Abstract
Gene expression is dynamically regulated by chromatin modifications on histone tails, such as acetylation. In general, histone acetylation promotes transcription, whereas histone deacetylation negatively regulates transcription. The interplay between histone acetyltranserases and histone deacetylases (HDACs) is pivotal for the regulation of gene expression required for long-term memory processes. Currently, very little is known about the role of individual HDACs in learning and memory. We examined the role of HDAC3 in long-term memory using a combined genetic and pharmacologic approach. We used HDAC3–FLOX genetically modified mice in combination with adeno-associated virus-expressing Cre recombinase to generate focal homozygous deletions of Hdac3 in area CA1 of the dorsal hippocampus. To complement this approach, we also used a selective inhibitor of HDAC3, RGFP136 [N-(6-(2-amino-4-fluorophenylamino)-6-oxohexyl)-4-methylbenzamide]. Immunohistochemistry showed that focal deletion or intrahippocampal delivery of RGFP136 resulted in increased histone acetylation. Both the focal deletion of HDAC3 as well as HDAC3 inhibition via RGFP136 significantly enhanced long-term memory in a persistent manner. Next we examined expression of genes implicated in long-term memory from dorsal hippocampal punches using quantitative reverse transcription-PCR. Expression of nuclear receptor subfamily 4 group A, member 2 (Nr4a2) and c-fos was significantly increased in the hippocampus of HDAC3–FLOX mice compared with wild-type controls. Memory enhancements observed in HDAC3–FLOX mice were abolished by intrahippocampal delivery of Nr4a2 small interfering RNA, suggesting a mechanism by which HDAC3 negatively regulates memory formation. Together, these findings demonstrate a critical role for HDAC3 in the molecular mechanisms underlying long-term memory formation.
Introduction
Transcription is thought to be a key step for long-term memory processes (Alberini, 2009). Transcription is promoted by specific chromatin modifications, such as histone acetylation, which modulate histone–DNA interactions (Kouzarides, 2007). Modifying enzymes, such as histone acetyltransferases (HATs) and histone deacetylases (HDACs), regulate the state of acetylation on histone tails. In general, histone acetylation promotes gene expression, whereas histone deacetylation leads to gene silencing. Numerous studies have shown that a potent HAT, cAMP response element-binding protein (CREB)-binding protein (CBP), is necessary for long-lasting forms of synaptic plasticity and long-term memory (for review, see Barrett and Wood, 2008).
In contrast, HDACs have been shown to be powerful negative regulators of long-term memory processes. Nonspecific HDAC inhibitors enhance synaptic plasticity as well as long-term memory (Levenson et al., 2004; Lattal et al., 2007; Vecsey et al., 2007; Bredy and Barad, 2008; Guan et al., 2009; Malvaez et al., 2010; Roozendaal et al., 2010). For example, HDAC inhibition can transform a learning event that does not lead to long-term memory into a learning event that does result in significant long-term memory (Stefanko et al., 2009). Furthermore, HDAC inhibition can also generate a form of long-term memory that persists beyond the point at which normal memory fails. HDAC inhibitors have been shown to ameliorate cognitive deficits in genetic models of Alzheimer's disease (Fischer et al., 2007; Kilgore et al., 2010). These demonstrations suggest that modulating memory via HDAC inhibition have considerable therapeutic potential for many cognitive disorders.
Currently, the role of individual HDACs in long-term memory formation remains primarily unexplored except for two recent studies. Kilgore et al. (2010) revealed that nonspecific HDAC inhibitors, such as sodium butyrate, inhibit class I HDACs (HDAC1, HDAC2, HDAC3, HDAC8) with little effect on the class IIa HDAC family members (HDAC4, HDAC5, HDAC7, HDAC9). This suggests that inhibition of class I HDACs is critical for the enhancement of cognition observed in many studies. Indeed, forebrain overexpression of HDAC2, but not HDAC1, negatively regulates memory formation (Guan et al., 2009). To date, no studies have examined the function of HDAC3 in memory formation. HDAC3 is the most highly expressed class I HDAC throughout the brain, including the hippocampus (Broide et al., 2007). HDAC3 alters gene expression as part of a large complex that contains corepressors, nuclear receptor corepressor 1 (NCoR) and silencing mediator for retinoid and thyroid-hormone receptors (SMRT), as well as class IIa HDACs, such as HDAC4 (Guenther et al. 2000; Li et al., 2000) (for review, see Karagianni and Wong, 2007). NCoR associates with HDAC3 through the deacetylase activation domain (DAD) of NCoR and a single amino acid substitution (Y478A) in the NCoR DAD results in a mutant protein that is unable to associate with or activate HDAC3 (Alenghat et al., 2008). In addition, class IIa HDACs may require interaction with HDAC3 for their HDAC activity (Fischle et al., 2002).
In this study, we examined the molecular and behavioral consequences of HDAC3 inhibition using a combined genetic and pharmacologic approach. Together, these strategies provide convergent findings demonstrating that HDAC3 is a critical negative regulator of long-term memory formation.
Materials and Methods
Subjects and surgical procedures.
HDAC3 floxed C57BL/6 mice were generated with loxP sites flanking exon 4 through exon 7 of the HDAC3 gene, a region required for the catalytic activity of the enzyme. These mice were generated in the laboratory of Dr. Mitch Lazar at the University of Pennsylvania and will be described in detail elsewhere. Briefly, targeted mutagenesis was performed in C57BL/6 embryonic stem cells and HDAC3–FLOX mice have been maintained on a C57BL/6 background. To generate a focal deletion, mice were infused with adeno-associated virus expressing Cre-recombinase (AAV2/1–Cre; Penn Vector Core, University of Pennsylvania, Philadelphia, PA) 2 weeks before behavioral procedures. Mice were anesthetized with isoflurane and placed in a digital Just For Mice stereotax (Stoelting). Virus (1.0 μl) was injected at a rate of 6 μl/h via an infusion needle positioned in the dorsal CA1 area of the hippocampus [anteroposterior (AP), −2.0 mm; mediolateral (ML), ±1.5 mm; dorsoventral (DV), −1.5 mm]. NCoR homozygous knock-in mice (referred to as DADm mice) were generated on C57BL/6 background using homologous recombination to incorporate a single amino acid substitution (Y478A) in the NCoR DAD. DADm mice are fully described by Alenghat et al. (2008). CBPKIX/KIX homozygous knock-in mice were generated as described previously (Kaspar et al., 2002). These mice carry a triple-point mutation in the phospho-CREB (KIX) binding domain of CBP. For the Nr4a2 knockdown experiment, SMART pool small interfering RNAs (siRNAs) (Dharmacon) targeted against Nr4a2 were prepared with jetSI (Polyplus Transfection) at a final concentration of 4 μm before injection. Intrahippocampal infusions of Nr4a2 siRNA or RNA-induced silencing complex (RISC)-free control siRNA were performed similarly to the infusion procedure above. These surgeries were performed on hippocampal AAV–Cre-infused HDAC3flox/flox and HDAC3+/+ mice 2 d before training. Immunohistochemistry and quantitative reverse transcription-PCR were used to confirm focal deletions and siRNA knockdown, respectively, and lack of either was used as criteria for exclusion from those experimental groups. For all other experiments, C57BL/6J male mice were acquired from The Jackson Laboratory. Mice were anesthetized with isoflurane, and bilateral cannulae (Plastics One) aimed at the dorsal hippocampus were stereotaxically implanted (AP, −1.7 mm; ML, ±1.2 mm; DV, −1.5 mm). For all experiments, mice were 8–12 weeks old and had access to food and water ad libitum in their home cages. Lights were maintained on a 12 h light/dark cycle, with all behavioral testing performed during the light portion of the cycle. All experiments were conducted according to National Institutes of Health guidelines for animal care and use and were approved by the Institutional Animal Care and Use Committee of the University of California, Irvine.
Drugs.
RGFP136 [N-(6-(2-amino-4-fluorophenylamino)-6-oxohexyl)-4-methylbenzamide] was provided by Repligen Corporation and has been described previously by Rai et al. (2010). Drug was dissolved in DMSO and diluted in a vehicle of 20% glycerol, 20% PEG 400, 20% propylene glycol, and 100 mm sodium acetate, pH 5.4. The final DMSO concentration was no greater than 10%, and the same concentration of DMSO was included in vehicle injections. For experiments, doses were 1.25 ng/side (0.5 μl volume) for intrahippocampal infusion and 30 or 150 mg/kg subcutaneously (10 ml/kg volume) for systemic administration.
