Abstract
The actin filament system is fundamental to cellular functions including regulation of shape, motility, cytokinesis, intracellular trafficking and tissue organization. Tropomyosins (Tm) are highly conserved components of actin filaments which differentially regulate filament stability and function. The mammalian Tm family consists of four genes; αTm, βTm, γTm and δTm. Multiple Tm isoforms (>40) are generated by alternative splicing and expression of these isoforms is highly regulated during development. In order to further identify the role of Tm isoforms during development, we tested the specificity of function of products from the γTm gene family in mice using a series of gene knockouts. Ablation of all γTm gene cytoskeletal products results in embryonic lethality. Elimination of just two cytoskeletal products from the γTm gene (NM1,2) resulted in a 50% reduction in embryo viability. It was also not possible to generate homozygous knockout ES cells for the targets which eliminated or reduced embryo viability in mice. In contrast, homozygous knockout ES cells were generated for a different set of isoforms (NM3,5,6,8,9,11) which were not required for embryogenesis. We also observed that males hemizygous for the knockout of all cytoskeletal products from the γTm gene preferentially transmitted the minus allele with 80–100% transmission. Since all four Tm genes are expressed in early embryos, ES cells and sperm, we conclude that isoforms of the γTm gene are functionally unique in their role in embryogenesis, ES cell viability and sperm function.
Key words: cytoskeleton, actin, tropomyosin, redundancy, isoforms
Introduction
Tropomyosin (Tm), an actin-associated protein, forms a head-to-tail continuous polymer positioned along the major groove of the actin filament providing stability and flexibility to the filaments.1–4 The mammalian Tm gene family consists of four genes, α, β, γ and δTm (Tpm1–4). Over 40 Tm isoforms are generated by alternative splicing. They can be classified as high molecular weight (HMW, 284 amino acids) or low molecular weight (LMW, 248 amino acids) and the majority are referred to as cytoskeletal isoforms, due to their prevalence in non-muscle cells.5
Tm isoforms are known to associate with actin filaments with different affinities and allow or inhibit the interaction of various actin binding proteins with the filaments.6,7–14 In addition, distinct Tm isoforms show specific subcellular localizations in several cell types both in vitro and in vivo where they can directly impact on the pool of filamentous actin.15,16 Together the above findings strongly implicate Tm isoforms in the regulation of actin filament dynamics and the spatial specialization of these filaments.5,14 Recent studies in yeast have demonstrated that this is an evolutionary ancient strategy to specialize actin filament function.17–19
Tm isoform expression is tissue specific and the repertoire of isoforms expressed is altered throughout development and with differentiation,20 strongly suggesting that each individual or group of isoforms is required to perform a specific biological function. Indeed, knockout (KO) mouse models of any of the α, β or γTm genes are embryonic lethal indicating that gene products from each of these genes are essential for embryonic survival and cannot be compensated by products from alternative Tm genes.21–23 Initial studies profiling the expression of Tm isoforms in undifferentiated embryonic stem (ES) cells, differentiated embryo bodies (EBs) together with developing mouse embryos demonstrate that the isoforms expressed early in mouse development are primarily LMW Tm products derived from the δ and γTm genes although ES cells cultured in vitro also express HMW Tms derived from the α and βTm genes.22,24,25
The γTm gene, one of the most highly alternatively spliced genes, produces at least 11 distinct cytoskeletal isoforms by the use of two alternative internal exons, 6a and 6b, and three alternative carboxy-terminal exons, 9a, 9c and 9d.26 We have previously demonstrated that although deletion of all cytoskeletal products from the γTm gene is lethal for embryonic development,22 deletion of exon 9c-containing isoforms (Tm5NM4 and NM7) or exon 9d-containing isoforms (Tm5NM1 and NM2) leads to viable animals.27,28
The importance of Tm gene products from the γTm gene during mammalian development has not been fully explored. We now extend our previous studies and evaluate the functional specificity of subsets of γTm isoforms in mice. Knockout constructs of the carboxy-terminal exons, 9a/b, 9c and 9d from the γTm gene were used to assess the viability of ES cells and mice. While ablation of all γTm gene cytoskeletal products by deletion of the amino-terminal coding exon 1b results in non-viable mice, elimination of exon 9c-containing isoforms (Tm5NM4,7) showed no impact on embryo viability whereas deletion of exon 9d (Tm5NM1,2) results in partial lethality. The viability of ES cells in vitro was also found to be comprised such that neither exon 1b nor 9d homozygous KO ES cells were generated. However, we were able to obtain homozygous KO ES cells for exon 9a/b (Tm5NM3,5,6,8,9,11). Since all four Tm family genes are expressed in early embryos, ES cells and sperm, we conclude that isoforms of the γTm gene perform specific functions in ES cell viability and embryogenesis that cannot be compensated by the other Tm genes. Furthermore, our data indicate that sperm lacking cytoskeletal Tm products of the γTm gene have a selective advantage.
Results
Knockout of γTm gene exon 1b is embryonic lethal.
As outlined above, all cytoskeletal isoforms encoded by the γTm gene incorporate exon 1b as the first coding exon (Fig. 1A). In order to study the effects on mouse function of deleting amino-terminal exon 1b, the γTm gene exon 1b was targeted in 129/SvJ ES cells according to Hook et al.22 (Fig. 1B). Mice carrying the exon 1b deletion (Δ1b+/−) were generated from two 129/SvJ ES cell lines (line#1 = 129 − Tpm3tm1(Δ1b)Pgun) (line#2 = 129 − Tpm3tm2(Δ1b)Pgun). Initial observations demonstrated that the KO of γTm exon 1b was embryonic lethal for both 129/SvJ lines. A summary of the breeding data obtained from mice hemizygous for the targeted deletion (Δ1b+/−) derived from ES cell lines Δ1b lines#1 and #2 is shown in Table 1. The observed numbers of wildtype (+/+), hemizygous (+/−) and homozygous (−/−) offspring do not fit expected Mendelian ratios for both of the 129/SvJ-derived Δ1b mouse lines. A preference for the transmission of the KO allele compared to the WT allele is observed in both 129/SvJ lines but most pronounced in the Δ1b line#2. No Δ1b−/− offspring were detected in either Δ1b lines#1 and #2 on the 129/SvJ background. The Δ1b line#2 mice on the 129/SvJ background were backcrossed onto a C57BL/6 background for five generations using a speed congenic breeding strategy. N5 generation Δ1b+/− mice (B6-Tpm3tm2(Δ1b)Pgun) were bred in order to determine whether null pups could be generated on the C57BL/6 background. In a similar result to the Δ1b lines#1 and #2 on the 129/SvJ background, no homozygous offspring could be detected in the C57BL/6 background (Table 1). This result confirms that homozygous loss of exon 1b on two different mouse backgrounds is lethal. However, we have observed that the ratio for +/+ and Δ1b+/− on the C57BL/6 background now meets expected Mendelian ratios being 1:2 for +/+ to +/− (Table 1). There appears to be no transmission ratio distortion on the C57BL/6 background.
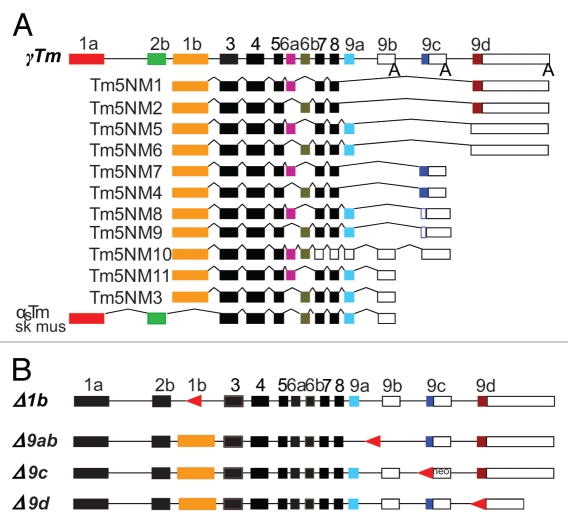
Figure 1.
