This work describes the development of a robust and quantitative system for elucidating the sequences and trans-acting factors required for de novo telomere formation in Arabidopsis thaliana. We determine that genetic redundancy and the nonhomologous end-joining factors Ku and Lig4 facilitate higher levels of de novo telomere formation.
Abstract
Conversion of a double-strand break into a telomere is a dangerous, potentially lethal event. However, little is known about the mechanism and control of de novo telomere formation (DNTF). DNTF can be instigated by the insertion of a telomere repeat array (TRA) into the host genome, which seeds the formation of a new telomere, resulting in chromosome truncation. Such events are rare and concentrated at chromosome ends. Here, we introduce tetraploid Arabidopsis thaliana as a robust genetic model for DNTF. Transformation of a 2.6-kb TRA into tetraploid plants resulted in a DNTF efficiency of 56%, fivefold higher than in diploid plants and 50-fold higher than in human cells. DNTF events were recovered across the entire genome, indicating that genetic redundancy facilitates recovery of DNTF events. Although TRAs as short as 100 bp seeded new telomeres, these tracts were unstable unless they were extended above a 1-kb size threshold. Unexpectedly, DNTF efficiency increased in plants lacking telomerase, and DNTF rates were lower in plants null for Ku70 or Lig4, components of the nonhomologous end-joining repair pathway. We conclude that multiple competing pathways modulate DNTF, and that tetraploid Arabidopsis will be a powerful model for elucidating the molecular details of these processes.
INTRODUCTION
The natural ends of chromosomes are distinguished from double-strand DNA breaks (DSBs) because they are packaged into telomeres, specialized nucleoprotein complexes assembled on a terminal array of short DNA repeats (TTTAGGG in Arabidopsis thaliana and most plants). The telomere repeat array (TRA) is mostly comprised of double-strand DNA but terminates in a short G-rich single-strand 3′ protrusion termed the G-overhang (Verdun and Karlseder, 2007). Telomere length is maintained in a dynamic range by opposing processes that shorten or extend the TRA (Shore and Bianchi, 2009). Incomplete DNA replication, nucleolytic degradation, and recombination events cause telomeres to shorten (Crabbe et al., 2004; Ferreira et al., 2004), whereas the telomerase extends the TRA (Collins, 2006). In vertebrates, the TRA is protected by a six-member protein complex termed shelterin (de Lange, 2005); however, in Arabidopsis and budding yeast (Saccharomyces cerevisiae), the predominant end protection complex is Cdc13/CTC1, Stn1, and Ten1 (Bertuch and Lundblad, 2006; Surovtseva et al., 2009). Loss of core telomere-capping components is highly deleterious, triggering DNA damage checkpoints, chromosome end-joining reactions, and widespread genome instability (Nugent et al., 1996; van Steensel et al., 1998; Baumann and Cech, 2001; Puglisi et al., 2008; Surovtseva et al., 2009).
Chromosomal DSBs are typically resolved by nonhomologous end-joining (NHEJ) and homologous recombination. They can also be subjected to “chromosome healing” in which telomerase adds telomere repeats at the break site to establish a new telomere (Pennaneach et al., 2006; Murnane, 2010). This latter process protects the nascent terminus from subsequent repair activities, but de novo telomere formation (DNTF) is perilous because it leads to deletion of the acentric distal chromosome fragment. Terminal deletions and DNTF are associated with several genetic disorders, including α-thalassemia and some forms of mental retardation (Wilkie et al., 1990; Flint et al., 1994), as well as cancer (Lee and Myung, 2009).
DNTF is best understood in budding yeast. In this setting, DNTF requires the telomere-capping protein Cdc13 and the telomerase accessory factor Est1, which collaborate in the recruitment and/or activation of telomerase following resection of the 5′ end of the TRA by the Mre11/Rad50/Xrs2 nuclease (Diede and Gottschling, 2001; Larrivée et al., 2004). In the absence of a TRA, telomerase is essential for the establishment of a new telomere (Pennaneach and Kolodner, 2004). However, if the break occurs adjacent to a TRA, telomerase is dispensable, and DNTF relies on double-strand telomere binding proteins, which presumably assist with the formation of a protective cap on the new telomere (Negrini et al., 2007). The KU70/80 heterodimer also plays an important, but enigmatic, role in yeast DNTF. Although Ku is an essential component of the NHEJ DSB repair pathway, it is also required to protect telomeres from end-joining reactions (DuBois et al., 2002). In addition, Ku binds the telomerase RNA subunit, and this association is essential for DNTF at sites lacking a TRA (Stellwagen et al., 2003).
DNTF has been studied in vertebrates (Barnett et al., 1993; Hanish et al., 1994; Bae and Baumann, 2007), but much less is known about the process. The DNTF assay involves transgenic introduction of a TRA, which results in chromosome truncation when the nontelomeric region of the construct integrates into an internal region of the chromosome and the TRA is recognized and established as a bona fide telomere. DNTF is supported by a TRA as short as 250 bp and requires the vertebrate telomere repeat sequence (TTAGGG) (Hanish et al., 1994; Okabe et al., 2000). DNTF is promoted by a double-strand telomeric DNA binding component of shelterin, TRF1. As in yeast, conversion of the TRA into a functional telomere does not require telomerase (Okabe et al., 2000; Gao et al., 2008). Finally, DNTF events are rare and are preferentially recovered near endogenous telomeres (Diede and Gottschling, 1999; Gao et al., 2008; Fortin et al., 2009), presumably reflecting the aneuploidy associated with chromosome truncation.
The ability of transgenic TRAs to acquire telomere function in plants was first demonstrated by Agrobacterium tumefaciens-mediated transformation of immature embryos from maize (Zea mays; Yu et al., 2006). In subsequent work, maize chromosomes were truncated using TRAs to create plant minichromosomes (Yu et al., 2007). Although the frequency of DNTF events was low, a whole-arm truncation was recovered in a spontaneous tetraploid event (Yu et al., 2007). This finding suggests that a major barrier to studying DNTF could be overcome by genetically buffering chromosome truncation.
Synthetic Arabidopsis tetraploids have served as models for understanding the effects of ploidy on plant development for some 40 years (Santos et al., 2003). Notably, large deletions that are lethal in the gametophytic generation of diploid Arabidopsis can be propagated in tetraploid (4X) plants (Vizir and Mulligan, 1999). Here, we introduce tetraploid Arabidopsis as a robust new model for DNTF. We observe a remarkably high rate of telomere truncation events, occurring throughout the entire Arabidopsis genome. In addition to testing the effects of cis-acting sequence requirements for DNTF, we exploit the genetic tractability of Arabidopsis to explore the contribution of telomerase and NHEJ components on DNTF. Unexpectedly, we find that although telomerase is required to sustain new telomeres over the long term, it is inhibitory to DNTF. By contrast, Ku and LIG4 are essential for efficient DNTF. These findings argue that multiple competing pathways influence the formation of new telomeres in higher plants.
