Abstract
Oocyte competence is a key factor limiting female fertility, yet the underlying molecular mechanisms that contribute to oocyte competence remain unclear. The objective of this study was to elucidate specific genes whose function contributes to oocyte competence. We observed that 6 of 20 target genes examined were differentially expressed between adult (more competent) and prepubertal (less competent) porcine in vitro matured (IVM) oocytes. These genes were the cholesterol synthesis related gene HMG-CoA reductase (HMGCR), fatty acid oxidation genes acyl-CoA synthetase long-chain family member 3 (ACSL3) and long-chain acyl-CoA dehydrogenase (ACADL), glycolytic genes fructose 1,6 bisphosphate aldolase (ALDOA) and lactate dehydrogenase C (LDHC), and tumor necrosis factor-α (TNF). These 6 genes, as well as 3 other genes (porcine endogenous retrovirus (PERV), transcribed loci 10 (TL10), serine/arginine-rich splicing factor 1 (SRSF1)), were further analyzed by comparing transcript abundance in IVM and in vivo matured (VVM) prepubertal and adult porcine oocytes. Among these 9 target genes, five were differentially expressed between IVM and VVM prepubertal oocytes, while eight genes were differentially expressed between IVM and VVM adult oocytes. None was differentially expressed between VVM prepubertal and adult oocytes. A functional study of TNF demonstrated that depletion of endogenous TNF decreased oocyte competence and TNFAIP6 expression in cumulus cells, while TNF in IVM medium regulated TNFAIP6 expression in cumulus cells. Differential expression of the genes identified in this study suggests that these genes may be functionally relevant to oocyte competence.
Keywords: oocyte, porcine, gene expression, in vitro maturation
INTRODUCTION
Oocyte quality, or developmental competence, affects embryonic development, fetal growth, and even health of the offspring (Eppig and O'Brien 1998). Improving and predicting oocyte quality in vitro is critical for successful application of assisted reproductive technologies to both reproductive medicine and animal agriculture. Pigs are important to biomedical research, because they are physiologically more similar to humans than the mouse (Lunney 2007). Thus, a better understanding of porcine oocyte competence could improve in vitro oocyte maturation not only in pigs but also in other mammalian species, including humans, resulting in better quality oocytes and a significant positive effect on human medicine, agricultural animal production and biomedical applications.
During maturation, the oocyte must undergo a series of cellular events, such as reorganization of cytoplasmic organelles, synthesis and storage of transcripts and proteins, and changes in cellular metabolism (Krisher 2004). These cellular events, collectively referred to as cytoplasmic maturation, are required for successful subsequent embryonic development (Eppig 1996). During oogenesis, oocytes accumulate a larger than necessary amount of transcripts that are important for oocyte maturation and early embryo development (Sasaki et al. 1999). Transcription and storage of most transcripts occurs during the early stages of oocyte growth. Transcription is then generally silenced as the germinal vesicle breaks down during resumption of meiosis (Brevini-Gandolfi et al. 1999). After transcriptional silencing, oocytes rely on stored transcripts to both continue meiosis and complete the initial cleavage divisions after fertilization, until the maternal zygotic transition (Hodgman et al. 2001; Stebbins-Boaz et al. 1996). Thus synthesis and storage of maternal transcripts prior to the onset of transcriptional silencing are essential for the oocyte to acquire developmental competence (De La Fuente and Eppig 2001). During oocyte meiotic maturation, degradation of transcripts also occurs; almost half of the existing transcripts undergo some degree of deadenylation, and about 30% of the total mRNA is degraded (Paynton et al. 1988). This degradation of mRNA during the transition from the germinal vesicle (GV) to the metaphase II (MII) stage of meiosis is a selective process based on gene function and stage of oocyte development (Su et al. 2007). Thus, either aberrant degradation or maintenance of transcripts during oocyte maturation could also be deleterious to oocyte quality.
Oocyte competence is progressively achieved as females approach puberty. The difference in developmental competence between oocytes derived from prepubertal and adult animals has been confirmed in bovine (Khatir et al. 1996; Revel et al. 1995) and ovine (O'Brien et al. 1996) species. In pigs, oocytes derived from prepubertal animals have decreased meiotic maturation, increased polyspermy and compromised embryonic development in vitro (Marchal et al. 2001). Nuclear transfer studies in cattle demonstrate that using enucleated, prepubertal oocytes as the recipient cytoplast result in decreased blastocyst formation compared with enucleated adult oocytes (Mermillod et al. 1998; Salamone et al. 2001). By comparing the relative abundance of transcripts between GV stage ovine oocytes derived from prepubertal and adult animals, seven developmentally important genes (Na+K+ ATPase, p34cdc2, glucose-transporter I, activin, zona occludens protein 2, poli(A)polymerase, E-cadherin) related to developmental competence were identified (Leoni et al. 2007). These results indicate that the decreased developmental competence of prepubertal derived oocytes is likely associated with incomplete cytoplasmic maturation, and may be specifically reflected in altered transcript abundance.
Elucidation of an aberrant transcript profile in prepubertal oocytes could lead to identification of important cellular mechanisms involved in oocyte competence. Previous microarray analysis in our laboratory detected a total of 999 genes that were differentially expressed in prepubertal versus adult MII stage oocytes (Paczkowski and Krisher, unpublished results). Further studies of target genes suggested by the microarray, as well as additional genes in these target pathways, may enhance our understanding of molecular mechanisms important to oocyte quality. The objective of this experiment was to elucidate specific porcine genes whose functions have been shown to influence oocyte competence in other species. Expression of 20 selected target genes were compared between adult (more competent) and prepubertal (less competent) oocytes by quantitative real-time PCR (qPCR), which revealed a select subset of differentially expressed mRNAs related to a narrow range of cellular functions. This selective set of differentially expressed mRNAs was further validated in a second model system using in vivo matured (VVM, more competent) and in vitro matured (IVM, less competent) oocytes. Furthermore, we performed a functional study to better understand the relationship of one selected gene, TNF, on oocyte competence.
Results
Experiment 1. Comparative gene expression following IVM of oocytes derived from prepubertal and adult animals
Transcript abundance of 20 candidate genes was compared between oocytes derived from adult and prepubertal animals by quantitative real-time PCR (qPCR). These genes are related to apoptosis (MAPK8, TNF, CAPS8 and CASP3), cholesterol synthesis (HMGCR, SREBF1), fatty acid oxidation (ACSL3, ACADL), the pentose phosphate pathway (G6PD, PGD), glycolysis (HK1, ALDOA, PGK1, ENO1, PKM2, LDHC, PDHA1), the citric acid cycle (IDH3B, ATP5G1), and calcium homeostasis (ATP2B1). Among these genes, TNF, ACSL3, ACADL, ALDOA and LDHC showed a significantly higher expression in oocytes from prepubertal animals. Only HMGCR was highly (P<0.05) expressed in adult derived oocytes (Figure 1).
Figure 1.
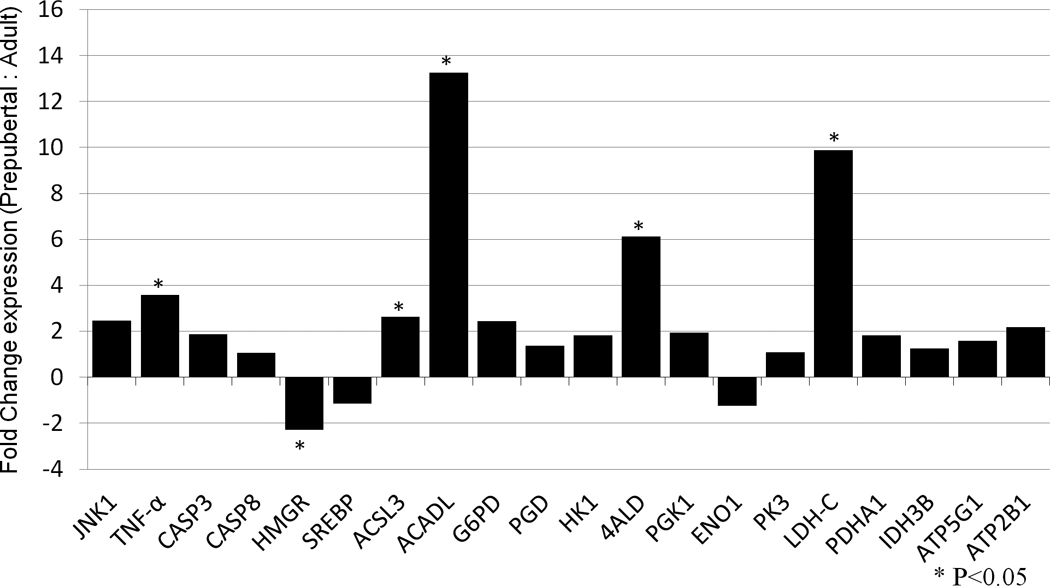
Expression of 20 target genes in oocytes derived from prepubertal compared to adult pigs, determined by qPCR analyses and relative to GAPDH. Bars above the x-axis represent genes that were upregulated in prepubertal oocytes; bars below x-axis represent genes that were upregulated in adult oocytes. * represents a significant difference in gene expression between prepubertal and adult oocytes (P <0.05).