Schematic representation of the γTm gene and targeted deletions of both amino- and carboxy-terminal exons. (A) The entire γTm gene showing alternatively spliced variants.3,26 All cytoskeletal products contain the exon 1b promoter, with choice of either exon 6a or 6b, and variations of the carboxy-terminal exon 9. Open boxes represent UTRs, lines represent introns and A represents poly A tail. (B) The γTm gene structure showing targeted deletions of exons 1b, 9a + 9b, 9c and 9d summarized as −1b, −9ab, −9c and −9d. Arrow-heads (in red) represent the LoxP sites remaining following exon deletions.
Table 1.
The Δ1b−/− genotype is embryonic lethal
| Background | KO line | +/+ | +/− | −/− |
| 129/SvJ | Δ1b line#1 (n = 230) | 50 | 180 | 0 |
| 129/SvJ | Δ1b line#2 (n = 104) | 6 | 98 | 0 |
| C57BL/6 | Δ1b line#2 (n = 152) | 50 | 102 | 0 |
Results of crossing Δ1b +/− mice on two genetic backgrounds, 129Sv/J and C57BL/6. Expected Mendelian ratios of 1:2:1 for +/+:+/−:−/− were not observed.
Deletion of γTm gene exon 1b is favored on the 129/SvJ background.
Our observation of the high number of Δ1b+/− line#1 and #2 mice on the 129/SvJ background (Table 1) indicates a possible transmission ratio distortion. In order to determine whether this can be attributed to altered production of gametes during female oogenesis or male spermatogenesis, the Δ1b line#2 on the 129/SvJ background was further assessed by breeding either Δ1b+/− males or Δ1b+/− females with 129/SvJ WT mice (Table 2). These data indicate a significant (p < 0.05) skewing of 85% transmission of the Δ1b ‘- allele’ from the Δ1b+/− male mice. The Δ1b+/− female mice transmitted the Δ1b ‘- allele’ at a significant (p < 0.05) higher (61.5%) frequency than the expected 50% (Table 2). We conclude that the transmission ratio distortion is common to both male and female hemizygous mice on the 129/SvJ background, but the effect is more pronounced in the males. Based on the 15% ‘+ allele’ and 85% ‘- allele’ transmission observed for sperm, we can predict that outcomes for +/+ and +/− would show a similar trend toward a higher skewing of +/− offspring as reported in Table 1.
Table 2.
Preferential transmission of the Δ1b negative allele
| Mouse line 129/SvJ background | Total | +/+ (% + allele transmission) | +/− (% - allele transmission) |
| Δ1b +/− ♂ X wt +/+ ♀* |
92 | 14 (15%) |
78 (85%) |
| Δ1b +/− ♀ X wt +/+ ♂* |
78 | 30 (38.5%) |
48 (61.5%) |
| Expected % | 50 | 50 |
p < 0.05 by χ2 test when compared with 1:1 distribution. Male or female mice with the Δ1b +/− genotype were crossed with wild type mice and the resulting genotype of the progeny determined. The expected Mendelian ratios of 1:1 for +/+:+/− were not observed.
Tropomyosin products from the four Tm genes (α, β, γ and δ) are expressed in mouse sperm.
Next we addressed the question of whether the male transmission ratio distortion that was observed on the 129/SvJ background, but not on the C57BL/6 background, was due to a difference between the Tm gene products expressed in mouse sperm from these two strains. Western blot analysis of proteins extracted from +/+ adult male mouse sperm tissue was performed on both 129/SvJ and C57BL/6 backgrounds. Using antibodies specific to detect products from each of the four Tm gene families (αTm, βTm, γTm and δTm), blots were exposed to antibodies γ/9d, α/9d and δ/9d. The results in Figure 2A–C show that high and low molecular weight Tm gene products are expressed in mouse sperm on both 129/SvJ and C57BL/6 mouse backgrounds and that all four genes express products in mouse sperm (Tm2, Tm6-αTm, Tm1-βTm, Tm5NM1/2-γTm and Tm4-δTm gene).
Figure 2.
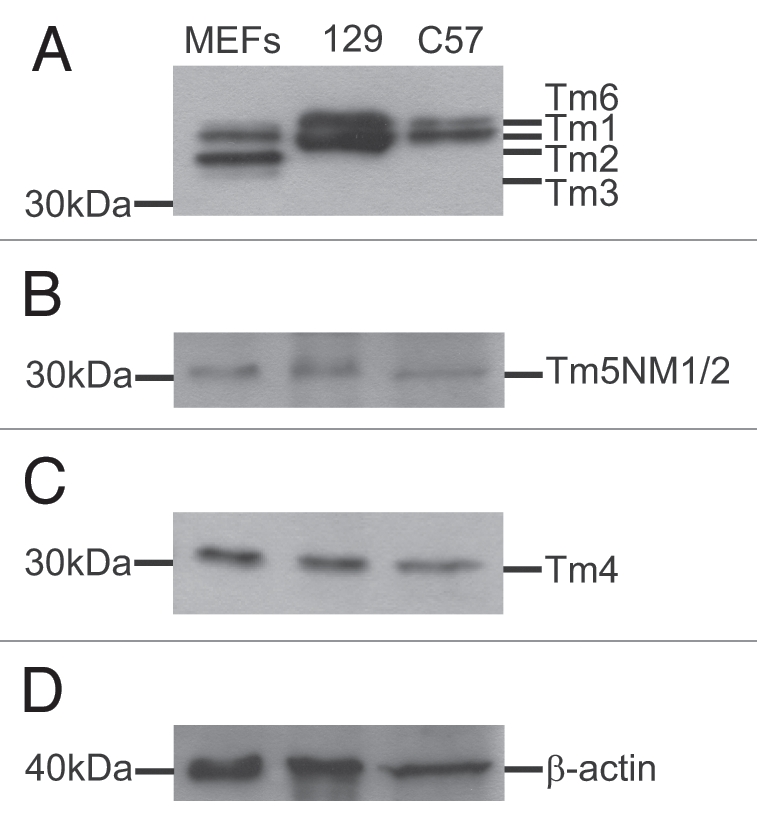
Western blot analysis of mouse sperm. Total mouse sperm protein (10 µg) from WT 129/SvJ and C57BL/6 mouse lines and primary mouse embryonic fibroblasts (MEF) control protein (10 µg for Part A; 1 µg for Parts B–D) were analyzed with antibodies for the presence of high and low molecular weight Tm gene products. Blots were probed with (A) α/9d, (B) γ/9d and (C) δ/9d which recognize products from α, β, γ and δ Tm genes. (D) β actin control blot.
Knockout of γTm gene exon 9d causes partial embryonic lethality on both 129/SvJ and C57BL /6 backgrounds.
Unlike our finding for the lethality caused by removing the γTm gene amino-terminal exon 1b, we have previously reported that removal of the carboxy-terminal exon 9c (129-Tpm3tm1(neo;Δ9c)Pgun) and exon 9d (129-Tpm3tm1(neo;Δ9d)Pgun, B6-Tpm3tm2(Δ9d)Pgun) results in viable animals.27,28 Subsequent breeding analysis of the Δ9d line is shown in Table 3. The expected Mendelian ratios were not achieved, with significantly fewer (p < 0.05) than expected Δ9d−/− mice generated. This phenomenon occurs in both 129/SvJ and C57BL/6 backgrounds. However, this result is not seen in the γTm gene Δ9c−/− on a 129/SvJ background (Table 3) where the expected and observed percentages of +/+, +/− and −/− mice are not significantly different.
