Abstract
Abiotic and biotic stresses are major limiting factors of crop yields and cause billions of dollars of losses annually around the world. It is hoped that understanding at the molecular level how plants respond to adverse conditions and adapt to a changing environment will help in developing plants that can better cope with stresses. Acquisition of stress tolerance requires orchestration of a multitude of biochemical and physiological changes, and most of these depend on changes in gene expression. Research during the last two decades has established that different stresses cause signal-specific changes in cellular Ca2+ level, which functions as a messenger in modulating diverse physiological processes that are important for stress adaptation. In recent years, many Ca2+ and Ca2+/calmodulin (CaM) binding transcription factors (TFs) have been identified in plants. Functional analyses of some of these TFs indicate that they play key roles in stress signaling pathways. Here, we review recent progress in this area with emphasis on the roles of Ca2+- and Ca2+/CaM-regulated transcription in stress responses. We will discuss emerging paradigms in the field, highlight the areas that need further investigation, and present some promising novel high-throughput tools to address Ca2+-regulated transcriptional networks.
The sessile nature of plants necessitates their adaptation to continuously changing and often unfavorable environmental conditions. These include many abiotic stresses that arise from an excess or deficit of water, temperature, and light in the physical environment and biotic stresses imposed by other organisms, such as bacteria, viruses, fungi, and insects (Boyer, 1982; Hadiarto and Tran, 2010; Miller et al., 2010; Winfield et al., 2010). It is estimated that hundreds of billions of dollars of crop losses around the world are due to abiotic and biotic stresses (Dhlamini et al., 2005), and much of the genetic potential for crop yield is not realized due to the effects of environmental stresses (Boyer, 1982). Plants have developed sophisticated mechanisms to perceive and respond to various stresses so that they adapt to their environment. Plants exhibit extraordinary plasticity in many of their growth and developmental processes in response to changes in their environment. Elucidation of mechanisms by which plants recognize and respond to various stresses is of great interest to plant biologists not only to elucidate fundamental principles in cellular signaling mechanisms but also to apply that knowledge to generate plants that can be grown under adverse environmental conditions. With the expected climate change during this century, understanding plant responses to changing environmental conditions is even more important. Climate change is anticipated to have many negative impacts on agriculture due to elevated temperature, salinity, unpredictable rains and floods in some places, and prolonged drought in other parts of the world (Pachauri and Reisinger, 2007; Reynolds, 2010). In recent years, considerable progress has been made in understanding the effects of stresses at the molecular level and how those changes may contribute to stress tolerance. Research in this area has uncovered several signaling pathways involving various messengers that participate in stress adaptation. Numerous studies indicate that Ca2+, a key messenger in regulating many growth and developmental processes, plays a crucial role in stress signaling. Several reviews have presented a comprehensive overview of Ca2+ role in various aspects of plant growth and development (Poovaiah and Reddy, 1993; Zielinski, 1998; Reddy, 2001; Snedden and Fromm, 2001; Sanders et al., 2002; Harper et al., 2004; Reddy and Reddy, 2004; Bouché et al., 2005; Hepler, 2005). Here, we focus primarily on the role of Ca2+- and Ca2+/calmodulin (CaM)-regulated gene expression in stress signaling. For those aspects of calcium signaling in plants that are not covered, the reader is referred to other recent reviews (Kim et al., 2009; DeFalco et al., 2010; Dodd et al., 2010; Galon et al., 2010a; Kudla et al., 2010).
CHANGES IN CELLULAR Ca2+ LEVELS IN RESPONSE TO ABIOTIC AND BIOTIC STRESS SIGNALS
Plant response to external cues can involve molecular, biochemical, physiological, and/or morphological changes, which must be balanced to achieve optimal plant growth and productivity. Signals perceived by cells are relayed by secondary messengers, such as Ca2+ ions, cyclic nucleotide monophosphates, inositol polyphosphates, nitric oxide, and other small molecules. The role of Ca2+ as one of the nutrients and as a key ion in maintaining the structural rigidity of the cell walls as well as in membrane structure and function has been known for a long time (Hepler, 2005). During the last three decades, numerous studies have shown that Ca2+ is an important messenger in eliciting responses to diverse signals, including many biotic and abiotic signals (Reddy, 2001; Hepler, 2005; McAinsh and Pittman, 2009; DeFalco et al., 2010). It appears that plants use Ca2+ as a messenger more than any other known messengers in plants and animals. This is evident from the fact that nearly all signals (developmental, hormonal, and stresses) cause changes in cellular Ca2+, primarily in the cytosol and, in some cases, in the nucleus and other organelles.
Several excellent reviews on signal-induced changes in the cytosolic Ca2+ concentration, [Ca2+]cyt, have been published recently (Lecourieux et al., 2006; Mazars et al., 2009; McAinsh and Pittman, 2009); hence, this aspect is covered only briefly here. [Ca2+]cyt is in the nanomolar range (100 to 200 nm), while in the cell wall and organelles, it is in the millimolar range (Trewavas and Malhó, 1998; Knight, 2000; Reddy, 2001). At higher concentrations, Ca2+ can chelate negatively charged molecules in the cell and hence can cause cytotoxicity. Therefore, to maintain low [Ca2+]cyt, cells actively pump Ca2+ to the apoplast or organelles. Using a variety of approaches to monitor free Ca2+ in the cytosol and other cellular compartments, it has been shown that many abiotic stresses (cold, heat, salt, drought, osmotic stress, mechanical stimuli such as touch and wind, oxidative stress, ozone, and hypoxia) rapidly elevate cellular Ca2+, mostly [Ca2+]cyt but in some cases nuclear Ca2+ ([Ca2+]nuc) or organellar Ca2+ ([Ca2+]org) (Knight et al., 1991, 1992, 1996, 1999; Biyaseheva et al., 1993; Price et al., 1994; Subbaiah et al., 1994a, 1998; Campbell et al., 1996; Levine et al., 1996; McAinsh et al., 1996; Russell et al., 1996; Taylor et al., 1996; Knight et al., 1997; Legué et al., 1997; Takahashi et al., 1997; Gong et al., 1998; Clayton et al., 1999; van Der Luit et al., 1999; Pei et al., 2000). Biotic stresses (pathogens, defense elicitors, and insect feeding) also cause changes in cellular calcium levels (Tavernier et al., 1995; Jabs et al., 1997; Zimmermann et al., 1997; Xu and Heath, 1998; Blume et al., 2000; Grant et al., 2000; Heath, 2000; Lecourieux et al., 2002; Gust et al., 2007; Maffei et al., 2007; Ranf et al., 2008; Ma et al., 2009a). Furthermore, changes in Ca2+ levels are specific to a given stress in terms of where the changes take place in the cell (e.g., cytosol, nucleus, organelles, or localized region within the cell), the magnitude and duration of Ca2+ elevation, and whether a single Ca2+ transient or multiple spikes occur, in which case the duration of spikes, the number of spikes, and the lag time between the spikes vary depending on the stimulus (Johnson et al., 1995; Allen et al., 2000, 2001; Tracy et al., 2008; Mazars et al., 2009; McAinsh and Pittman, 2009). These spatial and temporal patterns of cellular Ca2+ changes that are characteristic for a particular stimulus are termed Ca2+ signatures (Webb et al., 1996) and are thought to elicit specific and appropriate physiological responses to a given signal. For instance, cold and wind can initiate specific Ca2+ signals that are spatially and temporally distinct (van Der Luit et al., 1999). Moreover, different cell types in a tissue generate different Ca2+ signatures to a particular stimulus. Also, studies suggest that Ca2+ elevation in response to different stimuli may be generated by distinct mechanisms. Plants have developed elaborate mechanisms that involve Ca2+ channels, pumps, and exchangers (carriers), all of which control Ca2+ entry into and out of the cell and cellular compartments to maintain Ca2+ homeostasis and to bring rapid signal-specific changes in cellular Ca2+ in response to signals. Depending on the type of signal or the type of cell, internal and/or external Ca2+ stores could be involved in raising [Ca2+]cyt (Dodd et al., 2010; Kudla et al., 2010).
Signal-induced Ca2+ changes in plant nuclei have been reported (van Der Luit et al., 1999; Mazars et al., 2010) but not studied as extensively as signal-induced [Ca2+]cyt. Thus, nuclei have the potential to generate a Ca2+ signature (Xiong et al., 2004; Lecourieux et al., 2005; Mazars et al., 2009, 2010). In vitro studies with plant nuclei indicate that Ca2+ does not pass through nuclear pores passively and requires energy (Nicotera et al., 1989; Pauly et al., 2000). However, an in vivo study with animal cells indicates that Ca2+ can freely diffuse through nuclear pores at low concentrations but not above 300 nM, indicating that [Ca2+]cyt change may influence [Ca2+]nuc levels under certain condition but not others (al-Mohanna et al., 1994). Plant nuclei are also capable of generating Ca2+ changes that are not dependent on [Ca2+]cyt changes, suggesting that [Ca2+]nuc and [Ca2+]cyt levels can be regulated independently (Pauly et al., 2000; McAinsh and Pittman, 2009; Mazars et al., 2010). The mechanisms and the channels involved in signal-induced changes in [Ca2+]nuc have not been identified (Mazars et al., 2009, 2010). Therefore, the regulation of transcription by Ca2+ in plants may occur through processes controlled in the cytosol and in the nucleus or by a combination of both. For instance, studies on stress gene regulation in tobacco (Nicotiana tabacum) showed that wind-induced expression of one CaM isoform is regulated by a Ca2+-signaling pathway in the nucleus, while expression of a cold shock–induced isoform is regulated by a pathway in the cytoplasm (van Der Luit et al., 1999). Although the effect of individual stresses on cellular Ca2+ levels has been extensively studied, the effect of combinations of stresses that plants are subjected to normally has not been investigated in any detail. To understand the effects of multiple stresses, it will be necessary to investigate the type of Ca2+ signatures elicited by a combination of stresses. Calcium signatures elicited by a combination of stresses are likely to be different from those evoked by individual stresses.
DECODING OF Ca2+ SIGNATURE
Decoding complex signal-specific Ca2+ signatures is accomplished by myriad Ca2+ binding proteins in plants that function as Ca2+ sensors (Day et al., 2002; Boonburapong and Buaboocha, 2007). These Ca2+ binding proteins are thought to sense changes in cellular Ca2+ ([Ca2+]cyt and/or [Ca2+]nuc) and regulate downstream signaling events, thereby eliciting a physiological response that is appropriate for a signal. The majority of Ca2+ sensors are proteins with one or more highly conserved Ca2+ binding helix-turn-helix structures known as EF-hands (Nakayama et al., 2000; Day et al., 2002). In Arabidopsis thaliana, there are ~250 EF-hand–containing putative Ca2+ sensors, which represent ~1% of the predicted proteome (Day et al., 2002) (http://www.Arabidopsis.org/browse/genefamily/ef-hand.jsp), many of which have not been characterized or tested for Ca2+ binding. The number of EF-hands in different Ca2+ sensors ranges from one to six. Several one, two, and three EF-hand–containing proteins that were tested for Ca2+ binding showed Ca2+ binding at physiological concentrations (Reddy et al., 2004; I.S. Day, T. Brauch, D. Connor, and A.S.N. Reddy, unpublished data). The EF-hand–containing proteins can be broadly grouped into two major groups: sensor relays and sensor responders (Sanders et al., 2002; Kudla et al., 2010). For the most part, sensor relays do not have any known enzymatic or other functional domains. Rather, upon binding Ca2+, they interact with other proteins and regulate their activities. CaMs, CaM-like proteins (CMLs), and calcineurin B-like proteins (CBLs) fall into this group, with one exception (CaM7; discussed later) (Luan et al., 2002; Reddy and Reddy, 2004; McCormack et al., 2005; Luan, 2009; DeFalco et al., 2010). CaMs/CMLs interact with diverse proteins, whereas CBLs interact with a family of protein kinases called CBL-interacting protein kinases (CIPKs) (Chinnusamy et al., 2004; Luan et al., 2009; Weinl and Kudla, 2009; Batistic et al., 2010). On the other hand, sensor responders contain, in addition to one or more EF-hands, a catalytic or functional domain whose activity is regulated by Ca2+ binding to EF-hand motifs. Responders include Ca2+-dependent protein kinases (CDPKs; also called CPKs), Ca2+- and Ca2+/CaM-dependent protein kinases (CCaMKs), some DNA or lipid binding proteins, and a few enzymes (Day et al., 2002; Yang and Poovaiah, 2003; Harper and Harmon, 2005). Many calcium sensors are coded by multiple genes, and expression of many of these is induced by stresses (DeFalco et al., 2010). CCaMK is present in legumes, maize (Zea mays), tobacco, and other plants but not in Arabidopsis (DeFalco et al., 2010). In legumes, it plays an important role in nodule morphogenesis through transmission of Nod factor–induced (a signaling molecule from nitrogen-fixing rhizobial bacteria) Ca2+ transients (Gleason et al., 2006; Tirichine et al., 2006).
CaMs, a group of well-characterized Ca2+ sensors, and CMLs are implicated in a large number of diverse cellular processes, including many plant stress responses (Zielinski, 1998; Bouché et al., 2005). When bound to Ca2+, they relay the signal by binding to other proteins resulting in activation or inactivation of interacting proteins. Over 300 proteins that interact with CaMs and CMLs have been identified in plants (Reddy et al., 2002b; Zhang and Lu, 2003; Bouché et al., 2005; Popescu et al., 2007). In fact, among all known protein–protein interactions in plants, CaMs have the most interacting partners (Lee et al., 2010). Many of the CaM binding proteins (CBPs) identified using protein microarrays need further validation using other in vitro and in vivo approaches. A major challenge is to test experimentally the biological significance of these interactions. It is possible that not all interactions found using protein microarray and screening approaches are physiologically relevant. For example, the interactors may not be expressed in the same cell or may be localized to different compartments. The specificity of Ca2+ signaling is thought to be dependent on the interplay between Ca2+ signatures and Ca2+-sensing proteins. Different CaM proteins exhibit differential expression and are likely to show differential affinity to Ca2+ and to their target proteins (Reddy et al., 1999, 2004; McCormack et al., 2005; Popescu et al., 2007). In addition to EF-hand–containing Ca2+ binding proteins, there are other proteins that do not contain this motif (e.g., annexins and C2 domain–containing proteins) but bind Ca2+ (Clark and Roux, 1995; Reddy and Reddy, 2004; Laohavisit and Davies, 2011). Annexins are likely to function as responders as they function as enzymes and contain other domains. Several studies suggest that they are important regulators of plant stress responses (reviewed in Laohavisit and Davies, 2011). These multifunctional proteins can undergo stimulus-dependent (e.g., salt) Ca2+-mediated relocation from the cytosol to membranes (Lee et al., 2004), where they exert their enzymatic functions (e.g., peroxidase activity) or create Ca2+-permeable transport pathways (Jami et al., 2008; Laohavisit et al., 2009).
IMPACTS OF CELLULAR Ca2+ CHANGES ON GENE EXPRESSION
Expression of the right genes in the right cells/tissues at the right time is not only key to growth and development but also to environmental responses. Since the 1980s, it has been shown that almost all stresses, including seemingly innocuous signals such as touch and wind, regulate gene expression in plants (Braam and Davis, 1990; Thomashow, 1999; van Der Luit et al., 1999; Zhu, 2002; Shinozaki and Yamaguchi-Shinozaki, 2007; Hirayama and Shinozaki, 2010). From numerous global studies on gene expression in response to stresses, it is evident that reprogramming of the transcriptome is an important aspect of stress signaling and adaptation. However, molecular mechanisms by which stresses regulate gene expression and the role of stress-regulated genes in stress adaptation are just beginning to be uncovered. The changes in the transcriptome are primarily established by changes in gene expression, which are regulated by transcription factors (TFs) (Latchman, 1997; Brivanlou and Darnell, 2002). Soon after the discovery in the 1980s that Ca2+ functions as a messenger in plants, it was proposed that Ca2+ is likely to regulate gene expression (Poovaiah and Reddy, 1987). Due to the fact that most signal-specific changes in cellular Ca2+ occur rapidly (in seconds to minutes) and precede observed changes in signal-induced changes in gene expression, it is likely that some of these changes are mediated by Ca2+ (Hu et al., 2004; Kaplan et al., 2006; Lecourieux et al., 2006; McAinsh and Pittman, 2009). Several studies have now demonstrated that elevated levels of [Ca2+]cyt or [Ca2+]nuc modulate gene expression (Braam, 1992; van Der Luit et al., 1999; Kaplan et al., 2006).
