Abstract
Background and Aims
Recent papers indicated that epigenetic control is involved in transitions in bud dormancy, purportedly controlling gene expression. The present study aimed to identify genes that are differentially expressed in dormant and non-dormant Castanea sativa buds.
Methods
Two suppression subtractive hybridization cDNA libraries were constructed to characterize the transcriptomes of dormant apical buds of C. sativa, and buds in which dormancy was released.
Key Results
A total of 512 expressed sequence tags (ESTs) were generated in a forward and reverse subtractive hybridization experiment. Classification of these ESTs into functional groups demonstrated that dormant buds were predominantly characterized by genes associated with stress response, while non-dormant buds were characterized by genes associated with energy, protein synthesis and cellular components for development and growth. ESTs for a few genes involved in different forms of epigenetic modification were found in both libraries, suggesting a role for epigenetic control in bud dormancy different from that in growth. Genes encoding histone mono-ubiquitinase HUB2 and histone acetyltransferase GCN5L were associated with dormancy, while a gene encoding histone H3 kinase AUR3 was associated with growth. Real-time RT-PCR with a selection of genes involved in epigenetic modification and stress tolerance confirmed the expression of the majority of investigated genes in various stages of bud development, revealing a cyclical expression pattern concurring with the growth seasons for most genes. However, senescing leaves also showed an increased expression of several of the genes associated with dormancy, implying pleiotropy. Furthermore, a comparison between these subtraction cDNA libraries and the poplar bud dormancy transcriptome and arabidopsis transcriptomes for seed dormancy and non-dormancy indicated a common basis for dormancy in all three systems.
Conclusions
Bud dormancy and non-dormancy in C. sativa were characterized by distinct sets of genes and are likely to be under different epigenetic control.
Keywords: Bud dormancy, Castanea sativa, epigenetic modification, gene expression, leaf development, seed dormancy, senescence, subtraction library
INTRODUCTION
Perennial plants, such as Castanea sativa trees, can be distinguished by their ability to suspend and resume growth recurrently in response to environmental or seasonal conditions. The recurrent transitions of meristems into and out of dormancy are of primary significance to plant productivity and survival. In tree research, dormancy was referred to as ‘absence of visible growth in any plant structure containing a meristem’ (Lang et al., 1987). Dormancy was more recently re-defined as ‘the inability to initiate growth from meristems and other organs and cells with the capacity to resume growth under favourable conditions’ (Rohde and Bhalerao, 2007). Dormancy is crucial because it affects plant productivity, adaptability and distribution (Chuine and Beaubien, 2001). Perennial trees growing in temperate regions of the world are well adapted to the seasonal cycle, having a dormancy period in response to winter conditions when temperatures are low and the photoperiod is short.
Chestnut trees are found in temperate forests of Asia Minor and Mediterranean countries and their bud burst is driven mainly by temperature (Chuine, 2000; Kramer et al., 2000). Bud burst of many trees is often dependent on exposure to a particular duration of cool temperatures (chilling) to release dormancy, followed by an optimal temperature to permit growth in the spring (Martin, 1991). Trees use environmental cues to time growth-dormancy transitions in order to balance maximal growth and timely protection of their meristems against frost damage. Changes in the length of the growing season are expected with global warming. Between 1981 and 1991, the length of the growing season has increased by 5 d per °C temperature rise on average, or by 12 d at high latitudes (Zhang et al., 2004). Ecological and economical consequences have been monitored and reported (Kramer, 1995; Visser and Holleman, 2001; Howe et al., 2003) but processes related to the transition of dormancy are still poorly understood at the molecular level. This lack of knowledge might be due to inaccessibility of the tissues in which dormancy is imposed, time of sampling, or poor amenability to molecular methods. However, furthering insight in the molecular processes that underlie dormancy will improve understanding of this growth cessation as well as any predetermination of growth ability for the next season. Dormancy intensity can vary with time and conditions, e.g. dormancy release and imposition is not simultaneous in all the individual tree buds, apparently controlled by epigenetic mechanisms (Santamaría et al., 2009). Molecular changes take place in the morphologically identical dormant structures when the physiology changes (Arora et al., 2003; Cadman et al., 2006). Excellent reports cover physiological, hormonal and the few molecular aspects of dormancy in woody plants (Arora et al., 2003; Tanino, 2004; Böhlenius et al., 2006; Horvath et al., 2008; Ruttink et al., 2007; Rinne et al., 2011). However, reports on molecular and genetic components of signalling networks that regulate dormancy are still limited. The arabidopsis ABSCISIC ACID-INSENSITIVE3 (ABI3) protein plays a crucial role during late seed development and has an additional function in the vegetative meristem, particularly during growth-arresting conditions. Its homologue in poplar, a species well adapted to cold winters, is expressed in buds during natural bud set (Rohde et al., 2002). Similarly a microarray experiment on poplar bud set shows the major change in the expression 3–4 weeks after the onset of short days (Ruttink et al., 2007). An important group of genes involved in the acquisition of dormancy are the DAM (DORMANCY ASSOCIATED MADS-BOX) genes which are differentially regulated co-ordinately with endodormancy in buds of several perennial plant species (Horvath et al., 2010; Jiménez et al., 2010; Li et al., 2010). Derory et al. (2006) have recently published some differentially expressed genes during six bud flushing stages in sessile oak (Quercus petraea). Winter reportedly disrupts the circadian clock in chestnut (Ramos et al., 2005) and leafy spurge (Horvath et al., 2008). mRNA levels of CsTOC1 and CsLHY are cycled daily in chestnut seedlings and adult plants but during dormancy these genes are constitutively expressed. This alteration also affects CsPRR5, CsPRR7 and CsPRR9, components of the circadian oscillator feedback network (Ibañez et al., 2008). In poplar it has been reported that reducing the expression of PttTOC1, PttLHY1 and PttHLY2 by RNAi leads to a shortened internal period of clock-controlled genes expression rhythms which results in a shorter day-length requirement to induce growth arrest (Ibáñez et al., 2010). The association of epigenetic control with bud burst by chromatin remodelling has been reported (Horvath et al., 2003). Opposite patterns for acetylated H4 histone and genomic DNA methylation in dormant and non-dormant chestnut tree buds have been observed, providing evidence for different forms of epigenetic control during transition between different phases of dormancy that are likely to control related gene expression (Santamaría et al., 2009). However, differences in gene expression related to bud dormancy changes in line with changes in the seasons have not yet been reported in C. sativa.
The objective of the present study was to identify genes involved in C. sativa bud dormancy and non-dormancy using a transcriptome approach. Two suppression subtractive hybridization libraries were constructed, enriched for transcripts associated with dormancy or the absence of dormancy upon release. Suppression subtractive hybridization libraries, or subtraction libraries, form a reflection of the transcriptome of the studied condition. Functional classification was performed and the functional groups from the C. sativa libraries were compared with previously published datasets for dormant and non-dormant poplar buds and arabidopsis seeds. Real-time RT-PCR was used to verify expression of a selection of genes using several bud dormancy stages and other tissues.
MATERIALS AND METHODS
Plant material
Apical buds for the suppression subtractive hybridization were sampled in 2006 from 20 3-year-old Castanea sativa Mill. trees growing in Grado Asturias (experimental station ‘La Mata’). These trees had been produced from seed and grown under ambient conditions in 15-cm-diameter pots with soil containing peat/perlite. Bud collection was performed during bud burst in spring on 7 April (B01-03), 15 April (B08) and 23 April (B09-10) and during bud set in the autumn on 25 October (B93), 1 November (B99) and 10 November (B00) at three different time points in the day (1000 h, 1500 h and 2000 h) and pooled to avoid any effect of the circadian clock. To confirm gene expression during different bud dormancy stages and in several more tissues the same trees were sampled in 2008 following the same sampling strategy. Three biological replicate samples were collected for six apical bud development stages, four leaf development stages, as well as roots and stems (Fig. 1 and Table 1).