Immunohistochemistry.
Two weeks after mice were infused with AAV–Cre or 2 h after RGFP136 hippocampal infusion, mice were anesthetized deeply with sodium pentobarbital (100 mg/kg, i.p.) and perfused transcardially with ice-cold PBS, pH 7.4, followed by ice-cold 4% paraformaldehyde in PBS, pH 7.4, using a peristaltic perfusion pump (Thermo Fisher Scientific). The brains were removed, postfixed overnight at 4°C, and then transferred to 30% sucrose for 48 h at 4°C. Brains were frozen and cryocut to 20 μm coronal slices, and sections were stored in 0.1 m PBS. Floating sections were rinsed in 0.1% Triton X-100 (Thermo Fisher Scientific) in PBS, rinsed in PBS, and then blocked for 1 h at room temperature in 8% normal goat serum (NGS) (Jackson ImmunoResearch) with 0.3% Triton X-100 in PBS. Sections were rinsed in PBS and incubated overnight at 4°C in 2% NGS and 0.3% Triton X-100 in PBS with primary antibody. The sections were then rinsed in PBS and incubated for 2 h at room temperature with goat anti-rabbit IgG–FITC secondary antibody (1:1000; Millipore Bioscience Research Reagents). Sections were rinsed again in PBS and mounted on slides using ProLong Gold antifade reagent with DAPI (Invitrogen). Primary antibodies used were HDAC3 (1:1000; Millipore Corporation), HDAC2 (1:1000; Abcam), HDAC4 (1:500; Abcam), and acetyl-histone-H4K8 primary antibody (1:1000; Cell Signaling Technology).
Images were acquired on an Olympus BX51 microscope using a 4× or 20× objective, CCD camera (QImaging), and QCapture Pro 6.0 software (QImaging). All treatment groups were represented on each slide, and all images were acquired using the same exposure time. Immunolabeling was quantified using NIH ImageJ software by selecting the cell layer from comparable 20× images and calculating the optical density of immunofluorescence.
Quantitative real-time RT-PCR.
Quantitative real-time RT-PCR was performed to examine nuclear receptor subfamily 4 group A member 2 (Nr4a2) and c-fos expression. Tissue was collected by taking 1 mm punches from dorsal hippocampal slices in the area of the focal deletion in HDAC3flox/flox mice as confirmed by immunohistochemistry for HDAC3 and equivalent regions in HDAC3+/+ mice. RNA was isolated using RNeasy minikit (Qiagen). cDNA was made from 200 ng of total RNA using the Transcriptor First Strand cDNA Synthesis kit (Roche Applied Science). Primers were derived from the Roche Universal ProbeLibrary: Nr4a2 left primer, 5′-ttgcagaatatgaacatcgaca-3′; Nr4a2 right primer, 5′-gttccttgagcccgtgtct-3′; probe, ttctcctg; c-Fos left primer, 5′-ggggcaaagtagagcagcta-3′; c-Fos right primer, 5′-agctccctcctccgattc-3′; probe, atggctgc (both Nr4a2 and c-Fos probes are conjugated to the dye FAM); glyceraldehyde-3-phosphate dehydrogenase (GAPDH) left primer, 5′-atggtgaaggtcggtgtga-3′; right primer, 5′-aatctccactttgccactgc-3′; probe, tggcggtattgg (GAPDH probe is conjugated to Lightcycler Yellow 555). The non-overlapping dyes and quencher on the reference gene allow for multiplexing in the Roche LightCycle 480 II machine (Roche Applied Science). All values were normalized to GAPDH expression levels. Analysis and statistics were performed using the Roche proprietary algorithms and REST 2009 software based on the Pfaffl method (Pfaffl, 2001; Pfaffl et al., 2002).
Object recognition.
Training and testing for location-dependent object recognition memory (OLM) and novel object recognition memory (ORM) were performed as described previously (Roozendaal et al., 2010). Before training, mice were handled 1–2 min for 5 d and were habituated to the experimental apparatus 3 min/d for 3 consecutive days in the absence of objects. The experimental apparatus was a white rectangular open field (30 × 23 × 21.5 cm). During the training trial, mice were placed in the experimental apparatus with two identical objects (either 100 ml beakers, 2.5 cm diameter, 4 cm height; or large blue Lego blocks, 2.5 × 2.5 × 5 cm) and were allowed to explore these objects for 3 min, which does not result in short or long term memory (Stefanko et al., 2009). During the 24 h or 7 d retention test, mice were placed in the experimental apparatus for 5 min. For ORM, one copy of the familiar object (A3) and a new object (B1) were placed in the same location as during the training trial. For location-dependent OLM, one copy of the familiar object (A3) was placed in the same location as during the training trial, and one copy of the familiar object (A4) was placed in the middle of the box. All combinations and locations of objects were used in a balanced manner to reduce potential biases attributable to preference for particular locations or objects. All training and testing trials were videotaped and analyzed by individuals blind to the treatment condition and the genotype of subjects. A mouse was scored as exploring an object when its head was oriented toward the object within a distance of 1 cm or when the nose was touching the object. The relative exploration time was recorded and expressed by a discrimination index [DI = (tnovel − tfamiliar)/(tnovel + tfamiliar) × 100].
Statistics.
Datasets with only two groups were analyzed by independent samples t test. Datasets with four groups, such as the HDAC3–FLOX and Nr4a2 siRNA experiment, were analyzed by two-way ANOVA, and separate one-way ANOVAs were used to make specific comparisons when significant interactions were observed. Student–Newman–Keuls and least significant different post hoc tests were performed when appropriate. Simple planned comparisons were made using Student's t tests with α levels held at 0.05.
Results
Generation of focal HDAC3 deletion
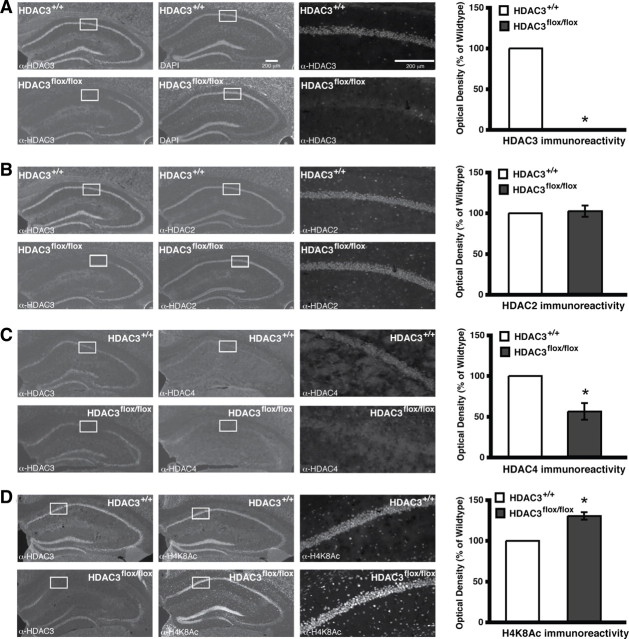
The overall goal of this study was to begin to understand the role of HDAC3 in long-term memory function. Focal deletions of HDAC3 allow for a detailed regional and task-selective behavioral analysis without developmental consequence. HDAC3flox/flox and HDAC3+/+ mice received bilateral intrahippocampal infusions of AAV–Cre recombinase (1 μl/side). AAV serotype 2/1 was used, which has the viral genome of serotype 2 and packaged in coat proteins from serotype 1 for efficient transduction of dorsal hippocampal pyramidal neurons (Burger et al., 2004). This viral infusion does not alter neuronal morphology indicated by intact nuclei visualized by DAPI staining but does lead to a complete, focal deletion of HDAC3 as demonstrated by loss of immunoreactivity in the dorsal hippocampus (Fig. 1A, bottom left).
Figure 1.
