The maize gene opaque5 encodes a monogalactosyldiacylglycerol synthase. Mutant analysis reveals that galactolipids with five double bonds are needed for thylakoid membrane function and cannot be substituted by the prevalent species with six double bonds. Galactolipid reduction results in altered starch granules, indicating a connection between amyloplast membranes and starch biosynthesis.
Abstract
The maize (Zea mays) opaque5 (o5) locus was shown to encode the monogalactosyldiacylglycerol synthase MGD1. Null and point mutations of o5 that affect the vitreous nature of mature endosperm engendered an allelic series of lines with stepwise reductions in gene function. C18:3/C18:2 galactolipid abundance in seedling leaves was reduced proportionally, without significant effects on total galactolipid content. This alteration in polar lipid composition disrupted the organization of thylakoid membranes into granal stacks. Total galactolipid abundance in endosperm was strongly reduced in o5- mutants, causing developmental defects and changes in starch production such that the normal simple granules were replaced with compound granules separated by amyloplast membrane. Complete loss of MGD1 function in a null mutant caused kernel lethality owing to failure in both endosperm and embryo development. The data demonstrate that low-abundance galactolipids with five double bonds serve functions in plastid membranes that are not replaced by the predominant species with six double bonds. Furthermore, the data identify a function of amyloplast membranes in the development of starch granules. Finally, the specific changes in lipid composition suggest that MGD1 can distinguish the constituency of acyl groups on its diacylglycerol substrate based upon the degree of desaturation.
INTRODUCTION
Maize (Zea mays) mutations that cause opaque kernel phenotypes are of interest because of their effects on seed texture and nutritional quality traits. The phenotype is defined by kernels that appear opaque to transmitted light, as opposed to the partially translucent appearance of normal vitreous seeds. At least 18 opaque gene loci are known, some of which are involved in storage protein metabolism (Thompson and Larkins, 1994; Hunter et al., 2002). The opaque2 (o2) locus, for example, encodes a transcription factor that activates genes coding for α-zein proteins, thus changing the constituency of protein bodies made in the endoplasmic reticulum (ER) and present in mature endosperm after desiccation (Kodrzycki et al., 1989; Schmidt et al., 1990). Another example is floury1, which encodes a membrane protein involved in targeting an α-zein to the interior of protein bodies (Holding et al., 2007). Macromolecular assembly of protein bodies is highly complex (Holding and Larkins, 2009), possibly explaining why opaque loci are identified frequently. In many instances, however, the cause of the phenotype appears to be unrelated to protein composition. For example, altered starch composition conferred by waxy1 mutations causes kernel opacity (reviewed in Coe et al., 1988). Similarly, mutations of the o5 locus condition opaque kernels without notable changes in total protein content, zein versus nonzein ratio, or amino acid composition (Hunter et al., 2002).
In this study, o5 was found to code for a glycosyl transferase that generates monogalactosyldiacylglycerol (MGDG). MGDG and digalactosyldiacylglycerol (DGDG) are the primary nonprotein components of plastid membranes, providing both structural and functional organization. Together, these galactolipids account for as much as 80% of total polar lipid content in plastids (Block et al., 1983; Dörmann and Benning, 2002; Hölzl and Dörmann, 2007). Galactolipid-based membranes are conserved features of organisms that undergo oxygenic photosynthesis, including Chloroplastida and cyanobacteria. Direct functions in photosynthesis are possible because specific galactolipids appear as ligands in three dimensional structures of photosynthetic complexes (Jordan et al., 2001; Loll et al., 2007). In addition, owing to its nonbilayer forming properties, MGDG is likely to provide membrane structure in areas of tight turns and high protein packing density (Webb and Green, 1991; Bruce, 1998; Lee, 2000; Dörmann and Benning, 2002). Nonphotosynthetic plastid membranes also contain MGDG and DGDG in high abundance, so galactolipid function is not restricted to light harvesting or electron transfer. Most higher-plant MGDG has a C18/C18 molecular structure, with an 18:3 fatty acid at both the sn-1 and sn-2 positions of the glycerol backbone. Some plants also contain MGDG with C18/C16 structures (Heinz, 1977; Maréchal et al., 2000). The C18/C18 species are thought to be derived from the ER lipid assembly pathway using fatty acids exported from the plastid, whereas C18/C16 lipids arise from the organellar lipid assembly pathway (Benning, 2008).
The enzyme MGDG synthase (MGD) (UDPgalactose: 1,2-diacylglycerol 3-β-d-galactosyl-transferase; EC 2.4.1.46), located in plastid envelope membranes, catalyzes transfer of a galactosyl residue from UDPgalactose to the sn-3 position of diacylglycerol (DAG) (Dörmann and Benning, 2002; Benning and Ohta, 2005). MGD activity has been studied most extensively in Arabidopsis thaliana, spinach (Spinacia oleracea), and cucumber (Cucumis sativus) (Joyard and Douce, 1987; Shimojima et al., 1997; Maréchal et al., 2000). MGD enzymes in these plants are divided into type A and type B classes based on amino acid sequences as well as enzymatic and physiological properties. The type A enzyme (MGD1) is targeted to the inner plastid envelope membrane, whereas the two type B enzymes (MGD2 and MGD3) are localized to outer envelope membranes (Maréchal et al., 2000; Awai et al., 2001). MGD1 is expressed in all plant tissues and is the most abundant form in chloroplasts, whereas MGD2 and MGD3 are detected primarily in nonphotosynthetic tissues, including flowers and roots (Awai et al., 2001; Kobayashi et al., 2004). MGD2 and MGD3 are induced by phosphate starvation, suggesting they function in specific conditions, whereas MGD1 appears to serve a general role in galactolipid biosynthesis based on its constitutive expression pattern (Awai et al., 2001; Kobayashi et al., 2004).
In Arabidopsis, the hypomorphic point mutation mgd1-1 conditioned chlorotic plants that exhibit ~50% reduction in chlorophyll (Jarvis et al., 2000). MGDG content was reduced to ~42% of normal, although total leaf lipid content was similar to the wild type. Chloroplasts were underdeveloped in the mutant, with reduced numbers of starch granules. The null allele mgd1-2 conditioned a near absence of both MGDG and DGDG as well as disruption of the photosynthetic plastid membranes, leading to a loss of photosynthetic ability for the plant (Kobayashi et al., 2007). Embryo development also was impaired in mgd1-2 mutants and the seeds were white, indicating that galactolipids have an essential role in embryogenesis (Kobayashi et al., 2007). These results indicate that neither MGD2 nor MGD3 compensates for loss of MGD1 in Arabidopsis.
This study characterized the effects of reducing or eliminating MGD1 function in leaves and endosperm of maize, a C4 monocot. The finding that mutations affecting MGD1 cause an opaque endosperm phenotype reveals a connection between membrane structure and storage compound metabolism in nonphotosynthetic plastids. Additionally, a hypomorphic o5- allele specifically affects abundance of minor MGDG or DGDG species with five double bonds in the two acyl groups combined. This seemingly subtle change in lipid structure, with essentially no reduction in the total galactolipid content, is physiologically significant. The data further reveal that acyl group identity can be a factor for MDG1 enzyme activity. In an o5 null mutant, as was observed in Arabidopsis, maize embryo development is aborted at an early stage, indicating that galactolipid function is essential in nonphotosynthetic and photosynthetic tissue.
RESULTS
Multiple Alleles of the o5 Locus and Resultant Phenotypes
The mutation o5-Ref, which causes visually distinctive opaque kernel and pale green virescent seedling phenotypes (Figures 1A and 1B), was first described by Robertson in 1967 (www.maizegdb.org). Other mutations that cause opaque kernels were subjected to complementation tests by crossing heterozygous mutants to homozygous o5-Ref plants. In three instances, ~50% of the progeny kernels were opaque, thus identifying new o5- alleles. These mutations, termed o5-PS3038, o5-5288, and o5-313328, condition various kernel and seedling phenotypes (summarized in Supplemental Table 1 online).
Figure 1.
Kernel and Seedling Phenotypes.
(A) o5-Ref kernel phenotype. Mature kernels from a self-pollinated ear of an o5-Ref/+ heterozygote were photographed under reflected light (black background) or backlit by a transilluminator (white background).
(B) Seedling phenotypes. Kernels of the indicated genotype were planted in the greenhouse, and seedlings were photographed 7 d after emergence.
(C) o5-PS3038 kernel phenotype. A portion of a self-pollinated ear from an o5-PS3038/+ heterozygote is shown. Homozygous mutant kernels are variable in size, often shriveled, floury in texture, and carotenoid deficient.
Both o5-Ref and o5-313328 condition pale-green, virescent seedlings that subsequently acquire full green color and develop apparently normally through maturity. Homozygous kernels bearing either mutation are opaque and slightly pale compared with yellow kernels of the congenic wild-type inbred W64A but germinate normally and exhibit near-normal morphology. The o5-5288 allele causes a stronger phenotype with white seedlings that fail to acquire green color and die 7 to 14 d postemergence. The kernel morphology of o5-5288 homozygotes is white and shrunken. The mutation o5-PS3038 is the most severe in this allelic group because it conditions defective kernels that fail to germinate. Mutant kernels observed on a segregating ear from a self-pollinated o5-PS3038/+ heterozygote are white and frequently severely shrunken (Figure 1C). The latter two mutations must be maintained as heterozygotes because homozygous individuals are nonviable.
Another allelic combination examined in this study is the o5-Ref/o5-PS3038 compound heterozygote. Such kernels are lighter in color and slightly collapsed at the crown compared with homozygous o5-Ref kernels. Seedlings grown from these kernels remain chlorotic and die within 2 weeks of emergence (Figure 1B), similar to o5-5288 homozygotes.
Molecular Characterization of the o5 Locus
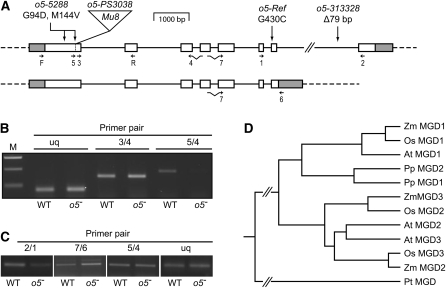
Transposon tagging was used to identify the o5 locus. As described previously, a Mu8 transposable element insertion was found tightly linked to the o5 locus (Yi et al., 2009). Briefly, an o5-PS3038/+ heterozygote was crossed to an o5-Ref homozygote to generate a population segregating 1:1 for o5-PS3038/o5-Ref mutant and +/o5-Ref normal offspring. As expected, approximately one-half of the kernels on such ears displayed the opaque phenotype. DNA isolated from mutant and normal seedlings was subjected to a high-throughput transposon display method called MuTA. A unique amplification product containing a Mu8 terminus and flanking genomic DNA was identified in all four o5-PS3038/o5-Ref mutant DNA samples and was absent in all four +/o5-Ref normal samples. The sequence of this product matched exactly to a published EST (GenBank accession number EE189213). Gene-specific primers from this sequence were used in combination with a Mu-end primer to amplify that specific Mu insertion. Forty-eight additional segregants from the cross were thus analyzed, and perfect cosegregation was observed, indicating this Mu8 insertion is linked to, and potentially causal of, the o5-PS3038 mutation (Yi et al., 2009).