Fig. 1.
Development stages of buds, leaves and control tissues of C. sativa, used for gene expression studies. The codes refer to the phenology stages according to the extended general BBCH scale (Hack et al., 1992) adapted for chestnut trees (Table 1).
Table 1.
Chestnut tree phenology stages according to the extended BBCH general scale
| Code | Tissue | Development stage | Sampling date |
|---|---|---|---|
| B93 | Buds | Leaves turn colour | 19 September 2008 |
| L93 | Leaves | Leaves turn colour | 19 September 2008 |
| L95a | Green leaves | 50 % of leaves have dropped | 23 September 2008 |
| L95b | Brown leaves | 50 % of leaves have dropped | 23 September 2008 |
| B99 | Buds | Winter or vegetative rest | 25 November 2008 |
| B00 | Buds | Winter dormancy | 26 January 2009 |
| B01-03 | Buds | Swelling of buds | 13 March 2009 |
| B08 | Buds | Opening of buds | 27 March 2009 |
| B09-10 | Buds | Growth of new green leaves | 27 March 2009 |
| L11-12 | Leaves | Start of leaf development | 14 April 2009 |
| S | Stem | 50 % of final stem length was reached | 14 April 2009 |
| R | Root | 50 % of final stem length was reached | 14 April 2009 |
Suppression subtractive hybridization
Total RNA was isolated from dormant (D) and non-dormant (ND) buds using the hot borate protocol with optimized PVP concentrations: 4·3 mg mL−1 for ND buds and 14·3 mg mL−1 for D buds (Santamaría et al., 2010). The RNA was used to generate two subtraction libraries, one enriched with dormancy-associated gene transcripts (D library) and the other enriched with gene transcripts associated with the absence of dormancy (ND library), using the Clontech PCR-Select™ cDNA Subtraction Kit (Clontech, Saint-Germain-en-Laye, France). For this purpose, total RNA from the samples was used both as ‘tester’ and ‘driver’ in two reciprocal experiments, to create both a forward and reverse library. The subtracted and amplified cDNAs were inserted into pGEM-T Easy and transformed to JM109 high-efficiency competent cells (Promega, Southampton, UK).
DNA sequencing and sequence analysis
Sequencing was performed on an ABI 3700 automated sequencer (Perkin-Elmer, Foster City, CA, USA) at the DNA Synthesis and Sequencing Facility, Macrogen (Seoul, Korea). EST homologies were analysed with BLASTN and BLASTX, querying the National Center for Biotechnology (NCBI) database and the populus database (Sterky et al., 2004). Each EST was evaluated by scoring the chance of random alignment with a database entry [E(xpect)Value], the highest percentage identity for a set of aligned segments to the same subject sequence [Max(imum)Identity].
Most ESTs showed high sequence similarities to plant genes (E-value < 0·05); for only three ESTs similarity was insufficient. Unique sequences were deposited in the NCBI dbEST database (GenBank accession nos. HO847068–HO847579; Supplementary Data Tables S1–S3, available online). Functional classification and categorization of genes were carried out using the Munich Information Centre for Protein Sequences (MIPS) database (http://mips.helmholtz-muenchen.de/proj/funcatDB/search_main_frame.html; Schoof et al., 2002). Functional groups were considered to be over-represented if a 1·5-fold or greater difference in EST abundance between the two subtraction libraries was observed that had a binomial probability (P) of 0·10 or smaller.
Real-time RT-PCR
Total RNA was extracted from buds, leaves, roots and stems using the hot borate protocol. Total RNA was purified using the RNeasy Plant Mini Kit (Qiagen, Crawley, UK) and each of the biological replicates was reverse transcribed using the Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics, Burgess Hill, UK). Primers for real-time PCR were generated using the DNASTAR software package PrimerSelect (Lasergene) and ordered from Invitrogen. The PCR conditions were 40 cycles with 10 s at 95 °C, 10 s at the specified annealing temperature (Table 2) and 30 s at 72 °C. PCR reactions were performed in 20 µL LightCycler®Capillaries using LightCycler® FastStart DNA Master SYBR® Green I. The reactions were carried out in a LightCycler 2·0 PCR thermal cycler and results were analysed using the LightCycler Software 4·05 (Roche Diagnostics). The MgCl2 concentration was optimized separately for each primer pair (Table 2). For negative controls, 1 µL of water was used instead of cDNA, as well as an RNA sample without reverse transcription (no-RT). Primer efficiency was tested using a standard curve for each gene. Subsequent to ampification, a melting curve analysis was performed to verify gene-specific amplification.
Table 2.
Gene-specific primer sequences, annealing temperatures and MgCl2 concentrations used for real-time RT-PCR
| Gene | EST # | Product length (bp) | Forward primer sequence | Reverse primer sequence | Annealing temp. (°C) | MgCl2 (mm) |
|---|---|---|---|---|---|---|
| GCN5L | K-D4-A01 | 300 | ACTGGCATCTGGGCGTGTG | ATGAAGCGGGCAATCGTGAC | 60·0 | 3 |
| HUB2 | K-D7-D01 | 433 | AGCCCCATGACGCACTGT | GCCTCGGTAAGCTCCAAAAC | 55·0 | 3 |
| GOLS | K-D8-G07 | 244 | AGATGATTTACCTAGACGGAGACA | AAAGAAGCCAGCATTGAAGTAT | 53·4 | 5 |
| RADSAM | L1-2–40 | 596 | CCGGTTCAATCTGGGAGTG | TTAGTGCAAGGCGAACATTTAG | 60·0 | 3 |
| SAMS | K-G4-C06 | 240 | AGGGCCATTGCTCAGGTCTCT | CCAGGTGAAGTCAGGGTCGTC | 60·0 | 1·5 |
| AUR3 | K-G2-H03 | 174 | AACGCATTTTCTTGATTCTCG | ATATACGTGGCAGCTTTGTTCTCG | 60·0 | 3 |
| HSPTF | K-G5-A04 | 167 | CGAGTTGCCCGAGTTTCTTTC | GGCTTTTGCACCATTGGACTATTA | 55·0 | 3 |
| HSPTF90 | K-G4-D02 | 136 | ACTTTGGTGAGGTCCTTGGTAGAG | AGGGAAATTGGGTAGCTGATAAAC | 55·0 | 3 |
| ACTIN | 188 | TCCATCATGAAGTGCGATGT | AACCTCCGATCCAGACACTG | 54·8 | 4·5 |
The absence of genomic DNA was confirmed by the no-RT control. Analysis of dissociation curves was performed to check gene-specific amplification. Agarose gel electrophoresis of the PCR products was used to verify amplicon size. Reactions were performed in duplicate for each biological replicate. Relative expression values for each target gene were expressed as fold-change using the ΔΔCt method (Livak and Schmittgen, 2001). Actin was tested and found to be not differentially expressed; therefore, relative expression values were normalized for actin and relative to the expression at stage B00 (Table 1).
Data analysis
Changes in gene expression among tissues and stages were analysed by analysis of variance (ANOVA) in Genstat 11 using the ΔΔCt values. Duncan's multiple range test was used to compare samples post-hoc.
RESULTS
Library construction and sequence analysis
Two subtraction libraries enriched for C. sativa bud transcripts associated with non-dormancy and dormancy were constructed, referred to here as the ND library and the D library, respectively. The lengths of the partial cDNAs varied from 89 to 1151 bp with an average length of 530 bp. In total, 806 sequences were generated. After sequencing, this initial set of 806 sequences was reduced to 512 unique consensus EST sequences (unigenes) comprising 150 contigs and 362 singletons. Only one of the ESTs was observed in both libraries, which is indicative of the high quality; this EST was removed from the genelist and analysis. The number of ESTs forming each contig varied between two and 22. The ND library contained 187 singletons and 67 contigs, while the D library contained 175 singletons and 83 contigs. The percentage redundancy was 26 % for the ND library and 32 % for the D library. After BLAST analysis and annotation a functional classification was performed according to a modified MIPS scheme.
