Abstract
Most core components of the neurotransmitter release machinery have homologues in other types of intracellular membrane traffic, likely underlying a universal mechanism of intracellular membrane fusion. However, no clear similarity between Munc13s and protein families generally involved in membrane traffic has been reported, despite the essential nature of Munc13s for neurotransmitter release. This crucial function was ascribed to a minimal Munc13 region called the MUN domain, which likely participates in SNARE complex assembly and is also found in CAPS. We have now used comparative sequence and structural analyses to study the structure and evolutionary origin of the MUN domain. We found weak, yet significant sequence similarities between the MUN domain and a set of protein subunits from several related vesicle tethering complexes, such as Sec6 from the exocyst complex and Vps53 from the GARP complex. Such an evolutionary relationship allows structure prediction of the MUN domain and suggests functional similarities between MUN domain-containing proteins and multisubunit tethering complexes such as exocyst, COG, GARP and Dsl1p. These findings further unify the mechanism of neurotransmitter release with those of other types of intracellular membrane traffic, and in turn support a role for tethering complexes in SNARE complex assembly.
Keywords: Munc13; CAPS; MUN domain; multisubunit tethering complexes exocyst, COG, GARP and Dsl1p complex; homology inference and structure prediction
Introduction
Extensive studies in diverse membrane compartments of organisms ranging from yeast to humans have shown that most types of intracellular membrane traffic are governed by members of several protein families, indicating the existence of a conserved core machinery for intracellular membrane fusion. 1 These conserved proteins include N-ethylmaleimide sensitive factor (NSF)/Sec18p, soluble NSF attachment proteins (SNAPs)/Sec17p, SNAP receptors (SNAREs), Sec1/Munc18 (SM) proteins, and small GTPases from the Rab family. 2 Correspondingly, the machinery that controls the release of neurotransmitters by Ca2+-triggered synaptic vesicle exocytosis contains members from each one of these families, and in addition includes specialized components that confer the exquisite regulation of this type of membrane fusion, which is critical for brain function. 3 Proteins from the Munc13/Unc13 family have commonly been considered as examples of such specialized factors, since they have not been related to any protein families generally involved in membrane traffic. However, genetic deletion of these proteins leads to abrogation of all forms of neurotransmitter release (spontaneous, sucrose-induced and Ca2+-evoked release) ,4; 5; 6 and the severity of this phenotype suggests an essential function that raises some critical questions: Is this function specific to synaptic vesicle fusion? Or is this function conserved in other types of intracellular membrane traffic? And, if so, which proteins perform this function in other systems? It is also intriguing that most types of intracellular membrane traffic depend on so-called tethering complexes such as exocyst, COG, GARP, Dsl1p complex, TRAPP and HOPS,7; 8; 9 but no such complexes have been clearly linked to neurotransmitter release.
The two major Munc13 isoforms in the mammalian brain are Munc13-1 and Munc13-2.10 They share a common domain content with two or three C2 domains (named C2A, C2B and C2C) and a diacylglycerol-binding C1 domain. Sequence analyses revealed the presence of two conserved regions between C2B and C2C in Munc13-related proteins (Munc13 homology domains (MHD) 1 and 2).11 A later study showed that the homologous region in Munc13 homologues extends beyond the MHD1 and MHD2 domains to include most of the region between C2B and C2C. 12 This extended homologous region (named the MUN domain) forms an independent folding unit and is required for vesicle priming. 12 This crucial function of the Munc13 MUN domain in neurotransmitter release likely involves a role in promoting assembly of SNARE complexes, since the Munc13-1 MUN domain binds to SNARE complexes and syntaxin-1/SNAP-25 heterodimers. 13; 14 MUN domains are also present in the Ca2+-dependent activator protein for secretion (CAPS), which may have a related role in vesicle priming, 15 and in distantly related proteins from fungi and plants. 11 Secondary structural predictions and fold recognition exercises suggested that the MUN domain mainly consists of alpha-helices and may form helical repeats. 12 The 3-dimensional structure and evolutionary origin of the MUN domain remain unknown.
With rapidly increasing numbers of protein sequences and structures in current databases and the development of powerful similarity search and structure prediction tools, more subtle homologous relationships between protein families can be revealed by comparative sequence and structural analysis. In this study, we show that the MUN domain is homologous to a set of protein subunits in several related multisubunit tethering complexes, including exocyst, COG, GARP and Dsl1p complex. These findings uncover what is likely to be a key missing link between the mechanisms of neurotransmitter release and those of other types of intracellular membrane traffic, allowing structure prediction of the MUN domain, and suggesting functional similarities between Munc13, CAPS and tethering complexes.