Intrahippocampal AAV2/1–Cre infusion in HDAC3flox/flox mice results in a complete, focal deletion of HDAC3 that correlates with increased histone acetylation. Images are 4×, except the right panels, which are 20× magnifications of the regions boxed in white. Histograms depict quantification of optical density as a percentage of wild type. A, Representative images showing DAPI labeling and HDAC3 immunoreactivity in hippocampi of AAV2/1–Cre infused HDAC3+/+ and HDAC3flox/flox mice. HDAC3 labeling is found throughout CA1, CA3, and the dentate gyrus, and no immunoreactivity is found in the AAV2/1–Cre infusion site of HDAC3flox/flox mice. *p < 0.05. B, Representative images showing HDAC2 immunoreactivity in hippocampus is unchanged in AAV2/1–Cre-infused HDAC3flox/flox mice. C, However, HDAC4 immunoreactivity is decreased in the region of the HDAC3 deletion. *p < 0.05. D, Furthermore, acetylation at H4K8 is increased specifically in the AAV2/1–Cre infusion site of HDAC3flox/flox mice. *p < 0.05.
Next, we examined immunoreactivity for other HDACs. HDAC2, another class I HDAC member, has been implicated in learning and memory (Guan et al., 2009), and it is part of a corepressor complex with HDAC1 (Laherty et al., 1997). We also chose HDAC4, a class IIa HDAC that can bind to HDAC3 in a corepressor complex (Grozinger and Schreiber, 2000; Fischle et al., 2002). HDAC3 deletion did not alter the expression of HDAC2 (Fig. 1B, bottom middle). In contrast, HDAC4 had reduced nuclear expression in the region of the HDAC3 deletion (F(1,6) = 7.53; p = 0.03) (Fig. 1C, bottom middle). These results suggest that deletion of HDAC3 has no observable effect, using immunohistochemical analysis, on expression of HDAC2; however, it has a significant effect on the expression of HDAC4.
To determine whether deletion of HDAC3 affected histone acetylation, acetylation of histone H4, lysine 8 (H4K8Ac) was examined. Acetylation at this site has been shown to increase after the dissociation of the NCoR/HDAC3 complex from promoter regions and consequently leads to an increase in transcriptional activity (Guenther et al., 2000; Li et al., 2000; Wang et al., 2010). Indeed, there was an observed increase in H4K8Ac in the region of HDAC3 deletion (F(1,5) = 7.18; p = 0.04) (Fig. 1D). These findings suggest that HDAC3, perhaps together with HDAC4, controls acetylation of H4K8 involved in transcriptional regulation (Agalioti et al., 2002).
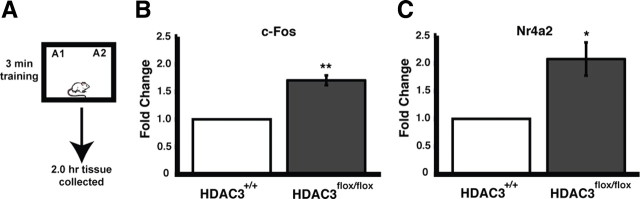
The absence of HDAC3, decreased expression of HDAC4, and the increase in histone acetylation suggested that gene expression would be increased in the region of the focal homozygous deletion of HDAC3. To test this, we examined expression of two immediate early genes, c-fos and Nr4a2, 2 h after object recognition training. Transcription of immediate early genes initiated by patterned synaptic activity is necessary for synaptic plasticity and long-term memory (for review, see Alberini, 2009). We have shown in a previous study that HDAC inhibition in the hippocampus maintained the expression of Nr4a2 at 2 h, beyond the point at which it would normally be expressed during memory consolidation (Vecsey et al., 2007). HDAC3flox/flox and HDAC3+/+ mice received bilateral intrahippocampal AAV–Cre infusions 2 weeks (for optimal gene deletion; data not shown) before training. During training, mice were placed in an arena with two identical objects for a subthreshold 3 min training session (Fig. 2A), which does not result in long-term memory (Stefanko et al., 2009). Tissue was collected by taking 1 mm punches from dorsal hippocampal slices in the area of the focal deletion in HDAC3flox/flox mice (n = 3) as confirmed by immunohistochemistry for HDAC3 and equivalent regions in HDAC3+/+ mice (n = 3). c-fos expression was significantly increased in the area of the focal deletion of HDAC3flox/flox mice compared with wild-type littermates (t(4) = 6.81; p = 0.002) (Fig. 2B). Similarly, Nr4a2 expression in the dorsal hippocampus was twofold greater in HDAC3flox/flox compared with HDAC3+/+ mice after training (t(4) = 4.05; p = 0.015) (Fig. 2C). Gene expression was also measured in naive controls that received hippocampal AAV–Cre infusions to determine potential basal differences (data not shown). Naive handled HDAC3flox/flox mice had significantly greater c-fos expression than wild-type mice (t(9) = 2.30; p < 0.05), yet basal Nr4a2 expression was unchanged (t(6) = 0.33; p = 0.75). Thus, Nr4a2 is differentially induced in the HDAC3flox/flox mice, in which training triggers greater gene expression but basal levels are unchanged compared with HDAC3+/+ mice. Together, these data reveal that HDAC3flox/flox mice have enhanced histone acetylation and gene expression in the focal deletion compared with wild-type controls.
Figure 2.
c-fos and Nr4a2 expression are increased in the area of focal homozygous deletion of Hdac3 in HDAC3flox/flox mice. A, Mice received subthreshold training (3 min) in an environment with two identical objects. B, Two hours after training, quantitative RT-PCR shows that c-fos expression is significantly increased in the dorsal hippocampus of HDAC3flox/flox mice compared with wild-type littermates (n = 3 per group; **p < 0.003). C, In addition, training induced greater Nr4a2 expression in the dorsal hippocampus of HDAC3flox/flox mice compared with wild-type littermates (n = 3 per group; *p < 0.02).
Deletion of HDAC3 in dorsal hippocampus leads to enhanced long-term memory for object location
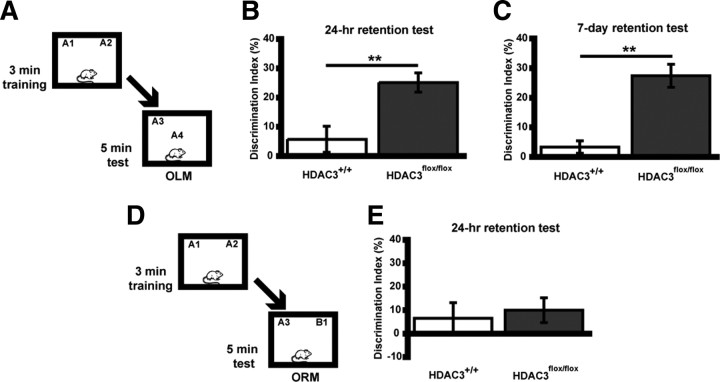
We hypothesized that the observed increases in histone acetylation and training-induced expression of genes implicated in long-term memory would likely result in enhanced performance in a behavioral paradigm used to assess learning and memory. Previous studies have shown that HDAC inhibition enhances memory such that a subthreshold learning event that would not result in long-term memory is transformed into an event leading to long-term memory (Stefanko et al., 2009). To test whether deletion of HDAC3 affects learning and memory in a similar manner, HDAC3flox/flox and HDAC3+/+ mice received bilateral intrahippocampal AAV–Cre infusions 2 weeks (for optimal gene deletion and protein clearance) before training. During training, mice were placed in an arena with two identical objects for a 3 min training session, which does not result in long-term memory (Stefanko et al., 2009), and then tested 24 h later in the same arena with one familiar object moved to a novel location (OLM) (Fig. 3A). Wild-type mice did not exhibit significant discrimination (n = 8; DI = 4.7 ± 3.0%), confirming that 3 min was a subthreshold training period. In contrast, HDAC3flox/flox mice displayed significant memory for object location, evident by a significantly greater discrimination index (n = 8; DI = 25.1 ± 3.3%; t(14) = 3.51; p = 0.003) (Fig. 3B). Groups did not differ in total exploration time of the two objects during either the training or retention test (data not shown). These results demonstrate that HDAC3 is a negative regulator of long-term memory in the dorsal hippocampus.
Figure 3.
Focal homozygous gene deletion of Hdac3 in the dorsal hippocampus leads to enhanced memory for object location (OLM), which persists at least 7 d but not for object recognition (ORM). A, Mice received subthreshold training (3 min) in an environment with two identical objects and received a retention test 24 h or 7 d later in which one object is moved to a new location. Schematic describes methods for B and C. B, HDAC3flox/flox mice exhibited significant long-term memory for object location 24 h after subthreshold training (n = 8 per group; **p < 0.005). C, In a different set of mice, the persistence of this enhanced memory was examined. HDAC3flox/flox mice displayed a significant preference for the novel object location compared with HDAC3+/+ mice during a 7 d retention test (n = 9 per group; **p < 0.001). D, Mice received subthreshold training (3 min) in an environment with two identical objects and received a retention test 24 h later in which one object is replaced with a novel one (ORM). Schematic describes methods for E. E, Neither HDAC3+/+ or HDAC3flox/flox mice exhibited significant preference for the novel object (n = 8 per group).