The genomic sequence adjacent to the Mu insertion associated with o5-PS3038 is within the locus that codes for a protein annotated as MGDG synthase type A (GenBank accession number EU954700), referred to here as MGD1. The transcribed region of this gene is located on chromosome 7.02 between nucleotides 117,701,235 and 117,708,502 of the B73 reference genome version 2, which as expected is near or at the inferred position of the o5 locus (Andorf et al., 2010). Mu8 is inserted into the first exon, 441 bases 3′ to the presumed initiation codon (Figure 2A). The effect of this mutation on the MGD1 mRNA level was examined by RT-PCR amplification of RNA from wild-type and mutant kernels from an ear of a self-pollinated o5-PS3038/+ heterozygote. Wild-type samples yielded an obvious amplification product when the PCR primers flanked the insertion site, whereas only a trace signal was evident in the mutant kernels (Figure 2B). Product from mutant kernels was most likely derived from pericarp tissue, which is maternal and, thus, heterozygous for a wild-type o5 allele. A primer pair located 3′ to the Mu insertion generated an obvious PCR product in both mutant and wild-type kernels. This suggests spurious transcript initiation from within the Mu element, as has been described previously for other Mu-induced mutations (Barkan and Martienssen, 1991).
Figure 2.
Characterization of the o5 Locus.
(A) Locus structure. Introns are shown as solid lines, exons as boxes, and untranslated regions as gray boxes. Mutation sites are indicated. Small arrows indicate the locations of oligonucleotide primers used in RT-PCR analyses in (B) and (C). The figure is drawn to scale except for intron 7, which is ~3.1 kb. The top diagram maps the transcript that appears most frequently in EST data, and the bottom diagram maps an alternative transcript that fails to remove intron 7 and uses a different 3′ end processing site.
(B) Transcript accumulation in o5-PS3038 kernels. MGD1 mRNA was detected by RT-PCR in wild-type (WT) and o5-PS3038/o5-PS3038 kernels (designated o5-) harvested from a segregating ear at 15 DAP. Primers locations are shown in (A). The ubiquitin (uq) gene transcript was used as a positive control. M, molecular weight standards.
(C) Transcript accumulation in o5-313328 kernels. Analysis as in (B).
(D) Phylogenetic relationship of MGD genes. The o5 gene product (shown as Zm MGD1) was aligned with MGD proteins from maize, rice, Arabidopsis, the moss Physcomitrella patens, and the diatom P. tricornutum. GenBank protein accession numbers are listed in Methods.
The gene encoding MGD1 was examined in three other o5- allelic mutants (see Supplemental Table 1 online). MGD1 cDNA was amplified by RT-PCR and sequenced from homozygous o5-Ref, o5-5288, and o5-313328 plants as well as from wild-type inbreds W64A, Oh43, and B73. Single nucleotide polymorphisms were identified that caused amino acid substitutions specifically in o5-Ref and o5-5288 mutants. In o5-Ref, a G/T transversion at nucleotide 1511 (numbering according to GenBank accession number BT042676) would change the residue at position 430 from a Gly to a Cys. This Gly is invariant in MGD coding sequences among vascular plants, moss, and species as divergent as the diatom Phaeodactylum tricornutum (see Supplemental Figure 1 online) (Botté et al., 2005). cDNA clones from o5-5288 seedlings contained two substitutions in MGD1 relative to the wild type. A G/A transition at nucleotide 504 would cause substitution of the Gly at residue 94 by Asp, and an A/G transition at nucleotide 653 changes the invariant Met codon at position 144 to a Val. Invariant residues are usually essential for protein function, indicating that the nonconservative amino acid substitutions in o5-Ref and o5-5288 are likely to impair MGD1 enzyme activity.
Changes in the gene encoding MGD1 were also detected in plants carrying o5-313328 (see Supplemental Table 1 online). Homozygotes for o5-313328 did not display any differences from the wild type in the full-length MGD1 cDNA sequence; however, genomic DNA contained a deletion of 79 bp in intron 7 that was not detected in any wild-type line. RT-PCR with primers spanning the seventh intron consistently generated decreased amounts of amplification product compared with normal, whereas primers that amplified an alternatively spliced and presumably nonfunctional transcript exhibited an increased signal (Figures 2A and 2C). This suggests that splicing of intron 7 is less efficient than normal owing to the deletion associated with o5-313328.
In summary, plants carrying four independent o5- alleles were altered in the gene encoding MGD1. Together with the fact MGD1 coding sequences could not be separated from the o5 locus, the data demonstrate that o5 codes for MGD1.
Bioinformatic Analyses
Genomics data (http://www.plantgdb.org and http://www.maizeGDB.org) revealed that the transcribed region of o5 spans ~7267 bp, including eight exons and seven introns (Figure 2A). The longest identified mature transcript (GenBank accession number BT042676) comprises 2188 bp with 1589 bp of coding sequence and noncoding regions of 223 and 374 bp at the 5′ and 3′ ends, respectively. The full-length protein product is predicted to contain 529 amino acid residues, including a 53-residue plastid transit peptide predicted by the TargetP algorithm (Emanuelsson et al., 2007). The predicted molecular mass of mature MGD1 is 51.7 kD.
The full-length amino acid sequence contains a conserved glycosyl transferase (GT) domain classified as family GT28 according to the Carbohydrate Active Enzymes database (Cantarel et al., 2009), whose members are predicted to exhibit the conserved GT-B structural fold (Lairson et al., 2008). This structure comprises residues 151 to 482 of the full-length MGD1 sequence, so an amino terminal extension of ~98 residues is attached to the catalytic domain in the mature protein. The first half of the GT domain is further classified as an MGDG synthase domain, denoted as structure PF06925 in the Pfam database (Finn et al., 2010). Maize MGD1 is 86% identical to Arabidopsis MGD1 from residues 137 to 519 of the full-length sequence, with no gaps in the alignment. Sequence identity between maize and Arabidopsis MGD1 from residues 1 to 136 is much less but still significant, with 32% identical amino acids over 120 aligned residues, including four gaps.
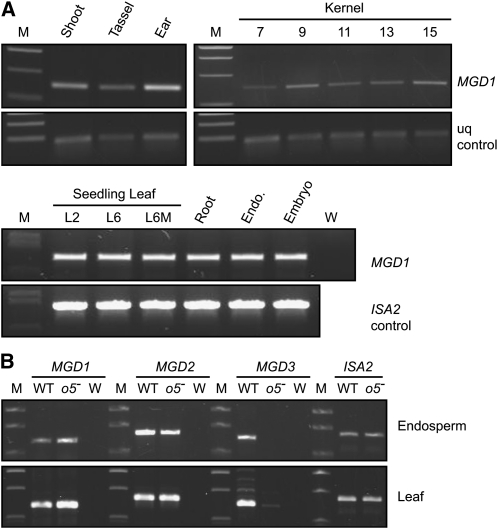
o5 is one of three loci in the maize genome coding for predicted MGDG synthases. The other two genes, mdg2 and mdg3, are located on chromosomes 5 and 1, respectively. MGD1 groups phylogenetically with type A MGD synthases, whereas MGD2 and MGD3 are related more closely to type B enzymes (Figure 2D). National Center for Biotechnology Information (NCBI) annotation of the MGD2 and MGD3 genes is discrepant between maize and rice (Oryza sativa), hence, the apparent nonconcordance in the phylogeny. As a type A MGD synthase, maize MGD1 would be predicted to be expressed widely throughout the plant. This was tested by RT-PCR amplification of MGD1 mRNA from seedling shoot, root, juvenile and adult leaf tissue, tassel, ear, kernels at various developmental stages, endosperm harvested 20 d after pollination (DAP), and 20 DAP embryo. Signals were evident in all tissues tested (Figure 3A), supporting classification of MGD1 as a type A enzyme.
Figure 3.
RT-PCR Analysis of Transcript Levels.
(A) Expression of MGD1 mRNA in wild-type tissues. RNA from the indicated tissues was amplified using primers specific for the MGD1 transcript. The ubiquitin (uq) or ISA2 transcripts were used as controls. M, molecular weight standards. W, no template control. Shoot and root were from seedlings. Ear tissue was from early development when the organ was ~2 mm in length. Whole kernels were analyzed at the indicated days after pollination. L2, leaf from two-leaf seedlings; L6, youngest leaf of six-leaf seedlings; L6M, mature seedling leaf defined as the outer one-third of the third leaf of six-leaf seedlings. Endosperm and embryo were from kernels harvested 20 DAP. In the top panel, primers 3 and 4 were used to amplify cDNA, and in the bottom panel, the primers were F and R. The locations of the primers are shown in Figure 2A.
(B) Expression of MGD1, MGD2, and MGD3 mRNAs in wild-type and o5-Ref tissues. Total RNA from endosperm or leaf was amplified using primers specific for the indicated transcript. The ISA2 transcript provided a positive control. M, molecular weight standards; WT, wild-type inbred W64A; o5-, plants homozygous for o5-Ref; W, no template control.