MIPS classification
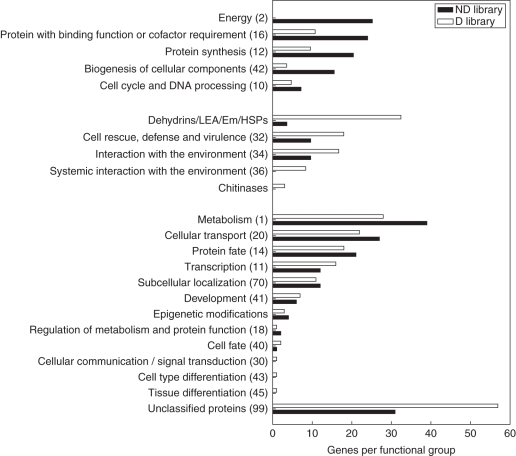
Of all ESTs, 86 % were annotated while for only three ESTs insufficient homology was retrieved for accurate identification. A further 13 % of the sequences showed similarity to proteins whose classification is not yet clear or unclassified. A modified MIPS classification was used to characterize the similarities and differences between the two subtraction libraries, based on functional groups of sequences (Fig. 2). Among the biological processes a high proportion of functionally assigned unigenes fell into the following classes Metabolism (67 sequences); Cellular Transport, Transport Facilities and Transport Routes (49 sequences); and Protein Fate (39 sequences); together accounting for 30 % of the assignable unigenes (Supplementary Data Table S1, available online). No over-representation was found for these classes in either library relative to the other library. Other assigned classes without over-representation in either of the two subtraction libraries relative to the other library and with substantial EST numbers were Transcription; Subcellular Localization; and Development.
Fig. 2.
Frequency distribution for the different classes of the modified MIPS classification with greater representation of the ND library (top) and the D library (middle), and for classes without over-representation (bottom); as well as frequencies for sequences without classification or homology. Numbers in brackets refer to the MIPS class identifier.
In the ND library there was a greater representation relative to the D-library of ESTs in the classes: Energy; Protein with Binding Function or Cofactor Requirement; Protein Synthesis; Biogenesis of Cellular Components; Cell Cycle and DNA Processing. The functional group Cell Cycle and DNA Processing contained five cyclin-associated genes, of which four were present in the ND library. The functional group Biogenesis of Cellular Components contained five ESTs homologous to tubulins that were all found in the ND library, and eight ESTs homologous to histones of which seven ESTs were present in the ND library and only one EST in the D library. The functional group Protein with Binding Factor or Cofactor Requirement contained eight ESTs homologous to laccases; seven ESTs for these lignin biosynthesis enzymes were present in the ND library and only one EST in the D library. In total, 77 ESTs fell in the five functional groups with over-representation in the ND library, while only 24 ESTs belonging to these functional groups were found in the D library. In addition, three ESTs with homology to aquaporins were found in the ND library and only one in the D library, included in the functional group Cellular Transport. Together, these results indicated higher transcription during bud burst of genes involved in growth, division and differentiation of cells.
In contrast, ESTs with greater representation in the D library fell into the classes: Cell Rescue, Defence and Virulence; Interaction with the Environment; Systemic Interaction with the Environment. Furthermore, 27 ESTs with homology to dehydrins, late embryogenesis abundant (LEA) proteins, Em proteins and heat shock proteins (HSPs) were found in the D library while only three ESTs in this group were found in the ND library. A small number of three ESTs with homology to chitinases were found, all of them in the D library. In total, 66 ESTs belonged to these five functional groups over-represented in the D library, and only 22 ESTs belonging to these functional groups were found in the ND library. Many of the ESTs in dormant buds were associated with stress, with greater representation in the D library (63 ESTs) across the MIPS classes Cell Rescue, Defence and Virulence (15 ESTs); Interaction with the Environment (14 ESTs); Systemic Interaction with the Environment (seven ESTs); and dehydrins, LEA proteins and Em proteins (27 ESTs), depending on the kind of stress perceived. Conversely, only 22 stress response-associated ESTs (eight, eight, three and three ESTs, respectively) were found in the ND library for these classes. Dormant buds contained a lower water content (P < 0·001), which might explain the higher number of stress-associated ESTs in the D library (Supplementary Data Fig. S1). ESTs with homology to different genes involved in epigenetic modifications were found in the D library (three) and the ND library (four). This suggested that epigenetic modifications control expression of genes involved in dormancy and growth.
Confirmation of gene expression
The EST BLAST analysis and functional classification allowed the identification of putative gene function. The presence of an EST in one of the two subtraction libraries is purportedly the result of higher expression of that gene under one of the conditions used for the suppression subtractive hybridization. To confirm differential expression in dormant and non-dormant buds a number of ESTs were selected that have a function consistent with epigenetic modifications or drought tolerance, two important events for bud development. The selected ESTs were orthologues of the following arabidopsis genes: a histone mono-ubiquitination 2 (HUB2; D library), a GCN5-like gene (GCN5L; D library), a histone H3 kinase (AUR3; ND library), S-adenosyl-l-methionine synthetase (SAMS; ND library) and a radical SAM protein (RADSAM; ND library). HUB2 catalyses the histone H2 mono-ubiquitination which plays a key role in seed dormancy possibly by regulating ABA levels, ABA sensitivity and other mechanisms (Liu et al., 2007; Chinnusamy et al., 2008). The CsHUB2 sequence was very similar to AtHUB2 and the putative amino acid sequence of both AtHUB2 and CsHUB2 contained a Structural Maintenance of Chromosomes (SMC) domain (TIGR02169; Supplementary Data Fig. S2). GCN5L function has not yet been described in plants, but its structure resembles that of GCN5, which plays a role in the regulation of histone acetylation and regulation of gene expression in response to light (Benhamed et al., 2006). In humans a GCN5L gene was held responsible for acetylation of H3K14 (Meyer et al., 2008). The putative CsGCN5L sequence contained a GCN5-like protein 1 domain (Supplementary Data Fig. S2). AUR3 is a histone serine kinase specific for H3S10 with transcripts and proteins most abundant in tissues containing dividing cells (Demidov et al., 2005). The putative CsAUR3 sequence is highly similar to the arabidopsis AUR3 sequence (Supplementary Data Fig. S2) as well as two other AUR genes of arabidopsis (data not shown) and contains a conserved protein kinase domain PKc. Radical SAM proteins catalyse diverse reactions, including unusual methylations, through generation of a radical species by reductive cleavage of S-adenosylmethionine (Sofia et al., 2001). Substrates for the radical SAM proteins are not nucleophilic, as opposed to conventional DNA methyltransferases: they are either electrophilic or sites normally considered to be unreactive. The putative amino acid sequence of CsRADSAM contained a radical SAM protein domain as well as a 2-methylthioadenine synthetase domain (Supplementary Data Fig. S2). SAMS is involved in DNA methylation and its gene expression is modified by ethylene induction (Yang and Hoffman, 1984). Most of the putative amino acid sequence contained a methionine adenosyltransferase domain (Supplementary Data Fig. S2). In addition to these genes involved in epigenetic control, the orthologues of galactinol synthase (GOLS; D library) and two heat shock protein transcription factors (HSPTF and HSPTF90; both ND library), purportedly playing an upstream role in tolerance to drought and heat stress (Yamada and Nishimura, 2008; Riechmann et al., 2000), were used for confirmation of expression. GOLS is highly expressed in seeds and in desiccation-tolerant parts of resurrection plants (Taji et al., 2002). Conserved domain analysis confirmed cDNA sequence analysis (Supplementary Data Fig. S2).