Sequence similarity searches and structure prediction for the MUN domain
Previous studies have not revealed homologous relationships between the MUN domain and known protein domain families. PSI-BLAST 16 searches using the MUN domain from mouse Munc13-1 (gene identification number (gi): 158706507, residues 859-1531) as the query found weak yet significant sequence similarities (e-value < 0.001) to Sec6 and Vps53, which are subunits of the tethering complexes exocyst and GARP (Golgi-associated retrograde protein), respectively. PSI-BLAST searches from Sec6 and Vps53 homologues also found MUN domains with significant e-values (<0.001). Using Munc13s and other MUN domain-containing proteins as queries, the profile-profile comparison-based program HHsearch 17 also consistently found Pfam 18 families of subunits from tethering complexes, such as exocyst, COG (conserved oligomeric Golgi), GARP and the Dsl1p complex, as top hits to the regions corresponding to the MUN domain. For example, the MUN domain from mouse Munc13-1 found Sec6 (pfam entry: PF06046) with a probability score of 98; the MUN domain-containing protein gi|22326641 from Arabidopsis thaliana found exocyst subunits Sec6 (score: 99.6), Sec3 (pfam entry: PF09763; score: 93.9), Exo70 (pfam entry: PF03081; score: 93.0) and Sec15 (pfam entry: PF04091; score: 90.5), Dsl1p complex subunit RINT1_TIP1 (pfam entry: PF04437; score: 86.7), and COG subunit COG4 (pfam entry: PF08318; score: 77.9).
Multisubunit tethering complexes mediate initial interactions between vesicles and the target membrane. 19 Although their exact functions remain unclear, they could be important to ensure membrane fusion specificity, and possibly catalyze key reactions in the fusion events. Both sequence analyses 9; 20 and 3D structure determinations 21; 22 have revealed distant similarities between many subunits of four member tethering complexes: exocyst (consisting of Sec3, Sec5, Sec6, Sec10, Sec15, Exo70 and Exo84; functioning in exocytosis), COG (consisting of COG1, COG2, COG3, COG4, COG5, COG6, COG7 and COG8; functioning in the Golgi) , GARP (consisting of Vps51, Vps52, Vps53 and Vps54; functioning in endosome to Golgi transport), and Dsl1p complex (consisting of Tip20p/RINT-1, Sec20 and Dsl1p/ZW10; functioning in Golgi to ER retrograde transport). Three other multisubunit tethering complexes TRAPI, TRAPII, and HOPS are evolutionarily unrelated to exocyst/COG/GARP/Dsl1p. 20
The structures of Exo70 of the exocyst complex 23; 24; 25 and Tip20p of the Dsl1p complex 26 revealed a similar arrangement of four domains (A-D) forming right-handed superhelices. In Exo70, these four domains are arranged in a linear way and adopt a rod-shaped structure. In Tip20p, the different orientation between the four domains results in a curved, hook-like structure. Structures of the C-terminal regions of exocyst subunits Sec6 27 and Sec15 28 have resemblance to domains C and D of Exo70 and Tip20p, while structures of the C-terminal region of Exo84 25 and N-terminal region of Dsl1p 26 show similarities to domains A and B of Exo70 and Tip20p. Another protein with a similar structure is the cargo binding domain of the yeast myosin V, 29 which has a related function in tethering yeast secretary vesicles to actin filaments. In the structural classification of proteins (SCOP) database, 30 the superhelical motif of Exo70 is classified in the ‘cullin-repeat like’ superfamily of the ‘alpha-alpha superhelix’ fold.
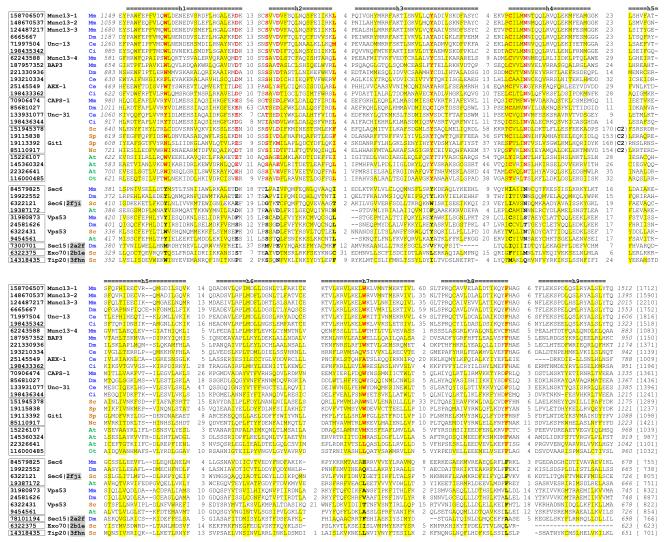
Multiple sequence alignments 31 of MUN domains, Sec6, Vps53, Sec15 and Tip20p suggested the presence of domains A, B, C and D in MUN domains (supplementary Figure1). The regions corresponding to domains C and D in MUN domains are more conserved than regions corresponding to domains A and B. Figure 1 shows the alignment of domains C and D in MUN domains and representative tethering complex subunits (Sec6, Vps53, Exo70, Sec15 and Tip20p). Secondary structure predictions and hydrophobicity patterns suggested the presence of nine major helices in MUN domains corresponding to the ones observed in domains C and D of Tip20p (the last helix is not present in the yeast Exo70 with solved structure).