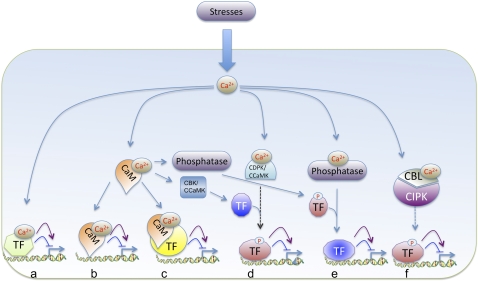
Figure 1 illustrates various ways signal-induced cellular Ca2+ levels can regulate gene expression either directly or indirectly through Ca2+ sensors. First, activated Ca2+ sensors can directly bind to cis-elements in the promoters of specific genes and induce or repress their expression (Figures 1a and 1b). Second, activated Ca2+ sensors can bind to DNA binding proteins and activate or inactivate them, thereby resulting in activation or repression of gene expression (Figure 1c). Finally, elevated cellular Ca2+ can activate Ca2+-regulated protein kinases (CDPK, CaM binding protein kinase [CBK], and CCaMK) or phosphatases, which in turn phosphorylate/dephosphorylate specific DNA binding proteins and regulate gene expression (Figures 1d to 1f). Several Ca2+ sensors (e.g., CaMs, CDPK3, and CDPK4) are localized to the nucleus, whereas others are translocated to the nucleus in response to stresses (e.g., At-CDPK2 in response to osmotic stress; Mc-CDPK1 in response to salt stress), suggesting a role for these proteins in nuclear functions (Dauwalder et al., 1986; Schuurink et al., 1996; Rodríguez-Concepción et al., 1999; Patharkar and Cushman, 2000; Dammann et al., 2003; Raichaudhuri et al., 2006). It is possible that a given signal-induced signature may modulate gene expression using one, a combination, or all of these pathways. As discussed below, there is evidence in support of Ca2+ regulation of gene expression using most of these pathways and a role for altered gene expression in stress responses.
Figure 1.
Signal-Induced Elevation of Cellular Calcium ([Ca2+]cyt and/or [Ca2+]nuc) Can Regulate Transcription by Different Mechanisms.
Elevated Ca2+ levels result in its binding to a Ca2+ sensor, which directly binds to specific DNA sequences and modulates gene expression (a and b). Activated calcium sensors (Ca2+/CaM or Ca2+/CML) interact with DNA binding proteins and modulate their activity resulting in altered transcription (c). Finally, an elevated level of calcium activates a protein kinase (CDPK, CBK, and/or CCaMK) either directly or through CaM or a protein phosphatase, which in turn phosphorylates or dephosphorylates a TF, respectively, resulting in activation or repression of transcription (d to f). Solid arrows indicate pathways with experimental evidence. Pathways lacking evidence are represented by broken arrows. Purple arrows indicate activation of gene expression; blue lines with a horizontal line represent repression.
Cellular Ca2+ levels have been shown to change expression of genes involved in stress responses. Elevated extracellular Ca2+ increased the expression of several genes, including those that encode calcium sensors (Braam, 1992). Furthermore, heat or cold shock induction of some genes is dependent on external calcium (Braam, 1992; Polisensky and Braam, 1996). The expression of some isoforms of CaM by cold and wind also requires changes in cellular Ca2+ (van Der Luit et al., 1999). Global studies of changes in gene expression in response to Ca2+ manipulations have revealed numerous target genes that are affected by Ca2+ signaling. To identify Ca2+-responsive genes in plants, Kaplan et al. (2006) generated specific [Ca2+]cyt transients in Arabidopsis seedlings and linked them to early transcriptome changes. Bioinformatic analysis revealed 230 Ca2+-responsive genes, of which 162 were upregulated and 68 downregulated. These included known early stress-responsive genes as well as genes of unknown function. A highly significant occurrence of a consensus sequence comprising two cis-elements that had previously been linked to abscisic acid (ABA) signaling, the ABA-responsive element (ABRE; CACGTG[T/C/G]) and its coupling element ([C/A]ACGCG[T/C/G]), in the upstream region of the upregulated genes was observed. Based on these data, it was concluded that, at least for some specific Ca2+ transients, ABREs function as Ca2+-responsive cis-elements. Kinetic analysis of some Ca2+-responsive genes showed they reached their maximal expression levels rapidly, within 30 min following the stimulus treatment (Kaplan et al., 2006).
CALCIUM AND Ca2+/CaM BINDING TFs
CaM Binding Transcription Factors
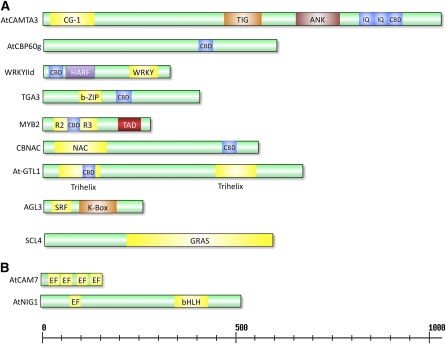
The recent release (TAIR10) of the Arabidopsis genome annotation has 27,416 protein coding genes (http://www.Arabidopsis.org/index.jsp). Among them, >2000 proteins (>7% of the total proteome) are identified as putative DNA binding TFs that are classified into 58 families according to their DNA binding domain and other conserved motifs (Table 1; Zhang et al., 2011) (http://planttfdb.cbi.pku.edu.cn/index.php?sp=At). About half of them belong to plant-specific families (Riechmann et al., 2000). Functions of many of these are yet to be discovered. In vitro screening of expression libraries with labeled CaM and probing of protein chips containing partial proteomes of Arabidopsis representing mostly TFs and signaling proteins (protein kinases, protein degradation–related proteins, heat shock proteins, CaMs/CMLs, and RNA binding proteins) with CaMs/CMLs resulted in identification of many CBPs (Reddy et al., 2002b, 2011; Yang and Poovaiah, 2003; Bouché et al., 2005; Popescu et al., 2007). Among them, over 90 CBPs fall into 10 families of DNA binding proteins (Reddy et al., 2002b; Popescu et al., 2007) (Table 1). All members in some families (e.g., CAMTAs) are CaM binding, whereas only certain members in other families (e.g WRKYs and Mybs) were found to interact with CaM or CMLs (see Supplemental Table 1 online). The domain organization of one representative member for each of the CaM binding TF families is presented in Figure 2A. The main properties of these CaM binding TFs are discussed below.
Table 1.
Arabidopsis TFs (2016) Are Grouped into 58 Families (Zhang et al., 2011)
| AP2 (25) | ARF (32) | ARR-B (16) | B3/ABI3/VP1 (71) | BBR/BPC (11) | BES1 (8) |
| C2H2 (104) | C3H (56) | CAMTA (6)a | CO-like (19) | CPP (10) | DBB (12) |
| Dof (43) | E2F/DP (12) | EIL (6) | ERF (132) | FAR1 (20) | G2-like (55) |
| GATA (31) | GRAS (36) | GRF (9) | GeBP (23) | HB-PHD (2) | HBother (8) |
| HDZIP (56) | HRT-like (2) | HSF (25) | LBD (43) | LFY (1) | LSD (6) |
| M-type (73) | MIKC (66) | MYB (159) | MYB_related (85) | NAC (135) | NF-X1 (2) |
| NF-YA (15) | NF-YB (19) | NF-YC (15) | NZZ/SPL (1) | Nin-like (14) | RAV (6) |
| S1Fa-like (4) | SAP (1) | SBP (18) | SRS (12) | STAT (2) | TALE (23) |
| TCP (28) | Trihelix (26) | VOZ (2) | WOX (17) | WRKY (89) | Whirly (4) |
| YABBY (7) | ZF-HD (17) | bHLH (194) | bZIP (101) | CBP60 (8)b |
AP2/ERF is subdivided into RAV, AP2, and ERF; HB is subdivided into HD-ZIP, TALE, WOX, HB-PHD, HB-other; MADS is subdivided into M type and MIKC. The number of TFs in each family is indicated in parentheses. Families with CaM binding TFs are indicated in bold. NF-YA is also called CCAAT-HAP2; NF-YB includes CCAAT-HAP3 and CCAAT-DR1; NF-YC is also called CAAT-HAP5.
In the plantTFDB2.0, seven CAMTAs are shown in error. The correct number is six. Two isoforms of CAMTA1 (AT5G09410) are indicated as two separate TFs.
Members of CBP60 family were recently identified as DNA binding proteins (Zhang et al., 2010) and were not included in the Zhang et al. (2011) table.
Figure 2.
Schematic Diagram of Ca2+ or Ca2+/CaM Binding DNA Proteins Showing Various Domains.
(A) Ca2+/CaM binding TFs. One representative member in each CaM binding TF family is shown. For a list of all Ca2+/CaM binding TFs, see Supplemental Table 1 online.
(B) DNA binding proteins that bind directly to Ca2+. CG-1, DNA binding domain; TIG, a nonspecific DNA binding domain; ANK, ankyrin repeats that are implicated in protein–protein interaction; IQ, Ca2+-independent CaM binding domain; CBD, CaM binding domain; HARF, conserved domain composed of these residues; WRKY, conserved domain containing these residues; R2R3, Myb DNA binding domain; TAD, transcription activation domain; NAC, conserved domain present in NAM-ATF-CUC proteins; SRF, serum response factor; K-Box, protein–protein interaction domain; GRAS, conserved domain in GAI-RGA-SCR proteins; EF, calcium binding motif; bHLH, basic helix-loop-helix domain. All proteins are drawn to scale. Numbers on the scale indicate the length of the protein in amino acids.
Calmodulin binding transcription activators (CAMTAs; also referred to as signal-responsive proteins or ethylene-induced CaM binding proteins), were first discovered in plants in a screen for CaM binding proteins (Reddy et al., 2000; Yang and Poovaiah, 2000, 2002; Bouché et al., 2002). This family of TFs is highly conserved and possesses multiple domains. CAMTAs are characterized by a CG-1 DNA binding domain at the N terminus, a TIG domain (an immunoglobulin-like fold found in some TFs) involved in nonspecific DNA binding, several ankyrin repeats that are implicated in protein–protein interaction, a Ca2+-dependent CaM binding domain, and Ca2+-independent CaM binding domains called the IQ motif (Figure 2A) (Bouché et al., 2002; Yang and Poovaiah, 2002; Finkler et al., 2007; Du et al., 2009). CAMTAs have also been identified in the genomes of other multicellular organisms, including mammals, flies, and worms (Han et al., 2006; Song et al., 2006). In Arabidopsis, there are six CAMTAs (CAMTA1 to CAMTA6), whose transcript levels are highly responsive (up- or downregulated) to diverse stresses (Reddy et al., 2000; Yang and Poovaiah, 2000, 2002). CAMTA transcript levels are induced upon cold and heat treatment (CAMTA1 and CAMTA3-6) as well as salinity (CAMTA1-4 and CAMTA6) (Yang and Poovaiah, 2002). Furthermore, CAMTA expression responds to phytohormones and secondary messengers known to mediate plant responses to biotic and abiotic stress, such as abscisic acid (CAMTA2 and CAMTA4-6), methyl jasmonate (CAMTA1, 3, and 4), ethylene (CAMTA1, 3, and 4), H2O2 (CAMTA2-6), salicylic acid (CAMTA2 and CAMTA4-6), and auxin (CAMTA1) (Yang and Poovaiah, 2002; Galon et al., 2010a). All CAMTAs are induced upon wounding (Yang and Poovaiah, 2002). The patterns of induction to multiple chemical and physical stimuli suggest the involvement of individual CAMTAs in multiple signal transduction pathways and stress responses. For example, CAMTAs have been shown to be involved in auxin signaling in growth and development as well as in stress response and may link the two pathways (Galon et al., 2010b). CAMTA1 repressor lines and camta1 mutants showed enhanced responsiveness to auxin, suggesting that in wild-type plants, enhanced expression of CAMTA1 in response to stresses suppresses the plant’s responsiveness to auxin (Galon et al., 2010b). The DNA cis-element that binds to CAMTA3 was identified as (G/A/C)CGCG(C/G/T) (Yang and Poovaiah, 2002). The core CGCG sequence was first identified as a binding site for a TF isolated from a parsley (Petroselinum crispum) cDNA library, giving the name CG-1 to the DNA binding domain of proteins interacting with this motif (da Costa e Silva, 1994). Later, analysis of the cis-element for this family revealed the existence of two core CAMTA binding motifs, CGCG and CGTG, the CGCG core–containing consensus motif is (A/C)CGCG(C/G/T) and the CGTG core–containing consensus motif is (A/C)CGTGT (Galon et al., 2008, 2010a; Doherty et al., 2009; Du et al., 2009; Kim et al., 2009). Studies with loss-of-function CAMTA3 mutants suggest that depending on the context it acts either as a positive or a negative regulator of transcription (Doherty et al., 2009; Du et al., 2009) and that its transcription repressor activity is dependent on CaM binding. A transient expression study with protoplasts indicated that the Ca2+/CaM complex functions as a negative regulator of the activity of the rice (Oryza sativa) CAMTA/SR protein Os-CBT (Choi et al., 2005).
An important group of TFs that participate in plant responses to stress belongs to the large MYB family, which contains functionally diverse proteins (Dubos et al., 2010). Several members of the MYB class of TFs were found to bind Ca2+/CaM (Popescu et al., 2007) (see Supplemental Table 1 online), and the DNA binding activity of one of these TFs is enhanced by Ca2+/CaM (Yoo et al., 2005). The MYB family of TFs is characterized by the structurally conserved DNA binding domain termed the MYB domain, which encompasses up to four imperfect repeats (R) of ~52 amino acids (Figure 2). Based on the number of repeats, plant MYB proteins are grouped into four classes: 4R-MYB, 3R-MYB, MYB-related (containing only a single or a partial MYB repeat), and two-repeat R2R3-MYB (Stracke et al., 2001; Dubos et al., 2010). The R2R3-MYB subfamily is the most common in plants (Stracke et al., 2001). A soybean (Glycine max) CaM, Gm-Cam4, was reported to mediate the Ca2+ signaling response by activating an R2R3-MYB2 TF (Yoo et al., 2005). Gm-CaM1 and Gm-Cam4 were shown to differentially regulate the DNA binding activity of AtMYB2 (At2g47190) (Yoo et al., 2005).
The third class of TFs with Ca2+/CaM-regulated members is the WRKY family. This group of TFs shares a characteristic DNA binding domain containing an almost invariant WRKY motif and an atypical Zn2+ finger structure (Figure 2) (Eulgem and Somssich, 2007). Based on the number of WRKY boxes (conserved amino acid sequence WRKYGQK) and type of zinc finger and function, WRKYs are grouped into three families: I, II, and III. The group II members are further divided into five subgroups (IIa, IIb, IIc, IId, and IIe). WRKY7 was the first WRKY TF reported to bind CaM in a Ca2+-dependent manner (Park et al., 2005). WRKY7 is a member of the WRKYIId subfamily, and all members of this subfamily (WRKY11, WRKY15, WRKY17, WRKY21, WRKY39, and WRKY74) were found to interact with Ca2+/CaM (Park et al., 2005) (see Supplemental Table 1 online). The WRKY TFs bind specifically to the W-box DNA cis-element (C/T)TGAC(C/T) (Eulgem and Somssich, 2007). A global analysis of Ca2+/CaM binding proteins in Arabidopsis using protein microarrays identified several additional WRKYs (WRKY43, 45, 50, and 53) that were shown to interact with different isoforms of CaM in a Ca2+-dependent manner (Popescu et al., 2007). The interactions between CaMs and WRKY43 and WRKY53 were confirmed by coimmunoprecipitation assays (Popescu et al., 2007).
TGA3, a member of a family of basic leucine zipper (bZIP) TFs, was identified as a CaM binding protein that binds the promoter of CaM3, and CaM binding of recombinant TGA enhanced its binding to the promoter (Szymanski et al., 1996). The bZIP family TFs contain a basic region for binding DNA and a leucine zipper dimerization domain (Jakoby et al., 2002) (Figure 2). A protein microarray-based Ca2+/CaM binding protein assay by Popescu et al. (2007) identified 18 possible bZIP family members as CaM binding, and the interaction of TGA3 with Ca2+/CaM binding was verified by coimmunoprecipitation (Popescu et al., 2007). Other bZIP family members, including the ABA-responsive TFs ABF1, 2, 3, and 4, which participate in the response to abiotic stresses, may be regulated by Ca2+ through their phosphorylation by CDPKs (reviewed in Galon et al., 2010a), which are known to be involved in stress responses (Cheng et al., 2002; Lee and Rudd, 2002; Reddy and Reddy, 2004). For example, CPK4 and CPK11 are stimulated by ABA signaling and phosphorylate ABF1 and ABF4 in vitro (Zhu et al., 2007). ABF2 has also been shown to bind CaM (Popescu et al., 2007).
A plant-specific family of CaM binding proteins called the CBP60s was first isolated from maize (Zea mays; Reddy et al., 1993) and then from tobacco (Lu and Harrington, 1994), Arabidopsis (Reddy et al., 2002b), and bean (Phaseolus vulgaris; Ali et al., 2003). In Arabidopsis, there are eight members in this family, and all but one bind CaM (Reddy et al., 2002b; Wang et al., 2009a; Zhang et al., 2010). Some of them have their CaM binding domain at the C terminus (Reddy et al., 1993, 2002b; Lu and Harrington, 1994) and others at the N terminus (Wang et al., 2009a; Zhang et al., 2010) (Figure 2). They are differentially expressed in response to biotic stresses and elicitors of plant defense (Ali et al., 2003; Wang et al., 2009a). Recently, two members of this family were reported to bind DNA and regulate expression of specific genes (Zhang et al., 2010).
