Abstract
Over the last decades, cancer research has focused on tumor suppressor genes and oncogenes. Genes in other cellular pathways has received less attention. Between 0.5% to 1% of the mammalian genome encodes for proteins that are tethered on the cell membrane via a glycosylphosphatidylinositol (GPI)-anchor. The GPI modification pathway is complex and not completely understood. Prion (PrP), a GPI-anchored protein, is infamous for being the only normal protein that when misfolded can cause and transmit a deadly disease. Though widely expressed and highly conserved, little is known about the functions of PrP. Pancreatic cancer and melanoma cell lines express PrP. However, in these cell lines the PrP exists as a pro-PrP as defined by retaining its GPI anchor peptide signal sequence (GPI-PSS). Unexpectedly, the GPI-PSS of PrP has a filamin A (FLNA) binding motif and binds FLNA. FLNA is a cytolinker protein, and an integrator of cell mechanics and signaling. Binding of pro-PrP to FLNA disrupts the normal FLNA functions. Although normal pancreatic ductal cells lack PrP, about 40% of patients with pancreatic ductal cell adenocarcinoma express PrP in their cancers. These patients have significantly shorter survival time compared with patients whose cancers lack PrP. Pro-PrP is also detected in melanoma in situ but is undetectable in normal melanocyte, and invasive melanoma expresses more pro-PrP. In this review, we will discuss the underlying mechanisms by which binding of pro-PrP to FLNA disrupts normal cellular physiology and contributes to tumorigenesis, and the potential mechanisms that cause the accumulation of pro-PrP in cancer cells.
Keywords: prion protein, filamins, cancer
Filamins
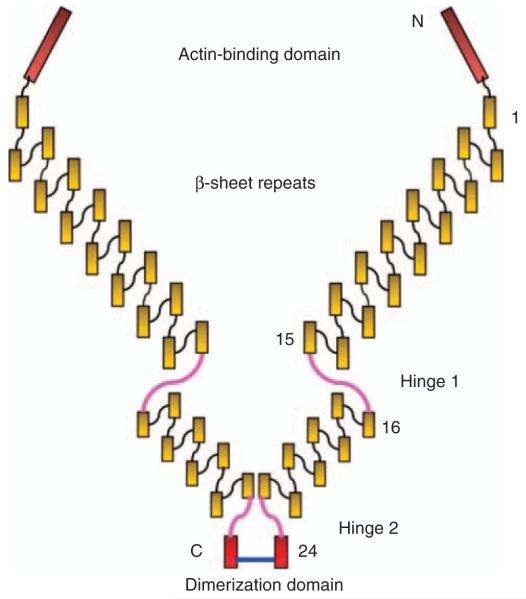
Filamins (FLNs) are cytolinkers that connect cell surface receptors to the cytoskeleton (Stossel et al., 2001; Feng and Walsh, 2004). Of the three FLNs, filamin A (FLNA) is the most abundantly and widely expressed, followed by filamin B. Expression of filamin C is restricted to myocytes in the heart, and in striated muscle. Native FLNs are homodimers with a relative molecular mass of about 280 kDa. At the N-terminus of each subunit, there is an actin-binding domain. FLNs bind and organize actin filaments creating an orthogonal actin network, which is important for maintaining cell morphology, membrane integrity, cell–cell and cell–matrix interactions. After the actin-binding domain, there are 24 β-sheets immunoglobulin-like domains. Between domains 15 and 16 and domains 23 and 24, there are two hinge regions, which permit high-angle branching of the actin filaments. Domain 24 is the self-association domain (Figure 1). The three FLNs share a high degree of homology, and have similar function in binding and organizing actin filaments. However, each FLN also has its unique binding partners. The functional diversity of the three FLNs is vividly illustrated in individuals with mutations in FLNA, FLNB or FLNC (Krakow et al., 2004; Robertson, 2005; Kley et al., 2007). Mutation in each gene has distinct phenotypes.
Figure 1.
Drawing of a dimeric FLNA: each monomeric FLNa contains an actin-binding domain (ABD) followed by 24 β-sheet immunoglobulin-linked domains, intersperse between these are two hinge regions, domain 24 is a self-association, dimerization domain.
FLNA
In this review we focus on FLNA. FLNA is located on the X chromosome. In male, FLNA deficiency due to a null mutation is embryonic lethal. In female, it causes ventricular heterotopia, a disease of abnormal neuronal migration (Fox et al., 1998; Lu and Sheen, 2005; Robertson, 2005). On the other hand, point mutations in the same gene cause a spectrum of diseases in multiple organ systems, such as skeletal and connective tissues (Robertson, 2005). In the genetically engineered mouse model, Flna deficiency (Flna−/−) is embryonic lethal. The embryo shows severe structural defects in the heart, and widespread vascular anomalies (Feng et al., 2006; Hart et al., 2006).
As a cytolinker protein, FLNA interacts with a plethora of proteins with diverse functions. These proteins include, cell surface receptors, cytoplasmic adapter proteins, signal transducing molecules and transcription factors (Stossel et al., 2001; Feng and Walsh, 2004; Robertson, 2005). Binding of FLNA regulates the transit, internalization or trafficking of many cell surface glycoproteins, such as glycoprotein Ibα (Williamson et al., 2002), dopamine D2 and D3 receptors (Lin et al., 2001; Li et al., 2002), furin (Liu et al., 1997), opioid receptor (Onoprishvili et al., 2003), calcitonin receptor (Seck et al., 2003), calcium-sensing receptor (Awata et al., 2001), cystic fibrosis transmembrane conductance regulator (Thelin et al., 2007) and caveolin-1 (Sverdlov et al., 2009).
Integrin is one of the best-characterized binding partners of FLNA (Sharma et al., 1995; Glogauer et al., 1998; Loo et al., 1998; Kiema et al., 2006). Integrins are bidirectional, allosteric cell surface molecules that are important in sensing and responding to the extracellular cues (Hynes, 2002; Ginsberg et al., 2005; Luo et al., 2007). Binding of FLNA to integrin β chain regulates cell spreading, migration and survival (Calderwood et al., 2001; Kim et al., 2008). FLNA modulates cell spreading and migration by competing with talin for integrin binding; an increase in FLNA binding inhibits cell migration (Calderwood et al., 2001). The function of FLNA is further fine-tuned by an auto-inhibitory domain on FLNA, which modulates the binding of FLNA to its ligands (Lad et al., 2007). FLNA also serves as a platform for the assembly of adapter and signaling molecules, such as Trio, Ral, Lnk, Ror 2, TRAF2 and SMADs. These molecules are important in many signal-transducing pathways. (Ohta et al., 1999; Bellanger et al., 2000; He et al., 2000; Leonardi et al., 2000; Sasaki et al., 2001; Nomachi et al., 2008).
FLNA function is also regulated by post-translational modifications, such as proteolytic cleavage and phosphorylation. FLNA fragments have been detected in the nucleus (Loy et al., 2003). In androgen-dependent prostate cancer cell lines, a small 90 kDa FLNA fragment is translocated into the nucleus where it binds the androgen receptor, and modulates its activity (Wang et al., 2007). Phosphorylation of FLNA by Ca2+/calmodulin-dependent protein kinase II causes cytoskeletal reorganization and endothelial barrier dysfunction (Borbiev et al., 2001). FLNA is also a substrate of PKCα (Tigges et al., 2003). Phosphorylation of FLNA by PKA renders FLNA more resistant to cleavage by calpain (Gorlin et al., 1990). Cyclin B1/Cdk-1 phosphorylates FLNA and regulates its ability to bind actin (Cukier et al., 2007). In response to EGF, FLNA is phosphorylated by ribosomal S6 kinase (Woo et al., 2004); phosphorylation is required for membrane ruffling (Vadlamudi et al., 2002). FLNA is also phosphorylated in a caveolin- and PI3 kinase-dependent manner to promote cell migration (Ravid et al., 2008). In lymphocytes, p56lck controls phosphorylation of FLNA and regulates focal adhesion kinase (Goldmann, 2002).
FLNA in cancer
Whether FLNA anomaly contributes to tumorigenesis has not been studied in detail. In prostate cancer, cancer metastasis correlates with cytoplasmic localization of FLNA (Bedolla et al., 2009). In melanoma cells, FLNA regulates the intracellular trafficking and degradation of the EGF receptor (Herlyn, 2006; Fiori et al., 2009). Immunohistochemical staining of melanoma biopsies show that in the dermis there are more FLNA positive tumor cells than in the epidermis (Bouffard et al., 1994). More recently, it is reported that FLNA is cleaved by Wnt5a-activated calpain 1, which then causes cytoskeleton remodeling, and enhances melanoma cell motility (O’Connell et al., 2009). FLNA is required for an efficient recombination DNA double strand break repair, suggesting that FLNA has a role in the maintenance of genomic stability (Yue et al., 2009). FLNA is upregulated in lung cancer cells undergoing epithelial-mesenchymal transition (Keshamouni et al., 2006). Squamous-cell carcinoma during cancer-stromal cell interaction also express a higher level of FLNA (Kamochi et al., 2008). These processes are important in tumor growth and metastasis.
Uhlen et al. (2005) compared the expression pattern of FLNA in different cancer tissues with their corresponding normal tissues by immunohistochemcial staining. Some noticeable differences were observed. For example, FLNA was undetectable in normal colon glandular cells, but ~50% of the colorectal cancer had moderate to high levels of FLNA. In normal pancreas, exocrine ductal cells had low to moderate levels of FLNA, the expression of FLNA was increased in pancreatic cancer.
A secreted variant of FLNA was detected in the plasma of patients with breast carcinoma and high-grade astrocytoma (Alper et al., 2009). FLNA was also overexpressed in peripheral cholangiocarcinomas (Guedj et al., 2009). In gene profiling studies, FLNA was overexpressed in human glioblastomas (Sun et al., 2006), salivary gland adenoid cystic carcinoma (Frierson et al., 2002) and in pancreatic cancer (Logsdon et al., 2003). On the other hand, FLNA was under expressed in human bladder cancer (Sanchez-Carbayo et al., 2006), gliomas (Bredel et al., 2005), colon adenocarcinoma (Notterman et al., 2001), lung carcinomas (Bhattacharjee et al., 2001) and renal carcinomas (Yusenko et al., 2009).
Despite its involvement in many cellular activities, FLNA is dispensable for cell-autonomous survival. Many cell types in Flna−/− mouse are able to migrate and function properly (Feng et al., 2006; Hart et al., 2006). Some human melanoma cell lines, such as M2 and M3 do not express FLNA. These cells lack actin fiber bundles and are less mobile in vitro (Byers et al., 1991). This phenotype is reversed in the A7 cells, which are derived from M2 cells after the transfection of an expression plasmid that encodes human FLNA (Cunningham et al., 1992). Since then this pair of isogenic cell lines has been used extensively to study the functionality of FLNA. Biological responses, such as cell spreading and migration, signal transduction and apoptosis observed in A7 cells, but not in M2 cells, have been attributed solely to FLNA function (Liu et al., 1997; Leonardi et al., 2000; Awata et al., 2001; Feng et al., 2003; He et al., 2003; Thelin et al., 2007; Zhu et al., 2007).
It is unlikely that anomaly in FLNA can initiate tumorigenesis. Patients with FLNA mutations do not have a higher incidence of cancers. On the other hand, disrupted FLNA function may contribute to the biology of cancers; modulating the functionality of growth factor receptors or signal transducing molecules, and provides the tumor cells with a growth advantage. Anomaly in FLNA may also modulate the functionality of adhesion molecules, which then facilitate the spreading and migration of cancer cells, giving rise to more aggressive cancers.
From scrapie, Creutzfeldt-Jakob disease, and kuru to prion
Scrapie is a form of transmissible spongiform encephalopathy (TSE) in sheep and goats, and is endemic in United Kingdom ever since the 1750s (Greig, 1950). First reported in 1920, Creutzfeldt-Jakob disease is a subacute spongiform encephalopathy in human (Creutzfeldt, 1920). Over the years and because of its rarity, Creutzfeldt-Jakob disease received little attention until the 1957 when Gajdusek and Zigas (1957) reported a new disease, Kuru. Kuru means to tremble in the Fore language of the East Highlanders of Papua New Guinea. Thus, the word Kuru describes vividly the clinical symptoms of the disease. A major advance in the understanding of Kuru was the serendipitous discovery that the spongiform histopathology, as seen in the brain of Kuru affected patients, was very similar to those found in scrapie. Subsequently, Gajdusek (2008) demonstrated that Creutzfeldt-Jakob disease and kuru are TSE in humans. It was thought that Kuru is transmitted because of the practice of cannibalism.
For decades, the etiology of the TSE remained elusive until 1982, when Prusiner (1982) isolated and characterized the infectious pathogen. They named the pathogen proteinaceous infectious particle or scrapie prion (PrPSc; Bolton et al., 1982; Prusiner, 1982). Subsequently, it was found that PrPSc was an aberrant, misfolded isoform of a highly conserved, and widely expressed normal cellular protein (Basler et al., 1986). Based on this finding, Prusiner proposed that the central event in the pathogenesis of prion disease is the conversion of a normal cellular prion protein, PrP, into an abnormal, pathogenic conformer, PrPSc (Prusiner, 1998). The accumulation of PrPSc then causes pathology in the brain. A familiar form of human TSE was later found to be caused by a mutation in the prion gene, PRNP (Hsiao and Prusiner, 1990; Goldfarb et al., 1994). Since then the term prion disease has been used synonymously with TSE. The next advance in prion research came from findings, which showed that mice, genetically engineered to lack the normal PrP, Prnp−/−, were resistant to PrPSc infection (Bueler et al., 1993), adding important support for the concept of prion pathogenesis.
