The authors show that genes can be separated into distinct low or high expression abundance groups. Histone marks reveal that this switch-like transition from low to high expression goes hand-in-hand with a change in chromatin status.
Keywords: bimodal, ChIP-seq, expression levels, RNA-FISH, RNA-seq
Abstract
The expression level of a gene is often used as a proxy for determining whether the protein or RNA product is functional in a cell or tissue. Therefore, it is of fundamental importance to understand the global distribution of gene expression levels, and to be able to interpret it mechanistically and functionally. Here we use RNA sequencing (RNA-seq) of mouse Th2 cells, coupled with a range of other techniques, to show that all genes can be separated, based on their expression abundance, into two distinct groups: one group comprised of lowly expressed and putatively non-functional mRNAs, and the other of highly expressed mRNAs with active chromatin marks at their promoters. These observations are confirmed in many other microarray and RNA-seq data sets of metazoan cell types.
Introduction
Expression level is frequently used as a way of characterizing gene function, by northern blotting, PCR, microarrays, and, more recently, RNA-sequencing (Wang et al, 2009a; RNA-seq). Therefore, it is a central issue in molecular biology to know how many transcripts are expressed in a cell at what levels. This question was studied very early in the history of molecular biology using methods such as reassociation kinetics (Hastie and Bishop, 1976), which indicated the existence of distinct abundance classes, and recently, we pointed out that separate peaks are visible in the abundance distributions of a number of microarray data sets (Hebenstreit et al, 2011). At the same time, microarrays or RNA-seq data have been described as displaying broad, roughly lognormal distributions of expression levels with no clear separation into discrete classes (Hoyle et al, 2002; Lu and King, 2009; Ramskold et al, 2009). There are several reasons for this: many samples are heterogeneous in terms of cell type (Hebenstreit and Teichmann, 2011) or are based on a previous generation of less sensitive microarrays, many are from unicellular organisms rather than animals, and finally, data processing and plotting methods can obscure the presence of distinct abundance classes. Here, we provide experimental and computational support for two overlapping major mRNA abundance classes. Our findings hold for metazoan data sets including human, mouse and Drosophila sources.
Results and discussion
We initially based our analysis on murine Th2 cells (Zhu et al, 2010), as these cells can be obtained in large quantities ex vivo and can be prepared as a pure and homogeneous cell population. Furthermore, there is a well-characterized set of genes whose proteins are known to be expressed and functional in Th2 cells, as well as a set of genes known to be not expressed in these cells (Supplementary Table S1 lists the genes we used in our study, Supplementary Figure S1 shows expression of two marker proteins in the cells).
We generated Th2 poly(A)+ RNA-seq data for two biological replicates and calculated gene expression levels using the standard measure of Reads Per Kilobase per Million (RPKM; Mortazavi et al, 2008, Supplementary Table S2 gives the number of reads and mappings we obtained). The expression levels of the biological replicates are highly correlated (r2=0.94, Supplementary Figure S2). We then calculated the mean RPKMs of the two samples for all genes and log2 transformed these values.
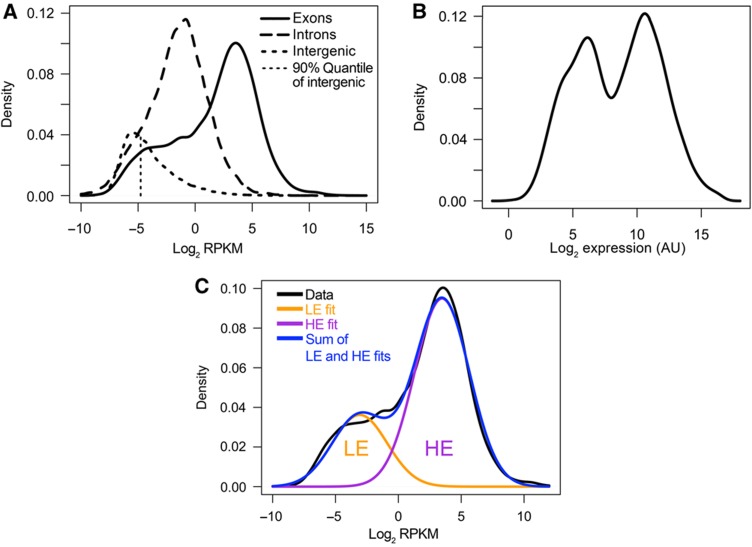
Displaying the distribution of all gene expression levels as a kernel density estimate (KDE) reveals an interesting structure: the majority of genes follow a normal distribution, which is centered at a value of ∼4 log2 RPKM (∼16 RPKM), whereas the remaining genes form a shoulder to the left of this main distribution (Figure 1A, solid line). This was conserved under different KDE bandwidths (Supplementary Figure S3, left panel) or different histogram representations (Supplementary Figure S3, right panel). As genes with zero reads cannot be included on the log scale, we prepared an alternative version of Figure 1A, where we assigned low RPKM values to these. This helps to illustrate the fraction of zero-read genes (Supplementary Figure S4). As a comparison, we studied microarray data for the same cell type from a recent publication (Wei et al, 2009). The correlation between the microarray and the RNA-seq data was very good and highly statistically significant (Pearson r2=0.83, Spearman ρ=0.84, Supplementary Figure S5). Surprisingly, displaying the distribution of microarray expression levels results in a clearly bimodal distribution (Figure 1B). Again, the appearance of the distribution was insensitive to the KDE bandwidth choice or histogram bin size (Supplementary Figure S6). The bimodality was conserved when alternative normalization and processing schemes were used, independent of KDE bandwidths (Supplementary Figure S7).