Figure 1. Multiple sequence alignment of domain C and domain D of the MUN domains.
This alignment was constructed by the PROMALS3D program 31 with manual adjustment according to available 3D structures and secondary structure predictions. The proteins are identified by their NCBI gene identification (gi) numbers. Their grouping is suggested by boxes around the gi numbers. Common names for some experimentally characterized proteins follow the gi numbers. PDB ids for sequences with known structures are shaded and shown after the common names. Non-polar residues in positions with mainly hydrophobic residues are shaded in yellow. Conserved residues in the MUN domain-containing proteins are shown in red and bold letters, and corresponding residues in proteins from vesicle tethering complexes are shown in black and bold letters. Starting and ending residues numbers (italic), as well as sequence lengths (in brackets), are shown. Insertion regions between the aligned blocks are replaced by the numbers of residues. The long insertion regions with the C2 domains in fungi MUN domain-containing proteins are indicated. Consensus secondary structure predictions for the MUN domains are shown above the alignment and marked from h1 to h9 for the nine major helices in domain C and domain D of tethering complex subunits such as Exo70 and Tip20p. Species names are represented by two letter abbreviations and colored as follows: metazoan: blue; fungi: orange; and plants: green. Species name abbreviations are: At: Arabidopsis thaliana; Ce: Caenorhabditis elegans; Ci: Ciona intestinalis; Dm: Drosophila melanogaster; Mm: Mus musculus; Nc: Neurospora crassa; Ot: Ostreococcus tauri; Sc: Saccharomyces cerevisiae; Sp: Schizosaccharomyces pombe.
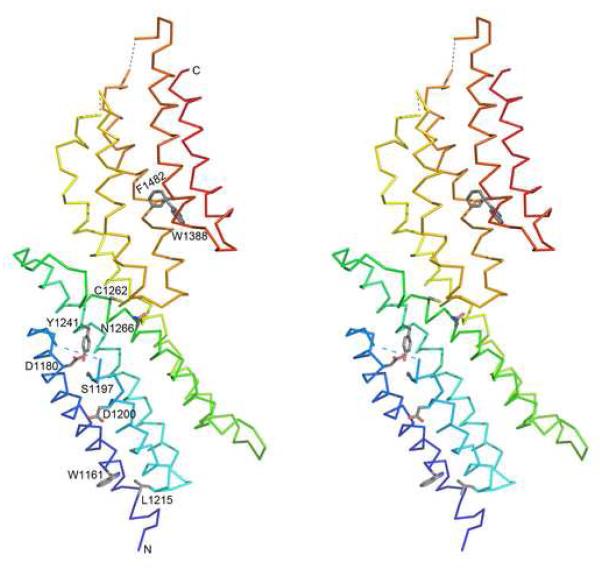
A structural model of the C-terminal region (domain C and domain D) of the MUN domain from mouse Munc13-1 was generated according to the Sec6 structure (pdb id: 2fji) by MODELLER.32 We calculated sequence conservation 33 for the MUN domains and mapped the top 10 most conserved residues (Figure 1, red columns) onto this structural model (Figure 2). These conserved residues form several spatial clusters. Residues W1161 and L1215 are two interacting large hydrophobic residues. In the structure of Sec6 with only domains C and D (pdb id: 2fji), these two residues are half buried. In the structure of Tip20 where all four domains are present (pdb id: 3fhn), residues in these two positions are buried in the interface of the domain B and domain C, and thus contribute to the interaction and orientation between these two domains. Residues D1180, S1197 and Y1241 are buried in the core of domain C. They are close to each other and may form specific interactions using their polar side chains. Conserved negatively charged residues occupy the position of the buried D1180 in both MUN domains and Sec6, but not in other related tethering complex subunits (Figure 1). The residue D1200 in helix 2 is a conserved exposed polar residue that could be important for mediating MUN domain interactions with other proteins. C1262 and N1266 in domain C are located at the interface between domain C and domain D. N1266 is also conserved in the Sec6 family. In the Sec6 structure, the side chain of this Asn residue is hydrogen-bonded to the main chain atoms in the turn region of helix 6 and helix 7 in domain D. This interaction could be important for maintaining the relative orientation between domains C and D. Residues W1388 and F1482 are two interacting large hydrophobic residues in the structural core of domain D.