PrP
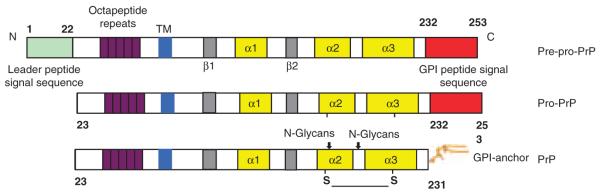
PrP is a relatively small glycoprotein and is tethered to the outer membrane leaflet using a glycosylphosphatidylinositol (GPI) anchor. The synthesis, processing and transit of PrP to the cell surface are multifaceted, cell-context dependent, and not completely understood (Hegde and Rane, 2003; Campana et al., 2005). PrP is synthesized as a pre-pro-PrP polypeptide of 253 amino acids (Figure 2). Residues 1–22 at the N-terminus are the leader peptide sequence. The last 22 amino acids at the C-terminus, from residues 232 to 253 are the GPI anchor peptide signal sequence (GPI-PSS). Both sequences are removed in the endoplasmic reticulum, and thus are absent from the mature PrP. Nuclear magnetic resonance studies reveal that the N-terminal half of the PrP is flexible and lacks noticeable secondary structure. On the other hand, the C-terminal region contains a well-defined globular domain with two β-sheets and three short α-helixes (Donne et al., 1997). This region also has two potential N-linked glycosylation sites and a disulfide bridge.
Figure 2.
Diagrammatic drawings of PrP and its processing from pre-pro-PrP to pro-PrP and to a mature, N-glycosylated and GPI-anchored PrP. Residues 1–22 contain the leader peptide sequence. Residues 232–253 contain the GPI-peptide signal sequence.
At the N-terminal end of PrP there is a highly conserved glycosaminoglycan binding motif, KKRPK (Caughey et al., 1994; Pan et al., 2002). Glycosaminoglycan participates in cell migration and adhesion, and serves as a coreceptor for growth factors. At the N-terminus there is also a highly conserved octapeptide repeat region, which binds divalent cations, such as Cu2+ and Zn2+ (Millhauser, 2007; Davies and Brown, 2008). PrP also has a large number of other binding partners. Some of these molecules are: laminin receptor (Simoneau et al., 2003), selectins (Li et al., 2007), glypican-1 (Mani et al., 2003), caveolin-1 (Mouillet-Richard et al., 2000), N-CAM (Schmitt-Ulms et al., 2001), dystroglycan (Keshet et al., 2000), heat shock proteins (Edenhofer et al., 1996), stress-inducible protein (Zanata et al., 2002) and Grb2 (Spielhaupter and Schatzl, 2001). PrP also binds to lipids (Mahfoud et al., 2002) and nucleic acids (Gabus et al., 2001). It is difficult to conceive how a relatively small protein can interacts with so many partners, and mediates diverse cellular processes that take place in different cellular compartments.
To identify proteins that are physically close to PrP in a more physiological setting, Schmitt-Ulms et al. (2004) carry out an in vivo crosslinking experiment followed by next-neighbor chemical analysis. It is found that in a normal mouse brain PrP is located in a submicrodomain on the cell membrane. Many of the PrP neighboring proteins are also GPI-anchored proteins, such as contactin-1 and LSAMP. Other neighboring proteins have immunoglobulin or fibronectin type III-like motifs. These proteins include N-CAM-2, MOG, L1cam and PGRL. As these proteins are adhesion molecules, it is postulated that PrP may participate in regulating cell–cell interaction in vivo. Consistent with this view is the recent finding that PrP is important in organizing the epithelial cell junctions in the small intestine (Morel et al., 2008).
On the cell surface PrP resides in lipid rafts and participates in signal transduction (Mouillet-Richard et al., 2000; Taylor and Hooper, 2006). PrP has a role in apoptosis in a cell context, as well as pathway-dependent manner. In some cell types, PrP functions as a pro-apoptotic mediator (Paitel et al., 2004). In other cell types, PrP functions as an anti-apoptotic mediator (Kuwahara et al., 1999; Bounhar et al., 2001; Chiarini et al., 2002). A recent study suggests that whether PrP is neuro-protective depends on the underlying pathogenesis (Steele et al., 2009). PrP has a pro-apoptotic role during endoplasmic reticulum stress, and an anti-apoptotic role during oxidative stress-induced cell death (Anantharam et al., 2008). PrP is also detected in the cytoplasm, nucleus, and is present in blood as well as other body fluids, such as milk (Franscini et al., 2006) and urine (Narang et al., 2005). Despite all the functions that are attributed to PrP, Prnp−/− mouse is apparently normal without overt aberrant phenotype (Bueler et al., 1993).
The GPI anchor
In mammalian cells, there are more than 100 GPI-anchored proteins with diverse functions (Ikezawa, 2002; Sharom and Lehto, 2002). It is not known why some proteins are GPI anchored whereas others are not; some proteins can exist either as a GPI anchored protein or a transmembrane protein. The biosynthesis of the GPI anchors and their attachment to proteins are intricate, protein-context and cell-context dependent (Maeda et al., 2006; Kinoshita et al., 2008). At least 25 genes are important in this pathway. The common core structure of the GPI anchor is synthesized in a stepped mechanism in the ER. The first step is the transfer of N-acetyl-glucosamine (GlcNAc) from UDP-GlcNAc to phosphatidylinositol to yield GlcNAc-phosphatidylinositol. This reaction is catalyzed by α1-6 GlcNAc transferase complex. The second step is the de-N-acetylation and generation of GlcN-phosphatidylinositol. Three mannose residues are then added sequentially. The last step in the GPI anchor modification pathway is the attachment of the already assembled GPI structure en bloc to the newly synthesized pro-protein in a transamidase reaction. This reaction is mediated by a protein oligomer comprising of five highly conserved proteins. The site of the proteolytic cleavage is referred to the ω site. The ω residue for mammalian proteins is confined to the amino acids glycine, serine, cysteine, alaine, aspartic acid and asparagine (Maeda et al., 2006; Kinoshita et al., 2008). Other than the ω residue, there is no other obvious motif in the GPI-PSS that signals the transamidase reaction; any hydrophobic sequences can function as a substrate in the transamidation reaction.
In general, the GPI-PSS contains 15–25 small hydrophobic amino acids, similar to a typical transmembrane domain. It is interesting that substitution of a single amino acid at the ω site of Qa2, a normally GPI-anchored protein, prevents its GPI anchor modification (Waneck et al., 1988). Nonetheless, Qa2 is still present on the cell surface as an integral membrane protein using the GPI-PSS as a surrogate transmembrane domain.
Does the GPI-PSS simply function as an inert substrate for the transamidase reaction, so that the protein is GPI-anchored? Stanners and colleagues do not think so. They propose that the GPI-PSS contains cryptic biological information that specifies the addition of a particular functional GPI-anchor, which ultimately regulates the functionality of the mature protein. This hypothesis is based on the finding that exchanging the GPI-PSS of N-CAM for the GPI-PSS of carcinoembryonic antigen (CEA) generates a mature protein with a N-CAM external domain, but with CEA-like biological properties (Screaton et al., 2000; Naghibalhossaini et al., 2007; Nicholson and Stanners, 2007). Different GPI-PSSs also affected the oligomerization of GPI-anchored proteins and their placement on either the basal side or epical side of the membrane (Paladino et al., 2008). Interestingly, although the coding region of the human PRNP and other mammalian Prnp is about 85% conserved, their GPI-PSS is almost 100% conserved (Table 1). On the other hand, their N-terminal peptide sequence, which is also discarded before maturation, is much less conserved. The significance of this conservation is not known.
Table 1.
The GPI-PSS of PrP is highly conserved
| Human | L | F | S | S | P | P | V | I | L | L | I | S | F | L | I | F | L | I | V | G |
| Goat | L | F | S | S | P | P | V | I | L | L | I | S | F | L | I | F | L | I | V | G |
| Sheep | L | F | S | S | P | P | V | I | L | L | I | S | F | L | I | F | L | I | V | G |
| C. Hamster | L | F | S | S | P | P | V | I | L | L | I | S | F | L | I | F | L | I | V | G |
| S. Hamster | L | F | S | S | P | P | V | I | L | L | I | S | F | L | I | F | L | M | V | G |
| Mouse | L | F | S | S | P | P | V | I | L | L | I | S | F | L | I | F | L | I | V | G |
| Rat | L | F | S | S | P | P | V | I | L | L | I | S | F | L | I | F | L | I | V | G |
| Rabbit | L | F | S | S | P | P | V | I | L | L | I | S | F | L | I | F | L | I | V | G |
| Cattle | L | F | S | S | P | P | V | I | L | L | I | S | F | L | I | F | L | I | V | G |
As GPI-anchored proteins are involved in different cellular activities, any malfunction in the GPI-anchored modification pathway is likely to contribute to tumorigenesis. Some components of the transamidase complex, such as PIG-T, PIG-U, PIG-S and GPAA1 are upregulated in bladder, breast, head and neck cancers (Jiang et al., 2007; Nagpal et al., 2008). Increased GPAA1 expression is also associated with hepatocarcinoma’s poor cellular differentiation and poor prognosis (Ho et al., 2006). However, the underlying mechanisms by which aberrant expressions of these genes contribute to tumorigenesis are not known. Worthy of note is the fact that many of the genes essential for GPI anchor modification are located in chromosomal regions, which are reported to be linked to, amplified, deleted or mutated in different human cancers (Table 2). Amplification or deletion in these regions may have impacted the integrity of the genes essential for the GPI-modification pathway. Aberrant expression of any one of these genes potentially can alter the physiology of many of the GPI-anchored proteins, and thus can have global impact on cellular physiology.
Table 2.
Chromosomal regions, which contain genes of the GPI-anchor pathway that have been reported to be linked to, amplified, deleted or mutated in human cancers
| Location | Genes | Functions | Linked to, amplified or deleted in cancers |
|---|---|---|---|
| 1p31.1 | PIG-K | A member of the cysteine protease family that catalyzes the transfer of a fully assembled GPI to protein |
Colon cancer (Daley et al., 2008) Pancreatic cancer (Kimmelman et al., 2008) Breast cancer (Hoggard et al., 1995; White et al., 1998) Mesothelioma (Lindholm et al., 2007) |
| 1p36.11 | PIG-V | Adds the second mannose to the GPI core | Meningioma (Guan et al., 2008) Sporadic chordoma (Miozzo et al., 2000) |
| 1q22 | DPM-3 | Stabilizes the dolichol-phosphatemannosesynthase complex, DPM-1, 2 and 3 |
Hepatocellular carcinoma (Wong et al., 2003; Skawran et al., 2008) Cervical Cancer (Narayan et al., 2007) Retinoblastoma (Zielinski et al., 2005) Lung adenocarcinoma (Goeze et al., 2002) |
| 1q23.2 | PIG-M | Transfers the first mannose to the GPI on the lumenal side of ER |
Sarcoma (Kresse et al., 2005) Neuroblastoma (Hirai et al., 1999) |
| 2p26-p21 | PIG-F | Transfers ethanolamine-phosphate to the third mannose in the GPI |
Ovarian neuroendocrine carcinoma (Mhawech-Fauceglia et al., 2008) Breast cancer (Argos et al., 2008) Colon cancer (Bisgaard et al., 2001) |
| 3q29 | PIG-X | A GPI mannosyl-transferase subunit that synthesize the core GPI structure in the ER |
Prostate cancer (Kim et al., 2007a) Head and neck squamous cell carcinoma (Lin et al., 2006) Adrenocortical carcinoma (Dohna et al., 2000) |
| 4p16.3 | PIG-G | Transfers ethanolamine phosphate to the GPI second mannose |
Prostate Cancer (Castro et al., 2009) Bladder cancer (Sibley et al., 2000) Breast cancer (Shivapurkar et al., 1999) |
| 8q24.3 | GPAA-1 | GPI anchor attachment protein 1 functions in GPI anchoring at the GPI transfer step |
Esophageal cancers (van Duin et al., 2007) Prostate cancer (Kim et al., 2007a) Colon cancer (Douglas et al., 2004; Tenesa et al., 2008) Cervical cancer (Narayan et al., 2007) Hepatocellular carcinoma (Ho et al., 2006) Breast cancer (Wu et al., 2006) |
| 9p13.3 | PIG-O | Transfer ethanolamine-phosphate to the third mannose in the GPI |
Olfactory neuroblastoma (Guled et al., 2008) Pancreatic cancer (Aguirre et al., 2004) |
| 9q34.13 | DPM-2 | Essential for the ER localization and stable expression of DPM1 |
Pancreatic cancer (Loukopoulos et al., 2007; Amundadottir et al., 2009) |
| 14q11–24 | PIG-H | Transfers GlcNAc to PI on the cytoplasmic side of the ER | Lung Cancer (Abujiang et al., 1998) Pancreatic cancer (Aguirre et al., 2004; Bashyam et al., 2005) |
| 15q21–22 | PIG-B | A subunit of the dolichol-P-mannose dependent mannosyltransferases |
Pancreatic cancer (Jonson et al., 1999) |
| 16p13.3 | PIG-Q | A subunit of the complex that catalyzes the transfer of GlcNAc from UDP-GlcNAc to PI |
Cervical carcinoma (Choi et al., 2007) Breast cancer (Naylor et al., 2005) Hepatocellular carcinoma (Wang et al., 2001; Katoh et al., 2005) Pancreatic cancer (Nowak et al., 2005) |
| 17p12 | PIG-L | An enzyme in the ER that catalyzes the second step of GPI biosynthesis, which is the de-N-acetylation of N-acetylglucosaminyl-phosphatidylinositol (GlcNAc-PI) |
Ovarian cancer (Kim et al., 2007b) Prostate cancer (Stanford et al., 2009) Lung adenocarcinoma (Goeze et al., 2002; Nakanishi et al., 2009) Osteosarcoma (Hulsebos et al., 1997; van Dartel et al., 2002; Selvarajah et al., 2008) Melanoma (Peris et al., 1995) Head and neck cancer (Adamson et al., 1994; Li et al., 1994) Breast cancer (Devilee et al., 1990) |
| 17p13.1- p12 |
MPDU1 | Required for the synthesis of lipid-linked sugars and GPI | Colon cancer (Daley et al., 2008) Gastric cancer (Yu et al., 2008) Pancreatic cancer (Aguirre et al., 2004) |
| 17p32.2 | PIG-S | A unit of transamidase transfers assembled GPI to proteins |
Breast, ovary, lung liver and uterus cancers (Nagpal et al., 2008) |
| 17q12 | PIG-W | An enzyme that acts in the third step of the GPI biosynthesis and acylates the inositol ring of PI |
Ovarian cancer (Dimova et al., 2009) Prostate cancer (Sun et al., 2008; Helfand et al., 2009) Gastric cancer (Maqani et al., 2006; Myllykangas et al., 2008) Breast cancer (Letessier et al., 2006; Thomassen et al., 2009) Lung cancer (Ma et al., 2006) Pancreatic cancer (Aguirre et al., 2004; Bashyam et al., 2005) |
| 18q21.33 | PIG-N | Transfer phospho-ethanoamine to the first mannose of the GPI |
Osteosarcoma (Johnson-Pais et al., 2003) Gallbladder and bile duct cancer (Saito et al., 2006) Colon cancer (Tenesa et al., 2008) |
| 20q11.22 | PIG-U | A subunit of the transamidase that attaches GPI anchor to proteins |
Melanoma (Brown et al., 2008) Thyroid cancer (Ishihara et al., 2008) Bladder cancer (Guo et al., 2004) Breast cancer (Wu et al., 2006) |
| 20q12–13 | PIG-T | A subunit of the GPI transamidase | Breast cancer (Guan et al., 1994; Larramendy et al., 2000; Vanden Bempt et al., 2006; Wu et al., 2006) Esophageal carcinoma (Fujita et al., 2003) Gastric cancer (Hidaka et al., 2003) |
| 20q13.13 | DPM-1 | The catalytic unit of the DPM1p, 2p, 3p complex | Cervical cancer (Scotto et al., 2008) Breast cancer (Mastracci et al., 2006) |
| 21q22.2 | PIG-P | Transfers GlcNAc from UDP-GlcNAc to PI | Prostate cancer (Torring et al., 2007; Ishkanian et al., 2009) |
Abbreviations: GlcNAc, N-acetyl-glucosamine; GPI, glycosylphosphatidylinositol; PI, phosphatidylinositol.