Table 3.
The Δ9d−/− genotype is partially embryonic lethal
| Mouse line | +/+ | +/− | −/− |
| Δ9d (n = 153)* 129/SvJ background |
43 | 87 | 23 |
| Observed % | 28.1 | 56.9 | 15 |
| Δ9d (n = 176)* C57BL/6 background |
52 | 105 | 19 |
| Observed % | 29.5 | 59.7 | 10.8 |
| Δ9c (n = 132) 129/SvJ background |
37 | 63 | 32 |
| Observed % | 28 | 47.8 | 24.2 |
| Expected % | 25 | 50 | 25 |
p < 0.05 by χ2 test when compared with 1:2:1 distribution. Results of crossing Δ9d +/− mice on two genetic backgrounds, 129Sv/J and C57BL/6.
γTm gene exon 9d homozygous deletions are not viable in vitro.
We have previously reported that elimination of both copies of the 1b-exon of the γTm gene in ES cells is not possible.22 Because of the partial lethality of the Δ9d−/− genotype, we evaluated whether it was possible to eliminate both copies of the 9d exon in vitro. In order to generate Δ9d−/− cells for in vitro analysis we attempted to re-target the WT allele in our Δ9d+/− ES cells by reintroducing the Δ9d targeting construct. There are three possible outcomes from the re-targeting: (1) random integration of the targeting vector leaving one WT allele and one KO allele, (2) re-targeting of the originally targeted locus resulting in one WT allele and one 9d KO neo allele or (3) targeting of the WT allele to yield one 9d KO neo allele and one 9d KO allele (Fig. 3).
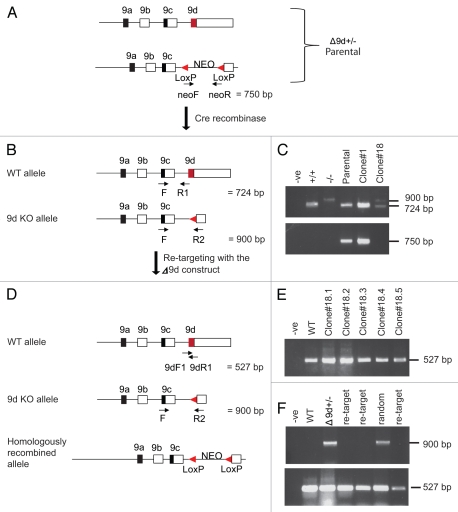
Figure 3.
Design and screening of −9d ES cells. (A) The −9d+/− homologously recombined (parental) allele containing the Neomycin cassette. An oligonucleotide primer set designed to the Neomycin cassette is also shown. (B) Following Cre-mediated recombination to remove the Neomycin cassette the expected −9d KO allele is shown with oligonucleotide primer sets common to both WT and −9d KO alleles. (C) Genomic DNA isolated from ES cell clones screened by PCR analysis. PCR analysis of outcomes from the three primer set F, R1 and R2 is shown in the upper part, with outcomes from the Neomycin primer set shown in the lower part. -ve, no template control; +/+, WT C57BL/6 ES cell DNA; −/−, genomic DNA from Δ9d deleted mouse line; Parental, homologously recombined −9d+/− clone containing the NEO cassette; Clone#1, clone containing the NEO cassette; Clone#18, clone with NEO cassette removed. (D) Following re-targeting of the −9d+/− ES clone (#18) with the −9d targeting construct, the possible outcomes from this re-targeting are WT (due to random integration), re-targeting back to the original 9d KO locus and targeting of the alternative allele. Oligonucleotide primer sets designed to detect the presence of exon 9d are shown. (E) Genomic DNA isolated from ES cell clones screened by PCR analysis using 9dF1 and 9dR1 primers. -ve, no template control; WT, C57BL/6 ES cell DNA; #1-#5 sample of the 330 clones. The predicted 527 bp fragment for exon 9d is shown. (F) The 330 ES cell clones were also analyzed by PCR for random and original locus re-targeting using F and R2 primers (upper part) and 9dF1 and 9dR1 primers (bottom part). -ve, no template control; WT, C57BL/6 ES cell DNA, −9d+/− ES Clone#18; re-target, clone where −9d targeting construct has re-inserted into the original targeted locus; random, clone where −9d targeting construct has re-inserted randomly. The 900 bp fragment for knockout of exon 9d and the 527 bp fragment for presence of exon 9d is shown.
Firstly, a Δ9d+/− parental ES cell clone (Fig. 3A) was subjected to Cre-mediated recombination to remove the Neomycin cassette (Fig. 3B). A total of 96 clones were picked and analyzed for the absence of the Neomycin cassette. The three primers used to confirm the absence of the Neomycin cassette were F and R1 which detected a 724 bp band corresponding to the WT allele and F and R2 detected the 900 bp KO allele band minus the Neomycin cassette (Fig. 3B and C upper part). A secondary confirmation for the absence of the Neomycin cassette was also performed using primers designed to the Neomycin cassette (neoF and neoR, Fig. 3A). One ES clone (#18) was found to contain the 900 bp exon 9d KO allele and was also lacking the Neomycin cassette (Fig. 3C lower part).
The Δ9d+/− ES clone (#18) was re-electroporated with the Δ9d targeting construct. The possible outcomes from this re-targeting are no change (due to random integration), re-targeting back to the original 9d KO locus and targeting of the alternative allele (Fig. 3D). Following selection, a total of 330 clones were picked and analyzed for the complete absence of exon 9d using primer set 9dF1 and 9dR1 (Fig. 3D). All ES cell clones analyzed contained the 527 bp band corresponding to the presence of exon 9d (Fig. 3E depicting some of the clones). As no Δ9d−/− ES cells could be found, the 330 Δ9d+/− ES cell clones were also analyzed by PCR using the F and R2 primers (Fig. 3D) in order to identify random integration and re-targeting of the original locus. Figure 3F (upper part) depicts 3 of the clones where re-targeting has occurred back into the original locus as determined by the absence of a 900 bp exon 9d KO band. The presence of the 900 bp exon 9d KO band in 1 clone is representative of the random integration seen for most of the clones analyzed. The presence of an exon 9d WT allele fragment of 527 bp (Fig. 3F lower part) was also used as a control in the PCR to check integrity of the DNA for each of the clones assayed. The data summarized in Table 4 shows that 324 clones were found to be random recombinants with 6 re-targeted back into the originally targeted locus. In a similar finding, Table 4 also shows a summary of the analysis performed in order to generate Δ1b−/− ES cells with 57 clones having re-targeted the construct back into the original locus and the remaining found to be random recombinants with no Δ1b−/− ES cells generated.22 We conclude that both exon 1b (common to products Tm5NM1-11) and exon 9d (common to products Tm5NM1,2) are essential for ES cell viability in vitro.
Table 4.
Neither Δ1b−/− nor Δ9d−/− ES cells are viable
| ES line | # Samples | Re-targeted original locus | Random insertions | Homozygous knockouts |
| Δ1b | 397 | 51 | 346 | 0 |
| Δ9d | 330 | 6 | 324 | 0 |
| Δ9ab | 48 | 1 | 46 | 1 |
Note, Δ1b data has been previously reported in Hook.22
γTm gene exons 9a + 9b homozygous deletions are viable in vitro.