Experiment 2. Comparative gene expression in IVM and VVM oocytes derived from prepubertal and adult animals
Using gene expression profiles from our prepubertal-adult model (Experiment 1), as well as a few candidate genes suggested by previous microarray and qPCR studies (Paczkowski and Krisher, unpublished results), we selected a panel of 9 differentially expressed genes that are associated with oocyte competence. These genes are porcine endogenous retrovirus (PERV), transcribed loci 10 (TL10), serine/arginine-rich splicing factor 1 (SRSF1), tumor necrosis factor-alpha (TNF), HMG-CoA reductase (HMGCR), acyl-CoA synthetase long-chain family member 3 (ACSL3), long-chain acyl-CoA dehydrogenase (ACADL), fructose 1,6 bisphosphate aldolase (ALDOA) and lactate dehydrogenase C (LDHC). Expression patterns of these 9 competence-related genes were then compared in IVM and VVM oocytes obtained from both prepubertal and adult animals. The comparison between IVM prepubertal and adult MII oocytes validated 55.6% (5/9) of genes that were determined to be differentially expressed by previous qPCR analyses in this model of oocyte competence. The comparison between IVM and VVM prepubertal oocytes determined that 55.6% (5/9) of our target genes were differentially expressed, and the comparison between IVM and VVM adult oocytes revealed that 88.9% (8/9) of the target genes were differentially expressed between more and less competent oocytes. None of the genes examined were differentially expressed between VVM prepubertal and adult oocytes (Table 1).
Table 1.
Summary of gene symbol, accession number, primer sequences and amplicon product length for the target genes examined.
| Gene Symbol |
Accession Number |
Forward Primer Sequence |
Reverse Primer Sequence |
Product Length |
|---|---|---|---|---|
| GAPDH | AF017079 | acatcaagaaggtggtgaag | attgtcgtaccaggaaatgag | 151 |
| PERV | AF038599 | tggtttgagggatggttca | acgggctttggtactgttgt | 190 |
| SRSF1 | NM_001038007 | tctgcatgtcctctgtgtga | tctaatgctcccaactgcaa | 211 |
| TL10 | CN162638 | aggagcttgtatccccatgt | ccctttcccagcttcataga | 230 |
| MAPK8 | AK230496 | atgaagcatcctgctcacct | cctcgcataccctcaaactt | 176 |
| TNF | NM_001031779 | cccattcaggctcaaacaat | gcctggactacatcccacat | 185 |
| CASP3 | AJ583707 | tgctttgtggaatcaagcac | cccaactgaccaaatgtcaa | 233 |
| CASP8 | AF069648.1 | tgttgcgagagcaatttagc | ttggacaaacacctccatga | 153 |
| HMCGR | BP436947 | gtgaaaaggagcgtgagctt | tgaatgtttcagtcgccaac | 106 |
| SREBF1 | NM_214157 | cactttctgacccgcttctt | gcttgctccaagaggtgttc | 236 |
| ACSL3 | NM_001143698 | ctggtgacagatgccttcaa | aggaatggagtttgcctcac | 184 |
| ACADL | NM_213897 | cttatgtggatgcccgagtt | tagtgttccctccctttcca | 183 |
| G6PD | XM_583628 | ccagaatctcatggtgctga | gatgacacaggcgatgttgt | 199 |
| PGD | CN159092 | aactgtcccacacacactgg | tcttcaccgcactgtcca | 172 |
| HK1 | BX915001 | aaggcagaagccacctaatg | gaaacggacgccactaaact | 166 |
| ALDOA | CA780251 | gctgagtgcagagaagtgtg | gccaggttattccaaggag | 233 |
| PGK1 | AY677198.1 | cttggactgtggtcctgaga | gcacagcaggtagcagtgtc | 204 |
| ENO1 | CK462922 | cacacatggctccagacact | tccctgtgatgtctcactgc | 154 |
| PKM2 | CN166623 | cttccttcatcccttgcttg | tagtacctgtgccgtgatgc | 224 |
| LDHC | U95378.1 | cctcttgggctattggactg | cctcctcctcagcattcaag | 198 |
| PDHA1 | X52990.1 | ggattgctctggcctgtaag | gctctctccacagacgttcc | 181 |
| IDH3B | DQ507860 | tgcagtggaccagagaagag | ccaatgacagcctcagtgaa | 220 |
| ATP5G1 | CF367963 | ttgtgtgatggtcaggaagc | caacctcctcttcgtcctgt | 158 |
| ATP2B1 | NM_214352.1 | cttgcttcggaaaccttacg | ccgtgcatttatttcgttga | 241 |
| TNFAIP6 | M31165 | tcataactccatatggcttgaac | tcttcgtactcatttgggaagcc | 391 |
Experiment 3. Validation of the relationship of TNF to oocyte competence
Experiment 3.1. The effect of TNF during IVM on oocyte nuclear maturation, developmental competence and TNFAIP6 expression in cumulus cells
There was no significant difference in the percentage of mature oocytes between those matured in the presence of 0 ng/ml, 0.1 ng/ml or 1 ng/ml TNF (Table 2). After in vitro fertilization (IVF), no significant difference in the percentage of cleaved embryos or blastocyst development, nor in blastocyst cell number, was found between treatment groups (Table 2). The level of TNFAIP6 mRNA in cumulus cells increased dramatically from zero to 24 hours post maturation (hpm), and then declined at 44 hpm, in all TNF treatments (P<0.05). Expression of TNFAIP6 was significantly increased after treatment with both 0.1 ng/ml and 1 ng/ml TNF compared to control (0 ng/ml) at 24 hpm, although there was no significant difference between 0.1 ng/ml and 1 ng/ml TNF treatment at this time (Figure 2). At 44 hpm, TNFAIP6 mRNA was significantly increased in 0.1 ng/ml TNF compared with control and 1 ng/ml treatment, but there was no significant difference in TNFAIP6 expression between 1 ng/ml and control at this time (Figure 2).
Table 2.
Comparative expression level of competence-related genes in in vivo or in vitro matured oocytes derived from prepubertal or adult pigs.*
| Gene Symbol |
In vitro prepubertal vs. adult |
Prepubertal in vitro vs. in vivo |
Adult in vitro vs. in vivo |
In vivo prepubertal vs. adult |
||||
|---|---|---|---|---|---|---|---|---|
| PERV | 0.821 | nsd | 0.384 | nsd | 0.244 | down | 0.522 | nsd |
| TL10 | 4.035 | up | 1.786 | up | 0.209 | down | 0.473 | nsd |
| SRSF1 | 3.259 | up | 1.357 | nsd | 0.166 | down | 0.398 | nsd |
| TNF | 4.899 | up | 1.87 | up | 0.272 | down | 0.713 | nsd |
| HMCGR | 0.827 | nsd | 0.461 | down | 0.439 | nsd | 0.766 | nsd |
| ACSL3 | 1.963 | up | 1.287 | nsd | 0.432 | down | 0.659 | nsd |
| ACADL | 1.23 | nsd | 0.54 | down | 0.222 | down | 0.506 | nsd |
| ALDOA | 1.55 | nsd | 1.38 | nsd | 0.428 | down | 0.48 | nsd |
| LDHC | 2.176 | up | 1.374 | up | 0.299 | down | 0.473 | nsd |
In each column, gene expression was compared between two groups, stated in the column heading. The numbers in each column showed the relative gene expression level of the first group normalized to that of the second group. ‘Up’ means that the gene was upregulated in the first group compared to the second group of the comparison, and likewise ‘down’ means the gene was down regulated in the first group compared to the second group; ‘nsd’ not significantly different.