Pancreatic cancer and prion
Because PrP is associated with cellular survival, proliferation and differentiation, aberrant PrP function may also contribute to tumorigenesis. PrP is overexpressed in human gastric cancers and contributes to resistance to chemotherapeutic agents (Liang et al., 2009). Expression microarray studies find that PRNP is overexpressed in human colorectal cancers (Antonacopoulou et al., 2008). PRNP is 1 of the 25 genes that is overexpressed in a panel of human pancreatic ductal adenocarcinoma (PDAC) cell lines (Han et al., 2002). However, the mechanism by which PrP overexpression promotes tumorigenesis is not clear. We have been studying the normal biology of PrP (Zanusso et al., 1998; Li et al., 2000). Based on an earlier report, which showed that PRNP is overexpressed in PDAC cell lines (Han et al., 2002), we investigated whether PrP was expressed in PDAC cell lines.
PDAC is the fourth leading cause of cancer deaths in the US and is one of the cancers with the poorest prognosis (Jemal et al., 2007). The overall median survival for all PDAC is 6 months and the 5-year survival rate is <5%. This bleak outcome reflects the aggressiveness and highly metastatic nature of the tumor, the lack of an early diagnostic marker, as well as the inefficacy of the treatment regimens. Pervasive genomewide aberrations impede the identification of the culprit genes in the etiology of human PDAC. Nonetheless, over the last two decades, there is significant progress in elucidating the molecular mechanisms underlying PDAC carcinogenesis. Many oncogenes, tumor suppressor genes and DNA mismatch repair genes are implicated in the etiology of PDAC (Li et al., 2004; Hezel et al., 2006; Hruban and Zamboni, 2009). The most common genetic lesions in human PDAC are mutations in KRAS, TP53, DPC4 (SMAD4) and CDNK2A (p16ink4a). It is now generally accepted that the KRAS mutation is one of the earliest and critical genetic lesion in the development of PDAC; about 90% of the PDAC cases have mutation in KRAS. In all, ~90% of the PDAC cases also have inactivation of CDNK2A. On the other hand, about half of the PDAC cases have mutations in TP53 or DPC4 (Hezel et al., 2006; Shi et al., 2008). The contribution of these genes to human PDAC development is supported by transgenic animal studies, which show that involvement by more than one of these genes is essential for the initiation and progression of PDAC (Hezel et al., 2006; Hruban et al., 2006; Wescott and Rustgi, 2008; Ottenhof et al., 2009). In addition, other growth factor genes, signal transducing molecules, transcription factors, as well as cell adhesion molecules are also implicated in the progression of human PDAC.
Garcea et al. (2005) examined evidence from published reports focused on molecular markers in PDAC, their correlation with tumor stage and grade, response to chemotherapy and long-term survival. The investigated markers included p53, p21, p16, p27, SMAD4, K-ras, cyclin D1, Bax, Bcl-2, epidermal growth factor receptor, epidermal growth factor, c-erbB2, HB-EGF, transforming growth factor-β, FGF, MMP, uPA, cathepsin, heparanase, E-cadherin, laminins, integrins, TMSF, CD44, cytokines, angiogenesis, vascular endothelial growth factor, interleukin-8 and β-catenin. These markers were previously reported as important in the pathogenesis of human PDAC. These authors concluded that for the most part, the evidence regarding the application of this panel of markers as prognostic indicators in PDAC was conflicting. Therefore, we need to continue the search for individual marker(s) or a group of markers that distinguish aggressive from less aggressive PDAC.
Fatal attraction between pro-PrP and FLNA
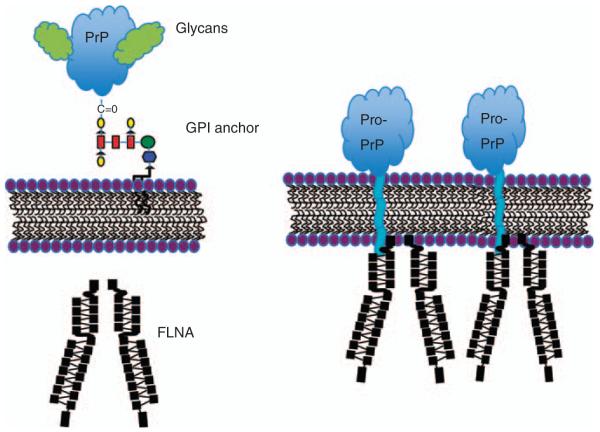
All PDAC cell lines (n = 7) studied express varying levels of PrP (Li et al., 2009). Further biochemical studies reveal that in contrast to PrP in normal cells, the PrP in the PDAC cell lines is unglycosylated, and exists as pro-PrP, retaining its normally cleaved GPI-PSS (Figure 2). This conclusion is based on the following findings: (1) PrP in the PDAC cell lines has a relative molecular mass of 26 kDa, a normal glycosylated, and GPI-anchored PrP has a relative molecular mass of 33–39 kDa; (2) treatment of PrP from PDAC cell lines with N-link glycosidase does not change its mobility in SDS–polyacrylamide gel electrophoresis, indicating that it does not contain N-linked glycan; (3) PrP from the PDAC cell lines is resistant to phopsholipase C, which cleaves the GPI anchor, indicating that the PrP from PDAC cell lines lacks the GPI-anchor; (4) the GPI-anchor protects a GPI-anchored protein from carboxypeptidase digestion, but PrP from the PDAC cell lines is susceptible to carboxypeptidase, indicating that the C-terminal of PrP lacks the GPI-anchor; (5) GPI-anchored PrP is normally find in lipid rafts, a micro-domain on the cell surface. However, PrP in the PDAC cell lines is not longer present in lipid raft; (6) a polyclonal antiserum that is specific for the GPI-PSS of PrP reacts with pro-PrP in the PDAC cell lines. Additional studies reveal that pro-PrP is readily detected on the cell surface of the PDAC cell line, using the GPI-PSS as a surrogate transmembrane domain (Figure 3).
Figure 3.
(a) Normal glycosylated and GPI anchored PrP on the cell surface; (b) pro-PrP on the cell surface using the GPI-PSS as a surrogate transmembrane domain and binds to FLNA just underneath the inner membrane leaflet. The sizes of the PrP and FLNA are not proportional. The size of PrP is approximately corresponding to two FLNA immunoglobulin domains.
Unexpectedly, we found that the GPI-PSS of pro-PrP has a FLNA binding motif. It is interesting to note that despite the fact that the GPI-PSS is discarded after the attachment of the GPI anchor, the FLNA binding motif on pro-PrP is highly conserved among mammalian PrPs (Table 1). Each immunoglobulin-like domain of FLNA has seven β-sheet strands (A–G). Most of the FLNA binding partners interact with the C and D strands of FLNA (Kiema et al., 2006; Nakamura et al., 2006, 2007). The atomic structure of the interface between the FLNA immunoglobulin-like domain and some of the common FLNA binding partners have been resolved (Kiema et al., 2006; Nakamura et al., 2006, 2007). These FLNA binding partners share a conserved hydrophobic amino-acid motif, consisting of multiple hydrophobic, nonpolar amino acids contacting residues.
First, binding of FLNA to pro-PrP is demonstrated by in vitro pull-down assays using recombinant FLNA and recombinant pro-PrP. FLNA only binds recombinant pro-PrP, but not mature PrP that lacks the GPI-PSS. Second, by co-immunoprecipitation, PrP copurifies with FLNA and vice versa in the PDAC cell lysates. Third, by immuno-fluorescent staining and confocal microscopic analysis, PrP and FLNA colocalize in the PDAC cell lines. Fourth, the co-immunoprecipitation of PrP and FLNA in PDAC cell lysate is competitively inhibited with a synthetic peptide corresponding to the GPI-PSS of PrP.
More recent studies using recombinant pro-PrP and FLNA individual domains reveal that pro-PrP binds to multiple domains, such as 10, 16, 17, 18, 20, 21 or 23, but not domains, 1–8, 11, 19, 22 or 24 of FLNA (Li et al., 2010). However, whether in vivo one FLNA dimer can simultaneously bind more than one pro-PrP is not known.
The GPI-PSS of PrP is comprised of 22 amino acids. Site-specific mutagenesis studies further reveal that the last five residues of the GPI-PSS are essential in binding FLNA. Therefore, the GPI-PSS is long enough to traverse the cell membrane. Furthermore, replacing residues 246 (Phe) and 250 (Leu) of the GPI-PSS to polar amino acids, such as tryptophan or tyrosine, completely eliminate the FLNA binding activity (Li et al., 2010). These results are consistent with earlier findings, which showed that the FLNA binding pocket could accommodate seven to nine amino acids and that nonpolar amino acids are important in the binding (Kiema et al., 2006; Nakamura et al., 2006, 2007).
The FLNA binding motif is present only on the GPI-PSS of PrP because it is absent in 14 other GPI-anchored proteins. Pro-PrP is undetectable in cells with normal GPI-anchored PrP, indicating the transit from pro-PrP to PrP is either very rapid or the removal of the pro-PrP is very efficient in these cells. Worthy of note is that it is reported that carbonic anhydrase, a normally GPI-anchored protein also exists as a pro-protein in a PDAC cell line (Fanjul et al., 2004, 2007). As the FLNA binding motif is absent on the GPI-PSS of carbonic anhydrase, therefore, even if carbonic anhydrase exists as pro-protein it will not be able to bind FLNA. On the other hand, there are more than one hundred GPI-anchored proteins, we cannot rule out the possibility that some other pro-proteins may be able to bind FLNA. A few GPI-anchored proteins, such as glypican-1 and CD24, have been reported to be important in tumorigenesis (Kayed et al., 2006; Fang et al., 2010). However, in these cases, it is not clear whether it is the protein portion of the molecule, the GPI anchor, or both that are critical. It is almost certain that GPI anchored proteins can contribute to tumor biology by numerous ways.
The reason the PrP GPI-PSS is not removed in the PDAC cell line is not because of a global defect in the GPI anchor modification machinery in the PDAC cell lines. Another GPI-anchored protein, CD55, is GPI-anchored in the PDAC cell lines. It is also not because of a mutation in the PRNP. We sequenced six of the seven PDAC cell lines and did not detect any mutation in the coding region of PRNP. Worthy of note is that the GPI-PSS of PrP is intrinsically inefficient in accepting the lipid anchor (Chen et al., 2001). In an in vitro GPI anchor modification assay comparing the efficiency of nine different GPI-PSSs, the GPI-PSS of PrP is by far the least efficient in accepting the GPI anchor—14% for pro-PrP versus 50% for pro-CD50 (Chen et al., 2001). Hence, a slight defect in the GPI assembly pathway will have a more dramatic effect on PrP than other GPI-anchored protein, such as CD55. As has been discussed earlier some of the genes that are essential in the GPI anchor modification pathways may contribute to human cancers, including pancreatic cancer (Table 1). Any defect in these genes will cause the accumulation of pro-PrP. In addition, slight defects in the ER quality control system or in the proteasome degradation pathway may also contribute to the accumulation of pro-PrP in these cells. On the other hand, as not all GPI anchored proteins are affected in the PDAC cell lines, these defects have to be somewhat restricted.
FLNA is an integrator of mechanical and chemical signaling events. Binding of pro-PrP to FLNA is likely to have widespread effects on the PDAC cells. Binding to pro-PrP may physically relocate FLNA from its normal environment, rendering it unable to interact with its regular binding partners. Alternatively, binding of pro-PrP may also compete for binding sites on FLNA that are normally reserved for other molecules. To study the effects of pro-PrP expression, we used small hairpin RNA to downregulate the expression of PrP in the PDAC cells. Although downregulation of PrP in the PDAC cell lines does not alter the expression levels of FLNA, it does alter the spatial distribution of FLNA in these cells. In control cells, FLNA is present just underneath the inner membrane leaflets. In PrP downregulated cells, it appears that FLNA is disconnected from the inner membrane leaflets and is more concentrated in the cytosol (Figure 3). This interpretation is further supported by biochemical approaches. In PrP downregulated cells, much less FLNA is copurified with membrane bound pro-PrP.
Downregulation of PrP in the PDAC cell lines also drastically alters the organization of actin filaments. Accordingly, the levels of proteins that are important in regulating actin polymerization and depolymerization, such as p-cofilin, LIMK-1 and LIMK-2 are also affected. These biochemical changes result in behavior changes of the PDAC cells. Compared with control cells, PrP downregulated PDAC cell lines have reduced in vitro proliferation and invasiveness. When the level of PrP is reduced, the growth of the PDAC cell lines as xenograft in nude mice is also greatly reduced. Collectively, these results suggest that the expression of pro-PrP modulates the functions of FLNA, and provides the PDAC cells with a growth advantage.