Effects of o5 Mutations on Lipid Constituents
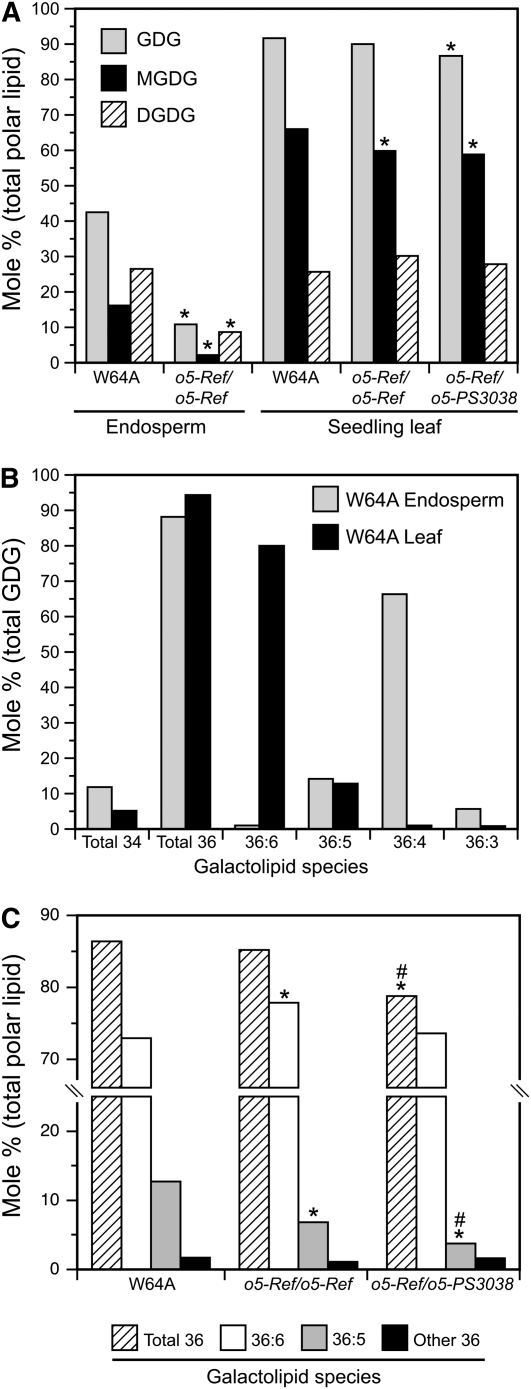
Polar lipids from seedling leaves and developing endosperm were characterized to test whether MGD function is altered in o5- mutants and to further define the effects of these mutations. Organic extracts were analyzed by mass spectrometry to identify and quantify individual phospholipids and galactolipids (Welti and Wang, 2004) (complete data set in Supplemental Data Set 1 online). The molar percentage of total galactosyldiacylglycerol (GDG) was ~42% in wild-type endosperm, whereas in seedlings, the GDG content was ~92% (Figure 4A). The distribution between MGDG and DGDG differed between the tissues, with monogalactosyl species constituting approximately one-third of total GDG in endosperm and two-thirds in seedling leaves (Figure 4A). The great majority of GDG in both endosperm and leaf were C18/C18 species (Figure 4B), and thus are expected to be derived from the ER assembly pathway (Benning, 2008). Among the C18/C18 GDG species, there were major differences between wild-type tissues in the degree of desaturation of the acyl groups. Endosperm contained predominantly 36:4 lipid, with ~13% 36:5 species and essentially no 36:6 lipids. Leaf tissue, conversely, contained predominantly 36:6 GDG species and very little 36:4 lipid. The 36:5 GDG content was similar in leaf and endosperm (Figure 4B).
Figure 4.
Lipid Content.
(A) Galactolipid species abundance in leaf and endosperm of the wild type and o5- mutants. MGDG, DGDG, and total GDG abundance is plotted as the molar percentage of total polar lipid. Values are averages of independent biological replicates (W64A endosperm, n = 4; o5-Ref/o5-Ref endosperm, n = 5; all seedling leaf tissue, n = 3; sd ≤ 2.1%). Asterisks indicate significant differences from the wild type (P < 0.01; Student's t test). Differences between o5-Ref/o5-Ref and o5-Ref/o5-PS3038 were not observed at this level of significance.
(B) Degree of desaturation in galactolipids of wild-type leaf and endosperm. Galactolipid species abundance is plotted as the molar percentage of total galactolipid. Independent replicates were as in (A) (sd ≤ 0.6%).
(C) Comparison of the degree of desaturation in galactolipids of wild-type and o5- mutant leaves. Galactolipid species abundance is plotted as the molar percentage of total polar lipid. Independent replicates were as in (A) (sd ≤ 1.6% for all samples and ≤0.6% for 36:5 species). Asterisks indicate significant differences from the wild-type value, and pound signs indicate significant differences from the o5-Ref/o5-Ref value (P < 0.01; Student's t test).
Total GDG content in endosperm was reduced approximately fourfold in homozygous o5-Ref mutants compared with the wild type (Figure 4A), supporting the conclusion that o5 codes for MGDG synthase. Both MGDG and DGDG abundance were reduced by the mutation. The distribution between 36:4 and 36:5 GDG was essentially unchanged in wild-type and o5-Ref endosperm (see Supplemental Data Set 1 online).
In contrast with endosperm, the total GDG level in leaves was not significantly altered in o5-Ref/o5-Ref homozygotes and was only slightly reduced when one copy of o5-Ref was present along with one copy of the null allele o5-PS3038 (Figure 4A). Thus, reduced total GDG level is not likely to be the cause of the leaf phenotypes. However, significant changes were noted in the abundance of 36:5 GDG species between the wild type and mutants. The total abundance of C18/C18 GDG was the same in W64A and o5-Ref leaves; however, the mutant had approximately one-half the wild-type amount of 36:5 GDG (Figure 4C). Heteroallelic o5-Ref/o5-PS3038 leaves had one-quarter of the wild-type 36:5 GDG content. Thus, the magnitude of the effect was proportional to dosage of partially functional alleles at the o5 locus. This effect was observed for both 36:5 MGDG and 36:5 DGDG (see Supplemental Data Set 1 online).
Proportionality to functional gene dosage, and accordingly to phenotypic severity, was observed only for 36:5 GDG content. Other changes in polar lipid abundance were observed; however, these were quantitatively small and did not correlate with phenotype. For example, MGDG abundance was reduced slightly but significantly in both mutant lines compared with the wild type (Figure 4A). This is not the cause of the leaf phenotypes because the total MGDG level was the same in o5-Ref/o5-Ref and o5-Ref/o5-PS3038 plants, although they display very different leaf phenotypes. Changes in phospholipid content between mutant and wild-type leaves also were observed (see Supplemental Data Set 1 online); however, these are minor species comprising in total <10% of the polar lipids. Proportionality to gene dosage or phenotype was not observed for any change in phospholipid content.
Expression of MGD Genes in Endosperm and Leaf
The greater effect of o5- mutations on total GDG level in endosperm than leaf could be explained by a compensation mechanism in the latter tissue. One possibility is that mgd2 or mgd3 expression is induced in o5- mutant leaf tissue but not in endosperm. This was investigated by examining the levels of transcripts coding for MGD using RT-PCR. MGD1, MGD2, and MGD3 transcripts were all present in both leaf and endosperm of wild-type plants (Figure 3B). The MGD1 mRNA level was not altered in either tissue by o5-Ref, consistent with identification of that allele as a point mutation. In both leaf and endosperm, MGD2 mRNA was present at the same levels in o5-Ref mutants as in the wild type. MGD3 mRNA was strongly reduced in concentration as a result of o5-Ref in both leaf and endosperm. The reason o5-Ref would affect MGD3 expression is not clear; however, the data taken together are inconsistent with the hypothesis that increased expression of MGD2 or MGD3 mRNA is the reason that o5-Ref has relatively little effect in leaf. The possibility remains that MGD2 functions in leaf but not in endosperm, despite the fact that the mRNA is present in both tissues.
Effects of o5 Mutations on Kernel Development
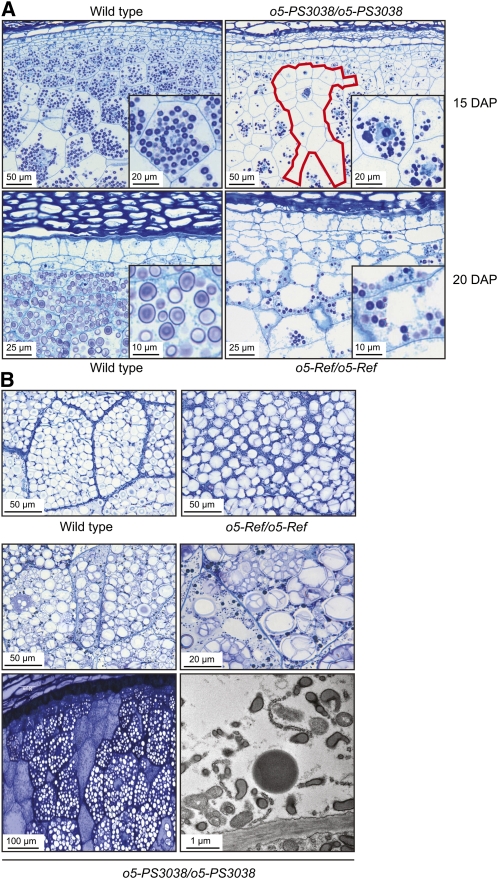
Endosperm characteristics were examined to investigate the nature of the opaque kernel phenotype. Homozygous o5-Ref mutants are viable, and mature kernels appeared normal except for the loss of translucency typical of wild-type maize seed (Figure 1A). Starchy endosperm tissue at mid-development (20 DAP) was abnormal, with highly vacuolated cells containing decreased numbers of starch grains that were noticeably smaller than the wild type (Figure 5A). At maturity, starch granule size in o5-Ref endosperm was similar to the wild type, although they appeared less densely packed and often compound (Figure 5B).
Figure 5.
Endosperm Development.
(A) Histological sections of immature endosperm. Wild-type and mutant kernels were collected at 15 DAP from a self-pollinated o5-PS3038/+ plant and at 20 DAP from a self-pollinated o5-Ref/+ plant. A cluster of cells in o5-PS3038/o5PS3038 endosperm largely devoid of starch grains is outlined in red.
(B) Histological sections of mature endosperm. Mature kernels were collected from the plants described in (A) and either fixed for histological analysis by light microscopy or embedded, thin-sectioned, and analyzed by TEM (bottom right panel). The TEM image depicts a cell from a region of o5-PS3038/o5PS3038 endosperm devoid of starch granules (bottom left panel).
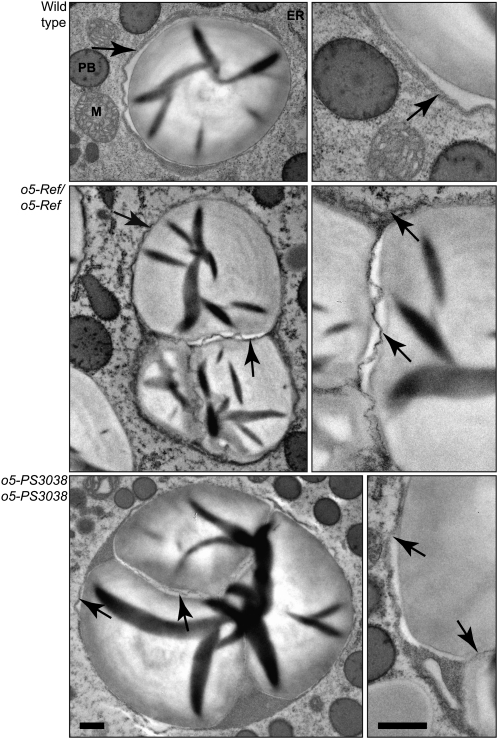
Homozygous o5-PS3038 mutants exhibit a defective kernel phenotype in which the seeds are small, white, collapsed to varying degrees, and have poorly developed, floury endosperms (Figure 1C). Homozygous mutant endosperm cells at 15 DAP had greatly decreased cellular contents and displayed a variety of cytological abnormalities, such as amoeboid nuclei and undefined globular structures (Figure 5A). At maturity, starch granules were evident in many cells, although they often showed highly irregular morphologies, including a dramatic enlargement and high degree of compounding (Figure 5B). Also of note were contiguous zones of cells devoid of starch grains or other recognizable structures (Figures 5A and 5B). These vacant zones, observed at both mid-development and maturity, were not seen in wild-type endosperm. Transmission electron microscopy (TEM) analysis revealed that cells in these zones were filled with many small vesicles and other bodies of unknown composition (Figure 5B).