RESULTS
Efficient Production and Recovery of DNTF Events in Tetraploid Arabidopsis
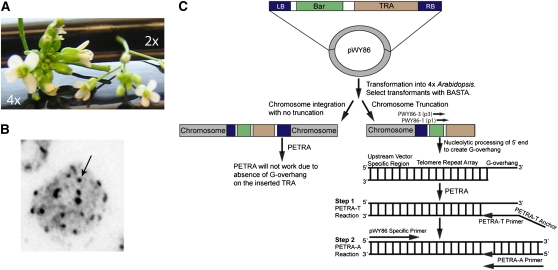
To investigate whether tetraploid Arabidopsis could serve as a model for DNTF, we generated tetraploids by treating the meristem of 7- to 14-d-old seedlings with a 0.1% colchicine solution (Henry et al., 2005). Potential tetraploids were initially screened by examining flower size (Figure 1A), which increases with ploidy (Henry et al., 2005). Plants grown from seeds of enlarged flowers were then subjected to a cytological screen, which revealed the presence of ~18 chromomeres (Figure 1B; Yu et al., 2006). Although it was not possible to definitively determine chromosome number in this assay, the data are consistent with tetraploidy.
Figure 1.
The Arabidopsis DNTF System.
(A) Increased flower size in tetraploid Arabidopsis. Flowers from tetraploid (4X) and diploid (2X) Arabidopsis are shown.
(B) Light microscopy of 4X nuclei. Ploidy was confirmed by counting chromomeres (arrow), the heterochromatic region near centromeres. In this image, 18 distinct chromomeres are visible.
(C) Schematic diagram depicting the pWY86 vector system and the PETRA assay. A T-DNA construct containing the BAR gene (for Basta selection), LB (left border), RB (right border), and a TRA in pWY86 is transformed into 4X Arabidopsis. The T-DNA can be fully integrated (left) or result in chromosome truncation and DNTF (right). Transformants are screened by PETRA using PWY86-1 (p1) or PWY86-3 (p3) and the PETRA-T primer, which will amplify products with an accessible G-overhang, reflecting DNTF.
A TRA-bearing construct previously used to seed DTNF in maize, pWY86 (Yu et al., 2006), was transformed into wild-type diploid (2X) and tetraploid (4X) Arabidopsis (Columbia ecotype; Figure 1C). For each construct, ~100 transgenic lines containing pWY86 (in some cases, from two independent transformation events) were selected for Basta (glufosinate) resistance and then screened for DNTF using a modification of the PCR assay, primer extension telomere rapid amplification (PETRA; Heacock et al., 2004; Figure 1C). In PETRA, the forward primer anneals to the G-overhang, a critical feature of a functional telomere, whereas the reverse primer binds a unique subtelomeric sequence. To detect DTNF, two PETRA reactions were performed with reverse primers that bind adjacent to each other 46 bp (PWY86-1) or 350 bp (PWY86-3) upstream of the TRA in the pWY86 construct (Figure 1C). Staggered products are generated when T-DNA insertion leads to DNTF rather than integration at an internal site in the chromosome (Figures 1C and 2A). The endogenous telomere on the left arm of chromosome 3 (3L) was monitored as a control.
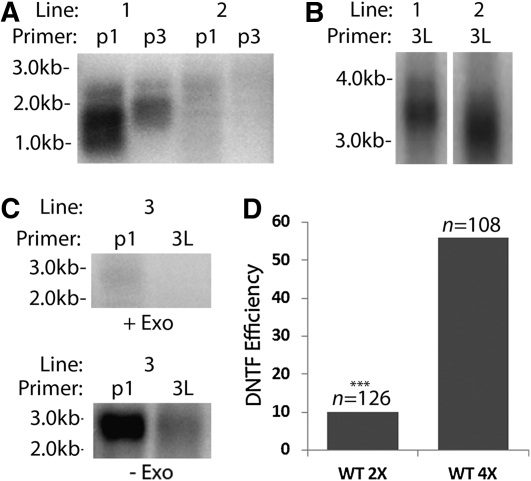
Figure 2.
Detection of DNTF Events by PETRA.
(A) PETRA results for 4X wild type carrying a 2.6-kb transgenic TRA. Two PETRA reactions with primers p1 or p3 (Figure 1C) were performed for each plant transformed with pWY86. Line 1 shows the staggered size products expected for a plant with DNTF. Line 2 shows nonspecific background bands. Molecular weight markers are indicated.
(B) PETRA results for the endogenous 3L telomere from two transformants.
(C) PETRA products are dependent on the presence of a 3′ overhang. PETRA was performed on transformants treated with T4 DNA polymerase, which has a strong 3′ to 5′ exonuclease activity (+exo), for 30 min (top) or untreated (−exo; bottom). Results using pWY86 primer (1) or a primer specific to the endogenous 3L telomere (e3L) are shown.
(D) Comparison of DNTF efficiency in 2X versus 4X Arabidopsis. Efficiency was calculated by dividing the number of DNTF events by the total number of lines screened (n). *** indicates a statistically significant difference (P > 0.0001) in the DNTF rate compared with 4X wild type transformed with a 2.6-kb TRA.
PETRA products were obtained from two independent transformation experiments using 4X wild-type Arabidopsis (T1 = 35 of 60 [58%], T2 = 26 of 48 [54%]; Figure 2D) for an average DNTF frequency of 56%. The congruence of these two data points and other results in duplicate transformation experiments (see Supplemental Table 1 online) demonstrated that the DNTF assay was reproducible. In contrast to results with 4X plants, only 10% of the transgenic events characterized in 2X plants led to DNTF (12 of 126). The elevated incidence of DNTF in 4X plants did not reflect an increased frequency of T-DNA insertion. DNA gel blot analysis using a probe against the BAR gene (Figure 1C) showed the same number of T-DNA insertions in 2X and 4X transformants (see Supplemental Figure 1B online).
Consistent with telomerase action at or near the chromosome break site, PETRA products were heterogeneous in length (Figure 2A), resembling those generated from the endogenous 3L telomere (Figure 2B). Furthermore, the nascent telomeres acquired a G-overhang because pretreatment of the reaction with T4 DNA polymerase, which harbors 3′ to 5′ exonuclease activity, blocked PETRA product synthesis (Figure 2C). Finally, DNA gel blot analysis of lines bearing potential truncation events confirmed DNTF (see Supplemental Figure 1 online).