A single member of the NAC TF family is known to interact with Ca2+/CaM. This family comprises a large group of plant-specific TFs with over 130 NACs in Arabidopsis. These proteins have a conserved N-terminal NAC domain (originally found in no apical meristem, ATAFs [Arabidopsis transcription activation factor] and cup-shaped cotyledon), whereas the C-terminal domain is highly variable (Figure 2). The CaM binding NAC protein (CBNAC) is a Ca2+-dependent CaM binding transcriptional repressor, and its repressor activity is enhanced by binding to Ca2+/CaM (Kim et al., 2007; Yoon et al., 2008). Its DNA cis-element is a GCTT core sequence flanked on both sides by other frequently repeating sequences (TTGCTTANNNNNNAAG) (Kim et al., 2007).
GT element binding proteins or GTLs are TFs that have one or two trihelix motifs (Figure 2), which bind the DNA cis-element GGTTAA (Smalle et al., 1998; Nagano et al., 2001). One GTL family member, At-GTL1 (AT1g33240), was identified as a Ca2+/CaM interacting protein in a screen of expression libraries using labeled recombinant CaM (Yoo et al., 2007) (see Supplemental Table 1 online).
MADS box proteins are a family of TFs characterized by the presence of a conserved ~60–amino acid N-terminal DNA binding motif (MADS box domain) that generally binds the consensus sequence CC(A/T)6GG (known as CArG motif) (Figure 2) (Shore and Sharrocks, 1995). The protein microarray probed with CaM/CMLs identified 25 members of the MADS box family proteins (e.g., AGL1, AGL3, and AGL8) (Popescu et al., 2007).
In addition, four scarecrow-like TFs (e.g., SCL4) and two NAM TFs were also found to bind CaM/CMLs (Popescu et al., 2007) (see Supplemental Table 1 online). The interactions of most of these TFs and Ca2+/CaM were based on protein array studies and need to be verified experimentally using other approaches.
In addition to these CaM binding proteins that interact with DNA, there are other CBPs that are involved in gene regulation but function as corepressors. For instance, a corepressor involved in auxin-regulated gene expression, IAA31, was identified as a Ca2+/CaM-interacting protein, and this interaction was confirmed by immunoprecipitation (Popescu et al., 2007), suggesting potential regulation of this TF by Ca2+/CaM. Some studies suggest that chromatin modifications involving DNA methylation and covalent modifications of histones, and chromatin remodeling, which require ATP hydrolysis, play a role in stress-induced reprogramming of the transcriptome (Walley and Dehesh, 2010). A CaM-activated nuclear NTPase has been reported in plants (Chen et al., 1987). It would be interesting to test if chromatin remodeling is altered in loss-of-function mutants of this CaM binding protein.
Ca2+ Binding TFs
As discussed above most TFs regulated by Ca2+ are Ca2+/CaM binding. However, there are at least two TFs that directly bind Ca2+. Arabidopsis NaCL-INDUCED GENE (NIG), a salt stress–responsive gene, encodes the first known plant TF involved in direct Ca2+ binding (Kim and Kim, 2006). NIG1 contains an EF-hand–like Ca2+ binding motif at its N-terminal region and a basic helix-loop-helix DNA binding domain at its C-terminal region (Figure 2B). NIG1 binds to the CANNTG motif, known as the E-box. There are other EF-hand proteins that have a DNA binding domain (Day et al., 2002), but their functions are not known.
Another TF that binds Ca2+ and has direct function in transcriptional regulation is At-CaM7. Normally Ca2+/CaMs do not act on their own directly; rather, they interact with other proteins either activating or deactivating their function. CaM7 appears unusual in that it directly interacts with promoters of genes involved in seedling development (Kushwaha et al., 2008). CaM7 binds to the Z-/G-box (ATACGTGT/CACGTG) in the promoters of light-regulated genes, thereby modulating their expression and photomorphogenesis. However, the effect of Ca2+ binding on CaM7 DNA binding activity has not been investigated. Given that not many Ca2+ sensors are tested for their DNA binding activity, it is likely that other Ca2+ sensors may also bind to specific DNA sequences. Future systematic biochemical analyses of other EF-hand–containing proteins will likely lead to identification of additional Ca2+ binding TF.
CALCIUM- AND Ca2+/CaM-REGULATED GENE EXPRESSION IN BIOTIC STRESS RESPONSES
In addition to many well-characterized defense signaling components (Chisholm et al., 2006; Boller and He, 2009), it is also well-established that pathogens cause substantial ion fluxes across membranes. In fact, these ion fluxes, which occur within a few minutes of plant–pathogen interaction, seem to precede and to be required for the activation of defense responses (Hu et al., 2004; Lecourieux et al., 2006). Among pathogen-induced ion fluxes, the involvement of Ca2+ signaling pathways in plant–microbe interactions has been relatively well studied. It is now well established that plant–microbe interactions that involve disease-causing microbes as well as beneficial symbiotic bacteria that induce nitrogen fixing nodules and arbuscular mycorhizal fungi induce Ca2+ signatures (Shaw and Long, 2003; Lecourieux et al., 2006; Kosuta et al., 2008). Transient changes in [Ca2+]cyt and/or [Ca2+]nuc are considered one of the early events that occur in response to microbes and microbe-associated elicitors (Lecourieux et al., 2006). A variety of proteinaceous elicitors (cryptogein, pep-13, elf18, and flg22) and nonproteinaceous elicitors (fungal oligosaccharides, glucans, and bacterial lipopolysaccharides), live plant pathogens, and interaction of plant resistance proteins with pathogen avirulence (Avr) factors induce Ca2+ signatures in cultured plant cells and intact leaves (Tavernier et al., 1995; Jabs et al., 1997; Zimmermann et al., 1997; Xu and Heath, 1998; Blume et al., 2000; Grant et al., 2000; Heath, 2000; Lecourieux et al., 2002; Gust et al., 2007; Ranf et al., 2008; Ma et al., 2009a).
The pathogen-induced Ca2+ signatures are generated by the coordinated action of Ca2+ influx through various types of channels on the plasma membrane and through pumps and cotransporters on various organelles (Kudla et al., 2010). Some recent studies have shown that plasma membrane–localized cyclic nucleotide gated channels (CNGCs), a family of CaM and cyclic nucleotide binding ion channels involved in uptake of Ca2+ and other cations, are some of the channels likely responsible for pathogen-induced changes in [Ca2+]cyt (Ali et al., 2007; Ma and Berkowitz, 2007; Ma et al., 2008, 2009b). However, little is known about the mechanisms by which pathogens/elicitors regulate CNGC activity (Ma et al., 2009a). A CaM binding endoplasmic reticulum–localized Ca2+-ATPase in tobacco also plays an important role in microbial/pathogen-associated molecular pattern (MAMP/PAMP)–induced Ca2+ changes. Silencing of this ATPase altered the MAMP-induced [Ca2+]cyt and [Ca2+]nuc signature and accelerated pathogen- and elicitor-induced cell death (Zhu et al., 2010). Other channels, pumps, and transporters are also likely to be involved in pathogen-induced changes in [Ca2+]cyt. Numerous studies have unequivocally demonstrated that the same elicitors that induce a Ca2+ signature also induce defense related genes at the transcriptional level (DeFalco et al., 2010). The role of cellular Ca2+ in plant immunity is further supported by genetic studies using mutants that are defective in Ca2+ channels and pumps. In the defense no death1 mutant of Arabidopsis, which lacks CNGC2 and shows no inward Ca2+ current in the presence of cAMP, hypersensitive response (i.e., plant cell death at the site of infection) to avirulent bacterial pathogen is impaired (Clough et al., 2000; Ali et al., 2007). Similarly, mutations in other CNGCs (CNGC4, CNGC11, and CNGC12) showed altered defense responses (Balagué et al., 2003; Yoshioka et al., 2006). Inactivation of an endoplasmic reticulum–localized Ca2+ pump or two vacuolar Ca2+ pumps results in altered plant defense responses (Boursiac et al., 2010; Zhu et al., 2010). What is not yet fully elucidated is how pathogen-induced Ca2+ signatures are translated to reprogramming of the transcriptome and altering defense responses. A role for several members of major groups of Ca2+ sensor proteins in plant–pathogen interaction has been described; this topic has been reviewed recently (DeFalco et al., 2010; Kudla et al., 2010) and therefore will not be discussed here. Our discussion will focus only on the role of Ca2+- and Ca2+/CaM-regulated gene expression in plant defense responses.
CaMs in Plant Defense
Using different plants such as soybean, Arabidopsis, and tobacco, a significant role for various CaM isoforms in plant defense has now been established (Harding et al., 1997; Heo et al., 1999; Chiasson et al., 2005; Takabatake et al., 2007; Zhu et al., 2010). These studies provide evidence that CaM might be one of the key players in transducing the pathogen-induced Ca2+ increase to downstream components of defense signaling. Early investigations on the role of CaM in plant–pathogen interactions have mainly used various CaM antagonists. However, conclusions from such studies were questioned because of the nonspecific effects of these drugs. Later, several studies provided genetic evidence for the role of CaM in plant defense responses. For example, in transgenic tobacco cells expressing a mutant CaM (VU-3) in which Lys at position 115 is changed to Arg making it hyperactive, the basal level of active oxygen species was greater than in control cells, and in addition, in response to cellulase, harpin, incompatible bacteria, and mechanical stress, these mutant CaM-expressing cells exhibited greater production of active oxygen species (Harding et al., 1997), providing indirect evidence that CaM is involved in plant defense responses. In a follow-up study, using cells and intact leaves of the VU-3 transgenic tobacco plants inoculated with incompatible Pseudomonas syringae pv syringae 61, cell death was shown to be accelerated in transgenic tobacco plants (Harding and Roberts, 1998). Silencing of specific pathogen-induced CaM isoforms in tomato (Solanum lycopersicum) resulted in enhanced susceptibility to virulent necrotrophic bacteria and fungi, suggesting the involvement of specific CaM isoforms in basal defense against necrotrophic pathogens (Takabatake et al., 2007). A CML in Arabidopsis (CML43) and tomato (APR134) is induced by pathogens, and silencing of this gene in tomato compromised immune response, while its overexpression in Arabidopsis accelerated hypersensitive response (Chiasson et al., 2005). These reports provided initial hints that CaM may contribute to plant defense responses. However, a direct effect on the expression of plant defense marker genes (e.g., PR) and on the resistance level of the VU-3 transgenic tobacco was not provided. Direct evidence for the involvement of CaM in plant defense responses was elegantly demonstrated by overexpression studies with soybean CaMs (Heo et al., 1999; Park et al., 2004). These studies demonstrated that the expression of SCaM4 and SCaM5 in transgenic tobacco and Arabidopsis leads to spontaneous lesions, increased PR gene expression, and enhanced resistance to bacterial, fungal, and viral pathogens. Furthermore, only the divergent SCaM4 and SCaM5, but not the conserved CaMs, were induced in response to pathogens, probably contributing to the specificity of defense-associated Ca2+ signaling. Interestingly, silencing of a tobacco CaM, Nb-CaM1, suppressed the tobacco mosaic virus p50-induced HR in tobacco cells but not the Cf9-Avr9 or Pto-AvrPto and Pst DC3000-induced cell death, suggesting that CaM can provide specificity to different pathogens (Zhu et al., 2010). Collectively, these studies demonstrate that CaMs play a critical role in plant defense.
As discussed above CaMs and CMLs generally regulate cellular processes indirectly by interacting with other proteins in a Ca2+-dependent manner and modulating their activity. Therefore, for CaMs to function in plant defense signaling pathways, they must be regulating the activity of genes that are associated with plant defense. Recent studies clearly show that CaMs interact with specific TFs to regulate gene expression, including the expression of defense genes (see below). In addition, it is likely that CaMs and CMLs may bind directly to specific promoters in a Ca2+-dependent manner and regulate their expression, since Arabidopsis CaM7 can bind to specific sequences in DNA directly and regulate expression (Kushwaha et al., 2008). Given the high identity of amino acid sequences of CaMs in plants, it will be interesting to see if other CaMs or CaM-like proteins also display DNA binding activity. While this study revealed a novel role for CaM in transcription, no study so far has shown a direct role for CaM in regulating plant defense genes. Instead, a majority of the studies have focused on investigating the role of CaM binding to TFs that regulate transcription of defense genes.
The Role of CaM Binding TFs in Plant Immunity
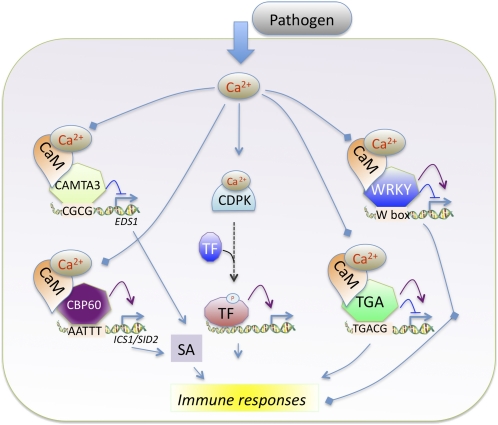
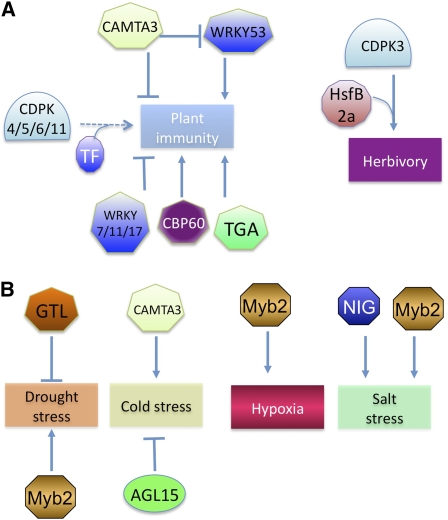
Among the CaM-interacting TF families, members of the CAMTA, WRKY, and bZIP (TGA) families and a novel family of CaM binding TFs (CBP60s) play a role in biotic stresses by modulating the expression of defense genes (Figure 3). Expression of two CBP60s in bean is induced in response to incompatible pathogens and elicitors of plant defense responses (salicylic acid [SA], jasmonic acid, hydrogen peroxide, and a fungal elicitor), suggesting a role for CaM binding TFs in plant defense (Ali et al., 2003). SA, a key defense hormone, is required for inducing local and systemic acquired resistance (immunity at the whole-plant level acquired after a local infection) in plants against diverse pathogens. Upon infection by pathogens, plants induce SA synthesis by activating the expression of Isochorismate Synthase 1 (ICS1)/SA Induction Deficient 2 (SID2), a key enzyme in SA biosynthesis (Wildermuth et al., 2001). Similarly, the expression of EDS1, one of the key genes in SA biosynthesis, is also upregulated in response to pathogens (Falk et al., 1999; Vlot et al., 2009). How plants regulate the expression of ICS1 or EDS1 in response to pathogens was not known. However, recent genetic studies have shown that two CaM binding TFs function as either a negative (CAMTA3) (Du et al., 2009) or a positive regulator (CBP60g) of EDS1 or ICS1/SID2, which are involved in SA biosynthesis and control its levels (Zhang et al., 2010). Mutants that lack CAMTA3 had elevated levels of SA and H2O2 and showed spontaneous lesions and constitutive activation of plant immune responses, including activation of SA biosynthetic and defense genes and increased resistance to a bacterial (P. syringae) and a fungal pathogen (Galon et al., 2008; Du et al., 2009), suggesting that CAMTA3 negatively regulates SA accumulation and pathogen defense.
Figure 3.
Diagram Illustrating the Known Roles of Ca2+ and Ca2+/CaM Binding TFs in Regulating Expression of Genes Involved in Plant Immunity.
Purple arrows indicate activation of gene expression; blue lines with a horizontal line indicate repression. Pathways lacking evidence are represented by broken arrows. Diamonds at the lines’ end indicate that the effect of Ca2+/CaM binding on TFs function is not known. See text for details.
Nonexpresser of PR gene1 (NPR1) is a critical signaling component downstream of SA. In the camta3 npr1 double mutant, constitutive expression of PR genes as well as disease resistance are similar to that in the camta3 mutant, suggesting that the activation of defense response is not mediated through NPR (G.S. Ali and A.S.N. Reddy, unpublished data). It has been shown that the DNA binding region in CAMTA3 binds the CGCG element in EDS1 and represses its expression (Du et al., 2009) (Figure 3). Binding of Ca2+/CaM to CAMTA3 was found to be necessary to negatively regulate EDS1 expression and plant immunity. This was demonstrated by the inability of an At-CAMTA3 mutant form, which does not bind CaM, to rescue the mutant phenotypes of atcamta3 (Du et al., 2009). These observations suggest that under normal conditions, resting Ca2+ levels are sufficient to maintain the binding of CaM to At-CAMTA3 for suppressing EDS1 expression. Although this study provided evidence that CaM is involved in regulating the activity of a defense-associated TF, it did not link the pathogen-induced Ca2+ signature to transcription. Choi et al. (2005) studied the function of the rice CAMTAs (Os-CBTs) in Arabidopsis protoplasts using synthetic promoters and found that Ca2+/CaM suppressed the CAMTA-mediated transcription activation. Constitutive activation of plant defense responses in camta3 mutants is temperature dependent (Du et al., 2009). Further studies are needed to address this temperature-dependent regulation of plant immunity. Also, the effect of Ca2+/CaM binding on interaction of CAMTA3 or other CAMTAs with DNA is not known and needs further investigation.