Figure 1.
Distribution of gene expression levels. (A) Kernel density estimates of RPKM distributions of RNA-seq data within exons, introns and intergenic regions as indicated. The fragments used to estimate intron and intergenic RPKM were based on randomizations using the same length distribution as the exonic parts of genes. The 90% quantile of the intergenic distribution is indicated. (B) Kernel density estimate of expression level distribution of microarray data (Wei et al, 2009). (C) Expectation-maximization-based curve fitting of RNA-seq data of A.
Visual inspection of both microarray and RNA-seq data thus reveals two overlapping main components of the distribution of gene expression levels. Quantifying this by curve fitting confirms a good fit to two distributions: the goodness-of-fit (measured by Akaike Information criterion, AIC (Akaike, 1974), Bayesian Information Criterion, BIC (Schwarz, 1978) or Likelihood ratio tests (Casella and Berger, 2001)) shows strong increases for both microarray and RNA-seq data when two-component models are fit by expectation-maximization (compared to single- or more-component models; Supplementary Figure S8). We designate the two groups of genes as the lowly expressed (LE) and highly expressed (HE) genes (Figure 1C), because we will present evidence below that the LE genes are expressed rather than simply being experimental background. Our findings are not limited to Th2 cells and hold for virtually all recently published metazoan RNA-seq data sets (e.g., Marioni et al, 2008; Mortazavi et al, 2008; Mudge et al, 2008; Wang et al, 2008; Supplementary Figure S9; Cloonan et al, 2008; and Supplementary Figure S10A and B) and all microarray data sets (e.g., Cui et al, 2009; GNF Atlas 3 (Chintapalli et al, 2007); FlyAtlas (Lattin et al, 2008); and Supplementary Figure S11) we have studied. The existence of further minor groups of genes cannot be excluded, but is not clear at this point due to the diverse curve-fitting results for the different data sets if higher-order (more than two components) models are considered.
The difference between the microarray and RNA-seq distributions is explained by the fact that the microarrays yield a signal for all genes, part of which is due to cross-hybridization of oligonucleotide probes if the gene is not strongly expressed. RNA-seq on the other hand yields a signal for a gene only if at least one sequencing read is found. The accuracy of RNA-seq is biased toward longer and more highly expressed genes, e.g., 5% of all genes account for 50% of all reads in our data as well as in other data sets (Mortazavi et al, 2008; Oshlack and Wakefield, 2009; Bullard et al, 2010).
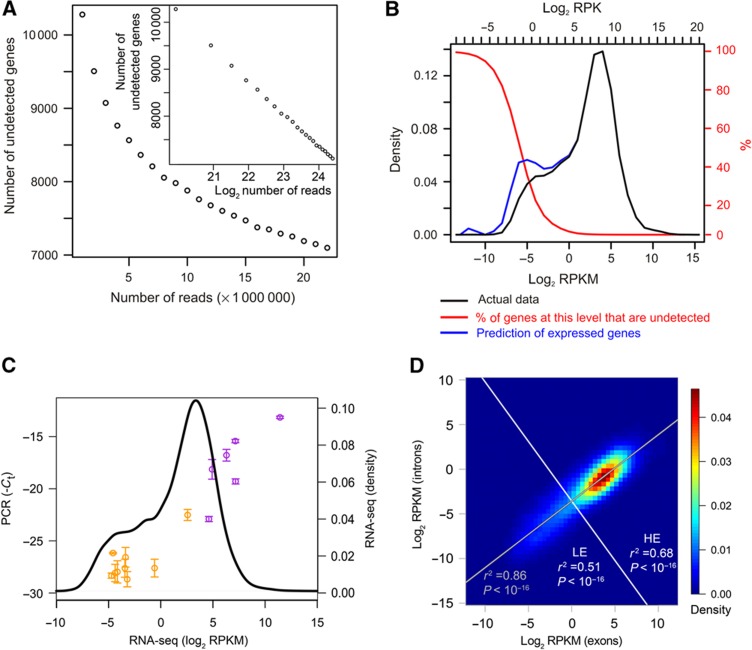
To explore how this accuracy bias affects the shape of the LE distribution, we studied the RNA-seq detection limit. We first plotted the number of genes with zero reads as a function of the total number of reads (taking subsets of the total reads). The number of genes without reads decreases slowly, with no change in slope and hence no indication of reaching a plateau. Even at a total of 25 million reads, ∼30% of all genes are undetected (Figure 2A). We further estimated the numbers of genes remaining undetected at each expression level by assuming Poisson-distributed read numbers (Jiang and Wong, 2009) and by determining the expected frequency of zeroes. This confirms the sensitivity drop at the lower end of the LE peak (Figure 2B). Extrapolating the numbers of expressed genes including the undetected ones reveals an emerging LE peak (Figure 2B). Thus, the smaller portion of LE genes in the RNA-seq data compared with the microarray data is at least partially due to the RNA-seq detection limit, although this only becomes a problem for genes at less than approximately −3 to −4 log2 RPKM. It should be noted that these low expression levels correspond to an absence of transcripts in the majority of cells, as we demonstrate further below.
Figure 2.
