Abstract
In Drosophila, the global increase in transcription from the male X chromosome to compensate for its monosomy is mediated by the male-specific lethal (MSL) complex consisting of five proteins and two non-coding RNAs, roX1 and roX2. After an initial sequence-dependent recognition by the MSL complex of 150–300 high affinity sites, the spread to the majority of the X-linked genes depends on local MSL-complex concentration and active transcription. We have explored whether any additional RNA species are associated with the MSL complex. No additional roX RNA species were found, but a strong association was found between a spliced and poly-adenylated msl2 RNA and the MSL complex. Based on our results, we propose a model in which a non-chromatin-associated partial or complete MSL-complex titrates newly transcribed msl2 mRNA and thus regulates the amount of available MSL complex by feedback. This represents a novel mechanism in chromatin structure regulation.
INTRODUCTION
Dosage compensation in Drosophila provides one of the best examples of chromosome-wide gene regulation (1,2). The compensation mechanism must fulfil two requirements. It must balance the relative expression between sex chromosomes and autosomes and also equalize the transcriptional activities of the two X chromosomes in the homogametic sex with that of the single X chromosome in the heterogametic sex. Examination of X-linked and autosomal gene expression in Drosophila, Caenorhabditis elegans and mammals has revealed that X-linked genes are expressed, on average, twice as strongly as autosomal genes (3–5), implying that dosage compensation involves compensation of X chromosome expression not only between the two sexes, but also in relation to the expression of autosomes. Thus, X inactivation in mammals might be considered as a response in females to the overexpression of the X chromosome. This overexpression is required by males because they have just one X chromosome.
The male restricted 2-fold increase of X chromosome gene expression in Drosophila fulfils both compensation requirements described above. This seems to be mediated by a general buffering mechanism for all monosomic regions, resulting in an ∼1.4-fold expression increase (6–8) followed by an ∼1.35-fold expression increase restricted to the X chromosome, mediated by the MSL complex (7–10). In Drosophila melanogaster, two non-coding RNAs, roX1 and roX2, have been shown to be essential components of the dosage compensation system. Together with at least five MSL proteins, roX1 and roX2 form male-specific lethal (MSL) complexes that ‘paint’ the male X chromosome, and mediate acetylation of H4 at lysine 16 on the male X chromosome, which partly explains its subsequent hypertranscription (2,11). Recent data suggest that the MSL complex, in addition, have intrinsic properties that constrain the activation potential of MOF (males absent on the first) to end up with the required 2-fold activation (12). The prevailing model of the targeting process is that the MSL complex is recruited to a set of 150–300 high-affinity MSL recognition elements (MRE) in a sequence-depending mechanism (13). Subsequent spreading to neighboring genes with lower affinities depends on the local MSL-complex concentration (14), X chromosome location (15) and active transcription (16,17). The sequence dependence of the spreading remains poorly understood. Irrespective of differences in transcript levels of genes and the transcription requirements of different cells, a 2-fold transcriptional up-regulation is required.
MSL2 protein is the limiting component for the formation of the MSL complex. In females, translation of msl2 mRNA is inhibited by the binding of the Sex lethal (Sxl) protein in both the 5′ and the 3′-UTRs (18–20). Overexpression of msl2 is toxic to females and becomes lethal for females and toxic to males in combination with the overexpression of msl1, again highlighting the requirement for correct concentrations of the MSL complex (12,19,21,22). Furthermore, association studies suggest that MSL2 levels are tuned to match those of the other MSL-complex components (23).
Although the roX genes are important for correct targeting of the MSL complex, in roX1 roX2 double mutants escaping males are recovered (22,24,25). This contrasts with the absence of escaper males when the msl genes are mutated. The partial viability of roX1 roX2 double mutants could be explained by the presence of additional roX RNA species (26,27), even though unsaturated screens have failed to isolate such species (27,28). It has also been hypothesized that MSL complex may bind to additional RNAs produced by cryptic transcription as a mediator of spreading (26).
Here we have used RNA-immunoprecipitation (RIP) followed by tiling array analysis to determine the association between the MSL complex and RNA moieties. We found no additional ncRNA associated to the complex. However, we observed a strong association between the msl2 mRNA and a non-chromatin-associated MSL complex. These data suggest a novel mechanism in chromatin structure regulation in which a non-chromatin-associated form of a chromatin regulatory complex attracts an intrinsic rate-limiting mRNA and thus regulates the amounts of the complex available for chromatin targeting by feedback.
MATERIALS AND METHODS
Immunostaining and in situ hybridization of polytene chromosomes
Polytene chromosomes from the salivary glands of third-instar larvae were prepared and stained essentially as previously described (29). Salivary glands were fixed in 2% formaldehyde in PBS, 0.1% Triton X-100, 0.2% NP-40 for 40 s followed by 2–3 min in 50% acetic acid, 1% formaldehyde. Polytene chromosomes were squashed as previously described (30). The slides were washed for 30 min in 1× PBS, 0.1% Triton X-100, transferred to blocking solution (0.1 M maleic acid, 0.15 M NaCl, 1% Boehringer blocking reagent) and incubated for 30 min at room temperature. The slides were then incubated overnight at 4°C with primary antibody raised against MSL2 (1:300), MSL3 (1:2000) or MLE (1:2000). The slides were washed 2× 10 min in 0.1 M maleic acid, 0.15 M NaCl, 0.3% Tween-20 and blocked for 30 min. Donkey anti-rabbit and anti-goat (Jackson Laboratories) conjugated with Cy3 or FITC (diluted 1:400) were used as secondary antibodies and incubated at room temperature for 2 h. The squashes were counterstained with DAPI (1 µg/ml) and washed 2× 10 min before mounting with Vectashield (Vector). Preparations were analyzed using a Zeiss Axiophot microscope equipped with a KAPPA DX20C charge-coupled device camera. Images were assembled and merged electronically using Adobe Photoshop. For in situ hybridization, the slides were prepared as above and hybridized and washed essentially as previously described (31). The slides were hybridized against a DIG-labeled antisense RNA probe against roX1 (GH10432) or msl2 (GH22488). The DIG-labeled probe was detected using sheep anti-Digoxigenin (0.4 µg/ml, Roche) followed by donkey–anti-sheep conjugated with AlexaFluor555 (Molecular Probes, diluted 1:500) and DAPI counterstaining. roX1 roX2 double mutant males were selected as non-GFP males from a yw roX1ex6 Df(1)roX252 P[w+4Δ4.3]/ FM7i,P[w+mC ActGFP]JMR3 stock. Females and males overexpressing msl2 were selected from a w; P[w+ hsp83:msl2] msl3/TM6B stock.