Arabidopsis CBP60g, a member of plant-specific CBPs, positively affects the expression of ICS1 (Figure 3), which is the source for a majority (>90%) of pathogen-induced SA (Wildermuth et al., 2001). Previously, gene expression analyses have shown induction of some CBP60s in bean by bacterial pathogens and defense elicitors (Ali et al., 2003), suggesting that they may play a role in plant defense. Recently, At-CBP60g was implicated in PAMP-triggered immunity and accumulation of SA, suggesting that CBP60g plays a positive role in plant immunity (Wang et al., 2009a; Zhang et al., 2010). Interestingly, At-CBP60g displays DNA binding activity, and it preferentially binds to a DNA sequence that contains AATTTT, which is present in the promoter of ICS1. Genome-wide bioinformatic analyses showed that the AATTTT motif is overrepresented in the promoters of genes induced by flg22 or Pst DC3000 (AvrRPM1), suggesting that in addition to regulating the expression of ICS1, CBP60g might also affect the expression of other defense-related genes (Zhang et al., 2010). Mutants that abolish CaM binding activity of CBP60g did not complement the mutant phenotype, suggesting that binding of CaM to CBP60g is essential for its function (Wang et al., 2009a). However, it remains to be seen how CaM binding affects its ability to interact with the ICS1 promoter. One possibility is that pathogen-induced changes in the binding affinity of Ca2+/CaM to CBP60g might lead to induction of ICS1. Taken together, experiments with CAMTA3 and CBP60g show that activated CaM (i.e., Ca2+-loaded CaM) can affect defense gene expression both positively and negatively and that pathogen-induced changes in Ca2+ might lead to altered affinity of CaM to these proteins, thereby changing their activity. Both AtCAMTA3 and CBP60g have several homologs in plants; if and how they affect plant defense responses remains to be investigated.
TGA is another family of CaM binding TFs whose members physically interact with NPR1, an important regulator of systemic acquired resistance (Szymanski et al., 1996; Kesarwani et al., 2007). There are eight TGA genes in Arabidopsis. A triple knockout mutant (tga2 tga5 tga6) is impaired in PR gene activation and shows no systemic acquired resistance, confirming their role in disease resistance. Reverse genetic approaches using single, double, and triple knockout lines suggest that a majority of them play a positive role in PR gene expression and resistance, whereas TGA2 might be a negative regulator of PR genes and resistance (Kesarwani et al., 2007). Translocation of NPR1 to the nucleus after pathogen attack leads to the stable binding of TGA2 to specific elements in the promoter of defense-associated genes and activation of gene expression (Figure 3) (Fan and Dong, 2002). How the activity of TGAs is affected by Ca2+/CaM is unknown, and similar to so many other CaM binding TFs, there is a great need for understanding the role of Ca2+ signaling in modulating the activity of these TFs.
There are other CaM binding TFs, such as several members of the WRKY, MYB, and NAC families, which have been demonstrated to play a role in plant defense responses. Besides the fact that they bind CaM, a functional significance of CaM binding to these proteins in plant defense is not known. The expression of Ca2+/CaM binding WRKY7 is regulated by flagellin and other PAMPs (Thilmony et al., 2006). Individual WRKY TFs are known to either positively or negatively regulate plant immunity (Figure 3) (Pandey and Somssich, 2009). The CaM binding subgroup of WRKYs likewise can be separated as positive or negative regulators of plant defense (Figure 3). As with CAMTA3, At-WRKY7 loss-of-function mutants exhibit enhanced resistance to Pst, while plants overexpressing WRKY7 showed an increased susceptibility to the pathogen, suggesting a negative regulatory role for WRKY7 in plant defense responses against bacterial pathogens (Kim et al., 2006). Supporting this, expression of the defense gene PR1 and SA accumulation is increased in wrky7 loss-of-function mutants and suppressed in overexpression lines (Kim et al., 2006). The Ca2+/CaM binding Arabidopsis WRKY11 and WRKY17 genes were also induced by the bacterium Pst (Journot-Catalino et al., 2006). Similar to wrky7 mutants, loss-of-function wrky11 mutant has increased resistance to Pst, and wrky11 wrky17 double mutants showed further enhancement of resistance, suggesting that they negatively regulate plant immunity. Expression analyses revealed that both WRKY11 and 17 modulate transcriptional activity and that some target genes were specific to each WRKY, while others were redundant (Journot-Catalino et al., 2006). By contrast, a loss-of-function mutant of WRKY53 showed enhanced disease susceptibility, whereas its overexpression lead to enhanced resistance against Pst and other pathogens, suggesting that it plays a positive role in plant immunity (Prasad et al., 2009; Murray et al., 2007; Hu et al., 2008). Several studies suggest that expression of positive and negative regulators is fine-tuned and is dependent on the stage of disease and lifestyle of pathogens with a majority of positive WRKYs becoming active in the early stages of disease, whereas a majority of negative regulators becoming active during the later stages of disease. The Ca2+/CaM binding role in modulating WRKY TFs remains to be discovered.
The expression of a plant-specific CaM binding protein (pathogen-induced CaM binding protein [PICBP]) with four CaM binding domains is induced in response to pathogens in Arabidopsis and bean (Ali et al., 2003; Reddy et al., 2003) and is constitutively expressed in the Arabidopsis accelerated cell death2-2 mutant (Reddy et al., 2003). Furthermore, the hrp1 mutant of P. syringae pv tabaci and elicitors of plant defense, such as SA and hydrogen peroxide–induced PICBP expression in bean (Ali et al., 2003; Reddy et al., 2003), suggest a role for PICBP in Ca2+-mediated defense signaling and cell death. This protein, coded by a single gene in Arabidopsis and potato (Solanum tuberosum), contains multiple nuclear localization signals and is localized to the nucleus (Reddy et al., 2002a, 2003) (see Supplemental Figure 1 online), suggesting that it affects a nuclear process; whether it regulates any aspect of gene expression remains to be studied.
Substantial experimental evidence points to a pivotal role for CDPKs in plant defense (Figure 3) (Ma and Berkowitz, 2007; Boudsocq et al., 2010). In Arabidopsis, several CDPKs, CDPK4, 5, 6, and 11, are implicated in a PAMP (flg22)-induced transcriptional reprogramming of plant defense genes (Boudsocq et al., 2010). Phosphorylation of specific TFs by these CDPKs is thought to regulate gene expression. However, the identity of TFs phosphorylated by any of these CDPKs is not known. CDPKs have been shown to phosphorylate a membrane-localized NADPH oxidase, which affects production of reactive oxygen species (Kobayashi et al., 2007), which in turn are known to induce expression and activation of defense-related TFs. An indication that CDPKs might regulate gene expression through TFs comes from microarray analyses where it was shown that CDPKs affect the expression of many genes (Boudsocq et al., 2010). It is likely that they accomplish this by modulating phosphorylation of TFs (Boudsocq et al., 2010). How activated CDPKs affect transcription of defense genes remains unexplored, and given the central role of CDPKs in relaying Ca2+ signaling, new discoveries are expected in the near future. Since CDPKs carry out Ca2+-dependent phosphorylation, it is possible that phosphorylation of TFs with known defense functions likely mediates pathogen-induced Ca2+ signatures.
There are other Ca2+/CaM binding proteins and Ca2+ binding proteins that are not TFs but function in plant disease resistance. For example, barley (Hordeum vulgare) MLO, a membrane protein, acts as a repressor of defense responses, and CaM binding activity is necessary for repressing the defense response (Kim et al., 2002a, 2002b). Cotton (Gossypium hirsutum) annexin ANN1 has been shown to inhibit callose synthase activity in a Ca2+-dependent manner, suggesting a role in pathogen response (Andrawis et al., 1993; Shin and Brown, 1999).
Fungal Pathogens
The expression of two Ca2+/CaM binding members (WRKY11 and WRKY17) of the WRKY IId family (Park et al., 2005) was enhanced by chitin treatment (Libault et al., 2007). WRKY11 (At4G31550) is expressed as three alternative transcripts whose expression varied over time (Libault et al., 2007). As chitin is a component of fungal cell walls, these WRKY TFs may be involved in fungal defense. The role of Ca2+/CaM in this role has not been investigated. CAMTA3 has also been shown to be a negative regulator of fungal resistance. Loss-of-function mutants showed increased resistance to the fungal pathogen Botrytis cinerea (Galon et al., 2008).
Herbivory
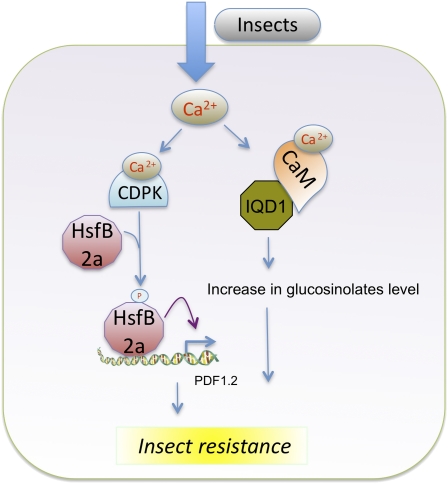
Insect feeding and isolated insect-derived elicitors are also known to lead to a Ca2+ signature (Maffei et al., 2007). Although a direct connection between CaM binding and any herbivory-associated transcription regulator is not known, the fact that herbivory leads to an elevated Ca2+ level suggests that Ca2+/CaMs may bind to TFs and regulate responses to herbivory. Studies with IQD1, which binds CaM in a Ca2+-dependent manner, have shown that it controls the levels of glucosinolates (GSs), which play an important role in herbivory, by regulating the expression of several genes involved in GS metabolism (Figure 4) (Levy et al., 2005). The loss-of-function iqd1 mutants have reduced GS, whereas overexpression lines showed increased GS and reduced herbivory (Levy et al., 2005). Since IQD1 has several nuclear localization signals and localizes to the nucleus (see Supplemental Figure 1 online), it is possible that IQD1 regulates gene expression by binding to DNA but this remains to be tested. Ca2+ can indirectly regulate transcription of herbivory-associated genes through CDPKs, and there is some evidence for this (Figure 4). A screen of cdpk mutants for herbivory-associated genes following insect attack revealed that cdpk3 and cdpk13 had lower transcript levels of the plant defensin gene PDF1.2 compared with the wild type (Kanchiswamy et al., 2010). Several TFs, such as HsfB2a, ERF1, and ATL2, which are involved in regulating the expression of the herbivory-associated marker PDF1.2, are phosphorylated by CDPKs (Kanchiswamy et al., 2010). In in vitro studies, CDPK3 was able to phosphorylate several TFs (including ERF1, HsfB2a, and CZF1/ZFAR1) in the presence of Ca2+, whereas CDPK13 phosphorylated only HsfB2a in the presence or absence of Ca2+. HsfB2a codes for a heat shock protein known for its involvement in stress responses. Interestingly, CDPK3- or CDPK13-derived phosphorylation of HsfB2a promotes PDF1.2 transcriptional activation, suggesting a role for these CDPKs in transcription regulation as a result of herbivory attack (Kanchiswamy et al., 2010) (Figure 4). An interesting picture that emerges from the biotic stress studies is that the Ca2+ signaling pathway in plant defense responses is fairly complex; it seems to involve an intricate network of interactions of Ca2+ sensors with positive and negative regulators of defense genes.
Figure 4.
Ca2+ and Ca2+/CaM Binding Proteins’ Role in Expression of Genes Controlling Herbivory.
IQD1, IQ-domain 1, a Ca2+/CaM binding region; HsfB2a, Heat shock factor B2a. Purple arrow indicates activation of gene expression. See text for details.
CALCIUM- AND Ca2+/CaM-REGULATED GENE EXPRESSION IN ABIOTIC STRESS RESPONSES
Drought
Drought is one of the most prevalent abiotic stresses and can seriously limit plant growth and survival (Blum, 1996). Physiologically, plants adapt to drought by increasing the efficiency of water uptake from soil, retaining water within cells, and/or by regulating water loss through stomata via transpiration (Yang et al., 2010a). Microarray analyses have shown that several hundred genes respond to water deficiency in a specific temporal and spatial expression pattern (Seki et al., 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). Water stress activates signaling cascades involving protein kinases/phosphatases (e.g., RPK1, SRK2C, CDPKs, and ABI1) and TFs (e.g., AREBs and DREBs) and upregulates production of chaperones and molecules involved in osmoprotectant metabolism. The phytohormone ABA plays an important role in cellular signaling in abiotic stresses, such as drought and salinity. ABA synthesis is induced under conditions of water stress and the increased level of ABA signals for stomatal closure in guard cells and induces expression of drought stress–related genes that encode proteins contributing to dehydration tolerance. Promoters of many ABA-responsive genes bear cis-acting elements known as ABRE (PyACGTGGC) (Uno et al., 2000). ABA-inducible transcription typically requires the existence of more than two ABREs or the combination of an ABRE with a coupling element at appropriate positions in the promoter regions (Yoshiba et al., 1999; Uno et al., 2000). Many of the Ca2+-regulated genes have these elements, suggesting that ABA may regulate ABA-responsive genes through cellular Ca2+ changes (Kaplan et al., 2006). Additionally, MYC and MYB recognition sites mediate ABA signaling for some stress-inducible genes (Urao et al., 1993; Abe et al., 1997, 2003). Besides ABA-dependent pathways, drought responses are also mediated by ABA-independent signaling pathways, such as those mediated by the DREB proteins (Agarwal et al., 2006; Yamaguchi-Shinozaki and Shinozaki, 2006; Seki et al., 2007).
Some members of the bZIP family are induced by drought, respond to ABA signaling, and activate expression of genes containing ABRE elements (Jakoby et al., 2002). These are known as ABFs or AREBs (ABRE binding factors or ABRE binding proteins) (Jakoby et al., 2002). Arabidopsis ABF2/AREB1, ABF4/AREB2, and ABF3 are upregulated by ABA, dehydration, and salinity stress. ABF2/AREB1 is a CaM binding TF (Popescu et al., 2007) and thus likely responds directly to changes in cellular Ca2+ levels. Single and multiple mutants of ABF2, 3, and 4 display reduced survival under drought conditions (Fujita et al., 2005; Yoshida et al., 2010). ABA-dependent phosphorylation by SnRK2 (SNF1-related protein kinase 2) confers full activity to AREB/ABF factors, which then induce expression of abiotic ABA-responsive genes in a cooperative manner (Uno et al., 2000; Fujita et al., 2005; Mizoguchi et al., 2010; Yoshida et al., 2010). The types of genes that AREB/ABF factors upregulate under water stress include regulatory proteins (phosphatases, kinases, and TFs) and functional genes encoding late embryogenesis abundant (LEA) proteins (Yoshida et al., 2010). In tomato, the drought- and salinity-induced SlAREB1 and SlAREB2 confer tolerance to water stress by activating genes encoding oxidative stress–related proteins, lipid transfer proteins, transcription regulators, and LEA proteins (Orellana et al., 2010).
The CaM binding protein MYB2 in Arabidopsis functions as a transcriptional activator under drought stress (Abe et al., 2003). MYB2 is transcriptionally induced by dehydration, and this upregulation is reversed upon rehydration (Urao et al., 1993). Additionally, upregulation of MYB2 mRNA is detected upon salt stress and treatment with ABA (Urao et al., 1993). Transgenic plants overexpressing MYB2 exhibit higher sensitivity to ABA and display enhanced ABA-induced expression of the dehydration-responsive gene rd22 (Abe et al., 2003). In vitro, MYB2 binds the MYB recognition element in the promoter of rd22, and in Arabidopsis leaf protoplasts it activates the transcription of a reporter gene driven by a 67-bp region of the rd22 promoter (Urao et al., 1993; Abe et al., 1997, 2003).
The GT-2 LIKE1 (GTL1) TF, a CaM binding member of the GTL family, is a negative regulator of drought resistance. An At-GTL1 loss-of-function mutant, gtl1, enhanced the capacity of plants to survive drought through reduced transpiration (Yoo et al., 2007, 2010). Lower density of stomata on the abaxial surface and high expression of STOMATAL DENSITY AND DISTRIBUTION1 (SDD1) was attributed to reduced transpiration (Yoo et al., 2010). GTL1 expression is downregulated by dehydration stress. GTL1 binds the promoter of SDD1 and represses its expression (Yoo et al., 2010). The expression of the Ca2+/CaM binding NAC TF, CBNAC, is upregulated upon exposure to a combination of drought and heat stress (Rizhsky et al., 2004), suggesting a role for CBNAC in stress responses. However, the mechanism of its involvement is not known.