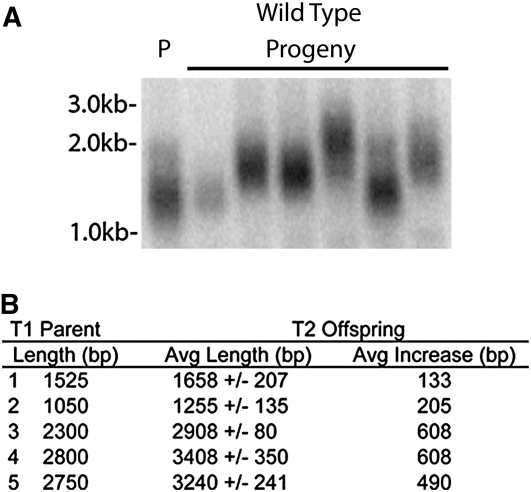
Telomeres in Arabidopsis (Columbia ecotype) typically span 2 to 5 kb (Shakirov and Shippen, 2004). To investigate whether new telomeres were subjected to the same length regulation as endogenous telomeres, PETRA was performed in the next plant generation (Figure 3A). Wild-type lines (4X) transformed with a 950-bp TRA showed on average a 650-bp increase in telomere length upon DNTF (Table 1). These telomeres were elongated further in the next generation (T2; Figures 3A and 3B). Telomeres generally increased in size, reaching up to 3.4 kb in T2 (Figure 3B). Conversely, new telomeres established using a 2.6-kb TRA seed, which falls within the wild-type range of endogenous Arabidopsis telomeres, did not increase in length (Table 1). Thus, the de novo telomeres formed in 4X Arabidopsis mimic endogenous telomeres in their architecture and length regulation.
Figure 3.
New Telomeres Function Like Native Telomeres.
(A) Parent–progeny analysis of new telomeres. PETRA results are shown for first- (T1) and second- (T2) generation 4X wild-type transformants carrying a 950-bp TRA. P denotes parent. Results for six progeny (T2) lines are shown.
(B) Compilation of PETRA product lengths in T1 4X wild-type parents and their T2 progeny transformed with the 950-bp TRA. Progeny from five parent lines were analyzed by PETRA. Six progeny were analyzed per parent.
Table 1.
Length of De Novo Telomeres Generated with Different TRAs
| TRAs Recovered |
||
| Initial TRA (bp) | Range (bp) | Average (bp) |
| 2600 | 950–4100 | 2562 ± 574 |
| 950 | 850–2925 | 1608 ± 359 |
| 800 | 850–3200 | 1458 ± 423 |
| 700 | 650–2950 | 1361 ± 334 |
| 400 | 825–1912 | 1436 ± 366 |
| 200 | 900–2900 | 1732 ± 243 |
| 100 | 650–1900 | 1142 ± 251 |
DNTF Events Occur at Random Sites in the Arabidopsis Genome
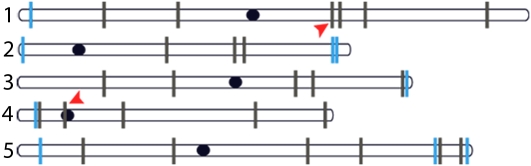
We mapped the sites of DNTF in 4X and 2X Arabidopsis using thermal asymmetric interlaced (TAIL)-PCR (Sessions et al., 2002). We randomly selected 10 diploid and 30 tetraploid DNTF lines and obtained PCR products for nine of 10 2X lines and 25 of 30 4X lines. In all the 2X lines, a new telomere formed close to the endogenous chromosome end (Figure 4, blue bars). By contrast, the 25 chromosome truncation events mapped in 4X plants were widely distributed throughout the entire genome (Figure 4, gray bars), consistent with random integration of T-DNA (Kim et al., 2007). Strikingly, several DNTF events resulted in large deletions, including the loss of 20 Mb from the right arm of chromosome 1 and a truncation event within the centromere-flanking region of chromosome 4 (Figure 4, arrowheads; Kumekawa et al., 2001). Whether this centromere truncation affects chromosome segregation is unknown. The elevated incidence of DNTF and recovery of large chromosomal deletions indicate that tetraploid Arabidopsis can be exploited as a robust model for DNTF.
Figure 4.
New Telomeres Form Throughout the Tetraploid Arabidopsis Genome.
Insertion sites for DNTF events with the 2.6-kb TRA were determined for 2X and 4X wild-type plants by TAIL-PCR. Insertions were mapped along the five nonhomologous chromosomes (2X, blue lines; 4X, black lines). Red arrowheads denote DNTF events resulting in a 20-Mb deletion and a centromere-adjacent telomere truncation. Dark ovals represent centromeres.
DNTF Requires a Properly Oriented TRA Containing 100 bp of TTTAGGG Repeats
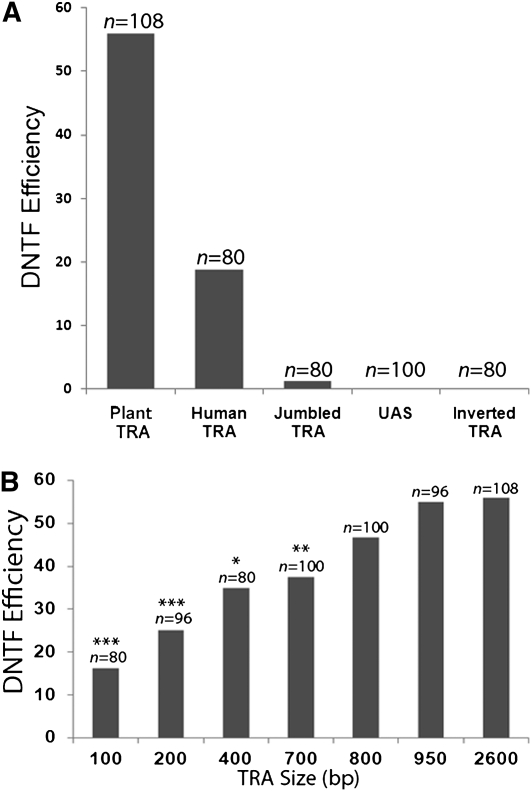
We investigated the sequence requirements for DNTF. Orientation proved to be important because 4X Arabidopsis plants transformed with a 900-bp TRA in an inverted orientation failed to promote DNTF (Figure 5A). Altering the repeat sequence was also detrimental (Figure 5). Only 1% (one of 80) of plants transformed with a 500-bp jumbled TRA (TTGATGG)n showed DNTF. Similarly, 600 bp of the yeast consensus upstream activating sequence (UAS) repeat (CGGAGGAGAGTCTTCCG) did not produce any DNTF events. In addition, DNTF was detected in ~18% (15/80) of plants transformed with a 750-bp TRA consisting of the vertebrate telomere repeat (TTAGGG)n, in contrast to 37% (37 of 100) in plants transformed with 700 bp of the Arabidopsis repeat (Figures 5A and 5B).
Figure 5.
DNTF Efficiency Is Dependent on the Sequence and Length of the TRA.
(A) Wild-type tetraploid plants were transformed with arrays consisting of Arabidopsis (TTTAGGG, 950 bp), human (TTAGGG, 750 bp), jumbled (TGGTTGAT, 500 bp), or UAS (CGGAGGAGAGTCTTCCG, 600 bp) repeat sequences cloned into the pBGW plasmid. The frequency of DNTF was determined by PETRA using primers targeting the pBGW plasmid backbone.
