-to-human transmission had been documented. Although the impact of this pandemic has been mild in terms of overall mortality, discerning the molecular factors enabling this virus’s success is crucial for future detection of potential pandemic threats. To this end, we identified positions in the HA protein sequence separating pH1N1 from classical swine strains. Our study revealed residues in the receptor-binding pocket that may have altered binding to the host cell receptor.
Swine hosts were shown to be susceptible to infection by both avian and human influenza viruses (37), owing to the presence of both avian and human receptors in the pig trachea (38). Therefore, reassortment between avian and human viruses may occur upon coinfection (39). Alternatively, viruses with preferential binding specificity to avian receptors may mutate (as a result of the human receptors’ presence in the pig trachea) and eventually acquire the ability to efficiently bind the human receptor. Indeed, our analysis showed that the residues detected as discriminating between the swine and pH1N1 strains are located at a mean distance of 10.2 Å from the receptor, and the residues discriminating between the human-seasonal and pH1N1 strains are located at a mean distance of 19.4 Å from the receptor. An independent t test with a confidence level of 0.95 showed that the difference between these means was indeed significant (p = 0.04). These results emphasize and support the probable role of the former identified positions in increasing binding to the human host. Therefore, these sites may represent milestones in the adaptation of the swine virus to humans. In this context it is noteworthy that four residues appeared in both analyses (residues 132, 171, 206, and 225, Fig. 5).
Fig. 5.
Positions detected as discriminating both between human circulating H1N1 and pH1N1 strains and between swine and pH1N1 strains. The receptor-binding domain of the HA protein from the human A/California/04/2009 H1N1 strain (PDB ID code 3lzg) is shown in cartoon representation. Front (A) and side (B) views of the RBD with the identified positions presented as all-atom spheres. Positions detected as discriminating between human circulating and pH1N1 are in blue, and those detected as discriminating between swine and pH1N1 are in green. Residues identified in both analyses are colored in pink. For clarity, only the overlapping positions are numbered. It is evident that residues discriminating between swine and pH1N1 strains are mostly around the receptor-binding pocket.
We validated the significance of residues 133A and 149, identified in our analysis. A hemagglutination assay showed that the introduction of mutation R133AK or of mutation R149K into the HA of A/swine/NC/18161/02 viruses had the effect of decreasing binding to chicken, goose, guinea pig, and human (type O) erythrocytes as compared with the parental strain (Table S3). Furthermore, we compared the pathogenesis of our two swine and two pandemic mutants with that of their parental strain in the DBA/2J mouse model. Both mutants were more virulent than their parental strain, whereas the mutated pandemic strains were less. Furthermore, two mutants were successfully generated for positions 171 and 132 (ranks 2 and 4) but did not alter erythrocyte binding patterns or pathogenicity in mice (Table S4). The consequences of such mutations in the human host, however, are still unclear, and further studies are needed to better address the question. Nevertheless the fact that such mild substitutions (i.e., lysine to arginine) caused significant phenotypic effects confirm the biological significance of HA positions 133A and 149 and validate the computational method presented here.
After this paper was submitted for publication, Ye et al. reported that the insertion of mutations D131E and S186P into the pandemic strain A/California/04/ 09/(H1N1) increased the virus’s pathogenicity in BALB/c mice (40). Indeed, our analysis detected these positions as discriminating between swine H1N1 and pH1N1 (Table 2), and D131 and S186 were identified as characteristic of the pandemic strain. Interestingly, the amino acids we predicted to be swine H1N1 characteristics (131E and 186P) were shown to generate the pathogenic strain studied by Ye et al. (40). An additional study published after the submission of this paper (41) disclosed two positions in the HA, namely, 200 and 227, as affecting receptor binding of the pandemic strain. Residue 200 is indeed a highly ranked position in our study (Table 2), and 227 was detected by our algorithm, albeit with a lower rank.
We suggest that the basis for the difference in antigenicity between the pH1N1 and seasonal human H1N1 strains is associated with the detected sites presented in our study. We suggest signature sites in the HA protein that may enable the efficient host cell binding, infection, and transmission of the strain in the human population. Bearing in mind that HA is the main antigen on the viral surface and is responsible for the first step in the viral infection (42), our study is a significant step toward providing testable predictions for positions that may contribute to the elusiveness of this previously undescribed viral strain.