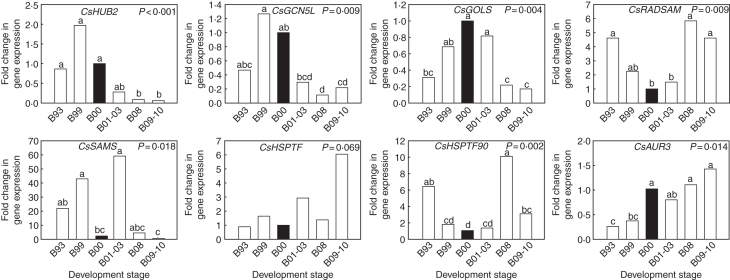
Six stages of bud development were used for confirmation and to further investigate expression of the selected ESTs from the two subtraction libraries, from early bud formation at the end of the growing season to bud burst at the start of the next growing season (Table 1). For CsHUB2, CsGCN5L and CsGOLS that were found in the D library higher expression was detected during the dormancy stages B99 and B00 than in the non-dormant stages B08 and B09-10 (P ≤ 0·009). This formed part of a transient increase in gene expression observed in the buds between the end of the growing season (B93) and the start of the next growing season (B09-10; Fig. 3), coinciding with bud set and bud burst. Three of the ESTs that were found in the ND library (representing CsRADSAM, CsHSPTF90 and CsAUR3) showed lower expression in one of the dormant stages B99 or B00 than in one of the non-dormant stages B08 or B09-10 (Fig. 3; P ≤ 0·014). The expression pattern of CsRADSAM displayed a negative reciprocal pattern of expression to that of CsHUB2, CsGCN5L and CsGOLS, displaying a transient decrease. CsHSPTF90 showed a similar transient decrease, with a peak when buds start to open (B08) and a second decrease at the start of the growing season (B09-10). CsHUB2, CsGCN5L, CsGOLS, CsRADSAM and CsHSPTF90 all displayed a cyclical pattern of expression that coincided with but set and bud burst. For CsAUR3 a steady increase in expression was observed, reaching a maximum during bud burst. Expression of CsHSPTF, found in the ND library, appeared higher in non-dormant buds B09-10 than in dormant buds B00. This result confirmed the result of the subtraction library, although it was not significant by a small margin (Fig. 3; P = 0·069). The only EST for which no confirmation was found was CsSAMS, which was found in the ND library, showing a strongly fluctuating expression pattern with higher expression during early bud development B93, vegetative rest B99 and during swelling of the buds at the end of the winter season B01-03. Although significant, these results for CsSAMS were incongruous with bud set and bud burst.
Fig. 3.
Relative gene expression of HUB2, GCN5L, GOLS, RADSAM, SAMS, HSPTF, HSPTF90 and AUR3 during six dormancy stages of C. sativa buds, assessed by real-time RT-PCR. Expression is relative to actin and normalized for stage B00 (highlighted in black; n = 3). Identical letters above the bars indicate no significant differences between stages in a Duncan's multiple range test, following a significant ANOVA test result. Development stages are described in Table 1.
Gene expression during leaf development
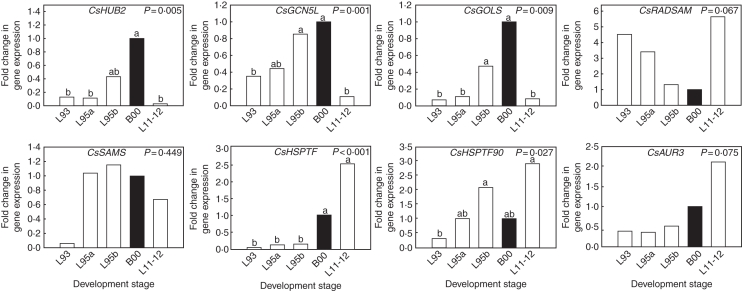
Since buds produce leaves at the start of the growing season, expression of the eight selected ESTs was also studied in leaves to see if trends in transcript abundance during induction and release from bud dormancy persisted during the different stages of subsequent growth. The hypothesis was tested that the genes represented in the D library are expressed low in the various stages of leaf development compared with dormant buds, while the genes represented in the ND library are expressed high. The three ESTs found in the D library, CsHUB2, CsGCN5L and CsGOLS, showed significant differences (P ≤ 0·009) in expression between the different leaf development stages, including young leaves and senescing leaves (Fig. 4). Young leaves L11-12 exhibited lower expression than dormant buds B00 for each of these genes, confirming association of transcript abundance with the presence of dormancy; and expression increased again in older and senescing leaves (stages L93 and L95). Expression of CsHSPTF (P < 0·001) was higher in young leaves L11-12 than in dormant buds B00 confirming association of transcript abundance with the absence of dormancy. The expression pattern of CsHSPTF90 was similar (P = 0·027); however, expression of CsHSPTF in older leaf development stages was low, while that of CsHSPTF90 was low as ageing leaves turned colour (stage L93), but increased again in senescing leaves (stage L95). Although expression of CsRADSAM and CsAUR3 appeared higher in young leaves (L11-12) than in dormant buds (B00) these differences were not significant. Expression of CsSAMS also did not differ significantly between the different leaf development stages. Although the hypothesis was accepted for four genes (CsHUB2, CsGCN5L, CsGOLS and CsHSPTF), this was only valid for young leaves in the case of the dormancy genes, and for senescing leaves in the case of CsHSPTF. Senescing leaves did not always demonstrate a clearly different expression level than dormant buds for the dormancy genes, which indicated that these genes play a pleiotropic role in these different development stages.
Fig. 4.
Relative gene expression of HUB2, GCN5L, GOLS, RADSAM, SAMS, HSPTF, HSPTF90 and AUR3 during four stages of C. sativa leaf development, assessed by real-time RT-PCR. Expression is relative to actin and normalized for stage B00 (highlighted in black; n = 3). Identical letters above the bars indicate no significant differences between stages in a Duncan's multiple range test, following a significant ANOVA test result. Development stages are described in Table 1.
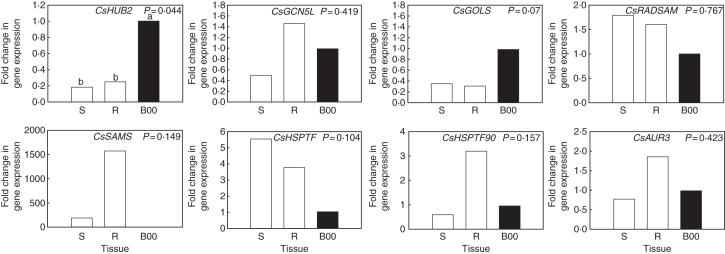
Expression of the selected genes was not restricted to buds and leaves. Similar levels of transcript abundance were observed in stem and root tissue for CsGCN5L, CsGOLS, CsRADSAM, CsHSPTF, CsHSPTF90 and CsAUR3 (Fig. 5). Expression of CsSAMS appeared 100-fold higher in stems and 1500-fold higher in roots than in dormant buds, although this was not significant. Expression of CsHUB2 was 5-fold lower in root and stem tissue than in dormant buds (B00). Gene expression in all investigated tissues is summarized in Supplementary Data Table S4.
Fig. 5.
Relative gene expression of HUB2, GCN5L, GOLS, RADSAM, SAMS, HSPTF, HSPTF90 and AUR3 in the stem and root tissue of C. sativa assessed by real-time RT-PCR. Expression is relative to actin and normalized for stage B00 (highlighted in black; n = 3). Identical letters above the bars indicate no significant differences between tissues in a Duncan's multiple range test, following a significant ANOVA test result.