Wild-type and mutant endosperm and starch granules were further examined by TEM (Figure 6). Normal tissue revealed characteristic simple granules surrounded individually by amyloplast envelope. The compound nature of starch in mature o5-PS3038 mutant endosperm was clearly evident in TEM, with internal membranes separating individual granules packed together within a single amyloplast. Entire compound granules were surrounded on the exterior with plastid envelope. Each particle within a single plastid displayed concentric growth rings, demonstrating compound granules derived from multiple initiation events rather than fractured simple granules. Growth rings in mutant starch grains appeared to be irregularly spaced compared with the uniform rings in wild-type granules. Mature o5-Ref/o5-Ref endosperm observed in TEM also exhibited numerous compound granules but less frequently than in the null mutant. These also were surrounded on the exterior by amyloplast envelope, with interior membranes separating individual particles (Figure 6). Most granules in o5-Ref tissue were close to the wild type in size and growth ring spacing.
Figure 6.
TEM Visualization of Mature Starch Grains in Endosperm Tissue.
Kernels on self-pollinated ears of o5-Ref/+ or o5-PS3038/+ heterozygotes were harvested at maturity, fixed, thin-sectioned, and analyzed by TEM. Dark-staining areas within granules are folds that arise during sectioning. Arrows indicate amyloplast envelope or internal membrane. Bars = 0.5 μm and apply uniformly within each column. Organelles are indicated: M, mitochondria; PB, protein bodies.
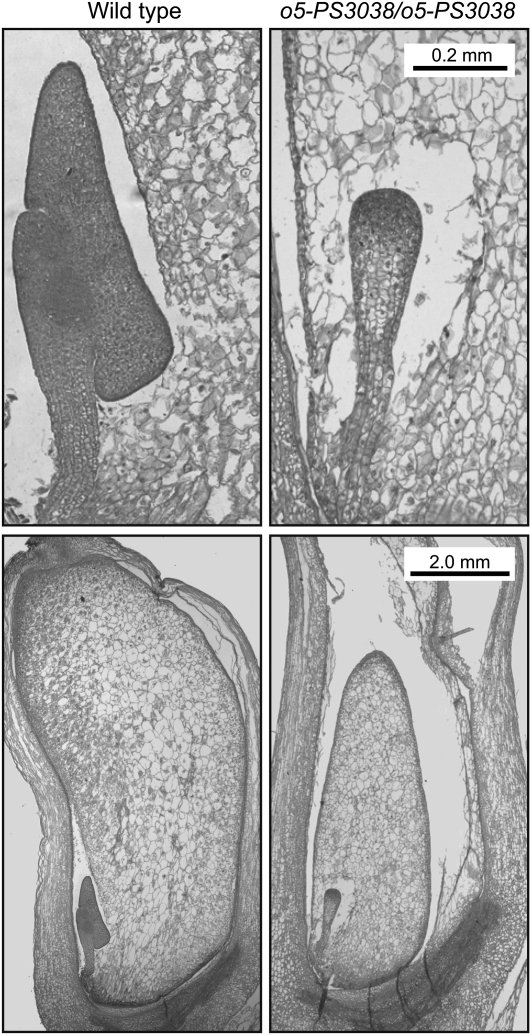
Embryo development was also severely affected in homozygous o5-PS3038 mutants. Longitudinal sections of developing kernels harvested 15 DAP revealed that the mutant embryos attained a relatively normal transition stage morphology but failed to progress further (Figure 7). Wild-type embryos had established an axis, shoot apical meristem, and scutellum by this point.
Figure 7.
Embryo Development.
Wild-type and homozygous o5-PS3808 kernels harvested from a segregating ear at 15 DAP were collected, and longitudinal sections were prepared such that both endosperm and embryo were included. Tissue sections were stained with Safranin O and fast green. Bars apply to both images in each row.
Effects of o5 Mutations on Starch Biosynthesis
Morphological abnormalities of starch granules in o5- mutants prompted more detailed investigation of starch biosynthesis. Consistent with fewer and smaller starch granules in o5-Ref endosperm at mid-development compared with the wild type (Figure 5A), the total starch content measured at 20 DAP was significantly reduced in the mutant (P = 0.02; Table 1). Water-soluble polysaccharide was elevated in the mutant at this stage by approximately sixfold compared with the wild-type level. By contrast, at maturity when o5-Ref starch granules are generally similar in size to the wild type (Figure 5B), neither the total starch content nor the water-soluble polysaccharide level was significantly altered in the mutant (Table 1). Amylose content was the same in mature starch granules from wild-type or o5-Ref/o5-Ref endosperm (Table 1).
Table 1.
Endosperm Carbohydrate Content
| Genotype | 20 DAP |
Mature |
|||
| Starch | WSP | Starch | WSP | Amylose (%) | |
| +/+ | 697 ± 74 | 4.0 ± 1.1 | 598 ± 51 | 3.3 ± 1.1 | 22 |
| o5-Ref/o5-Ref | 502 ± 45 | 23.1 ± 2.0 | 563 ± 27 | 3.0 ± 1.0 | 25 |
Starch and water-soluble polysaccharide (WSP) content is expressed as mg/g dry weight. Values are the mean ± sd from three biological replicates. Amylose content is expressed as the percentage of total carbohydrate in purified starch.
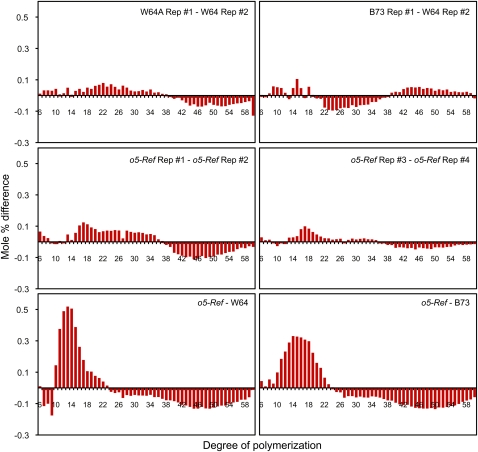
Glucan chain length distribution in amylopectin from o5-Ref and wild-type endosperm was determined using fluorophore-assisted carbohydrate electrophoresis (Morell et al., 1998; O'Shea et al., 1998). Biological replicates of starch from kernels harvested 20 DAP showed that the measured chain length distribution was highly reproducible for both wild-type and o5-Ref homozygotes (Figure 8). Compared with B73 or W64A wild-type starch, o5-Ref starch had an elevated frequency of linear chains with degrees of polymerization in the 12 to 21 range. The chain lengths that are increased in frequency constitute the crystalline lamellae of amylopectin (Buléon et al., 1998).
Figure 8.
Amylopectin Chain Length Distribution.
Total starch from the indicated genotypes was debranched with commericial isoamylase, and mole percentage of linear chains of each degree of polymerization was normalized to the total number of chains from DP5 to DP48. Plots show the difference between the mole percentage of each chain length for the two indicated lines. The designations #1, #2, etc., indicate independent biological replicates of the same genotype. For comparison between genotypes (bottom row), the average values for four independent replicates of the mutant and two independent replicates of each wild-type inbred line were compared.
[See online article for color version of this figure.]
The assembly state of isoamylase-type starch debranching enzymes (ISA) was analyzed because mutations affecting these enzymes change amylopectin chain length frequency in the same size range that is altered in o5-Ref mutants (Dinges et al., 2001). Three bands of ISA activity are observed in extracts of 20 DAP wild-type endosperm after native-PAGE, electrophoretic transfer to a starch-containing gel, and then staining with I2/KI (Kubo et al., 2010). Endosperm of o5-Ref kernels reproducibly exhibited a fourth band of ISA activity (Figure 9). The levels of ISA1 and ISA2 protein determined by immunoblot analysis using isoform-specific IgG fractions were the same in o5-Ref and wild-type endosperm. Multiple ISA activity bands are known to result from different assembly states of the ISA1 and ISA2 proteins in multimeric complexes in both maize and rice (Utsumi and Nakamura, 2006; Kubo et al., 2010). The data suggest that the change in membrane lipid composition caused by o5-Ref results in altered assembly of one or more of these ISA complexes.
Figure 9.
Starch Modifying Enzymes.
Extracts of 20 DAP endosperm were separated by native-PAGE and analyzed by zymogram. Differential color staining in I2/KI revealed changes in starch structure owing to specific enzyme activities. Blue bands result from ISA complexes and red bands from starch branching enzymes. ISA form I is known to be an ISA1 homomeric complex, whereas forms II and III are ISA1/ISA2 heteromers. The asterisk indicates the position of an ISA activity observed reproducibly in o5-Ref endosperm that is not seen in the wild type.
Mutations of o5 Disrupt Chloroplast Structure
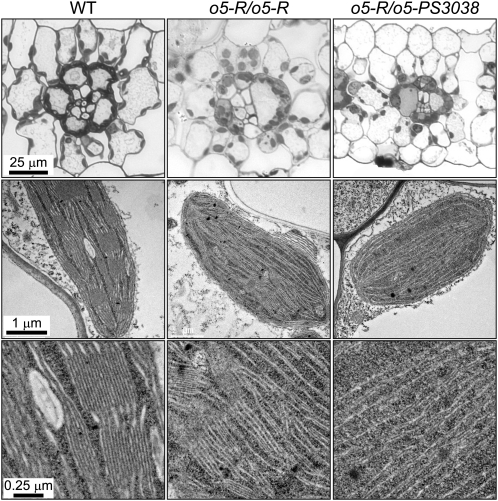
Further microscopy characterization of photosynthetic tissue was undertaken to investigate the nature of the pale-green or colorless seedling phenotype observed in viable o5-Ref homozygotes or seedling-lethal o5-Ref/o5-PS3038 compound heterozygotes (Figure 1B). The o5-Ref mutant leaves in cross section appear similar to the wild type with regard to the number of chloroplasts present in mesophyll and bundle sheath cells. The chloroplasts appear less tightly appressed to the periphery of mutant bundle sheath cells than in the wild type (Figure 10). The number of chloroplasts present in the more severely affected o5-Ref/o5-PS3038 seedlings is noticeably reduced from the wild type, particularly in mesophyll cells.
Figure 10.
Effects of o5- Mutations on Leaf and Chloroplast Morphology.
Seedling leaves were fixed and sectioned for either light microscopy or TEM. The top row shows chloroplasts as dark staining bodies that appear contiguous in the bundle sheath cells. Individual mesophyll chloroplasts visualized by TEM are shown in the bottom two rows. Bars apply uniformly within each row. WT, wild type.
TEM observation of individual chloroplasts revealed defects in membrane assembly that increased in severity proportional to the reduction in 36:5 galactolipid content (Figure 10). Highly organized granal stacks of thylakoid membranes were clearly observed in chloroplasts of wild-type mesophyll cells. Chloroplasts in o5-Ref/o5-Ref mesophyll had less extensive grana, and this effect was more severe in o5-Ref/o5-PS3038 plastids where internal membrane structure appeared sparse and even less organized (Figure 10). Normal bundle sheath chloroplasts also possess an organized thylakoid membrane system although they lack grana and are the major sites of transient leaf starch accumulation. The thylakoids in chloroplasts of o5-Ref/o5-PS3038 bundle sheath cells showed decreased laminar structure, and starch grains were rare and much smaller (see Supplemental Figure 2 online).