Computational Methods
An elaborate description of the methodology is provided in SI Text S5. Briefly, H1N1 HA sequences from swine and human hosts were collected from the NCBI Influenza Database (14). Duplicate sequences and partial sequences (less than 80% of full length) were removed from the data. We aligned sequences using the MUSCLE program (43) and visually inspected alignments to verify their quality. The main analysis was limited to the RBD (positions 114–268, H3 numbering) (44). Two datasets were created from these sequences: pH1N1 sequences versus prior circulating human strains, and pH1N1 sequences versus classical swine strains. The first dataset consisted of 706 pH1N1 sequences and 852 prior circulating human strains, excluding the 1918 historical strain. The second dataset consisted of 245 swine sequences and 782 pH1N1 sequences. In each dataset, sequences were labeled according to the group they belonged to (“pH1N1,” “swine,” or “human circulating” strains).
We used JBoost (http://jboost.sourceforge.net/) and the above datasets to identify positions in HA that distinguish pH1N1 isolates from human circulating H1N1 isolates, as well as positions that distinguish pH1N1 from swine H1N1 isolates (Fig. S3).
Supplementary Material
Acknowledgments.
We acknowledge Aaron Arvey and Maya Schushan for fruitful discussions. We also thank the St. Jude Hartwell Center for Biotechnology and Bioinformatics. This work was supported by the Specific Targeted Research Project project “EuroFlu,” funded by the FP6 European Commission Program (SP5B-CT-2007-044098 to N.B.-T.) R.W. was supported in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract HHSN266200700005C. D.M. was supported by the Edmond J. Safra Bioinformatics program at Tel Aviv University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014854108/-/DCSupplemental.
References
- 1.Dawood FS, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans: Novel swine-origin influenza A (H1N1) virus investigation team. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Pandemic (H1N1) 2009—update 101 ( http://www.who.int/csr/don/2010_05_21. 2010.
- 3.Sinha NK, Roy A, Das B, Das S, Basak S. Evolutionary complexities of swine flu H1N1 gene sequences of 2009. Biochem Biophys Res Commun. 2009;390:349–351. doi: 10.1016/j.bbrc.2009.09.060. [DOI] [PubMed] [Google Scholar]
- 4.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hay AJ, Gregory V, Douglas AR, Lin YP. The evolution of human influenza viruses. Philos Trans R Soc Lond B Biol Sci. 2001;356:1861–1870. doi: 10.1098/rstb.2001.0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kendal AP, Goldfield M, Noble GR, Dowdle WR. Identification and preliminary antigenic analysis of swine influenza-like viruses isolated during an influenza outbreak at Fort Dix, New Jersey. J Infect Dis. 1977;136(Suppl):S381–S385. doi: 10.1093/infdis/136.supplement_3.s381. [DOI] [PubMed] [Google Scholar]
- 7.Myers KP, Olsen CW, Gray GC. Cases of swine influenza in humans: A review of the literature. Clin Infect Dis. 2007;44:1084–1088. doi: 10.1086/512813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinde V, et al. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N Engl J Med. 2009;360:2616–2625. doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- 9.Childs RA, et al. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat Biotechnol. 2009;27:797–799. doi: 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maines TR, et al. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science. 2009;325:484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garten RJ, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh Y, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freund Y, Mason L. The alternating decision tree algorithm; Proceedings of the 16th International Conference on Machine Learning; San Francisco: Morgan Kaufmann Publishers; 1999. pp. 124–133. [Google Scholar]