Figure 2. Stereo diagrams of the structural model of domain C and domain D of the MUN domain from mouse Munc13-1.
α-Helices are labeled in accordance with Figure 1. Sidechains of conserved residues (highlighted in red letters in Figure 1) are shown as sticks. N- and C-termini are marked. Long loops are omitted with dotted lines connecting the end points. These diagrams are made by program PyMOL.
Sequence grouping, phyletic distribution and domain architecture of MUN domain-containing proteins
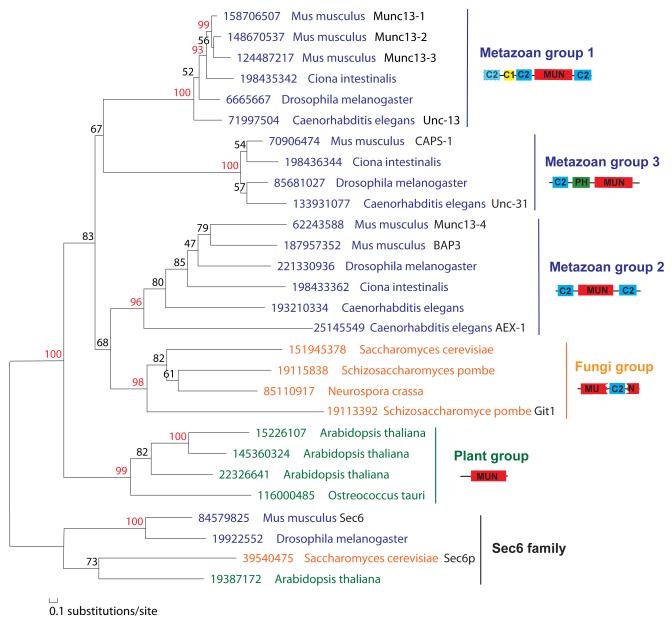
Phylogenetic analysis using MOLPHY 34 suggested clear separation of five major groups of MUN domain-containing proteins with distinct domain contents and phyletic distributions (Figure 3). We also found divergent MUN domain-containing proteins in Dictyostelium discoideum and Trichomonas vaginalis, but not in other protists. On the other hand, tethering complexes exocyst and GARP subunits have a wider species distribution 20 than MUN domain-containing proteins. MUN domain-containing proteins probably originate from duplication of one exocyst/GARP subunit, e.g., Sec6 or Vps53. Addition of C2 domains in metazoan and yeast MUN domain-containing proteins suggested that they have specialized roles in regulating Ca2+-dependent exocytosis. Indeed, the function of exocyst in neural cells does not seem to overlap with synaptic vesicle exocytosis. 35
Figure 3. Phylogenetic tree of representative MUN domains and Sec6 proteins (as an out group) by MOLPHY.
34 Sequence groups are marked on the right with domain architecture diagrams shown. The dashed outline of the first C2 domain in metazoan group 1 suggests that it is absent in some proteins. JTT amino acid substitution model 54 was used. Only positions with gap fraction less than 50% were selected for tree building. The local estimates of bootstrap percentages were obtained by the RELL method, 55 as implemented in the program ProtML of MOLPHY (-R option). Branch support values above 90 are shown in red numbers. A similar tree supporting the separation of five major groups of MUN domains were obtained by using the PHYML program 56 with the JTT amino acid substitution model and with rate variation across sites modeled by a discrete gamma-distribution.
Metazoan group 1 includes experimentally characterized mammalian Munc13-1, Munc13-2, Munc13-3 and the Unc-13 protein from Caenorhabditis elegans. These proteins are expressed in the nervous systems and are important for priming synaptic vesicles for fusion. Proteins in this group are detected only in metazoan species, such as vertebrates, insects, and the base groups Ciona and Trichoplax. They have a C1 domain that is absent in the other four groups. The MUN domain is bounded by two C2 domains (C2B and C2C in Munc13-1). An additional N-terminal C2 domain (C2A) is present in mouse Munc13-1 and ubiquitous Munc13-2, but is absent from mouse Munc13-3. Proteins from lower metazoan species such as Ciona and Trichoplax also lack the N-terminal C2A domain, suggesting that the addition of C2A in mouse Munc13-1 and Munc13-2 is a late evolutionary event.