(B) TRAs of various lengths were transformed into wild-type tetraploid plants and DNTF events were scored by PETRA. Efficiency was calculated as described in Figure 2D. *** indicates a P value < 0.0001, ** P < 0.001, * P < 0.01.
To establish an optimal TRA length for DNTF, derivatives of pWY86 with varying amounts of TTTAGGG repeats were transformed into 4X Arabidopsis (Figure 5B). Previous studies indicate that 1 kb represents a critical length threshold for Arabidopsis telomeres (Heacock et al., 2004). Below this size, telomeres begin to be recruited into end-joining reactions. In tert mutants (which harbor a null mutation in the catalytic subunit of telomerase), the smallest TRA detected with an intact G-overhang is ~300bp (Heacock et al., 2004). In 4X wild type, even the smallest TRA we tested, corresponding to 100 bp, initiated DNTF, albeit at a substantially reduced rate relative to a 900- or 950-bp TRA (16% versus 54%, respectively; Figure 5B; see Supplemental Table 1 online). New telomeres recovered from plants bearing a 100-bp TRA ranged from 650 to 1.9 kb (average = 1.1 kb; Table 1), indicating that up to 1.8 kb of telomere repeats was added to the nascent terminus in a single plant generation. A similar trend was observed with other TRAs shorter than 1 kb (Table 1).
A small number of the plants transformed with the 100-bp TRA contained new telomeres substantially shorter than 1 kb (see Supplemental Figure 2A, lines 1 and 4, online). We suspected that such telomeres would be prone to end-joining reactions. Telomere fusion (TF)-PCR (Heacock et al., 2004) was performed using primers to test for sister chromatid fusions with the new telomere, as well as fusions between the TRA and endogenous telomere 3L (see Supplemental Figure 2B online). TF-PCR products were generated in reactions with lines 1 and 4, but not when new telomeres were longer than 1 kb (see Supplemental Figure 2B, lines 2 and 3, online). As expected, the endogenous 3L telomere did not engage in extensive end-joining reactions with the new TRA (see Supplemental Figure 2B, 2 + 3L, online). We conclude that nascent telomeres must exceed a critical 1-kb size threshold to avert end-to-end chromosome fusions.
Inactivation of Telomerase Increases the Frequency of DNTF
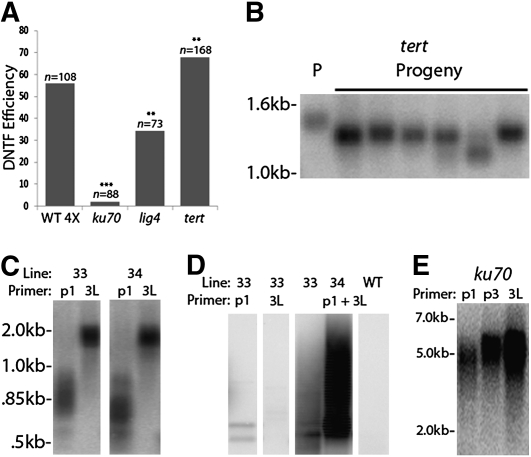
We used a genetic approach to examine the role of telomerase in converting a TRA into a functional telomere. Second generation (G2) tert mutants were made tetraploid and then transformed in the next generation with pWY86. The frequency of DNTF was monitored in two separate transformation experiments. On average, 72% of the transformants displayed DNTF (T1 = 68 of 96 [70%], T2 = 53 of 72 [74%]), a statistically higher fraction than 4X wild-type plants (56%; P ≤ 0.01; Figure 6A). DNA gel blot analysis confirmed that the average transgene copy number was the same (or lower) in tert compared with wild-type transformants (see Supplemental Figure 3 online). Thus, the elevated frequency of DNTF in 4X tert mutants does not reflect increased T-DNA integration but rather argues that telomerase is inhibitory to DNTF.
Figure 6.
Establishment of a New Telomere Is Dependent on Ku and Lig4, but Not on Telomerase.
(A) DNTF efficiency was monitored in the 4X mutant Arabidopsis lines shown. Efficiency was calculated as in Figure 2D.
(B) PETRA results for DNTF in a T1 tert-deficient parent (P) and several T2 progeny.
(C) PETRA results for two 4X tert lines (33 and 34) showing new telomeres shorter than 1 kb. PETRA products for the endogenous 3L telomere are shown for comparison.
(D) TF-PCR results for lines 33, 34, and wild type. Reactions were conducted with a single primer (p1 or e3L) or with both primers.
(E) PETRA results for a 4X ku70 transformant using p1, p3, or e3L.
As for endogenous telomeres, telomerase was needed to maintain the nascent telomere once it is established. In first generation (G1) tert mutants, endogenous telomeres are ~1 kb shorter than wild type and then decline by ~200 to 300 bp each plant generation thereafter (Riha et al., 2001). Similarly, in 4X tert transformants (G3 for tert), the average TRA associated with a nascent telomere was ~1.5 kb (Table 2), corresponding to the loss of 1.1 kb from the 2.6-kb pWY86 TRA. In the next generation, the new telomere decreased in size by ~200 bp (Figure 6B).
Table 2.
Size Variation for De Novo Telomeres in Different Genetic Backgrounds
| TRA Recovered |
|||
| Genotype | Initial TRA (bp) | Range (bp) | Avg (bp) |
| Wild type | 2600 | 950–4100 | 2562 ± 574 |
| G4 tert | 2600 | 650–2050 | 1529 ± 460 |
| G4 tert (endogenous) | ND | 670–2600 | 1466 ± 657 |
| lig4 | 2600 | 600–3050 | 1508 ± 722 |
ND, not determined.
Like 4X wild-type transformants, a subset of the de novo telomeres formed with the 2.6-kb TRA in 4X tert were significantly shorter than 1 kb (Figure 6C). Only a low level of sister fusions were detected with newly formed telomeres (Figure 6D, p1 alone). Consistent with previous analysis of tert mutants, no fusions were detected with the 3L telomere control (3L alone; Heacock et al., 2004). Unexpectedly, however, TF-PCR products were observed in reactions targeting the new telomere and 3L (p1 + 3L). We conclude that a short, unstable TRA in a telomerase-negative setting is capable of recruiting a fully capped and functional telomere into an end-joining event.
NHEJ Machinery Is Required for DNTF in Arabidopsis
DNTF would appear to be in direct competition with DNA repair pathways because factors necessary to authenticate the T-DNA as a telomere seed must displace or compete with components required for T-DNA integration. The situation may be more complex, given studies in yeast showing that Ku, a core component of NHEJ machinery, promotes telomerase recruitment at DSBs and at the same time protects natural telomeres from chromosome fusion (Stellwagen et al., 2003; Wang et al., 2009). To test how NHEJ components influence new telomere formation in a multicellular eukaryote, we transformed pWY86 with a 2.6-kb TRA into tetraploid plants carrying a null mutation in KU70 or DNA ligase IV (LIG4).