In the next experiment, we tested the persistence of long-term memory induced by HDAC3 deletion. Previously, we demonstrated that novel object recognition after 10 min training is evident 24 h later, but this memory fails when tested after 7 d (Stefanko et al., 2009). Mice received a 3 min training period, followed 7 d later by a retention test. As shown in Figure 3C, HDAC3flox/flox mice (n = 9; DI = 27.4 ± 4.0%) had a greater preference for object in the new location compared with wild-type mice (t(16) = 5.30; p < 0.001). Groups did not differ in total exploration time of the two objects during either the training or retention test (data not shown). These results suggest that HDAC3 deletion leads to long-term memory formation that is persistent and lasts beyond the point at which normal long-term memory fails.
We next examined whether the focal HDAC3 deletion affected long-term memory in a standard novel object recognition task (ORM) (Fig. 3D). In this task, there is no change in context or object location, but one of the familiar objects is replaced with a novel object. The dorsal hippocampus has been shown to encode information regarding context and location (O'Keefe, 1999; Fanselow, 2000; Maren and Holt, 2000; Smith and Mizumori, 2006); however, other brain regions, such as insular cortex, are important for long-term memory for the object itself (Balderas et al., 2008; Roozendaal et al., 2010). This distinct neural circuitry for the ORM and OLM tasks can reveal the specificity of our treatment. Figure 3E shows that, after subthreshold training (3 min), both HDAC3flox/flox (n = 8) and HDAC3+/+ (n = 8) mice spent similar amounts of time with both the familiar and novel objects on test day (t(14) = 0.40; p = 0.70). Groups did not differ in total exploration time of the two objects during either the training or retention test (data not shown). Together, the data in Figure 3 suggest that focal deletion of HDAC3 in the dorsal hippocampus results in a selective enhancement of long-term memory for the object location (Fig. 3B) but not the object itself (Fig. 3E).
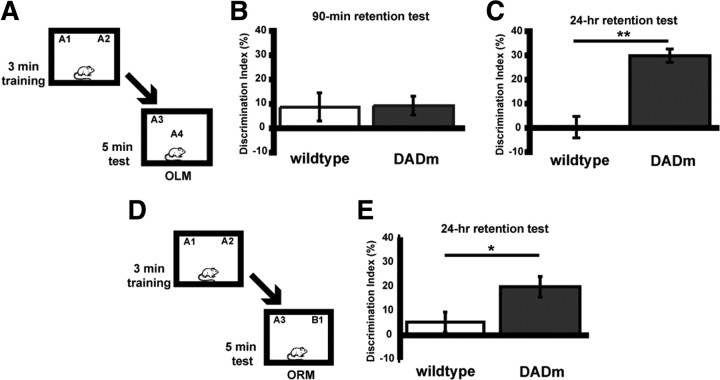
To further support the role of HDAC3 as a negative regulator of long-term memory formation, we also examined genetically modified NCoR homozygous knock-in mice. These mice carry a single amino acid substitution (Y478A) in the DAD of NCoR that disrupts its binding to HDAC3 (Alenghat et al., 2008) (these mice are referred to as DADm mice). Mice were subjected to a subthreshold training period (3 min) and tested for short-term (90 min) memory for object location (Fig. 4A). Short-term memory is a distinct form of memory that does not require transcription (for review, see Alberini, 2009). As shown in Figure 4B, DADm (n = 11) and wild-type (n = 13) mice showed a similar lack of preference for the novel object location after a 90 min retention test (t(22) = 0.08; p = 0.94). In a different set of mice to examine long-term memory at 24 h (mice were also given a 3 min training period), DADm mice (n = 10) exhibited significant preference for the novel location of the familiar object compared with wild-type controls (n = 9; t(17) = 3.52; p = 0.003) (Fig. 4C). Groups did not differ in total exploration time of the two objects during either the training or retention test (data not shown). These results are consistent with findings that HDAC inhibition enhances long-term memory, but short-term memory is unaffected (Stefanko et al., 2009).
Figure 4.
Loss of HDAC3/NCoR interaction enhances long-term OLM and ORM formation but has no effect on short-term memory. A, Mice received subthreshold training (3 min) in an environment with two identical objects and received a retention test 90 min (B) or 24 h (C) later in which one object is moved to a new location. B, Subthreshold training did not result in significant short-term memory by either genotype when tested 90 min later (n = 11–13 per group). C, However, DADm mice showed a significant preference for the novel object location 24 h after training compared with wild types (n = 9–10 per group; **p < 0.005). D, Mice received subthreshold training (3 min) in an environment with two identical objects and received a retention test 24 h later in which one object is replaced with a novel one (ORM). Schematic describes methods for E. E, DADm mice showed a significant preference for the novel object itself 24 h after training compared with wild types (n = 12–18 per group; *p < 0.05).
We next examined whether loss of NCoR/HDAC3 interactions affected long-term memory in the ORM task. Because these are traditional knock-in mice, mutant NCoR is present in all cells expressing NCoR. Thus, we predicted that DADm mice would exhibit enhanced memory in the ORM task (Fig. 4D) as well. Figure 4E shows that, after subthreshold training (3 min), DADm mice (n = 18) showed a greater preference for the novel object than the wild-type mice (n = 12) on test day (t(28) = 2.19; p = 0.04). These data suggest that brain regions mediating ORM, such as insular and perirhinal cortex, are also regulated by NCoR/HDAC3. Thus, disruption of the interaction between NCoR and HDAC3, which is sufficient to abrogate HDAC3 activity, results in similar effects as HDAC inhibition by enhancing long-term, but not short-term, memory.
The HDAC inhibitor RGFP136 affects HDAC4 expression and histone acetylation
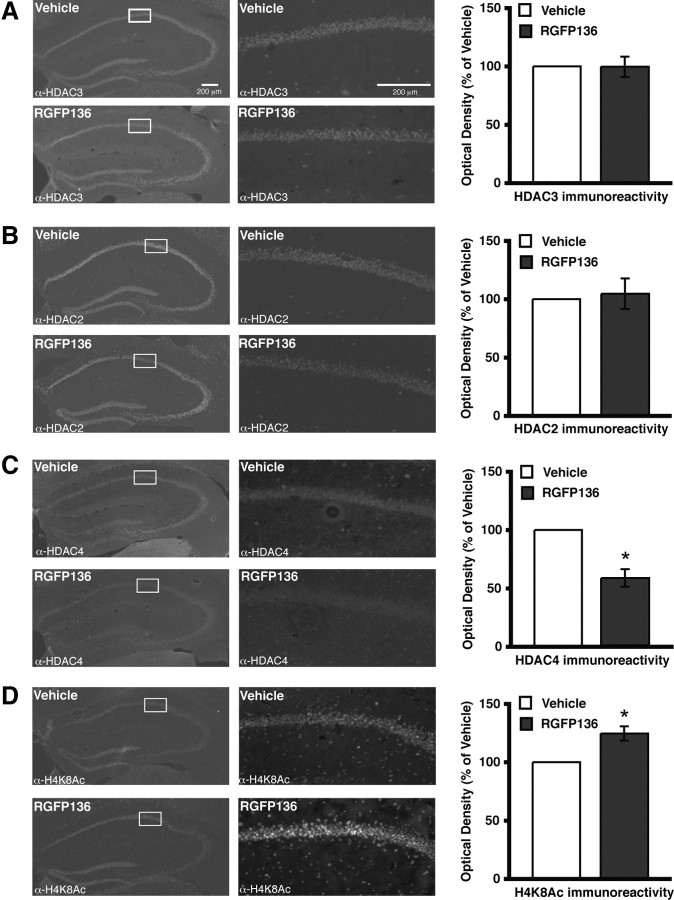
A new pimelic diphenylamide HDAC inhibitor, RGFP136, has been characterized as a class I HDAC inhibitor with greatest inhibition of HDAC3 (Rai et al., 2010). Using this compound, we tested whether acute inhibition of HDAC3 produced similar changes to that observed in the HDAC3flox/flox mice with respect to HDAC2–HDAC4 expression as well as histone acetylation. Brains from C57BL/6 mice with bilateral hippocampal cannulae were collected 2 h after 0.5 μl infusions of RGFP136 (1.25 ng/side) or vehicle. The drug infusion does not alter neuronal morphology compared with vehicle as visualized by DAPI staining (data not shown). HDAC3 nuclear immunoreactivity is similar in drug-infused mice compared with vehicle-treated mice (Fig. 5A, bottom right). HDAC2 expression is unchanged (Fig. 5B). Similar to results obtained from HDAC3flox/flox mice shown in Figure 1C, there is a loss of HDAC4 expression (F(1,5) = 8.35; p = 0.03) (Fig. 5C). To determine the effect of RGFP136 on histone acetylation, H4K8Ac was examined. As predicted, RGFP136 infusion resulted in an increase of immunoreactivity for H4K8Ac compared with vehicle (F(1,6) = 6.60; p = 0.04) (Fig. 5D). These findings mirror the results from HDAC3flox/flox mice (Fig. 1) and further support that HDAC3 may be involved in HDAC4 expression and inhibition of HDAC3 results in increased histone acetylation.