Immunostaining and in situ hybridization of S2-cells
Schneider’s line 2 cells (ATCC CRL-1963) were grown as a suspension culture at 25°C in Erlenmeyer flasks to a density of 1 × 107 cells/ml in Drosophila SFM medium (Invitrogen), supplemented with 100 U/ml Penicillin G, 100 µg/ml Streptomycin sulfate and 2 mM of l-glutamine. 0.2 × 107 cells were placed on Polysine slides (VWR) and incubated at room temperature for 1 h. The cells were fixed for 10 min with 2% formaldehyde in PBS and permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature. The slides were washed twice in 70% ethanol before dehydration at 80, 95 and 99.5% ethanol and then air dried. The slides were hybridized and washed as described above using DIG-labeled antisense RNA probes against roX1 (GH10432), roX2 (GH18991), msl2 (GH22488) or CkIIβ cDNA and immunostained in parallel with anti-MSL1 (1:500).
RIP-chip
For each sample treatment condition, we used 300 ml cultured D. melanogaster Schneider’s line 2 cells. The cells were pelleted, washed twice in 100 ml 10 mM HEPES pH 7.4, 140 mM NaCl and resuspended in 50 ml of lysis buffer (20 mM HEPES pH 7.4, 3 mM MgCl2, 0.1% Triton X-100, 1 mM dithiothreitol, 0.5 mM PMSF, 10 U/ml RNasin and 0.5× complete protease inhibitor cocktail, Roche). The cells were allowed to swell on ice for 10 min and then homogenized on ice with 30 strokes of a Dounce homogenizer. The nuclei were pelleted at 2 000g for 5 min and used either as native samples (non-cross-linked) or after formaldehyde cross-linking. For the cross-linked sample (FA), the pelleted nuclei were resuspended in 50 ml lysis buffer and cross-linked by adding formaldehyde to a final concentration of 0.5% and incubated for 10 min at room temperature. The cross-linking was stopped by adding glycine (final concentration 0.125 M), the nuclei were washed once in lysis buffer and resuspended in 2 ml of sonication buffer (20 mM HEPES pH 7.4, 10% glycerol, 0.1 M NaCl, 1 mM MgCl2, 0.1% Triton X-100, 1 mM dithiothreitol, 0.5 mM PMSF, 10 U/ml RNasin and 0.5× Protease inhibitor cocktail) and sonicated using a Bioruptor (Diagenode) for 4 min at high setting (30 s ON, 30 s OFF). For the native samples the pelleted nuclei were resuspended in 2 ml sonication buffer and sonicated for 1, 3 or 6 min (N1, N3 and N6, respectively). Cellular debris was removed by centrifugation at 14 000g for 25 min at 4°C and the supernatants were used for immunoprecipitations.
For immunoprecipitation, 1–2 mg nuclear extracts were incubated with 5 µl anti-MSL2, 5 µl anti-MOF or no antibody for 45 min at 4°C, with agitation. The antibody complexes were precipitated by incubation with 75 µl of Dynabeads conjugated to protein-A (Invitrogen) for 30 min at 4°C, with agitation. The supernatant was removed and the beads were washed twice with PBS (150 mM NaCl), 0.1% Triton X-100, 32 U/ml RNasin, 0.5× Protease inhibitor cocktail and twice in PBS (300 mM NaCl), 0.1% Triton X-100, 32 U/ml RNasin, 0.5× Protease inhibitor cocktail. The cross-linking in FA samples was reversed by adding 200 µl 0.45 M LiCl to the beads and incubating for 3–4 h at 65°C. RNA was isolated using TRI-reagent (Ambion), followed by purification using an RNeasy kit (Qiagen) according to the instructions by the suppliers. The RNA samples were concentrated and reverse-transcribed into cDNA using random primers with an ImPromII first-strand synthesis kit (Promega). The single-stranded cDNA was purified with a QIAquick PCR purification Kit (Qiagen) then the purified cDNA samples were amplified using a WGA2 GenomePlex Complete whole genome amplification kit (Sigma), according to the manufacturer’s recommendations.
For tiling array analysis, amplified DNA samples were fragmented, labeled and hybridized to an Affymetrix Drosophila Genome 2.0 array. The signal intensity data were analyzed with the Affymetrix Tiling Analysis Software (v. 1.1.02) using 90 bp bandwidth and perfect match only. For absolute amount analysis (transcript profile), the bandwidth was set to zero. RNA enrichment ratios for all genes in nuclear extract N6 were calculated as the average enrichment ratio value of all probes located within exons of each gene. Total amounts of nuclear gene transcript were calculated from the corresponding input sample as the average value of all probes located within exons of each gene. Only genes with at least 10 probes within exons were included. For the comparison of our RIP profiles to MSL-complex binding profile the ChIP-chip data for MSL1, MSL3 and MOF from (32) were downloaded from the ArrayExpress database (accession number M-EXP-1508). The signal intensity data were analyzed with the Affymetrix Tiling Analysis Software (v. 1.1.02) using parameters: 300 bp bandwidth and perfect match only. The data were converted to D. melanogaster genomic release 5 using the Coordinates Converter Tool at FlyBase (33).
RIP-qPCR
Three biological replicates of native nuclear preps from Schneider’s line 2 cells (ATCC CRL-1963) were prepared as described above. The nuclei were washed once in lysis buffer and fractionation into nucleoplasm and chromatin was done essentially as described in (34). The pelleted nuclei were resuspended in 2 ml nuclear lysis buffer (20 mM HEPES pH 7.9, 3 mM EDTA, 10% glycerol, 150 mM KAc, 1.5 mM MgCl2, 0.1% Triton X-100, 1 mM dithiothreitol, 0.5 mM PMSF, 20 U/ml RNasin and 1 × Protease inhibitor cocktail) and centrifuged at 14 000g for 30 min at 4°C. The supernatant was collected as the nucleoplasmic fraction. The pelleted chromatin was resuspended in 2.8 ml of nuclear lysis buffer and solubilized by sonication for 6 min at high setting (30 s ON, 30 s OFF). Cellular debris was removed by centrifugation at 14 000g for 20 min at 4°C and the supernatants were collected as the chromatin fractions.
For immunoprecipitation, 5–10 mg nucleoplasmic and corresponding cytoplasmic extracts, respectively, were incubated with 5 µl anti-MSL2 (replicate A) or 2 µl anti-MSL2 serum (replicate B and C) for 45 min at 4°C with agitation. The antibody complexes were then precipitated as described above. RNA was isolated using TRI-reagent (Ambion), followed by DNase treatment using TURBO DNA-free kit (Ambion) and purification using an RNeasy kit (Qiagen) according to the manufacturer’s instruction. For replicate C, poly(A)+ RNA was isolated using Dynabeads Oligo (dT)25 (Invitrogen) in line with the instruction by the suppliers. The RNA samples were reverse-transcribed into cDNA using iScript cDNA synthesis kit (Bio-Rad).
The RIP cDNA and the corresponding total cDNA were quantified by real-time PCR using SYBR green supermix (Bio-Rad). The primer pairs used were: roX1 (5′-TACCGCTCTCTTTCGGGACTTG-3′, 5′-TCCATCACTCTCTATCGGGCTG-3′), roX2 (5′-GGCCATCGAAAGGGTAAATTG-3′, 5′-ACTGTCCGTAAGACAATTCAAC-3′), msl2 (5′-CTGGACACGAATAGTGAAGCC-3′, 5′-TTGCAGCAATCCCAGCATC-3′), actin (5′-CAGCCAGCAGTCGTCTAATC-3′, 5′-ACAACCAGAGCAGCAACTTC-3′) and Rpl32 (5′-CGATGTTGGGCATCAGATAC-3′, 5′-CCCAAGATCGTGAAGAAGC-3′). The relative enrichment levels were calculated in relation to actin cDNA in each replicate.