Interaction between pro-PrP and FLNA in melanoma cell line A7
As discussed earlier human melanoma cell lines, such as M2 and M3 cells do not express FLNA. These cells lack actin fiber bundles and are less mobile in vitro when compared with cells with FLNA. The deficiency is restored in A7 cell, which was derived from M2 cell by transfection of a plasmid encoding human FLNA. We posit if this pair of tumor cell lines expresses pro-PrP, it will provide an excellent model system to further study the interacting between pro-PrP and FLNA.
We find that both M2 and A7 cells express comparable levels of PrP. Furthermore, as in the PDAC cell lines, the PrP exists as pro-PrP (Li et al., 2010). In A7 cells, pro-PrP copurifies and colocalizes with FLNA. Binding of cell surface pro-PrP to FLNA in A7 cells provides stability to cell surface pro-PrP. Pro-PrP on the cell surface of A7 cells has a much longer half-life than PrP on the cell surface of M2 cells, which lack FLNA.
As in PDAC cells, reducing PrP expression in A7 cells also does not change the expression level of FLNA, but the spatial distribution of FLNA is noticeably altered. In control cells, FLNA concentrates at the leading edges. In PrP downregulated A7 cells, FLNA is retracted from the inner membrane leaflet. In control A7 cells, the actin filaments are well organized. In PrP downregulated A7 cells, the actin filaments are dis-organized. Accordingly, the levels of p-cofilin and LIMK1 are reduced in PrP downregulated A7 cells, but not in similarly downregulated M2 cells. These results suggest that the effect of reduced p-cofilin in PrP downregulated cells depends on the binding of pro-PrP to FLNA. If the effects were simply due to a reduction in PrP, we would have observed reduced p-cofilin level in PrP downregulated M2 cells.
A7 cells are used to demonstrate the role FLNA has in cell spreading and migration (Cunningham et al., 1992). We find that reducing PrP expression greatly diminishes the spreading and migration of A7 cells. Therefore, the enhanced cell spreading and migration observed in A7 cells are because of the binding of pro-PrP to FLNA.
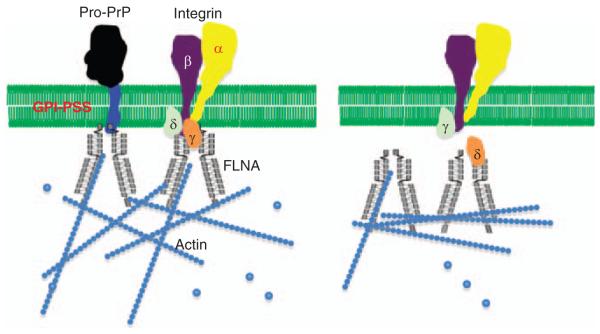
One of the best-characterized FLNA binding partners is the integrin β chain, such as β1 and β7. Integrins regulate cell spreading and migration (Hynes, 2002; Ginsberg et al., 2005; Luo et al., 2007). Therefore, in A7 cells, PrP, FLNA and integrin β1 may coexist in a trimeric complex. We find that in A7 cells, Although FLNA copurifies with PrP, and integrin β1 copurifies with FLNA, integrin β1 does not copurify with PrP. Therefore, in A7 cells, FLNA/PrP and FLNA/integrin β1 exist in two distinct complex, one containing FLNA and PrP, the other containing FLNA and integrin β1 (Figure 4). However, these two complexes appear to be functionally linked, because of the fact that the amount of FLNA copurified with integrin β1 is greatly reduced in PrP downregulated A7 cells. In PrP downregulated A7 cells, FLNA is dissociated from the inner membrane leaflets. This spatial change may be the reason that less integrin β1 is bound to FLNA in PrP downregulated A7 cells. Results of immunofluorescent staining of integrin β1 and FLNA in PrP downregulated A7 cells are consistent with this hypothesis. In control cells, integrin β1 and FLNA colocalize at the leading edges. In PrP downregulated A7 cells, cell surface integrin β1 is no longer associated with FLNA. FLNA appears to retract from the inner membrane leaflet. It is known that the motif on integrin β1 that binds FLNA is located at the N-terminus, proximal to the inner membrane leaflet (Loo et al., 1998). We propose that in A7 cells pro-PrP pulls FLNA closer to the inner membrane leaflet, enabling FLNA to bind to the short cytoplasmic tail of integrin β1 more effectively. We further propose that when PrP is downregulated, FLNA dissociates from the inner membrane leaflet, moving away from the β-chain (Figure 4). This disconnect is likely to affect the bidirectional functionality of integrin β1.
Figure 4.
A minimum drawing model on the interplay between pro-PrP, FLNA and integrin β1 in A7 cells. Presence of pro-PrP pulls FLNA closer to the inner membrane leaflet, allowing it to interact with integrin β1. When the level of pro-PrP is reduced, FLNA is retracted from the inner membrane leaflet rendering it unable to bind integrin β1. γ and δ are proteins that are normally associated with either FLNA or integrins.
Focal adhesion kinase is a critical component of the integrin-signaling cascade, and a regulator human melanoma cell migration (Akasaka et al., 1995; Tomar and Schlaepfer, 2009). Our finding that downregulation of PrP in A7 cells also reduces the levels of p-focal adhesion kinase is consistent with the view that less p-focal adhesion kinase correlates with reduced cell migration. Integrin signaling is complicated, a much more detailed biochemical analysis will be needed to further identify the upstream and downstream molecules that are altered in PrP downregulated A7 cells.
Although our studies on M2 and A7 cells suggest that binding of pro-PrP interferes with the interaction between FLNA and integrin β1, it remains to be determined whether this association also occurs in other cancers that express pro-PrP, FLNA and integrin. Furthermore, much more detailed biochemical study is required to identify the altered downstream and upstream signaling events as a consequence of binding of pro-PrP to FLNA.
Prevention of binding of pro-PrP to FLNA modulates cell spreading and migration
To further support our hypothesis that binding of pro-PrP to FLNA is important in cell migration, we took advantage of a recent finding. We found that a pentapeptide, KKRPK, had cell penetrating capacity (Yin et al., 2008). We hypothesized that if we added a KKRPK motif to the N-terminus of the PrP GPI-PSS, the peptide might be able to enter cells, and competed for the binding of pro-PrP to FLNA.
The KKRPK-GPI-PSS peptide is not toxic. It does not alter the expression of PrP, FLNA or integrin β1 in A7 cells. However, when A7 cells are incubated with the peptide, their spreading and migration are significantly reduced. Thus, binding of pro-PrP to FLNA indeed occurs in living cells, is important for A7 cell spreading and migration, and can be inhibited by the KKRPK-GPI-PSS. Although the synthetic peptide also blocks the copurification of FLNA and pro-PrP in cell lysate, it is possible that in a live cell the peptide may also interfere with other cellular processes that are unrelated to the binding of FLNA to pro-PrP. Nonetheless, our finding provides ‘proof of principle’ that interaction between pro-PrP and FLNA may be a potential target for therapeutic intervention in tumor cells that express both pro-PrP and FLNA.
Expression of pro-PrP in the PDAC correlates with shorter survival
To investigate whether our findings in cell lines have clinical implications, we determine whether PrP is expressed in normal human pancreas, in pancreas with pancreatitis, in pancreas with pre-neoplastic lesions, such as PanIN-1, -2, and -3, or in pancreas with PDAC. In normal human pancreas, only islet cells show PrP immunoreactivity. Neither acinar nor ductal epithelial cells stain for PrP. PrP is also undetectable in the duct cells in chronic pancreatitis, PanIN-1, and PanIN-2. About 13% PanIN-3 specimens show weak PrP staining. Among the 83 PDAC cases examined, 34 show strong PrP staining. The PDAC tumor cells also react with the anti-GPI-PSS antiserum indicating the presence of the GPI-PSS. Thus, as in the PDAC cell lines, PrP exists as pro-PrP in human PDAC lesions (Li et al., 2009).
Most importantly, the expression of PrP is associated with poorer clinical outcome. Patients with intra-tumoral PrP have a median survival time of 360 days, whereas patients without PrP expression in their tumors have a mean survival time of >1000 days. This association is independent of other factors, such as age and gender of the patient, as well as the size or differentiation stage of the tumor. Therefore, expression of PrP divides the PDAC patients into two groups with drastic difference in their survival rates. PrP expression is not essential for PDAC initiation because only 41% of the PDAC cases have detectable PrP. However, the presence of PrP is associated with poorer clinical outcome. Therefore, similar to PDAC cell lines, PDAC tumors with PrP have a growth advantage and are more aggressive.
Although all PDAC cell lines (n = 7) express PrP, only 41% of the tumor biopsies have PrP. One possibility is that during in vitro culture PrP bearing tumor cells are preferentially expanded. This view is consistent with our finding that PrP positive cells have growth advantage. So far, we have only studied the expression of PrP in resectable PDAC, if we examine PrP expression in nonresectable or in autopsies pancreatic cancer tissues we would observe a high proportion of the tumors that have PrP.
In normal pancreas only islet cells express detectable PrP; neither ductal cells nor acinar cells have detectable PrP. However, we can’t rule out the possibility that these cells may express PrP, but at a level that is beyond the detection limit of immunohistochemcial staining. As the anti-PrP GPI-PSS does not react with normal islet cells, therefore, the PrP in islet cell is a normal GPI-anchored PrP. It is interesting to note that in contrast to PrP, FLNA is undetectable in islet cells, but is detectable in low level in ductal cells, and the level is upregulated in PDAC (Uhlen et al., 2005). This finding is also consistent with an early report, which shows that FLNA is upregulated in human PDAC (Logsdon et al., 2003). At present time, the underlying mechanisms that cause the upregulation of PrP, FLNA and accumulation of pro-PrP in pancreatic ductal cells are not known.
Relationship between PrP expression and other genetic lesions in human PDAC
The four most extensively studied genetic lesions in human PDAC were mutations in K-RAS, SMAD4, TP53 and inactivation of CDNK2A (Li et al., 2004; Hezel et al., 2006; Hruban and Zamboni, 2009). Recently, we investigated whether there was a correlation between the expression of PrP and TP53 mutation. In normal pancreas, p53 was undetectable. Mutations in the TP53 caused the accumulation of p53, and thus p53 became detectable (Boschman et al., 1994). When stained with anti-PrP mAb, most of the immunoreactivity in tumor cells was detected in the cytosol, and the nucleus was by and large negative. In contrast, when stained with the anti-p53 mAb, most of the p53 immunoreactivity was detected in the nucleus (not shown). Two-color staining clearly showed the colocalization of PrP and p53 in the same tumor cells. Interestingly, in the 34 PrP positive tumor samples, 22 of the tumors (65%) also had detectable p53. On the other hand, in PrP negative tumors, only 12 out of the 49 (24%) PrP negative tumors had p53. These results suggested that tumor cells with TP53 mutations preferentially expressed PrP. Alternatively, expression of PrP might favor the accumulation of p53. So far, we have shown a correlation between PrP expression and TP53 mutation. The underlying mechanisms that link TP53 mutation and pro-PrP expression still need to be investigated. Mutation in p53 can result in gain of novel functions, which then affects numerous cellular pathways (Brosh and Rotter, 2009). A recent study finds that mutant p53 regulates cellular invasion by promoting integrin recycling (Muller et al., 2009).
Expression of pro-PrP in human melanoma
The progression from melanocyte to melanoma is complex and not completely understood (Herlyn, 2006). By immunohistochemical staining, normal human melanocytes do not react with either the anti-PrP mAb or the anti-PrP GPI-PSS specific antibody, indicating that normal melanocytes do not express PrP (Li et al., 2010). In contrast, both the anti-PrP mAb and anti-PrP GPI-PSS antibody react with melanoma in situ and invasive melanoma in the dermal component express the highest levels of PrP. These results suggest that expression of PrP may be important in the neoplastic transformation of melanocyte and invasion of human melanoma. However, whether expression of pro-PrP has diagnostic or prognostic value will require studying a much larger cohort of melanoma patients.
Future directions
It is a certainty that interaction between pro-PrP and FLNA does not occur in all tumor cells. Human neuroblastoma cell lines (n = 2) express neither PrP nor FLNA (not shown). Small-cell lung carcinoma cell lines (n = 3) express FLNA, but not PrP (not shown). On the other hand, leukemia cell lines, such as Jurket express a normal GPI-anchored PrP (Li et al., 2003). In addition to melanoma and PDAC, human hepatocarcinoma cell lines (n = 5) express both pro-PrP and FLNA. It is not known why some tumor cell lines express pro-PrP and FLNA although others do not. The current concept divides genes involved in tumorigenesis into driver genes and passenger genes (Liu, 2008). PrP is clearly not a driver, but more likely a passenger with deadly potential.
Over the last two decades, cancer biologists have focused on oncogenes and tumor suppressor genes. Recently, metabolic enzymes involved in diverse physiological functions, such as circadian clock regulation, intermediate metabolism, fucosylation or lipid synthesis, have emerged as critical factors in tumorigenesis (Deberardinis et al., 2008; Miyoshi et al., 2008; Sahar and Sassone-Corsi, 2009; Yan et al., 2009; Nomura et al., 2010). Enzymes involved in the GPI-anchor modification pathway may also have a role in the tumorigenesis of some human cancers (Table 2).
To fully elucidate the contributions of PrP to tumor biology some additional questions need to be addressed. For example, what is the mechanism that causes the upregulation of PrP in some tumor cells, but not all? What are the mechanisms that cause the failure to remove the GPI-PSS on PrP, or the accumulation of pro-PrP? Finally, as for the FLNA binding site on the GPI-PSS of PrP—is this by design or it is just an unfortunate blunder?
Acknowledgements
We thank all the collaborators who have contributed to these studies, and Pearl Ling for careful reading of the paper, suggestion and editorial assistant. Start-up funds from the Department of Pathology to WX. Supported by R21-CA133559-01 from NIH and a pilot Grant from SDRC#5P30AR039750 from NIH.
Footnotes
Conflict of interest The authors declare no conflict of interest.