Figure 2.
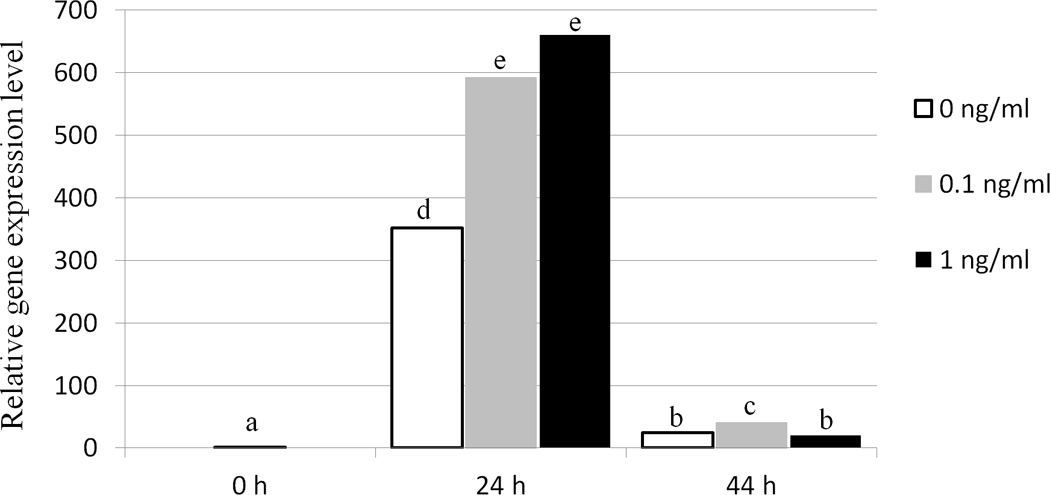
Relative expression of TNFAIP6 in porcine cumulus cells treated with different concentrations of TNF (0 ng/ml, 0.1 ng/ml, 1 ng/ml) during in vitro maturation, as determined by qPCR analysis. Data were normalized to TNFAIP6 expression at 0 h post maturation. Columns with different superscript letters differ significantly between time points and TNF treatments (P <0.05).
Experiment 3.2. The effect of anti-TNF antibody during IVM on oocyte nuclear maturation, developmental competence and TNFAIP6 expression in cumulus cells
The percentage of mature oocytes was significantly decreased after addition of 100 µg/ml anti-TNF during maturation (Table 3). In addition, percentages of cleaved embryos and blastocyst development were also significantly decreased, although blastocyst cell number was not different (Table 3). Embryonic development was also examined after IVM with nonspecific immunoglobulin (IgG) or anti-TNF antibody; no significant difference in the percentage of cleaved embryos was observed between either treatment group and control, although treatment with anti-TNF resulted in reduced embryonic cleavage compared to treatment with IgG. Blastocyst formation, both as a percentage of total oocytes and cleaved embryos, was significantly decreased following anti-TNF treatment during IVM when compared with control, whereas no differences were observed between controls and IgG treatment (Table 4). Although treatment with IgG significantly decreased expression of TNFAIP6 in cumulus cells when compared with control at 24 hpm, a further significant reduction in TNFAIP6 expression was observed following treatment with anti-TNF. At 44 hpm TNFAIP6 mRNA was not significantly different between any treatment group (Figure 3).
Table 3.
Effects of TNF during in vitro maturation on meiotic maturation and subsequent embryonic development of porcine oocytes following IVF/IVC*.
| TNF concentration |
Percentage of oocytes reaching to MII (n) |
Embryonic cleavage (n) |
Blastocyst development (n) |
Blastocyst/cleaved § | Blastocyst cell number (n) |
|---|---|---|---|---|---|
| 0 ng/ml | 66.1 ± 6.1% (62) | 57.8 ± 2.9% (288) | 23.0 ± 2.7% (288) | 39.4 ± 4.0% | 45.0 ± 2.3 (65) |
| 0.1 ng/ml | 62.5 ± 6.5% (58) | 64.2 ± 3.0% (249) | 25.8 ± 2.8% (249) | 39.7 ± 3.9% | 43.1 ± 2.8 (63) |
| 1 ng/ml | 50.0 ± 7.3% (48) | 63.2 ± 2.7% (279) | 26.1 ± 3.7% (279) | 38.2 ± 4.8% | 55.3 ± 2.8 (72) |
Data are reported as mean ± SEM. No significant differences (P > 0.05) were found in percentage of oocytes reaching to MII and embryonic development in all treatment groups.
Percentage of blastocysts is calculated from the cleaved embryos.
Table 4.
Effects of anti-TNF during in vitro maturation on oocyte meiotic maturation and subsequent embryonic development following IVF/IVC.
| Anti-TNF concentration |
Meiotic maturation (n) |
Cleaved embryos (n) |
Blastocyst development (n) | Blastocyst/cleaved § | Blastocyst cell number (n) |
|---|---|---|---|---|---|
| 0 µg/ml | 56.6 ± 6.9%a (53) | 53.8 ± 2.7%a (291) | 17.7 ± 2.0%a (291) | 33.4 ± 3.7%a | 42.9 ± 2.4 (51) |
| 100 µg/ml | 31.4 ± 8.0%b (35) | 39.2 ± 4.2%b (119) | 9.9 ± 2.6%b (119) | 24.2 ± 6.9%b | 51.1 ± 8.9 (12) |
Different superscripts within a column denote a significant difference, P < 0.05. Data are reported as mean ± SEM.
Percentage of blastocysts is calculated from the cleaved embryos.
Figure 3.
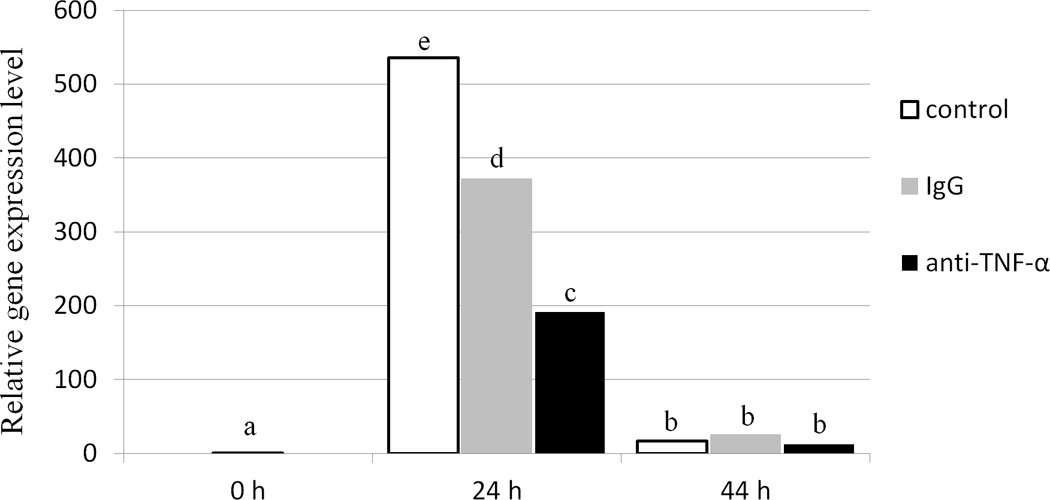
Relative expression of TNFAIP6 in porcine cumulus cells treated with 100 µg/ml of either IgG or anti-TNF during in vitro maturation, as determined by qPCR analysis. Data were normalized against the TNFAIP6 expression level at 0 h post maturation. Bars with different superscripts differ significantly between time points and TNF treatments (P <0.05).