A transcriptome comparison between poplar and C. sativa buds
A transcriptome analysis was recently described for the induction of bud dormancy in poplar (Ruttink et al., 2007). The ESTs for C. sativa buds were compared with the genelists for poplar, comparing the induction of bud dormancy in poplar with the release from bud dormancy in C. sativa. Of the 196 poplar genes orthologous to the C. sativa ESTs in the D library, 55 showed concurring increasing expression with induction of dormancy, while 27 showed decreasing and 114 unaltered expression. Conversely, of the 210 poplar genes orthologous to the C. sativa ESTs in the ND library, 78 showed concurring decreasing expression with induction of dormancy, while 38 showed an increasing and 94 unaltered expression (Supplementary Data Tables S2 and S3). These data indicated a relatively small similarity in transcriptome composition of the D (28 %) and ND libraries (37 %) between C. sativa and poplar. However, the real-time PCR data suggested a high reliability of both C. sativa subtraction libraries. Furthermore, for the seven confirmed ESTs six poplar orthologues were available with expression data; of these, only two poplar orthologues (GOLS and AUR3) showed a similar expression pattern in C. sativa while four orthologues (GCN5L, HUB2, HSPTF and HSPTF90) were not significant in poplar. This difference between genelists and gene expression patterns in poplar and C. sativa buds possibly highlights the difference between induction of and release from bud dormancy.
A transcriptome comparison between arabidopsis seeds and C. sativa buds
Like buds, seeds can cycle through dormant states and gene expression for these different states of dormancy has been studied in arabidopsis seeds. The functional classification of the dormant and non-dormant C. sativa buds in this paper was compared with the functional classification of dormant and non-dormant arabidopsis seeds, using the arabidopsis sets of 442 genes associated with dormancy and 779 genes associated with non-dormancy (Cadman et al., 2006). Several similarities between both analyses were observed. Similar to dormant C. sativa buds, a reduced number of genes were observed in dormant arabidopsis seeds associated with the classes Protein Synthesis and Energy. Moreover, similar to dormant seeds, a higher number of genes associated with stress response were found in the D library than in the ND library. A difference was formed by genes involved in the class Cell Cycle and DNA Processing, for which in arabidopsis seeds a similar number of genes were found in dormant and non-dormant seeds; as well as the class Transcription, for which in arabidopsis seeds over-representation was observed in dormant seeds.
Of the 442 arabidopsis genes with higher expression in dormant seeds, orthologues of ten specific genes were found in the C. sativa D library. These were predominantly stress-associated genes and included a dormancy-associated gene (DRM1) and a second dormancy-associated gene (AT2G33830), as well as a NAD+ ADP-ribosyltransferase, an LEA protein, two Em-like proteins (GEA6 and GEA1), two heat-shock proteins (HSP70 and HSP101), a shoot gravitropism gene (SGR2) and a galactinol synthase gene (GOLS2) (Supplementary Data Table S5). Conversely, five orthologous genes were found in the C. sativa D library with higher expression in non-dormant aradidopsis seeds, indicating some differences between the two systems. These were an extracellular dermal glycoprotein, a potassium transporter (KUP3), a cysteine proteinase, a dehydrin (COR47) and a 60S ribosomal protein (L27AB). Of the 779 arabidopsis genes with higher expression in non-dormant seeds, 13 orthologous genes were found in the C. sativa ND library, predominated by the functional groups Metabolism and Biogenesis of Cellular Components. This group contained three tubulin genes (TUA4, TUA6 and TUB1) as well as genes encoding a histone H2A, a cysteine proteinase (RD19A), a gibberellin response protein (GASA4), a serine carboxipeptidase S10, a hydroxymethylbilane synthase, a xyloglucan:xyloglucosyl transferase (XTH33), 2-dehydro-3-deoxyphosphoheptonate aldolase 1 (DHS2), glyceraldehyde-3-phosphate dehydrogenase (GAPC), a kinase interacting family protein and a SPIRAL-like protein. No discrepancy was found for orthologues in the C. sativa ND library that displayed higher expression in the dormant arabidopsis seeds. Of these 28 orthologous shared genes between arabidopsis seeds and C. sativa buds, 25 were also shared with poplar buds exposed to short days to induce bud dormancy, but only ten showed a similar expression pattern in association with dormancy (Ruttink et al., 2007; Supplementary Data Table S5). This lower similarity in gene expression between the C. sativa and poplar buds is likely to be due to the difference in sampling strategy, as concluded above. Overall, comparison between C. sativa bud dormancy and arabidopsis seed dormancy transcriptomes indicated strong similarities as well as some discrepancies.
DISCUSSION
Bud dormancy is fundamental to the study of plant developmental processes and its regulation is of significant economic importance to fruit and horticultural industries. Gaining knowledge of the genes involved in bud burst and bud set generates insight in this process, enables the development of markers and offers the potential to control bud dormancy for scientific and applied purposes. This paper reports a functional gene classification during dormant and non-dormant stages and a collection of differentially expressed genes in dormant and non-dormant C. sativa buds, generated with suppression subtractive hybridization and validated with real-time RT-PCR. Suppression subtractive hybridization is efficient in generating cDNA libraries highly enriched for differentially expressed genes of both high and low abundance (Diatchenko et al., 1996). Several recent studies have also used similar approaches or whole-genome transcriptomics to study the dormancy process in other systems (Pnueli et al., 2002; Rohde et al., 2002, 2007; Schrader et al., 2004; Horvath et al., 2005, 2006, 2008; Cadman et al., 2006; Finch-Savage et al., 2007; Mazzitelli et al., 2007; Rohde and Bhalerao, 2007; Ruttink et al., 2007; Mathiason et al., 2009; Ophir et al., 2009). Comparing the lists of genes and functions from different studies using different dormancy systems allows the identification of shared pathways of common importance. The results from this report were compared with the poplar bud dormancy study and the arabidopsis seed dormancy study (Cadman et al., 2006; Ruttink et al., 2007).
Bud dormancy is associated with stress tolerance
Dormant buds of C. sativa were characterized by a high representation of transcripts involved in cell rescue, defence and virulence, interaction with the environment, low temperature stress and dehydratation protection. Different proteins were reported to accumulate during water loss to protect and establish cellular structures during dehydration. These include the LEA proteins (Ingram and Bartels, 1996; Phillips et al., 2002) and HSPs (Wehmeyer and Vierling, 2000). Dehydrins and Em proteins (members of LEAs) may also protect the cells during dehydration. In buds from oak (Derory et al., 2006) and poplar (Ruttink et al., 2007) LEA gene expression was also higher during the dormant stage. The expression across different dormant states of arabidopsis seeds shows higher expression of genes encoding LEA and Em proteins as well as HSPs compared with non-dormant states (Cadman et al., 2006). Certain HSPs are induced in response to low temperatures, and recent data suggest that they are involved in cold acclimation in C. sativa (Ramos et al., 2005). HSPTFs are transcription factors that control the expression of heat-shock proteins. CsHSPTF and CsHSPTF90 found in the ND library are likely to induce HSPs during bud burst and subsequent leaf growth when required by the conditions. Gene expression for CsHSPTF90 was higher during bud burst, while for CsHSPTF expression appeared higher during subsequent early bud growth (Fig. 3). During early leaf development transcript abundance for both HSPTFs indeed appeared higher (Fig. 4), supporting a role for HSPTFs subsequent to bud burst.
Stress tolerance and drought tolerance are of quintessential importance for the survival of buds. Galactinol synthase has been described to play an important role in this (Taji et al., 2002). Galactinol synthase is a key regulatory enzyme in the biosynthesis of the raffinose family of oligosaccharides (RFOs) and has been associated with bud dormancy as well as leaf senescence. Galactinol synthase was expressed from 1 week after the onset of short days and remained expressed throughout the dormant period in poplar buds (Rohde and Bhalerao, 2007). Galactinol synthase is highly expressed in dormant buds from sessile oak (Derory et al., 2006), in seeds and in desiccation-tolerant parts of resurrection plants (Taji et al., 2002). RFOs are thought to play a role in desiccation tolerance of seeds and galactinol synthase was reported to be involved in drought and heat-stress tolerance (Pukacka and Wójkiewicz, 2002; Taji et al., 2002; Zhao et al., 2003). CsGOLS expression was observed to be higher in dormant buds than in non-dormant buds. Together, these data strongly indicated that dormant buds of C. sativa contain multiple mechanisms to protect against the adverse effects of dehydration. The results from the C. sativa D library were in agreement with published data.