DISCUSSION
o5 Encodes a Type A MGDG Synthase
The combined genetic and biochemical analyses presented here make clear that the maize o5 gene encodes a type A MGD synthase. A specific transposon within the gene that codes for MGD1 was genetically inseparable from a defined o5- mutation. Furthermore, plants carrying any of three additional independent o5- alleles also contained specific mutant variants at the locus encoding MGD1. Accumulation of direct and indirect products of MGD1 (i.e., MGDG and DGDG, respectively) is strongly reduced in endosperm tissue in mutants carrying lesions in o5. Taken together, these data demonstrate that MGD1 defects are the causative agents of the observed phenotypes in o5- mutants.
Galactolipids in Maize Endosperm and Leaf
Wild-type leaf and endosperm varied considerably in galactolipid content in three aspects. First, seedling leaves contained >90% GDG as a molar percentage of total polar lipids compared with 42% of total in endosperm. Second, MGDG abundance relative to DGDG was different in the two tissues, with approximately one-third of the total GDG as monogalactosyl forms in endosperm and two-thirds in leaf. Finally, the degree of desaturation was very different. About 67% of endosperm GDG was a 36:4 species (i.e., 18:2 fatty acyl groups at both the sn-1 and sn-2 positions), and this tissue was essentially devoid of 36:6 species. By contrast, leaf comprised ~80% 36:6 species, whereas 36:4 GDG was detected at only a trace level. These endosperm characteristics are consistent with previous characterization of potato (Solanum tuberosum) amyloplast membranes, which showed DGDG in excess of MGDG, and preponderance of 18:2 fatty acids relative to 18:3 species in all galactolipids (Fishwick and Wright, 1980).
Maize leaf GDG content was high relative to that reported for other plant species (e.g., 60 to 70% in Arabidopsis leaves) (Dörmann et al., 1995; Jarvis et al., 2000). This discrepancy could be explained by differences in developmental stage, inherent species characteristics, or possibly the use of mass spectrometry as the detection method in this study. Regardless, GDGs are the major polar lipids present in both maize and Arabidopsis leaves. Maize GDG has ~95% C18/C18 species thought to derive from the eukaryotic pathway of GDG biosynthesis. Previous reports indicated that maize was devoid of C18/C16 GDG species (Mongrand et al., 1998 and references therein); however, the use of mass spectrometry now reveals that a minor amount of GDG biosynthesis proceeds through the prokaryotic pathway in both maize leaves and endosperm.
Unique Effects of o5- Mutations on Leaf Galactolipids
This study characterized the effects of disrupted MGD1 function in maize, a monocot species that uses the C4 photosynthetic pathway. The o5- alleles affected lipid content in ways that were not seen previously in analysis of mgd1 mutants in Arabidopsis (Jarvis et al., 2000; Kobayashi et al., 2007), thus providing insight into functions of specific lipid species. In mutant maize leaves, the reduction in total GDG content was much less severe than that observed in Arabidopsis, even though in both instances chlorotic phenotypes were observed. Notably, there was no significant difference in total GDG content between W64A and homozygous o5-Ref leaves. In o5-Ref/o5-PS3038 compound heterozygotes, which are severely chlorophyll deficient, the total GDG decrease was 5% of the wild-type value. This compares to a total GDG decrease of 20% in hypomorphic Arabidopsis mgd1-1 mutants. With regard to MGDG specifically, mutant maize seedlings were reduced by <11% compared with the 42% reduction seen in Arabidopsis. Thus, the data reveal that photosynthetic development can be impaired as the result of less severe and more specific changes in the GDG profile than have been observed previously.
Further analysis revealed that 36:5 galactolipids are specifically required for normal chloroplast structure and function and that this requirement cannot be substituted by 36:6 species. There was clear correspondence between the level of MGD1 function and 36:5 GDG content. The o5-Ref/o5-Ref line with two copies of the hypomorphic point mutation showed 50% reduction in 36:5 GDG compared with the wild type, whereas o5-Ref/o5-PS3038, with one copy of the hypomorphic allele and one null allele, was reduced 75%. Successively greater reduction in 36:5 GDG corresponded to the increasing phenotypic severity of the two mutant lines. No other parameters of lipid content exhibited such correspondence. Taken together, the data are strong evidence that 36:5 GDG deficiency causes the photosynthetic defects.
The specific reduction in 36:5 galactolipids, and not 36:6 lipids, indicates molecular recognition of acyl group structure on the DAG acceptor substrate by MGD synthase(s). Galactolipid precursors with 36 carbons in the acyl groups are provided from the ER lipid assembly pathway with alkene groups already in place, prior to conversion to DAG in the plastid and subsequent glycosylation by MGD1 (Benning, 2008). Thus, reduction in 36:5 GDG content but not 36:6 GDG most likely is explained at the level of the galactosyl transferase. The alternative is that o5- mutations affect fatty acyl desaturases in the ER, which is implausible given the enzymatic function of MGD1 and its location in the inner plastid envelope. Both MGDG and DGDG were reduced in 36:5 frequency to the same extent, suggesting that DGDG synthase activity is not affected by the desaturation state of the DAG substrate. This observation is consistent with the fact that MGDG produced by MGD1 in the inner envelope is the substrate of DGD1 and DGD2 located in the outer envelope membrane (Kobayashi et al., 2007; Benning, 2008).
The relatively small change in galactolipids in o5-Ref/o5-Ref or o5-Ref/o5-PS3038 leaves could result from inherent properties of the G430C mutant MGD1 enzyme and/or by compensatory activity of another MGD synthase(s). MGD1-G430C could be altered in enzymatic rate or substrate binding but not devoid of function, so that MGDG synthesis is reduced but not eliminated. This is likely because seedlings with two copies of o5-Ref apparently attain full photosynthetic capacity, whereas those with only one copy remain chlorotic and are inviable, and because 36:5 GDG content increases with two copies of the mutant allele relative to only one copy. These observations are consistent with the hypothesis that MGD1-G430C is impaired in recognition of 36:5 DAG as a substrate but not 36:6 DAG. Based on molecular modeling studies, G430 is predicted to be located within the active site of MGD1, near the sites involved in binding to both the UDPGal donor and the DAG acceptor substrate (Botté et al., 2005). Thus, perturbation of DAG substrate selection owing to the G430C mutation is plausible.
The possibility also exists that MDG2 could be active in maize seedling leaves so that loss of MGD1 activity has only a small effect on total MGDG synthesis. According to this hypothesis, the MGD2 enzyme would have reduced activity toward 36:5 DAG substrates relative to 36:6 DAG. MGD2 mRNA is present in seedling leaves, at the same level in either o5- or wild-type plants. MGD3 can be ruled out as a compensating enzyme because the corresponding transcript is not detected in mutant leaves. MGD2 activity cannot fully explain the slight changes in GDG levels, however, because such compensation should occur to the same extent regardless of whether one or two copies of o5-Ref are present. Compensation clearly does not occur in Arabidopsis because an mgd1 null mutation caused essentially complete absence of GDG in seedlings even though MDG2 and MDG3 transcripts were present (Kobayashi et al., 2007). Thus, partially reduced activity of the mutant MGD1 encoded by o5-Ref is responsible for the specific lipid phenotype, at least to some extent if not entirely.
Galactolipid Function in Chloroplasts
The o5-Ref/o5-PS3038 seedlings exhibited altered chloroplast structure attributed to deficiency of 36:5 GDG. The observed morphological defects in the plastid are more severe than those caused by the hypomorphic allele mgd1-1 in Arabidopsis (Jarvis et al., 2000) despite much smaller changes in MGDG or total GDG content. In the dicot, the amount of thylakoid membrane was substantially decreased, although the remaining membranes formed morphologically normal granal stacks. By contrast, o5-Ref/o5-PS3038 mesophyll thylakoid membrane structure was drastically altered such that normal grana were lost. An intermediate degree of thylakoid disorganization was observed in o5-Ref/o5-Ref mesophyll, supporting the conclusion that 36:5 GDG content is the determining factor. The extensive granal stacks of mesophyll thylakoid membrane are enriched in photosystem II complexes, and assembly of these complex structures apparently requires a minimal amount of 36:5 MDGD and/or 36:5 DGDG. Formation of grana is not the only membrane process that requires 36:5 GDG, however, because loss of integrity in the internal membranes of bundle sheath chloroplasts, as well as mesophyll plastids, was evident in o5- seedlings. Mesphophyll and bundle sheath chloroplasts in C4 plants such as maize exhibit discrete structural features and carry out different biochemical functions. Bundle sheath chloroplasts lack grana and are specialized for carbon fixation in the stroma by the Calvin cycle (reviewed in Waters and Langdale, 2009). Thus, there appears to be a generalized requirement for 36:5 lipids in thylakoid membrane biogenesis that is not restricted to any particular aspect of photosynthesis.
A possible explanation for the function of 36:5 GDG in chloroplasts is that they have a structural organization that differs from that of 36:6 GDG, and lipids with these different properties are necessary for proper thylakoid structure and/or assembly of protein complexes therein. Although most membrane lipids are organized in a bilayer structure, MGDG typically assumes a nonbilayer, reverse hexagonal phase organization (HII) (Cullis and de Kruijff, 1979). Such nonbilayer structures are thought to be particularly suited for tight turns in membranes and/or for packing of membrane around densely populated protein complexes (Simidjiev et al., 2000; Dörmann and Benning, 2002; Hölzl and Dörmann, 2007). Previous research showed that the extent of desaturation affects the propensity of MGDG to adopt the HII structure, such that fewer double bonds favor bilayer formation. Notably, changes from five or more double bonds to less than five can significantly affect the bilayer/HII distribution (Gounaris et al., 1983). Thus, the effects of o5- mutations on 36:5 GDG abundance may have changed the abundance and/or location of bilayer and nonbilayer structures within the thylakoids. This in turn could explain the disorganized membranes, loss of granal stacks, and reduced chlorophyll in o5- leaves.
Galactolipid Function in Amyloplasts
The results of this study provide strong support for the conclusion that a functional connection exists between starch biosynthesis and the structure of internal amyloplast membranes. In contrast with leaves, total GDG content was strongly reduced in endosperm of o5-Ref homozygotes, to ~25% of normal, and all species of MGDG and DGDG were significantly decreased. Two explanations for the quantitatively different effects between leaf and endosperm can be considered. First, MGD2 or MGD3 activity may compensate for loss of MGD1 in leaf, but not in endosperm. This is unlikely, however, because there are no differences in MGD2 or MGD3 mRNA levels between endosperm and leaf in the wild type or o5- mutants and because, as noted previously, o5-Ref gene dosage clearly determines significant components of the leaf lipid profile. A second explanation could again be that the mutant MGD1 protein differentially recognizes DAG substrates based on degree of desaturation. In contrast with the leaf, endosperm contains virtually no 36:6 GDG species, whereas 36:4 GDG is predominant. If MGD1-G430C is less active against 36:5 DAG than 36:6 DAG, as proposed for leaf, it might be even less active using 36:4 DAG as the substrate. This could explain why the decrease in total GDG is much more severe in endosperm than in leaf.