Cold
Most temperate plants exhibit cold acclimation (i.e., increased tolerance to freezing by prior exposure to low nonfreezing temperatures), which involves changes in gene expression (Fowler and Thomashow, 2002; Kreps et al., 2002). The C-repeat binding factors CBF1, 2, and 3, which are also called DREB1B, 1C, and 1A, respectively, are TFs that induce the expression of a large number of genes (CBF regulon) involved in cold acclimation (Riechmann et al., 2000; Maruyama et al., 2004; Sakamoto et al., 2004; Vogel et al., 2005). Several lines of evidence have implicated Ca2+ in cold acclimation. For example, a rapid increase in [Ca2+]cyt is required for cold induction of KIN1, a member of the CBF regulon (Knight et al., 1996). Also, cold acclimation was prevented by the administration of Ca2+ chelators and Ca2+ channel blockers (Monroy et al., 1997). Recent research has revealed a role for CAMTAs in cold acclimation (Doherty et al., 2009). Doherty et al. (2009) analyzed the promoter region of the three CBFs and found seven conserved DNA motifs (CM1-7). The CM2 motif found in CBF2 matched the CAMTA core DNA binding motif and the tested CAMTAs (1, 2, 3, and 5) were able to bind the CBF2 promoter. Mutant studies revealed that camta3 plants had a reduction of cold-induced accumulation of CBF2 transcripts (Doherty et al., 2009). Many cold-responsive genes contain the CAMTA binding sequence CGCG and therefore may be transcriptionally regulated by CAMTA proteins upon exposure to cold (Doherty et al., 2009). Although camta3 mutants did not show a cold stress–related phenotype, the camta1 camta3 double mutant was impaired in freezing tolerance, suggesting a likely functional redundancy between these two genes.
Several MADS box gene family members in Arabidopsis bind CaM (Popescu et al., 2007) and are likely directly regulated by cellular Ca2+ levels. Some of these CaM binding proteins are downregulated upon exposure to cold (AGL3, AGL8, AGL15, and AGL32) (Hannah et al., 2005), raising the possibility that they may be transcriptional regulators mediating cold stress responses. Many of these TFs are implicated in floral development and flowering (Dornelas et al., 2011). Furthermore, several wheat (Triticum aestivum) genes associated with flower development are implicated in abiotic stress responses (Tardif et al., 2007). It would be interesting to investigate whether these TFs are involved in integrating cold stress with flowering. Additionally, as some of the MADS proteins are expressed not only in flowers, but in other aboveground vegetative organs, a more general role in cold stress responses is a possibility that would require further investigation (Huang et al., 1995; Teper-Bamnolker and Samach, 2005). As in Arabidopsis, rice expression profiling during desiccation, cold, and salt stress revealed differential expressions of several MADS box genes (Arora et al., 2007). Interestingly, the CaM binding AGL15 directly binds to and modulates expression of several genes encoding proteins in the MYB, WRKY, ABF, and AP2-domain families that are implicated in stress responses (Hill et al., 2008; Zheng et al., 2009; Dubos et al., 2010). It has been shown that AGL15 negatively regulates the TF CBF2 (Hill et al., 2008). Seedlings constitutively expressing AGL15 show downregulation of CBF2 compared with wild-type plants, suggesting an AGL15 role in cold stress responses.
An indirect regulation of cold acclimation by Ca2+/CaM was revealed in a study of a Ca2+/CaM-regulated receptor kinase CRLK1 (Yang et al., 2010b). Ca2+/CaM binding to CRLK1 upregulates its activity. Later work showed that CRLK1 interacts with MEKK1, a mitogen-activated protein (MAP) kinase kinase kinase family member, both in vitro and in planta (Yang et al., 2010c). In crlk1 mutants, the cold-triggered MAP kinase activation was abolished, and the cold-induced expression levels of genes involved in MAP kinase signaling were altered.
Salt
Over 6% of land (>800 million hectares) throughout the world is affected by salinity, a major factor limiting crop productivity (Munns and Tester, 2008). In response to salt stress, plants activate various signaling pathways, including those involving Ca2+ that promote adequate cellular responses (Zhu, 2002). A multitude of genes are activated upon exposure to salinity, including ion channels, receptors, signaling molecules, and genes involved in producing compatible solutes (e.g., osmoprotectants Gly betaine and Pro) (Tuteja, 2007). The phytohormone ABA plays an important role in adaptation of plants to salinity.
The salt-overly-sensitive (SOS) pathway is important in the decoding of salt stress–mediated Ca2+ signatures (Chinnusamy et al., 2004; Mahajan et al., 2008). The SOS3 (CBL4)/SOS2 (CIPK24) complex regulates ion homeostasis by modulating the expression and activity of SOS1, a plasma membrane–localized Na+/H+ antiporter, which exports Na+ to the apoplast (Chinnusamy et al., 2004). CaM is also proving to be a major sensory molecule in the Ca2+-induced response to salt stress (Bouché et al., 2005). Some members of the CAMTA family of TFs are upregulated in response to salt stresses as well as other abiotic and biotic stresses (Yang and Poovaiah, 2002). Plant lines expressing At-CAMTA1:β-glucuronidase (GUS) and exposed to NaCl showed an increase in GUS expression in leaves with increasing salt concentration (Galon et al., 2010b), implicating CAMTA1 in salt stress response. The spatial expression of AtCAMTA1:GUS in response to NaCl stress was different from that resulting from heat shock.
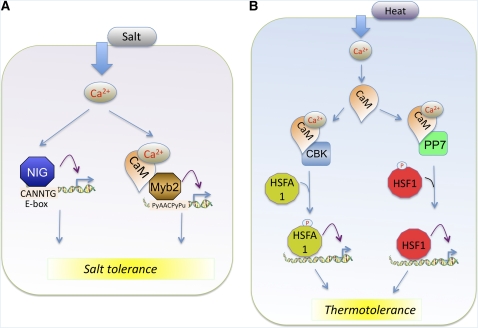
Also, a divergent type of soybean CaM isoform (Gm-CaM4), which is induced by salt, has been reported to mediate salt-induced Ca2+ signaling by activating the R2R3-type MYB2 TF (Yoo et al., 2005). MYB2 is an upstream regulator of a set of salt- and dehydration-responsive genes. CaM binds to At-MYB2 in a Ca2+-dependent manner, which enhances the DNA binding activity of MYB2 and its transcriptional activation (Figure 5A). A closely related CaM (Gm-CaM1) inhibits its DNA binding activity. Overexpression of Gm-CaM4 leads to constitutive expression of salt- and dehydration-responsive genes, including the Pro-synthesizing enzyme P5CS1 (Δ1-pyrroline-5-carboxylate synthetase-1), which confers salt tolerance by facilitating Pro accumulation (Yoo et al., 2005).
Figure 5.
Regulation of Genes Involved in Abiotic Stresses by Ca2+ and Ca2+/CaM Binding TFs.
Salt (A) and heat (B). Purple arrows indicate activation of gene expression. NIG, NaCl-inducible gene 1, an EF-hand–containing TF; PP7, Ca2+-CaM binding protein phosphatase 7; HSF, heat shock factor. See text for details.
One of the EF-hand–containing proteins predicted to contain a bHLH DNA binding domain (Day et al., 2002) was isolated as a salt-induced gene (Kim and Kim, 2006). This Ca2+ binding TF, named At-NIG1 (NaCl-induced gene), binds to the canonical E-box element (CANNTG) found in the promoter region of several salt stress–responsive genes (Figure 5A). Although the biological role of Ca2+ binding on the activity of NIG1 remains unclear, NIG1 loss-of-function mutant plants are more sensitive to salt stress and abscisic acid (Kim and Kim, 2006). Phosphorylation of NIG1 may be involved, as the motifs RXX(S/T) and KXX(S/T) of the salt stress response kinase SOS2 (Liu et al., 2000) are present in the NIG1 protein. There are a few other EF-hand–containing proteins with a DNA binding motif (Day et al., 2002), the functions of which have not been studied.
Another group of plant-specific CaM binding proteins, At-BT1-5 (Arabidopsis BTB and TAZ domain proteins 1-5) that respond to H2O2 and SA as well as to different abiotic stresses were reported (Du and Poovaiah, 2004). These proteins have been shown to interact with transcriptional regulators (Du and Poovaiah, 2004). At-BT1 and 2 localize to the nucleus and cytosol, whereas At-BT3, 4, and 5 are cytosolic (Robert et al., 2009). At-BT proteins participate in Arabidopsis development, metabolic processes, and hormone signaling (Ren et al., 2007; Robert et al., 2009; Mandadi et al., 2009), and a role in abiotic stress has been suggested as they exhibit differential expression upon exposure to salt (At-BT2, 3, and 4) and cold stress (At-BT2, 3, and 5) (Du and Poovaiah, 2004; Mandadi et al., 2009). Furthermore, At-BT1, 2, and 5 are induced in plants overexpressing the salt tolerance–conferring TF At-bZIP60, suggesting involvement of these At-BT proteins in protecting plants from salt stress (Fujita et al., 2007).
Expression of a nuclear CaM binding protein (CaMBP25) is induced by several abiotic stresses (dehydration, low temperature, or high salinity). Plants overexpressing CaMBP25 showed increased sensitivity to salt and osmotic stresses, whereas antisense lines were more tolerant to these stresses, suggesting it functions as a negative regulator of these stress responses (Perruc et al., 2004). Whether these nuclear CBPs function in regulating gene expression remains to be seen. Extracellular ATP also plays a regulatory role in plant stress adaptation by modulating expression of stress-related genes. A rise in extracellular ATP is observed in response to biotic and abiotic stresses. Increased extracellular ATP causes an elevation in [Ca2+]cyt in Arabidopsis as well as upregulation of transcription of MPK3, a kinase known to be involved in salt, cold, and touch responses (Demidchik et al., 2009).
Heat
Plants are constantly exposed to fluctuations in temperature. Heat stress can disturb cellular homeostasis, impair plant growth, and even cause plant death. Plant thermotolerance can be achieved through accumulation of heat shock proteins, whose transcription is tightly regulated by TFs. Heat-induced elevation of cellular Ca2+ results in changes in expression of several genes, including Ca2+ sensors (Braam, 1992; Zhang et al., 2009). A CaM3 knockout mutant in Arabidopsis displayed impaired thermotolerance, whereas CaM3 overexpression increased plant thermotolerance. Expression of heat shock genes was increased in CaM3-overexpressing lines and reduced in knockout plants (Zhang et al., 2009). A loss-of-function mutant of a CaM binding phosphatase (PP7) reduced tolerance to heat and its overexpression, which increased heat shock proteins expression and conferred thermotolerance (Liu et al., 2007). Since this phosphatase interacts with a heat shock TF (HSF1), it is likely that Ca2+/CaM through PP7 modulates the activity of HSF1. A CaM binding protein kinase (CBK) also positively regulates thermotolerance (Liu et al., 2007). This kinase, which regulates transcription of heat shock proteins, interacts with a heat shock TF (At-HSFA1a) and phosphorylates it. These results suggest that Ca2+, through CaM binding kinases and phosphatases, modulates the activity of heat shock TFs, resulting in changes in heat shock proteins (Figure 5B). CAMTA1, which is induced by auxin, has also been implicated in the heat shock response. CAMTA1:GUS plants that were exposed to heat shock showed expression in leaf trichomes at the leaf base and in the root cortex (Galon et al., 2010b), while the promoter of an auxin-induced gene showed no expression. This suggests that CAMTA1 responds to heat shock through promoter elements that are different from those responding to auxin.
Mechanical Stress
Mechanical stress such as touch and wind alter plant growth. Mechanical stimuli in plants lead to a general stress response (Walley and Dehesh, 2010). It is well established that mechanical stimulation causes an increase of [Ca2+]cyt (Knight et al., 1991, 1992; Monshausen et al., 2009). Calcium changes are thought to link the mechanical stimulus to plant growth responses. Expression of several genes, including CaM and CaM-related genes, is induced by mechanical stimuli (Braam and Davis, 1990; Braam et al., 1997; van Der Luit et al., 1999; Walley and Dehesh, 2010). A bioinformatic analysis of the data obtained in a transcriptome study following mechanical stress identified an overrepresented cis-element in the promoter of rapid wound-responsive genes (Walley et al., 2007). This element, CGCGTT, termed the rapid stress response element (RSRE), contains the core CAMTA cis-element CGCG. Several stress-related genes with the CGCG core sequences have been shown to bind CAMTAs; hence, it is likely that RSRE-containing genes are also regulated by this family of TFs (Doherty et al., 2009; Du et al., 2009) and that CAMTAs may be involved in the rapid response to stresses including wounding (Walley et al., 2007; Walley and Dehesh, 2010). Fusion of the RSRE to a reporter gene was found to be sufficient for activation of the reporter gene in response to several abiotic and biotic stresses.
Ca2+/CaM has also been shown to be involved in the response to mechanical wounding through a MAP kinase (MPK) pathway (Takahashi et al., 2011). Arabidopsis MPK8, a component of the wound signaling pathway that negatively regulates ROS accumulation by controlling expression of the Rboh D gene, is known to be phosphorylated and activated by a MAP kinase kinase, MKK3. However, full activation of MPK8 was shown to require Ca2+-dependent binding of CaM4 (Takahashi et al., 2011).
Flooding
Flooding affects vegetation in river plains and cultivated lands and is a major concern for food production. The primary stress experienced by plants growing in flooded soils is limited oxygen availability. Plants respond to low oxygen stress by shifting the carbohydrate metabolism in root cells from an oxidative to a fermentative pathway to allow limited energy production while facing limited oxygen supply (reviewed in Subbaiah and Sachs, 2003). In maize, 20 anaerobic proteins (ANPs) get selectively synthesized under hypoxia and end up accounting for more than 70% of the total translation. Most ANPs are enzymes of glycolysis or sugar-phosphate metabolism and help flooded plants survive under adverse conditions. For example, maize mutants that are null for the ANP alcohol dehydrogenase (ADH) only survive a few hours of anoxia, while most maize genotypes survive up to 3 d of anaerobic treatment at 27°C (Subbaiah and Sachs, 2003).
Genes encoding ANPs are rapidly induced upon even mild oxygen deprivation and are downregulated upon reoxygenation. In maize seedlings and cultured cells, gene expression in response to low oxygen is signaled by an elevation of [Ca2+]cyt (Subbaiah et al., 1994a, 1994b). Intracellular Ca2+ levels increase within minutes after oxygen depletion in cultured cells and decrease within seconds after reoxygenation (Subbaiah et al., 1994a). The observed Ca2+ increase occurs by mobilization from intracellular stores (Subbaiah et al., 1994a). Anoxic activation of genes encoding ADH1 and sucrose synthase1 is suppressed upon blocking Ca2+ channels with ruthenium red, an inhibitor of organellar Ca2+ fluxes (Subbaiah et al., 1994a, 1994b). In Arabidopsis, the hypoxia-induced CaM binding TF AtMYB2 activates ADH1 expression under low oxygen conditions by binding to the GT-motif (5′-TGGTTT-3′) in the ADH1 promoter (Hoeren et al., 1998). In rice, the protein kinase CIPK15 contributes to low oxygen tolerance by modulating the SnRK1A kinase-dependent sugar-sensing cascade, which regulates plant sugar and energy production (Lee et al., 2009).
Ca2+ AND Ca2+/CaM IN POSTTRANSCRIPTIONAL GENE REGULATION
The examples discussed above are focused on Ca2+- and Ca2+/CaM-regulated gene expression at the transcriptional level. Recent studies suggest that posttranscriptional regulation of gene expression at the level of mRNA transport, pre-mRNA splicing, mRNA turnover, and small RNAs (small interfering RNAs, microRNAs, trans-acting small interfering RNAs, and other noncoding RNAs) also play an important role in stress-induced reprogramming of the transcriptome (Iida et al., 2004; Lee et al., 2006; Palusa et al., 2007; Reddy, 2007; Chinnusamy et al., 2010; Walley and Dehesh, 2010; Walley et al., 2010; Yao et al., 2010). It is possible that Ca2+ may regulate gene expression at the posttranscriptional level. It is known that alternative splicing of pre-mRNAs is significantly impacted by stresses (Iida et al., 2004; Reddy, 2007). The fact that there are several RNA binding proteins that interact with CaM suggests their role in RNA metabolism (Reddy et al., 2002b; Delaney et al., 2006; Popescu et al., 2007). Furthermore, alternative splicing of one of the splicing regulators is changed by cellular Ca2+ manipulations (S.G. Palusa and A.S.N. Reddy, unpublished data). In Arabidopsis, the small subunit of the cleavage and polyadenylation specificity factor binds CaM, and this interaction inhibits its RNA binding activity in a Ca2+-dependent manner, suggesting that signals through cellular Ca2+ changes can regulate polyadenylation and functions associated with this process (Delaney et al., 2006).