Figure 5.
Intrahippocampal RGFP136 infusions increases histone acetylation. Images on left are 4×, and 20× magnifications of the regions boxed in white are on the right. Histograms depict quantification of optical density as a percentage of vehicle. A, HDAC3 immunoreactivity is unaltered in area of infusion 2 h after RGFP136 treatment compared with vehicle. B, Representative images show HDAC2 immunoreactivity in dorsal hippocampus is also unchanged by drug treatment. C, However, HDAC4 nuclear immunoreactivity is decreased in the region of the RGFP136 infusion. *p < 0.05. D, Furthermore, acetylation at H4K8 is increased in RGFP136-infused mice compared with those treated with vehicle. *p < 0.05.
RGFP136 treatment leads to enhanced long-term memory for object recognition and location
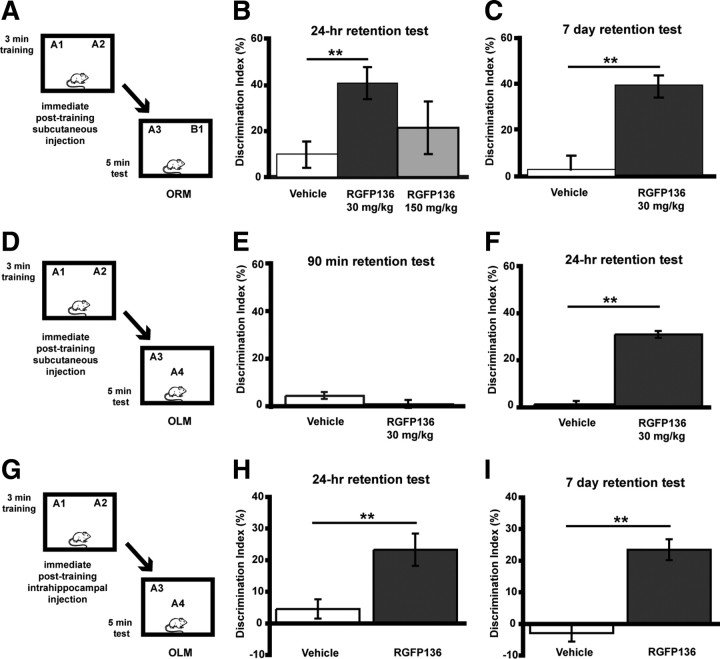
Next, we examined the ability of RGFP136 to modulate long-term memory. Mice were given a subthreshold 3 min training period (Fig. 6A), followed immediately by subcutaneous injection of RGFP136 (30 or 150 mg/kg) or vehicle. As shown in Figure 6B, mice receiving the 30 mg/kg dose (n = 9; DI = 40.7 ± 7.1%) exhibited a significantly greater preference for the novel object than vehicle-treated controls (n = 7; DI = 12.4 ± 5.7%; F(1,14) = 0.48; p = 0.01). Because animals treated with 150 mg/kg RGFP136 did not exhibit significantly enhanced preference for the novel object (n = 9; DI = 21.5 ± 11.5%; F(1,14) = 4.83; p = .530), only the low dose (30 mg/kg) was used to test the persistence of memory for the familiar object 7 d after the initial exposure in a different set of mice. Mice treated with 30 mg/kg RGFP136 immediately after a 3 min training period and tested 7 d later (n = 10; DI = 38.8 ± 4.6%) showed significant preference for the novel object (t(17) = 2.06, p < 0.05) compared with vehicle controls (n = 9; DI = 2.5 ± 6.3%) (Fig. 6C). Groups did not differ in total exploration time of the two objects during either the training or retention test (data not shown). Next, we examined OLM after RGFP136 systemic delivery. Animals were tested for short-term memory 90 min after training. No significant preference for the object in the novel location was evident in either the RGFP136 or vehicle-treated groups (RGFP136, n = 10, DI = 0.5 ± 1.4%; vehicle, n = 9, DI = 4.1 ± 1.4%; t(17) = 1.835; p > 0.05) (Fig. 6E). In a different set of mice to examine long-term memory at 24 h (mice were also given a 3 min training period), mice treated with posttraining RGFP136 (30 mg/kg, s.c.) exhibited significant preference for the object in the novel location compared with vehicle controls (F(1,17) = 192.21; p < 0.001) (Fig. 6F). These findings mirror the effects in HDAC3flox/flox mice as well as a recent study using a general HDAC inhibitor (Stefanko et al., 2009), in which enhanced memory is demonstrated in long-term, but not short-term, memory tests.
Figure 6.
The HDAC inhibitor RGFP136 enhances long-term memory for ORM and OLM. A, Mice received subthreshold training (3 min) in an environment with two identical objects immediately followed by subcutaneous injection of RGFP136 and received a retention test 24 h (B) or 7 d (C) later in which one object is replaced with a novel one. B, Mice treated with the 30 mg/kg dose exhibited a significant preference for the novel object than vehicle-treated controls, whereas 150 mg/kg treatment resulted in memory no different from vehicle (n = 7–9 per group; two-way ANOVA, **p < 0.01). C, In a different set of mice, the persistence of this enhanced memory was examined. Mice receiving subcutaneous injection of RGFP136 (30 mg/kg) exhibited significantly increased exploration of the novel object compared with vehicle-treated mice during a 7 d retention test (n = 9–10 per group; **p < 0.01). D, Mice received subthreshold training (3 min) in an environment with two identical objects immediately followed by a subcutaneous injection of RGFP136 (30 mg/kg) or vehicle and received a retention test 90 min (E) or 24 h (F) later in which one object is moved to a new location. E, Subthreshold training did not result in significant short-term memory after RGFP136 (30 mg/kg) or vehicle injections when tested 90 min later. F, Mice treated with the 30 mg/kg RGFP136 exhibited significant preference for the object in the novel location compared with vehicle-treated controls (n = 9–10 per group; **p < 0.001). G, Mice received subthreshold training (3 min) in an environment with two identical objects followed immediately by intrahippocampal infusion of RGFP136 and received a retention test 24 h (H) or 7 d (I) later in which one object is moved to a new location. H, Intrahippocampal RGFP136 treatment led to significant preference for the novel object location 24 h after subthreshold training (n = 7 per group; **p < 0.01). I, In a different set of mice, the persistence of this enhanced memory was examined. Mice receiving intrahippocampal RGFP136 also displayed a significant preference for the novel object location compared with vehicle-treated mice during a 7 d retention test (n = 8 per group; **p < 0.001).