RESULTS
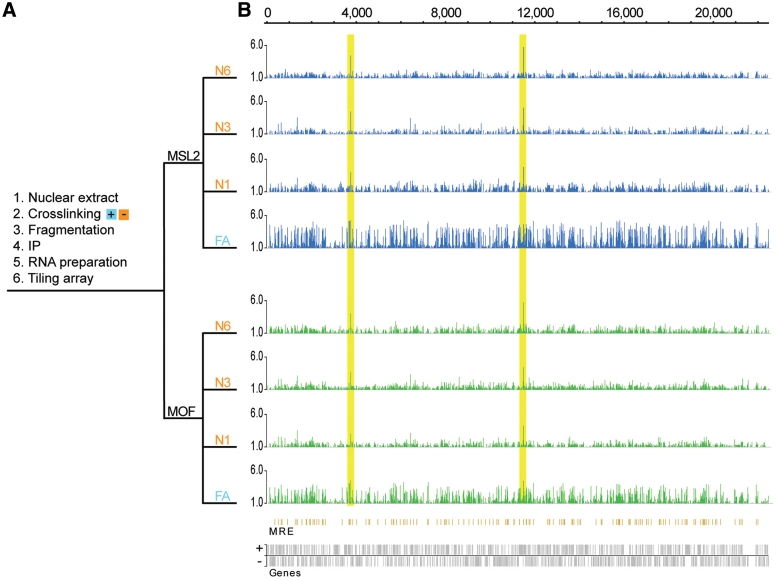
To determine the potential association of the MSL complex with RNA moieties in an unbiased procedure, we performed RIP experiments on nuclear extracts from S2 cells, with and without cross-linking, followed by tiling-array analysis (RIP-chip) (Figure 1). For the RIP experiment, we chose MSL2 as a representative of the MSL-complex scaffold (consisting of MSL1 and MSL2) required for the complex to bind DNA and MOF as a representative of the complex members required for spreading (1,35).
Figure 1.
MSL-complex associates with roX1 and roX2 RNAs transcribed from the X chromosome. (A) Schematic outline of the RIP method. N1, N3 and N6 correspond to native nuclear extracts sonicated for 1, 3 and 6 min, respectively. FA corresponds to formaldehyde-cross-linked nuclear extract. (B) The tiling array results are computed as the ratio between the RIP value and the value of the corresponding nuclear input RNA preparation; MSL2-RIPs (blue), MOF-RIPs (green). Ratios calculated between the RIP values and a MOCK-RIP (instead of input) yield comparable results (data not shown). The plots show the mean enrichment ratios obtained using a bandwidth of 90 bp. Numbers on the x-axis denote chromosomal position along the X chromosome in kilobase. The y-axis shows the RIP enrichment as the log2-ratio. In the resulting profile, enrichments >1 (which correspond approximately to the top 1% of binding) are shown. Genes expressed from left to right are shown above the horizontal line and genes expressed in the opposite direction are shown below the line. The high peaks within the yellow boxes are roX1 and roX2, respectively. Below the enrichment plots, the MRE sites are indicated (orange) as previously characterized by (13). No significant correlation of immunoprecipitated RNAs to MREs was found.
MSL-complex associates with roX1 and roX2 RNAs from the X chromosome
We first investigated the enrichment of RNAs transcribed from the X chromosome in the MSL2- and MOF-RIPs. We found that, as predicted, roX1 and roX2 are highly enriched in the complex (Figure 1). We found no evidence of any additional X-linked non-coding RNA associated with the MSL complex when roX1 and roX2 are present (Figure 1). Neither did we find any evidence that the MSL-complex attracts any cryptic RNA species produced at the MREs. The overall enrichment levels of the X chromosome and the autosomes are comparable (Supplementary Figure S1). In the formaldehyde fixed samples (FA), X chromosome transcripts in general were enriched compared with transcript on other chromosomes. We therefore compared our RIP-chip data with available ChIP-chip data representing the MSL-complex binding profile. The results showed that the RIP from the native nuclear extract is highly enriched in roX RNAs while the RIP experiment from the formaldehyde cross-linked nuclear extract is in addition enriched in transcripts from genes targeted by the MSL complex (Figure 2A–C). This is probably a consequence of cross-linked chromatin fragments including RNA moieties linked by active transcription.
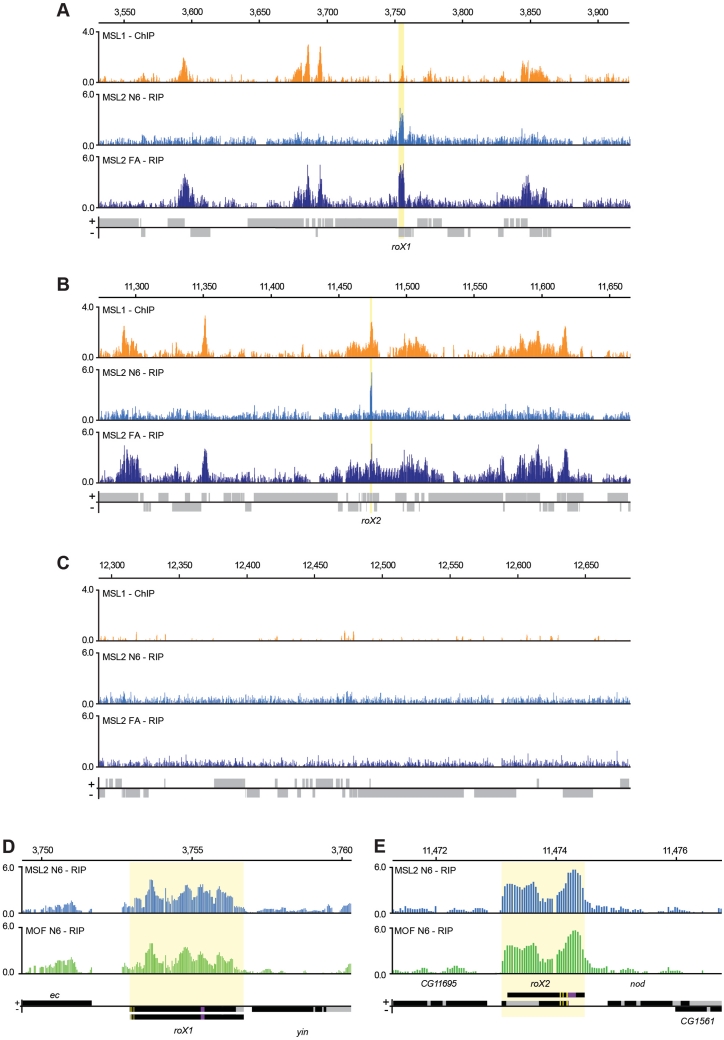
Figure 2.