References
- Abujiang P, Mori TJ, Takahashi T, Tanaka F, Kasyu I, Hitomi S, et al. Loss of heterozygosity (LOH) at 17q and 14q in human lung cancers. Oncogene. 1998;17:3029–3033. doi: 10.1038/sj.onc.1202230. [DOI] [PubMed] [Google Scholar]
- Adamson R, Jones AS, Field JK. Loss of heterozygosity studies on chromosome 17 in head and neck cancer using microsatellite markers. Oncogene. 1994;9:2077–2082. [PubMed] [Google Scholar]
- Aguirre AJ, Brennan C, Bailey G, Sinha R, Feng B, Leo C, et al. High-resolution characterization of the pancreatic adenocarcinoma genome. Proc Natl Acad Sci USA. 2004;101:9067–9072. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akasaka T, van Leeuwen RL, Yoshinaga IG, Mihm MC, Jr, Byers HR. Focal adhesion kinase (p125FAK) expression correlates with motility of human melanoma cell lines. J Invest Dermatol. 1995;105:104–108. doi: 10.1111/1523-1747.ep12313396. [DOI] [PubMed] [Google Scholar]
- Alper O, Stetler-Stevenson WG, Harris LN, Leitner WW, Ozdemirli M, Hartmann D, et al. Novel anti-filamin-A antibody detects a secreted variant of filamin-A in plasma from patients with breast carcinoma and high-grade astrocytoma. Cancer Sci. 2009;100:1748–1756. doi: 10.1111/j.1349-7006.2009.01244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, Arslan AA, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharam V, Kanthasamy A, Choi CJ, Martin DP, Latchoumycandane C, Richt JA, et al. Opposing roles of prion protein in oxidative stress- and ER stress-induced apoptotic signaling. Free Radic Biol Med. 2008;45:1530–1541. doi: 10.1016/j.freeradbiomed.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonacopoulou AG, Grivas PD, Skarlas L, Kalofonos M, Scopa CD, Kalofonos HP. POLR2F, ATP6V0A1 and PRNP expression in colorectal cancer: new molecules with prognostic significance? Anticancer Res. 2008;28:1221–1227. [PubMed] [Google Scholar]
- Argos M, Kibriya MG, Jasmine F, Olopade OI, Su T, Hibshoosh H, et al. Genomewide scan for loss of heterozygosity and chromosomal amplification in breast carcinoma using single-nucleotide polymorphism arrays. Cancer Genet Cytogenet. 2008;182:69–74. doi: 10.1016/j.cancergencyto.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awata H, Huang C, Handlogten ME, Miller RT. Interaction of the calcium-sensing receptor and filamin, a potential scaffolding protein. J Biol Chem. 2001;276:34871–34879. doi: 10.1074/jbc.M100775200. [DOI] [PubMed] [Google Scholar]
- Bashyam MD, Bair R, Kim YH, Wang P, Hernandez-Boussard T, Karikari CA, et al. Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia. 2005;7:556–562. doi: 10.1593/neo.04586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler K, Oesch B, Scott M, Westaway D, Walchli M, Groth DF, et al. Scrapie and cellular PrP isoforms are encoded by the same chromosomal gene. Cell. 1986;46:417–428. doi: 10.1016/0092-8674(86)90662-8. [DOI] [PubMed] [Google Scholar]
- Bedolla RG, Wang Y, Asuncion A, Chamie K, Siddiqui S, Mudryj MM, et al. Nuclear versus cytoplasmic localization of filamin A in prostate cancer: immunohistochemical correlation with metastases. Clin Cancer Res. 2009;15:788–796. doi: 10.1158/1078-0432.CCR-08-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger JM, Astier C, Sardet C, Ohta Y, Stossel TP, Debant A. The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat Cell Biol. 2000;2:888–892. doi: 10.1038/35046533. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgaard ML, Jager AC, Dalgaard P, Sondergaard JO, Rehfeld JF, Nielsen FC. Allelic loss of chromosome 2p21–16.3 is associated with reduced survival in sporadic colorectal cancer. Scand J Gastroenterol. 2001;36:405–409. doi: 10.1080/003655201300051252. [DOI] [PubMed] [Google Scholar]
- Bolton DC, McKinley MP, Prusiner SB. Identification of a protein that purifies with the scrapie prion. Science. 1982;218:1309–1311. doi: 10.1126/science.6815801. [DOI] [PubMed] [Google Scholar]
- Borbiev T, Verin AD, Shi S, Liu F, Garcia JG. Regulation of endothelial cell barrier function by calcium/calmodulin-dependent protein kinase II. Am J Physiol Lung Cell Mol Physiol. 2001;280:L983–L990. doi: 10.1152/ajplung.2001.280.5.L983. [DOI] [PubMed] [Google Scholar]
- Boschman CR, Stryker S, Reddy JK, Rao MS. Expression of p53 protein in precursor lesions and adenocarcinoma of human pancreas. Am J Pathol. 1994;145:1291–1295. [PMC free article] [PubMed] [Google Scholar]
- Bouffard D, Duncan LM, Howard CA, Mihm MC, Jr, Byers HR. Actin-binding protein expression in benign and malignant melanocytic proliferations. Hum Pathol. 1994;25:709–714. doi: 10.1016/0046-8177(94)90305-0. [DOI] [PubMed] [Google Scholar]
- Bounhar Y, Zhang Y, Goodyer CG, LeBlanc A. Prion protein protects human neurons against Bax-mediated apoptosis. J Biol Chem. 2001;276:39145–39149. doi: 10.1074/jbc.C100443200. [DOI] [PubMed] [Google Scholar]
- Bredel M, Bredel C, Juric D, Harsh GR, Vogel H, Recht LD, et al. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res. 2005;65:8679–8689. doi: 10.1158/0008-5472.CAN-05-1204. [DOI] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- Brown KM, Macgregor S, Montgomery GW, Craig DW, Zhao ZZ, Iyadurai K, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- Byers HR, Etoh T, Doherty JR, Sober AJ, Mihm MC., Jr Cell migration and actin organization in cultured human primary, recurrent cutaneous and metastatic melanoma. Time-lapse and image analysis. Am J Pathol. 1991;139:423–435. [PMC free article] [PubMed] [Google Scholar]
- Calderwood DA, Huttenlocher A, Kiosses WB, Rose DM, Woodside DG, Schwartz MA, et al. Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat Cell Biol. 2001;3:1060–1068. doi: 10.1038/ncb1201-1060. [DOI] [PubMed] [Google Scholar]
- Campana V, Sarnataro D, Zurzolo C. The highways and byways of prion protein trafficking. Trends Cell Biol. 2005;15:102–111. doi: 10.1016/j.tcb.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Castro P, Creighton CJ, Ozen M, Berel D, Mims MP, Ittmann M. Genomic profiling of prostate cancers from African American men. Neoplasia. 2009;11:305–312. doi: 10.1593/neo.81530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B, Brown K, Raymond GJ, Katzenstein GE, Thresher W. Binding of the protease-sensitive form of PrP (prion protein) to sulfated glycosaminoglycan and congo red. J Virol. 1994;68:2135–2141. doi: 10.1128/jvi.68.4.2135-2141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Knez JJ, Merrick WC, Medof ME. Comparative efficiencies of C-terminal signals of native glycophosphatidylinositol (GPI)-anchored proproteins in conferring GPI-anchoring. J Cell Biochem. 2001;84:68–83. doi: 10.1002/jcb.1267. [DOI] [PubMed] [Google Scholar]
- Chiarini LB, Freitas AR, Zanata SM, Brentani RR, Martins VR, Linden R. Cellular prion protein transduces neuroprotective signals. EMBO J. 2002;21:3317–3326. doi: 10.1093/emboj/cdf324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YW, Bae SM, Kim YW, Lee HN, Kim YW, Park TC, et al. Gene expression profiles in squamous cell cervical carcinoma using array-based comparative genomic hybridization analysis. Int J Gynecol Cancer. 2007;17:687–696. doi: 10.1111/j.1525-1438.2007.00834.x. [DOI] [PubMed] [Google Scholar]
- Creutzfeldt HG. Über eine eigenartige herdförmige Erkrankung des Zentralnervensystems. Vorlaüfige Mitteilung. Zeitschrift für die gesamte Neurologie und Psychiatrie. 1920;57:1–18. [Google Scholar]
- Cukier IH, Li Y, Lee JM. Cyclin B1/Cdk1 binds and phosphorylates Filamin A and regulates its ability to cross-link actin. FEBS Lett. 2007;581:1661–1672. doi: 10.1016/j.febslet.2007.03.041. [DOI] [PubMed] [Google Scholar]
- Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, et al. Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 1992;255:325–327. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- Daley D, Lewis S, Platzer P, MacMillen M, Willis J, Elston RC, et al. Identification of susceptibility genes for cancer in a genomewide scan: results from the colon neoplasia sibling study. Am J Hum Genet. 2008;82:723–736. doi: 10.1016/j.ajhg.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies P, Brown DR. The chemistry of copper binding to PrP: is there sufficient evidence to elucidate a role for copper in protein function? Biochem J. 2008;410:237–244. doi: 10.1042/BJ20071477. [DOI] [PubMed] [Google Scholar]
- Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devilee P, Cornelisse CJ, Kuipers-Dijkshoorn N, Jonker C, Pearson PL. Loss of heterozygosity on 17p in human breast carcinomas: defining the smallest common region of deletion. Cytogenet Cell Genet. 1990;53:52–54. doi: 10.1159/000132893. [DOI] [PubMed] [Google Scholar]
- Dimova I, Orsetti B, Negre V, Rouge C, Ursule L, Lasorsa L, et al. Genomic markers for ovarian cancer at chromosomes 1, 8 and 17 revealed by array CGH analysis. Tumori. 2009;95:357–366. doi: 10.1177/030089160909500315. [DOI] [PubMed] [Google Scholar]
- Dohna M, Reincke M, Mincheva A, Allolio B, Solinas-Toldo S, Lichter P. Adrenocortical carcinoma is characterized by a high frequency of chromosomal gains and high-level amplifications. Genes Chromosomes Cancer. 2000;28:145–152. [PubMed] [Google Scholar]
- Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, Cohen FE, et al. Structure of the recombinant full-length hamster prion protein PrP(29-231): the N terminus is highly flexible. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas EJ, Fiegler H, Rowan A, Halford S, Bicknell DC, Bodmer W, et al. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res. 2004;64:4817–4825. doi: 10.1158/0008-5472.CAN-04-0328. [DOI] [PubMed] [Google Scholar]
- Edenhofer F, Rieger R, Famulok M, Wendler W, Weiss S, Winnacker EL. Prion protein PrPc interacts with molecular chaperones of the Hsp60 family. J Virol. 1996;70:4724–4728. doi: 10.1128/jvi.70.7.4724-4728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Zheng P, Tang J, Liu Y. CD24: from A to Z. Cell Mol Immunol. 2010;7:100–103. doi: 10.1038/cmi.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanjul M, Alvarez L, Hollande E. Expression and subcellular localization of a 35-kDa carbonic anhydrase IV in a human pancreatic ductal cell line (Capan-1) J Histochem Cytochem. 2007;55:783–794. doi: 10.1369/jhc.6A7112.2007. [DOI] [PubMed] [Google Scholar]
- Fanjul M, Alvarez L, Salvador C, Gmyr V, Kerr-Conte J, Pattou F, et al. Evidence for a membrane carbonic anhydrase IV anchored by its C-terminal peptide in normal human pancreatic ductal cells. Histochem Cell Biol. 2004;121:91–99. doi: 10.1007/s00418-003-0616-2. [DOI] [PubMed] [Google Scholar]
- Feng S, Resendiz JC, Lu X, Kroll MH. Filamin A binding to the cytoplasmic tail of glycoprotein Ibalpha regulates von Will-ebrand factor-induced platelet activation. Blood. 2003;102:2122–2129. doi: 10.1182/blood-2002-12-3805. [DOI] [PubMed] [Google Scholar]
- Feng Y, Chen MH, Moskowitz IP, Mendonza AM, Vidali L, Nakamura F, et al. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc Natl Acad Sci USA. 2006;103:19836–19841. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- Fiori JL, Zhu TN, O’Connell MP, Hoek KS, Indig FE, Frank BP, et al. Filamin A modulates kinase activation and intracellular trafficking of epidermal growth factor receptors in human melanoma cells. Endocrinology. 2009;150:2551–2560. doi: 10.1210/en.2008-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JW, Lamperti ED, Eksioglu YZ, Hong SE, Feng Y, Graham DA, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- Franscini N, El Gedaily A, Matthey U, Franitza S, Sy MS, Burkle A, et al. Prion protein in milk. PLoS One. 2006;1:e71. doi: 10.1371/journal.pone.0000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frierson HF, Jr, El-Naggar AK, Welsh JB, Sapinoso LM, Su AI, Cheng J, et al. Large scale molecular analysis identifies genes with altered expression in salivary adenoid cystic carcinoma. Am J Pathol. 2002;161:1315–1323. doi: 10.1016/S0002-9440(10)64408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Sakakura C, Shimomura K, Nakanishi M, Yasuoka R, Aragane H, et al. Chromosome arm 20q gains and other genomic alterations in esophageal squamous cell carcinoma, as analyzed by comparative genomic hybridization and fluorescence in situ hybridization. Hepatogastroenterology. 2003;50:1857–1863. [PubMed] [Google Scholar]
- Gabus C, Auxilien S, Pechoux C, Dormont D, Swietnicki W, Morillas M, et al. The prion protein has DNA strand transfer properties similar to retroviral nucleocapsid protein. J Mol Biol. 2001;307:1011–1021. doi: 10.1006/jmbi.2001.4544. [DOI] [PubMed] [Google Scholar]
- Gajdusek DC. Review. Kuru and its contribution to medicine. Philos Trans R Soc Lond B Biol Sci. 2008;363:3697–3700. doi: 10.1098/rstb.2008.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajdusek DC, Zigas V. Degenerative disease of the central nervous system in New Guinea; the endemic occurrence of kuru in the native population. N Engl J Med. 1957;257:974–978. doi: 10.1056/NEJM195711142572005. [DOI] [PubMed] [Google Scholar]