Sensitivity of RNA-seq. (A) Detection of genes in dependency of the total read numbers on linear scale and log2 scale (inset). Random subsets of the total reads for the two RNA-seq replicates were taken and the number of genes with zero reads were plotted versus the total read numbers used. The figure represents an average of five independent subsets for each data point. (B) Prediction of genes remaining undetected due to Poisson statistics underlying RNA-seq. The theoretically expected fraction of genes remaining undetected (red, y axis on the right side of the figure in red) was determined for each expression level. This was used to infer the expressed genes including the undetected ones (blue) from the actual expression data (black, bins indicated by tick marks across top). In addition to the RPKM scale, the reads per kilobase (RPK) scale (without normalization to the total number of mapped reads) is shown on top, which was used for the calculation of the (integer-) Poisson statistic and which, in contrast to the RPKM scale, depends on the total number of sequencing reads. (C) RT–PCR for the genes are listed in Supplementary Table S1. The RNA-seq expression levels of the genes are plotted versus the negative threshold cycles (Ct) of the PCRs. The plot is overlaid (with the same x axis scaling) upon the kernel density estimate of the RNA-seq expression level distribution (black line) to show the positions of the genes in the total expression distribution. Genes either in the LE peak of the RNA-seq distribution or which have been previously characterized as not expressed in Th2 cells are shown in orange. Genes known to be expressed are shown in purple. Error bars indicate s.e.m. from three independent biological replicates. Please refer to Supplementary Tables S1 and S6 for details of genes and PCR primers. (D) Correlation of RPKM within exons and introns based on the RNA-seq data from Figure 1A. Correlation and significance of correlation were calculated for the whole distribution (gray) or for LE and HE genes separately. Division into LE and HE was performed along a line (white) perpendicular to a fitted trendline (gray), centered at Exon RPKM=1. The data points are shown as 2D kernel density estimate.
To further confirm that the LE genes correspond to low expression and not experimental noise, we performed real-time PCRs. We tested amplification by exon-spanning primers of a set of genes that are known to be expressed or not expressed in Th2 cells, plus five random genes that we detected between −3.7 and −5 log2 RPKM in the RNA-seq experiment (Supplementary Table S1). We were able to successfully PCR-amplify all genes with high specificity. The expressed genes map to the HE peak, while almost all unexpressed genes map to the LE peak, if we align the PCR results with the microarray/RNA-seq data (Figure 2C).
We also tested the extent to which genomic DNA can be detected in our polyA-purified mRNA sample, as proposed by Ramskold et al (2009) as a means of quantifying experimental background. We randomly selected intergenic fragments with the same length distribution as genes, 10 kb away from genes. The resulting RPKM distribution contains a high number of zero-RPKM fragments (79%), while the majority of non-zero fragments peaks slightly left of the LE shoulder (Figure 1A). The 90% quantile of this intergenic background distribution is at −4.97 log2 RPKM, which means that we can be quite confident (with probability >90%) that genes with an RPKM value above this level are truly expressed rather than representing experimental background noise (Figure 1A). Further, the overlap between the intergenic and the normalized LE fit is small (Supplementary Figure S12). We cannot rule out that detection of intergenic DNA corresponds to transcription as well, which would make the case for transcription of LE genes even stronger.
Analysis of the strand-specific mRNA-sequencing data of ES cells by Cloonan et al (2008) yields similar conclusions. The poly(A)-purification protocol selects for reads that are antisense to genes (the antisense reads correspond to mRNA). In the distribution of ‘sense’ reads (corresponding to antisense transcripts in genic regions), more than 50% of genic regions have zero reads. This distribution is unimodal and shifted by ∼2 log2 RPKM with respect to the LE distribution, and overlaps almost perfectly with the distribution of reads in intergenic regions (Supplementary Figure S10A).
We next determined the distribution of RPKM within introns, again using fragments with the same length distribution as transcripts (please note that our intronic read densities are not enriched at 5′ or 3′ ends of the intronic regions (Supplementary Figure S13)). The resulting intronic distribution is significantly higher than the intergenic background (two-sided Wilcoxon rank-sum test, P<2.2 × 10−16) and peaks at roughly −1 log2 RPKM (Figure 1A). Introns thus have one- to two orders of magnitude lower read density than exons. This suggests that we are detecting incompletely processed transcripts at a low but significant and uniform level across the whole range of transcript abundances.
As introns are one- to two orders of magnitude longer than exons, introns should be detectable with roughly the same accuracy as exons, if the full-length set of introns of a gene is used. If we plot the RPKM in exonic regions versus the RPKM in intronic regions for each gene, there is significant correlation (r2=0.86, P<2.2 × 10−16) across the whole spectrum of expression levels. Calculating the correlation for LE and HE genes separately yields only slightly lower correlations among LE genes compared with HE genes, and both correlations are highly significant (Figure 2D). This provides evidence that confirms that LE genes are transcribed rather than experimental background: there would not be such a high correlation between introns and exons, particularly in the low-abundance region, if their detection were due to noise.