The remaining carboxy-terminal exon that had not previously been targeted is exon 9a. This exon is used by both cytoskeletal and muscle products from the γTm gene. Although skeletal muscle products are expressed at later stages of embryo development,29 we addressed the question of whether the removal of exons 9a and 9b could be achieved in vitro. We first targeted C57BL/6 ES cells to yield a Δ9ab+/− ES cell line (Tpm3tm1(Δ9ab)Pgun) (Fig. 4). Next we were able to generate Δ9ab−/− ES cells using successive rounds of electroporation (Table 4). The exon 9ab targeting construct (Fig. 4A) was introduced into C57BL/6 ES cells and EcoRV digestion was used to determine homologously recombined alleles. Southern blot analysis of ES cell DNA following EcoRV digestion shows the expected 5.5 kb band for the WT allele and a 3.1 kb band for the recombined allele (Fig. 4B). A homologously recombined Δ9ab clone was subjected to CMV-Cre recombinase followed by selection in ganciclovir to eliminate clones containing the Thymidine Kinase-Neomycin cassette. The resulting outcomes from this Cre-mediated recombination are an unchanged WT allele plus either a floxed (flanked by LoxP sites) exon 9a + 9b (Δ9abFlox) allele or complete deletion of all sequences between the most distant LoxP sites (Δ9ab+/−) (Fig. 4C). Oligonucleotide primer sets common to the WT and both altered alleles were used in PCR analysis to show either the floxed or complete deletion of exon 9a + 9b (Fig. 4D). Using the primer set 9723F and 9b3′R, a WT allele of 315 bp and a floxed allele of 405 bp is shown in the Flox sample (Fig. 4D and upper part). Note that the fully deleted exon 9a + 9b clone only has the 315 bp band. Further confirmation of the absence of exon 9a + 9b was carried out using primer set 9724F and 9b3′R (Fig. 4E) which yielded a 400 bp fragment (Fig. 4D and lower part). A fragment of 2.6 kb for the WT allele was not determined in this PCR assay and is shown below in a subsequent long range PCR assay.
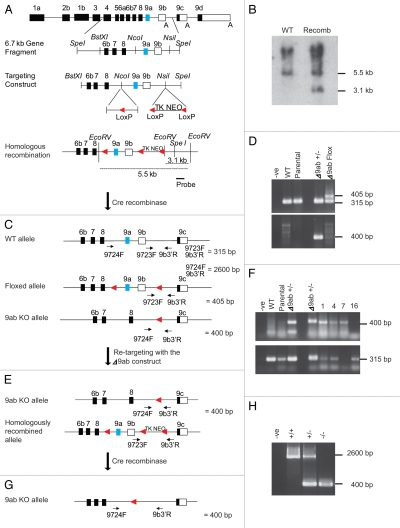
Figure 4.
Design and screening of −9ab ES cells. (A) The γTm gene showing the subcloned 6.7 kb SpeI fragment containing exons 9a and 9b with flanking intron sequence. The targeting construct with a LoxP sequence and a cassette containing LoxP—pGK Thymidine Kinase—pGK NeomycinR—LoxP. A 1.2 kb probe located outside the construct region is shown. (B) Southern blot analysis of C57BL/6 ES cell DNA following EcoRV digestion shows the expected 5.5 kb band for the WT allele and a 3.1 kb band for the recombined allele (Recomb) following probing with the 1.2 kb probe. (C) Following Cre recombinase and Gancyclovir selection the resulting alleles expected are WT, a floxed exon 9a + 9b or a full 9ab KO allele. Oligonucleotide primer sets common to the WT and both altered alleles are shown. (D) Genomic DNA isolated from ES cell clones was screened by PCR analysis. WT: +/+, Parental: homologously recombined clone containing exons 9a + b and the TKNEO cassette, −9ab+/−: clone lacking exons 9a + 9b and −9abFlox: clone containing a floxed exon 9a + 9b allele. PCR analysis outcomes from the primer set 9723F and 9b3′R is shown in the upper part and primer set 9724F and 9b3′R lower part. (E) Following re-targeting of the −9ab+/− clone with the −9ab construct the expected outcomes are a 9ab KO and homologously recombined alleles. (F) Genomic DNA isolated from ES cell clones was screened by PCR analysis using primer sets 9724F and 9b3−R to yield a 400 bp band and 9723F and 9b3′R to yield a 315 bp band. (G) Following the second round of Cre recombination and selection the expected outcome will be two −9ab−/− alleles. An oligonucleotide primer set common to both WT allele and the fully deleted exon 9a + 9b allele was used in order to confirm any −9ab−/− ES cell clones. (H) Genomic DNA isolated from ES cell clones were screened by PCR analysis. A single −9ab−/− ES cell clone was determined by long range PCR against both WT+/+ and −9ab+/− controls. The expected 2.6 kb fragment for the WT allele and a 400 bp fragment for the deleted exons 9a and 9b is shown.
An ES cell clone containing the complete deletion of exon 9a + 9b allele (Δ9ab+/−) was re-targeted with the Δ9ab construct in order to generate a double KO. The possible outcomes following this re-targeting are a homologously recombined allele and 9ab KO allele (Fig. 4E). Using primer sets as described in Figure 4C, one clone out of 48 was found by PCR analysis to be targeted with the 9ab construct on the alternative allele (Fig. 4F). Clone#7 contains the 400 bp fragment corresponding to the 9ab KO allele and lacks the 315 bp fragment that would have been amplified if an intact WT allele was present. In contrast, clone#16 represents a recombination back into the original locus due to the absence of the 9ab KO allele (400 bp) and presence of the 315 bp WT allele (Fig. 4F and Table 4). The doubly targeted clone#7 was again subjected to Cre recombinase to yield the fully 9ab KO alleles (Fig. 4G). A resulting viable Δ9ab−/− ES cell clone was genotyped by long range PCR, the expected 2.6 kb fragment for the WT allele and a 400 bp fragment for the deleted exons 9a and 9b is shown (Fig. 4H). An oligonucleotide primer set designed to amplify exon 9a was also used to confirm that exons 9a and 9b had been successfully deleted (data not shown). From these results we can conclude that exons 9a + 9b (common to products Tm5NM3,5,6,8,9,11) are not essential for ES cell viability.
Discussion
Critical cellular functions such as cellular trafficking, cell division and motility are directly regulated by the actin cytoskeleton. To control the stability and dynamics of actin filaments during these processes the communication and interaction of a large number of actin-associated proteins with the actin filaments are required. Our recent studies have shown that members of the Tm protein family are central regulators of actin filament dynamics and structure and can define distinct actin filament populations (reviewed in ref. 5). The results presented in this study using a range of different Tm KO mouse models shows that Tm gene products from different Tm genes are not functionally redundant supporting the idea of unique cellular functions of Tm isoforms from different genes. Furthermore, we demonstrate that Tms play a role in sperm function prior to or during fertilization, skewing transmission with a selection advantage for alleles lacking all cytoskeletal γTm gene products.
Transmission ratio distortion by deletion of γTm gene products.
Breeding data from the Δ1b+/− matings and Δ1b+/− cross with WT animals are indicative of a distorted transmission ratio based on impairment of both sperm and oocyte function. Our results also show a minor but significant distortion of transmission ratio through the female germ line. The involvement of female gamete production in transmission ratio distortion has previously been shown in a KO mouse model with a deletion of the Src homology 2 domain-containing adapter protein SHB.30 The large skewing of the transmission ratio in the offspring of the Δ1b+/− males mated with Δ1b+/+ females however is suggestive of a more prominent impairment of sperm function which provides a selection advantage for sperm lacking any cyoskeletal γTm gene products. The subsequent analysis of breeding data using the Δ9d−/− and Δ9c−/− γTm gene mice demonstrates that this selection advantage is not due to isoforms containing either of these two exons. It is possible that products containing exons 9a/b may be responsible for the strong phenotype observed in the Δ1b−/− γTm gene mice which will be the subject for further studies using the Δ9a/b−/− γTm gene mouse line. Alternatively, the phenotype may reflect a failure to be able to produce any cytoskeletal products from the γTm gene.