- Garcea G, Neal CP, Pattenden CJ, Steward WP, Berry DP. Molecular prognostic markers in pancreatic cancer: a systematic review. Eur J Cancer. 2005;41:2213–2236. doi: 10.1016/j.ejca.2005.04.044. [DOI] [PubMed] [Google Scholar]
- Ginsberg MH, Partridge A, Shattil SJ. Integrin regulation. Curr Opin Cell Biol. 2005;17:509–516. doi: 10.1016/j.ceb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Glogauer M, Arora P, Chou D, Janmey PA, Downey GP, McCulloch CA. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J Biol Chem. 1998;273:1689–1698. doi: 10.1074/jbc.273.3.1689. [DOI] [PubMed] [Google Scholar]
- Goeze A, Schluns K, Wolf G, Thasler Z, Petersen S, Petersen I. Chromosomal imbalances of primary and metastatic lung adenocarcinomas. J Pathol. 2002;196:8–16. doi: 10.1002/path.1009. [DOI] [PubMed] [Google Scholar]
- Goldfarb LG, Brown P, Cervenakova L, Gajdusek DC. Genetic analysis of Creutzfeldt-Jakob disease and related disorders. Philos Trans R Soc Lond B Biol Sci. 1994;343:379–384. doi: 10.1098/rstb.1994.0032. [DOI] [PubMed] [Google Scholar]
- Goldmann WH. p56(lck) Controls phosphorylation of filamin (ABP-280) and regulates focal adhesion kinase (pp125(FAK)) Cell Biol Int. 2002;26:567–571. doi: 10.1006/cbir.2002.0900. [DOI] [PubMed] [Google Scholar]
- Gorlin JB, Yamin R, Egan S, Stewart M, Stossel TP, Kwiatkowski DJ, et al. Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J Cell Biol. 1990;111:1089–1105. doi: 10.1083/jcb.111.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig JR. Scrapie in sheep. J Comp Pathol. 1950;60:263–266. doi: 10.1016/s0368-1742(50)80024-3. [DOI] [PubMed] [Google Scholar]
- Guan XY, Meltzer PS, Dalton WS, Trent JM. Identification of cryptic sites of DNA sequence amplification in human breast cancer by chromosome microdissection. Nat Genet. 1994;8:155–161. doi: 10.1038/ng1094-155. [DOI] [PubMed] [Google Scholar]
- Guan Y, Hata N, Kuga D, Yoshimoto K, Mizoguchi M, Shono T, et al. Narrowing of the regions of allelic losses of chromosome 1p36 in meningioma tissues by an improved SSCP analysis. Int J Cancer. 2008;122:1820–1826. doi: 10.1002/ijc.23297. [DOI] [PubMed] [Google Scholar]
- Guedj N, Zhan Q, Perigny M, Rautou PE, Degos F, Belghiti J, et al. Comparative protein expression profiles of hilar and peripheral hepatic cholangiocarcinomas. J Hepatol. 2009;51:93–101. doi: 10.1016/j.jhep.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Guled M, Myllykangas S, Frierson HF, Jr, Mills SE, Knuutila S, Stelow EB. Array comparative genomic hybridization analysis of olfactory neuroblastoma. Mod Pathol. 2008;21:770–778. doi: 10.1038/modpathol.2008.57. [DOI] [PubMed] [Google Scholar]
- Guo Z, Linn JF, Wu G, Anzick SL, Eisenberger CF, Halachmi S, et al. CDC91L1 (PIG-U) is a newly discovered oncogene in human bladder cancer. Nat Med. 2004;10:374–381. doi: 10.1038/nm1010. [DOI] [PubMed] [Google Scholar]
- Han H, Bearss DJ, Browne LW, Calaluce R, Nagle RB, Von Hoff DD. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002;62:2890–2896. [PubMed] [Google Scholar]
- Hart AW, Morgan JE, Schneider J, West K, McKie L, Bhattacharya S, et al. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006;15:2457–2467. doi: 10.1093/hmg/ddl168. [DOI] [PubMed] [Google Scholar]
- He HJ, Kole S, Kwon YK, Crow MT, Bernier M. Interaction of filamin A with the insulin receptor alters insulin-dependent activation of the mitogen-activated protein kinase pathway. J Biol Chem. 2003;278:27096–27104. doi: 10.1074/jbc.M301003200. [DOI] [PubMed] [Google Scholar]
- He X, Li Y, Schembri-King J, Jakes S, Hayashi J. Identification of actin binding protein, ABP-280, as a binding partner of human Lnk adaptor protein. Mol Immunol. 2000;37:603–612. doi: 10.1016/s0161-5890(00)00070-5. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Rane NS. Prion protein trafficking and the development of neurodegeneration. Trends Neurosci. 2003;26:337–339. doi: 10.1016/S0166-2236(03)00143-7. [DOI] [PubMed] [Google Scholar]
- Helfand BT, Loeb S, Meeks JJ, Fought AJ, Kan D, Catalona WJ. Pathological outcomes associated with the 17q prostate cancer risk variants. J Urol. 2009;181:2502–2507. doi: 10.1016/j.juro.2009.01.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlyn M. Molecular targets in melanoma: strategies and challenges for diagnosis and therapy. Int J Cancer. 2006;118:523–526. doi: 10.1002/ijc.21605. [DOI] [PubMed] [Google Scholar]
- Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- Hidaka S, Yasutake T, Kondo M, Takeshita H, Yano H, Haseba M, et al. Frequent gains of 20q and losses of 18q are associated with lymph node metastasis in intestinal-type gastric cancer. Anticancer Res. 2003;23:3353–3357. [PubMed] [Google Scholar]
- Hirai M, Yoshida S, Kashiwagi H, Kawamura T, Ishikawa T, Kaneko M, et al. 1q23 gain is associated with progressive neuroblastoma resistant to aggressive treatment. Genes Chromosomes Cancer. 1999;25:261–269. doi: 10.1002/(sici)1098-2264(199907)25:3<261::aid-gcc8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Ho JC, Cheung ST, Patil M, Chen X, Fan ST. Increased expression of glycosyl-phosphatidylinositol anchor attachment protein 1 (GPAA1) is associated with gene amplification in hepatocellular carcinoma. Int J Cancer. 2006;119:1330–1337. doi: 10.1002/ijc.22005. [DOI] [PubMed] [Google Scholar]
- Hoggard N, Hey Y, Brintnell B, James L, Jones D, Mitchell E, et al. Identification and cloning in yeast artificial chromosomes of a region of elevated loss of heterozygosity on chromosome 1p31.1 in human breast cancer. Genomics. 1995;30:233–243. doi: 10.1006/geno.1995.9882. [DOI] [PubMed] [Google Scholar]
- Hruban RH, Adsay NV, Albores-Saavedra J, Anver MR, Biankin AV, Boivin GP, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66:95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- Hruban RH, Zamboni G. Pancreatic cancer. Special issue—insights and controversies in pancreatic pathology. Arch Pathol Lab Med. 2009;133:347–349. doi: 10.5858/133.3.347. [DOI] [PubMed] [Google Scholar]
- Hsiao K, Prusiner SB. Inherited human prion diseases. Neurology. 1990;40:1820–1827. doi: 10.1212/wnl.40.12.1820. [DOI] [PubMed] [Google Scholar]
- Hulsebos TJ, Bijleveld EH, Oskam NT, Westerveld A, Leenstra S, Hogendoorn PC, et al. Malignant astrocytoma-derived region of common amplification in chromosomal band 17p12 is frequently amplified in high-grade osteosarcomas. Genes Chromosomes Cancer. 1997;18:279–285. [PubMed] [Google Scholar]
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Ikezawa H. Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol Pharm Bull. 2002;25:409–417. doi: 10.1248/bpb.25.409. [DOI] [PubMed] [Google Scholar]
- Ishihara T, Tsuda H, Hotta A, Kozaki K, Yoshida A, Noh JY, et al. ITCH is a putative target for a novel 20q11.22 amplification detected in anaplastic thyroid carcinoma cells by array-based comparative genomic hybridization. Cancer Sci. 2008;99:1940–1949. doi: 10.1111/j.1349-7006.2008.00900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishkanian AS, Mallof CA, Ho J, Meng A, Albert M, Syed A, et al. High-resolution array CGH identifies novel regions of genomic alteration in intermediate-risk prostate cancer. Prostate. 2009;69:1091–1100. doi: 10.1002/pros.20959. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- Jiang WW, Zahurak M, Zhou ZT, Park HL, Guo ZM, Wu GJ, et al. Alterations of GPI transamidase subunits in head and neck squamous carcinoma. Mol Cancer. 2007;6:74. doi: 10.1186/1476-4598-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Pais TL, Nellissery MJ, Ammerman DG, Pathmanathan D, Bhatia P, Buller CL, et al. Determination of a minimal region of loss of heterozygosity on chromosome 18q21.33 in osteosarcoma. Int J Cancer. 2003;105:285–288. doi: 10.1002/ijc.11070. [DOI] [PubMed] [Google Scholar]
- Jonson T, Gorunova L, Dawiskiba S, Andren-Sandberg A, Stenman G, ten Dijke P, et al. Molecular analyses of the 15q and 18q SMAD genes in pancreatic cancer. Genes Chromosomes Cancer. 1999;24:62–71. doi: 10.1002/(sici)1098-2264(199901)24:1<62::aid-gcc9>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Kamochi N, Nakashima M, Aoki S, Uchihashi K, Sugihara H, Toda S, et al. Irradiated fibroblast-induced bystander effects on invasive growth of squamous cell carcinoma under cancer-stromal cell interaction. Cancer Sci. 2008;99:2417–2427. doi: 10.1111/j.1349-7006.2008.00978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H, Shibata T, Kokubu A, Ojima H, Loukopoulos P, Kanai Y, et al. Genetic profile of hepatocellular carcinoma revealed by array-based comparative genomic hybridization: identification of genetic indicators to predict patient outcome. J Hepatol. 2005;43:863–874. doi: 10.1016/j.jhep.2005.05.033. [DOI] [PubMed] [Google Scholar]
- Kayed H, Kleeff J, Keleg S, Siang X, Penzel R, Giese T, et al. Correlation of glypican-1 expression with TGF-beta, BMP, and activin receptors in pancreatic ductal adenocarcinoma. Int J Oncol. 2006;29:1139–1148. [PubMed] [Google Scholar]
- Keshamouni VG, Michailidis G, Grasso CS, Anthwal S, Strahler JR, Walker A, et al. Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. J Proteome Res. 2006;5:1143–1154. doi: 10.1021/pr050455t. [DOI] [PubMed] [Google Scholar]
- Keshet GI, Bar-Peled O, Yaffe D, Nudel U, Gabizon R. The cellular prion protein colocalizes with the dystroglycan complex in the brain. J Neurochem. 2000;75:1889–1897. doi: 10.1046/j.1471-4159.2000.0751889.x. [DOI] [PubMed] [Google Scholar]
- Kiema T, Lad Y, Jiang P, Oxley CL, Baldassarre M, Wegener KL, et al. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Kim H, Sengupta A, Glogauer M, McCulloch CA. Filamin A regulates cell spreading and survival via beta1 integrins. Exp Cell Res. 2008;314:834–846. doi: 10.1016/j.yexcr.2007.11.022. [DOI] [PubMed] [Google Scholar]
- Kim JH, Dhanasekaran SM, Mehra R, Tomlins SA, Gu W, Yu J, et al. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res. 2007a;67:8229–8239. doi: 10.1158/0008-5472.CAN-07-1297. [DOI] [PubMed] [Google Scholar]
- Kim SW, Kim JW, Kim YT, Kim JH, Kim S, Yoon BS, et al. Analysis of chromosomal changes in serous ovarian carcinoma using high-resolution array comparative genomic hybridization: potential predictive markers of chemoresistant disease. Genes Chromosomes Cancer. 2007b;46:1–9. doi: 10.1002/gcc.20384. [DOI] [PubMed] [Google Scholar]
- Kimmelman AC, Hezel AF, Aguirre AJ, Zheng H, Paik JH, Ying H, et al. Genomic alterations link Rho family of GTPases to the highly invasive phenotype of pancreas cancer. Proc Natl Acad Sci USA. 2008;105:19372–19377. doi: 10.1073/pnas.0809966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Fujita M, Maeda Y. Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress. J Biochem. 2008;144:287–294. doi: 10.1093/jb/mvn090. [DOI] [PubMed] [Google Scholar]
- Kley RA, Hellenbroich Y, van der Ven PF, Furst DO, Huebner A, Bruchertseifer V, et al. Clinical and morphological phenotype of the filamin myopathy: a study of 31 German patients. Brain. 2007;130:3250–3264. doi: 10.1093/brain/awm271. [DOI] [PubMed] [Google Scholar]
- Krakow D, Robertson SP, King LM, Morgan T, Sebald ET, Bertolotto C, et al. Mutations in the gene encoding filamin B disrupt vertebral segmentation, joint formation and skeletogenesis. Nat Genet. 2004;36:405–410. doi: 10.1038/ng1319. [DOI] [PubMed] [Google Scholar]
- Kresse SH, Berner JM, Meza-Zepeda LA, Gregory SG, Kuo WL, Gray JW, et al. Mapping and characterization of the amplicon near APOA2 in 1q23 in human sarcomas by FISH and array CGH. Mol Cancer. 2005;4:39. doi: 10.1186/1476-4598-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara C, Takeuchi AM, Nishimura T, Haraguchi K, Kubosaki A, Matsumoto Y, et al. Prions prevent neuronal cell-line death. Nature. 1999;400:225–226. doi: 10.1038/22241. [DOI] [PubMed] [Google Scholar]
- Lad Y, Kiema T, Jiang P, Pentikainen OT, Coles CH, Campbell ID, et al. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO J. 2007;26:3993–4004. doi: 10.1038/sj.emboj.7601827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larramendy ML, Lushnikova T, Bjorkqvist AM, Wistuba II, Virmani AK, Shivapurkar N, et al. Comparative genomic hybridization reveals complex genetic changes in primary breast cancer tumors and their cell lines. Cancer Genet Cytogenet. 2000;119:132–138. doi: 10.1016/s0165-4608(99)00226-5. [DOI] [PubMed] [Google Scholar]
- Leonardi A, Ellinger-Ziefelbauer H, Franzoso G, Brown K, Siebenlist U. Physical and functional interaction of filamin (actin-binding protein-280) and tumor necrosis factor receptor-associated factor 2. J Biol Chem. 2000;275:271–278. doi: 10.1074/jbc.275.1.271. [DOI] [PubMed] [Google Scholar]