Unexpectedly, 4X lig4 transformants showed a statistically significant decrease in DNTF events relative to 4X wild-type plants, 26 of 73 (36%) versus 61 of 108 (56%) (Figure 6A). The value does not reflect decreased integration of pWY86 since T-DNA integration does not require LIG4 or KU (Friesner and Britt, 2003). Moreover, we assayed DNTF in 4X lig4 transformants resistant to BASTA and thus bearing an integrated T-DNA. The average length of a new telomere in 4X lig4 mutants was 1.5 kb, 1.1 kb shorter than the 2.6-kb TRA in pWY86 (Table 2). Because LIG4 does not make a significant contribution to telomere maintenance in Arabidopsis (Heacock et al., 2007), the data suggest that the TRA was subjected to nucleolytic digestion or deletional recombination prior to becoming a fully capped telomere.
An even more dramatic decrease in DNTF was observed in ku70 4X mutants. Only 2% (two of 88) of the transformants formed new telomeres (Figure 6A). In Arabidopsis, endogenous telomeres are grossly extended in the absence of Ku (Bundock et al., 2002; Riha et al., 2002). PETRA revealed that the new telomeres formed in these two lines were elongated to approximately the same extent as the endogenous 3L telomere, with the addition of ~2.5 kb in one generation (Figure 6E). These results show that if a telomere can form in the absence of Ku, it is subjected to the same length regulation as endogenous telomeres. We conclude that NHEJ components directly or indirectly promote DNTF at chromosome breaks and, further, that Ku plays an additional, specialized role in new telomere formation.
DISCUSSION
Conversion of a DSB into a fully capped telomere by DNTF is a potentially lethal event, leading to gene loss and genome instability. Thus, cells must evolve mechanisms to strictly control DNTF. Emerging data from budding yeast reveal that the ATR ortholog, Mec1, promotes genome integrity by negatively regulating DNTF as part of the DNA damage response (Makovets and Blackburn, 2009; Lydeard et al., 2010; Zhang and Durocher, 2010). Factors that modulate DNTF in multicellular eukaryotes are largely unexplored, but as this study illustrates, the flowering plant Arabidopsis is poised to fill this gap in understanding. For example, unlike vertebrate models, null mutations in ATR or telomere-capping proteins are viable in Arabidopsis (Watson and Riha, 2010). Furthermore, as demonstrated here, tetraploid Arabidopsis has the necessary genome-buffering capacity to reveal fundamental insights into the mechanism of DNTF.
Remarkably, up to one-half of the 4X Arabidopsis transformants we tested acquired truncated chromosomes capped by new telomeres. This represents a fivefold increase in DNTF events relative to diploid Arabidopsis and a much higher frequency than in yeast (<1%; Kramer and Haber, 1993), human embryonic fibroblasts (<2%; Barnett et al., 1993), or maize (~9%; Yu et al. 2006). The incidence of DNTF events in 4X Arabidopsis is comparable to mammalian cancer cell lines (40–60%), which are characterized by abrogated cell cycle checkpoints and rampant aneuploidy (Nigg, 2001). In normal diploids, including Arabidopsis, DNTF events are almost uniformly recovered near chromosome ends (Barnett et al., 1993; Yu et al., 2006). By contrast, DNTF events arise throughout the entire 4X Arabidopsis genome, an outcome that does not reflect an increased T-DNA integration in tetraploid plants. Rather, the genetic redundancy of the tetraploid genome appears to provide a less stringent filter for telomere-mediated chromosome truncation.
A Critical Length Threshold for New Telomeres
Arabidopsis can establish a telomere with a TRA of only 100 bp. DNTF events may actually occur with little to no TRA seed sequence. Arabidopsis telomerase extends primers lacking any complementarity to the telomerase RNA template in vitro by aligning the 3′ terminus at a “default” position within the RNA (Fitzgerald et al., 2001). Moreover, in yeast and mammals, telomeres can be formed in the complete absence of a TRA (Flint et al., 1994) via microhomology between the nucleotides at the 3′ terminus of the break site and the telomerase RNA template (Stellwagen et al., 2003). DTNF at the α-globin locus in humans and in mouse embryonic stem cells is thought to proceed by such a mechanism (Wong et al., 1997; Sprung et al., 1999).
By exploiting a PCR strategy devised to follow the fate of individual Arabidopsis telomeres, we discovered that the TRA must be elongated above a critical 1-kb length threshold to establish a fully capped telomere that prohibits end-joining reactions. The average length of new telomeres formed with a 100-bp TRA was 1.1 kb. Moreover, telomere fusion events were detected with TRAs shorter than 1 kb, whereas longer TRAs were immune. The molecular switch underlying this pivotal length threshold is unknown, but it may represent the minimal TRA required to recruit a sufficient number of telomere-capping proteins or to assemble into a protective secondary structure such as a t-loop.
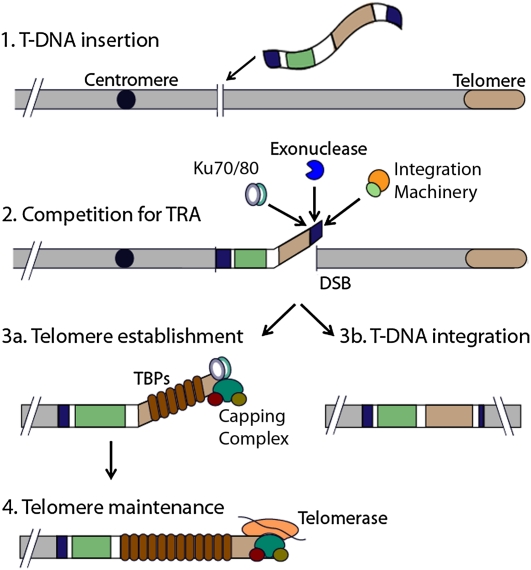
Competition for the Nascent Chromosome Terminus
Once a TRA is exposed, we speculate that it is engaged by multiple competing pathways (Figure 7). As in mammalian cells (Okabe et al., 2000), we found that telomerase is not required to establish a new telomere when the break occurs adjacent to a TRA (Figure 7, steps 1–3). However, telomerase is needed to maintain the integrity of the new telomere in successive generations (Figure 7, step 4). Successful de novo telomeres formed from short TRAs are extended past 1 kb in the first generation and then brought into the wild-type range in the second generation after transformation. Intriguingly, DNTF occurred at an even higher frequency in telomerase-deficient 4X plants than in wild-type 4X. One explanation is that telomerase interferes with DNTF by competing with proteins needed to form a protective cap and thus stabilize the nascent terminus (Figure 7, steps 2 and 3a).
Figure 7.
Model for DNTF Establishment and Maintenance in Arabidopsis.