Reduced GDG content in endosperm membranes had a clear effect on starch, which is likely the reason for the opaque kernel phenotype. At mid-development, granules were smaller in o5-Ref and o5-PS3038 homozygous endosperm than in the wild type, suggesting a delay in initiation and/or slower growth rate. Irregular growth ring spacing was observed in mutants, which could reflect altered granule growth processes or may be a consequence of distorted granule shape. Compound granules were frequent in mature mutant endosperm. In the null mutant, almost every granule was compound, and in some instances, there appeared to be 10 or more separate initiation events within the compound granules. Distinct initiation events, as opposed to fractured granules, were demonstrated by TEM observation of separate sets of concentric growth rings.
Little is known about initiation of starch granules in plants in general and endosperm in particular. One clue comes from observations that granule initiation in maize, oat (Avena sativa), or barley (Hordeum vulgare) endosperm occurs in plastids containing internal thylakoids, possibly within pockets of such membranes (Buttrose, 1960; Badenhuizen, 1969). Based on such ultrastructural data, association with thylakoid-like membranes within amyloplasts has been proposed as a functional aspect of starch polymer synthesis, granule formation, and growth (Salema and Badenhuizen, 1967; Barlow and Sargent, 1978). Direct evidence addressing this hypothesis is lacking; however, this study provides some support by showing a drastic switch in granule complexity when membrane lipids are altered. In normal maize, granules are first observed in endosperm within elongated, irregularly shaped amyloplasts containing multiple starch particles. Granules grow as development proceeds, and during this period, the amyloplasts divide such that each starch particle is surrounded individually by a double membrane (Badenhuizen, 1969; Shannon et al., 2009). The switch to compound granules in endosperm reduced in GDG content could arise from changes in the amyloplast division process such that membranes are unable to completely separate the growing starch particles. In this instance, outer envelope membrane would surround multiple starch grains, separated from each other by internal plastid membrane. Compound granules are the norm in rice endosperm, where internal membrane is thought to form separate compartments within a single amyloplast, each containing an individual starch particle (Yun and Kawagoe, 2010). Growth of the particles results in a compound granule within each plastid, similar to those observed here in o5- mutants.
Another interpretation is that abnormal thylakoid membrane structure could affect the temporal or spatial details of granule initiation during the early stages of amyloplast development, so that later separation into individual simple granules does not occur normally. Internal membrane structure may also influence or organize the coordinated activities of starch biosynthetic enzymes. This hypothesis is proposed as an explanation of the alteration in amylopectin fine structure observed to result from decreased GDG content (Figure 8). Amylopectin structure is determined by the combined activities of starch synthases, starch branching enzymes, and starch debranching enzymes (Myers et al., 2000; Ball and Morell, 2003; Zeeman et al., 2007). The functions of one or more of these enzymes could be affected by the structure of amyloplast thylakoids, thus resulting in alterations in granule gross morphology, initiation, growth rate, and/or fine structure. Abnormal assembly of ISA complexes in o5-Ref endosperm is consistent with this hypothesis. The specific connection between GDG content, amyloplast thylakoid structure, and starch biosynthesis currently is not known, although the data of this study indicate clearly that such a connection does exist.
The possibility must be considered that lipid composition indirectly affects starch granule formation. For example, o5- mutations may impact the plastid import systems, and disrupted protein or metabolite transport could secondarily cause defects in granule production. This explanation is unlikely, however, because amyloplasts in o5-Ref mutants appear to be fully functional in terms of starch level, and mature kernels are viable without germination defects. Altered protein import into amyloplasts would have global effects on metabolism, including starch biosynthesis, and significant changes in metabolite transfer would have similarly broad effects, including decreased total starch level. Indirect effects of MGD1 deficiency owing to changes in gene expression should also be considered. As with all opaque mutants studied, numerous changes in gene expression at the level of mRNA accumulation were found in o5-Ref endosperm (Hunter et al., 2002). Several genes that could affect starch biosynthesis either directly or indirectly are included in the set of ~90 genes that were observed to have at least a twofold difference in mRNA level. Altered gene expression could result from signals derived from galacto- or phospholipid levels changed as a result of MGD1 deficiency and/or from altered metabolite levels.
Galactolipid Function in Embryo and Endosperm Development
Plastids are known to be crucial for various developmental processes, and nearly one-third of known Arabidopsis embryo-defective mutants affect plastid genes (Hsu et al., 2010; Inaba and Ito-Inaba, 2010). The phenotypes of o5- plants bear striking parallels to maize etched1 (et1) mutants. Like o5, weak et1 alleles cause virescent seedlings that are initially pale but then become normal dark green (Sangeetha and Reddy, 1991; Scanlon et al., 1994; da Costa e Silva et al., 2004). Strong alleles cause defective endosperm development and embryo arrest, whereas heteroallelic combinations cause albino seedlings that die soon after germination. A striking aspect of et1 endosperm defects is sectors of cells devoid of starch. Like o5, the primary defects of et1 mutants appear to affect plastid function. The et1 locus encodes a zinc ribbon protein that is targeted to plastids and appears to function in transcription (da Costa e Silva et al., 2004). Thus, functionally deficient plastids, impaired through distinct mechanisms, lead to highly similar plant and kernel phenotypes in o5- and et1- mutants.
Photosynthesis is thought to be critical for embryo development in many species, particularly dicots where embryos become green for much of seed development (Tschiersch et al., 2011). The physiological benefit of embryo photosynthesis is unclear, but one hypothesis is that the oxygen produced is critical to support respiration and maintain appropriate redox status in the hypoxic cellular environment of developing seeds. The ability to rescue Arabidopsis mgd1-2 null mutant embryos with exogenous Suc would suggest that photosynthetic dysfunction is primarily responsible for the embryo lethality (Kobayashi et al., 2007). However, normal embryo development and seed germination in nonphotosynthetic albino mutants of Arabidopsis and maize (Prikryl et al., 2008; Liu et al., 2010) would suggest this explanation is not adequate. Furthermore, impaired photosynthesis seems an implausible explanation for the defects in o5- maize endosperm development. Cereal grains have a single layer of photosynthetic collenchyma cells in the pericarp, but the endosperm does not contain chlorophyll (Tschiersch et al., 2011). As such, on segregating ears, the homozygous o5-PS3038 mutant endosperms are surrounded by photosynthetically competent, genetically heterozygous maternal pericarp tissue, yet display defects. Therefore, a disruption of critical nonphotosynthetic plastid functions must be responsible for the mutant endosperm phenotype in maize.
As mentioned, assembly and function of several photosynthetic protein complexes require specific galactolipid interactions (Liu et al., 2004; Sakurai et al., 2006; Hölzl and Dörmann, 2007; Jones, 2007; Loll et al., 2007; Domonkos et al., 2008; Zhou et al., 2009). As such, it is likely that membrane composition could also affect plastid membrane protein complexes that function in processes other than photosynthesis. Plastids are centers for many metabolic processes, notably fatty acid biosynthesis (Joyard et al., 2010) and carotenoid biosynthesis (Lopez et al., 2008), which also involve integral membrane protein complexes. Several key hormones, including gibberellins (Yamaguchi, 2008), abscisic acid (Nambara and Marion-Poll, 2005), brassinosteroids (Boutté and Grebe, 2009), and cytokinins (Sakakibara, 2006), derive from plastidic isoprenoid metabolism. Perturbations in this system could readily explain the pleiotropic effects of mgd1 mutations on development as well as the observed carotenoid deficiency. Other integral membrane protein complexes include the plastid protein import machinery (Kessler and Schnell, 2009). Thus, membrane composition could potentially affect multiple critical protein complexes involved in plastid processes that are required for cellular function and plant viability. The disruption of such other functions likely results in embryo lethality of mutants entirely lacking MGD1 such as o5-PS3038.
METHODS
Plant Materials
The mutations o5-PS3038, o5-5288, and o5-313328 were isolated independently from self-pollinated ears of active Mutator lines in nontargeted mutagenesis experiments. The mutations were backcrossed into the B73 and/or W64A genetic backgrounds for at least three generations, and o5-Ref was backcrossed into the same inbreds for more than five generations. Nonviable mutations were maintained in the heterozygous state, and such plants were identified as backcross parents by appearance of ~25% opaque kernels on self-pollinated ears. Plants carrying o5-Ref were maintained in the homozygous state after backcrossing.
Kernels were collected from self-pollinated o5-Ref/+ or o5-PS3038/+ ears in the B73 genetic background segregating 25% homozygous mutant kernels. Controls were wild-type sibling kernels from the same segregating ears, with genotypes of either o5-/+ or +/+. Kernels from such ears were harvested at 15 or 20 DAP and used immediately for light microscopy analysis of endosperm and embryo tissue. The same tissues were used for comparison of MGD1 transcript levels between wild-type and o5-PS3038/o5-PS3038 mutant kernels. Mature kernels were used for TEM analysis.
Developing kernels used for analysis of enzymes, starch structures, transcript abundance, or polar lipid content were harvested from field-grown plants at 20 DAP, immediately frozen in liquid N2, and stored at −80° until use. Where indicated, endosperm and embryo tissue was separated by dissection after thawing kernels on ice. Greenhouse-grown seedlings provided leaf, root, and shoot tissue for analysis of transcript levels and leaf tissue for polar lipid quantification.
Phylogenetic Analysis
Related sequences were identified in the NCBI protein database by performing a BLASTp search (Camacho et al., 2009). Selected sequences were aligned using ClustalW (Larkin et al., 2007), followed by manual editing, and the output alignment is shown in Supplemental Figure 1 online. Conserved residues in the output alignment file were shaded using the Boxshade program (http://www.ch.embnet.org/software/BOX_form.html). The phylogenetic tree was produced with MEGA4 using default parameters of the unweighted pair group method with arithmetic mean (UPGMA) algorithm, although neighbor joining gave an identical topology (Tamura et al., 2007). GenBank protein accessions used in the analysis are as follows: Zm MGD1 (the product of o5), NP_001142118; Os MGD1, Q69QJ7; At MGD1, NP_194906; Pp MGD1, XP_001758690; Pp MGD2, XP_001755870; Pt MGD, XP_002181685; Os MGD2, Q6UTZ2; Zm MGD3, NP_001170057; Zm MGD2, NP_001147778; Os MGD3, Q0DWQ1; At MGD3, NP_565352; and At MGD2, NP_568394.