- Letessier A, Sircoulomb F, Ginestier C, Cervera N, Monville F, Gelsi-Boyer V, et al. Frequency, prognostic impact, and subtype association of 8p12, 8q24, 11q13, 12p13, 17q12, and 20q13 amplifications in breast cancers. BMC Cancer. 2006;6:245. doi: 10.1186/1471-2407-6-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong P, Pan T, Xiao F, Yin S, Chang B, et al. Normal cellular prion protein is a ligand of selectins: binding requires Le(X) but is inhibited by sLe(X) Biochem J. 2007;406:333–341. doi: 10.1042/BJ20061857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yu S, Nakamura F, Yin S, Xu J, Petrolla AA, et al. Binding of pro-prion to filamin A disrupts cytoskeleton and correlates with poor prognosis in pancreatic cancer. J Clin Invest. 2009;119:2725–2736. doi: 10.1172/JCI39542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yu S, Nakamura F, Pentikainen O, Singh N, Yin S, et al. Pro-prion binds filamin A facilitating its interaction with integrin β1 and contributes to melanomagenesis. J Biol Chem. 2010 doi: 10.1074/jbc.M110.147413. (e-pub ahead of print 21 July 2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- Li M, Li C, Weingarten P, Bunzow JR, Grandy DK, Zhou QY. Association of dopamine D(3) receptors with actin-binding protein 280 (ABP-280) Biochem Pharmacol. 2002;63:859–863. doi: 10.1016/s0006-2952(01)00932-7. [DOI] [PubMed] [Google Scholar]
- Li R, Liu T, Wong BS, Pan T, Morillas M, Swietnicki W, et al. Identification of an epitope in the C terminus of normal prion protein whose expression is modulated by binding events in the N terminus. J Mol Biol. 2000;301:567–573. doi: 10.1006/jmbi.2000.3986. [DOI] [PubMed] [Google Scholar]
- Li R, Liu T, Yoshihiro F, Trat-Lehmann M, Obrenovich M, Kuekrek H, et al. On the same cell type GPI-anchored normal cellular prion and DAF protein exhibit different biological properties. Biochem Biophys Res Com. 2003;303:446–451. doi: 10.1016/s0006-291x(03)00354-1. [DOI] [PubMed] [Google Scholar]
- Li X, Lee NK, Ye YW, Waber PG, Schweitzer C, Cheng QC, et al. Allelic loss at chromosomes 3p, 8p, 13q, and 17p associated with poor prognosis in head and neck cancer. J Natl Cancer Inst. 1994;86:1524–1529. doi: 10.1093/jnci/86.20.1524. [DOI] [PubMed] [Google Scholar]
- Liang J, Ge F, Guo C, Luo G, Wang X, Han G, et al. Inhibition of PI3K/Akt partially leads to the inhibition of PrP(C)-induced drug resistance in gastric cancer cells. FEBS J. 2009;276:685–694. doi: 10.1111/j.1742-4658.2008.06816.x. [DOI] [PubMed] [Google Scholar]
- Lin M, Smith LT, Smiraglia DJ, Kazhiyur-Mannar R, Lang JC, Schuller DE, et al. DNA copy number gains in head and neck squamous cell carcinoma. Oncogene. 2006;25:1424–1433. doi: 10.1038/sj.onc.1209166. [DOI] [PubMed] [Google Scholar]
- Lin R, Karpa K, Kabbani N, Goldman-Rakic P, Levenson R. Dopamine D2 and D3 receptors are linked to the actin cytoskeleton via interaction with filamin A. Proc Natl Acad Sci USA. 2001;98:5258–5263. doi: 10.1073/pnas.011538198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm PM, Salmenkivi K, Vauhkonen H, Nicholson AG, Anttila S, Kinnula VL, et al. Gene copy number analysis in malignant pleural mesothelioma using oligonucleotide array CGH. Cytogenet Genome Res. 2007;119:46–52. doi: 10.1159/000109618. [DOI] [PubMed] [Google Scholar]
- Liu ET. Functional genomics of cancer. Curr Opin Genet Dev. 2008;18:251–256. doi: 10.1016/j.gde.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Liu G, Thomas L, Warren RA, Enns CA, Cunningham CC, Hartwig JH, et al. Cytoskeletal protein ABP-280 directs the intracellular trafficking of furin and modulates proprotein processing in the endocytic pathway. J Cell Biol. 1997;139:1719–1733. doi: 10.1083/jcb.139.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, et al. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- Loo DT, Kanner SB, Aruffo A. Filamin binds to the cytoplasmic domain of the beta1-integrin. Identification of amino acids responsible for this interaction. J Biol Chem. 1998;273:23304–23312. doi: 10.1074/jbc.273.36.23304. [DOI] [PubMed] [Google Scholar]
- Loukopoulos P, Shibata T, Katoh H, Kokubu A, Sakamoto M, Yamazaki K, et al. Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: identification of genetic indicators that predict patient outcome. Cancer Sci. 2007;98:392–400. doi: 10.1111/j.1349-7006.2007.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy CJ, Sim KS, Yong EL. Filamin-A fragment localizes to the nucleus to regulate androgen receptor and coactivator functions. Proc Natl Acad Sci USA. 2003;100:4562–4567. doi: 10.1073/pnas.0736237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Sheen V. Periventricular heterotopia. Epilepsy Behav. 2005;7:143–149. doi: 10.1016/j.yebeh.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Gao M, Lu Y, Feng X, Zhang J, Lin D, et al. Gain of 1q25–32, 12q23–24.3, and 17q12–22 facilitates tumorigenesis and progression of human squamous cell lung cancer. J Pathol. 2006;210:205–213. doi: 10.1002/path.2050. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Ashida H, Kinoshita T. CHO glycosylation mutants: GPI anchor. Methods Enzymol. 2006;416:182–205. doi: 10.1016/S0076-6879(06)16012-7. [DOI] [PubMed] [Google Scholar]
- Mahfoud R, Garmy N, Maresca M, Yahi N, Puigserver A, Fantini J. Identification of a common sphingolipid-binding domain in Alzheimer, prion, and HIV-1 proteins. J Biol Chem. 2002;277:11292–11296. doi: 10.1074/jbc.M111679200. [DOI] [PubMed] [Google Scholar]
- Mani K, Cheng F, Havsmark B, Jonsson M, Belting M, Fransson LA. Prion, amyloid beta-derived Cu(II) ions, or free Zn(II) ions support S-nitroso-dependent autocleavage of glypican-1 heparan sulfate. J Biol Chem. 2003;278:38956–38965. doi: 10.1074/jbc.M300394200. [DOI] [PubMed] [Google Scholar]
- Maqani N, Belkhiri A, Moskaluk C, Knuutila S, Dar AA, El-Rifai W. Molecular dissection of 17q12 amplicon in upper gastro-intestinal adenocarcinomas. Mol Cancer Res. 2006;4:449–455. doi: 10.1158/1541-7786.MCR-06-0058. [DOI] [PubMed] [Google Scholar]
- Mastracci TL, Shadeo A, Colby SM, Tuck AB, O’Malley FP, Bull SB, et al. Genomic alterations in lobular neoplasia: a microarray comparative genomic hybridization signature for early neoplastic proliferationin the breast. Genes Chromosomes Cancer. 2006;45:1007–1017. doi: 10.1002/gcc.20368. [DOI] [PubMed] [Google Scholar]
- Mhawech-Fauceglia P, Odunsi K, Dim D, Nowak N, Lele S, Cheney RT, et al. Array-comparative genomic hybridization analysis of primary endometrial and ovarian high-grade neuroendocrine carcinoma associated with adenocarcinoma: mystery resolved? Int J Gynecol Pathol. 2008;27:539–546. doi: 10.1097/PGP.0b013e31816bcda4. [DOI] [PubMed] [Google Scholar]
- Millhauser GL. Copper and the prion protein: methods, structures, function, and disease. Annu Rev Phys Chem. 2007;58:299–320. doi: 10.1146/annurev.physchem.58.032806.104657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miozzo M, Dalpra L, Riva P, Volonta M, Macciardi F, Pericotti S, et al. A tumor suppressor locus in familial and sporadic chordoma maps to 1p36. Int J Cancer. 2000;87:68–72. [PubMed] [Google Scholar]
- Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. 2008;143:725–729. doi: 10.1093/jb/mvn011. [DOI] [PubMed] [Google Scholar]
- Morel E, Fouquet S, Strup-Perrot C, Thievend C Pichol, Petit C, Loew D, et al. The cellular prion protein PrP(c) is involved in the proliferation of epithelial cells and in the distribution of junction-associated proteins. PLoS One. 2008;3:e3000. doi: 10.1371/journal.pone.0003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillet-Richard S, Ermonval M, Chebassier C, Laplanche JL, Lehmann S, Launay JM, et al. Signal transduction through prion protein. Science. 2000;289:1925–1928. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- Myllykangas S, Junnila S, Kokkola A, Autio R, Scheinin I, Kiviluoto T, et al. Integrated gene copy number and expression microarray analysis of gastric cancer highlights potential target genes. Int J Cancer. 2008;123:817–825. doi: 10.1002/ijc.23574. [DOI] [PubMed] [Google Scholar]
- Naghibalhossaini F, Yoder AD, Tobi M, Stanners CP. Evolution of a tumorigenic property conferred by glycophosphatidyl-inositol membrane anchors of carcinoembryonic antigen gene family members during the primate radiation. Mol Biol Cell. 2007;18:1366–1374. doi: 10.1091/mbc.E06-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal JK, Dasgupta S, Jadallah S, Chae YK, Ratovitski EA, Toubaji A, et al. Profiling the expression pattern of GPI transamidase complex subunits in human cancer. Mod Pathol. 2008;21:979–991. doi: 10.1038/modpathol.2008.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179:1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Pudas R, Heikkinen O, Permi P, Kilpelainen I, Munday AD, et al. The structure of the GPIb-filamin A complex. Blood. 2006;107:1925–1932. doi: 10.1182/blood-2005-10-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Matsumoto S, Iwakawa R, Kohno T, Suzuki K, Tsuta K, et al. Whole genome comparison of allelic imbalance between noninvasive and invasive small-sized lung adenocarcinomas. Cancer Res. 2009;69:1615–1623. doi: 10.1158/0008-5472.CAN-08-3218. [DOI] [PubMed] [Google Scholar]
- Narang HK, Dagdanova A, Xie Z, Yang Q, Chen SG. Sensitive detection of prion protein in human urine. Exp Biol Med (Maywood) 2005;230:343–349. doi: 10.1177/153537020523000508. [DOI] [PubMed] [Google Scholar]
- Narayan G, Bourdon V, Chaganti S, Arias-Pulido H, Nandula SV, Rao PH, et al. Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: identification of candidate amplified and overexpressed genes. Genes Chromosomes Cancer. 2007;46:373–384. doi: 10.1002/gcc.20418. [DOI] [PubMed] [Google Scholar]
- Naylor TL, Greshock J, Wang Y, Colligon T, Yu QC, Clemmer V, et al. High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 2005;7:R1186–R1198. doi: 10.1186/bcr1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson TB, Stanners CP. Identification of a novel functional specificity signal within the GPI anchor signal sequence of carcinoembryonic antigen. J Cell Biol. 2007;177:211–218. doi: 10.1083/jcb.200701158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomachi A, Nishita M, Inaba D, Enomoto M, Hamasaki M, Minami Y. Receptor tyrosine kinase Ror2 mediates Wnt5a-induced polarized cell migration by activating c-Jun N-terminal kinase via actin-binding protein filamin A. J Biol Chem. 2008;283:27973–27981. doi: 10.1074/jbc.M802325200. [DOI] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
- Nowak NJ, Gaile D, Conroy JM, McQuaid D, Cowell J, Carter R, et al. Genome-wide aberrations in pancreatic adenocarcinoma. Cancer Genet Cytogenet. 2005;161:36–50. doi: 10.1016/j.cancergencyto.2005.01.009. [DOI] [PubMed] [Google Scholar]
- O’Connell MP, Fiori JL, Baugher KM, Indig FE, French AD, Camilli TC, et al. Wnt5A activates the calpain-mediated cleavage of filamin A. J Invest Dermatol. 2009;129:1782–1789. doi: 10.1038/jid.2008.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. The small GTPase RalA targets filamin to induce filopodia. Proc Natl Acad Sci USA. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoprishvili I, Andria ML, Kramer HK, Ancevska-Taneva N, Hiller JM, Simon EJ. Interaction between the mu opioid receptor and filamin A is involved in receptor regulation and trafficking. Mol Pharmacol. 2003;64:1092–1100. doi: 10.1124/mol.64.5.1092. [DOI] [PubMed] [Google Scholar]
- Ottenhof NA, Milne AN, Morsink FH, Drillenburg P, Ten Kate FJ, Maitra A, et al. Pancreatic intraepithelial neoplasia and pancreatic tumorigenesis: of mice and men. Arch Pathol Lab Med. 2009;133:375–381. doi: 10.5858/133.3.375. [DOI] [PubMed] [Google Scholar]
- Paitel E, Sunyach C, da Costa C Alves, Bourdon JC, Vincent B, Checler F. Primary cultured neurons devoid of cellular prion display lower responsiveness to staurosporine through the control of p53 at both transcriptional and post-transcriptional levels. J Biol Chem. 2004;279:612–618. doi: 10.1074/jbc.M310453200. [DOI] [PubMed] [Google Scholar]
- Paladino S, Lebreton S, Tivodar S, Campana V, Tempre R, Zurzolo C. Different GPI-attachment signals affect the oligomerisation of GPI-anchored proteins and their apical sorting. J Cell Sci. 2008;121:4001–4007. doi: 10.1242/jcs.036038. [DOI] [PubMed] [Google Scholar]