CONCLUSIONS AND PERSPECTIVES
Transcriptional reprogramming is a unifying common principle among the stress-induced responses. Plants have exploited Ca2+ more extensively than any other messenger in regulating stress responses. This is evident by elaborate mechanisms that maintain Ca2+ homeostasis and elicit stress-specific Ca2+ signatures. More importantly, plants have myriad Ca2+ sensors that either directly regulate cellular processes or interact with other proteins in a Ca2+-dependent manner and regulate their activity/function to elicit cellular responses. About 3% of the Arabidopsis proteome is involved in Ca2+ signaling. Although significant progress has been made in elucidating Ca2+-mediated events involved in signal-induced responses in general, the roles of Ca2+- and Ca2+/CaM-regulated TFs in stress regulation are just beginning to be understood.
Limited studies on CaM binding TFs are revealing interesting paradigms. These studies suggest that Ca2+- and Ca2+/CaM- regulated TFs play a key role in suppressing inappropriate activation of plant defense and stress adaptation responses (Figure 6). This is exemplified with studies on CAMTA3 and some WRKYs, which suppress plant defense, and GT-2, which suppresses drought resistance. It is also evident that Ca2+- and Ca2+/CaM-regulated TFs function both as positive and negative regulators of stress responses. For instance, CAMTA3 functions as a negative regulator of plant immunity, whereas CBP60, TGAs, and specific CDPKs function as positive regulators in disease resistance. A given CaM binding TF can function in multiple stresses. Furthermore, in response to some stresses, a CAM binding TF can function as a positive regulator of transcription, whereas the same TF acts as a negative regulator of gene expression in response to other stresses. This is illustrated by CAMTA3, which acts as a negative regulator of bacterial and fungal resistance and positive regulator of cold stress responses (Figure 6). This suggests that whether a particular TF functions as an activator or a repressor depends on the gene and other proteins that interact with the promoter of that gene.
Figure 6.
Overview of Negative and Positive Regulation of Biotic and Abiotic Stresses by Ca2+ and Ca2+/CaM Binding TFs.
Biotic (A) and abiotic (B) stresses. Positive regulation is indicated by an arrow, and negative regulation is indicated by lines with a horizontal line. Pathways lacking evidence are represented by broken arrows.
Functions of only a few CaM binding TFs have been studied so far. Functional studies with other Ca2+- and Ca2+/CaM-regulated TFs are necessary to fully understand their roles in stress responses. In addition, it is necessary to decipher Ca2+-regulated transcriptional networks involved in a particular stress and in a combination of stresses. This will require application of emerging tools and the use of novel approaches that combine system level analyses with conventional genetic and biochemical studies. To elucidate Ca2+/CaM-regulated signaling networks, it is necessary to identify direct targets of each Ca2+- and Ca2+/CaM-regulated TF globally and integrate this data into networks maps. Chromatin immunoprecipitation combined with next generation sequencing should allow genome-wide identification of direct targets of TFs regulated by Ca2+ or Ca2+/CaM (Johnson et al., 2007; Schmidt et al., 2009). Chromatin immunoprecipitation, coupled with transcriptome analyses performed with RNA-seq (Wang et al., 2009b) or microarrays using an approach similar to Wang et al. (2006) should help identify the consequences of TF binding to direct targets (i.e., induction or repression of gene expression) and also identify secondary effects of TFs on gene expression. Integration of the data generated from such studies for each TF with physiological responses observed with mutants will pave the way to construct Ca2+/CaM-regulated transcriptional networks and their impact on stress responses. Such studies should help identify key nodes and commonalities among different stress signaling pathways and lead to unraveling of the choreography of Ca2+-mediated signal transcriptional networks. A recent study revealed a significant overlap in transcriptional responses to multiple individual stress conditions (Narsai et al., 2010). Identification of Ca2+/CaM-regulated TFs that regulate a core set of genes that respond to several abiotic treatments could be particularly useful in developing plants that are tolerant to multiple stresses. In addition, these analyses will also shed light on why plants often respond oppositely to biotic and abiotic stresses and hopefully lead to devising strategies for uncoupling biotic from abiotic stress responses in developing multistress resistant varieties.
Cellular Ca2+ changes and Ca2+-mediated alterations in gene expression in response to a combination of stresses that mimic natural environments and the effect of such changes at the molecular levels are least studied and require much further research (Mittler, 2006; Mittler and Blumwald, 2010). This is critical for developing plants that are stress tolerant under field conditions. Taking advantage of natural variation by comparing ecotypes that are tolerant to different stresses to stress-sensitive ecotypes to analyze Ca2+ signaling events might prove useful in identifying critical Ca2+-mediated pathways important for stress adaptation. In vivo interaction and dynamics of CaM binding TFs with activated CaM/CMLs in normal and stressed plants using live-cell imaging methods, such as bimolecular fluorescence complementation, fluorescence resonance energy transfer, fluorescence lifetime imaging microscopy, and analysis of protein complexes associated with CaMs/CMLs, are needed to obtain further insights into mechanistic aspects of Ca2+/CaM-regulated gee expression.
As discussed above, significant progress has been made in understanding the biological roles of some CaM binding TFs. However, in only a few cases is the effect of Ca2+ or Ca2+/CaM in regulating their DNA binding activity known. Furthermore, mechanisms of regulation of TFs by Ca2+ and Ca2+/CaM are completely unknown, and this is certainly an area that requires further investigation in the future.
It is hoped that the next decade will address many of the unanswered questions pertinent to Ca2+-mediated transcriptional networks and mode of regulation of CaM binding TFs. A thorough understanding of Ca2+- and Ca2+/CaM-regulated transcriptional networks will aid in developing new crop varieties with enhanced tolerance to environmental stresses.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Diagram of Two Nuclear CaM Binding Proteins Implicated in Biotic (PICBP and IQD1) and Abiotic Stress (CaMBP25).
Supplemental Table 1. CaM Binding Transcription Factors in Arabidopsis.
Supplementary Material
Acknowledgments
We thank current and former members in the Reddy laboratory and our collaborators who have contributed to calcium research. Research on signaling mechanisms in our laboratory is/was funded by grants from the from the National Science Foundation, the National Aeronautics and Space Administration, the Department of Energy, the U.S. Department of Agriculture, the Defense Advanced Research Projects Agency, and the Office of Naval Research to A.S.N.R. We thank Abdel-Ghany Salah for his comments on the manuscript.
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P.K., Agarwal P., Reddy M.K., Sopory S.K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 25: 1263–1274 [DOI] [PubMed] [Google Scholar]
- Ali G.S., Reddy V.S., Lindgren P.B., Jakobek J.L., Reddy A.S. (2003). Differential expression of genes encoding calmodulin-binding proteins in response to bacterial pathogens and inducers of defense responses. Plant Mol. Biol. 51: 803–815 [DOI] [PubMed] [Google Scholar]
- Ali R., Ma W., Lemtiri-Chlieh F., Tsaltas D., Leng Q., von Bodman S., Berkowitz G.A. (2007). Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G.J., Chu S.P., Harrington C.L., Schumacher K., Hoffmann T., Tang Y.Y., Grill E., Schroeder J.I. (2001). A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature 411: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Allen G.J., Chu S.P., Schumacher K., Shimazaki C.T., Vafeados D., Kemper A., Hawke S.D., Tallman G., Tsien R.Y., Harper J.F., Chory J., Schroeder J.I. (2000). Alteration of stimulus-specific guard cell calcium oscillations and stomatal closing in Arabidopsis det3 mutant. Science 289: 2338–2342 [DOI] [PubMed] [Google Scholar]
- al-Mohanna F.A., Caddy K.W., Bolsover S.R. (1994). The nucleus is insulated from large cytosolic calcium ion changes. Nature 367: 745–750 [DOI] [PubMed] [Google Scholar]
- Andrawis A., Solomon M., Delmer D.P. (1993). Cotton fiber annexins: A potential role in the regulation of callose synthase. Plant J. 3: 763–772 [DOI] [PubMed] [Google Scholar]
- Arora R., Agarwal P., Ray S., Singh A.K., Singh V.P., Tyagi A.K., Kapoor S. (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagué C., Lin B., Alcon C., Flottes G., Malmström S., Köhler C., Neuhaus G., Pelletier G., Gaymard F., Roby D. (2003). HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O., Waadt R., Steinhorst L., Held K., Kudla J. (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61: 211–222 [DOI] [PubMed] [Google Scholar]
- Biyaseheva A.E., Molotkovskii Y.G., Mamonov L.K. (1993). Increase of free Ca2+ in the cytosol of plant protoplast in response to heat stress as related to Ca2+ homeostasis. Russ. J. Plant Physiol. 40: 540–544 [Google Scholar]
- Blum A. (1996). Crop responses to drought and the interpretation of adaptation. Plant Growth Regul. 20: 135–148 [Google Scholar]
- Blume B., Nürnberger T., Nass N., Scheel D. (2000). Receptor-mediated increase in cytoplasmic free calcium required for activation of pathogen defense in parsley. Plant Cell 12: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., He S.Y. (2009). Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonburapong B., Buaboocha T. (2007). Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 7: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N., Scharlat A., Snedden W., Bouchez D., Fromm H. (2002). A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 277: 21851–21861 [DOI] [PubMed] [Google Scholar]
- Bouché N., Yellin A., Snedden W.A., Fromm H. (2005). Plant-specific calmodulin-binding proteins. Annu. Rev. Plant Biol. 56: 435–466 [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M.R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S.H., Sheen J. (2010). Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursiac Y., Lee S.M., Romanowsky S., Blank R., Sladek C., Chung W.S., Harper J.F. (2010). Disruption of the vacuolar calcium-ATPases in Arabidopsis results in the activation of a salicylic acid-dependent programmed cell death pathway. Plant Physiol. 154: 1158–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J.S. (1982). Plant productivity and environment. Science 218: 443–448 [DOI] [PubMed] [Google Scholar]
- Braam J. (1992). Regulated expression of the calmodulin-related TCH genes in cultured Arabidopsis cells: induction by calcium and heat shock. Proc. Natl. Acad. Sci. USA 89: 3213–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J., Davis R.W. (1990). Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell 60: 357–364 [DOI] [PubMed] [Google Scholar]
- Braam J., Sistrunk M.L., Polisensky D.H., Xu W., Purugganan M.M., Antosiewicz D.M., Campbell P., Johnson K.A. (1997). Plant responses to environmental stress: Regulation and functions of the Arabidopsis TCH genes. Planta 203(suppl): S35–S41 [DOI] [PubMed] [Google Scholar]
- Brivanlou A.H., Darnell J.E., Jr. (2002). Signal transduction and the control of gene expression. Science 295: 813–818 [DOI] [PubMed] [Google Scholar]
- Campbell A.K., Trewavas A.J., Knight M.R. (1996). Calcium imaging shows differential sensitivity to cooling and communication in luminous transgenic plants. Cell Calcium 19: 211–218 [DOI] [PubMed] [Google Scholar]
- Chen Y.-R., Datta N., Roux S.J. (1987). Purification and partial characterization of a calmodulin-stimulated nucleoside triphosphatase from pea nuclei. J. Biol. Chem. 262: 10689–10694 [PubMed] [Google Scholar]
- Cheng S.H., Willmann M.R., Chen H.C., Sheen J. (2002). Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129: 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiasson D., Ekengren S.K., Martin G.B., Dobney S.L., Snedden W.A. (2005). Calmodulin-like proteins from Arabidopsis and tomato are involved in host defense against Pseudomonas syringae pv. tomato. Plant Mol. Biol. 58: 887–897 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Schumaker K., Zhu J.K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 55: 225–236 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J.K., Sunkar R. (2010). Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 639: 39–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Choi M.S., et al. (2005). Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J. Biol. Chem. 280: 40820–40831 [DOI] [PubMed] [Google Scholar]
- Clark G.B., Roux S.J. (1995). Annexins of plant cells. Plant Physiol. 109: 1133–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton H., Knight M.R., Knight H., McAinsh M.R., Hetherington A.M. (1999). Dissection of the ozone-induced calcium signature. Plant J. 17: 575–579 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Fengler K.A., Yu I.C., Lippok B., Smith R.K., Jr, Bent A.F. (2000). The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 97: 9323–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa e Silva O. (1994). CG-1, a parsley light-induced DNA-binding protein. Plant Mol. Biol. 25: 921–924 [DOI] [PubMed] [Google Scholar]
- Dammann C., Ichida A., Hong B., Romanowsky S.M., Hrabak E.M., Harmon A.C., Pickard B.G., Harper J.F. (2003). Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol. 132: 1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwalder M., Roux S.J., Hardison L. (1986). Distribution of calmodulin in pea seedlings: Immunocytochemical localization in plumules and root apices. Planta 168: 461–470 [PubMed] [Google Scholar]
- Day I.S., Reddy V.S., Shad Ali G., Reddy A.S. (2002). Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol. 3: RESEARCH0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco T.A., Bender K.W., Snedden W.A. (2010). Breaking the code: Ca2+ sensors in plant signalling. Biochem. J. 425: 27–40 [DOI] [PubMed] [Google Scholar]
- Delaney K.J., Xu R., Zhang J., Li Q.Q., Yun K.Y., Falcone D.L., Hunt A.G. (2006). Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol. 140: 1507–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V., et al. (2009). Plant extracellular ATP signalling by plasma membrane NADPH oxidase and Ca2+ channels. Plant J. 58: 903–913 [DOI] [PubMed] [Google Scholar]
- Dhlamini Z., Spillane C., Moss J.P., Ruane J., Urquia N., Sonnino A. (2005). Status of Research and Applications of Crop Biotechnologies in Developing Countries: Preliminary Assessment. (Rome: Food and Agriculture Organization of the United Nations; ), pp. 1–53 [Google Scholar]
- Dodd A.N., Kudla J., Sanders D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Doherty C.J., Van Buskirk H.A., Myers S.J., Thomashow M.F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornelas M.C., Patreze C.M., Angenent G.C., Immink R.G. (2011). MADS: The missing link between identity and growth? Trends Plant Sci. 16: 89–97 [DOI] [PubMed] [Google Scholar]
- Du L., Ali G.S., Simons K.A., Hou J., Yang T., Reddy A.S., Poovaiah B.W. (2009). Ca(2+)/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457: 1154–1158 [DOI] [PubMed] [Google Scholar]
- Du L., Poovaiah B.W. (2004). A novel family of Ca2+/calmodulin-binding proteins involved in transcriptional regulation: interaction with fsh/Ring3 class transcription activators. Plant Mol. Biol. 54: 549–569 [DOI] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I.E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Falk A., Feys B.J., Frost L.N., Jones J.D., Daniels M.J., Parker J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W., Dong X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkler A., Ashery-Padan R., Fromm H. (2007). CAMTAs: Calmodulin-binding transcription activators from plants to human. FEBS Lett. 581: 3893–3898 [DOI] [PubMed] [Google Scholar]
- Fowler S., Thomashow M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M.M., Seki M., Hiratsu K., Ohme-Takagi M., Shinozaki K., Yamaguchi-Shinozaki K. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Mizukado S., Fujita Y., Ichikawa T., Nakazawa M., Seki M., Matsui M., Yamaguchi-Shinozaki K., Shinozaki K. (2007). Identification of stress-tolerance-related transcription-factor genes via mini-scale Full-length cDNA Over-eXpressor (FOX) gene hunting system. Biochem. Biophys. Res. Commun. 364: 250–257 [DOI] [PubMed] [Google Scholar]
- Galon Y., Aloni R., Nachmias D., Snir O., Feldmesser E., Scrase-Field S., Boyce J.M., Bouché N., Knight M.R., Fromm H. (2010b). Calmodulin-binding transcription activator 1 mediates auxin signaling and responds to stresses in Arabidopsis. Planta 232: 165–178 [DOI] [PubMed] [Google Scholar]
- Galon Y., Finkler A., Fromm H. (2010a). Calcium-regulated transcription in plants. Mol. Plant 3: 653–669 [DOI] [PubMed] [Google Scholar]
- Galon Y., Nave R., Boyce J.M., Nachmias D., Knight M.R., Fromm H. (2008). Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 582: 943–948 [DOI] [PubMed] [Google Scholar]
- Gleason C., Chaudhuri S., Yang T., Muñoz A., Poovaiah B.W., Oldroyd G.E. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Gong M., van der Luit A.H., Knight M.R., Trewavas A.J. (1998). Heat-shock-induced changes in intracellular Ca2+ level in tobacco seedlings in relation to thermotolerance. Plant Physiol. 116: 429–437 [Google Scholar]
- Grant M., Brown I., Adams S., Knight M., Ainslie A., Mansfield J. (2000). The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 23: 441–450 [DOI] [PubMed] [Google Scholar]