To test the effect of site-specific delivery of RGFP136 on long-term memory, we examined its ability to modulate memory for object location (OLM) (Fig. 6G). Mice fitted with bilateral hippocampal cannulae received either 0.5 μl infusions of RGFP136 (n = 7; 1.25 ng/side) or vehicle (n = 7) immediately after a subthreshold 3 min training period. Mice treated with RGFP136 showed a greater preference for object in the novel location 24 h later than controls treated with vehicle (t(12) = 3.16; p = 0.008) (Fig. 6H). In addition, a separate group of mice were given a subthreshold 3 min training period and then a 7 d OLM retention test. RGFP136-treated mice (n = 8) showed significantly greater preference for object in the novel location than vehicle-treated controls (n = 8; t(14) = 6.10; p < 0.001) (Fig. 6I). Groups did not differ in total exploration time of the two objects during either the training or retention test (data not shown). We next examined whether intrahippocampal delivery of RGFP136 affected long-term memory in a standard novel object recognition task (ORM; data not shown). Both RGFP136 (DI = 0.3 ± 4.2%) and vehicle-treated mice (DI = 0.9 ± 6.7%) did not show a preference for the novel object 24 h later (p > 0.05), demonstrating that site-specific delivery only enhances hippocampus-dependent long-term memory. In summary, RGFP136 treatment leads to similar effects on long-term memory when delivered to the dorsal hippocampus as the HDAC3 dorsal hippocampus deletions.
RGFP136 enhancement of long-term memory for object location requires CBP
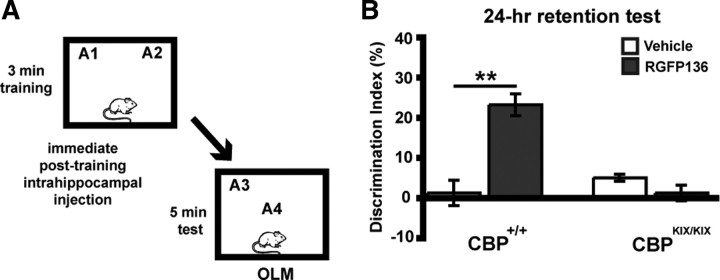
HDAC3 is found in the nucleus and cytoplasm in which it can regulate transcription of genes as well as perform other nontranscriptional functions (e.g., deacetylate nonhistone proteins) (for review, see Karagianni and Wong, 2007). To test whether the enhancements in memory formation observed in Figure 6 may be attributable to the transcription of genes necessary for long-term memory, we used genetically modified CBP mutant mice carrying a triple-point mutation in the phospho-CREB (KIX) binding domain of CBP (CBPKIX/KIX mice) (Kaspar et al., 2002). Previously, we have demonstrated that HDAC inhibition, by either sodium butyrate or trichostatin A, enhances hippocampal synaptic plasticity via a CREB/CBP interaction (Vecsey et al., 2007). To see whether RGFP136 is acting via a similar mechanism, we infused this drug into the dorsal hippocampus of CBPKIX/KIX and CBP+/+ mice after a 3 min subthreshold training period and tested the effects on OLM (Fig. 7A). We found overall effects of genotype (F(1,24) = 17.12; p < 0.001), drug treatment (F(1,24) = 17.15; p < 0.001), as well as interaction of genotype × drug treatment (F(1,24) = 34.41; p < 0.001). RGFP136-treated CBP+/+ mice (n = 5) showed significantly greater preference for object in the novel location than vehicle-treated controls (n = 6; p < 0.001) (Fig. 7B). However, RGFP136 had no effect on novel location preference in the CBPKIX/KIX mice (n = 8 for vehicle and n = 9 for RGFP136; p = 1.0). Groups did not differ in total exploration time of the two objects during either the training or retention test (data not shown). These results indicate that RGFP136 enhances long-term memory through a CBP-dependent mechanism.
Figure 7.
The HDAC inhibitor RGFP136 requires CBP to enhance long-term memory of OLM. A, Mice received subthreshold training (3 min) in an environment with two identical objects immediately followed by intrahippocampal infusions of RGFP136 (1.25 ng/side) or vehicle (0.5 μl/side) and received a retention test 24 h later in which one object is moved to a new location. B, Wild-type CBP+/+ mice that received intrahippocampal RGFP136 immediately after training showed significant long-term memory for the object location compared with vehicle-treated mice. CBPKIX/KIX mice showed no effect of drug treatment (n = 5–9 per group; two-way ANOVA, **p < 0.001).
Deletion of HDAC3 in dorsal hippocampus requires Nr4a2 expression to enhance long-term memory
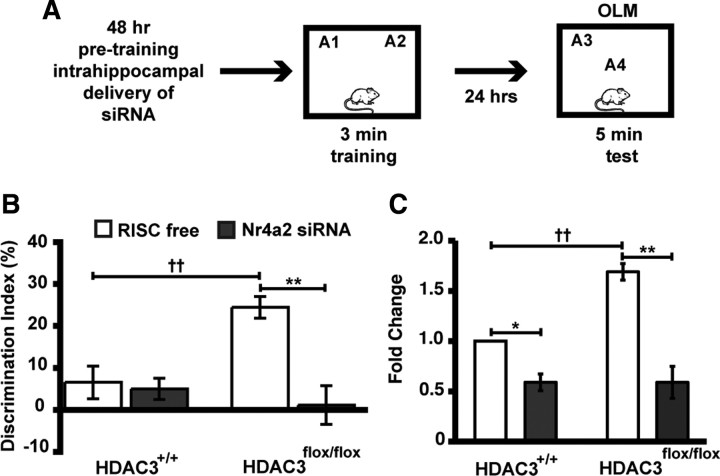
Nr4a2 is a CREB-dependent gene implicated in long-term memory (Peña de Ortiz et al., 2000; von Hertzen and Giese, 2005; Colón-Cesario et al., 2006; Vecsey et al., 2007). We showed in Figure 2 that subthreshold training induced greater Nr4a2 gene expression in the dorsal hippocampi of HDAC3flox/flox mice compared with wild-type controls. To determine whether this increase in Nr4a2 expression is necessary for enhanced long-term memory in HDAC3flox/flox mice, we infused siRNA targeting Nr4a2 48 h before training (Fig. 8A). We found an overall effect of siRNA treatment (F(1,35) = 11.98; p = 0.001), genotype (F(1,35) = 5.08; p = 0.03), and an interaction of genotype × siRNA treatment (F(1,35) = 7.31; p = 0.005). HDAC3flox/flox mice infused with RISC-free siRNA (n = 10) demonstrated significant preference for the object in the novel location, which was blocked by Nr4a2 siRNA treatment (n = 10; p < 0.001) (Fig. 8B). HDAC3+/+ mice did not display preference for the novel location after either RISC-free or Nr4a2 siRNA treatment (n = 10 for RISC free and n = 9 for Nr4a2 siRNA; p = 1.0). Two hours after testing, brains were collected to determine levels of Nr4a2 mRNA in the dorsal hippocampus. Significant effects were found for genotype (F(1,8) = 11.24; p = 0.01), siRNA treatment (F(1,8) = 56.45; p < 0.001), and an interaction of genotype × siRNA treatment (F(1,8) = 10.82; p = 0.01). Figure 8C shows that the infusion of Nr4a2 siRNA significantly decreased Nr4a2 expression in wild-type and HDAC3flox/flox mice compared with RISC-free siRNA-infused controls (wild-type, p = 0.02; HDAC3flox/flox, p < 0.001). In addition, HDAC3flox/flox mice treated with RISC-free siRNA also demonstrated an increased induction of Nr4a2 mRNA after the long-term memory test (p = 0.002 vs HDAC3+/+ RISC free). This enhancement posttest is similar to increases seen after training (Fig. 2B). These data yield a potential mechanism for the negative regulation of long-term memory by HDAC3.
Figure 8.
Nr4a2 siRNA attenuates the long-term memory enhancement observed in HDAC3flox/flox mice. A, At 48 h after infusions of Nr4a2 or RISC-free siRNA, HDAC3flox/flox and HDAC3+/+ mice received subthreshold training (3 min) in an environment with two identical objects and received a retention test 24 h later in which one object is moved to a new location. B, HDAC3flox/flox mice infused with RISC free (n = 10) exhibited significant memory for object location compared with HDAC3+/+ mice (††p = 0.001), which was blocked by Nr4a2 siRNA treatment (n = 9–10 per group; **p < 0.001). C, At 2 h after testing, quantitative RT-PCR shows that Nr4a2 siRNA treatment significantly reduced Nr4a2 expression in both HDAC3flox/flox and HDAC3+/+ mice (n = 3 per group; **p < 0.001 and *p < 0.05 vs respective RISC-free siRNA controls). HDAC3flox/flox mice also exhibited an increased induction of Nr4a2 mRNA after the long-term memory test (††p = 0.002 vs HDAC3+/+ RISC free).