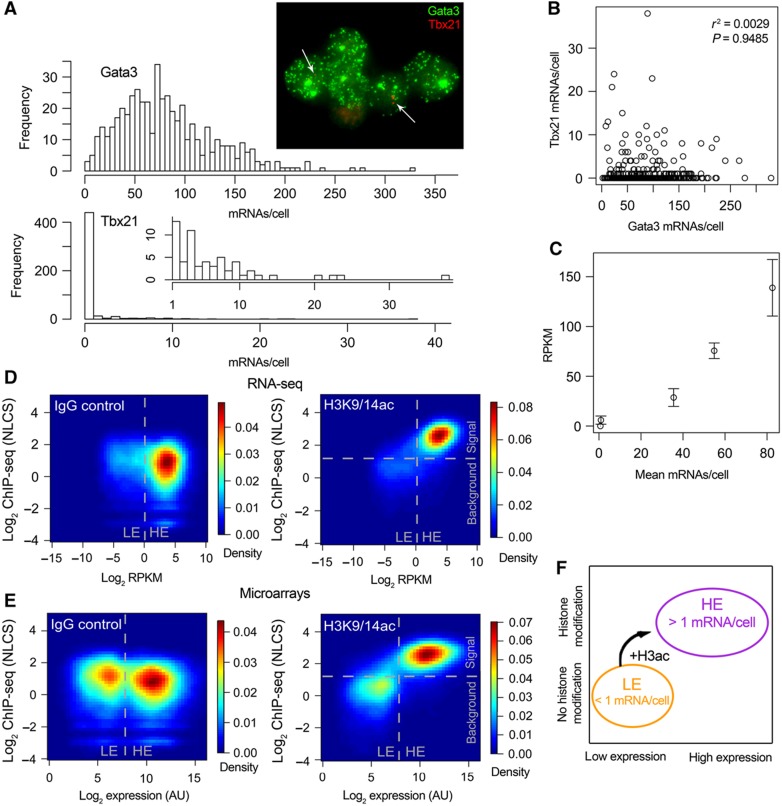
We next studied gene expression using a single-cell approach by performing single-molecule RNA-fluorescence in situ hybridization (FISH; Raj et al, 2008) for five genes that are expressed at different levels according to the literature and our RNA-seq data. The distributions of mRNA numbers per cell were very broad for expressed genes (e.g., Gata3), while low mRNA numbers from ‘not-expressed’ genes (e.g., Il2) were still detected (Figure 3A). All genes had Fano factors (σ2/μ) larger than 4, indicating that they had extra-Poisson variation (a Poisson random variable would have σ2/μ=1) and therefore burst-like transcription (Raj and van Oudenaarden, 2009) (Supplementary Table S3). Importantly, cells expressing Tbx21 were not anticorrelated with cells expressing Gata3 (Figure 3B), meaning that we do not have a sub-population of Th1 cells in our Th2 cell populations. This demonstrates that LE expression is not due to a contaminating cell type, as the same cells express groups of genes at HE and others at LE levels.
Figure 3.
(A) Distribution of mRNA numbers among single cells. Histograms for Gata3 and Tbx21 (with an inset histogram starting from 1 instead of 0 to better illustrate higher expressions) and a sample fluorescence microscopy image are shown. Tbx21 transcripts are marked with white arrows to ease identification. (B) Correlation between Gata3 and Tbx21 expression. Correlation coefficient and significance are inset. (C) Plot of mean mRNA numbers per cell versus RNA-seq RPKM of five genes. Error bars indicate s.e.m. from two RNA-seq biological replicates. (D, E) 2D kernel density estimates of gene expression level versus ChIP-seq signal for each gene for RNA-seq (D) and microarray (E) data. Divisions between background and signal for the ChIP-seq component were determined by curve fitting with the software EpiChIP (Hebenstreit et al, 2011) and are indicated. Divisions between LE and HE groups of genes are indicated. (F) Scheme summarizing the results.
It should be further noted that the LE/HE groups cannot theoretically result from a mixture of different cell types. Mixing of different cell types leads to gene expression levels for each gene that are an average across cell types. Hence, such distributions will become more unimodal, not less so (following the central limit theorem).
As the RPKM as measured by RNA-seq should be proportional to the mean mRNA numbers per cell, we can use the RNA-FISH results to estimate how our RPKM values translate into mRNA numbers. We find that one RPKM corresponds to an average of roughly one transcript per cell in our Th2 data set (Figure 3C). Please note that the value of one RPKM/one transcript on average per cell serves as an estimate only as it is based on a limited number of data points. See Supplementary Figure S14 for log-transformed versions of Figure 3A–C.
To study the nature of the LE and HE groups in more detail, we prepared Th2 ChIP-seq data for the activating H3K9/14 acetylation histone modification (Roh et al, 2005; Wang et al, 2009b) (H3K9/14ac) and one IgG control. We calculated the histone-modification level at each gene by identifying a globally enriched window around the transcription start sites of genes, and using reads in this window as a measure of each gene's modification level, normalized by the total reads (giving the normalized locus-specific chromatin state, NLCS, as used in Hebenstreit et al (2011)). Thus, we were able to plot histone-modification levels of each gene against expression levels from the RNA-seq or microarray data using a heatmap representation (Figure 3D, RNA-seq and Figure 3E, microarrays). Supplementary Figure S15 is an alternative version of this figure, where we randomly assigned low RPKM values to the zero-read genes.
This strikingly confirms the two groups of gene expression levels, as there is a very good agreement between LE genes and absence of histone marks on one hand, and HE genes and presence of H3K9/14ac marks on the other hand (Figure 3D–E). This is seen for both the microarrays as well as the RNA-seq data. This extends previous findings of the relationship between H3K9/14ac and transcriptional activation by revealing an on/off-type of correlation between this histone mark and the LE/HE groups of genes. It should be noted that there is a very weak correlation within the LE and HE groups. The strongest correlation is within the RNA-seq HE group with a correlation coefficient r2=0.29 in log space and r2=0.097 on linear space.
As the LE group of genes is still expressed at low levels and contains at least five genes that are characterized as not expressed and non-functional in Th2 cells, it seems likely that the HE group of genes represents the active and functional transcriptome of cells. This is supported by SILAC proteomics data (Graumann et al, 2008), which is available for the embryonic stem cell data we presented earlier (Supplementary Figure S10) and which indicates protein expression of HE genes only (Supplementary Figure S10C). The tight correlation recently observed between RNA and protein levels in three mammalian cell lines also supports this (Lundberg et al, 2010).