Entire roX1 and roX2 mRNAs are associated with a complete or partial MSL complex. (A–C) In formaldehyde cross-linked extracts, actively transcribed RNAs are immunoprecipitated by chromatin-associated proteins. Comparison of a ChIP-chip profile representing the MSL complex (MSL1-ChIP) and the MSL2 RIP-chip profiles from the native nuclear extract (MSL2 N6-RIP) and formaldehyde-cross-linked nuclear extract (MSL2 FA-RIP). Representative 400 kb regions from the X chromosome including roX1 (A) and roX2 (B) are compared to a representative 400 kb region from chromosome 3 R (C). The plots show the mean enrichment ratios obtained using a bandwidth of 300 bp for the MSL1 ChIP-chip and a bandwidth of 90 bp for the MSL2 RIP-chip. Numbers on the x-axis denote chromosomal position along the chromosome in kb. The y-axis shows the ChIP and RIP enrichments, respectively, as the log2 ratio. Genes expressed from left to right are shown above the horizontal line and genes expressed in the opposite direction are shown below the line. The roX1 and roX2 loci are indicated by yellow boxes. The MSL1 ChIP data is from (32). (D and E) High resolution enrichment profiles of MSL2-RIP and MOF-RIP at the rox1 (D) or roX2 (E) locus. Exons are indicated in black and introns in grey. The different transcript forms of roX1 and roX2 are indicated. The described DNAse hypersensitive regions and roX-boxes suggested to be of functional importance (36,51–53) are indicated as deep-purple and yellow boxes, respectively. We observed a general enrichment of the entire mRNA and no obvious specificity to parts of the RNAs.
A detailed analysis of the roX1- and roX2-precipitated RNAs shows that the entire roX1 and roX2 RNAs are immunoprecipitated by antibodies against both MOF and MSL2 (Figure 2D–E and Supplementary Figure S2). A higher enrichment of roX2 compared to roX1 was observed in all conditions except in the formaldehyde cross-linked sample, suggesting that roX2 is more stably bound to the complex in S2 cells.
The MSL complex is strongly associated with nuclear msl2 mRNA
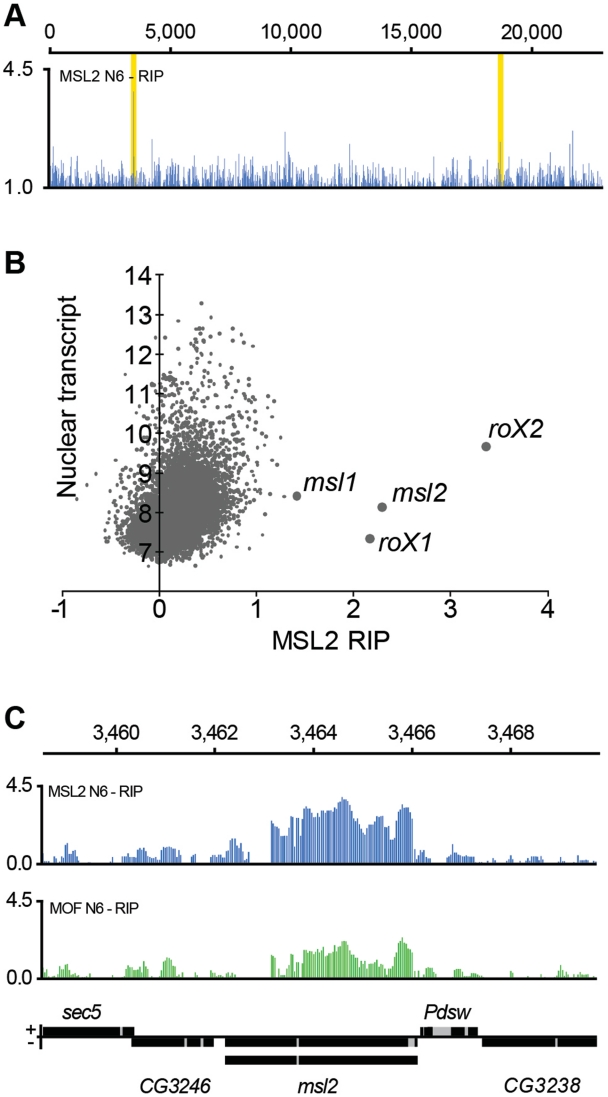
It is well established that the roX genes can exert their activity in trans as well as in cis (24). We, therefore, asked whether any autosomal RNA has a stable association with the MSL complex. Visual inspection of enrichment curves readily identified a second target with a comparable enrichment ratio to roX1 and roX2, namely the msl2 transcript itself (Figure 3A). A ranking of all nuclear transcripts according to their mean enrichment ratio showed that msl2 binding is indeed comparable with the roX enrichments. Furthermore, msl1 also scored highly in this ranking, although much lower than roX1, roX2 and msl2 (Figure 3B and Supplementary Figure S3). It should be stressed that not only the enrichment ratio but also the absolute amount of msl2 RNA pulled down with the MSL complex are comparable to roX levels (Figure 3B and Supplementary Figure S3). We asked whether the msl2 and msl1 RNAs were pulled down by free MSL2 protein, or by MSL2 associated with the MSL complex. Notably, roX1 and roX2 RNAs are enriched at similar levels in both MSL2-RIP and MOF-RIP. In contrast, msl2 RNA is enriched in both RIPs but at higher levels in the MSL2-RIP than in the MOF-RIP (compare Figure 2D–E with Figure 3C). The results suggest that msl2 RNA is associated with a less stable MSL complex. Still, since the MOF-RIP also shows high enrichment of msl2 and msl1 RNA, we conclude that msl2 and msl1 RNAs are associated with a partial or complete nuclear MSL complex (Figure 3C and Supplementary Figure S4). We have not found any significant sequence identities or predicted secondary structure similarities between msl2 and roX1 or roX2.
Figure 3.
MSL complex is strongly associated with nuclear msl2 mRNA. (A) High resolution enrichment profiling along chromosome 2 L shows that msl2 is enriched in the MSL2-RIP at similar ratios to roX1 and roX2. The high peaks within the yellow boxes correspond to msl2 (left box) and msl1 (right box). (B) Average calculated enrichment levels of genes (x-axis) plotted against the average amount of nuclear transcript (y-axis, log2 scale) for all genes with at least 10 probes within exons. roX1, roX2 and msl2 clearly ordinate as highly enriched. Note that the absolute amount of msl2 RNA pulled down is similar to the absolute amounts of the roX RNAs. (C) The entire msl2 mRNA is associated with a complete or partial MSL complex. High resolution enrichment profiles of MSL2-RIP (blue) and MOF-RIP (green) at the msl2 locus. The msl2 gene is transcribed from right to left and the different splice forms are indicated. Numbers on the x-axis denote chromosomal position along the X chromosome in kilobase. The y-axis shows the RIP enrichment as the log2 ratio.
msl2 RNA is bound by a non-chromatin-associated MSL complex
We considered two plausible models to account for a potential function for the strong association of a nuclear complete or partial MSL complex with the msl2 RNA. First, msl2 may aid the correct targeting of the MSL complex, by functioning similarly to roX. Second, since the local concentration of MSL complex is important for spreading and therefore for the correct regulation of most X-linked genes, we can hypothesize that the non-chromatin bound fraction of the MSL complex titrates the msl2 RNA. A combination of these two models is also possible. Recalling that the roX RNAs coat the male X chromosome, co-localizing with MSL complex, we performed in situ hybridization with RNA probes against roX1 and msl2. As previously shown, roX1 decorates the male X chromosome in a similar pattern to the MSL complex. However, we did not observe chromatin-associated msl2 RNA in the wild-type (Figure 4A). Previously, a co-transcriptional assembly of the MSL complex at the roX loci has been suggested (1,27,36). In contrast, the MSL complex is not found at the msl2 locus in published ChIP-chip analysis (Supplementary Figure S5). This indicates that the association of MSL complex with msl2 RNA is not co-transcriptional.