Metazoan group 2 is typified by the mammalian Munc13-4 proteins. Munc13-4 is important for calcium-dependent exocytosis of dense core granules in various types of cells. 36; 37 Munc13-4 orthologs are also restricted to the metazoan species including the base groups such as Ciona and Trichoplax. They have similar domain architecture to metazoan group 1, with the MUN domain sandwiched by two C2 domains; however, they lack the C1 domain. Many species such as mouse, Drosphila, C. elegans and Trichoplax have two Munc13-4-like proteins. In mouse, the BAI-associated protein 3 (BAP3) 38 is closely related to Munc13-4. In C. elegans, AEX-1 is a divergent copy of Munc13-4. AEX-1 is found in postsynaptic muscles and retrogradely regulates presynaptic neural activity. 39; 40
Metazoan group 3 contains the CAPS proteins 41. These proteins are also restricted to the animal kingdom, including deep branches such as Ciona and Trichoplax. CAPS proteins are expressed in the nervous system and are critical for the priming and refilling of reliable pools in dense core vesicle exocytosis. 42; 43; 44 Recent data have also suggested a role in synaptic vesicle priming. 15 They have one C2 domain and one Pleckstrin homology (PH) domain N-terminal to the MUN domain. The PH domain in CAPS-1 binds phosphatidylinositol 4,5-bisphosphate (PIP2) and is responsible for membrane localization of CAPS. 45; 46 A PH domain is also present in exocyst subunits Sec3 47 and Exo84.48 Interestingly, Sec3 also binds PIP2 47 and its PH domain is located in the N-terminal region responsible for this binding.
Fungi group consists of Munc13 orthologs from fungi which have been annotated as hypothetical proteins or “C2 domain containing proteins”. They have one C2 domain inserted inside the MUN domain. Such an insertion is located between domain C and domain D (after helix 4, Figure 1). The functions of fungi MUN domain-containing proteins are unclear. Most fungi species have only one MUN domain-containing protein. One exception is Schizosaccharomyces pombe, which has two such proteins. The divergent one (Git1) is important for glucose activation of adenylate cyclase. 49
Plant group consists of Munc13 orthologs from plants and green algae. MUN domains in these proteins have been classified as DUF810 (DUF: domain of unknown function) in the Pfam database. Lineage-specific expansion is observed in this group. For example, Arabidopsis thaliana has five copies of these proteins. These proteins are about 1000 residues long, with the MUN domains located at the C-termini. The N-terminal regions about 350 residues mainly consist of helices. No C2 domains or other known domains are found in this group of proteins. The functions of plant MUN domain-containing proteins await experimental studies.
Implications of functional similarity between MUN domain-containing proteins and tethering complexes
More than fifteen years ago, research from diverse systems led to the general belief that most types of intracellular membrane traffic are governed by similar core machineries with conserved members from a few of protein families. 50 This belief has been reinforced over the years, and it is now clear that membrane traffic at most intracellular compartments involves SNAREs, SM proteins, NSF/Sec18p, SNAPs/Sec17p and small GTPases such as Rabs. Correspondingly, members from each one of these families have been implicated in neurotransmitter release. 3 However, it was puzzling that Munc13/Unc13s play such crucial roles in neurotransmitter release, 4; 5; 6 and yet they had not been found to have any similarities with factors generally involved in other forms of membrane traffic. Conversely, traffic at most membrane compartments was known to involve at least one multisubunit tethering complex such as exocyst, COG, GARP, Dsl1p complex, TRAPP or HOPS, but no such complex had been implicated in neurotransmitter release. Our analyses now uncover a similarity between distinct subunits of several of these complexes (exocyst, COG, GARP and Dsl1p complex) and the MUN domain, which is at the heart of the key function of Munc13s in vesicle priming. These results allowed us to predict the structure of the MUN domain and suggest that the function of Munc13s may be at least partially related to those of tethering complexes, further unifying the mechanism of neurotransmitter release with those of other forms of intracellular membrane traffic.