Bud burst is associated with the cell cycle
The C. sativa ND library was characterized by cDNAs for proteins involved in growth (cyclins, histones and tubulins) and water relations, energy and metabolism. Dormancy release resulted in the up-regulated transcription of cyclins and histone genes (Devitt and Stafstrom, 1995; Horvath et al., 2002; Freeman et al., 2003). In underground buds of leafy spurge, histone H3 and a tubulin were shown to be differentially expressed in adventitious shoot buds after breaking paradormancy (Anderson and Horvath, 2001; Horvath et al., 2002). β-Tubulin, which has been suggested as a marker for monitoring dormancy in tree buds (Bergervoet et al., 1999), is a basic structural unit of microtubules whose function is required for cell division and cell elongation. High expression of three H2A genes was observed in non dormant arabidopsis seeds, as well as in onion bulbs where the peak in expression indicated the time of dormancy break (Carter et al., 1999; Cadman et al., 2006). In general, high expression of genes involved in cell cycle and translation was observed in non-dormant systems.
Aquaporins are a class of membrane-bound water channel proteins that facilitate water transport across membranes (Maurel, 1997). ESTs with similarity to an aquaporin gene were found to be down-regulated at the dormancy to growth-phase transition in Rubus idaeus buds (Mazzitelli et al., 2007). The over-representation of aquaporins in the ND library of C. sativa concurs with the published literature and with the increased water content of buds associated with growth in this development stage (Supplementary Data Fig. S1).
Laccases are widespread in plants, and are involved in the biosynthesis of lignin. In fungi, laccases are involved in lignin degradation, development-associated pigmentation, detoxification and pathogenesis; in bacteria, laccases are related to endospore coat protein biosynthesis (Sharma and Kuhad, 2008). A large number of laccases were found in the C. sativa ND library, putatively playing a role in lignin synthesis in the growing tissues. This result is consistent with the increased expression of laccase genes after chemical induction of dormancy release in grape buds (Ophir et al., 2009). The results from the ND library identified many genes involved in active growth, which is in agreement with the literature.
A comparison of dormancy in C. sativa buds with poplar buds and arabidopsis seeds
A relatively small similarity in transcriptome composition was observed between the C. sativa D and ND libraries and the poplar genesets. Various explanations are conceivable. Firstly, the two species are phylogenetically too distant to display the same genesets. This seems unlikely given the greater similarity between the transcriptomes of C. sativa bud dormancy and arabidopsis seed dormancy. Not only were the studied tissues different; the two species are also located in different phylogenetic clades. Secondly, cDNA subtraction libraries may not be a proper reflection of the differences in transcriptomes. This too seems unlikely, since several publications have reported reliable results with this technique (e.g. Diatchenko et al., 1996). Thirdly, induction of and release from bud dormancy may not be entirely comparable, and only a limited number of genes could have an expression pattern that mirrors dormancy changes. Surprisingly, this last explanation seems most probable since the number of confirmed genes with dissimilar expression patterns between poplar and C. sativa was high (four out of six).
It has been hypothesized that seed and bud dormancy involve similar processes (Rohde et al., 2002). This hypothesis was tested by comparing the C. sativa D library with the arabidopsis seed dormancy transcriptome, which was characterized by whole-genome transcriptomics. The absence of dormancy in both C. sativa buds and arabidopsis seeds was strongly associated with energy and protein synthesis (Cadman et al., 2006). Conversely, both arabidopsis seed dormancy and C. sativa bud dormancy showed strong association with stress response, which is likely to prepare them for unfavourable conditions. A total of 28 putative orthologous genes were shared between C. sativa buds and arabidopsis seeds, of which 23 were correctly matched for the state of dormancy. The shared functional groups of proteins suggest that seed and bud dormancy use similar molecular pathways. Moreover, the high degree of sequence conservation of the shared orthologous genes underlines the importance of these molecular pathways in both dormancy systems. Seeds as well as buds are suitable structures to survive adverse conditions outside the growing season, thus contributing to the survival of the individual. Dormancy is of benefit since it results in scattering over time of sprouting or germination. Synchronization of these events with the start of the growing season inevitably requires an appropriate environmental cue to signal suitable conditions for growth resumption. Therefore, it is not surprising to detect the convergence of molecular pathways that underlie bud and seed dormancy, even in two different species.
A role for epigenetic modification in bud dormancy transitions
Epigenetic modifications control expression of targeted genes through chromatin remodelling. Two forms of epigenetic control, DNA methylation and H4 acetylation, have been reported to play a role in C. sativa bud burst and bud set (Santamaría et al., 2009). HUB1- and HUB2-mediated H2B monoubiquitination facilitates transcriptional control of targeted genes in arabidopsis, possibly through H3 hypermethylation, which controls flowering time (Cao et al., 2008). Both the hub1 and hub2 mutants displayed reduced seed dormancy (Liu et al., 2007), while early leaf and root growth were inhibited (Fleury et al., 2007). The latter was achieved by down-regulating cell proliferation through reduced expression of genes involved in the cell cycle and cytokinesis. A described role for histone H2B ubiquitination in these different developmental processes highlights the importance of this control mechanism in plant development. In C. sativa buds, HUB2 was found in the D library and the expression peaked during dormancy induction and leaf senescence. Therefore, it is possible that in C. sativa H2B monoubiquitination plays a role in bud dormancy, possibly by reducing the expression of genes involved in cell cycling (Fig. 2). AtAUR3 encodes a histone serine kinase (H3-S10 specific). Aurora-like kinases play a role in chromosome segregation and cytokinesis in yeast, plant and animal systems (Demidov et al., 2005). The transcripts and proteins of all three kinases are most abundant in tissues containing dividing cells (Gutierrez, 2009). An AUR3 orthologue was found in the ND library and gene expression was higher during bud burst and new leaves than during bud set. Thus, it is possible that phosphorylation of histone H3 serine plays a role in cell division upon bud burst in C. sativa. Expression of a GCN5-related gene (AT4G19985) is lower in dormant seeds of arabidopsis than in non-dormant seeds (Cadman et al., 2006). Therefore, it is plausible that completion of arabidopsis seed germination coincides with and is preceded by histone hyperacetylation. Acetylation of H3 has been described to depend on GCN5 (Wang et al., 1998). The hyperacetylated form of histone H3 was also associated with continued plant growth and delayed senescence, as was concluded from a study with the hda6 mutant with strongly reduced transcript levels for a histone deacetylase (Wu et al., 2008). It is therefore surprising that GCN5L, similar in structure to GCN5, was observed in the D library and that its expression was increased both in dormant buds and in senescing leaves of C. sativa. One possible explanation is that GCN5L and GCN5 play complementary roles in acetylation of subsets of histones, hence controlling different sets of genes associated with either the metabolically inactive state (GCN5L) or the growing phase (GCN5). Although results for CsGCN5L appeared inconsistent, this seems to reflect the strong lack of studies into GCN5L in plants. On the whole, different forms of epigenetic control seem closely linked to bud set and bud burst in C. sativa.