Gene ontology (GO) analysis of LE and HE genes in the Th2 cells supports the notion that HE comprises the functional transcriptome, as many T-cell-specific processes (e.g., GO:0050863, GO:0045582, GO:0042110) and housekeeping processes are enriched (Supplementary Table S4). On the other hand, many GO terms referring to differentiation of other cell types (e.g., ear development GO:0043583, neuron fate commitment GO:0048663) are enriched among the LE set of genes (Supplementary Table S5).
In conclusion, our data show that two large groups of genes can be discriminated based on the distribution of expression levels. RNA-FISH indicates that the boundary between the groups is found at an expression level of roughly one transcript per cell. In addition, H3K9/14ac marks are associated with the promoters of HE genes only (Figure 3F). It thus seems likely that the LE/HE groups reflect different transcription kinetics depending on the chromatin state or vice versa. The LE group is likely to correspond to ‘leaky’ expression, producing non-functional transcripts. The majority of LE genes are expressed at less than one copy per cell on average, and it would be interesting to know whether such stochastic expression has any function, e.g., in cell differentiation, or any deleterious effects. There may be a trade-off between the cost of repressing expression entirely and unwanted consequences of stochastic expression.
Regulation of gene expression is mostly mediated by transcription factor binding events at promoters and enhancers (Heintzman et al, 2009). Often, differential regulation induces only small changes in expression levels, probably serving to fine-tune expression and shifting genes within the HE group. Our data suggest that in addition to this, there is a key decision about whether a gene becomes ‘switched on’ and expressed, which coincides with a boost in both transcription and H3K9/14ac histone modification.
Materials and methods
Th2 cell differentiation culture
Spleens of C57BL/6 mice aged from 7 weeks to 4 months were removed and softly homogenized through a nylon mesh. The medium used throughout the cell cultures was IMDM supplemented with 10% FCS, 2 μM L-glutamine, penicillin, streptomycin and 50 μM β-mercaptoethanol. Cells were washed twice and purified by a Ficoll density gradient centrifugation. CD4+CD62L+ cells were isolated by a two-step MACS purification using the CD4+CD62L+ T Cell Isolation Kit II (Miltenyi Biotec). Cells were seeded into 24-well plates that had been coated with a mix of anti-CD3 (1 μg/ml, clone 145-2C11, eBioscience) and anti-CD28 (5 μg/ml, clone 37.51, eBioscience) antibodies overnight, at a density of 250 000 cells/ml and a total volume of 2 ml. The following cytokines and antibodies, respectively, were added to the Th2 culture: recombinant murine IL-4 (10 ng/ml, R&D Systems), and neutralizing IFN-γ (5 μg/ml, Sigma). Cells were cultured for 4–5 days at 37°C, 5% CO2. After this, cells were taken away from the activation stimulus, diluted 1:2 in fresh medium containing the same cytokine concentration as before. After 2–3 days of resting time, cells were directly crosslinked in formaldehyde for preparing ChIP-seq samples. For FACS stainings, cells were restimulated with phorbol dibutyrate and ionomycin (both used at 500 ng/ml, both from Sigma) for 4 h in the presence of Monensin (2 μM, eBioscience) for the last 2 h after the resting phase. For real-time PCRs, the cells were lysed in Trizol.
FACS staining
After restimulation, cells were washed in PBS and fixed in IC fixation buffer from the Foxp3 staining kit (eBioscience). Staining for intracellular transcription factor expression was carried out according to the eBioscience protocol, using Permeabilization buffer (eBioscience) and the following antibodies: anti-GATA3-Alexa647 (one test, TWAJ, eBioscience), anti-Tbx21-PE (1/400, clone eBio4B10, eBioscience). Stained cells were analyzed on a FACSCalibur (BD Biosciences) flow cytometer using Cellquest Pro and FlowJo software.
Real-time PCR
RNA of ∼106 cells was isolated with Trizol (Invitrogen) according to the manufacturer's protocol. cDNA was produced using Superscript III reverse transcriptase (Invitrogen), following the protocol supplied by the manufacturer. The cDNA was subjected to real-time PCR, using the SYBR green PCR master mix (Applied Biosystems) and a 7900 HT Real-Time PCR system (Applied Biosystems). The threshold cycles (Ct) were determined. The primer sequences used are listed in Supplementary Table S6 and were mostly obtained from ‘Primerbank’ (http://pga.mgh.harvard.edu/primerbank/; Spandidos et al, 2010).
RNA-seq data generation
Poly-(A)+ RNA was purified from ∼500 000 cells using the Oligotex kit (Qiagen). The manufacturer's protocol was slightly modified to include additional final elution steps resulting in a larger volume. After precipitation of RNA to concentrate it, first- and second-strand cDNA synthesis was performed using the Just cDNA kit (Stratagene), skipping the blunting step and directly proceeding to PCI extraction. Quality of the cDNA was tested by real-time PCR for a housekeeping gene. After this, the cDNA was sonicated for a total of 45 min using the Diagenode Bioruptor at maximum power settings, cycling 30 s sonications with 30 s breaks. After precipitation, the sample was processed using the ChIP-seq sample prep kit (Illumina) with a slightly modified protocol (PCR before gel extraction). Sequencing for 36 or 41 bp was carried out on an Illumina GAII genome analyzer. The data were deposited at Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/), accession number GSE28666.