After the first step of T-DNA insertion (Step 1), there is a competition for the TRA by Ku, T-DNA integration machinery, and exonucleases at the DSB formed at the 3′ end of the T-DNA (Step 2). T-DNA integration will occur if integration machinery out competes Ku (Step 3b). If Ku out competes the integration machinery, controlled exonucleolytic processing exposes the TRA (Step 2). A new telomere is established by association of double-strand telomere binding proteins and the Cdc13/CTC1, Stn1, and Ten1 capping complex (Step 3a). After telomere establishment, the new telomere must be maintained by telomerase (Step 4) above the critical 1-kb length threshold to prevent end-joining reactions.
We also exploited the genetic tractability of Arabidopsis to investigate how NHEJ machinery affects DNTF in a multicellular organism. Unexpectedly, we discovered that Ku and LIG4 directly or indirectly promote DNTF. NHEJ occurs by multiple routes in Arabidopsis, including a Ku- and LIG4-independent pathway. Neither of these factors is required for T-DNA integration (Gallego et al., 2003; van Attikum et al., 2003; Li et al., 2005; Heacock et al., 2007). The frequency of DNTF events in 4X lig4 mutants was significantly reduced, and the new telomere tracts were shorter than in 4X wild-type plants. Notably, in plants doubly deficient in LIG4 and TERT, extreme nucleolytic degradation is observed prior to end-joining (Heacock et al., 2007). Therefore, LIG4 may temporarily sequester the TRA from nucleolytic attack, providing additional time for telomere-capping proteins to engage the terminus (Figure 7, step 2).
Finally, we discovered that Ku plays a critical role in DNTF in Arabidopsis. Unlike LIG4, Ku is an essential component of the chromosome terminus, functioning in telomere length regulation and protection of the telomeric C-strand (Bertuch and Lundblad, 2003; Wang et al., 2009). Bertuch and colleagues propose a two-faced model for yeast Ku that explains its dual functions in NHEJ and telomere maintenance. In this model, Ku differentiates between telomeric and nontelomeric DNA based on the orientation of its two molecular faces (Ribes-Zamora et al., 2007). Accordingly, Ku interaction with the TRA on the T-DNA could define this region as a telomere instead of a double-strand break. In this view, full integration of the T-DNA into the chromosome would be favored over DNTF (Figure 7, steps 3a and 3b). In contrast to yeast, Ku does not associate with the TER1 telomerase RNP in Arabidopsis (Cifuentes-Rojas et al., 2011) and instead acts as a potent negative regulator of telomere length (Riha et al., 2002). Thus, Ku could potentially play an indirect role in promoting DNTF. For example, in the absence of Ku, the ultralong endogenous telomeres may sequester double-strand telomere binding proteins, preventing the transgenic TRA from being established as a functional telomere.
DNTF and Chromosome Engineering
Our findings indicate that natural polyploids can be manipulated as a platform for chromosome engineering and plant breeding through telomere truncation. Given the high frequency of DNTF and a semi-high–throughput method for identifying these events, it should now be possible to recover chromosome truncations at a desired location. With different selectable markers, a streamlined chromosome could be created by multiple truncation events and then reintroduced into a diploid by conventional genetic crosses (Yu et al., 2007) or through centromere-mediated genome elimination (Ravi and Chan, 2010). Finally, because truncated chromosomes can be transmitted to progeny, the consequences of such events could be examined over several generations.
METHODS
Arabidopsis thaliana Lines, Tetraploidization, and Transformation
The lig4, tert, and ku70 T-DNA insertion lines have been previously described (Riha et al., 2001, 2002; Heacock et al., 2007). Mutant lines (4X) were generated by the application of 2 mL of a 0.1% colchicine solution to the apical meristem of 7- to 14-d-old seedlings (second generation homozygous [G2] for the various T-DNA insertion). Seeds were collected from large flowers. Cytological confirmation of tetraploidy was performed by counting chromomeres of 4′,6-diamidino-2-phenylindole-stained nuclei (Yu et al., 2006). G3 (4X) plants were then transformed by the floral dip method (Zhang et al., 2006).
Plasmid Construction
The 2.6-kb and 950-, 800-, 700-, 400-, and 200-bp TRA derivatives of pWY86 were obtained by transforming pWY86 into Stbl2 cells (Invitrogen) and screening individual colonies by restriction digestion with EcoRI and BglII. A 100-bp TRA, 950-bp TRA, jumbled TRA, UAS, vertebrate TRA, and inverted TRA were constructed using a modified PCR reaction described in detail below and cloned into a Gateway entry vector. These constructs were then transferred to Agrobacterium tumefaciens destination vectors pBGW or pBWG by the Gateway clonase reaction (Karimi et al., 2002). The pWY86 and pBGW constructs were propagated in Invitrogen’s Stbl2 recombination-impaired cells and grown at 30°C instead of 37°C to minimize intrarepeat array recombination. Integrity of the constructs in A. tumefaciens was confirmed by extracting plasmid from a sample of the cells used to dip the plants and analyzing by restriction digestion.
Gateway Cloning of Repeat Arrays
TRAs were synthesized by performing a modified PCR reaction using two complementary oligonucleotide primers composed of a number of the desired repeats. Primers were designed to produce a single-strand overhang when annealed. The primers were resuspended to a stock concentration of 100 μM. The optimal oligonucleotide concentration to produce arrays of the correct size was determined (1 μL of oligo stock:20 μL of water; also in 1:40, 1:80, and 1:150 concentrations) in a reaction that contained phusion polymerase and reagents (New England Biolabs): 4 μL of 10 High Fidelity buffer (New England Biolabs), 1.2 μL of 2.5 mM deoxynucleotide triphosphates, 4 μL of oligo number 1 dilution, 4 μL of oligo number 2 dilution, 6.2 μL of water, and 0.2 μL of phusion polymerase. Cycling conditions were: (1) 98°C for 30 s, (2) 98°C for 10 s, (3) 58°C for 15 s, (4) 72°C for 5 s, (5) go to step 2 four more times, (6) 98°C for 10 s, (7) 58°C for 15 s, (8) 72°C for 5 s and add 2 s each cycle, (9) go to step 6 34 more times, and (10) 4°C hold. The PCR reaction produced a broad smear that varied in length depending on the primer concentration (lower concentrations produced high molecular weight products, whereas higher primer concentrations produced lower molecular weight products). After running a gel to determine which oligo concentration produced the desired product size range, a larger (three times for each reagent) reaction was set up with that primer concentration, and the products were run on a gel; the desired size range was extracted from the gel (Promega SV Wizard kit) and cloned using Invitrogen’s blunt end TOPO cloning kit (catalog no. K250020) with the pCR8/GW/TOPO vector, which contains Gateway attL1 and attL2 sites flanking the cloning site. The TOPO reaction was transformed into Stbl2 or Stbl4 recombination impaired cells (Invitrogen) and grown at 30°C (instead of 37°C) to minimize intrarepeat array recombination. After Escherichia coli transformation, the repeat array length and orientation were determined for individual clones by restriction analysis and sequencing. Depending on the orientation, the repeat array was moved into pBGW or pBWG via the Gateway clonase reaction. pBGW and pBWG are identical except for the orientation of the gateway cassette. Most of the repeat arrays recovered were less than 700 bp and were stable in the Stbl2 line and in A. tumefaciens.