Discussion
In this study, we tested 20 candidate genes that are related to oocyte competence in a prepubertal-versus-adult model. We identified 6 differentially expressed genes identified in other specie (TNF, HMGCR, ACSL3, ACADL, ALDOA, LDHC), plus 3 more genes (PERV, TL10, SRSF1) based on a previous study (Paczkowski and Krisher, unpublished results). We further tested expression of this panel of genes in both in vitro and in vivo matured oocytes derived from both adult and prepubertal females, to validate their importance in a second model system of oocyte competence. In addition, this model allowed us to evaluate how the expression of these genes is altered by the challenges of the in vitro environment. To our knowledge, this is the first time that two well-established models have been used in the same study to determine the relationship between gene expression and oocyte competence in pigs.
The 5 differentially expressed genes (TL10, SRSF1, TNF, ACSL3 and LDHC) identified in both experiments were all upregulated in less competent oocytes in the in vitro prepubertal versus adult model. Interestingly, in adult oocytes, in the in vitro-versus-in vivo matured model, where 8 out of 9 genes were differentially expressed, all genes were downregulated in less competent oocytes. We hypothesize that this discrepancy between up- and down-regulation in less competent oocytes between prepubertal and adult animals is due to differences in the maturity of cellular mechanisms. During the final phase of oocyte development, global transcriptional activity decreases, allowing the oocytes to progress through meiosis (Brevini-Gandolfi et al. 1999). This transcriptional silencing process is mediated by epigenetic mechanisms, including DNA methylation (Curradi et al. 2002). The genome-wide methylation status is significantly lower in IVM prepubertal oocytes than in IVM adult oocytes at the GV stage, indicating that regulation of gene expression mediated by DNA methylation is not fully established in prepubertal oocytes (Ptak et al. 2006). This could result in prepubertal oocytes being more transcriptionally active prior to re-initiation of meiosis, as well as inappropriately continuing some minor level of expression even during meiotic maturation. Even more significantly, transcripts may not be appropriately degraded during meiotic maturation due to the immature cellular mechanisms in IVM prepubertal oocytes. Thus, cellular mechanisms that control transcription and transcript degradation may not be functioning correctly in the immature oocyte, resulting in the observed up-regulation of genes in IVM oocytes compared to adult. It is important to realize that upregulation of gene expression in prepubertal oocytes does not necessarily imply improved oocyte quality.
In adult oocytes, the genome methylation pattern is well established and the transcriptional control system is mature. The down-regulation of target genes in less competent IVM adult oocytes may indicate either insufficient transcript storage before germinal vesicle breakdown (GVBD) in oocytes collected from abattoir-derived ovarian follicles not yet near ovulation, or excessive transcript degradation after GVBD due to the suboptimal in vitro culture environment.
In the prepubertal in vivo-in vitro matured model, in vitro culture environment may cause both up-regulation of genes due to immature cellular transcription control mechanisms, or down-regulation of genes due to the suboptimal environment. Therefore, genes were regulated in different directions in these oocytes. Regulation of specific genes may depend on how extensive the immature gene regulation mechanisms have been altered by the in vitro culture environment in the prepubertal oocyte. We did not compare gene expression patterns between IVM prepubertal oocytes and VVM adult oocytes, as both female age and maturation environment confound this comparison. Thus, it is difficult to draw meaningful conclusions from this comparison as both female age and maturation environment are contributing factors, making it impossible to determine which factor is responsible for the effects observed.
An important point to note is that the differences we observed in gene expression patterns between prepubertal and adult oocytes were only observed after maturation in the in vitro environment, not in vivo, indicating that the prepubertal oocyte is less able to handle the challenges of the in vitro environment, and that these stresses may then negatively alter gene expression and or transcript degradation when compared to adult oocytes.
Of our panel of 9 oocyte competence genes, 5 were further validated in the second experiment by again comparing gene expression levels between IVM prepubertal and adult oocytes. The discrepancies observed in qPCR results between experiments 1 and 2 may be attributed to the independent pools of cDNA utilized for qPCR analysis in these two independent experiments. A previous study of cumulus cells demonstrated that using qPCR analysis to validate microarray data could reach 60% (15 out of 25 target genes) validation rate in the original samples; however, only 8 of the 15 validated genes could be confirmed using independent cumulus cell samples (van Montfoort et al. 2008). Gene expression is variable between pools of cells, particularly when oocytes are pooled in large number and across several females. However, using independent pools of cDNA for qPCR analysis may actually increase the reliability of any consistently identified differences in transcript abundance between two treatment groups.
HMGCR is the rate limiting enzyme of the cholesterol biosynthetic pathway (Brown and Goldstein 1990). Cholesterol-enriched lipid rafts are present in membranes of mouse oocytes and pre-implantation embryos, and treating zygotes with a cholesterol-depleting drug prevents embryonic development (Comiskey and Warner 2007). Exposure to follicular fluid meiosis-activating sterol, an intermediate of cholesterol biosynthesis, during IVM can increase the quality of porcine oocytes (Faerge et al. 2006). These findings suggest that cholesterol is important in oocytes and embryos for supporting pre-implantation development. These results support previous findings that cholesterol synthesis is important to oocyte developmental potential (Faerge et al. 2006).
ACSL3 and ACADL are two important enzymes in the lipid β-oxidation pathway, while LDHC and ALDOA are two enzymes related to glycolysis. A recent study in mice demonstrated that lipid β-oxidation is essential for oocyte developmental competence and early embryo development (Dunning et al. 2010). Porcine oocytes, compared with other mammalian species, are characterized by a high lipid content (McEvoy et al. 2000), stored mainly as lipid droplets in the cytoplasm that are co-localized with mitochondria. Exposure to inhibitors of lipid β-oxidation during oocyte maturation results in developmental failure post IVF (Sturmey et al. 2006). Elevated glucose metabolism via glycolysis in oocytes has been correlated to improved developmental competence in cattle, cats and pigs (Herrick et al. 2006; Krisher and Bavister 1999; Spindler et al. 2000). Recently, our laboratory also demonstrated aberrant protein abundance of ALDOA and lactate dehydrogenase A (LDH-A) in prepubertal COCs, suggesting that these two genes may play a role in the mechanisms resulting in developmental competence (Paczkowski and Krisher 2010). These results substantiate previous work suggesting that fatty acid and glucose metabolism play an important role in oocyte competence (Krisher 2004; Sturmey et al. 2009).
TL10 represents a transcribed locus with unknown gene identification and functional annotation. SRSF1 is an essential sequence specific splicing factor involved in pre-mRNA splicing (Kim et al. 2009). In addition, SRSF1 also mediates post-splicing activities, such as mRNA nuclear export and translation (Michlewski et al. 2008). We currently have no information about how these genes function in the context of oocyte quality.
In the current study, TNF was upregulated in less competent oocytes in the in vitro prepubertal-adult model as well as the prepubertal in vivo-in vitro model. This suggests that high levels of TNF may be detrimental to oocyte competence. Exposure of porcine oocytes to high concentrations of TNF (5 ng/ml) in vitro decreased the percentage of meiotic maturation, whereas exposure to 10 or 100 ng/ml TNF resulted in a significant increase in the frequency of defective spindles or abnormal microfilament distribution (Ma et al. 2010). Despite the negative effects of TNF on oocytes in vitro, several in vivo studies suggested an important role of TNF during follicle development and oocyte growth (Johnson et al. 1999; Onagbesan et al. 2000). Our results demonstrate that some level of TNF is required for oocyte competence, as anti-TNF antibody specifically decreased embryonic development. Together, our findings suggest that TNF is an important component of oocyte maturation, but that excessive concentrations may be detrimental.