The evolutionary relationship established here suggests that insights into MUN domain function may be derived from what is known about tethering complexes, and vice versa. The overall concepts of tethering or docking refer to establishing long-range interactions between two membranes to bring them into proximity and facilitate their eventual fusion. Such long-range interactions can involve binding of the tethering complexes to proteins that reside in each membrane, including Rabs, coats or SNAREs, which provides a first level of specificity for membrane fusion. 7; 8; 9 Interestingly, recent studies have suggested a role for Unc13 in vesicle docking in C. elegans. 51 Conversely, docking was normal in central synapses of Munc13-1 knockout mice, and the strong impairment of release observed in these mice suggested a role in vesicle priming. 4 It is plausible that redundant docking mechanisms at central synapses may explain this potential contradiction 3; however, regardless of the potential role of Munc13/Unc13s in docking, it is most likely that these proteins play a role downstream of docking. Functional evidence strongly suggested that Munc13/Unc13s are involved in SNARE complex assembly, 52 which was further supported by the observation that the Munc13-1 MUN domain binds to SNARE complexes and syntaxin-1/SNAP-25 heterodimers. 13; 14 This observation suggests that tethering complexes may have a role not only in tethering per se, but also in downstream events that lead to SNARE complex assembly, a notion that has been supported for some tethering complexes. 7 Intriguingly, binding of the Munc13-1 MUN domain to SNARE complexes requires membrane anchoring of the complexes, suggesting that the MUN domain binds to both SNAREs and membranes. 13 Similarly, tethering complexes exocyst, COG, GARP, Dsl1p complex, and HOPS bind to SNAREs and/or membrane lipids. 9; 26
All these observations show that there are indeed common themes in the functions of the MUN domain and of tethering complexes, although it also seems clear that they play multiple roles and it is plausible that not all these roles may be shared among the MUN domain and different tethering complexes. It is also worth noting that there is considerable variability in the nature of tethering complexes. 19 HOPS and TRAPP are not evolutionarily related to exocyst, COG, GARP and Dsl1p complex; while these latter four complexes are clearly related given the homology between some of their subunits, they also exhibit clear differences in the number of subunits. It is likely that these differences arose because of specific regulatory requirements of fusion in the specific membrane compartments where they function. However, a common theme is that these complexes normally contain several subunits, raising the possibility that the Munc13 MUN domain may also function as part of a large complex that includes related proteins (e.g., CAPS).
Clearly, much research will be required to fully understand how MUN domains and tethering complexes function, but the connection established here leads to multiple predictions that can now be tested experimentally. For instance, it will be interesting to examine whether fragments of the MUN domain corresponding to the individual domains of tethering complexes of known structures can fold autonomously. Such fragments would be valuable to dissect which sequences of the MUN domain are involved in different types of interactions, including interactions with the SNAREs 13; 14 or putative intramolecular interactions with the Munc13-1 N-terminus 12 that could provide a link to RIMs and Rab3. 53 Moreover, it will also be interesting to investigate whether interactions mapped within the MUN domain are conserved in tethering complexes, or vice versa. These experiments will be crucial to further understand how conserved are the mechanisms of membrane docking and fusion at different membrane compartments.
Supplementary Material
Acknowledgements
We would like to thank Lisa Kinch for critical reading of the manuscript and helpful suggestions. This work was supported in part by NIH grants GM67165 (to NVG) and NS37200 (to JR), and by Welch foundation grants I1505 (to NVG) and I1304 (to JR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–43. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 2.Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–47. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 3.Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat Struct Mol Biol. 2008;15:665–74. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Augustin I, Rosenmund C, Sudhof TC, Brose N. Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature. 1999;400:457–61. doi: 10.1038/22768. [DOI] [PubMed] [Google Scholar]