RNA-seq data processing
Reads were mapped to the mouse genome (mm9) with Bowtie (Langmead et al, 2009) using the command options -m 1--best –strata--solexa1.3-quals, and were assigned to exons of RefSeq genes using custom perl scripts. We used the gene symbol as the primary identifier. Supplementary Table S2 shows the numbers of mapped reads. We further generated a library of splice junctions based on RefSeq genes, mapped unmapped reads to these and added the numbers of hits to the genes. The numbers of mapped reads per gene were corrected for mapability based on the ‘CRG’ tracks of the UCSC genome browser. RPKM were then calculated. In the case that multiple splice variants existed, the most highly expressed one was selected as representative for a gene's expression level. For generating the RPKM distributions of intergenic regions, we considered regions with a distance of at least 10 kb to any RefSeq or Ensembl gene. The distribution was based on random fragments of the same length distribution as gene lengths. Mapability was accounted for, and the randomization was performed 20 times. The same procedure was followed for determining the read distribution within introns (of RefSeq genes). To test for a possible RPKM bias in 5′ or 3′ ends of intronic regions, the introns of each gene were aligned, and if the intronic region was at least 6 kb in total, RPKM were separately determined for the most 5′ 2 kb, for the 2 kb in the center and for the most 3′ 2 kb. The full-length of introns was used (for the sake of higher sensitivity) for plotting RPKM of introns versus exons (as in Figure 2D). A trend line was calculated based on a least squares fit of the log2-transformed data. Division into LE and HE was made along a line perpendicular to the trendline, crossing at Exon RPKM=1. Correlations and significances calculated were based on Pearson's product moment correlation coefficient.
We prepared alternative versions of Figures 1A and 3D, where we assigned a random log2 RPKM value derived from a Normal distribution with μ=−12 and σ=1 to each gene without sequencing reads (Supplementary Figures S4 and S15).
Integration of the RNA-seq data with mircoarray- and histone-modification data was based on gene symbols.
The RNA-seq data of Cloonan et al (2008) was downloaded from the NCBI short-read archive (http://www.ncbi.nlm.nih.gov/sra/), accession number SRX003912. The reads were mapped to mm9 in colorspace format using Bowtie with similar settings as above. The mapped reads were separated into those sense and those antisense to RefSeq genes and processed similarly as described above. Read distributions in intergenic regions were determined as described above for our data.
RNA-seq data from Mudge et al (2008) was downloaded from GEO, accession number GSE12297. We used the processed data for ‘Cerebellar cortex 40 Control’ directly and performed no further calculations, except log transformation and kernel density estimation. The RNA-seq data for ‘skeletal muscle’ from Wang et al (2008) was downloaded from GEO (accession number GSE12946). We used the data that was mapped to the human genome (hg18), assigned it to RefSeq genes, and processed it similarly as described above. We further downloaded RNA-seq data from Marioni et al (2008) from the Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra/). The data for human liver tissue were used (accession numbers SRX000571 and SRX000604). The two files were concatenated, mapped to the human genome (hg18) with Bowtie and processed further as described above. Finally, RNA-seq data for mouse brain (Mortazavi et al, 2008) was downloaded from SRA (accession numbers SRX000350 and SRX001866). As described above, the two files were concatenated, mapped to the mouse genome (mm9) with Bowtie and processed further.
Kernel density estimation
Gene expression distributions were displayed as KDEs in most cases. These were calculated using the function ‘density()’ of the freely available statistical software package ‘R’ (http://www.r-project.org/). We used default settings of this function unless stated otherwise. This means a Gaussian kernel and that the ‘bandwidth equals 0.9 times the minimum of the standard deviation and the interquartile range divided by 1.34 times the sample size to the negative one-fifth power (corresponding to Silverman's ‘rule of thumb’, Silverman (1986), page 48, equation (3.31)), unless the quartiles coincide when a positive result will be guaranteed’ (R manual). For 2D kernel density estimations we used the function ‘kde2d()’ of the R library ‘MASS’ with the default bandwidth and a Gaussian kernel. This bandwidth is calculated based on a variation of above formula for the 1D case, where the factor 1.06 instead of 0.9 is used. Densities were estimated at 50 grid points in either direction and displayed as heatmaps.
RNA-seq data sensitivity analysis
The RNA-seq detection limit was explored by two different approaches. First, random subsets of different sizes were taken from the total reads we generated. The number of genes that remained undetected (zero reads) were plotted as a function of the subset size. The subsetting was performed five times for each subset size and the average number of zero-read genes was determined.
As a second approach, we determined the expected number of zero-read genes depending on the expression level. To this end, we calculated the expected number of reads for each gene in dependency of the expression level (as reads per kilobase, RPK, instead of RPKM which includes normalization by the total number of mapped reads) and gene length (the length distribution of all genes was used). The expected read number is generally assumed to be Poisson distributed (Jiang and Wong, 2009) and can be used as an estimator of the single parameter of a Poisson distribution, λ, which is equal to mean and variance of the distribution. Studying the probability density function of a Poisson distribution for a certain λ reveals the expected frequency of zeros, which corresponds to genes of a certain length that remain undetected at a certain RPK, despite being expressed. Assuming an equal distribution of gene lengths at all expression levels, we could thus sum up the proportion of zero-read genes for all gene lengths and thus obtain the total expected portion of undetected genes for all RPK levels. For instance, at RPK=1 we would expect two sequencing reads for a gene that is 2 kb long and one read for a 1 kb gene (giving the same expression level). As the actual read numbers vary according to a Poisson distribution, not all genes that are expressed at that level will have exactly one or two reads, respectively, but some will have more and some none at all. The Poisson distribution gives the expected portion of zeros, which would be 37% for the 1 kb gene and 13.5% for the 2 kb gene. Thus, if we detect 150 1-kb genes and 250 2-kb genes at RPK=1, we can estimate that a further 127 (=150/(1−0.37)−150+250/(1−0.135)−250) genes of the same lengths are expressed at the same level but remain undetected.