TAIL-PCR
To determine sites of T-DNA insertion or DNTF, TAIL-PCR was performed using the TAIL, modified TAIL, and high-efficiency TAIL methods (Sessions et al., 2002; Liu and Chen, 2007). Primer sequences are shown in Supplemental Table 2 online. TAIL-PCR products were cloned into the TOPO-TA 2.1 vector and were sequenced using the M13 forward primer (Invitrogen).
PETRA and TF-PCR
PETRA and TF-PCR were performed as described in Heacock et al. (2004) with the following modifications. The PETRA-T reaction was followed by multiple PETRA-A reactions using the PETRA-A primer and either of two primers specific to pWY86, PWY86#1 and pWY86#3, or either of two primers specific to pBGW, PBGW#3 and PBGW#4 (see Supplemental Table 2 online for primer sequences). PETRA on ku70 samples was performed as described previously, except that the extension time was increased to 4 min to amplify the longer telomeres.
Statistical Analysis of DNTF Efficiency
An online Fisher’s exact test (http://www.langsrud.com/fisher.htm) was used to compare wild-type tetraploid lines transformed with the 2.6-kb TRA against all other TRAs used and against all other genetic backgrounds. In two cases, duplicate transformation experiments were performed with the same construct (see Supplemental Table 1 online). No statistically significant difference was observed in these cases, and the results were combined to get an overall DNTF efficiency number for that particular construct.
DNA Gel Blot Analysis
Fifty micrograms of genomic DNA was digested with one of three enzymes (SwaI, SmaI, or HindIII). Digests were ethanol precipitated and then separated on a 1% agarose gel. This was followed by transfer to a nylon membrane (GE) and hybridization with a High Prime (Roche) internally labeled dsDNA probe (probe no. 2) at 65°C. Enzymes were picked so that a staggered product would be formed in the event of a truncation.
To determine equal number of insertion events per line, genomic DNA was digested with EcoRI and HindIII. Following ethanol precipitation, DNA gel blotting was performed, probing with a dsDNA probe specific for the BAR gene.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DNTF Detected by DNA Gel Blot Analysis.
Supplemental Figure 2. TRAs Shorter Than 1 kb Are Prone to End-Joining Reactions.
Supplemental Figure 3. 4X Tert Acquires the Same Number of T-DNA Insertions as 4X Wild Type.
Supplemental Table 1. DNTF Efficiencies in Different Genetic Backgrounds.
Supplemental Table 2. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Christina Cutting and Alexandra Villanova for technical assistance, the Birchler laboratory for providing the pWY86 plasmid, Luca Comai and Dr. B. Dilkes for tetraploid Arabidopsis seeds and advice on tetraploidization, and Craig Kaplan, Jeff Kapler, and the Shippen laboratory for helpful advice. This work was supported by grants from the National Institutes of Health (GM-065383) and the National Science Foundation (MCB-0349993) to D.E.S. and by a Ruth L. Kirschstein National Research Service Award (GM-800052) to J.C.L.
AUTHOR CONTRIBUTIONS
A.D.N. and J.C.L. designed and performed the research. A.D.N. and J.C.L. analyzed the data. A.D.N., J.C.L., and D.E.S. wrote the manuscript.
References
- Bae N.S., Baumann P. (2007). A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol. Cell 26: 323–334 [DOI] [PubMed] [Google Scholar]
- Barnett M.A., Buckle V.J., Evans E.P., Porter A.C., Rout D., Smith A.G., Brown W.R. (1993). Telomere directed fragmentation of mammalian chromosomes. Nucleic Acids Res. 21: 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P., Cech T.R. (2001). Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Bertuch A.A., Lundblad V. (2003). Which end: Dissecting Ku’s function at telomeres and double-strand breaks. Genes Dev. 17: 2347–2350 [DOI] [PubMed] [Google Scholar]
- Bertuch A.A., Lundblad V. (2006). The maintenance and masking of chromosome termini. Curr. Opin. Cell Biol. 18: 247–253 [DOI] [PubMed] [Google Scholar]
- Bundock P., van Attikum H., Hooykaas P. (2002). Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucleic Acids Res. 30: 3395–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rojas C., Kannan K., Tseng L., Shippen D.E. (2011). Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc. Natl. Acad. Sci. USA 108: 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Collins K. (2006). The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 7: 484–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L., Verdun R.E., Haggblom C.I., Karlseder J. (2004). Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306: 1951–1953 [DOI] [PubMed] [Google Scholar]
- de Lange T. (2005). Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- Diede S.J., Gottschling D.E. (1999). Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99: 723–733 [DOI] [PubMed] [Google Scholar]
- Diede S.J., Gottschling D.E. (2001). Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 11: 1336–1340 [DOI] [PubMed] [Google Scholar]
- DuBois M.L., Haimberger Z.W., McIntosh M.W., Gottschling D.E. (2002). A quantitative assay for telomere protection in Saccharomyces cerevisiae. Genetics 161: 995–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M.G., Miller K.M., Cooper J.P. (2004). Indecent exposure: When telomeres become uncapped. Mol. Cell 13: 7–18 [DOI] [PubMed] [Google Scholar]
- Fitzgerald M.S., Shakirov E.V., Hood E.E., McKnight T.D., Shippen D.E. (2001). Different modes of de novo telomere formation by plant telomerases. Plant J. 26: 77–87 [DOI] [PubMed] [Google Scholar]
- Flint J., Craddock C.F., Villegas A., Bentley D.P., Williams H.J., Galanello R., Cao A., Wood W.G., Ayyub H., Higgs D.R. (1994). Healing of broken human chromosomes by the addition of telomeric repeats. Am. J. Hum. Genet. 55: 505–512 [PMC free article] [PubMed] [Google Scholar]
- Fortin F., Beaulieu Bergeron M., Fetni R., Lemieux N. (2009). Frequency of chromosome healing and interstitial telomeres in 40 cases of constitutional abnormalities. Cytogenet. Genome Res. 125: 176–185 [DOI] [PubMed] [Google Scholar]
- Friesner J., Britt A.B. (2003). Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 34: 427–440 [DOI] [PubMed] [Google Scholar]
- Gallego M.E., Bleuyard J.Y., Daoudal-Cotterell S., Jallut N., White C.I. (2003). Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant J. 35: 557–565 [DOI] [PubMed] [Google Scholar]
- Gao Q., Reynolds G.E., Wilcox A., Miller D., Cheung P., Artandi S.E., Murnane J.P. (2008). Telomerase-dependent and -independent chromosome healing in mouse embryonic stem cells. DNA Repair (Amst.) 7: 1233–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanish J.P., Yanowitz J.L., de Lange T. (1994). Stringent sequence requirements for the formation of human telomeres. Proc. Natl. Acad. Sci. USA 91: 8861–8865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock M., Spangler E., Riha K., Puizina J., Shippen D.E. (2004). Molecular analysis of telomere fusions in Arabidopsis: Multiple pathways for chromosome end-joining. EMBO J. 23: 2304–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock M.L., Idol R.A., Friesner J.D., Britt A.B., Shippen D.E. (2007). Telomere dynamics and fusion of critically shortened telomeres in plants lacking DNA ligase IV. Nucleic Acids Res. 35: 6490–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry I.M., Dilkes B.P., Young K., Watson B., Wu H., Comai L. (2005). Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics 170: 1979–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim S.I., Veena, Gelvin S.B. (2007). Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J. 51: 779–791 [DOI] [PubMed] [Google Scholar]