- 5.Richmond JE, Davis WS, Jorgensen EM. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat Neurosci. 1999;2:959–64. doi: 10.1038/14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varoqueaux F, Sigler A, Rhee JS, Brose N, Enk C, Reim K, Rosenmund C. Total arrest of spontaneous and evoked synaptic transmission but normal synaptogenesis in the absence of Munc13-mediated vesicle priming. Proc Natl Acad Sci U S A. 2002;99:9037–42. doi: 10.1073/pnas.122623799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–82. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Pfeffer SR. Transport-vesicle targeting: tethers before SNAREs. Nat Cell Biol. 1999;1:E17–22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- 9.Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J Cell Sci. 2002;115:2627–37. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- 10.Brose N, Hofmann K, Hata Y, Sudhof TC. Mammalian homologues of Caenorhabditis elegans unc-13 gene define novel family of C2-domain proteins. J Biol Chem. 1995;270:25273–80. doi: 10.1074/jbc.270.42.25273. [DOI] [PubMed] [Google Scholar]
- 11.Koch H, Hofmann K, Brose N. Definition of Munc13-homology-domains and characterization of a novel ubiquitously expressed Munc13 isoform. Biochem J. 2000;349:247–53. doi: 10.1042/0264-6021:3490247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basu J, Shen N, Dulubova I, Lu J, Guan R, Guryev O, Grishin NV, Rosenmund C, Rizo J. A minimal domain responsible for Munc13 activity. Nat Struct Mol Biol. 2005;12:1017–8. doi: 10.1038/nsmb1001. [DOI] [PubMed] [Google Scholar]
- 13.Guan R, Dai H, Rizo J. Binding of the Munc13-1 MUN domain to membrane-anchored SNARE complexes. Biochemistry. 2008;47:1474–81. doi: 10.1021/bi702345m. [DOI] [PubMed] [Google Scholar]
- 14.Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. Accessory proteins stabilize the acceptor complex for synaptobrevin, the 1:1 syntaxin/SNAP-25 complex. Structure. 2008;16:308–20. doi: 10.1016/j.str.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jockusch WJ, Speidel D, Sigler A, Sorensen JB, Varoqueaux F, Rhee JS, Brose N. CAPS-1 and CAPS-2 are essential synaptic vesicle priming proteins. Cell. 2007;131:796–808. doi: 10.1016/j.cell.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–60. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 18.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kummel D, Heinemann U. Diversity in structure and function of tethering complexes: evidence for different mechanisms in vesicular transport regulation. Curr Protein Pept Sci. 2008;9:197–209. doi: 10.2174/138920308783955252. [DOI] [PubMed] [Google Scholar]
- 20.Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croteau NJ, Furgason ML, Devos D, Munson M. Conservation of helical bundle structure between the exocyst subunits. PLoS ONE. 2009;4:e4443. doi: 10.1371/journal.pone.0004443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munson M. Tip20p reaches out to Dsl1p to tether membranes. Nat Struct Mol Biol. 2009;16:100–2. doi: 10.1038/nsmb0209-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore BA, Robinson HH, Xu Z. The crystal structure of mouse Exo70 reveals unique features of the mammalian exocyst. J Mol Biol. 2007;371:410–21. doi: 10.1016/j.jmb.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamburger ZA, Hamburger AE, West AP, Jr., Weis WI. Crystal structure of the S.cerevisiae exocyst component Exo70p. J Mol Biol. 2006;356:9–21. doi: 10.1016/j.jmb.2005.09.099. [DOI] [PubMed] [Google Scholar]
- 25.Dong G, Hutagalung AH, Fu C, Novick P, Reinisch KM. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat Struct Mol Biol. 2005;12:1094–100. doi: 10.1038/nsmb1017. [DOI] [PubMed] [Google Scholar]
- 26.Tripathi A, Ren Y, Jeffrey PD, Hughson FM. Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex. Nat Struct Mol Biol. 2009;16:114–23. doi: 10.1038/nsmb.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sivaram MV, Furgason ML, Brewer DN, Munson M. The structure of the exocyst subunit Sec6p defines a conserved architecture with diverse roles. Nat Struct Mol Biol. 2006;13:555–6. doi: 10.1038/nsmb1096. [DOI] [PubMed] [Google Scholar]
- 28.Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat Struct Mol Biol. 2005;12:879–85. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- 29.Pashkova N, Jin Y, Ramaswamy S, Weisman LS. Structural basis for myosin V discrimination between distinct cargoes. EMBO J. 2006;25:693–700. doi: 10.1038/sj.emboj.7600965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–40. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 31.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–59. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 33.Pei J, Grishin NV. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17:700–12. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- 34.Adachi J, Hasegawa M. Computer Science Monographs (The Institute of Statistical Mathematics) No. 28. Tokyo: 1996. MOLPHY version 2.3, Programs for Molecular Phylogenetics Based on Maximum Likelihood. [Google Scholar]