Cumulus expansion is mediated by the production of an extracellular matrix comprised of a hyaluronan backbone, and is stabilized by various hyaluronan-binding proteins, including TNFAIP6 (Fulop et al. 1997). In our study, TNFAIP6 expression in cumulus cells increased dramatically at 24 hpm and was responsive to TNF treatment. This pattern of TNFAIP6 expression agrees with that in porcine preovulatory follicles reported by another group. Interestingly, the pattern of TNFAIP6 expression in cumulus cells during IVM is also similar to the pattern of in vivo TNF secretion by oocytes (Johnson et al. 1999). As suggested by several studies, cumulus cell TNFAIP6 expression may be a marker of oocyte competence (Assidi et al. 2010; Assidi et al. 2008; Tesfaye et al. 2009) Our functional study of TNF revealed a positive correlation between the concentration of TNF in IVM medium and TNFAIP6 mRNA expression in cumulus cells. Depletion of endogenous TNF decreased TNFAIP6 mRNA expression levels in cumulus cells and resulted in decreased meiotic maturation and embryonic development. These results suggest that oocyte derived TNF regulates the expression of TNFAIP6 in cumulus cells in vitro, and thus may indeed be reflective of oocyte quality. We hypothesize that a minimal concentration of endogenous oocyte TNF, reflected by a critical transcript level, may be important in regulating appropriate TNFAIP6 expression and function in cumulus cells. In this study, however, high levels of TNFAIP6 in cumulus cells, induced by adding exogenous TNF, did not result in increased oocyte competence. This suggests that a minimal level of TNFAIP6 expression in cumulus cells is required for oocyte competence, but that further increasing this level may not be beneficial. Thus, increased concentrations of TNF in poor quality oocytes, although detrimental, may be reflected by increased TNFAIP6 levels in cumulus cells, suggesting that higher TNFAIP6 expression is not necessarily better for oocyte quality.
The present study provides a molecular sketch of genes and metabolic pathways, including the cholesterol synthesis, fatty acid oxidation and glycolysis, that may be important regulators of oocyte quality in the pig. For one of these genes, TNF, we demonstrate functional importance. Additional functional studies of the remaining target genes in our panel may further elucidate the contribution of these genes and the pathways they participate in to oocyte quality. Ultimately, this information may result not only in a better understanding of oocyte competence and how better to support optimal competence during in vitro maturation, but also in the identification of biomarkers that could be used to predict oocyte quality.
Materials and Methods
Materials
All chemicals were purchased from Sigma Aldrich (St. Louis, MO, USA) unless specified otherwise.
Animals
All procedures were approved by the University of Illinois Institutional Animal Care and Use Committee, and have been judged to minimize discomfort, distress, injury and pain.
In vitro maturation
Ovaries of cycling adult sows and prepubertal gilts were collected from two local abattoirs (Momence Packing Co. and Indiana Packers Corp., respectively), and transferred to the laboratory at 30 − 34°C in 0.9% (w/v) NaCl. Prepubertal ovaries were confirmed by the absence of developed corpora lutea. Oocytes were aspirated from 3–8 mm follicles using an 18-gauge needle. Oocytes with several layers of unexpanded cumulus cells and evenly dark cytoplasm were selected and rinsed in Hepes-buffered synthetic oviductal fluid supplemented with 0.1% BSA (SOF-Hepes (Good and Frishette 1966)). Selected oocytes were matured in vitro in Purdue Porcine Medium modified for maturation (PPMmat) (Herrick et al. 2006; Herrick et al. 2003; Stroble et al. 2002) containing 2 mM glucose, 6 mM lactate, 0.2 mM pyruvate, 0.5 mM cysteamine, 1% recombinant human albumin (recombumin, G-MM, Vitrolife, Kungsbacka, Sweden), 0.2% fetuin, 50 ng/ml EGF, 0.01 units/ml each LH and FSH (Sioux Biochemicals, Sioux City, IA) for 42 to 44 h in 7% CO2 in humidified air at 38.7°C.
Assessment of Meiotic Status
After maturation, oocytes were denuded by vortexing for 3 minutes in SOF-Hepes with 0.01% (w/v) hyaluronidase. Denuded oocytes were mounted, fixed in chloroform:acetic acid:ethanol (1:3:6, v/v/v) for 72 h, stained with 1% (w/v) orcein in acetic acid, and examined for meiotic stage at 400 × magnification. Oocytes reaching telophase and metaphase II were considered mature.
In vitro fertilization (IVF) and embryo culture (IVC)
After maturation, oocytes were denuded as described above, washed twice in modified Tris-buffered medium (mTBM) (Abeydeera and Day 1997) supplemented with 2 mM caffeine and 0.2% (w/v) fraction V BSA and placed in 50 µl drops of mTBM under 10 ml embryo tested, washed mineral oil (20 oocytes/drop). Semen from the same boar was used throughout the experiments. Sperm preparation was performed by placing 500 µl of chilled, extended (1:5 dilution, Androhep EnduraGuard, Minitube of America Inc., Verona, WI, USA) semen, onto a gradient of 45%:90% Percoll (GE Healthcare Life Sciences, Uppsala, Sweden), and centrifuged for 20 min at 700 × g. The supernatant was removed and the remaining sperm pellet washed twice with 5.0 ml D-PBS by centrifuging for 5 min at 1000 × g. Sperm were then counted and diluted (1 × 106 sperm/ml) in mTBM (Abeydeera and Day 1997) and added (50 µl) to fertilization drops containing oocytes for a final sperm concentration of 5 × 105 sperm/ml. Gametes were co-incubated for 5 h in 6% CO2 in humidified air. Following co-incubation, putative zygotes were washed 3 times and cultured in 50 µl NCSU-23 medium (Long et al. 1999; Machaty et al. 1998; Petters and Wells 1993) (10 zygotes/drop) containing 0.4% crystallized BSA (MP Biomedicals, Solon, OH, USA) under 10 ml mineral oil in 6% CO2, 10% O2, balance N2 for 6 days, when embryonic cleavage and blastocyst development were determined. Blastocysts were stained with 0.01 mg/ml Hoechst 33342 to ascertain total blastocyst cell number under fluorescence at 400 × magnification.
In vivo matured ovulated oocyte collection
Adult and prepubertal pigs were synchronized then superstimulated prior to surgical oocyte retrieval. Superovulation maximizes the number of oocytes retrieved per female so that costs can be minimized. However, superovulation does influence oocyte quality (Baart et al. 2009; Foote and Ellington 1988).Therefore the oocytes used in this study, although in vivo matured, are somewhat different than naturally ovulated oocytes. It is possible that expression of our target genes may differ between superovulated and naturally ovulated in vivo matured oocytes. In spite of the physiologic differences reported in superovulated oocytes, such oocytes are still routinely used to produce normal offspring in both human medicine and agricultural animal production systems. Compared with IVM, superovulation has fewer effects on oocyte quality, and thus has been considered a more competent oocyte in this study. The animals’ estrous cycles were synchronized by feeding 15 mg/day of an oral progestin, altrenogest (Matrix; Intervet America Inc. Millsboro, DE, USA) for 14–18 days. The day following the final day of matrix feeding, females received 1000 IU of pregnant mares’ serum gonadotropin (Calbiochem, La Jolla, CA, USA) followed in 72 h by 1000 IU of human chorionic gonadotropin (hCG, Calbiochem). Surgical oocyte retrieval was performed 40 h post hCG injection. Pigs were restrained with a limp rope snare and injected with an initial intra-muscular injection of 3 ml anesthetic cocktail consisting of 5.9 mg/kg Ketamine, 1.47 mg/kg Telozol, 2.9 mg/kg Xylazine and 0.09 mg/kg atropine, followed by 7 ml intra-venous injection of the same solution via ear vein canula. Endotracheal tubes were inserted into the nares for connection to the anesthesia apparatus, supplying oxygen (2 L/min) and 5% halothane. A midline, lower abdominal incision was made through the body wall, the oviduct and part of the uterus exteriorized, and ovulated oocytes recovered via retrograde flush of the oviducts using approximately 20 ml warm SOF-Hepes. If necessary, oocytes were denuded via vortexing for 3 min in SOF-Hepes with 0.01% (w/v) hyaluronidase.
Quantitative PCR
Oocytes
Metaphase II stage oocytes were identified by extrusion of the first polar body before being frozen at −80°C with lysis buffer (Dynabeads mRNA DIRECT Kit, Invitrogen, Carlsbad, CA). Three separate pools of 20 oocytes each were collected for each treatment for qPCR analysis. Poly A+ RNA was extracted from the samples using the Dynabeads mRNA DIRECT Kit (Invitrogen).