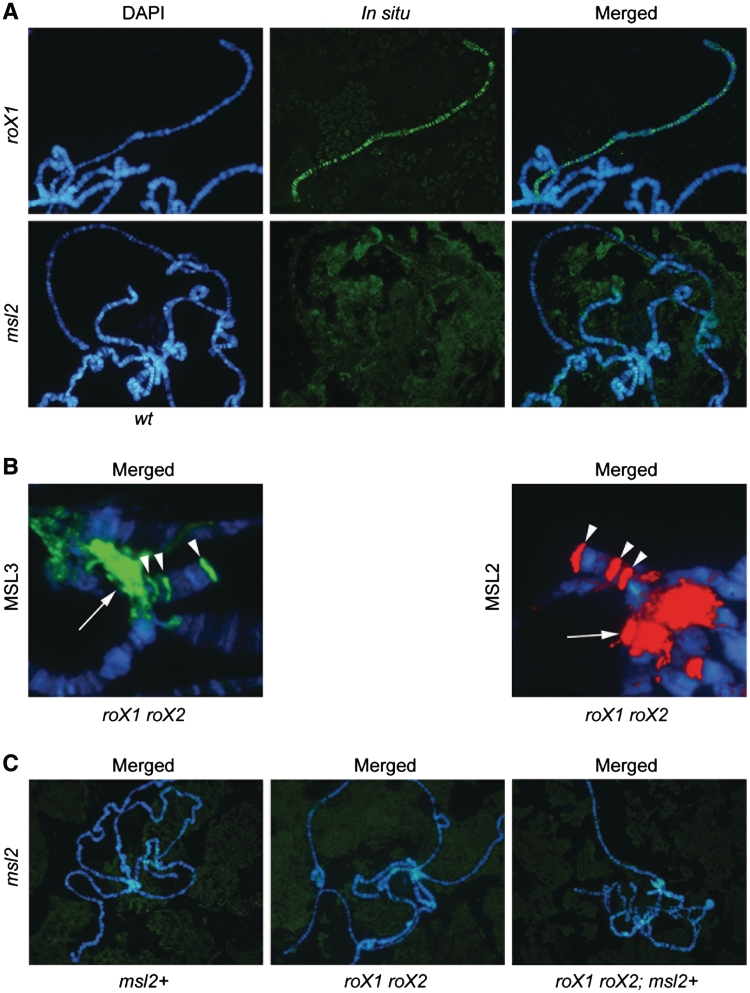
Figure 4.
In polytene chromosomes, msl2 RNA is not associated with the chromatin bound MSL complex. (A) In situ hybridization shows that roX1 decorates the male X chromosome (top row of images). No msl2 RNA associated with the wild-type male X chromosome is detected (bottom row of images). (B) In roX1 roX2 mutant males MSL2 and MSL3 target the chromocenter and three specific sites on the fourth chromosome. Merged images of immunostainings showing DAPI in blue, MSL3 in green and MSL2 in red. The chromocenter is indicated by an arrow and the specific sites on the fourth chromosome with arrowheads (Supplementary Figure S6). (C) Merged images from in situ hybridization with msl2 RNA antisense probe on males overexpressing msl2 (left panel), roX1 roX2 mutant males (middle) and roX1 roX2 mutant males overexpressing msl2 (right) (Supplementary Figure S8).
We speculated that a loss of roX1 roX2 and/or overexpression of msl2 may enhance an intrinsic property of the msl2 RNA to be included in the chromatin-associated MSL complex. In roX1 roX2 mutant males, MSL complex is still detected at a reduced number of sites on the X chromosome, but strong binding is also reproducibly found in the chromocenter and at a few specific sites on the fourth chromosome (Figure 4B and Supplementary Figure S6) (24,37). Notably, the specific sites on the fourth chromosome targeted by the MSL complex in roX1 roX2 mutants are also targeted weakly by the MLE protein alone in wild-type, both in males and females (Supplementary Figure S7). We hybridized roX1 roX2 mutant male larvae with and without overexpression of msl2 and wild-type males with overexpression of msl2. In none of these cases did we detect msl2 RNA linked to the X chromosome, in the chromocenter, or the highly specific sites on the fourth chromosome (Figure 4C and Supplementary Figure S8).
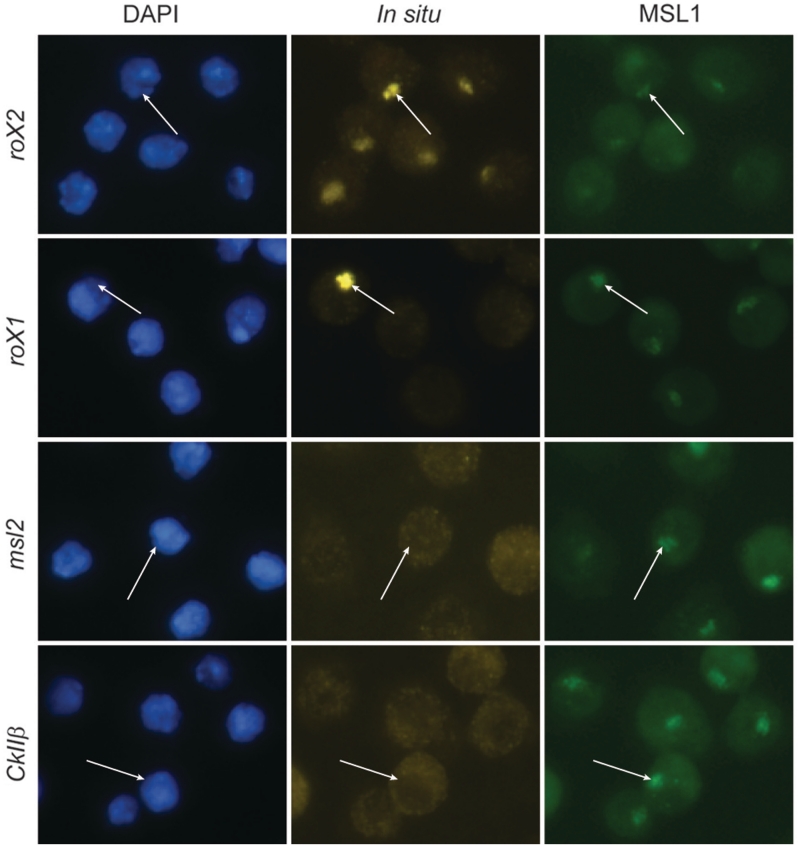
In the RIP experiments performed on S2 cells a strong enrichment of msl2 RNA in the MSL complex was detected. To test if msl2 is included in an X chromosome-associated MSL complex in S2 cells, we performed in situ hybridizations on these cells (Figure 5). As expected, roX2 RNAs co-localize with the MSL1 protein on the X chromosome. In agreement with the results from salivary glands we did not find any enrichment of msl2 RNA on the X chromosome in S2 cells, indicating that the msl2 RNA associates to a non-chromatin bound MSL complex. Strong roX2 hybridization was observed on the X chromosome territories in all cells. Interestingly, roX1 hybridization was as strong, but only detected in a small fraction of cells (<0.5%). This suggests that the observed low expression of roX1 in S2 cells is due to expression in few individual cells and not a general low expression level in all cells.