- 35.Murthy M, Garza D, Scheller RH, Schwarz TL. Mutations in the exocyst component Sec5 disrupt neuronal membrane traffic, but neurotransmitter release persists. Neuron. 2003;37:433–47. doi: 10.1016/s0896-6273(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 36.Feldmann J, Callebaut I, Raposo G, Certain S, Bacq D, Dumont C, Lambert N, Ouachee-Chardin M, Chedeville G, Tamary H, Minard-Colin V, Vilmer E, Blanche S, Le Deist F, Fischer A, de Saint Basile G. Munc13-4 is essential for cytolytic granules fusion and is mutated in a form of familial hemophagocytic lymphohistiocytosis (FHL3) Cell. 2003;115:461–73. doi: 10.1016/s0092-8674(03)00855-9. [DOI] [PubMed] [Google Scholar]
- 37.Pivot-Pajot C, Varoqueaux F, de Saint Basile G, Bourgoin SG. Munc13-4 regulates granule secretion in human neutrophils. J Immunol. 2008;180:6786–97. doi: 10.4049/jimmunol.180.10.6786. [DOI] [PubMed] [Google Scholar]
- 38.Shiratsuchi T, Oda K, Nishimori H, Suzuki M, Takahashi E, Tokino T, Nakamura Y. Cloning and characterization of BAP3 (BAI-associated protein 3), a C2 domain-containing protein that interacts with BAI1. Biochem Biophys Res Commun. 1998;251:158–65. doi: 10.1006/bbrc.1998.9408. [DOI] [PubMed] [Google Scholar]
- 39.Doi M, Iwasaki K. Regulation of retrograde signaling at neuromuscular junctions by the novel C2 domain protein AEX-1. Neuron. 2002;33:249–59. doi: 10.1016/s0896-6273(01)00587-6. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita M, Iwasaki K, Doi M. The non-neuronal syntaxin SYN-1 regulates defecation behavior and neural activity in C. elegans through interaction with the Munc13-like protein AEX-1. Biochem Biophys Res Commun. 2009;378:404–8. doi: 10.1016/j.bbrc.2008.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens DR, Rettig J. The Ca(2+)-dependent activator protein for secretion CAPS: do I dock or do I prime? Mol Neurobiol. 2009;39:62–72. doi: 10.1007/s12035-009-8052-5. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Schirra C, Stevens DR, Matti U, Speidel D, Hof D, Bruns D, Brose N, Rettig J. CAPS facilitates filling of the rapidly releasable pool of large dense-core vesicles. J Neurosci. 2008;28:5594–601. doi: 10.1523/JNEUROSCI.5672-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Speese S, Petrie M, Schuske K, Ailion M, Ann K, Iwasaki K, Jorgensen EM, Martin TF. UNC-31 (CAPS) is required for dense-core vesicle but not synaptic vesicle exocytosis in Caenorhabditis elegans. J Neurosci. 2007;27:6150–62. doi: 10.1523/JNEUROSCI.1466-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujita Y, Xu A, Xie L, Arunachalam L, Chou TC, Jiang T, Chiew SK, Kourtesis J, Wang L, Gaisano HY, Sugita S. Ca2+-dependent activator protein for secretion 1 is critical for constitutive and regulated exocytosis but not for loading of transmitters into dense core vesicles. J Biol Chem. 2007;282:21392–403. doi: 10.1074/jbc.M703699200. [DOI] [PubMed] [Google Scholar]
- 45.Grishanin RN, Klenchin VA, Loyet KM, Kowalchyk JA, Ann K, Martin TF. Membrane association domains in Ca2+-dependent activator protein for secretion mediate plasma membrane and dense-core vesicle binding required for Ca2+-dependent exocytosis. J Biol Chem. 2002;277:22025–34. doi: 10.1074/jbc.M201614200. [DOI] [PubMed] [Google Scholar]
- 46.Grishanin RN, Kowalchyk JA, Klenchin VA, Ann K, Earles CA, Chapman ER, Gerona RR, Martin TF. CAPS acts at a prefusion step in dense-core vesicle exocytosis as a PIP2 binding protein. Neuron. 2004;43:551–62. doi: 10.1016/j.neuron.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Orlando K, He B, Xi F, Zhang J, Zajac A, Guo W. Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J Cell Biol. 2008;180:145–58. doi: 10.1083/jcb.200704128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin R, Junutula JR, Matern HT, Ervin KE, Scheller RH, Brunger AT. Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. EMBO J. 2005;24:2064–74. doi: 10.1038/sj.emboj.7600699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kao RS, Morreale E, Wang L, Ivey FD, Hoffman CS. Schizosaccharomyces pombe Git1 is a C2-domain protein required for glucose activation of adenylate cyclase. Genetics. 2006;173:49–61. doi: 10.1534/genetics.106.055699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–3. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- 51.Weimer RM, Gracheva EO, Meyrignac O, Miller KG, Richmond JE, Bessereau JL. UNC-13 and UNC-10/rim localize synaptic vesicles to specific membrane domains. J Neurosci. 2006;26:8040–7. doi: 10.1523/JNEUROSCI.2350-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richmond JE, Weimer RM, Jorgensen EM. An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature. 2001;412:338–41. doi: 10.1038/35085583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dulubova I, Lou X, Lu J, Huryeva I, Alam A, Schneggenburger R, Sudhof TC, Rizo J. A Munc13/RIM/Rab3 tripartite complex: from priming to plasticity? EMBO J. 2005;24:2839–50. doi: 10.1038/sj.emboj.7600753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 55.Hasegawa M, Kishino H, Saitou N. On the maximum likelihood method in molecular phylogenetics. J Mol Evol. 1991;32:443–5. doi: 10.1007/BF02101285. [DOI] [PubMed] [Google Scholar]
- 56.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.