Interestingly, we saw the transmission ratio distortion only on the 129/SvJ mouse background but not after congenic back-crossing onto the C57BL/6 mouse strain. This observation is in agreement with a number of studies that show strain differences in fertilization ability for different mammalian species including mice, rats and hamsters.31–33 In particular a poor fertilizing ability in vitro has been reported for various substrains of C57BL/6 mice when compared to other inbred and outbred strains,34 an effect that has also been observed after artificial insemination,35 ruling out these observations to be in vitro artefacts. The selection advantage that we observe in the 129/SvJ strain may therefore be masked by the poor fertilizing ability of C57BL/6 mice. Tm gene products have been detected in a previous study in bovine spermatozoa.36 However the expression profile of individual Tm gene products in mouse sperm has not been studied so far. Our data demonstrates that Tm products from all Tm genes are expressed in mouse sperm and that there appears to be no differences in the expression of the products between 129/SvJ and C57BL/6 mouse lines.
To our knowledge this is the first indication for mouse sperm gaining a selection advantage due to the manipulation of the actin cytoskeleton.
Tropomyosin as a potential actin filament regulator in sperm function.
While microtubules are the major cytoskeletal component driving sperm motility via the motile force generation of the sperm flagellum, the actin cytoskeleton has been shown to be equally critical for sperm motility.37,38 Both actin filament destabilizing drugs and changes in the expression of endogenous actin associated proteins impairs sperm motility. Although some of the results from these studies are contradictory, possibly due to species differences or differences in experimental conditions—the actin filament destabilizing drug cytochalasin D inhibits sperm motility in macaque sperm,38 but not in guinea pig sperm,37—the results provide strong evidence for the involvement of actin filament stability and dynamics in sperm motility. The actin cytoskeleton furthermore controls sperm maturation, also known as capacitation and the formation of the acrosome complex prior to fertilization.39–41 Throughout these processes the actin cytoskeleton undergoes a complex re-organization with an initial increase in F-actin in the sperm head during capacitation followed by a fast disappearance of F-actin at the start of the zona pellucida-induced acrosome reaction.39 Interference with actin function using a monoclonal anti-actin antibody has been shown to inhibit the acrosome reaction.41 To mediate this re-organization a range of actin-associated proteins are involved such as the F-actin severing protein gelsolin and the actin motor protein myosinII.42,43 The activities of gelsolin and myosin II have previously been shown to be regulated by Tms.14,44 The subcellular compartmentalization of distinct actin filament populations in sperm may therefore be instrumental in regulating sperm motility and fertilization. A change in the Tm composition such as the absence of γTm gene products may facilitate the acrosome reaction and thereby lead to a positive selection of the γTm exon 1b KO allele in the 129/SvJ mouse background. Such a profound impact on sperm would be expected to result in a systematic elimination of the γTm gene. The fact that −/− individuals are not viable clearly prevents this outcome.
Functional redundancy does not occur between tropomyosin genes.
Tm gene isoforms do not appear to be redundant throughout evolutionary development. The elimination of various Tm genes in yeast and worms results in lethality indicating that Tm isoforms perform essential functions.4,45 The elimination of cytoskeletal Tm genes in Drosophila does lead to compensating mutations that result in substitution of an alternative Tm isoform.46–48 The observed embryonic lethality of Δ1b−/− γTm gene mice and reduction of viable Δ9d−/− γTm gene mice show that while the lack of a subset of Tm isoforms can be either fully or partially compensated by other products from the same gene,27,28 the total depletion of products from this gene cannot be compensated. Together with previous studies that show the requirement of products from the α, β and γTm gene for survival,21–23 our present data strongly support the idea that products from different Tm genes are not functionally redundant. The absence of lethality in the Δ9c−/− γTm gene mouse line in combination with the generation of Δ9ab−/− γTm gene ES cells demonstrate that these gene products have some intragenetic functional redundancy. While mice lacking products from the βTm and γTm gene show lethality due to impaired development of the heart, our screening for γTm KO ES cells are suggestive of a more severe defect of cellular function due to the lack of cytoskeletal γTm gene products since neither Δ9d−/− nor Δ1b−/− γTm gene ES cells could be generated. These findings stress the unique functional role of specific Tm products being generated from different Tm genes.
The ability to spatially and temporally regulate the expression of particular combinations of Tm isoforms through the use of different Tm genes and alternative splicing equips mammalian cells with a highly efficient tool to generate functional diversity of dynamic cytoskeletal structures. Our work provides strong evidence for the direct control of vital actin-cytoskeleton dependent cellular functions via the expression of individual sets of Tm proteins early in development.
Materials and Methods
Targeting constructs.
A summary of the γTm gene with targeted deletions for exons 1b, 9ab, 9c and 9d is shown in Figure 1B. The exon 1b targeted deletion (Tpm3tm1(Δ1b)Pgun) is described in Hook et al.22 Briefly, the 132 bp coding sequence of exon 1b as well as 64 bp of intronic sequence 3′ to exon 1b was deleted using a cassette containing LoxP—pGK Thymidine Kinase—pGK NeomycinR—LoxP (obtained from Allan Bradley) which was subsequently removed by Cre-mediated recombination. The exon 9c targeted deletion (Tpm3tm1(neo;Δ9c)Pgun) is described in Vrhovski et al.27 Briefly, the 100 bp coding sequence and 3′ UTR were replaced with a cassette containing LoxP—NeomycinR—LoxP. The exon 9d targeted deletion (Tpm3tm1(neo;Δ9d)Pgun) has been previously described and shown in Figure 3A.28 Briefly, the 100 bp coding sequence and 100 bp of 3′ UTR were replaced with a cassette containing LoxP—pGK NeomycinR—LoxP which was later removed in vivo using Cre-mediated recombination.
The exon 9ab targeted deletion (Tpm3tm1(Δ9ab)Pgun) has not been previously described. A genomic clone from the mouse γTm (TPM3) gene was isolated by screening a 129 strain mouse genomic library (λ DASH II Vector, Stratagene) with oligonucleotide probes directed towards γTm exon 9c as described in Vrhovski et al.27 A 6.7 kb SpeI fragment containing exons 9a and 9b with flanking intron sequence (Fig. 4A) was isolated and cloned into pBlueScript SK+ (Stratagene). In order to generate the floxed targeting construct, a 5.2 kb BstXI-SpeI fragment containing all exons from 6b–9b was subcloned out of the 6.7 kb fragment and cloned into pBlueScript SK+. Two oligos (64 bp) containing the LoxP sequence and restriction sites were annealed and inserted into the NcoI sites 5′ to exon 9a. A cassette containing LoxP—pGK Thymidine Kinase—pGK NeomycinR—LoxP was inserted in the NsiI site 3′ to exon 9b (Fig. 4A).
Generation of targeted ES cells.
The targeting of ES cells for exons 1b, 9c and 9d has been previously described in reference 22, 27 and 28. Briefly, 40 µg of linearized construct was transfected into mouse R1 129/SvJ embryonic stem (ES) cells by electroporation (0.24 kV, 500 µF, Time Constant = 10.0–12.0 ms). The 9d targeting construct was also transfected into mouse Bruce 4 C57BL/6 ES cells using the same conditions.