Discussion
The results from this study demonstrate that HDAC3 is a critical negative regulator of long-term memory formation. Focal homozygous gene deletion of HDAC3 resulted in not only loss of HDAC3 but also a significant decrease in HDAC4 expression. Neurons lacking HDAC3 had increased histone acetylation of histone H4 lysine 8 (H4K8Ac), which correlated with increased Nr4a2 and c-fos expression in the area of the focal HDAC3 deletion in the dorsal hippocampus. Focal homozygous deletion of HDAC3 in the dorsal hippocampus lead to facilitated long-term memory formation after a subthreshold training period. This subthreshold training period failed to yield long-term memory in control animals. The genetic approach to examine the role of HDAC3 in long-term memory formation was complemented with a pharmacological approach using an HDAC inhibitor shown to be more selective for HDAC3 than other class I HDACs. This compound, called RGFP136, when delivered to the dorsal hippocampus, resulted in decreased HDAC4 expression, increased H4K8Ac, and also significantly facilitated long-term memory formation via a CBP-dependent manner in the hippocampus. Our final experiment demonstrated that HDAC3 may modulate long-term memory formation via the expression of the immediate early gene and transcription factor Nr4a2. Together, these genetic and neuropharmacological approaches identify HDAC3 as a critical negative regulator of memory.
HDAC3 is expressed throughout the brain, with particularly strong gene expression in the hippocampus (Broide et al., 2007). However, no study to date has examined the role of HDAC3 in the brain. Previous in vitro studies have shown that HDAC3 and HDAC4 interact with each other in large complexes (Grozinger and Schreiber, 2000; Fischle et al., 2002). Interactions between HDAC3 and HDAC4 create a functional complex involved in transcriptional regulation. HDAC4 is considered to be in the “inactive state” until bound to HDAC3, an interaction necessary for its enzymatic activity (Fischle et al., 2002). A study by Lahm et al. (2007) supported previous findings that class IIa HDACs (HDAC4, HDAC5, HDAC7, and HDAC9) are inactive on acetylated substrates, thus distinguishing them from class I HDACs (HDAC1, HDAC2, HDAC3, and HDAC8). This has called into question the catalytic activity of class IIa HDACs. An equally reasonable idea is that the natural substrate of these enzymes has not been identified. In any case, the interaction between HDAC4 and HDAC3 is facilitated by corepressor proteins NCoR and SMRT, which form a large complex with HDACs and other proteins (Fischle et al., 2002). HDAC4 and HDAC3 bind independently to different domains of SMRT and NCoR, but the proximity allows for interactions of HDAC3 and HDAC4 proteins.
To complement our genetic and pharmacological approach to study HDAC3, we also used genetically modified NCoR mutant mice. These mice, referred to as DADm mice, carry a single amino acid substitution (Y478A) in the DAD of NCoR, which results in a mutant NCoR protein that is unable to associate with or activate HDAC3 (Guenther et al., 2001; Ishizuka and Lazar, 2005; Alenghat et al., 2008). When given a subthreshold training period, DADm homozygous knock-in mice exhibited significant long-term memory compared with wild-type littermates, which failed to show any long-term memory. These data support the idea that a functional complex between NCoR and HDAC3 is required to repress long-term memory formation.
Lahm et al. (2007) showed that a critical residue for HDAC3 activity is a tyrosine at amino acid 298, which if mutated to a histidine (Y298H) completely abrogates enzymatic function. Interestingly, HDAC4 and other class IIa enzymes normally have a histidine at this position, which provides a potential reason why HDAC4 has such poor enzymatic activity on traditional substrates. Commonly used HDAC inhibitors, such as valproic acid, sodium butyrate, phenylbutyrate, and suberoylanilide hydroxamic acid (SAHA), have been shown to greatly inhibit class I HDACs (HDAC1, HDAC2, HDAC3, HDAC8) with little effect on the class IIa HDAC family members (HDAC4, HDAC5, HDAC7, HDAC9) (Kilgore et al., 2010). This suggests that inhibition of class I HDACs are critical for the reported effects of HDAC inhibition, such as the enhancement of cognition. Indeed, HDAC2, which has been shown to negatively regulate memory formation, has been implicated as a specific target of commonly used HDAC inhibitors (Guan et al., 2009).
Recently, a new class of HDAC inhibitor called pimelic diphenylamide compounds has been identified (Chou et al., 2008; Xu et al., 2009; Rai et al., 2010). These inhibitors are slow-on/slow-off, competitive tight-binding inhibitors that specifically target class I HDACs, with the greatest inhibitory effect on HDAC3 (Chou et al., 2008; Xu et al., 2009). RGFP136, used in these studies, has an IC50 of 5.2 μm for HDAC1, 3.0 μm for HDAC2, and 0.4 μm for HDAC3 using purified recombinant HDACs. After systemic subcutaneous injection, the maximum drug concentration (Cmax) in the brain is ∼1.7 μm for a 30 mg/kg dose. This suggests that, after systemic administration, as in the data shown in Figure 6, RGFP136 is at a sufficient concentration in the brain to inhibit HDAC3 but perhaps not HDAC1 or HDAC2. Furthermore, the immunofluorescence data indicate that RGFP136 disrupts HDAC4 expression, with no effect on HDAC2 expression. Thus, although RGFP136 affects class I HDACs, the effects observed in this study are most likely via HDAC3. Behaviorally, when delivered site specifically to the dorsal hippocampus, RGFP136 transformed a learning event that does not result in long-term memory into an event that now does lead to long-term memory. Furthermore, this facilitation of long-term memory via RGFP136 resulted in persistent long-term memory observed 7 d later when normal long-term memory retrieval for object location fails. Subcutaneous injection of RGFP136 also facilitated long-term memory for object location (Fig. 6F) as well as long-term memory for a familiar object (Fig. 6B).
Importantly, we found that, in the hippocampus, RGFP136 requires CBP to facilitate long-term memory formation. CBPKIX/KIX mice, which contain a mutation in the phospho-CREB (KIX) binding domain of CBP (Kaspar et al., 2002), failed to exhibit significant long-term memory for object location when RGFP136 was delivered to the dorsal hippocampus (Fig. 7B). These results suggest that RGFP136 is functioning via a CBP-dependent mechanism to regulate transcription required for hippocampus-dependent long-term memory.
Despite the consistent enhancements of long-term memory by deletion of HDAC3, short-term memory was unaffected (Figs. 4B, 6E). A major difference between these forms of memory is that, in general, transcription is essential for the formation of long-term memory but not short-term memory. We and others have found that HDAC inhibition by sodium butyrate, trichostatin A (TSA), or SAHA have no effect on short-term memory (Korzus et al., 2004; Levenson et al., 2004; Yeh et al., 2004; Vecsey et al., 2007; Stefanko et al., 2009). Additionally, homozygous knock-in NCoR mice in this study exhibit enhanced long-term memory but not enhanced short-term memory. In contrast, Guan et al. (2009) found enhanced short-term memory in HDAC2 knock-out mice in which the knock-out is generated by crossing HDAC2–FLOX mice with nestin–Cre transgenic mice. Thus, the differences observed on short-term memory may be attributable to either functional differences between HDAC3 and HDAC2 or developmental/compensation effects in HDAC2 knock-out mice. In any case, it is still remarkable that traditional knock-out (HDAC2) or knock-in (NCoR) mice exhibit similar enhanced long-term memory phenotypes as acute disruption of HDAC activity by pharmacological manipulation.
A major finding in this study is the relationship of hippocampal HDAC3 deletion with increased Nr4a2 expression. Nr4a2 is a CREB-dependent gene that has been implicated in long-term memory (Peña de Ortiz et al., 2000; von Hertzen and Giese, 2005; Colón-Cesario et al., 2006; Vecsey et al., 2007). We have demonstrated previously that Nr4a2 expression is enhanced by the HDAC inhibitor TSA during memory consolidation (Vecsey et al., 2007). In this study, we also observed enhanced Nr4a2 expression in HDAC3flox/flox mice after learning (Fig. 2C). It has been suggested that HDACs may terminate the CREB-dependent transcription for this gene (Fass et al., 2003), and thus the removal of HDAC3 allows transcription to be maintained for a longer period. Activation of Nr4a2 is critical for the expression of long-term memory, as demonstrated by our behavioral study using siRNA (Fig. 8). HDAC3flox/flox mice with a homozygous deletion of HDAC3 in the dorsal hippocampus failed to exhibit enhanced long-term memory when Nr4a2 siRNA was infused into the area of HDAC3 deletion before training. These findings are in agreement with another study that found hippocampal infusions of Nr4a2 antisense, which did not affect c-fos expression, caused impaired long-term memory for a spatial discrimination task (Colón-Cesario et al., 2006). These data suggest a mechanism by which the loss of HDAC3 enhances long-term memory by allowing increased and/or prolonged CREB/CBP-dependent transcription of Nr4a2.