The data indicate that the observed differences in expression of CsHUB2, CsGCN5L, CsGOLS, CsRADSAM, CsHSPTF, CsHSPTF90 and CsAUR3 in apical buds and leaves are a reflection of the dormancy status and development in these tissues, and that the cDNA subtraction libraries are a good representation of the transcriptomes during bud dormancy and bud burst. However, some of these genes are likely to display pleiotropy, since their transcript abundance is also associated with leaf senescence. Transcriptome comparisons between arabidopsis seeds and C. sativa buds are consistent with a common basis for dormancy in both processes.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following tables and figures. Table S1: EST numbers per functional group in the D and ND libraries. Table S2: EST list of the D library, containing C. sativa data and sequence similarity to the best hit in a NCBI BLAST search, the best hit and sequence similarity in a populus DB BLAST search, closest arabidopsis homologue, and poplar gene expression data from Ruttink et al. (2007). Table S3: EST list of the ND library, containing C. sativa data and sequence similarity to the best hit in a NCBI BLAST search, the best hit and sequence similarity in a populus DB BLAST search, closest arabidopsis homologue, and poplar gene expression data from Ruttink et al. (2007). Table S4: a summary of gene expression in various C. sativa tissues. Table S5: genes found in the C. sativa D library as well as in the arabidopsis seed dormancy transcriptome; genes in the C. sativa D library and in the arabidopsis transcriptome associated with non-dormancy; and genes found in the C. sativa ND library as well as in the arabidopsis transcriptome associated with non-dormancy. Fig. S1: relative water content in buds with different developmental stages which are described in Table 1; the bar in the top right corner depicts the maximum LSD. Fig. S2: translated amino acid alignments of CsGCN5L1, CsHUB2, CsGOLS, CsRADSAM, CsSAMS, CsAUR3, CsHSPTF90 and CsHSPTF with truncated orthologous sequences from arabidopsis and other species.
ACKNOWLEDGEMENTS
We are grateful to SERIDA (Servicio Regional de Investigación y Desarrollo Agroalimentario) for donating all C. sativa plant material, particularly Dr Isabel Feito. This work was supported by an FPU fellowship to M.E.S. and the MICINN projects AGL2007-62907 and AGL2010-22351-C03-01.
LITERATURE CITED
- Anderson JV, Horvath DP. Random sequencing of cDNAs and identification of mRNAs. Weed Science. 2001;49:590–597. [Google Scholar]
- Arora R, Rowland LJ, Tanino K. Induction and release of bud dormancy in woody perennials: a science comes of age. Hortscience. 2003;38:911–921. [Google Scholar]
- Benhamed M, Bertrand C, Servet C, Zhou DX. Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. The Plant Cell. 2006;18:2893–2903. doi: 10.1105/tpc.106.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergervoet JHW, Jing HC, Van den Hout JWE, et al. Expression of beta-tubulin during dormancy induction and release in apical and axillary buds of five woody species. Physiologia Plantarum. 1999;106:238–245. [Google Scholar]
- Böhlenius H, Huang T, Charbonnel-Campaa L, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE. Gene expression profile of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. The Plant Journal. 2006;46:805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- Cao Y, Dai Y, Cui S, Ma L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. The Plant Cell. 2008;20:2586–2602. doi: 10.1105/tpc.108.062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CE, Partis MD, Thomas B. The expression of histon 2A in onion (Allium cepa) during the onset of dormancy, storage and emergence from dormancy. New Phytologist. 1999;143:461–470. doi: 10.1046/j.1469-8137.1999.00484.x. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Gong Z, Zhu JK. Abscisic acid-mediated epigenetic processes in plant development and stress responses. Journal of Integrative Plant Biology. 2008;50:1187–1195. doi: 10.1111/j.1744-7909.2008.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuine I. A unified model for budburst of trees. Journal of Theoretical Biology. 2000;207:337–347. doi: 10.1006/jtbi.2000.2178. [DOI] [PubMed] [Google Scholar]
- Chuine I, Beaubien EG. Phenology is a major determinant of tree species range. Ecology Letters. 2001;4:500–510. [Google Scholar]
- Demidov D, Van Damme D, Geelen D, Blattner FR, Houben A. Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. The Plant Cell. 2005;17:836–848. doi: 10.1105/tpc.104.029710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derory J, Léger P, Garcia V, et al. Transcriptome analysis of bud burst in sessile oak (Quercus petraea) New Phytologist. 2006;170:723–738. doi: 10.1111/j.1469-8137.2006.01721.x. [DOI] [PubMed] [Google Scholar]
- Devitt ML, Stafstrom JP. Cell cycle regulation during growth-dormancy cycles in pea axillary buds. Plant Molecular Biology. 1995;29:255–265. doi: 10.1007/BF00043650. [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau Y-FC, Campbell AP, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Sciences of the USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Cadman CSC, Toorop PE, Lynn JR, Hilhorst HWM. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. The Plant Journal. 2007;51:60–78. doi: 10.1111/j.1365-313X.2007.03118.x. [DOI] [PubMed] [Google Scholar]
- Fleury D, Himanen K, Cnops G, et al. The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. The Plant Cell. 2007;19:417–432. doi: 10.1105/tpc.106.041319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman D, Riou-Khamlichi C, Oakenfull EA, Murray JAH. Isolation, characterization and expression of cyclin and cyclin-dependent kinase genes in Jerusalem artichoke (Helianthus tuberosus L.) Journal of Experimental Botany. 2003;54:303–308. doi: 10.1093/jxb/erg047. [DOI] [PubMed] [Google Scholar]
- Gutierrez C. The arabidopsis cell division cycle. In: Last R, Chang C, Jander G, et al., editors. The Arabidopsis book. Rockville, MD: American Society of Plant Biologists; 2009. pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack H, Bleiholder H, Buhr L, et al. Einheitliche Codierung der phänologischen. Entwicklungsstadien mono- und dikotyler Pflanzen – Erweiterte BBCH-Skala, Allgemein. Nachrichtenblatt des Deutschen Pflanzenschutzdienstes. 1992;44:265–270. [Google Scholar]
- Horvath DP, Chao WS, Anderson JV. Molecular analysis of signals controlling dormancy and growth in underground adventitious buds of leafy spurge. Plant Physiology. 2002;128:1439–1446. doi: 10.1104/pp.010885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath DP, Anderson JV, Chao WS, Foley ME. Knowing when to grow: signals regulating bud dormancy. Trends in Plant Science. 2003;8:534–540. doi: 10.1016/j.tplants.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Horvath DP, Soto-Suárez M, Chao WS, Jia Y, Anderson JV. Transcriptome analysis of paradormancy release in root buds of leafy spurge (Euphorbia esula) Weed Science. 2005;53:795–801. [Google Scholar]
- Horvath DP, Anderson JV, Soto-Suárez M, Chao WS. Transcriptome analysis of leafy spurge (Euphorbia esula) crown buds during shifts in well-defined phases of dormancy. Weed Science. 2006;54:821–827. [Google Scholar]
- Horvath DP, Chao WS, Suttle JC, Thimmapuram J, Anderson JV. Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.) BMC Genomics. 2008;9:536. doi: 10.1186/1471-2164-9-536. doi:10.1186/1471-2164-9-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath DP, Sung S, Kim D, Chao W, Anderson J. Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Molecular Biology. 2010;73:169–179. doi: 10.1007/s11103-009-9596-5. [DOI] [PubMed] [Google Scholar]
- Howe GT, Aitken SN, Neale DB, Jermstad KD, Wheeler NC, Chen THH. From genotype to phenotype: unraveling the complexities of cold adaptation in forest trees. Canadian Journal of Botany. 2003;81:1247–1266. [Google Scholar]
- Ibañez C, Ramos A, Acebo P, et al. Overall alteration of circadian clock gene expression in the chestnut cold response. PLoS ONE. 2008;3:e3567. doi: 10.1371/journal.pone.0003567. doi:10.1371/journal.pone.0003567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez C, Kozarewa I, Johansson M, Ögren E, Rohde A, Eriksson ME. Circadian clock components regulate entry and affect exit of seasonal dormancy as well as winter hardiness in Populus trees. Plant Physiology. 2010;153:1823–1833. doi: 10.1104/pp.110.158220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annual Review of Plant Biology. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Jiménez S, Reighard GL, Bielenberg DG. Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Molecular Biology. 2010;73:157–167. doi: 10.1007/s11103-010-9608-5. [DOI] [PubMed] [Google Scholar]