The Δ9ab targeting construct was generated using a subcloned 5.2 kb BstXI-SpeI fragment containing all exons from 6b–9b. A 64 bp fragment containing the LoxP sequence and restriction sites was inserted into the NcoI sites 5′ to exon 9a. A cassette containing LoxP—pGK Thymidine Kinase—pGK NeomycinR—LoxP was inserted in the NsiI site 3′ to exon 9b. The Δ9ab targeting construct (40 µg) (Fig. 4A) was electroporated into Bruce 4 ES cells using the conditions above. ES cells were cultured for 10 days in KNOCKOUTTm DMEM/F-12 (Invitrogen, 10829-018) containing 10% FBS, 2 mM glutamine, non-essential amino acids (NEAA) (Invitrogen, 11140), 100 µM β-ME, 1,000 units/mL of ESGRO® (Millipore Corporation, ESG1107), with selection using 300 µg/mL G418 (Invitrogen, 10131). A total of 132 clones were picked and grown to confluence in 24-well tissue culture plates prior to harvesting cell pellets for DNA analysis. Genomic DNA was isolated and 10 µg was digested with EcoRV. Digested genomic DNA was subjected to Southern blot analysis using a 1.2 kb external probe 3′ to the region of homologous recombination (Fig. 4A). Homologous recombinant clones for exon 9ab were expanded and re-electroporated with 20 µg CMV-Cre recombinase (Stratagene, 10347-011), followed by selection in 1 µM Ganciclovir (Roche Products, R10047) to eliminate recombined clones containing the Thymidine Kinase-Neomycin cassette. Surviving ES cell clones were isolated, expanded and screened by PCR analysis for either floxed or complete deletion of exon 9a + 9b (Fig. 4C). PCR screening for products up to 1 kb were routinely performed using either EconoTaq Plus Green EconoTaq Plus Green (Lucigen, 30033-1) or Taq DNA polymerase (New England BioLabs, M0267L). A total of 35 cycles with annealing between 55–60°C were used to generate the products shown in Figure 4D (upper and lower parts). The primer set to determine the floxed allele is (9723F: 5′-CAT TTC TCG CTC ACT CGA GG-3′) and (9b3′R: 5′-TGT GCC ACA TTC ATA GCC CTG G-3′) and for the absence of exon 9a and 9b the primer set used was (9724F: 5′-CTG GAA TCT GCC ATC CAT CC-3′) and 9b3′R. PCR products were analyzed by agarose gel electrophoresis in Tris-acetate-EDTA. The primer set designed to amplify exon 9a is (9aF1: 5′-GAG CTC TAT GCC CAG AAA C-3′) and (9aR1: 5′-AGA GGT CAT GTC ATT GAG G-3′).
Generation and screening for double knockout Δ9d ES clones.
A Δ9d+/− ES cell clone containing the Neomycin cassette was electroporated with 20 µg CMV-Cre recombinase using the method above in order to remove the Neomycin cassette in vitro. The 96 clones picked were analyzed by a PCR assay previously described in reference 28, using three primers (F: 5′-GAG ATT TAG TGC GGA ACT-3′), (R1: 5′-GTG GGT CTC TGG CTT GCC TA-3′) and (R2: 5′-CCA ACA GGG TCC GTA AAC C-3′) for a total of 35 cycles with annealing at 50°C to determine a Δ9d+/− ES cell clone lacking the Neomycin cassette. As a secondary confirmation for the absence of the Neomycin cassette, a primer set (neoF: 5′-GCT ATT CGG CTA TGA CTA GGG-3′) and (neoR: 5′-GAA GGC GAT AGA AGG CGA TG-3′) was also used with the above PCR conditions. The resulting Δ9d+/− ES cell clone (Clone#18) (Fig. 3C) lacking the Neomycin cassette was re-electroporated with the exon 9d targeting construct described by Fath et al.28 followed by growth and selection in G418 as described above. A total of 330 clones were picked and analyzed for the complete absence of both copies of exon 9d. These ES clones were screened using the primer set (9dF1: 5′-CTG AAG TGC ACC AAA GAG G-3′) and (9dR1: 5′-GTC TCC CAC AGT GGA GCC T-3′) designed to the coding region of exon 9d. The 330 clones were further analyzed using the F and R2 primers (above) to determine whether the construct had been integrated either randomly or back into the originally targeted locus.
Generation and screening for double knockout Δ9ab ES clones.
A Δ9ab+/− ES cell clone containing the complete absence of exon 9a + 9b allele was re-electroporated with 40 µg of linearized construct as described above. One ES cell clone out of 48 was found by PCR analysis (using primers 9723F; 9724F and 9b3′R above) to be targeted with the 9ab targeting construct on the alternative allele. The doubly targeted clone was again subjected to Cre recombinase to remove the coding regions of exons 9a and 9b as well as the Thymidine Kinase-Neomycin cassette. Following the second round of Cre-recombination and selection, the 9724F and 9b3′R oligonucleotide primer set common to both WT allele and the fully deleted exon 9a + 9b allele was used in order to confirm the presence of any Δ9ab−/− ES cell clones. A single Δ9ab−/− ES cell clone was determined by long range PCR against both WT+/+ and Δ9ab+/− controls. PCR screening for products over 2 kb were performed using LongAmp Taq DNA polymerase (New England BioLabs, M0323S). A total of 35 cycles with annealing between 55–60°C were used to generate PCR products. The expected WT fragment of 2.6 kb and a deleted exons 9a and 9b fragment of 400 bp is shown (Fig. 4H). An oligonucleotide primer set designed to amplify exon 9a (9aF1: 5′-GAG CTC TAT GCC CAG AAA C-3′) and (9aR1: 5′-AGA GGT CAT GTC ATT GAG G-3′) were also used to confirm the absence of exon 9a coding region (data not shown).
Generation and screening of knockout mice.
All animal experimentation was performed in accordance with institutional guidelines and guidelines of the National Health and Medical Research Council, Canberra, Australia. Chimeric mice were generated for the targeted γTm exons 1b, 9c and 9d as previously described in reference 22, 27 and 28. Δ9ab−/− ES cells have so far been used for in vitro analysis in this study. Chimeric mice for exons 1b, 9c and 9d were bred against either 129/SvJ or C57BL/6 mice lines. Genomic DNA was isolated from either ear clip or tail tip mouse tissues and genotyped by PCR for the presence of both WT and KO alleles.
Δ1b+/− mice from the 129/SvJ background were also inter-crossed onto the C57BL/6 background via speed congenic breeding (Transgenic Animal Services Qld, Australia) for five generations. N5 generation Δ1b+/− mice were bred in order to generate null pups. Progeny were genotyped by PCR as previously described in reference 22. Briefly, oligonucleotide primers common to both alleles:- (9341: 5′-GGC TAC AAC GCC GAG CGG AG-3′) and (9342: 5′-CGG GGC TCG ATT CTT TCC AG-3′) were designed to regions either side of the exon 1b deletion to generate a WT fragment of 405 bp and a deleted exon fragment of 355 bp. The PCR products were analyzed on a 2% agarose gel in 1x Tris-acetate-EDTA.
Western blot analysis.