Figure 5.
In S2 cells, msl2 RNA is not associated with the chromatin bound MSL complex. Immunostaining and in situ hybridization show that roX2 (yellow) decorates the X chromosome identified by MSL1 (green). Note that roX1 decorates the X chromosome in a small fraction of cells (<0.5%). Nuclei are visualized with DAPI. Neither the msl2 RNA nor the negative control CkIIβ RNA are enriched on the X chromosome. In each row, the location of one X chromosome in a representative cell is indicated with an arrow.
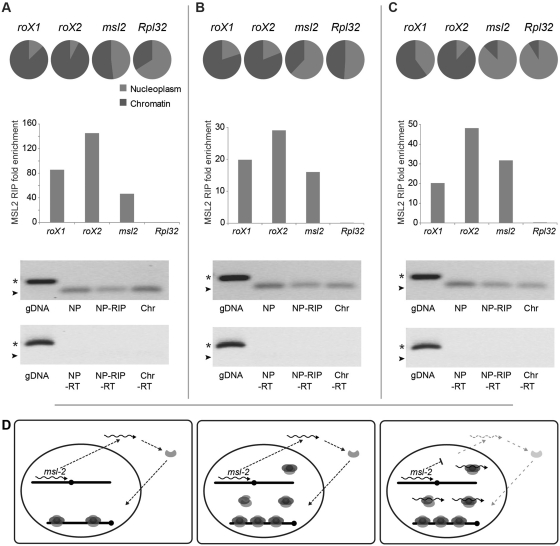
Next, we performed a crude fractionation of nuclei into a chromatin fraction and a nucleoplasmic fraction and determined the relative levels of roX1, roX2 and msl2 by qPCR. The results show that the majority of both roX1 and roX2 RNAs are located in the chromatin fraction while the msl2 transcripts are primarily found in the nucleoplasm (Figure 6A–C). MSL2-RIP experiments using the nucleoplasmic fractions show very strong enrichments of roX1, roX2 and msl2 RNA, verifying the nuclear RIP-chip data. Gel electrophoresis of the RIP experiment qPCR products shows that the MSL complex-associated msl2 RNAs are spliced. The results also show that the msl2 RNA associated to the MSL complex is poly-adenylated, since msl2 RNA is enriched to the same extent when using poly-adenylated RNA (Figure 6C) as when using total RNA (Figure 6A–B) in the analysis. To exclude that the enrichment of msl2 RNA in the nucleoplasm is caused by contaminating cytoplasm, we performed MSL2-RIP also on corresponding cytoplasmic fractions. Weak enrichments of roX and msl2 RNAs were detected in the cytoplasm but much lower than the enrichments found in corresponding nucleoplasm (Supplementary Figure S9). We conclude that a spliced poly-adenylated msl2 RNA associates with a nucleoplasmic complete or partial MSL complex.
Figure 6.
Spliced and poly-adenylated msl2 RNA is associated to a non-chromatin bound MSL complex. (A–C) Fractionation and RIP experiments of three biological replicates using total RNA extraction (A and B) and extracted poly(A)+ RNA (C). The circle diagrams specify the relative proportion of the RNAs in the nucleoplasm (gray) and the chromatin fractions (dark gray). The majority of the roX1 and roX2 RNAs are located in the chromatin fraction while msl2 and the negative control Rpl32 are mainly found within the nucleoplasm. As illustrated in the histograms, enrichment of msl2 RNA in the MSL complex is confirmed by RIP experiments on the nucleoplasmic fractions using MSL2 antibodies. Displayed are the fold enrichments relative to actin RNA. Immunoprecipitated msl2 in experiments A, B and C correspond to 3, 4 and 10% of input, respectively. Gel-electrophoretic analyses of amplified genomic DNA (gDNA), immunoprecipitated (NP RIP) and total RNA from the nucleoplasm (NP) and from chromatin (Chr) fractions show that msl2 RNAs associated to the MSL complex are spliced. The PCR product sizes of the spliced msl2 amplicon and the unspliced amplicon are indicated by arrowheads and stars, respectively. The minus reverse transcriptase controls (-RT) are shown below. (D) Model for feedback regulation of MSL complex via the association of a non-chromatin MSL complex with the msl2 mRNA. When free nuclear MSL complex is present this non-chromatin MSL-complex titrates msl2 RNA and thus reduces the amount of msl2 transcript available for export and translation, which in turn regulates the complex production as a feedback mechanism.
DISCUSSION
For many chromatin-associated factors, the relative concentrations direct their binding. Thus, correct targeting will strictly depend on the concentration of these protein complexes. It is well established that the MSL complex is restricted to males by a translational block caused by Sxl protein binding to the msl2 RNAs. However, it has been suggested that Sxl binds not only all msl2 transcripts, but also to a subset of msl1 transcripts and prevents their translation in females (19). This reflects the potential of these two RNAs to be regulated through specific protein interactions. These two RNAs encode the core of the dosage compensation complex. Here, we provide evidence that a non-chromatin bound fraction of the MSL complex has high affinity for nuclear msl2 mRNA and also affinity for nuclear msl1 mRNA. Based on this, we propose a novel mechanism for fine-tuning the MSL complex concentration by feedback.
We have used both formaldehyde fixed nuclear extracts and native nuclear extracts in our RIP experiments. In both these sample types, we detected strong enrichments of roX1, roX2 and msl2 transcripts. In the native extracts, we could not detect significant enrichment of RNA transcribed from genes targeted by the MSL complex nor any enrichment of RNA molecules transcribed from MREs. Although, we found robust enrichment of roX1 and roX2 we did not find any additional roX RNA that could explain the escape of male lethality seen in roX1 roX2 double mutants. It should be stressed that it is still possible that a roX-like encoding gene is harbored in the heterochromatic regions which are not fully covered by the tiling arrays. Alternatively, several different RNAs may substitute for roX1 and roX2 but with much lower affinity or the MSL complex can exert its function without an RNA molecule. In contrast, in the formaldehyde fixed sample we detected enrichments of transcripts from genes targeted by the MSL complex. We conclude that RIP using formaldehyde cross-linked extracts will be enriched in RNA species directly linked to the targeted protein as well as nascent RNA molecules linked via chromatin and active transcription. This is important to consider when using formaldehyde fixation in RNA immunoprecipitation experiments.