- Gust A.A., Biswas R., Lenz H.D., Rauhut T., Ranf S., Kemmerling B., Götz F., Glawischnig E., Lee J., Felix G., Nürnberger T. (2007). Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J. Biol. Chem. 282: 32338–32348 [DOI] [PubMed] [Google Scholar]
- Hadiarto T., Tran L.S. (2010). Progress studies of drought-responsive genes in rice. Plant Cell Rep. 30: 297–310 [DOI] [PubMed] [Google Scholar]
- Han J., Gong P., Reddig K., Mitra M., Guo P., Li H.S. (2006). The fly CAMTA transcription factor potentiates deactivation of rhodopsin, a G protein-coupled light receptor. Cell 127: 847–858 [DOI] [PubMed] [Google Scholar]
- Hannah M.A., Heyer A.G., Hincha D.K. (2005). A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 1: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding S.A., Oh S.H., Roberts D.M. (1997). Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J. 16: 1137–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding S.A., Roberts D.M. (1998). Incompatible pathogen infection results in enhanced reactive oxygen and cell death responses in transgenic tobacco expressing a hyperactive mutant calmodulin. Planta 206: 253–258 [Google Scholar]
- Harper J.F., Breton G., Harmon A. (2004). Decoding Ca(2+) signals through plant protein kinases. Annu. Rev. Plant Biol. 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Harper J.F., Harmon A. (2005). Plants, symbiosis and parasites: A calcium signalling connection. Nat. Rev. Mol. Cell Biol. 6: 555–566 [DOI] [PubMed] [Google Scholar]
- Heath M.C. (2000). Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol. 3: 315–319 [DOI] [PubMed] [Google Scholar]
- Heo W.D., Lee S.H., Kim M.C., Kim J.C., Chung W.S., Chun H.J., Lee K.J., Park C.Y., Park H.C., Choi J.Y., Cho M.J. (1999). Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc. Natl. Acad. Sci. USA 96: 766–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler P.K. (2005). Calcium: A central regulator of plant growth and development. Plant Cell 17: 2142–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Wang H., Perry S.E. (2008). A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 53: 172–185 [DOI] [PubMed] [Google Scholar]
- Hirayama T., Shinozaki K. (2010). Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 61: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Hoeren F.U., Dolferus R., Wu Y., Peacock W.J., Dennis E.S. (1998). Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 149: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Barlet X., Deslandes L., Hirsch J., Feng D.X., Somssich I., Marco Y. (2008). Transcriptional responses of Arabidopsis thaliana during wilt disease caused by the soil-borne phytopathogenic bacterium, Ralstonia solanacearum. PLoS One 3: e2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X.Y., Neill S.J., Cai W.M., Tang Z.C. (2004). Induction of defence gene expression by oligogalacturonic acid requires increases in both cytosolic calcium and hydrogen peroxide in Arabidopsis thaliana. Cell Res. 14: 234–240 [DOI] [PubMed] [Google Scholar]
- Huang H., Tudor M., Weiss C.A., Hu Y., Ma H. (1995). The Arabidopsis MADS-box gene AGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol. Biol. 28: 549–567 [DOI] [PubMed] [Google Scholar]
- Iida K., Seki M., Sakurai T., Satou M., Akiyama K., Toyoda T., Konagaya A., Shinozaki K. (2004). Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res. 32: 5096–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabs T., Tschöpe M., Colling C., Hahlbrock K., Scheel D. (1997). Elicitor-stimulated ion fluxes and O2- from the oxidative burst are essential components in triggering defense gene activation and phytoalexin synthesis in parsley. Proc. Natl. Acad. Sci. USA 94: 4800–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., Parcy F.; bZIP Research Group (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Jami S.K., Clark G.B., Turlapati S.A., Handley C., Roux S.J., Kirti P.B. (2008). Ectopic expression of an annexin from Brassica juncea confers tolerance to abiotic and biotic stress treatments in transgenic tobacco. Plant Physiol. Biochem. 46: 1019–1030 [DOI] [PubMed] [Google Scholar]
- Johnson C.H., Knight M.R., Kondo T., Masson P., Sedbrook J., Haley A., Trewavas A. (1995). Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science 269: 1863–1865 [DOI] [PubMed] [Google Scholar]
- Johnson D.S., Mortazavi A., Myers R.M., Wold B. (2007). Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Journot-Catalino N., Somssich I.E., Roby D., Kroj T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchiswamy C.N., et al. (2010). Regulation of Arabidopsis defense responses against Spodoptera littoralis by CPK-mediated calcium signaling. BMC Plant Biol. 10: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan B., Davydov O., Knight H., Galon Y., Knight M.R., Fluhr R., Fromm H. (2006). Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18: 2733–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarwani M., Yoo J., Dong X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.S., Park B.O., Yoo J.H., Jung M.S., Lee S.M., Han H.J., Kim K.E., Kim S.H., Lim C.O., Yun D.J., Lee S.Y., Chung W.S. (2007). Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. J. Biol. Chem. 282: 36292–36302 [DOI] [PubMed] [Google Scholar]
- Kim J., Kim H.Y. (2006). Functional analysis of a calcium-binding transcription factor involved in plant salt stress signaling. FEBS Lett. 580: 5251–5256 [DOI] [PubMed] [Google Scholar]
- Kim K.C., Fan B., Chen Z. (2006). Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol. 142: 1180–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.C., Chung W.S., Yun D.J., Cho M.J. (2009). Calcium and calmodulin-mediated regulation of gene expression in plants. Mol. Plant 2: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.C., et al. (2002b). Mlo, a modulator of plant defense and cell death, is a novel calmodulin-binding protein. Isolation and characterization of a rice Mlo homologue. J. Biol. Chem. 277: 19304–19314 [DOI] [PubMed] [Google Scholar]
- Kim M.C., Panstruga R., Elliott C., Müller J., Devoto A., Yoon H.W., Park H.C., Cho M.J., Schulze-Lefert P. (2002a). Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416: 447–451 [DOI] [PubMed] [Google Scholar]
- Knight H. (2000). Calcium signaling during abiotic stress in plants. Int. Rev. Cytol. 195: 269–324 [DOI] [PubMed] [Google Scholar]
- Knight H., Trewavas A.J., Knight M.R. (1996). Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H., Trewavas A.J., Knight M.R. (1997). Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 12: 1067–1078 [DOI] [PubMed] [Google Scholar]
- Knight H., Veale E.L., Warren G.J., Knight M.R. (1999). The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 11: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight M.R., Campbell A.K., Smith S.M., Trewavas A.J. (1991). Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526 [DOI] [PubMed] [Google Scholar]
- Knight M.R., Smith S.M., Trewavas A.J. (1992). Wind-induced plant motion immediately increases cytosolic calcium. Proc. Natl. Acad. Sci. USA 89: 4967–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Ohura I., Kawakita K., Yokota N., Fujiwara M., Shimamoto K., Doke N., Yoshioka H. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19: 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S., Hazledine S., Sun J., Miwa H., Morris R.J., Downie J.A., Oldroyd G.E. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps J.A., Wu Y., Chang H.S., Zhu T., Wang X., Harper J.F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J., Batistic O., Hashimoto K. (2010). Calcium signals: The lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha R., Singh A., Chattopadhyay S. (2008). Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 20: 1747–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laohavisit A., Davies J.M. (2011). Annexins. New Phytol. 189: 40–53 [DOI] [PubMed] [Google Scholar]
- Laohavisit A., Mortimer J.C., Demidchik V., Coxon K.M., Stancombe M.A., Macpherson N., Brownlee C., Hofmann A., Webb A.A., Miedema H., Battey N.H., Davies J.M. (2009). Zea mays annexins modulate cytosolic free Ca2+ and generate a Ca2+-permeable conductance. Plant Cell 21: 479–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman D.S. (1997). Transcription factors: An overview. Int. J. Biochem. Cell Biol. 29: 1305–1312 [DOI] [PubMed] [Google Scholar]
- Lecourieux D., Lamotte O., Bourque S., Wendehenne D., Mazars C., Ranjeva R., Pugin A. (2005). Proteinaceous and oligosaccharidic elicitors induce different calcium signatures in the nucleus of tobacco cells. Cell Calcium 38: 527–538 [DOI] [PubMed] [Google Scholar]
- Lecourieux D., Mazars C., Pauly N., Ranjeva R., Pugin A. (2002). Analysis and effects of cytosolic free calcium increases in response to elicitors in Nicotiana plumbaginifolia cells. Plant Cell 14: 2627–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourieux D., Ranjeva R., Pugin A. (2006). Calcium in plant defence-signalling pathways. New Phytol. 171: 249–269 [DOI] [PubMed] [Google Scholar]
- Lee B.H., Kapoor A., Zhu J., Zhu J.K. (2006). STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 18: 1736–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Rudd J.J. (2002). Calcium-dependent protein kinases: Versatile plant signalling components necessary for pathogen defence. Trends Plant Sci. 7: 97–98 [DOI] [PubMed] [Google Scholar]
- Lee K., Thorneycroft D., Achuthan P., Hermjakob H., Ideker T. (2010). Mapping plant interactomes using literature curated and predicted protein-protein interaction data sets. Plant Cell 22: 997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.W., Chen P.W., Lu C.A., Chen S., Ho T.H., Yu S.M. (2009). Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal. 2: ra61. [DOI] [PubMed] [Google Scholar]
- Lee S., Lee E.J., Yang E.J., Lee J.E., Park A.R., Song W.H., Park O.K. (2004). Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell 16: 1378–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legué V., Blancaflor E., Wymer C., Perbal G., Fantin D., Gilroy S. (1997). Cytoplasmic free Ca2+ in Arabidopsis roots changes in response to touch but not gravity. Plant Physiol. 114: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A., Pennell R.I., Alvarez M.E., Palmer R., Lamb C. (1996). Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr. Biol. 6: 427–437 [DOI] [PubMed] [Google Scholar]
- Levy M., Wang Q., Kaspi R., Parrella M.P., Abel S. (2005). Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 43: 79–96 [DOI] [PubMed] [Google Scholar]
- Libault M., Wan J., Czechowski T., Udvardi M., Stacey G. (2007). Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol. Plant Microbe Interact. 20: 900–911 [DOI] [PubMed] [Google Scholar]
- Liu H.T., Li G.L., Chang H., Sun D.Y., Zhou R.G., Li B. (2007). Calmodulin-binding protein phosphatase PP7 is involved in thermotolerance in Arabidopsis. Plant Cell Environ. 30: 156–164 [DOI] [PubMed] [Google Scholar]
- Liu J., Ishitani M., Halfter U., Kim C.S., Zhu J.K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97: 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Harrington H.M. (1994). Isolation of tobacco cDNA clones encoding calmodulin-binding proteins and characterization of a known calmodulin-binding domain. Plant Physiol. Biochem. 32: 413–422 [Google Scholar]
- Luan S. (2009). The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 14: 37–42 [DOI] [PubMed] [Google Scholar]
- Luan S., Lan W., Chul Lee S. (2009). Potassium nutrition, sodium toxicity, and calcium signaling: Connections through the CBL-CIPK network. Curr. Opin. Plant Biol. 12: 339–346 [DOI] [PubMed] [Google Scholar]
- Luan S., Kudla J., Rodríguez-Concepcíon M., Yalovsky S., Gruissem W. (2002). Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 14(suppl): S389–S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Berkowitz G.A. (2007). The grateful dead: Calcium and cell death in plant innate immunity. Cell. Microbiol. 9: 2571–2585 [DOI] [PubMed] [Google Scholar]
- Ma W., Qi Z., Smigel A., Walker R.K., Verma R., Berkowitz G.A. (2009b). Ca2+, cAMP, and transduction of non-self perception during plant immune responses. Proc. Natl. Acad. Sci. USA 106: 20995–21000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Smigel A., Tsai Y.C., Braam J., Berkowitz G.A. (2008). Innate immunity signaling: Cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol. 148: 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Smigel A., Verma R., Berkowitz G.A. (2009a). Cyclic nucleotide gated channels and related signaling components in plant innate immunity. Plant Signal. Behav. 4: 277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei M.E., Mithöfer A., Boland W. (2007). Before gene expression: Early events in plant-insect interaction. Trends Plant Sci. 12: 310–316 [DOI] [PubMed] [Google Scholar]
- Mahajan S., Pandey G.K., Tuteja N. (2008). Calcium- and salt-stress signaling in plants: Shedding light on SOS pathway. Arch. Biochem. Biophys. 471: 146–158 [DOI] [PubMed] [Google Scholar]
- Mandadi K.K., Misra A., Ren S., McKnight T.D. (2009). BT2, a BTB protein, mediates multiple responses to nutrients, stresses, and hormones in Arabidopsis. Plant Physiol. 150: 1930–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., Sakuma Y., Kasuga M., Ito Y., Seki M., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38: 982–993 [DOI] [PubMed] [Google Scholar]
- Mazars C., Bourque S., Mithöfer A., Pugin A., Ranjeva R. (2009). Calcium homeostasis in plant cell nuclei. New Phytol. 181: 261–274 [DOI] [PubMed] [Google Scholar]
- Mazars C., Thuleau P., Lamotte O., Bourque S. (2010). Cross-talk between ROS and calcium in regulation of nuclear activities. Mol. Plant 3: 706–718 [DOI] [PubMed] [Google Scholar]
- McAinsh M.R., Clayton H., Mansfield T.A., Hetherington A.M. (1996). Changes in stomatal behavior and guard cell cytosolic free calcium in response to oxidative stress. Plant Physiol. 111: 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh M.R., Pittman J.K. (2009). Shaping the calcium signature. New Phytol. 181: 275–294 [DOI] [PubMed] [Google Scholar]
- McCormack E., Tsai Y.C., Braam J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10: 383–389 [DOI] [PubMed] [Google Scholar]
- Miller G., Suzuki N., Ciftci-Yilmaz S., Mittler R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2006). Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11: 15–19 [DOI] [PubMed] [Google Scholar]
- Mittler R., Blumwald E. (2010). Genetic engineering for modern agriculture: Challenges and perspectives. Annu. Rev. Plant Biol. 61: 443–462 [DOI] [PubMed] [Google Scholar]
- Mizoguchi M., Umezawa T., Nakashima K., Kidokoro S., Takasaki H., Fujita Y., Yamaguchi-Shinozaki K., Shinozaki K. (2010). Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant Cell Physiol. 51: 842–847 [DOI] [PubMed] [Google Scholar]
- Monroy A.F., Labbé E., Dhindsa R.S. (1997). Low temperature perception in plants: Effects of cold on protein phosphorylation in cell-free extracts. FEBS Lett. 410: 206–209 [DOI] [PubMed] [Google Scholar]
- Monshausen G.B., Bibikova T.N., Weisenseel M.H., Gilroy S. (2009). Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21: 2341–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Murray S.L., Ingle R.A., Petersen L.N., Denby K.J. (2007). Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol. Plant. Microbe Interact. 20: 1431–1438 [DOI] [PubMed] [Google Scholar]
- Nagano Y., Inaba T., Furuhashi H., Sasaki Y. (2001). Trihelix DNA-binding protein with specificities for two distinct cis-elements: Both important for light down-regulated and dark-inducible gene expression in higher plants. J. Biol. Chem. 276: 22238–22243 [DOI] [PubMed] [Google Scholar]
- Nakayama S., Kawasaki H., Kretsinger R. (2000). Evolution of EF-hand proteins. Calcium Homeostasis, Carafoli E., Krebs J., (New York: Springer; ), pp. 29–58 [Google Scholar]
- Narsai R., Castleden I., Whelan J. (2010). Common and distinct organ and stress responsive transcriptomic patterns in Oryza sativa and Arabidopsis thaliana. BMC Plant Biol. 10: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotera P., McConkey D.J., Jones D.P., Orrenius S. (1989). ATP stimulates Ca2+ uptake and increases the free Ca2+ concentration in isolated rat liver nuclei. Proc. Natl. Acad. Sci. USA 86: 453–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana S., Yañez M., Espinoza A., Verdugo I., González E., Ruiz-Lara S., Casaretto J.A. (2010). The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 33: 2191–2208 [DOI] [PubMed] [Google Scholar]
- Pachauri R.K., Reisinger A. eds (2007). Climate Change 2007: Synthesis Report. In IPCC Fourth Assessment Report (AR4): Contribution of Working Groups I, II and III to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. (Geneva, Switzerland: Intergovernmental Panel on Climate Change; ), pp. 1–104 [Google Scholar]
- Palusa S.G., Ali G.S., Reddy A.S. (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 49: 1091–1107 [DOI] [PubMed] [Google Scholar]
- Pandey S.P., Somssich I.E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.Y., et al. (2004). Pathogenesis-related gene expression by specific calmodulin isoforms is dependent on NIM1, a key regulator of systemic acquired resistance. Mol. Cells 18: 207–213 [PubMed] [Google Scholar]
- Park C.Y., Lee J.H., Yoo J.H., Moon B.C., Choi M.S., Kang Y.H., Lee S.M., Kim H.S., Kang K.Y., Chung W.S., Lim C.O., Cho M.J. (2005). WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 579: 1545–1550 [DOI] [PubMed] [Google Scholar]
- Patharkar O.R., Cushman J.C. (2000). A stress-induced calcium-dependent protein kinase from Mesembryanthemum crystallinum phosphorylates a two-component pseudo-response regulator. Plant J. 24: 679–691 [DOI] [PubMed] [Google Scholar]
- Pauly N., Knight M.R., Thuleau P., van der Luit A.H., Moreau M., Trewavas A.J., Ranjeva R., Mazars C. (2000). Control of free calcium in plant cell nuclei. Nature 405: 754–755 [DOI] [PubMed] [Google Scholar]