Mouse sperm collected from the vas deferens and caudal epididymus of adult male mice. The sperm were washed twice in PBS and pelleted to remove seminiferous proteins. Mouse sperm protein was prepared by adding a 200 µL volume of RIPA buffer (10 mM Tris-HCl, pH 7.6; 2% SDS; 2 mM DTT) to each sperm pellet followed by sonication. A protein assay was performed using Precision Red (Cytoskeleton Inc., ADV02) to determine protein concentration. Total protein (10 µg) from mouse sperm and a control protein (either 1 µg or 10 µg) sample from primary mouse embryonic fibroblast (PMEF) line were denatured and reduced in sample buffer containing β-Mercaptoethanol. Proteins were analyzed by sodium dodecyl sulfate 12.5% polyacrylamide gel electrophoresis (SDS-PAGE) using 29:1 acrylamide:bis-acrylamide (3.3%C) (Bio-Rad Labs, 161-0156). Gels were transferred to PDVF membrane (Millipore Corporation, IPVH00010) via electroblotting techniques. The PVDF membranes were blocked in 5% skim milk powder in Tris buffered saline (TBS) pH 7.5 for 1 h at room temperature and washed in TBS with 0.1% Tween-20 (TBST) for 3 × 5 min.
The primary antibodies used for western analysis were:- mouse monoclonal γ9d, mouse monoclonal α/9d and rabbit antiserum WS4/9d. The γ9d antibody recognizes products from the γTm gene that contain the 9d exon.49 The α/9d mouse monoclonal recognizes products from both the α and βTm gene that contain the 9d exon (Tms 1, 2, 3, 5a, 5b and 6).50 The δ/9d rabbit polyclonal antiserum recognizes a single exon 9d-containing product from the δTm gene.51 Primary antibodies were diluted in TBS and were incubated with the PVDF membranes at room temperature for 2 h. The membranes were washed in TBST (as above) prior to adding secondary antibody at 1:5,000 (goat anti-mouse Ig or goat anti-rabbit Ig conjugated to horseradish peroxidase, Jackson ImmunoResearch, 115-035-003; 111-035-003) for 1 h at room temperature. The membranes were washed in TBST (as above) and protein bands were detected using a western Lightning kit (Perkin Elmer Life Sciences, NEL101001EA) followed by exposure to Fuji Super RX X-ray film (Fuji Film, 100NIF).
Statistical analysis.
The distribution of maternal and paternal allele for determining transmission ratio distortion were analyzed using a χ2. The null hypothesis was rejected at p < 0.05.
Acknowledgements
This work was supported by Australian National and Medical Research Council (NH&MRC) Grant #321705 (P.G. & G.S.), and funding from the Oncology Children's Foundation (P.G.). P.G. is a Principal Research Fellow of the National Health Medical Research Council (Grant 163626). We thank Drs. Stephen Palmer and Justine Stehn for critical reading of the manuscript.
Abbreviations
- Tm
tropomyosin
- ES
embryonic stem
- WT
wild type
- KO
knockout
- TK
thymidine kinase
- NEO
neomycin
- CMV
cytomegalovirus
- PCR
polymerase chain reaction
- kb
kilobase
- bp
base pair
- N
generation
- PMEF
primary mouse embryonic fibroblast
References
- 1.Cooper JA. Actin dynamics: tropomyosin provides stability. Curr Biol. 2002;12:523–525. doi: 10.1016/s0960-9822(02)01028-x. [DOI] [PubMed] [Google Scholar]
- 2.Lees-Miller JP, Helfman DM. The molecular basis for tropomyosin isoform diversity. Bioessays. 1991;13:429–437. doi: 10.1002/bies.950130902. [DOI] [PubMed] [Google Scholar]
- 3.Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–341. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Araya E, Berthier C, Kim E, Yeung T, Wang X, Helfman DM. Regulation of coiled-coil assembly in tropomyosins. J Struct Biol. 2002;137:176–183. doi: 10.1006/jsbi.2002.4463. [DOI] [PubMed] [Google Scholar]
- 5.Gunning P, O'Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- 6.Pittenger MF, Kistler A, Helfman DM. Alternatively spliced exons of the beta tropomyosin gene exhibit different affinities for F-actin and effects with nonmuscle caldesmon. J Cell Sci. 1995;108:3253–3265. doi: 10.1242/jcs.108.10.3253. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF) Cell Motil. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- 8.Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broschat KO, Weber A, Burgess DR. Tropomyosin stabilizes the pointed end of actin filaments by slowing depolymerization. Biochemistry. 1989;28:8501–8506. doi: 10.1021/bi00447a035. [DOI] [PubMed] [Google Scholar]
- 10.Burgess DR, Broschat KO, Hayden JM. Tropomyosin distinguishes between the two actin-binding sites of villin and affects actin-binding properties of other brush border proteins. J Cell Biol. 1987;104:29–40. doi: 10.1083/jcb.104.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa R, Yamashiro S, Matsumura F. Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin. Potentiation of protective ability of tropomyosins by 83 kDa nonmuscle caldesmon. J Biol Chem. 1989;264:7490–7497. [PubMed] [Google Scholar]
- 13.Wawro B, Greenfield NJ, Wear MA, Cooper JA, Higgs HN, Hitchcock-DeGregori SE. Tropomyosin regulates elongation by formin at the fast-growing end of the actin filament. Biochemistry. 2007;46:8146–8155. doi: 10.1021/bi700686p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryce NS, Schevzov G, Ferguson V, Percival JM, Lin JJ, Matsumura F, et al. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol Biol Cell. 2003;14:1002–1016. doi: 10.1091/mbc.E02-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin C, Schevzov G, Gunning P. Alternatively spliced N-terminal exons in tropomyosin isoforms do not act as autonomous targeting signals. J Struct Biol. 2010;170:286–293. doi: 10.1016/j.jsb.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 16.Schevzov G, Fath T, Vrhovski B, Vlahovich N, Rajan S, Hook J, et al. Divergent Regulation of the Sarcomere and the Cytoskeleton. J Biol Chem. 2008;283:275–283. doi: 10.1074/jbc.M704392200. [DOI] [PubMed] [Google Scholar]
- 17.Coulton AT, East DA, Galinska-Rakoczy A, Lehman W, Mulvihill DP. The recruitment of acetylated and unacetylated tropomyosin to distinct actin polymers ermits the discrete regulation of specific myosins in fission yeast. J Cell Sci. 2010;123:3235–3243. doi: 10.1242/jcs.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clayton JE, Sammons MR, Stark BC, Hodges AR, Lord M. Differential regulation of unconventional fission yeast myosins via the actin track. Curr Biol. 2010;20:1423–1431. doi: 10.1016/j.cub.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 19.Stark BC, Sladewski TE, Pollard LW, Lord M. Tropomyosin and myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Mol Biol Cell. 2010;21:989–1000. doi: 10.1091/mbc.E09-10-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schevzov G, O'Neill G. Tropomyosin gene expression in vivo and in vitro. Adv Exp Med Biol. 2008;644:43–59. [PubMed] [Google Scholar]
- 21.Blanchard EM, Iizuka K, Christe M, Conner DA, Geisterfer-Lowrance A, Schoen FJ, et al. Targeted ablation of the murine alpha-tropomyosin gene. Circ Res. 1997;81:1005–1010. doi: 10.1161/01.res.81.6.1005. [DOI] [PubMed] [Google Scholar]