We further used above calculation to estimate how the distribution of expression levels is affected by the sensitivity of RNA-seq. To this end, we binned the actual expression distribution into bins of size 1 on the log2 RPK scale and extrapolated the number of expressed genes by adding the inferred number of undetected genes to each bin.
Microarray data
Microarray data (Th2) of Wei et al (2009) were downloaded from GEO, accession number GSE14308. Either normalized (by the authors) microarray data were used (Figure 1B, Supplementary Figure S5, S6 and S8), in which case present (P) and absent (A) calls of the probesets were ignored, or custom processing schemes were applied to the raw data (Supplementary Figure S7 and S8). The mean of the two replicates of the microarray data was calculated for each probeset and was log2-transformed. These values were then linked to RefSeq genes based on the Affymetrix MOE430 2.0 build 27 annotations. If more than one probeset was mapping to a gene, the probeset with the highest intensity was chosen as representative of the gene's expression level.
We further downloaded microarray data for murine bone cells from the GNF Mouse GeneAtlas V3 (Lattin et al (2008); GEO, GSE10246) and processed them as described above. Similarly, the processed microarray data for two replicates of human Cd133+ cells (Cui et al, 2009) were downloaded from GEO, accession number GSE12646, and processed (using Affymetrix build 28 annotations for the Affymetrix U133A chip). Finally, we downloaded from GEO (accession number GSE7763) microarray data for Drosophila eye tissue from the FlyAtlas (Chintapalli et al, 2007). We mapped the probesets to genes using Affymetrix probe annotations (build 28) for GeneChip Drosophila Genome 2.0 and processed the data the same way as the other data sets.
Curve fitting
Curve fitting and/or clustering of the data into LE and HE sets by expectation-maximization was performed on the log2-transformed RNA-seq or microarray data using the R library ‘Mclust’. The log-likelihood values output by Mclust were used to calculate AIC (Akaike, 1974), BIC (Schwarz, 1978) and likelihood ratio statistics (Casella and Berger, 2001). The latter were calculated for the model with n components as the null model and the one with n+1 components as the alternative model (0<n<9). We approximated the test statistics with χ2 distributions and calculated the P-values with R.
SILAC data
Processed SILAC data for murine embryonic stem cells was downloaded from the Supplementary Material of Graumann et al (2008). Using UCSC table browser, we linked the protein expression data to the RNA-seq data of Cloonan et al (2008) by referencing the RefSeq protein ID provided by Graumann et al (2008) to the gene symbol, which we used as gene identifier for the RNA-seq data. A protein was regarded as expressed if it had a non-zero ‘MS intensity’ value.
GO analysis
Genes were clustered into LE and HE subsets by expectation-maximization using the R library Mclust. Enrichment analysis of ‘process’ GO terms was performed with the Generic Gene Ontology (GO) Term Finder (http://go.princeton.edu/cgi-bin/GOTermFinder; Boyle et al, 2004) using the combined LE/HE set of genes as the custom background. Bonferroni-adjusted P-values were used.
Single-molecule FISH
We performed single-molecule FISH on the Th2 cells and counted the mRNAs in individual cells as described previously (Raj et al, 2008). Briefly, harvested Th2 cells were fixed with 3.7% formaldehyde for 10 min, washed twice with PBS, and permeabilized in 70% ethanol. For hybridization, the samples were resuspended in 100 μl of hybridization solution containing labeled DNA probes in 2 × SSC, 1 mg/ml BSA, 10 mM VRC, 0.5 mg/ml Escherichia coli tRNA and 0.1 g/ml dextran sulfate, with 10–25% formamide, which varies for different probes, and incubated overnight at 30°C. The next day, the samples were washed twice by incubating in 1 ml of wash solution consisting of 10–25% formamide and 2 × SSC for 30 min. The sequences of the probes are available upon request.
Image acquisition
The samples were resuspended in glucose oxidase anti-fade solution, which contains 10 mM Tris (pH 7.5), 2 × SSC, 0.4% glucose, supplemented with glucose oxidase and catalase. Then 8 μl of cell suspension was sandwiched between two coverglasses, and mounted on glass slides using a silicone gasket. Images were taken using a Nikon TE2000 inverted fluorescence microscope equipped with a × 100 oil-immersion objective and a Princeton Instruments camera using MetaMorph software (Molecular Devices, Downington, PA). Stacks of images were taken automatically with 0.4 microns between the z-slices.
Image analysis
To segment the cells, a marker-guided watershed algorithm was used. Briefly, cell boundaries were obtained by running an edge detection algorithm on the bright-field image of the cells. To generate markers, the centroid of the region enclosed by individual cell boundaries is computed. A marker-guided watershed algorithm was then run on the distance transformation of the cell boundaries, using the markers located within the cell boundaries (Supplementary Figure S16). The resultant cell segmentation image was then manually curated for occasional mis-segmentations.
To quantify the number of RNA molecules in each cell, a log filter was run over each optical slice of an image stack to enhance signals. A threshold was taken on the resultant image stack to pick up mRNA spots. The locations of mRNA spots were then taken to be the regional maximum pixel value of each connected region (Supplementary Figure S17). The number of mRNA spots located within the cell boundaries of an individual cell was thus quantified.