RT-PCR
Plant tissues were frozen in liquid nitrogen and ground to a powder in a mortar and pestle. Total RNA was extracted with RNeasy plant mini kit (Qiagen). DNase-treated total RNA (3 μg) was reverse transcribed in a 20-μL reaction with Superscript III (Invitrogen) using oligo(dT) primers. Genomic DNA contamination was checked by PCR using ubiquitin primers on DNase-treated RNA as a template. The cDNA synthesis reaction (1 μL) was used as template in PCR reactions with gene-specific primers. Ubiquitin primers or ISA2 primers used in control reactions were previously described (Lee et al., 2009; Kubo et al., 2010). Primer sequences for amplification of MGD1 mRNA were as follows: OP5-F, 5′-ATGCCACGAGACTGACCTCT-3′; OP5-R, 5′-TGAGGCTGAT-GGACTACACG-3′; o5-1, 5′-GATGGAAGAATGTATGGGTGCTTG-3′; o5-2, 5′-TGGGCCAAACCAGTCGGCAACT-3′; o5-3, 5′-GGAGTTCGGCGACGACTACCAG-3′; o5-4, 5′-GTGACAAGCTTATGGAACCATGTTGG-3′; o5-5, 5′-GAAGGTGCTGATCCTCATGA-3′; o5-6, 5′-GGAGTAAAACAATATCTACGATGAGTTGC-3′; o5-7, 5′-GACCGAAGGATGAGCTGCGAAGAG-3′. Primers for amplification of MGD2 mRNA were as follows: MGD2-F, 5′-CATTCCACAGCACCTCGCCTA-3′; MGD2-R, 5′-CCACGACATATGGGACGTTC-3′. Primers for amplification of MGD3 mRNA were as follows: MGD3-F, 5′-CCTACTTGTACGCCAACGA-3′; MGD3-R, 5′-CCACGACATATGGGACGTTC-3′.
PCR conditions using primers o5-1 to o5-7 were 94°C for 4 min, followed by 31 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 60 s, and a final extension of 72° for 10 min. PCR conditions using primers OP5-F and OP5-R were 95°C for 2 min, followed by 30 cycles of 95°C for 30 s, 51°C for 30 s, 72°C for 60 s, and a final extension of 72°C for 5 min. Amplification of MGD2 mRNA used the same protocol as for the OP5-F/OP5-R pair except that the annealing temperature was 60°C and 35 cycles were performed. MGD3 mRNA was amplified using the same protocol as for the OP5-F/OP5-R pair except that the annealing temperature was 63°C. In all instances, the number of cycles was less than that necessary to reach a saturating level of signal observed as intensity of DNA bands after staining with ethidium bromide.
Lipid Analysis
Lipids were extracted from maize (Zea mays) seedling leaves or developing endosperm following the protocol specified by the Kansas Lipidomics Research Center for Arabidopsis leaf extractions (http://www.k-state.edu/lipid/ lipidomics/leaf-extraction.html). The abundance of specific lipids in the extracts was determined by electrospray ionization-quadrupole tandem mass spectrometry according to lipid profiling protocols established by the facility (http://www.k-state.edu/lipid/lipidomics/profiling.htm). These methods detect phosphatidylcholine, lysophosphatidylcholine, phosphatidylserine, phosphatidylethanolamine, lysophosphatidyl-ethanolamine, MGDG, DGDG, phosphatidylglycerol, phosphatidylinositol, and phosphatidic acid. Within each class defined by the head group, mass data specifically identify the total number of carbons and the total number of double bonds present in the two acyl groups combined.
Analyses of Carbohydrates and Starch Modifying Enzymes
Methods for determination of starch quantity in mature and developing endosperm tissue, starch extraction, determination of the linear chain length distribution within amylopectin by fluorescence-assisted carbohydrate electrophoresis, and determination of amylose content by Sepharose CL-2B chromatography have been previously described (Kubo et al., 2010 and references therein). Detection of ISA enzyme activities and other starch modifying enzyme activities by zymogram analysis was performed as previously described (Kubo et al., 2010).
Microscopy
Specimens for visualization of embryo development were dissected from a self-pollinated o5-PS3038/+ ear at 15 DAP. Kernels were fixed in formalin–acetic acid–alcohol and embedded in paraffin (Berlyn and Miksche, 1976). Microtome sections of 10-μm thickness were affixed to glass microscope slides, deparaffinized in xylene, stained with Safranin O and fast green, and mounted in Permount. Leaf and endosperm specimens for histological analyses were fixed in 2% paraformaldehyde and 3% gluteraldehyde buffered in 0.1 M cacodylate, pH 7.2. For light microscopy analyses of endosperm, samples were embedded in LR White resin and sectioned to 1 μm on a Leica EM UC6 ultramicrotome. Sections were stained with Periodic Acid Schiff’s and methylene blue. For light microscopy analyses of leaf tissue, specimens were embedded in Spurr's resin, sectioned to 1 μm, and stained with toluidine blue. Samples for TEM were also embedded in Spurr's resin and then postfixed with 1% osmium tetroxide, sectioned to 80 nm, affixed to grids, and stained with uranyl acetate and lead citrate. Histological specimens were photographed with an Olympus BX-60 microscope and a Jenoptik C-5 camera system. TEM examination was performed with a JEOL 2100 200 kV scanning and transmission electron microscope.
Accession Numbers
The GenBank/EMBL database accession number of the EST sequence used as a reference to identify the o5 gene is EE189213. The reference for the cDNA sequence used to identify the position of specific mutations is accession number BT042676, and a cDNA sequence originally annotating the o5 gene product is EU954700. The maize ubiquitin and ISA2 mRNA sequences used as controls for RT-PCR analyses are specified by accession numbers EU959403 and AY172633, respectively.
AUTHOR CONTRIBUTIONS
A.M.M., M.G.J., T.A.H.-B., and P.W.B. designed the research, performed research, analyzed the data, and wrote the article. Q.L. and G.Y. performed research. P.S.S. designed and performed the research.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of the Maize MGD Amino Acid Sequences with MGD Proteins from Other Species.
Supplemental Figure 2. Wild Type and Mutant Bundle Sheath Chloroplasts Visualized by TEM.
Supplemental Table 1. Allele and Phenotype Summary.
Supplemental Data Set 1. Lipid Profiling Raw Data.
Supplemental Data Set 2. Text File of Alignment of the MGD Amino Acid Sequences.
Supplementary Material
Acknowledgments
We acknowledge the assistance of the Iowa State University William Keck Metabolomics Facility for analysis of starch chain length distributions and the Iowa State University Microscopy and Nanoimaging Facility for assistance with microscopy. Harry T. Horner provided helpful discussions interpreting TEM images. We also acknowledge the assistance of the Kansas Lipodomics Research Center, Kansas State University, for lipid analyses.
References
- Andorf C.M., Lawrence C.J., Harper L.C., Schaeffer M.L., Campbell D.A., Sen T.Z. (2010). The Locus Lookup tool at MaizeGDB: Identification of genomic regions in maize by integrating sequence information with physical and genetic maps. Bioinformatics 26: 434–436 [DOI] [PubMed] [Google Scholar]
- Awai K., Maréchal E., Block M.A., Brun D., Masuda T., Shimada H., Takamiya K., Ohta H., Joyard J. (2001). Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98: 10960–10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhuizen N.P. (1969). The Biogenesis of Starch Granules in Higher Plants. (New York: Appleton-Century-Crofts; ). [Google Scholar]
- Ball S.G., Morell M.K. (2003). From bacterial glycogen to starch: Understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54: 207–233 [DOI] [PubMed] [Google Scholar]
- Barkan A., Martienssen R.A. (1991). Inactivation of maize transposon Mu suppresses a mutant phenotype by activating an outward-reading promoter near the end of Mu1. Proc. Natl. Acad. Sci. USA 88: 3502–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow P.W., Sargent J.A. (1978). The ultrastructure of the regenerating root cap of Zea mays L. Ann. Bot. (Lond.) 42: 791–799 [Google Scholar]
- Benning C. (2008). A role for lipid trafficking in chloroplast biogenesis. Prog. Lipid Res. 47: 381–389 [DOI] [PubMed] [Google Scholar]
- Benning C., Ohta H. (2005). Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J. Biol. Chem. 280: 2397–2400 [DOI] [PubMed] [Google Scholar]
- Berlyn G.P., Miksche J.P. (1976). Botanical Microtechnique and Cytochemistry. (Ames, IA: Iowa State University Press: ). [Google Scholar]
- Block M.A., Dorne A.J., Joyard J., Douce R. (1983). Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. II. Biochemical characterization. J. Biol. Chem. 258: 13281–13286 [PubMed] [Google Scholar]
- Botté C., Jeanneau C., Snajdrova L., Bastien O., Imberty A., Breton C., Maréchal E. (2005). Molecular modeling and site-directed mutagenesis of plant chloroplast monogalactosyldiacylglycerol synthase reveal critical residues for activity. J. Biol. Chem. 280: 34691–34701 [DOI] [PubMed] [Google Scholar]
- Boutté Y., Grebe M. (2009). Cellular processes relying on sterol function in plants. Curr. Opin. Plant Biol. 12: 705–713 [DOI] [PubMed] [Google Scholar]
- Bruce B.D. (1998). The role of lipids in plastid protein transport. Plant Mol. Biol. 38: 223–246 [PubMed] [Google Scholar]
- Buléon A., Colonna P., Planchot V., Ball S. (1998). Starch granules: Structure and biosynthesis. Int. J. Biol. Macromol. 23: 85–112 [DOI] [PubMed] [Google Scholar]
- Buttrose M.S. (1960). Submicroscopic development and structure of starch granules in cereal endosperms. J. Ultrastruct. Res. 4: 231–257 [DOI] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. (2009). BLAST+: Architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009). The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 37(Database issue): D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe E.H., Jr, Neuffer M.G., Hoisington D.A. (1988). The genetics of corn. In Corn and Corn Improvement, Sprague G.F., Dudley J.W., (Madison, WI: American Society of Agronomy; Crop Science Society of America; Soil Science Society of America; ), pp. 81–258 [Google Scholar]
- Cullis P.R., de Kruijff B. (1979). Lipid polymorphism and the functional roles of lipids in biological membranes. Biochim. Biophys. Acta 559: 399–420 [DOI] [PubMed] [Google Scholar]
- da Costa e Silva O., Lorbiecke R., Garg P., Müller L., Wassmann M., Lauert P., Scanlon M., Hsia A.P., Schnable P.S., Krupinska K., Wienand U. (2004). The Etched1 gene of Zea mays (L.) encodes a zinc ribbon protein that belongs to the transcriptionally active chromosome (TAC) of plastids and is similar to the transcription factor TFIIS. Plant J. 38: 923–939 [DOI] [PubMed] [Google Scholar]
- Dinges J.R., Colleoni C., Myers A.M., James M.G. (2001). Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol. 125: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domonkos I., Laczkó-Dobos H., Gombos Z. (2008). Lipid-assisted protein-protein interactions that support photosynthetic and other cellular activities. Prog. Lipid Res. 47: 422–435 [DOI] [PubMed] [Google Scholar]
- Dörmann P., Benning C. (2002). Galactolipids rule in seed plants. Trends Plant Sci. 7: 112–118 [DOI] [PubMed] [Google Scholar]