- Pan T, Wong BS, Liu T, Li R, Petersen RB, Sy MS. Cell-surface prion protein interacts with glycosaminoglycans. Biochem J. 2002;368:81–90. doi: 10.1042/BJ20020773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris K, Keller G, Chimenti S, Amantea A, Kerl H, Hofler H. Microsatellite instability and loss of heterozygosity in melanoma. J Invest Dermatol. 1995;105:625–628. doi: 10.1111/1523-1747.ep12323809. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Nat Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid D, Chuderland D, Landsman L, Lavie Y, Reich R, Liscovitch M. Filamin A is a novel caveolin-1-dependent target in IGF-I-stimulated cancer cell migration. Exp Cell Res. 2008;314:2762–2773. doi: 10.1016/j.yexcr.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Robertson SP. Filamin A: phenotypic diversity. Curr Opin Genet Dev. 2005;15:301–307. doi: 10.1016/j.gde.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- Saito S, Ghosh M, Morita K, Hirano T, Miwa M, Todoroki T. The genetic differences between gallbladder and bile duct cancer cell lines. Oncol Rep. 2006;16:949–956. [PubMed] [Google Scholar]
- Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Masuda Y, Ohta Y, Ikeda K, Watanabe K. Filamin associates with Smads and regulates transforming growth factor-beta signaling. J Biol Chem. 2001;276:17871–17877. doi: 10.1074/jbc.M008422200. [DOI] [PubMed] [Google Scholar]
- Schmitt-Ulms G, Hansen K, Liu J, Cowdrey C, Yang J, DeArmond SJ, et al. Time-controlled transcardiac perfusion cross-linking for the study of protein interactions in complex tissues. Nat Biotechnol. 2004;22:724–731. doi: 10.1038/nbt969. [DOI] [PubMed] [Google Scholar]
- Schmitt-Ulms G, Legname G, Baldwin MA, Ball HL, Bradon N, Bosque PJ, et al. Binding of neural cell adhesion molecules (N-CAMs) to the cellular prion protein. J Mol Biol. 2001;314:1209–1225. doi: 10.1006/jmbi.2000.5183. [DOI] [PubMed] [Google Scholar]
- Scotto L, Narayan G, Nandula SV, Arias-Pulido H, Subramaniyam S, Schneider A, et al. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: potential role in progression. Genes Chromosomes Cancer. 2008;47:755–765. doi: 10.1002/gcc.20577. [DOI] [PubMed] [Google Scholar]
- Screaton RA, DeMarte L, Draber P, Stanners CP. The specificity for the differentiation blocking activity of carcinoembryonic antigen resides in its glycophosphatidyl-inositol anchor. J Cell Biol. 2000;150:613–626. doi: 10.1083/jcb.150.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seck T, Baron R, Horne WC. Binding of filamin to the C-terminal tail of the calcitonin receptor controls recycling. J Biol Chem. 2003;278:10408–10416. doi: 10.1074/jbc.M209655200. [DOI] [PubMed] [Google Scholar]
- Selvarajah S, Yoshimoto M, Ludkovski O, Park PC, Bayani J, Thorner P, et al. Genomic signatures of chromosomal instability and osteosarcoma progression detected by high resolution array CGH and interphase FISH. Cytogenet Genome Res. 2008;122:5–15. doi: 10.1159/000151310. [DOI] [PubMed] [Google Scholar]
- Sharma CP, Ezzell RM, Arnaout MA. Direct interaction of filamin (ABP-280) with the beta 2-integrin subunit CD18. J Immunol. 1995;154:3461–3470. [PubMed] [Google Scholar]
- Sharom FJ, Lehto MT. Glycosylphosphatidylinositol-anchored proteins: structure, function, and cleavage by phosphatidylinositol-specific phospholipase C. Biochem Cell Biol. 2002;80:535–549. doi: 10.1139/o02-146. [DOI] [PubMed] [Google Scholar]
- Shi C, Daniels JA, Hruban RH. Molecular characterization of pancreatic neoplasms. Adv Anat Pathol. 2008;15:185–195. doi: 10.1097/PAP.0b013e31817bf57d. [DOI] [PubMed] [Google Scholar]
- Shivapurkar N, Sood S, Wistuba II, Virmani AK, Maitra A, Milchgrub S, et al. Multiple regions of chromosome 4 demonstrating allelic losses in breast carcinomas. Cancer Res. 1999;59:3576–3580. [PubMed] [Google Scholar]
- Sibley K, Bell S, Knowles MA. Redefining a critical region of LOH on 4p16.3 in bladder cancer. Genes Chromosomes Cancer. 2000;29:378–379. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1045>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Simoneau S, Haik S, Leucht C, Dormont D, Deslys JP, Weiss S, et al. Different isoforms of the non-integrin laminin receptor are present in mouse brain and bind PrP. Biol Chem. 2003;384:243–246. doi: 10.1515/BC.2003.027. [DOI] [PubMed] [Google Scholar]
- Skawran B, Steinemann D, Weigmann A, Flemming P, Becker T, Flik J, et al. Gene expression profiling in hepatocellular carcinoma: upregulation of genes in amplified chromosome regions. Mod Pathol. 2008;21:505–516. doi: 10.1038/modpathol.3800998. [DOI] [PubMed] [Google Scholar]
- Spielhaupter C, Schatzl HM. PrPC directly interacts with proteins involved in signaling pathways. J Biol Chem. 2001;276:44604–44612. doi: 10.1074/jbc.M103289200. [DOI] [PubMed] [Google Scholar]
- Stanford JL, FitzGerald LM, McDonnell SK, Carlson EE, McIntosh LM, Deutsch K, et al. Dense genome-wide SNP linkage scan in 301 hereditary prostate cancer families identifies multiple regions with suggestive evidence for linkage. Hum Mol Genet. 2009;18:1839–1848. doi: 10.1093/hmg/ddp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele AD, Zhou Z, Jackson WS, Zhu C, Auluck P, Moskowitz MA, et al. Context dependent neuroprotective properties of prion protein (PrP) Prion. 2009;3:240–249. doi: 10.4161/pri.3.4.10135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- Sun J, Purcell L, Gao Z, Isaacs SD, Wiley KE, Hsu FC, et al. Association between sequence variants at 17q12 and 17q24.3 and prostate cancer risk in European and African Americans. Prostate. 2008;68:691–697. doi: 10.1002/pros.20754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Sverdlov M, Shinin V, Place AT, Castellon M, Minshall RD. Filamin A regulates caveolae internalization and trafficking in endothelial cells. Mol Biol Cell. 2009;20:4531–4540. doi: 10.1091/mbc.E08-10-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Hooper NM. The prion protein and lipid rafts. Mol Membr Biol. 2006;23:89–99. doi: 10.1080/09687860500449994. [DOI] [PubMed] [Google Scholar]
- Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelin WR, Chen Y, Gentzsch M, Kreda SM, Sallee JL, Scarlett CO, et al. Direct interaction with filamins modulates the stability and plasma membrane expression of CFTR. J Clin Invest. 2007;117:364–374. doi: 10.1172/JCI30376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomassen M, Tan Q, Kruse TA. Gene expression meta-analysis identifies chromosomal regions and candidate genes involved in breast cancer metastasis. Breast Cancer Res Treat. 2009;113:239–249. doi: 10.1007/s10549-008-9927-2. [DOI] [PubMed] [Google Scholar]
- Tigges U, Koch B, Wissing J, Jockusch BM, Ziegler WH. The F-actin cross-linking and focal adhesion protein filamin A is a ligand and in vivo substrate for protein kinase C alpha. J Biol Chem. 2003;278:23561–23569. doi: 10.1074/jbc.M302302200. [DOI] [PubMed] [Google Scholar]
- Tomar A, Schlaepfer DD. Focal adhesion kinase: switching between GAPs and GEFs in the regulation of cell motility. Curr Opin Cell Biol. 2009;21:676–683. doi: 10.1016/j.ceb.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torring N, Borre M, Sorensen KD, Andersen CL, Wiuf C, Orntoft TF. Genome-wide analysis of allelic imbalance in prostate cancer using the Affymetrix 50 K SNP mapping array. Br J Cancer. 2007;96:499–506. doi: 10.1038/sj.bjc.6603476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Li F, Adam L, Nguyen D, Ohta Y, Stossel TP, et al. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat Cell Biol. 2002;4:681–690. doi: 10.1038/ncb838. [DOI] [PubMed] [Google Scholar]
- van Dartel M, Cornelissen PW, Redeker S, Tarkkanen M, Knuutila S, Hogendoorn PC, et al. Amplification of 17p11.2 approximately p12, including PMP22, TOP3A, and MAPK7, in high-grade osteosarcoma. Cancer Genet Cytogenet. 2002;139:91–96. doi: 10.1016/s0165-4608(02)00627-1. [DOI] [PubMed] [Google Scholar]
- van Duin M, van Marion R, Vissers KJ, Hop WC, Dinjens WN, Tilanus HW, et al. High-resolution array comparative genomic hybridization of chromosome 8q: evaluation of putative progression markers for gastroesophageal junction adenocarcinomas. Cytogenet Genome Res. 2007;118:130–137. doi: 10.1159/000108293. [DOI] [PubMed] [Google Scholar]
- Vanden Bempt I, Vanhentenrijk V, Drijkoningen M, De Wolf-Peeters C. Comparative expressed sequence hybridization reveals differential gene expression in morphological breast cancer subtypes. J Pathol. 2006;208:486–494. doi: 10.1002/path.1911. [DOI] [PubMed] [Google Scholar]
- Waneck GL, Stein ME, Flavell RA. Conversion of a PI-anchored protein to an integral membrane protein by a single amino acid mutation. Science. 1988;241:697–699. doi: 10.1126/science.3399901. [DOI] [PubMed] [Google Scholar]
- Wang G, Zhao Y, Liu X, Wang L, Wu C, Zhang W, et al. Allelic loss and gain, but not genomic instability, as the major somatic mutation in primary hepatocellular carcinoma. Genes Chromosomes Cancer. 2001;31:221–227. doi: 10.1002/gcc.1138. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kreisberg JI, Bedolla RG, Mikhailova M, deVere White RW, Ghosh PM. A 90 kDa fragment of filamin A promotes Casodex-induced growth inhibition in Casodex-resistant androgen receptor positive C4-2 prostate cancer cells. Oncogene. 2007;26:6061–6070. doi: 10.1038/sj.onc.1210435. [DOI] [PubMed] [Google Scholar]
- Wescott MP, Rustgi AK. Pancreatiic cancer: translating lessons from mouse models and hereditary syndromes. Cancer Prev Res (Phila Pa) 2008;1:503–506. doi: 10.1158/1940-6207.CAPR-08-0195. [DOI] [PubMed] [Google Scholar]
- White GR, Varley JM, Heighway J. Isolation and characterization of a human homologue of the latrophilin gene from a region of 1p31.1 implicated in breast cancer. Oncogene. 1998;17:3513–3519. doi: 10.1038/sj.onc.1202487. [DOI] [PubMed] [Google Scholar]
- Williamson D, Pikovski I, Cranmer SL, Mangin P, Mistry N, Domagala T, et al. Interaction between platelet glycoprotein Ibalpha and filamin-1 is essential for glycoprotein Ib/IX receptor anchorage at high shear. J Biol Chem. 2002;277:2151–2159. doi: 10.1074/jbc.M109384200. [DOI] [PubMed] [Google Scholar]
- Wong N, Chan A, Lee SW, Lam E, To KF, Lai PB, et al. Positional mapping for amplified DNA sequences on 1q21–q22 in hepatocellular carcinoma indicates candidate genes over-expression. J Hepatol. 2003;38:298–306. doi: 10.1016/s0168-8278(02)00412-9. [DOI] [PubMed] [Google Scholar]
- Woo MS, Ohta Y, Rabinovitz I, Stossel TP, Blenis J. Ribosomal S6 kinase (RSK) regulates phosphorylation of filamin A on an important regulatory site. Mol Cell Biol. 2004;24:3025–3035. doi: 10.1128/MCB.24.7.3025-3035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Guo Z, Chatterjee A, Huang X, Rubin E, Wu F, et al. Overexpression of glycosylphosphatidylinositol (GPI) transamidase subunits phosphatidylinositol glycan class T and/or GPI anchor attachment 1 induces tumorigenesis and contributes to invasion in human breast cancer. Cancer Res. 2006;66:9829–9836. doi: 10.1158/0008-5472.CAN-06-0506. [DOI] [PubMed] [Google Scholar]
- Yan H, Bigner DD, Velculescu V, Parsons DW. Mutant metabolic enzymes are at the origin of gliomas. Cancer Res. 2009;69:9157–9159. doi: 10.1158/0008-5472.CAN-09-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Fan X, Yu S, Li C, Sy MS. Binding of recombinant but not endogenous prion protein to DNA causes DNA internalization and expression in mammalian cells. J Biol Chem. 2008;283:25446–25454. doi: 10.1074/jbc.M800814200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhou H, Jin Y, Bai J, Yu Y, Geng J, et al. Three distinct regions of allelic deletion on chromosome 17 involved in sporadic gastric cancer. Hepatogastroenterology. 2008;55:1487–1491. [PubMed] [Google Scholar]
- Yue J, Wang Q, Lu H, Brenneman M, Fan F, Shen Z. The cytoskeleton protein filamin-A is required for an efficient recombinational DNA double strand break repair. Cancer Res. 2009;69:7978–7985. doi: 10.1158/0008-5472.CAN-09-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusenko MV, Kuiper RP, Boethe T, Ljungberg B, van Kessel AG, Kovacs G. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer. 2009;9:152. doi: 10.1186/1471-2407-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanata SM, Lopes MH, Mercadante AF, Hajj GN, Chiarini LB, Nomizo R, et al. Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neuroprotection. EMBO J. 2002;21:3307–3316. doi: 10.1093/emboj/cdf325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanusso G, Liu D, Ferrari S, Hegyi I, Yin X, Aguzzi A, et al. Prion protein expression in different species: analysis with a panel of new mAbs. Proc Natl Acad Sci USA. 1998;95:8812–8816. doi: 10.1073/pnas.95.15.8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu TN, He HJ, Kole S, D’Souza T, Agarwal R, Morin PJ, et al. Filamin A-mediated down-regulation of the exchange factor Ras-GRF1 correlates with decreased matrix metalloproteinase-9 expression in human melanoma cells. J Biol Chem. 2007;282:14816–14826. doi: 10.1074/jbc.M611430200. [DOI] [PubMed] [Google Scholar]
- Zielinski B, Gratias S, Toedt G, Mendrzyk F, Stange DE, Radlwimmer B, et al. Detection of chromosomal imbalances in retinoblastoma by matrix-based comparative genomic hybridization. Genes Chromosomes Cancer. 2005;43:294–301. doi: 10.1002/gcc.20186. [DOI] [PubMed] [Google Scholar]