Half of the mRNA from each sample was used to assess genomic DNA contamination by reverse transcriptase (RT) and mock-RT reactions. RNA samples (3 µl) were incubated with 1 µl dNTP (10 mM), 2 µl random hexamer primers (dN6; 100 ng/µl), and 4 µl water for 5 min at 65 °C. Random primed RNA samples were divided for RT and mock RT reactions. For RT reactions, 50% of random primed RNA (5 µl) was incubated with 2 µl 5× transcription buffer, 1 µl dithiothreitol (0.1 mM), 0.75 µl Moloney Murine Leukemia Virus (MMLV; 200 U/µl), 0.5 µl RNase Out (40 U/µl), and 0.75 µl water. Mock RT reactions were performed using the remaining 50% of random primed RNA as described above; however, MMLV and RNase Out were excluded from the samples. Reverse transcription was performed at 25 °C for 10 min, 37 °C for 50 min, and 70 °C for 10 min. cDNA samples were amplified for GAPDH with 0.5 µl dNTP (10 mM), 0.5 µl forward (20 µM) and 0.5 µl reverse (20 µM) primers, 2.5 µl 10× transcription buffer, 0.25 µl HotMaster Taq (5 U/µl) polymerase (Eppendorf, Hamburg, Germany), and 15.75 µl water. Samples were incubated at 94 °C for 1 min and 40 seconds, 40-cycles of 94 °C for 20 seconds, 58 °C for 10 seconds, and 65 °C for 30 seconds, followed by 65 °C for 2 min. PCR products were separated by gel electrophoresis and stained with ethidium bromide. The presence of a gel band in mock PCR samples indicated genomic DNA contamination. All samples displayed a positive RT reaction and did not contain genomic DNA.
For the remaining half of the mRNA, single strand and double strand cDNA were generated using the RT template switching SMART technology (Wang et al. 2000; Zhu et al. 2001) and DNA samples were purified using the Machery-Nagel PCR Clean-up Gel Extraction Kit (Machery-Nagel, Easton, PA). Linear amplification was performed using the MEGAscript High Yield Transcription Kit (Ambion, Austin, TX), followed by DNase treatment. Amplified mRNA samples were purified from template cDNA using the Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA) and single strand cDNA were generated by random primed cDNA synthesis. Primer design and qPCR were performed as previously described (Fleming-Waddell et al. 2007). Current gene annotations were obtained from the Genbank and primers were designed for qPCR using Primer3 (Rozen and Skaletsky 2000). Accession number, primer sequence, and product length of target genes are presented in Table 5. Information for primers for GAPDH, PERV, SRSF1, TL10 (Paczkowski and Krisher, unpublished results) and TNFAIP6 (Shimada et al. 2004) were previously reported. For the remaining genes, primers were tested on pools of 20 porcine oocytes and primer specificity was determined by melting curve analysis and gel electrophoresis. PCR products were cloned into pCR 2.1 TOPO vectors and transformed into One Shot TOP10 chemically competent E. coli. Plasmids were sequenced to confirm the identity of the transcript and quantified using a ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Target genes were analyzed using QuantiFast SYBR Green PCR reagents (Qiagen) on a MasterCycler Realplex2 (Eppendorf North America, Inc; Westbury, NY, USA). A standard curve was generated from serial dilutions of EcoRI digested plasmids (107 to 101 molecules) and the efficiency of the primers was calculated.
Table 5.
Effects of anti-TNF-α and IgG during porcine oocyte in vitro maturation on subsequent embryonic development following IVF/IVC.
| Treatment | Cleaved embryos (n) |
Blastocyst development (n) |
Blastocyst/cleaved | Blastocyst cell number (n) |
|---|---|---|---|---|
| Control | 49.7 ± 3.3%ab (115) | 11.1 ± 1.7%a(115) | 23.7 ± 3.9%a | 59.2 ± 6.3 (12) |
| 100 µg/ml IgG | 64.0 ± 5.6%a (81) | 8.6 ± 1.8%ab(81) | 14.3 ± 3.1%ab | 47.7 ± 3.6 (8) |
| 100 µg/ml anti TNF | 47.0 ± 3.9%b (91) | 3.0 ± 1.5%b(91) | 5.3 ± 2.7%b | 45.5 ± 3.0 (4) |
Different superscripts within a column denote significant difference, P < 0.05. Data are reported as mean ± SEM.
Cumulus cells
After removal from oocytes, cumulus cells were transferred to a 1.5 ml tube and centrifuged for 1 min at 700 × g. The supernatant was removed and the cumulus cells were frozen at −80°C for qPCR analysis. Total RNA was extracted from pooled samples of cumulus cells using the RNeasy kit (Qiagen) as recommended by the manufacturer, followed by DNase treatment and eluted in 30 µl water. After extraction, the total RNA concentration of cumulus cell samples was determined by using the Quant-iT RiboGreen RNA Assay Kit (Invitrogen). Since the number of cumulus cells varied between pooled samples, the lowest RNA concentration in all samples was used as the standard for normalization and an equal amount of RNA was used for all reverse transcription reactions. Reverse transcription was performed with SuperScript III (Invitrogen) and random hexamer primers (dN6; 100 ng/µl). Reactions were performed at 42°C for 15 min, 50°C for 50 min and at 70°C for 15 min for enzyme inactivation. Primer design and qPCR were performed as described above.
Experimental design
Experiment 1. The objective of this experiment was to compare transcript abundance of 20 candidate genes between prepubertal and adult MII oocytes derived following IVM to identify a ‘panel’ of candidate genes that are reflective of oocyte competence. Each treatment was replicated three times, with 20 oocytes per pool in each replicate.
Experiment 2. The objective of this experiment was to further examine a select subset of genes from Experiment 1 that were differentially expressed between IVM and VVM MII oocytes collected from both prepubertal and adult females (2×2 factorial; in vitro-versus-in vivo matured model). In vivo matured ovulated MII oocytes were collected from 6 prepubertal and 6 adult females. Transcript abundance of the selected genes was compared between these 4 treatment groups. In each treatment, oocytes were pooled in 3 separate replicates with 20 oocytes per replicate.
Experiment 3. The objective of this experiment was to validate the functional relationship of TNF to oocyte competence in adult oocytes, suggested by our gene expression analysis results. In Experiment 3.1, the effect of TNF supplementation during IVM on a) oocyte nuclear maturation and subsequent embryo development and b) TNFAIP6 mRNA expression in cumulus cells was examined. Adult oocytes were matured in PPMmat with TNF (0, 0.1 and 1 ng/ml). In each replicate, 10 oocytes were used to determine meiotic status after IVM, and 40–50 oocytes from the same pool were used for in vitro fertilization (IVF) and in vitro embryo culture (IVC) to assess embryonic development. This experiment was replicated 6 times. In addition, three separate pools of cumulus cells from 10 COCs were independently sampled from each treatment at 0 h, 24 h and 44 hpm to analyze TNFAIP6 expression. In Experiment 3.2, the effect of anti-TNF antibody during in vitro oocyte maturation on a) subsequent embryonic development and b) TNFAIP6 mRNA expression in cumulus cells was examined. Adult oocytes were matured in PPMmat or PPMmat with 100 µg/ml anti-TNF antibody. In each replicate, 10 oocytes were used to determine meiotic status after IVM, and 40–50 oocytes from the same pool were used for IVF and IVC to assess embryonic development. This experiment was replicated 4 times. To determine the specificity of antibody effects, oocytes were matured in PPMmat without antibody, and PPMmat with 100 µg/ml of either IgG or anti-TNF antibody. Cumulus cells from 10 COCs were sampled from each treatment at 0 h, 24 h and 44 hpm for qPCR analysis. The remaining oocytes in each pool were used for IVF and IVC to assess subsequent embryonic developmental. This experiment was replicated 3 times.