In summary, the experiments presented here demonstrate that HDAC3 is a critical negative regulator of long-term memory formation. RGFP136, a pimelic diphenylamide compound, represents a promising pharmacotherapeutic approach for cognitive impairments. RGFP136 and genetic manipulation of HDAC3 (via HDAC3flox/flox and DADm mice) had similar effects at the molecular and behavioral levels. It is likely that HDAC3 performs its role in memory processes via its interactions with NCoR as well as HDAC4. However, future studies are necessary to determine the exact nature of these interactions and their effects on acetylation, gene expression, and learning and memory.
Footnotes
This work was supported by the Whitehall Foundation, National Institute of Mental Health Grant R01MH081004, and National Institute on Drug Abuse Grant R01DA025922 (all to M.A.W.) and National Research Service Award Predoctoral Fellowship F31MH85494 (R.M.B.). The generation of the HDAC3–FLOXed mice was supported by National Institutes of Health Grant DK43806 (M.A.L.). We thank the Functional Genomics and Gene Targeting Core and the Transgenic Mouse Core of the Penn Diabetes and Endocrinology Research Center (National Institutes of Health Grant DK19525) for generating targeted embryonic stem cells and knock-in mice, respectively. We also thank Steven Blue, Theresa Lien, and Angelina Sylvain for their technical assistance.
J.R.R. and S.J. are employed by Repligen Corporation.
References
- Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bućan M, Ahima RS, Kaestner KH, Lazar MA. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderas I, Rodriguez-Ortiz CJ, Salgado-Tonda P, Chavez-Hurtado J, McGaugh JL, Bermudez-Rattoni F. The consolidation of object and context recognition memory involve different regions of the temporal lobe. Learn Mem. 2008;15:618–624. doi: 10.1101/lm.1028008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RM, Wood MA. Beyond transcription factors: the role of chromatin modifying enzymes in regulating transcription required for memory. Learn Mem. 2008;15:460–467. doi: 10.1101/lm.917508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Barad M. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction, and reconsolidation of conditioned fear. Learn Mem. 2008;15:39–45. doi: 10.1101/lm.801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1–11 in the rat brain. J Mol Neurosci. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- Burger C, Gorbatyuk OS, Velardo MJ, Peden CS, Williams P, Zolotukhin S, Reier PJ, Mandel RJ, Muzyczka N. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol Ther. 2004;10:302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Chou CJ, Herman D, Gottesfeld JM. Pimelic diphenylamide 106 is a slow, tight-binding inhibitor of class I histone deacetylases. J Biol Chem. 2008;283:35402–35409. doi: 10.1074/jbc.M807045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Cesario WI, Martínez-Montemayor MM, Morales S, Félix J, Cruz J, Adorno M, Pereira L, Colón N, Maldonado-Vlaar CS, Peña de Ortiz S. Knockdown of Nurr1 in the rat hippocampus: implications to spatial discrimination learning and memory. Learn Mem. 2006;13:734–744. doi: 10.1101/lm.407706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanselow MS. Contextual fear, gestalt memories, and the hippocampus. Behav Brain Res. 2000;110:73–81. doi: 10.1016/s0166-4328(99)00186-2. [DOI] [PubMed] [Google Scholar]
- Fass DM, Butler JE, Goodman RH. Deacetylase activity is required for cAMP activation of a subset of CREB target genes. J Biol Chem. 2003;278:43014–43019. doi: 10.1074/jbc.M305905200. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T, Lazar MA. The nuclear receptor corepressor deacetylase activating domain is essential for repression by thyroid hormone receptor. Mol Endocrinol. 2005;19:1443–1451. doi: 10.1210/me.2005-0009. [DOI] [PubMed] [Google Scholar]
- Karagianni P, Wong J. HDAC3: taking the SMRT-N-CoRrect road to repression. Oncogene. 2007;26:5439–5449. doi: 10.1038/sj.onc.1210612. [DOI] [PubMed] [Google Scholar]
- Kaspar BK, Vissel B, Bengoechea T, Crone S, Randolph-Moore L, Muller R, Brandon EP, Schaffer D, Verma IM, Lee KF, Heinemann SF, Gage FH. Adeno-associated virus effectively mediates conditional gene modification in the brain. Proc Natl Acad Sci U S A. 2002;99:2320–2325. doi: 10.1073/pnas.042678699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, Rumbaugh G. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfí A, Koch U, De Francesco R, Steinkühler C, Gallinari P. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattal KM, Barrett RM, Wood MA. Systemic or intrahippocampal delivery of histone deacetylase inhibitors facilitates fear extinction. Behav Neurosci. 2007;121:1125–1131. doi: 10.1037/0735-7044.121.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, Sanchis-Segura C, Vo D, Lattal KM, Wood MA. Modulation of chromatin modification facilitates extinction of cocaine-induced conditioned place preference. Biol Psychiatry. 2010;67:36–43. doi: 10.1016/j.biopsych.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maren S, Holt W. The hippocampus and contextual memory retrieval in Pavlovian conditioning. Behav Brain Res. 2000;110:97–108. doi: 10.1016/s0166-4328(99)00188-6. [DOI] [PubMed] [Google Scholar]
- O'Keefe J. Do hippocampal pyramidal cells signal non-spatial as well as spatial information? Hippocampus. 1999;9:352–364. doi: 10.1002/(SICI)1098-1063(1999)9:4<352::AID-HIPO3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Peña de Ortiz S, Maldonado-Vlaar CS, Carrasquillo Y. Hippocampal expression of the orphan nuclear receptor gene hzf-3/nurr1 during spatial discrimination learning. Neurobiol Learn Mem. 2000;74:161–178. doi: 10.1006/nlme.1999.3952. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai M, Soragni E, Chou CJ, Barnes G, Jones S, Rusche JR, Gottesfeld JM, Pandolfo M. Two new pimelic diphenylamide HDAC inhibitors induce sustained frataxin upregulation in cells from Friedreich's ataxia patients and in a mouse model. PLoS One. 2010;5:e8825. doi: 10.1371/journal.pone.0008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozendaal B, Hernandez A, Cabrera SM, Hagewoud R, Malvaez M, Stefanko DP, Haettig J, Wood MA. Membrane-associated glucocorticoid activity is necessary for modulation of long-term memory via chromatin modification. J Neurosci. 2010;30:5037–5046. doi: 10.1523/JNEUROSCI.5717-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Mizumori SJ. Learning-related development of context-specific neuronal responses to places and events: the hippocampal role in context processing. J Neurosci. 2006;26:3154–3163. doi: 10.1523/JNEUROSCI.3234-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanko DP, Barrett RM, Ly AR, Reolon GK, Wood MA. Modulation of long-term memory for object recognition via HDAC inhibition. Proc Natl Acad Sci U S A. 2009;106:9447–9452. doi: 10.1073/pnas.0903964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hertzen LS, Giese KP. Memory reconsolidation engages only a subset of immediate-early genes induced during consolidation. J Neurosci. 2005;25:1935–1942. doi: 10.1523/JNEUROSCI.4707-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Xia X, Weiss RE, Refetoff S, Yen PM. Distinct and histone-specific modifications mediate positive versus negative transcriptional regulation of TSHalpha promoter. PLoS One. 2010;5:e9853. doi: 10.1371/journal.pone.0009853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Soragni E, Chou CJ, Herman D, Plasterer HL, Rusche JR, Gottesfeld JM. Chemical probes identify a role for histone deacetylase 3 in Friedreich's ataxia gene silencing. Chem Biol. 2009;16:980–989. doi: 10.1016/j.chembiol.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh SH, Lin CH, Gean PW. Acetylation of nuclear factor-kappaB in rat amygdala improves long-term but not short-term retention of fear memory. Mol Pharmacol. 2004;65:1286–1292. doi: 10.1124/mol.65.5.1286. [DOI] [PubMed] [Google Scholar]