- 14.Bao YM, et al. The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorrell EM, Wan H, Araya Y, Song H, Perez DR. Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proc Natl Acad Sci USA. 2009;106:7565–7570. doi: 10.1073/pnas.0900877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tumpey TM, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 17.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 18.Xu R, et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei CJ, et al. Cross-neutralization of 1918 and 2009 influenza viruses: Role of glycans in viral evolution and vaccine design. Sci Transl Med. 2010;2:24ra21. doi: 10.1126/scitranslmed.3000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matrosovich MN, et al. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology. 1997;233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- 21.Suphaphiphat P, et al. Mutations at positions 186 and 194 in the HA gene of the 2009 H1N1 pandemic influenza virus improve replication in cell culture and eggs. Virol J. 2010;7:157. doi: 10.1186/1743-422X-7-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matrosovich M, et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medeiros R, Escriou N, Naffakh N, Manuguerra JC, van der Werf S. Hemagglutinin residues of recent human A(H3N2) influenza viruses that contribute to the inability to agglutinate chicken erythrocytes. Virology. 2001;289:74–85. doi: 10.1006/viro.2001.1121. [DOI] [PubMed] [Google Scholar]
- 25.Ito T, et al. Receptor specificity of influenza A viruses correlates with the agglutination of erythrocytes from different animal species. Virology. 1997;227:493–499. doi: 10.1006/viro.1996.8323. [DOI] [PubMed] [Google Scholar]
- 26.Boon AC, et al. Cross-reactive neutralizing antibodies directed against pandemic H1N1 2009 virus are protective in a highly sensitive DBA/2 mouse influenza model. J Virol. 2010;84:7662–7667. doi: 10.1128/JVI.02444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen GW, et al. Genomic signatures of human versus avian influenza A viruses. Emerg Infect Dis. 2006;12:1353–1360. doi: 10.3201/eid1209.060276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finkelstein DB, et al. Persistent host markers in pandemic and H5N1 influenza viruses. J Virol. 2007;81:10292–10299. doi: 10.1128/JVI.00921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miotto O, Heiny A, Tan TW, August JT, Brusic V. Identification of human-to-human transmissibility factors in PB2 proteins of influenza A by large-scale mutual information analysis. BMC Bioinformatics. 2008;9(Suppl 1):S18. doi: 10.1186/1471-2105-9-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen JE, Gardner SN, Vitalis EA, Slezak TR. Conserved amino acid markers from past influenza pandemic strains. BMC Microbiol. 2009;9:77. doi: 10.1186/1471-2180-9-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science. 2010;328:1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamuri AU, Dos Reis M, Hay AJ, Goldstein RA. Identifying changes in selective constraints: Host shifts in influenza. PLoS Comput Biol. 2009;5:e1000564. doi: 10.1371/journal.pcbi.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forsberg R, Christiansen FB. A codon-based model of host-specific selection in parasites, with an application to the influenza A virus. Mol Biol Evol. 2003;20:1252–1259. doi: 10.1093/molbev/msg149. [DOI] [PubMed] [Google Scholar]
- 34.Holmes EC. Virology. Helping the resistance. Science. 2010;328:1243–1244. doi: 10.1126/science.1190994. [DOI] [PubMed] [Google Scholar]
- 35.Katz J, et al. Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Wkly Rep. 2009;58:521–524. [PubMed] [Google Scholar]
- 36.Treanor J. Influenza vaccine—Outmaneuvering antigenic shift and drift. N Engl J Med. 2004;350:218–220. doi: 10.1056/NEJMp038238. [DOI] [PubMed] [Google Scholar]
- 37.Gambaryan AS, et al. Receptor-binding properties of swine influenza viruses isolated and propagated in MDCK cells. Virus Res. 2005;114:15–22. doi: 10.1016/j.virusres.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 38.Ito T, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma W, Kahn RE, Richt JA. The pig as a mixing vessel for influenza viruses: Human and veterinary implications. J Mol Genet Med. 2008;3:158–166. [PMC free article] [PubMed] [Google Scholar]
- 40.Ye J, et al. Variations in the hemagglutinin of the 2009 H1N1 pandemic virus: Potential for strains with altered virulence phenotype? PLoS Pathog. 2010;6:e1001145. doi: 10.1371/journal.ppat.1001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Vries RP, et al. Only two residues are responsible for the dramatic difference in receptor binding between swine and new pandemic H1 hemagglutinin. J Biol Chem. 2011;286:5868–5875. doi: 10.1074/jbc.M110.193557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. H5 avian and H9 swine influenza virus haemagglutinin structures: Possible origin of influenza subtypes. EMBO J. 2002;21:865–875. doi: 10.1093/emboj/21.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin T, et al. The hemagglutinin structure of an avian H1N1 influenza A virus. Virology. 2009;392:73–81. doi: 10.1016/j.virol.2009.06.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.