- Pei Z.M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G.J., Grill E., Schroeder J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734 [DOI] [PubMed] [Google Scholar]
- Perruc E., Charpenteau M., Ramirez B.C., Jauneau A., Galaud J.P., Ranjeva R., Ranty B. (2004). A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J. 38: 410–420 [DOI] [PubMed] [Google Scholar]
- Polisensky D.H., Braam J. (1996). Cold-shock regulation of the Arabidopsis TCH genes and the effects of modulating intracellular calcium levels. Plant Physiol. 111: 1271–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovaiah B.W., Reddy A.S.N. (1987). Calcium messenger system in plants. CRC Crit. Rev. Plant Sci. 6: 47–103 [DOI] [PubMed] [Google Scholar]
- Poovaiah B.W., Reddy A.S.N. (1993). Calcium and signal transduction in plants. CRC Crit. Rev. Plant Sci. 12: 185–211 [DOI] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Seay M., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2007). Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. USA 104: 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K.V.S.K., Ali G.S., Reddy A.S.N. (2009). WRKY53 transcription factor is a key component in Flg22 signaling. Poster 48, Plant Pathogen Interactions. In Plant Biology 2009, July 18–July 22, 2009, Honolulu, HI. American Society of Plant Biologists, Rockville, MD. Abstract no. P48069 [Google Scholar]
- Price A.H., Taylor A., Ripley S.J., Griffiths A., Trewavas A.J., Knight M.R. (1994). Oxidative signals in tobacco increase cytosolic calcium. Plant Cell 6: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichaudhuri A., Bhattacharyya R., Chaudhuri S., Chakrabarti P., Dasgupta M. (2006). Domain analysis of a groundnut calcium-dependent protein kinase: nuclear localization sequence in the junction domain is coupled with nonconsensus calcium binding domains. J. Biol. Chem. 281: 10399–10409 [DOI] [PubMed] [Google Scholar]
- Ranf S., Wünnenberg P., Lee J., Becker D., Dunkel M., Hedrich R., Scheel D., Dietrich P. (2008). Loss of the vacuolar cation channel, AtTPC1, does not impair Ca2+ signals induced by abiotic and biotic stresses. Plant J. 53: 287–299 [DOI] [PubMed] [Google Scholar]
- Reddy A.S. (2001). Calcium: Silver bullet in signaling. Plant Sci. 160: 381–404 [DOI] [PubMed] [Google Scholar]
- Reddy A.S. (2007). Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 58: 267–294 [DOI] [PubMed] [Google Scholar]
- Reddy A.S., Day I.S., Narasimhulu S.B., Safadi F., Reddy V.S., Golovkin M., Harnly M.J. (2002a). Isolation and characterization of a novel calmodulin-binding protein from potato. J. Biol. Chem. 277: 4206–4214 [DOI] [PubMed] [Google Scholar]
- Reddy A.S., Reddy V.S., Golovkin M. (2000). A calmodulin binding protein from Arabidopsis is induced by ethylene and contains a DNA-binding motif. Biochem. Biophys. Res. Commun. 279: 762–769 [DOI] [PubMed] [Google Scholar]
- Reddy A.S.N., Ben-Hur A., Day I.S. (2011). Experimental and computational approaches for the study of calmodulin interactions. Phytochemistry 72: 1007–1019. [DOI] [PubMed] [Google Scholar]
- Reddy A.S.N., Takezawa D., Fromm H., Poovaiah B.W. (1993). Isolation and characterization of two cDNAs that encode for calmodulin-binding proteins fron corn root tips. Plant Sci. 94: 109–117 [Google Scholar]
- Reddy V.S., Ali G.S., Reddy A.S. (2002b). Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J. Biol. Chem. 277: 9840–9852 [DOI] [PubMed] [Google Scholar]
- Reddy V.S., Ali G.S., Reddy A.S. (2003). Characterization of a pathogen-induced calmodulin-binding protein: mapping of four Ca2+-dependent calmodulin-binding domains. Plant Mol. Biol. 52: 143–159 [DOI] [PubMed] [Google Scholar]
- Reddy V.S., Day I.S., Thomas T., Reddy A.S. (2004). KIC, a novel Ca2+ binding protein with one EF-hand motif, interacts with a microtubule motor protein and regulates trichome morphogenesis. Plant Cell 16: 185–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V.S., Reddy A.S. (2004). Proteomics of calcium-signaling components in plants. Phytochemistry 65: 1745–1776 [DOI] [PubMed] [Google Scholar]
- Reddy V.S., Safadi F., Zielinski R.E., Reddy A.S.N. (1999). Interaction of a kinesin-like protein with calmodulin isoforms from Arabidopsis. J. Biol. Chem. 274: 31727–31733 [DOI] [PubMed] [Google Scholar]
- Ren S., Mandadi K.K., Boedeker A.L., Rathore K.S., McKnight T.D. (2007). Regulation of telomerase in Arabidopsis by BT2, an apparent target of TELOMERASE ACTIVATOR1. Plant Cell 19: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds M.P. (2010). Climate Change and Crop Production. (Cambridge, MA: CABI International; ). [Google Scholar]
- Riechmann J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rizhsky L., Liang H., Shuman J., Shulaev V., Davletova S., Mittler R. (2004). When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert H.S., Quint A., Brand D., Vivian-Smith A., Offringa R. (2009). BTB and TAZ DOMAIN scaffold proteins perform a crucial function in Arabidopsis development. Plant J., 58: 109–121 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M., Yalovsky S., Zik M., Fromm H., Gruissem W. (1999). The prenylation status of a novel plant calmodulin directs plasma membrane or nuclear localization of the protein. EMBO J. 18: 1996–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A.J., Knight M.R., Cove D.J., Knight C.D., Trewavas A.J., Wang T.L. (1996). The moss, Physcomitrella patens, transformed with apoaequorin cDNA responds to cold shock, mechanical perturbation and pH with transient increases in cytoplasmic calcium. Transgenic Res. 5: 167–170 [DOI] [PubMed] [Google Scholar]
- Sakamoto H., Maruyama K., Sakuma Y., Meshi T., Iwabuchi M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 136: 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D., Pelloux J., Brownlee C., Harper J.F. (2002). Calcium at the crossroads of signaling. Plant Cell 14(suppl.): S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D., Wilson M.D., Spyrou C., Brown G.D., Hadfield J., Odom D.T. (2009). ChIP-seq: Using high-throughput sequencing to discover protein-DNA interactions. Methods 48: 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurink R.C., Chan P.V., Jones R.L. (1996). Modulation of calmodulin mRNA and protein levels in barley aleurone. Plant Physiol. 111: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Seki M., Umezawa T., Urano K., Shinozaki K. (2007). Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 10: 296–302 [DOI] [PubMed] [Google Scholar]
- Shaw S.L., Long S.R. (2003). Nod factor elicits two separable calcium responses in Medicago truncatula root hair cells. Plant Physiol. 131: 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Brown R.M., Jr. (1999). GTPase activity and biochemical characterization of a recombinant cotton fiber annexin. Plant Physiol. 119: 925–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Shore P., Sharrocks A.D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229: 1–13 [DOI] [PubMed] [Google Scholar]
- Smalle J., Kurepa J., Haegman M., Gielen J., Van Montagu M., Van Der Straeten D. (1998). The trihelix DNA-binding motif in higher plants is not restricted to the transcription factors GT-1 and GT-2. Proc. Natl. Acad. Sci. USA 95: 3318–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden W., Fromm H. (2001). Calmodulin as a versatile calcium signal transducer in plants. New Phytol. 151: 35–66 [DOI] [PubMed] [Google Scholar]
- Song K., Backs J., McAnally J., Qi X., Gerard R.D., Richardson J.A., Hill J.A., Bassel-Duby R., Olson E.N. (2006). The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell 125: 453–466 [DOI] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Subbaiah C.C., Bush D.S., Sachs M.M. (1994a). Elevation of cytosolic calcium precedes anoxic gene expression in maize suspension-cultured cells. Plant Cell 6: 1747–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah C.C., Bush D.S., Sachs M.M. (1998). Mitochondrial contribution to the anoxic Ca2+ signal in maize suspension-cultured cells. Plant Physiol. 118: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah C.C., Sachs M.M. (2003). Molecular and cellular adaptations of maize to flooding stress. Ann. Bot. (Lond.) 91(Spec No): 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah C.C., Zhang J., Sachs M.M. (1994b). Involvement of intracellular calcium in anaerobic gene expression and survival of maize seedlings. Plant Physiol. 105: 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski D.B., Liao B., Zielinski R.E. (1996). Calmodulin isoforms differentially enhance the binding of cauliflower nuclear proteins and recombinant TGA3 to a region derived from the Arabidopsis Cam-3 promoter. Plant Cell 8: 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabatake R., Karita E., Seo S., Mitsuhara I., Kuchitsu K., Ohashi Y. (2007). Pathogen-induced calmodulin isoforms in basal resistance against bacterial and fungal pathogens in tobacco. Plant Cell Physiol. 48: 414–423 [DOI] [PubMed] [Google Scholar]
- Takahashi F., Mizoguchi T., Yoshida R., Ichimura K., Shinozaki K. (2011). Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Mol. Cell 41: 649–660 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Isobe M., Muto S. (1997). An increase in cytosolic calcium ion concentration precedes hypoosmotic shock-induced activation of protein kinases in tobacco suspension culture cells. FEBS Lett. 401: 202–206 [DOI] [PubMed] [Google Scholar]
- Tardif G., Kane N.A., Adam H., Labrie L., Major G., Gulick P., Sarhan F., Laliberté J.F. (2007). Interaction network of proteins associated with abiotic stress response and development in wheat. Plant Mol. Biol. 63: 703–718 [DOI] [PubMed] [Google Scholar]
- Tavernier E., Wendehenne D., Blein J.P., Pugin A. (1995). Involvement of free calcium in action of cryptogein, a proteinaceous elicitor of hypersensitive reaction in tobacco cells. Plant Physiol. 109: 1025–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.R., Manison N.F.H., Fernandez C., Wood J., Brownlee C. (1996). Spatial organization of calcium signaling involved in cell volume control in the fucus rhizoid. Plant Cell 8: 2015–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teper-Bamnolker P., Samach A. (2005). The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17: 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilmony R., Underwood W., He S.Y. (2006). Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 46: 34–53 [DOI] [PubMed] [Google Scholar]
- Thomashow M.F. (1999). PLANT COLD ACCLIMATION: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Tirichine L., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- Tracy F.E., Gilliham M., Dodd A.N., Webb A.A., Tester M. (2008). NaCl-induced changes in cytosolic free Ca2+ in Arabidopsis thaliana are heterogeneous and modified by external ionic composition. Plant Cell Environ. 31: 1063–1073 [DOI] [PubMed] [Google Scholar]
- Trewavas A.J., Malhó R. (1998). Ca2+ signalling in plant cells: The big network! Curr. Opin. Plant Biol. 1: 428–433 [DOI] [PubMed] [Google Scholar]
- Tuteja N. (2007). Mechanisms of high salinity tolerance in plants. Methods Enzymol. 428: 419–438 [DOI] [PubMed] [Google Scholar]
- Uno Y., Furihata T., Abe H., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T., Yamaguchi-Shinozaki K., Urao S., Shinozaki K. (1993). An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Der Luit A.H., Olivari C., Haley A., Knight M.R., Trewavas A.J. (1999). Distinct calcium signaling pathways regulate calmodulin gene expression in tobacco. Plant Physiol. 121: 705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot A.C., Dempsey D.A., Klessig D.F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Vogel J.T., Zarka D.G., Van Buskirk H.A., Fowler S.G., Thomashow M.F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Walley J.W., Coughlan S., Hudson M.E., Covington M.F., Kaspi R., Banu G., Harmer S.L., Dehesh K. (2007). Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. 3: 1800–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley J.W., Dehesh K. (2010). Molecular mechanisms regulating rapid stress signaling networks in Arabidopsis. J. Integr. Plant Biol. 52: 354–359 [DOI] [PubMed] [Google Scholar]
- Walley J.W., Kelley D.R., Savchenko T., Dehesh K. (2010). Investigating the function of CAF1 deadenylases during plant stress responses. Plant Signal. Behav. 5: 802–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Amornsiripanitch N., Dong X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Tsuda K., Sato M., Cohen J.D., Katagiri F., Glazebrook J. (2009a). Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog. 5: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. (2009b). RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb A.A.R., McAinsh M.R., Taylor J.E., Hetherington A.M. (1996). Calcium ions as intracellular second messengers in higher plants. Adv. Bot. Res. 22: 45–96 [Google Scholar]
- Weinl S., Kudla J. (2009). The CBL-CIPK Ca(2+)-decoding signaling network: Function and perspectives. New Phytol. 184: 517–528 [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Winfield M.O., Lu C., Wilson I.D., Coghill J.A., Edwards K.J. (2010). Plant responses to cold: Transcriptome analysis of wheat. Plant Biotechnol. J. 8: 749–771 [DOI] [PubMed] [Google Scholar]
- Xiong T.C., Jauneau A., Ranjeva R., Mazars C. (2004). Isolated plant nuclei as mechanical and thermal sensors involved in calcium signalling. Plant J. 40: 12–21 [DOI] [PubMed] [Google Scholar]
- Xu H., Heath M.C. (1998). Role of calcium in signal transduction during the hypersensitive response caused by basidiospore-derived infection of the cowpea rust fungus. Plant Cell 10: 585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yang S., Vanderbeld B., Wan J., Huang Y. (2010a). Narrowing down the targets: Towards successful genetic engineering of drought-tolerant crops. Mol. Plant 3: 469–490 [DOI] [PubMed] [Google Scholar]
- Yang T., Chaudhuri S., Yang L., Du L., Poovaiah B.W. (2010b). A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 285: 7119–7126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Poovaiah B.W. (2000). An early ethylene up-regulated gene encoding a calmodulin-binding protein involved in plant senescence and death. J. Biol. Chem. 275: 38467–38473 [DOI] [PubMed] [Google Scholar]
- Yang T., Poovaiah B.W. (2002). A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 277: 45049–45058 [DOI] [PubMed] [Google Scholar]
- Yang T., Poovaiah B.W. (2003). Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci. 8: 505–512 [DOI] [PubMed] [Google Scholar]
- Yang T., Shad Ali G., Yang L., Du L., Reddy A.S., Poovaiah B.W. (2010c). Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal. Behav. 5: 991–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Ni Z., Peng H., Sun F., Xin M., Sunkar R., Zhu J.K., Sun Q. (2010). Non-coding small RNAs responsive to abiotic stress in wheat (Triticum aestivum L.). Funct. Integr. Genomics 10: 187–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C.Y., Jin J.B., Miura K., Jin Y.H., Gosney M., Mickelbart M.V., Bressan R.A., Hasegawa P.M. (2007). Ca2+/CaM signaling through AtGTL1 mediates drought stress adaptation. In Plant Biology & Botany 2007 Joint Congress, July 7–July 11, 2007, Chicago, IL. American Society of Plant Biologists, Rockville, MD. Abstract no. M05001 [Google Scholar]
- Yoo C.Y., Pence H.E., Jin J.B., Miura K., Gosney M.J., Hasegawa P.M., Mickelbart M.V. (2010). The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22: 4128–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J.H., et al. (2005). Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in arabidopsis. J. Biol. Chem. 280: 3697–3706 [DOI] [PubMed] [Google Scholar]
- Yoon H.K., Kim S.G., Kim S.Y., Park C.M. (2008). Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol. Cells 25: 438–445 [PubMed] [Google Scholar]
- Yoshiba Y., Nanjo T., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Stress-responsive and developmental regulation of Delta(1)-pyrroline-5-carboxylate synthetase 1 (P5CS1) gene expression in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 261: 766–772 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Yoshioka K., Moeder W., Kang H.G., Kachroo P., Masmoudi K., Berkowitz G., Klessig D.F. (2006). The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell 18: 747–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Jin J., Tang L., Zhao Y., Gu X., Gao G., Luo J. (2011). PlantTFDB 2.0: Update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 48: D1114–D1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lu Y.T. (2003). Calmodulin-binding protein kinases in plants. Trends Plant Sci. 8: 123–127 [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhou R.G., Gao Y.J., Zheng S.Z., Xu P., Zhang S.Q., Sun D.Y. (2009). Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol. 149: 1773–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xu S., Ding P., Wang D., Cheng Y.T., He J., Gao M., Xu F., Li Y., Zhu Z., Li X., Zhang Y. (2010). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 107: 18220–18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Ren N., Wang H., Stromberg A.J., Perry S.E. (2009). Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S.Y., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Caplan J., Mamillapalli P., Czymmek K., Dinesh-Kumar S.P. (2010). Function of endoplasmic reticulum calcium ATPase in innate immunity-mediated programmed cell death. EMBO J. 29: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski R.E. (1998). Calmodulin and calmodulin-binding proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 697–725 [DOI] [PubMed] [Google Scholar]
- Zimmermann S., Nürnberger T., Frachisse J.M., Wirtz W., Guern J., Hedrich R., Scheel D. (1997). Receptor-mediated activation of a plant Ca2+-permeable ion channel involved in pathogen defense. Proc. Natl. Acad. Sci. USA 94: 2751–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.