- Kramer K. Phenotypic plasticity of the phenology of seven European tree species in relation to climatic warming. Plant, Cell & Environment. 1995;18:93–104. [Google Scholar]
- Kramer K, Leinonen I, Loustau D. The importance of phenology for the evaluation of impact of climate change on growth of boreal, temperate and Mediterranean forests ecosystems: an overview. International Journal of Biometeorology. 2000;44:67–75. doi: 10.1007/s004840000066. [DOI] [PubMed] [Google Scholar]
- Lang GA, Early JD, Martin GC, Darnell RL. Endo-, para-, and ecodormancy: physiological terminology and classifications for dormancy research. Hortscience. 1987;22:371–377. [Google Scholar]
- Li Z-M, Zhang J-Z, Mei L, Deng X-X, Hu C-G, Yao J-L. PtSVP, an SVP homolog from trifoliate orange (Poncirus trifoliata L. Raf.), shows seasonal periodicity of meristem determination and affects flower development in transgenic Arabidopsis and tobacco plants. Plant Molecular Biology. 2010;74:129–142. doi: 10.1007/s11103-010-9660-1. [DOI] [PubMed] [Google Scholar]
- Liu Y, Koornneef M, Soppe WJJ. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. The Plant Cell. 2007;19:433–444. doi: 10.1105/tpc.106.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martin GC. Bud dormancy in deciduous fruit trees. In: Steward FC, editor. Plant physiology: a treatise. New York, NY: Academic Press; 1991. pp. 183–225. [Google Scholar]
- Mathiason K, He D, Grimplet J, et al. Transcript profiling in Vitis riparia during chilling requirement fulfillment reveals coordination of gene expression patterns with optimized bud break. Functional and Integrative Genomics. 2009;9:81–96. doi: 10.1007/s10142-008-0090-y. [DOI] [PubMed] [Google Scholar]
- Maurel C. Aquaporins and water permeability of plant membranes. Annual Review of Plant Physiology and Plant Molecular Biology. 1997;48:399–429. doi: 10.1146/annurev.arplant.48.1.399. [DOI] [PubMed] [Google Scholar]
- Mazzitelli L, Hancock RD, Haupt S, et al. Co-ordinated gene expression during phases of dormancy release in raspberry (Rubus idaeus L.) buds. Journal of Experimental Botany. 2007;58:1035–1045. doi: 10.1093/jxb/erl266. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Donner AJ, Knuesel MT, York AG, Espinosa JM, Taatjes DJ. Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. The EMBO Journal. 2008;27:1447–1457. doi: 10.1038/emboj.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ophir R, Pang X, Halaly T, et al. Gene-expression profiling of grape bud response to two alternative dormancy-release stimuli expose possible links between impaired mitochondrial activity, hypoxia, ethylene-ABA interplay and cell enlargement. Plant Molecular Biology. 2009;71:403–423. doi: 10.1007/s11103-009-9531-9. [DOI] [PubMed] [Google Scholar]
- Phillips JR, Hilbricht T, Salamini F, Bartels D. A novel abscisic acid- and dehydration-responsive gene family from the resurrection plant Craterostigma plantagineum encodes a plastid-targeted protein with DNA-binding activity. Planta. 2002;215:258–266. doi: 10.1007/s00425-002-0755-z. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Hallak-Herr E, Rozenberg M, et al. Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. The Plant Journal. 2002;31:319–330. doi: 10.1046/j.1365-313x.2002.01364.x. [DOI] [PubMed] [Google Scholar]
- Pukacka S, Wójkiewicz E. Carbohydrate metabolism in Norway maple and sycamore seeds in relation to desiccation tolerance. Journal of Plant Physiology. 2002;159:273–279. [Google Scholar]
- Ramos A, Pérez-Solís E, Ibáñez C, et al. Winter disruption of the circadian clock in chestnut. Proceedings of the National Academy of Sciences of the USA. 2005;102:7037–7042. doi: 10.1073/pnas.0408549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Rinne PLH, Welling A, Vahala J, et al. Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-β-glucanases to reopen signal conduits and release dormancy in Populus. The Plant Cell. 2011;23:130–146. doi: 10.1105/tpc.110.081307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Bhalerao RP. Plant dormancy in the perennial context. Trends in Plant Science. 2007;12:217–223. doi: 10.1016/j.tplants.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Rohde A, Prinsen E, De Rycke R, Engler G, Van Montagu M, Boerjan W. PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. The Plant Cell. 2002;14:1885–1901. doi: 10.1105/tpc.003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Ruttink T, Hostyn V, Sterck L, Van Driessche K, Boerjan W. Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar. Journal of Experimental Botany. 2007;58:4047–4060. doi: 10.1093/jxb/erm261. [DOI] [PubMed] [Google Scholar]
- Ruttink T, Arend M, Morreel K, et al. A molecular timetable for apical bud formation and dormancy induction in poplar. The Plant Cell. 2007;19:2370–2390. doi: 10.1105/tpc.107.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría ME, Hasbún R, Valera MJ, et al. Acetylated H4 histone and genomic DNA methylation patterns during bud set and bud burst in Castanea sativa. Journal of Plant Physiology. 2009;166:1360–1390. doi: 10.1016/j.jplph.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Santamaría ME, Toorop PE, Rodríguez R, Cañal MJ. Dormant and non-dormant Castanea sativa Mill. buds require different polyvinylpyrrolidone concentrations for optimal RNA isolation. Plant Science. 2010;178:55–60. [Google Scholar]
- Schoof H, Zaccaria P, Gundlach H, et al. MIPS Arabidopsis thaliana Database (MAtDB): an integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Research. 2002;30:91–93. doi: 10.1093/nar/30.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader J, Moyle R, Bhalerao R, et al. Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. The Plant Journal. 2004;40:173–187. doi: 10.1111/j.1365-313X.2004.02199.x. [DOI] [PubMed] [Google Scholar]
- Sharma KK, Kuhad RC. Laccase: enzyme revisited and function redefined. Indian Journal of Microbiology. 2008;48:309–316. doi: 10.1007/s12088-008-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Research. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky F, Bhalerao RR, Unneberg P, et al. A Populus EST resource for plant functional genomics. Proceedings of the National Academy of Sciences of the USA. 2004;101:13951–13956. doi: 10.1073/pnas.0401641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, et al. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. The Plant Journal. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- Tanino KK. Hormones and endodormancy induction in woody plants. Journal of Crop Improvement. 2004;10:157–199. [Google Scholar]
- Visser ME, Holleman LJM. Warmer springs disrupt the synchrony of oak and winter moth phenology. Proceedings of the Royal Society of London Series B – Biological Sciences. 2001;268:289–294. doi: 10.1098/rspb.2000.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Liu L, Berger SL. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes & Development. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehmeyer N, Vierling E. The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiology. 2000;122:1099–1108. doi: 10.1104/pp.122.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Zhang L, Zhou C, Yu C-W, Chaikam V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. Journal of Experimental Botany. 2008;59:225–234. doi: 10.1093/jxb/erm300. [DOI] [PubMed] [Google Scholar]
- Yamada K, Nishimura M. Cytosolic heat shock protein 90 regulates heat shock transcription factor in Arabidopsis thaliana. Plant Signaling & Behavior. 2008;3:660–662. doi: 10.4161/psb.3.9.5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annual Review of Plant Physiology. 1984;35:155–189. [Google Scholar]
- Zhang X, Friedl MA, Schaaf CB, Strahler AH. Climate controls on vegetation phenological patterns in northern mid- and high latitudes inferred from MODIS data. Global Change Biology. 2004;10:1133–1145. [Google Scholar]
- Zhao TY, Meeley RB, Downie B. Aberrant processing of a maize GALACTINOL SYNTHASE transcript is caused by heat stress. Plant Science. 2003;165:245–256. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.