ChIP-seq data analysis
We used murine Th2 cell data for the H3K9/14ac histone modification and an IgG control from Hebenstreit et al (2011) (available on GEO, accession number GSE23092). The reads were mapped to the mouse genome (mm9) using Bowtie as for the RNA-seq analysis. Further steps of the analysis were performed using the software EpiChIP (http://epichip.sourceforge.net/index.html; Hebenstreit et al, 2011). Briefly, the mapped reads were assumed to be the ends of 200-bp-long fragments following the XSET method (Pepke et al, 2009). Then EpiChIP was used to identify an optimal sequence window with respect to gene coordinates for analysis of the histone-modification status at all (RefSeq) genes. The resulting window of −400 to +807 bp at transcriptional start sites was used to quantify the ChIP-seq signal for each gene (the area below the peaks within this window), which was normalized by the total (genomewide) area to yield ‘NLCS’ (Hebenstreit et al, 2011). These values were log2 transformed and displayed against the RNA-seq or microarray expression levels as two-dimensional density estimations. The threshold separating background from signal was determined with the curve-fitting function of EpiChIP. For the alternative version of Figure 3D (Supplementary Figure S15), we assigned a random log2 RPKM value derived from a normal distribution with μ=−3 and σ=1 to each gene without ChIP-seq sequencing reads.
Supplementary Material
Supplementary Figures S1–17, Supplementary Tables S1–3, S6
Acknowledgments
We thank Guilhem Chalancon and Joseph Marsh for reading the manuscript and making valuable suggestions, Ines de Santiago and Ana Pombo for helpful and interesting discussions, Lucy Colwell for her role in establishing a fruitful collaboration, and Jonathon Howard for reminding us of the importance of absolute numbers.
Author contributions: Experiments, with the exception of RNA-FISH, were carried out by DH. RNA-FISH staining and image processing were carried out by MF. Computational analyses were carried out by DH, with contributions from MG and VC. DH and SAT wrote the manuscript with contributions from MF and AVO.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akaike H (1974) New look at statistical-model identification. Ieee T Automat Contr Ac 19: 716–723 [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G (2004) GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, Dudoit S (2010) Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casella G, Berger RL (2001) Statistical Inference, 2nd edn. Pacific Grove, CA, USA: Duxbury Press [Google Scholar]
- Chintapalli VR, Wang J, Dow JA (2007) Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39: 715–720 [DOI] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, Robertson AJ, Perkins AC, Bruce SJ, Lee CC, Ranade SS, Peckham HE, Manning JM, McKernan KJ, Grimmond SM (2008) Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5: 613–619 [DOI] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K (2009) Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann J, Hubner NC, Kim JB, Ko K, Moser M, Kumar C, Cox J, Scholer H, Mann M (2008) Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5111 proteins. Mol Cell Proteomics 7: 672–683 [DOI] [PubMed] [Google Scholar]
- Hastie ND, Bishop JO (1976) The expression of three abundance classes of messenger RNA in mouse tissues. Cell 9: 761–774 [DOI] [PubMed] [Google Scholar]
- Hebenstreit D, Gu M, Haider S, Turner DJ, Lio P, Teichmann SA (2011) EpiChIP: gene-by-gene quantification of epigenetic modification levels. Nucleic Acids Res 39: e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebenstreit D, Teichmann S (2011) Analysis and simulation of gene expression profiles in pure and mixed cell populations. Physical Biology 8: 035013. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle DC, Rattray M, Jupp R, Brass A (2002) Making sense of microarray data distributions. Bioinformatics 18: 576–584 [DOI] [PubMed] [Google Scholar]
- Jiang H, Wong WH (2009) Statistical inferences for isoform expression in RNA-Seq. Bioinformatics 25: 1026–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, Sweet MJ (2008) Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, King RD (2009) An investigation into the population abundance distribution of mRNAs, proteins, and metabolites in biological systems. Bioinformatics 25: 2020–2027 [DOI] [PubMed] [Google Scholar]
- Lundberg E, Fagerberg L, Klevebring D, Matic I, Geiger T, Cox J, Algenas C, Lundeberg J, Mann M, Uhlen M (2010) Defining the transcriptome and proteome in three functionally different human cell lines. Mol Syst Biol 6: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Mudge J, Miller NA, Khrebtukova I, Lindquist IE, May GD, Huntley JJ, Luo S, Zhang L, van Velkinburgh JC, Farmer AD, Lewis S, Beavis WD, Schilkey FD, Virk SM, Black CF, Myers MK, Mader LC, Langley RJ, Utsey JP, Kim RW et al. (2008) Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in post-mortem cerebellum. PLoS ONE 3: e3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshlack A, Wakefield MJ (2009) Transcript length bias in RNA-seq data confounds systems biology. Biol Direct 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S, Wold B, Mortazavi A (2009) Computation for ChIP-seq and RNA-seq studies. Nat Methods 6: S22–S32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S (2008) Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5: 877–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A (2009) Single-molecule approaches to stochastic gene expression. Annu Rev Biophys 38: 255–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D, Wang ET, Burge CB, Sandberg R (2009) An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Zhao K (2005) Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev 19: 542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G (1978) Estimating dimension of a model. Ann Stat 6: 461–464 [Google Scholar]
- Silverman BW (1986) Density Estimation. London: Chapman and Hall [Google Scholar]
- Spandidos A, Wang X, Wang H, Seed B (2010) PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res 38: D792–D799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M (2009a) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Schones DE, Zhao K (2009b) Characterization of human epigenomes. Curr Opin Genet Dev 19: 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O’Shea JJ, Zhao K (2009) Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30: 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE (2010) Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol 28: 445–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–17, Supplementary Tables S1–3, S6