Statistical analysis
For oocyte qPCR results, the mRNA abundance of target genes was normalized to an internal control, GAPDH, for sample to sample comparisons. For cumulus cell qPCR results, in which the RNA could be quantified, the expression level of the target gene at 0 hpm was set at 1, and data from other time points were calculated accordingly. Relative expression ratios were obtained by the comparative threshold cycle method (Livak and Schmittgen 2001). Data were analyzed using the relative expression software tool, REST 2009 version 2.0.13 (Pfaffl et al. 2002). Percentages of maturation, embryonic cleavage, and blastocyst development, as well as blastocyst total cell number were analyzed using two-way ANOVA, with maturation treatment included in the model as a fixed factor and replicate as a random factor. Percentage data (maturation success, embryonic cleavage and blastocyst development) were arcsin transformed. Treatment differences were determined by Bonferroni or Fisher's LSD Least Significant Difference (LSD) multiple comparison test when treatment was significant. Significance was determined as P<0.05. Data are reported as mean ± SEM.
Acknowledgements
We thank Dr. Ryan Cabot and Dr. Zoltan Machaty for providing prepubertal ovaries, and Dr. Matthew Wheeler for helping with in vivo matured oocyte collection.
Funding for this project was provided by the University of Illinois, College of Agricultural, Consumer and Environmental Sciences.
Abbreviations used in the manuscript
- GV
germinal vesicle (GV)
- GVBD
germinal vesicle breakdown
- hpm
hours post maturation
- IVF
in vitro fertilization
- IVM
in vitro matured
- MII
metaphase II
- qPCR
quantitative real-time PCR
- VVM
in vivo matured
Footnotes
J. Ida was partially funded by National Center for Research Resources, NIH, T35 RR020292 grant.
References
- Abeydeera LR, Day BN. Fertilization and subsequent development in vitro of pig oocytes inseminated in a modified tris-buffered medium with frozen-thawed ejaculated spermatozoa. Biology of Reproduction. 1997;57(4):729–734. doi: 10.1095/biolreprod57.4.729. [DOI] [PubMed] [Google Scholar]
- Assidi M, Dieleman SJ, Sirard MA. Cumulus cell gene expression following the LH surge in bovine preovulatory follicles: potential early markers of oocyte competence. Reproduction. 2010;140(6):835–852. doi: 10.1530/REP-10-0248. [DOI] [PubMed] [Google Scholar]
- Assidi M, Dufort I, Ali A, Hamel M, Algriany O, Dielemann S, Sirard MA. Identification of potential markers of oocyte competence expressed in bovine cumulus cells matured with follicle-stimulating hormone and/or phorbol myristate acetate in vitro. Biol Reprod. 2008;79(2):209–222. doi: 10.1095/biolreprod.108.067686. [DOI] [PubMed] [Google Scholar]
- Baart EB, Macklon NS, Fauser BJ. Ovarian stimulation and embryo quality. Reprod Biomed Online. 2009;18 Suppl 2:45–50. doi: 10.1016/s1472-6483(10)60448-8. [DOI] [PubMed] [Google Scholar]
- Brevini-Gandolfi TA, Favetta LA, Mauri L, Luciano AM, Cillo F, Gandolfi F. Changes in poly(A) tail length of maternal transcripts during in vitro maturation of bovine oocytes and their relation with developmental competence. Mol Reprod Dev. 1999;52(4):427–433. doi: 10.1002/(SICI)1098-2795(199904)52:4<427::AID-MRD12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Atherosclerosis. Scavenging for receptors. Nature. 1990;343(6258):508–509. doi: 10.1038/343508a0. [DOI] [PubMed] [Google Scholar]
- Comiskey M, Warner CM. Spatio-temporal localization of membrane lipid rafts in mouse oocytes and cleaving preimplantation embryos. Dev Biol. 2007;303(2):727–739. doi: 10.1016/j.ydbio.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curradi M, Izzo A, Badaracco G, Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol. 2002;22(9):3157–3173. doi: 10.1128/MCB.22.9.3157-3173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente R, Eppig JJ. Transcriptional activity of the mouse oocyte genome: companion granulosa cells modulate transcription and chromatin remodeling. Dev Biol. 2001;229(1):224–236. doi: 10.1006/dbio.2000.9947. [DOI] [PubMed] [Google Scholar]
- Dunning KR, Cashman K, Russell DL, Thompson JG, Norman RJ, Robker RL. Beta-Oxidation Is Essential for Mouse Oocyte Developmental Competence and Early Embryo Development. Biol Reprod. 2010 doi: 10.1095/biolreprod.110.084145. [DOI] [PubMed] [Google Scholar]
- Eppig JJ. Coordination of nuclear and cytoplasmic oocyte maturation in eutherian mammals. Reprod Fertil Dev. 1996;8(4):485–489. doi: 10.1071/rd9960485. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, O'Brien MJ. Comparison of preimplantation developmental competence after mouse oocyte growth and development in vitro and in vivo. Theriogenology. 1998;49(2):415–422. doi: 10.1016/s0093-691x(97)00413-5. [DOI] [PubMed] [Google Scholar]
- Faerge I, Strejcek F, Laurincik J, Rath D, Niemann H, Schellander K, Rosenkranz C, Hyttel PM, Grondahl C. The effect of FF-MAS on porcine cumulus-oocyte complex maturation, fertilization and pronucleus formation in vitro. Zygote. 2006;14(3):189–199. doi: 10.1017/S0967199406003765. [DOI] [PubMed] [Google Scholar]
- Fleming-Waddell JN, Wilson LM, Olbricht GR, Vuocolo T, Byrne K, Craig BA, Tellam RL, Cockett NE, Bidwell CA. Analysis of gene expression during the onset of muscle hypertrophy in callipyge lambs. Anim Genet. 2007;38(1):28–36. doi: 10.1111/j.1365-2052.2006.01562.x. [DOI] [PubMed] [Google Scholar]
- Foote RH, Ellington JE. Is a superovulated oocyte normal? Theriogenology. 1988;29(1):111–123. [Google Scholar]
- Fulop C, Kamath RV, Li Y, Otto JM, Salustri A, Olsen BR, Glant TT, Hascall VC. Coding sequence, exon-intron structure and chromosomal localization of murine TNF-stimulated gene 6 that is specifically expressed by expanding cumulus cell-oocyte complexes. Gene. 1997;202(1–2):95–102. doi: 10.1016/s0378-1119(97)00459-9. [DOI] [PubMed] [Google Scholar]
- Good AE, Frishette WA. Crystals in dried smears of synovial fluid. JAMA. 1966;198(1):198–199. doi: 10.1001/jama.198.1.198. [DOI] [PubMed] [Google Scholar]
- Herrick JR, Brad AM, Krisher RL. Chemical manipulation of glucose metabolism in porcine oocytes: effects on nuclear and cytoplasmic maturation in vitro. Reproduction. 2006;131(2):289–298. doi: 10.1530/rep.1.00835. [DOI] [PubMed] [Google Scholar]
- Herrick JR, Conover-Sparman ML, Krisher RL. Reduced polyspermic fertilization of porcine oocytes utilizing elevated bicarbonate and reduced calcium concentrations in a single-medium system. Reprod Fertil Dev. 2003;15(4):249–254. doi: 10.1071/rd03001. [DOI] [PubMed] [Google Scholar]
- Hodgman R, Tay J, Mendez R, Richter JD. CPEB phosphorylation and cytoplasmic polyadenylation are catalyzed by the kinase IAK1/Eg2 in maturing mouse oocytes. Development. 2001;128(14):2815–2822. doi: 10.1242/dev.128.14.2815. [DOI] [PubMed] [Google Scholar]
- Johnson ML, Murdoch J, Van Kirk EA, Kaltenbach JE, Murdoch WJ. Tumor necrosis factor alpha regulates collagenolytic activity in preovulatory ovine follicles: relationship to cytokine secretion by the oocyte-cumulus cell complex. Biol Reprod. 1999;61(6):1581–1585. doi: 10.1095/biolreprod61.6.1581. [DOI] [PubMed] [Google Scholar]