We describe two potential functions for the strong association of a nuclear MSL complex with the msl2 RNA. First, msl2 may aid the correct targeting of the MSL complex, by functioning similarly to roX. This could have been a function in a more ancient form of the MSL complex and may in part have been retained during evolution. In favour of this model is the fact that roX1 roX2 double mutations are not completely male lethal, in contrast to msl mutations. The elevation of cellular levels of MSL1 and MSL2 that partially suppress roX1 roX2 male lethality, also supports this model. Second, since the local concentration of MSL complex is important for spreading and, therefore, also for the correct regulation of most X-linked genes, we can hypothesize that the non-chromatin bound fraction of the MSL complex titrates the msl2 RNA. Thus, this process would regulate the amount of MSL2 protein and assembled MSL complex, as a feedback mechanism.
Since the msl2 RNA is highly enriched in a nuclear but not chromatin-associated MSL complex, we hypothesize that this interaction functions as a feedback control that avoids elevated MSL-complex levels and compensates for overshooting (Figure 6D). It has been observed that ectopic expression of an msl2-GFP transgene led to reduction of the endogenous MSL2 protein levels (23). This is in line with what would be predicted from a feedback mechanism as hypothesized here. The classical example of self-mRNA targeting of proteins is from Escherichia coli, in which the ribosomal proteins bind to their mRNA and inhibit translation. The ribosome biogenesis can thus be regulated solely through the amount of rRNA which synthesis rate becomes rate limiting for ribosome assembly. As long as the process of ribosome assembly requires ribosomal proteins, the corresponding mRNAs will escape translation inhibition (38). Intrinsic association of RNA in chromatin regulation has been reported for the yeast SET1C histone methyl transferase complex. In this case, the association is linked to co-translational assembly of the complex (39).
The dosage compensation system is under strong evolutionary pressure to respond as new genes, chromosome regions or chromosome arms join the X chromosome and it has been shown that the MSL proteins are under adaptive evolution (40). Interestingly, strong positive selection was detected in MSL1 and MSL2 protein domains shown to be responsible for their specific targeting to the X chromosome (41,42). Positive selection has also been shown to act on the MSL binding sites (43). This means that the dosage compensation system is under constant pressure to adapt to emerging changes. The target sequences are under constant selection as are the targeting proteins, in particular MSL1 and MSL2. Thus, the concentration requirements will also need constant adjustments. A feedback module based on association of a rate limiting mRNA would provide an optimal target for evolution to act on in order to provide a dynamic fine-tuning of complex concentration, and therefore, correct targeting. This may explain the affinity of the MSL complex to both msl1 and msl2 RNAs which encode the core proteins for a functional complex. At limiting concentrations of freely diffusible proteins and protein complexes only the sites with highest affinity will bind while at high concentrations even sites with low affinity will bind. It should be stressed that chromatin-associated factors are often described to be highly sensitive to correct dose (44,45). This is observed for the Polycomb-group proteins involved in maintaining repression of homeotic genes (46,47) as well as for suppressors of variegation, important for heterochromatin formation (48–50). In both these cases many loci exhibit dosage effects, indicating strict dosage needs. As more RNA immunoprecipitation data are produced it is likely that the msl2 titration reported here is just one example of a general self-affinity module important for feedback regulation.
ACCESSION NUMBER
The microarray data reported in this article have been deposited at http://www.ncbi.nlm.nih.gov/geo/ (Accession: GSE20249).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Carl Tryggers and Philip Sörenssen Foundations (to P.S.); the Kempe Foundations (A.M.J. and A.A.); and the Swedish Research Council and Magnus Bergvalls Foundation (J.L.). Funding for open access charge: The Swedish Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Kuroda for antibodies, Y. Park and V. Meller for fly stocks, K. Ekström for technical assistance and members of the Larsson and Stenberg labs for stimulating discussions.
REFERENCES
- 1.Gelbart ME, Kuroda MI. Drosophila dosage compensation: a complex voyage to the X chromosome. Development. 2009;136:1399–1410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsson J, Meller VH. Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res. 2006;14:417–431. doi: 10.1007/s10577-006-1064-3. [DOI] [PubMed] [Google Scholar]
- 3.Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, Malley JD, Eastman PS, Oliver B. Global analysis of X-chromosome dosage compensation. J. Biol. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Oliver B. Dosage compensation goes global. Curr. Opin. Genet. Dev. 2007;17:113–120. doi: 10.1016/j.gde.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Stenberg P, Lundberg LE, Johansson AM, Rydén P, Svensson MJ, Larsson J. Buffering of segmental and chromosomal aneuploidies in Drosophila melanogaster. PLoS Genet. 2009;5:e100302. doi: 10.1371/journal.pgen.1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, Macalpine DM, Oliver B. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 2010;8:e1000320. doi: 10.1371/journal.pbio.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prestel M, Feller C, Becker PB. Dosage compensation and the global re-balancing of aneuploid genomes. Genome Biol. 2010;11:216. doi: 10.1186/gb-2010-11-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamada FN, Park PJ, Gordadze PR, Kuroda MI. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 2005;19:2289–2294. doi: 10.1101/gad.1343705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng X, Koya SK, Kong Y, Meller VH. Coordinated regulation of heterochromatic genes in Drosophila melanogaster males. Genetics. 2009;182:481–491. doi: 10.1534/genetics.109.102087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Straub T, Becker PB. Dosage compensation: the beginning and end of generalization. Nat. Rev. Genet. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- 12.Prestel M, Feller C, Straub T, Mitlöhner H, Becker PB. The activation potential of MOF is constrained for dosage compensation. Mol. Cell. 2010;38:815–826. doi: 10.1016/j.molcel.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 13.Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahlsveen IK, Gilfillan GD, Shelest VI, Lamm R, Becker PB. Targeting determinants of dosage compensation in Drosophila. PLoS Genet. 2006;2:e5. doi: 10.1371/journal.pgen.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorchakov AA, Alekseyenko AA, Kharchenko P, Park PJ, Kuroda MI. Long-range spreading of dosage compensation in Drosophila captures transcribed autosomal genes inserted on X. Genes Dev. 2009;23:2266–2271. doi: 10.1101/gad.1840409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol. Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Sass GL, Pannuti A, Lucchesi JC. Male-specific lethal complex of Drosophila targets activated regions of the X chromosome for chromatin remodeling. Proc. Natl Acad. Sci. USA. 2003;100:8287–8291. doi: 10.1073/pnas.1332749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bashaw GJ, Baker BS. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell. 1997;89:789–798. doi: 10.1016/s0092-8674(00)80262-7. [DOI] [PubMed] [Google Scholar]