- 22.Hook J, Lemckert F, Qin H, Schevzov G, Gunning P. Gamma tropomyosin gene products are required for embryonic development. Mol Cell Biol. 2004;24:2318–2323. doi: 10.1128/MCB.24.6.2318-2323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jagatheesan G, Rajan S, Wieczorek DF. Investigations into tropomyosin function using mouse models. J Mol Cell Cardiol. 2010;48:893–898. doi: 10.1016/j.yjmcc.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muthuchamy M, Pajak L, Howles P, Doetschman T, Wieczorek DF. Developmental analysis of tropomyosin gene expression in embryonic stem cells and mouse embryos. Mol Cell Biol. 1993;13:3311. doi: 10.1128/mcb.13.6.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clayton L, Johnson MH. Tropomyosin in preimplantation mouse development: identification, expression and organization during cell division and polarization. Exp Cell Res. 1998;238:450–464. doi: 10.1006/excr.1997.3854. [DOI] [PubMed] [Google Scholar]
- 26.Dufour C, Weinberger RP, Schevzov G, Jeffrey PL, Gunning P. Splicing of two internal and four carboxyl-terminal alternative exons in nonmuscle tropomyosin 5 pre-mRNA is independently regulated during development. J Biol Chem. 1998;273:18547–18555. doi: 10.1074/jbc.273.29.18547. [DOI] [PubMed] [Google Scholar]
- 27.Vrhovski B, Lemckert F, Gunning P. Modification of the tropomyosin isoform composition of actin filaments in the brain by deletion of an alternatively spliced exon. Neuropharmacology. 2004;47:684–693. doi: 10.1016/j.neuropharm.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 28.Fath T, Agnes Chan YK, Vrhovski B, Clarke H, Curthoys N, Hook J, et al. New aspects of tropomyosin-regulated neuritogenesis revealed by the deletion of Tm5NM1 and 2. Eur J Cell Biol. 2010;89:489–498. doi: 10.1016/j.ejcb.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 29.Sutherland CJ, Elsom VL, Gordon ML, Dunwoodie SL, Hardeman EC. Coordination of skeletal muscle gene expression occurs late in mammalian development. Dev Biol. 1991;146:167–178. doi: 10.1016/0012-1606(91)90457-e. [DOI] [PubMed] [Google Scholar]
- 30.Kriz V, Mares J, Wentzel P, Funa NS, Calounova G, Zhang XQ, et al. Shb null allele is inherited with a transmission ratio distortion and causes reduced viability in utero. Dev Dyn. 2007;236:2485–2492. doi: 10.1002/dvdy.21257. [DOI] [PubMed] [Google Scholar]
- 31.Archer JR, Self SJ, Winchester BG. Mechanism of infertility in t-complex mice. Genet Res. 1978;32:79–84. doi: 10.1017/s0016672300018541. [DOI] [PubMed] [Google Scholar]
- 32.Huck UW, Lisk RD. Determinants of mating success in the golden hamster (Mesocricetus auratus): I. Male capacity. J Comp Psychol. 1985;99:98–107. doi: 10.1037/0735-7036.99.1.98. [DOI] [PubMed] [Google Scholar]
- 33.Sharma OP, Hays RL. Heterospermic insemination and its effect on the offspring ratio in rats. J Reprod Fertil. 1975;45:533–535. doi: 10.1530/jrf.0.0450533. [DOI] [PubMed] [Google Scholar]
- 34.Parkening TA, Chang MC. Strain differences in the in vitro fertilizing capacity of mouse spermatozoa as tested in various media. Biol Reprod. 1976;15:647–653. doi: 10.1095/biolreprod15.5.647. [DOI] [PubMed] [Google Scholar]
- 35.Robl JM, Dziuk PJ. Penetration of mouse eggs at various intervals after insemination as influenced by concentration of sperm and strain of male. J Exp Zool. 1987;242:181–187. doi: 10.1002/jez.1402420209. [DOI] [PubMed] [Google Scholar]
- 36.Yagi A, Paranko J. Localization of actin, alpha-actinin and tropomyosin in bovine spermatozoa and epididymal epithelium. Anat Rec. 1992;233:61–74. doi: 10.1002/ar.1092330109. [DOI] [PubMed] [Google Scholar]
- 37.Azamar Y, Uribe S, Mujica A. F-actin involvement in guinea pig sperm motility. Mol Reprod Dev. 2007;74:312–320. doi: 10.1002/mrd.20578. [DOI] [PubMed] [Google Scholar]
- 38.Correa LM, Thomas A, Meyers SA. The macaque sperm actin cytoskeleton reorganizes in response to osmotic stress and contributes to morphological defects and decreased motility. Biol Reprod. 2007;77:942–953. doi: 10.1095/biolreprod.107.060533. [DOI] [PubMed] [Google Scholar]
- 39.Brener E, Rubinstein S, Cohen G, Shternall K, Rivlin J, Breitbart H. Remodeling of the actin cytoskeleton during mammalian sperm capacitation and acrosome reaction. Biol Reprod. 2003;68:837–845. doi: 10.1095/biolreprod.102.009233. [DOI] [PubMed] [Google Scholar]
- 40.Dvorakova K, Moore HD, Sebkova N, Palecek J. Cytoskeleton localization in the sperm head prior to fertilization. Reproduction. 2005;130:61–69. doi: 10.1530/rep.1.00549. [DOI] [PubMed] [Google Scholar]
- 41.Liu DY, Martic M, Clarke GN, Grkovic I, Garrett C, Dunlop ME, et al. An anti-actin monoclonal antibody inhibits the zona pellucida-induced acrosome reaction and hyperactivated motility of human sperm. Mol Hum Reprod. 2002;8:37–47. doi: 10.1093/molehr/8.1.37. [DOI] [PubMed] [Google Scholar]
- 42.Finkelstein M, Etkovitz N, Breitbart H. The role and regulation of sperm gelsolin prior to fertilization. J Biol Chem. doi: 10.1074/jbcM11 170951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sosnik J, Buffone MG, Visconti PE. Analysis of CAPZA3 localization reveals temporally discrete events during the acrosome reaction. J Cell Physiol. 2009;224:575–580. doi: 10.1002/jcp.22211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dabrowska R, Hinssen H, Galazkiewicz B, Nowak E. Modulation of gelsolin-induced actin-filament severing by caldesmon and tropomyosin and the effect of these proteins on the actin activation of myosin Mg(2+)-ATPase activity. Biochem J. 1996;315:753–759. doi: 10.1042/bj3150753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drees B, Brown C, Barrell BG, Bretscher A. Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J Cell Biol. 1995;128:383–392. doi: 10.1083/jcb.128.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tansey T, Schultz JR, Miller RC, Storti RV. Small differences in Drosophila tropomyosin expression have significant effects on muscle function. Mol Cell Biol. 1991;11:6337–6342. doi: 10.1128/mcb.11.12.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kreuz AJ, Simcox A, Maughan D. Alterations in flight muscle ultrastructure and function in Drosophila tropomyosin mutants. J Cell Biol. 1996;135:673–687. doi: 10.1083/jcb.135.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tetzlaff MT, Jackle H, Pankratz MJ. Lack of Drosophila cytoskeletal tropomyosin affects head morphogenesis and the accumulation of oskar mRNA required for germ cell formation. EMBO J. 1996;15:1247–1254. [PMC free article] [PubMed] [Google Scholar]
- 49.Lin JJ, Hegmann TE, Lin JL. Differential localization of tropomyosin isoforms in cultured nonmuscle cells. J Cell Biol. 1988;107:563–572. doi: 10.1083/jcb.107.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schevzov G, Vrhovski B, Bryce NS, Elmir S, Qiu MR, O'Neill GM, et al. Tissue-specific tropomyosin isoform composition. J Histochem Cytochem. 2005;53:557–570. doi: 10.1369/jhc.4A6505.2005. [DOI] [PubMed] [Google Scholar]
- 51.Hannan AJ, Gunning P, Jeffrey PL, Weinberger RP. Structural compartments within neurons: developmentally regulated organization of microfilament isoform mRNA and protein. Mol Cell Neurosci. 1998;11:289–304. doi: 10.1006/mcne.1998.0693. [DOI] [PubMed] [Google Scholar]