- Dörmann P., Hoffmann-Benning S., Balbo I., Benning C. (1995). Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7: 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Finn R.D., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38(Database issue): D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishwick M.J., Wright A.J. (1980). Isolation and characterization of amyloplast envelope membranes from Solanum tuberosum. Phytochemistry 19: 55–59 [Google Scholar]
- Gounaris K., Mannock D.A., Sen A., Brain A.P.R., Williams W.P., Quinn P.J. (1983). Polyunsaturated fatty acyl residues of galactolipids are involved in the control of bilayer/non-bilayer transitions in higher plant chloroplasts. Biochim. Biophys. Acta 732: 229–242 [Google Scholar]
- Heinz E. (1977). Enzymatic reactions in galactolipid biosynthesis. In Lipids and Lipid Polymers in Higher Plants, Tevini M., Lichtenthaler H.K., (Berlin, Heidelberg, and New York: Springer-Verlag; ), pp. 102–120 [Google Scholar]
- Holding D.A., Larkins B.A. (2009). Zein storage proteins. In Molecular Genetic Approaches to Maize Improvement, Kriz A.L., Larkins B.A., (Berlin: Springer-Verlag; ), pp. 269–286 [Google Scholar]
- Holding D.R., Otegui M.S., Li B., Meeley R.B., Dam T., Hunter B.G., Jung R., Larkins B.A. (2007). The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell 19: 2569–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölzl G., Dörmann P. (2007). Structure and function of glycoglycerolipids in plants and bacteria. Prog. Lipid Res. 46: 225–243 [DOI] [PubMed] [Google Scholar]
- Hsu S.C., Belmonte M.F., Harada J.J., Inoue K. (2010). Indispensable roles of plastids in Arabidopsis thaliana embryogenesis. Curr. Genomics 11: 338–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter B.G., Beatty M.K., Singletary G.W., Hamaker B.R., Dilkes B.P., Larkins B.A., Jung R. (2002). Maize opaque endosperm mutations create extensive changes in patterns of gene expression. Plant Cell 14: 2591–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T., Ito-Inaba Y. (2010). Versatile roles of plastids in plant growth and development. Plant Cell Physiol. 51: 1847–1853 [DOI] [PubMed] [Google Scholar]
- Jarvis P., Dörmann P., Peto C.A., Lutes J., Benning C., Chory J. (2000). Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc. Natl. Acad. Sci. USA 97: 8175–8179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.R. (2007). Lipids in photosynthetic reaction centres: Structural roles and functional holes. Prog. Lipid Res. 46: 56–87 [DOI] [PubMed] [Google Scholar]
- Jordan P., Fromme P., Witt H.T., Klukas O., Saenger W., Krauss N. (2001). Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411: 909–917 [DOI] [PubMed] [Google Scholar]
- Joyard J., Douce R. (1987). Galactolipid biosynthesis. In The Biochemistry of Plants: Lipids: Structure and Function, Stumpf P.K., (New York: Academic Press; ), pp. 215–274 [Google Scholar]
- Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. (2010). Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism. Prog. Lipid Res. 49: 128–158 [DOI] [PubMed] [Google Scholar]
- Kessler F., Schnell D. (2009). Chloroplast biogenesis: Diversity and regulation of the protein import apparatus. Curr. Opin. Cell Biol. 21: 494–500 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Awai K., Takamiya K., Ohta H. (2004). Arabidopsis type B monogalactosyldiacylglycerol synthase genes are expressed during pollen tube growth and induced by phosphate starvation. Plant Physiol. 134: 640–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Kondo M., Fukuda H., Nishimura M., Ohta H. (2007). Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc. Natl. Acad. Sci. USA 104: 17216–17221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodrzycki R., Boston R.S., Larkins B.A. (1989). The opaque-2 mutation of maize differentially reduces zein gene transcription. Plant Cell 1: 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Colleoni C., Dinges J.R., Lin Q., Lappe R.R., Rivenbark J.G., Meyer A.J., Ball S.G., James M.G., Hennen-Bierwagen T.A., Myers A.M. (2010). Functions of heteromeric and homomeric isoamylase-type starch-debranching enzymes in developing maize endosperm. Plant Physiol. 153: 956–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lairson L.L., Henrissat B., Davies G.J., Withers S.G. (2008). Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 77: 521–555 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lee A.G. (2000). Membrane lipids: It’s only a phase. Curr. Biol. 10: R377–R380 [DOI] [PubMed] [Google Scholar]
- Lee B.H., Johnston R., Yang Y., Gallavotti A., Kojima M., Travençolo B.A.N., Costa Lda.F., Sakakibara H., Jackson D. (2009). Studies of aberrant phyllotaxy1 mutants of maize indicate complex interactions between auxin and cytokinin signaling in the shoot apical meristem. Plant Physiol. 150: 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Gong Q., Ma Y., Li P., Li J., Yang S., Yuan L., Yu Y., Pan D., Xu F., Wang N.N. (2010). cpSecA, a thylakoid protein translocase subunit, is essential for photosynthetic development in Arabidopsis. J. Exp. Bot. 61: 1655–1669 [DOI] [PubMed] [Google Scholar]
- Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. (2004). Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428: 287–292 [DOI] [PubMed] [Google Scholar]
- Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2007). Lipids in photosystem II: Interactions with protein and cofactors. Biochim. Biophys. Acta 1767: 509–519 [DOI] [PubMed] [Google Scholar]
- Lopez A.B., Yang Y., Thannhauser T.W., Li L. (2008). Phytoene desaturase is present in a large protein complex in the plastid membrane. Physiol. Plant. 133: 190–198 [DOI] [PubMed] [Google Scholar]
- Maréchal E., Awai K., Block M.A., Brun D., Masuda T., Shimada H., Takamiya K., Ohta H., Joyard J. (2000). The multigenic family of monogalactosyl diacylglycerol synthases. Biochem. Soc. Trans. 28: 732–738 [PubMed] [Google Scholar]
- Mongrand S., Bessoule J.-J., Cabantous F., Cassagne C. (1998). The C16:3/C18:3 fatty acid balance in photosynthetic tissues from 468 plant species. Phytochemistry 49: 1049–1064 [Google Scholar]
- Morell M.K., Samuel M.S., O’Shea M.G. (1998). Analysis of starch structure using fluorophore-assisted carbohydrate electrophoresis. Electrophoresis 19: 2603–2611 [DOI] [PubMed] [Google Scholar]
- Myers A.M., Morell M.K., James M.G., Ball S.G. (2000). Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 122: 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56: 165–185 [DOI] [PubMed] [Google Scholar]
- O'Shea M.G., Samuel M.S., Konik C.M., Morell M.K. (1998). Fluorophore-assisted carbohydrate electrophoresis (FACE) of oligosaccharides: efficiency of labelling and high-resolution separation. Carbohydr. Res. 307: 1–12 [Google Scholar]
- Prikryl J., Watkins K.P., Friso G., van Wijk K.J., Barkan A. (2008). A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 36: 5152–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. (2006). Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57: 431–449 [DOI] [PubMed] [Google Scholar]
- Sakurai I., Shen J.-R., Leng J., Ohashi S., Kobayashi M., Wada H. (2006). Lipids in oxygen-evolving photosystem II complexes of cyanobacteria and higher plants. J. Biochem. 140: 201–209 [DOI] [PubMed] [Google Scholar]
- Salema R., Badenhuizen N.P. (1967). The production of reserve starch granules in the amyloplasts of Pellionia daveauana N. E. Br. J. Ultrastruct. Res. 20: 383–399 [DOI] [PubMed] [Google Scholar]
- Sangeetha H.G., Reddy A.R. (1991). Genetic and biochemical analysis of the Etched (et) mutant of Zea mays L. Maydica 36: 343–354 [Google Scholar]
- Scanlon M.J., James M.G., Stinard P.S., Myers A.M., Robertson D.S. (1994). Characterization of ten new mutations of the maize Etched-1 locus. Maydica 39: 301–308 [Google Scholar]
- Schmidt R.J., Burr F.A., Aukerman M.J., Burr B. (1990). Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc. Natl. Acad. Sci. USA 87: 46–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon J.C., Garwood D.L., Boyer C.D. (2009). Genetics and physiology of starch development. In Starch: Chemistry and Technology, Bemiller J.N., Whistler R.L., (London: Academic Press; ), pp. 23–82 [Google Scholar]
- Shimojima M., Ohta H., Iwamatsu A., Masuda T., Shioi Y., Takamiya K. (1997). Cloning of the gene for monogalactosyldiacylglycerol synthase and its evolutionary origin. Proc. Natl. Acad. Sci. USA 94: 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simidjiev I., Stoylova S., Amenitsch H., Jávorfi T., Mustárdy L., Laggner P., Holzenburg A., Garab G. (2000). Self-assembly of large, ordered lamellae from non-bilayer lipids and integral membrane proteins in vitro. Proc. Natl. Acad. Sci. USA 97: 1473–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thompson G., Larkins B.A. (1994). Characterization of zein genes and their regulation in maize endosperms. In The Maize Handbook, Freeling M., Walbot V., (Berlin: Springer-Verlag; ), pp. 639–646 [Google Scholar]
- Tschiersch H., Borisjuk L., Rutten T., Rolletschek H. (2011). Gradients of seed photosynthesis and its role for oxygen balancing. Biosystems 103: 302–308 [DOI] [PubMed] [Google Scholar]
- Utsumi Y., Nakamura Y. (2006). Structural and enzymatic characterization of the isoamylase1 homo-oligomer and the isoamylase1-isoamylase2 hetero-oligomer from rice endosperm. Planta 225: 75–87 [DOI] [PubMed] [Google Scholar]
- Waters M.T., Langdale J.A. (2009). The making of a chloroplast. EMBO J. 28: 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb M., Green B. (1991). Biochemical and biophysical properties of thylakoid acyl lipids. Biochim. Biophys. Acta 1060: 133–158 [Google Scholar]
- Welti R., Wang X. (2004). Lipid species profiling: a high-throughput approach to identify lipid compositional changes and determine the function of genes involved in lipid metabolism and signaling. Curr. Opin. Plant Biol. 7: 337–344 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yi G., Luth D., Goodman T.D., Lawrence C.J., Becraft P.W. (2009). High-throughput linkage analysis of Mutator insertion sites in maize. Plant J. 58: 883–892 [DOI] [PubMed] [Google Scholar]
- Yun M.-S., Kawagoe Y. (2010). Septum formation in amyloplasts produces compound granules in the rice endosperm and is regulated by plastid division proteins. Plant Cell Physiol. 51: 1469–1479 [DOI] [PubMed] [Google Scholar]
- Zeeman S.C., Smith S.M., Smith A.M. (2007). The diurnal metabolism of leaf starch. Biochem. J. 401: 13–28 [DOI] [PubMed] [Google Scholar]
- Zhou F., Liu S., Hu Z., Kuang T., Paulsen H., Yang C. (2009). Effect of monogalactosyldiacylglycerol on the interaction between photosystem II core complex and its antenna complexes in liposomes of thylakoid lipids. Photosynth. Res. 99: 185–193 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.