- Kramer K.M., Haber J.E. (1993). New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes Dev. 7(12A): 2345–2356 [DOI] [PubMed] [Google Scholar]
- Kumekawa N., Hosouchi T., Tsuruoka H., Kotani H. (2001). The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 4. DNA Res. 8: 285–290 [DOI] [PubMed] [Google Scholar]
- Larrivée M., LeBel C., Wellinger R.J. (2004). The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 18: 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Myung K. (2009). Faithful after break-up: Suppression of chromosomal translocations. Cell. Mol. Life Sci. 66: 3149–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Vaidya M., White C., Vainstein A., Citovsky V., Tzfira T. (2005). Involvement of KU80 in T-DNA integration in plant cells. Proc. Natl. Acad. Sci. USA 102: 19231–19236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.G., Chen Y. (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43: 649–650, 652, 654 passim [DOI] [PubMed] [Google Scholar]
- Lydeard J.R., Lipkin-Moore Z., Jain S., Eapen V.V., Haber J.E. (2010). Sgs1 and exo1 redundantly inhibit break-induced replication and de novo telomere addition at broken chromosome ends. PLoS Genet. 6: e1000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovets S., Blackburn E.H. (2009). DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat. Cell Biol. 11: 1383–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane J.P. (2010). Telomere loss as a mechanism for chromosome instability in human cancer. Cancer Res. 70: 4255–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S., Ribaud V., Bianchi A., Shore D. (2007). DNA breaks are masked by multiple Rap1 binding in yeast: Implications for telomere capping and telomerase regulation. Genes Dev. 21: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A. (2001). Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2: 21–32 [DOI] [PubMed] [Google Scholar]
- Nugent C.I., Hughes T.R., Lue N.F., Lundblad V. (1996). Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Okabe J., Eguchi A., Masago A., Hayakawa T., Nakanishi M. (2000). TRF1 is a critical trans-acting factor required for de novo telomere formation in human cells. Hum. Mol. Genet. 9: 2639–2650 [DOI] [PubMed] [Google Scholar]
- Pennaneach V., Kolodner R.D. (2004). Recombination and the Tel1 and Mec1 checkpoints differentially effect genome rearrangements driven by telomere dysfunction in yeast. Nat. Genet. 36: 612–617 [DOI] [PubMed] [Google Scholar]
- Pennaneach V., Putnam C.D., Kolodner R.D. (2006). Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol. Microbiol. 59: 1357–1368 [DOI] [PubMed] [Google Scholar]
- Puglisi A., Bianchi A., Lemmens L., Damay P., Shore D. (2008). Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 27: 2328–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi M., Chan S.W. (2010). Haploid plants produced by centromere-mediated genome elimination. Nature 464: 615–618 [DOI] [PubMed] [Google Scholar]
- Ribes-Zamora A., Mihalek I., Lichtarge O., Bertuch A.A. (2007). Distinct faces of the Ku heterodimer mediate DNA repair and telomeric functions. Nat. Struct. Mol. Biol. 14: 301–307 [DOI] [PubMed] [Google Scholar]
- Riha K., McKnight T.D., Griffing L.R., Shippen D.E. (2001). Living with genome instability: Plant responses to telomere dysfunction. Science 291: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Riha K., Watson J.M., Parkey J., Shippen D.E. (2002). Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21: 2819–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J.L., Alfaro D., Sanchez-Moran E., Armstrong S.J., Franklin F.C., Jones G.H. (2003). Partial diploidization of meiosis in autotetraploid Arabidopsis thaliana. Genetics 165: 1533–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov E.V., Shippen D.E. (2004). Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell 16: 1959–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Bianchi A. (2009). Telomere length regulation: Coupling DNA end processing to feedback regulation of telomerase. EMBO J. 28: 2309–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprung C.N., Reynolds G.E., Jasin M., Murnane J.P. (1999). Chromosome healing in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 96: 6781–6786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A.E., Haimberger Z.W., Veatch J.R., Gottschling D.E. (2003). Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 17: 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva Y.V., Churikov D., Boltz K.A., Song X., Lamb J.C., Warrington R., Leehy K., Heacock M., Price C.M., Shippen D.E. (2009). Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Bundock P., Overmeer R.M., Lee L.Y., Gelvin S.B., Hooykaas P.J. (2003). The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 31: 4247–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska A., de Lange T. (1998). TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Verdun R.E., Karlseder J. (2007). Replication and protection of telomeres. Nature 447: 924–931 [DOI] [PubMed] [Google Scholar]
- Vizir I.Y., Mulligan B.J. (1999). Genetics of gamma-irradiation-induced mutations in Arabidopsis thaliana: Large chromosomal deletions can be rescued through the fertilization of diploid eggs. J. Hered. 90: 412–417 [DOI] [PubMed] [Google Scholar]
- Wang Y., Ghosh G., Hendrickson E.A. (2009). Ku86 represses lethal telomere deletion events in human somatic cells. Proc. Natl. Acad. Sci. USA 106: 12430–12435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J.M., Riha K. (2010). Comparative biology of telomeres: Where plants stand. FEBS Lett. 584: 3752–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie A.O., Lamb J., Harris P.C., Finney R.D., Higgs D.R. (1990). A truncated human chromosome 16 associated with alpha thalassaemia is stabilized by addition of telomeric repeat (TTAGGG)n. Nature 346: 868–871 [DOI] [PubMed] [Google Scholar]
- Wong A.C., Ning Y., Flint J., Clark K., Dumanski J.P., Ledbetter D.H., McDermid H.E. (1997). Molecular characterization of a 130-kb terminal microdeletion at 22q in a child with mild mental retardation. Am. J. Hum. Genet. 60: 113–120 [PMC free article] [PubMed] [Google Scholar]
- Yu W., Han F., Gao Z., Vega J.M., Birchler J.A. (2007). Construction and behavior of engineered minichromosomes in maize. Proc. Natl. Acad. Sci. USA 104: 8924–8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Lamb J.C., Han F., Birchler J.A. (2006). Telomere-mediated chromosomal truncation in maize. Proc. Natl. Acad. Sci. USA 103: 17331–17336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Durocher D. (2010). De novo telomere formation is suppressed by the Mec1-dependent inhibition of Cdc13 accumulation at DNA breaks. Genes Dev. 24: 502–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.