- Khatir H, Lonergan P, Carolan C, Mermillod P. Prepubertal bovine oocyte: a negative model for studying oocyte developmental competence. Mol Reprod Dev. 1996;45(2):231–239. doi: 10.1002/(SICI)1098-2795(199610)45:2<231::AID-MRD17>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Kim DJ, Oh B, Kim YY. Splicing factor ASF/SF2 and transcription factor PPAR-gamma cooperate to directly regulate transcription of uncoupling protein-3. Biochem Biophys Res Commun. 2009;378(4):877–882. doi: 10.1016/j.bbrc.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Krisher RL. The effect of oocyte quality on development. J Anim Sci. 2004;82 E-Suppl:E14–E23. doi: 10.2527/2004.8213_supplE14x. [DOI] [PubMed] [Google Scholar]
- Krisher RL, Bavister BD. Enhanced glycolysis after maturation of bovine oocytes in vitro is associated with increased developmental competence. Mol Reprod Dev. 1999;53(1):19–26. doi: 10.1002/(SICI)1098-2795(199905)53:1<19::AID-MRD3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Leoni GG, Bebbere D, Succu S, Berlingeur F, Mossa F, Galioto M, Boglioli L, Ledda S, Naitana S. Relations between relative mRNA abundance and developmental competence of ovine oocytes. Molecular Reproduction and Development. 2007;74(2):249–257. doi: 10.1002/mrd.20442. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long CR, Dobrinsky JR, Johnson LA. In vitro production of pig embryos: comparisons of culture media and boars. Theriogenology. 1999;51(7):1375–1390. doi: 10.1016/S0093-691X(99)00081-3. [DOI] [PubMed] [Google Scholar]
- Lunney JK. Advances in swine biomedical model genomics. Int J Biol Sci. 2007;3(3):179–184. doi: 10.7150/ijbs.3.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CH, Yan LY, Qiao J, Sha W, Li L, Chen Y, Sun QY. Effects of tumor necrosis factor-alpha on porcine oocyte meiosis progression, spindle organization, and chromosome alignment. Fertil Steril. 2010;93(3):920–926. doi: 10.1016/j.fertnstert.2009.01.131. [DOI] [PubMed] [Google Scholar]
- Machaty Z, Day BN, Prather RS. Development of early porcine embryos in vitro and in vivo. Biol Reprod. 1998;59(2):451–455. doi: 10.1095/biolreprod59.2.451. [DOI] [PubMed] [Google Scholar]
- Marchal R, Feugang JM, Perreau C, Venturi E, Terqui M, Mermillod P. Meiotic and developmental competence of prepubertal and adult swine oocytes. Theriogenology. 2001;56(1):17–29. doi: 10.1016/s0093-691x(01)00539-8. [DOI] [PubMed] [Google Scholar]
- McEvoy TG, Coull GD, Broadbent PJ, Hutchinson JS, Speake BK. Fatty acid composition of lipids in immature cattle, pig and sheep oocytes with intact zona pellucida. Journal of Reproduction & Fertility. 2000;118(1):163–170. [PubMed] [Google Scholar]
- Mermillod P, Le Bourhis D, Lonergan P, Khatir H, Heyman Y. Assessment of cytoplasmic competence of prepubertal calf oocytes by use of nuclear transfer. Theriogenology. 1998;49(1):187–187. [Google Scholar]
- Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30(2):179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- O'BRIEN JK, Dwarte D, Ryan JP, Maxwell WM, Evans G. Developmental capacity, energy metabolism and ultrastructure of mature oocytes from prepubertal and adult sheep. Reprod Fertil Dev. 1996;8(7):1029–1037. doi: 10.1071/rd9961029. [DOI] [PubMed] [Google Scholar]
- Onagbesan OM, Mast J, Goddeeris B, Decuypere E. Effect of TNF-alpha on LH and IGF-I modulated chicken granulosa cell proliferation and progesterone production during follicular development. J Reprod Fertil. 2000;120(2):433–442. [PubMed] [Google Scholar]
- Paczkowski M, Krisher R. Aberrant protein expression is associated with decreased developmental potential in porcine cumulus-oocyte complexes. Mol Reprod Dev. 2010;77(1):51–58. doi: 10.1002/mrd.21102. [DOI] [PubMed] [Google Scholar]
- Paynton BV, Rempel R, Bachvarova R. Changes in state of adenylation and time course of degradation of maternal mRNAs during oocyte maturation and early embryonic development in the mouse. Dev Biol. 1988;129(2):304–314. doi: 10.1016/0012-1606(88)90377-6. [DOI] [PubMed] [Google Scholar]
- Petters RM, Wells KD. Culture of pig embryos. J Reprod Fertil. 1993 Suppl 48:61–73. [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak G, Matsukawa K, Palmieri C, Della Salda L, Scapolo PA, Loi P. Developmental and functional evidence of nuclear immaturity in prepubertal oocytes. Hum Reprod. 2006;21(9):2228–2237. doi: 10.1093/humrep/del184. [DOI] [PubMed] [Google Scholar]
- Revel F, Mermillod P, Peynot N, Renard JP, Heyman Y. Low developmental capacity of in vitro matured and fertilized oocytes from calves compared with that of cows. J Reprod Fertil. 1995;103(1):115–120. doi: 10.1530/jrf.0.1030115. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Salamone DF, Damiani P, Fissore RA, Robl JM, Duby RT. Biochemical and developmental evidence that ooplasmic maturation of prepubertal bovine oocytes is compromised. Biol Reprod. 2001;64(6):1761–1768. doi: 10.1095/biolreprod64.6.1761. [DOI] [PubMed] [Google Scholar]
- Sasaki R, Nakayama T, Kato T. Microelectrophoretic analysis of changes in protein expression patterns in mouse oocytes and preimplantation embryos. Biol Reprod. 1999;60(6):1410–1418. doi: 10.1095/biolreprod60.6.1410. [DOI] [PubMed] [Google Scholar]
- Shimada M, Yamashita Y, Ito J, Okazaki T, Kawahata K, Nishibori M. Expression of two progesterone receptor isoforms in cumulus cells and their roles during meiotic resumption of porcine oocytes. J Mol Endocrinol. 2004;33(1):209–225. doi: 10.1677/jme.0.0330209. [DOI] [PubMed] [Google Scholar]
- Spindler RE, Pukazhenthi BS, Wildt DE. Oocyte metabolism predicts the development of cat embryos to blastocyst in vitro. Mol Reprod Dev. 2000;56(2):163–171. doi: 10.1002/(SICI)1098-2795(200006)56:2<163::AID-MRD7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Hake LE, Richter JD. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J. 1996;15(10):2582–2592. [PMC free article] [PubMed] [Google Scholar]
- Stroble KA, Herrick JR, Conover ML, Krisher RL. Assessment of a novel media system for in vitro porcine embryo production. Biology of Reproduction. 2002;66 Suppl. 1:145. [Google Scholar]
- Sturmey RG, O'Toole PJ, Leese HJ. Fluorescence resonance energy transfer analysis of mitochondrial:lipid association in the porcine oocyte. Reproduction. 2006;132(6):829–837. doi: 10.1530/REP-06-0073. [DOI] [PubMed] [Google Scholar]
- Sturmey RG, Reis A, Leese HJ, McEvoy TG. Role of fatty acids in energy provision during oocyte maturation and early embryo development. Reproduction in Domestic Animals. 2009;44 Suppl 3:50–58. doi: 10.1111/j.1439-0531.2009.01402.x. [DOI] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Affourtit J, Eppig JJ. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;302(1):104–117. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesfaye D, Ghanem N, Carter F, Fair T, Sirard MA, Hoelker M, Schellander K, Lonergan P. Gene expression profile of cumulus cells derived from cumulus-oocyte complexes matured either in vivo or in vitro. Reprod Fertil Dev. 2009;21(3):451–461. doi: 10.1071/rd08190. [DOI] [PubMed] [Google Scholar]
- van Montfoort APA, Geraedts JPM, Dumoulin JCM, Stassen APM, Evers JLH, Ayoubi TAY. Differential gene expression in cumulus cells as a prognostic indicator of embryo viability: a microarray analysis. Molecular Human Reproduction. 2008;14(3):157–168. doi: 10.1093/molehr/gam088. [DOI] [PubMed] [Google Scholar]
- Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM. High-fidelity mRNA amplification for gene profiling. Nat Biotechnol. 2000;18(4):457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. BioTechniques. 2001;30(4):892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]