- 19.Kelley RL, Solovyeva I, Lyman LM, Richman R, Solovyev V, Kuroda MI. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell. 1995;81:867–877. doi: 10.1016/0092-8674(95)90007-1. [DOI] [PubMed] [Google Scholar]
- 20.Kelley RL, Wang J, Bell L, Kuroda MI. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature. 1997;387:195–199. doi: 10.1038/387195a0. [DOI] [PubMed] [Google Scholar]
- 21.Chang KA, Kuroda MI. Modulation of MSL1 abundance in female Drosophila contributes to the sex specificity of dosage compensation. Genetics. 1998;150:699–709. doi: 10.1093/genetics/150.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng X, Rattner BP, Souter S, Meller VH. The severity of roX1 mutations is predicted by MSL localization on the X chromosome. Mech. Dev. 2005;122:1094–1105. doi: 10.1016/j.mod.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Straub T, Neumann MF, Prestel M, Kremmer E, Kaether C, Haass C, Becker PB. Stable chromosomal association of MSL2 defines a dosage-compensated nuclear compartment. Chromosoma. 2005;114:352–364. doi: 10.1007/s00412-005-0020-x. [DOI] [PubMed] [Google Scholar]
- 24.Meller VH, Rattner BP. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 2002;21:1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menon DU, Meller VH. Imprinting of the Y chromosome influences dosage compensation in roX1 roX2 Drosophila melanogaster. Genetics. 2009;183:811–820. doi: 10.1534/genetics.109.107219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilik I, Akhtar A. roX RNAs: non-coding regulators of the male X chromosome in flies. RNA Biol. 2009;6:113–121. doi: 10.4161/rna.6.2.8060. [DOI] [PubMed] [Google Scholar]
- 27.Oh H, Park Y, Kuroda MI. Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes Dev. 2003;17:1334–1339. doi: 10.1101/gad.1082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujii S, Amrein H. Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J. 2002;21:5353–5363. doi: 10.1093/emboj/cdf556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsson J, Chen JD, Rasheva V, Rasmuson Lestander A, Pirrotta V. Painting of fourth, a chromosome-specific protein in Drosophila. Proc. Natl Acad. Sci. USA. 2001;98:6273–6278. doi: 10.1073/pnas.111581298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White RAH. Immunolabelling of Drosophila. In: Roberts DB, editor. Drosophila, a practical approach. Oxford: IRL Press; 1998. pp. 215–240. [Google Scholar]
- 31.Meller VH, Gordadze PR, Park Y, Chu X, Stuckenholz C, Kelley RL, Kuroda MI. Ordered assembly of roX RNAs into MSL complexes on the dosage-compensated X chromosome in Drosophila. Curr. Biol. 2000;10:136–143. doi: 10.1016/s0960-9822(00)00311-0. [DOI] [PubMed] [Google Scholar]
- 32.Kind J, Vaquerizas JM, Gebhardt P, Gentzel M, Luscombe NM, Bertone P, Akhtar A. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–828. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 33.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aygün O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl Acad. Sci. USA. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu. Rev. Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- 36.Kelley RL, Lee OK, Shim YK. Transcription rate of noncoding roX1 RNA controls local spreading of the Drosophila MSL chromatin remodeling complex. Mech. Dev. 2008;125:1009–1019. doi: 10.1016/j.mod.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng X, Meller VH. roX RNAs are required for increased expression of X-linked genes in Drosophila melanogaster males. Genetics. 2006;174:1859–1866. doi: 10.1534/genetics.106.064568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 39.Halbach A, Zhang H, Wengi A, Jablonska Z, Gruber IM, Halbeisen RE, Dehé PM, Kemmeren P, Holstege F, Géli V, et al. Cotranslational assembly of the yeast SET1C histone methyltransferase complex. EMBO J. 2009;28:2959–2970. doi: 10.1038/emboj.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachtrog D, Jensen JD, Zhang Z. Accelerated adaptive evolution on a newly formed X chromosome. PLoS Biol. 2009;7:e82. doi: 10.1371/journal.pbio.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levine MT, Holloway AK, Arshad U, Begun DJ. Pervasive and largely lineage-specific adaptive protein evolution in the dosage compensation complex of Drosophila melanogaster. Genetics. 2007;177:1959–1962. doi: 10.1534/genetics.107.079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez MA, Vermaak D, Bayes JJ, Malik HS. Species-specific positive selection of the male-specific lethal complex that participates in dosage compensation in Drosophila. Proc. Natl Acad. Sci. USA. 2007;104:15412–15417. doi: 10.1073/pnas.0707445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachtrog D. Positive selection at the binding sites of the male-specific lethal complex involved in dosage compensation in Drosophila. Genetics. 2008;180:1123–1129. doi: 10.1534/genetics.107.084244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veitia RA. Exploring the etiology of haploinsufficiency. BioEssays. 2002;24:175–184. doi: 10.1002/bies.10023. [DOI] [PubMed] [Google Scholar]
- 45.Edger PP, Pires JC. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009;17:699–717. doi: 10.1007/s10577-009-9055-9. [DOI] [PubMed] [Google Scholar]
- 46.Kennison JA, Russell MA. Dosage-dependent modifiers of homoeotic mutations in Drosophila melanogaster. Genetics. 1987;116:75–86. doi: 10.1093/genetics/116.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campbell RB, Sinclair DA, Couling M, Brock HW. Genetic interactions and dosage effects of Polycomb group genes of Drosophila. Mol. Gen. Genet. 1995;246:291–300. doi: 10.1007/BF00288601. [DOI] [PubMed] [Google Scholar]
- 48.Henikoff S. Dosage-dependent modification of position-effect variegation in Drosophila. BioEssays. 1996;18:401–409. doi: 10.1002/bies.950180510. [DOI] [PubMed] [Google Scholar]
- 49.Tartof KD, Bishop C, Jones M, Hobbs CA, Locke J. Towards an understanding of position effect variegation. Dev. Genet. 1989;10:162–176. doi: 10.1002/dvg.1020100306. [DOI] [PubMed] [Google Scholar]
- 50.Locke J, Kotarski MA, Tartof KD. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics. 1988;120:181–198. doi: 10.1093/genetics/120.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kageyama Y, Mengus G, Gilfillan G, Kennedy HG, Stuckenholz C, Kelley RL, Becker PB, Kuroda MI. Association and spreading of the Drosophila dosage compensation complex from a discrete roX1 chromatin entry site. EMBO J. 2001;20:2236–2245. doi: 10.1093/emboj/20.9.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park Y, Mengus G, Bai X, Kageyama Y, Meller VH, Becker PB, Kuroda MI. Sequence-specific targeting of Drosophila roX genes by the MSL dosage compensation complex. Mol. Cell. 2003;11:977–986. doi: 10.1016/s1097-2765(03)00147-3. [DOI] [PubMed] [Google Scholar]
- 53.Park SW, Kuroda MI, Park Y. Regulation of histone H4 Lys16 acetylation by predicted alternative secondary structures in roX noncoding RNAs. Mol. Cell. Biol. 2008;28:4952–4962. doi: 10.1128/MCB.00219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.