Abstract
The caudal homeobox (cdx) gene family is critical for specification of caudal body formation and erythropoiesis. In zebrafish, cdx4 expression is controlled by the Wnt pathway, but the molecular mechanism of this regulation is not fully understood. Here, we provide evidence that Tcf3 suppresses cdx4 expression through direct binding to multiple sites in the cdx4 gene regulatory region. Tcf3 requires corepressor molecules such as Groucho (Gro)/TLE and HDAC1 for activity. Using zebrafish embryos and cultured mammalian cells, we show that the transcription factor E4f1 derepresses cdx4 by dissociating corepressor proteins from Tcf3 without inhibiting its binding to cis-regulatory sites in the DNA. Further, the E3 ubiquitin ligase Lnx2b, acting as a scaffold protein irrespective of its enzymatic activity, counteracts the effects of E4f1. We propose that the modulation of Tcf3 repressor function by E4f1 assures precise and robust regulation of cdx4 expression in the caudal domain of the embryo.
Keywords: caudal body domain, Cdx4, E4f1, Tcf3, Wnt signalling
Introduction
The process of anteroposterior (AP) axis specification is comparatively well understood in amphibians and fish where a caudal-to-rostral gradient of Wnt activity is instrumental in AP patterning (MacDonald et al, 2009). Canonical Wnt signalling stabilizes cytosolic β-catenin, which translocates into the nucleus and binds to LEF/TCF transcription factors to mediate target gene expression. Four evolutionary well-conserved factors (Lef1/LEF1, Tcf3/TCF7L1, Tcf4/TCF7L2 and Tcf7/TCF1) are involved in Wnt/β-catenin signalling. Although LEF/TCF family members exhibit some functional redundancy (Galceran et al, 1999; Nagayoshi et al, 2008), individual LEF/TCF factors often behave differently during embryonic development. In Xenopus, XTcf3 establishes early dorsal polarity while XLef1 has important roles in mesoderm patterning (Brannon et al, 1997; Roël et al, 2002). In zebrafish, Lef1 regulates brain neurogenesis (Lee et al, 2006; Bonkowsky et al, 2008) and pectoral fin outgrowth in cooperation with Tcf7 (Nagayoshi et al, 2008), and Tcf3 specifies anterior neural tissues (Kim et al, 2000; Dorsky et al, 2003).
The distinctive functional differences between LEF/TCF family members seem to be due to their intrinsic properties as transcriptional activators or repressors. LEF/TCF factors act as repressors when bound to corepressor proteins such as Gro/TLE and HDAC, and this repression is relieved by β-catenin recruitment (Cavallo et al, 1998; Brantjes et al, 2001; Hovanes et al, 2001; Hurlstone and Clevers, 2002; MacDonald et al, 2009). However, Lef1 cannot substitute for Tcf3 repressor function in AP patterning (Kim et al, 2000; Dorsky et al, 2003), suggesting that the primary embryonic function of Lef1 is transcriptional activation in the presence of β-catenin (Gat et al, 1998; Hovanes et al, 2001). In contrast, Tcf3 function involves mostly or entirely cooperation with corepressors to act as a transcriptional repressor in embryonic domains in which Wnt signals are limiting (Kim et al, 2000; Dorsky et al, 2003; Gribble et al, 2009). In the developing zebrafish and mouse embryo, AP body patterning largely depends on the transcriptional repressor function of Tcf3 (Kim et al, 2000; Dorsky et al, 2003; Merrill et al, 2004). This is most clearly indicated by the fact that the tcf3a mutant headless (hdl) shows anterior truncations due to overall posteriorization, similar to embryos with hyperactivated Wnt signalling (Kim et al, 2000). In contrast, when Wnt signalling is inhibited, zebrafish embryos show truncated tails reminiscent of the loss-of-function phenotype of cdx1a and cdx4 (Shimizu et al, 2005).
The evolutionary conserved Cdx/caudal homeobox transcription factors, known Wnt target genes (Shimizu et al, 2005; Pilon et al, 2006, 2007), have critical roles in patterning of caudal structures, early endoderm specification, gut AP patterning, establishment of the intestinal epithelium and haematopoiesis, at least in part by regulating expression of certain Hox genes (Isaacs et al, 1998; van den Akker et al, 2002; Davidson et al, 2003; Bansal et al, 2006; Cheng et al, 2008; Flores et al, 2008; Chen et al, 2009; Faas and Isaacs, 2009; Young and Deschamps, 2009; Young et al, 2009; Gao and Kaestner, 2010). Aberrant expression of human CDX2 is frequently detected in AML and paediatric ALL patients, and seems to be causative of leukaemia development via altering HOX gene expression (Scholl et al, 2007; Riedt et al, 2009; Thoene et al, 2009). Additionally, Cdx genes function as key factors during haematopoiesis, and ectopic expression of Cdx4 can trigger leukaemogenesis in mice (Bansal et al, 2006; Wang et al, 2008). Thus, the expression of Cdx genes must be tightly regulated during development, but the mechanism leading to the induction of Cdx genes in a precise pattern in the embryo is not fully understood.
In zebrafish, Cdx4 is the major factor that governs caudal tissue specification and primitive erythropoiesis (Davidson et al, 2003). Here, we provide evidence that Tcf3 in cooperation with Gro/TLE and HDAC1 suppresses cdx4 expression through direct binding to the cdx4 gene regulatory region. We show that E4f1, previously characterized as a transcriptional repressor of cyclin A2 (Fajas et al, 2001), derepresses cdx4 by disrupting a complex between corepressor proteins and Tcf3 while leaving Tcf3 bound to its cognate sites in the cdx4 regulatory region. As a further mechanism in precise modulation of cdx4 expression, we find that the multi-PDZ domain-containing E3 ubiquitin ligase Lnx2b (one of the zebrafish homologues of Ligand of Numb protein X-2; Nie et al, 2002; Ro and Dawid, 2009; Ro and Dawid, 2010) counteracts E4f1 function by stabilizing the Tcf3–Gro/TLE–HDAC1 repressor complex. These observations introduce a novel mechanism that modulates Tcf3 repressor activity and through this the output of the Wnt signalling pathway in AP patterning. We propose that this mechanism contributes to the establishment of the precise cdx4 expression domain in the zebrafish embryo, assuring normal development of the caudal body region and erythropoiesis.
Results
E4f1 is a positive factor in tail development
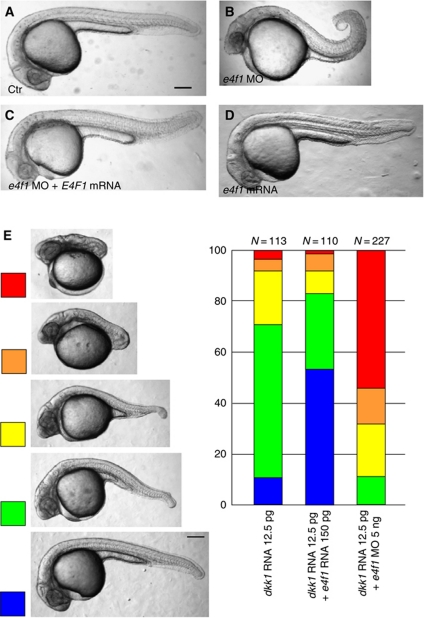
Our interest in the function of E4f1 started from the observations that it is a binding partner of Lnx2b (previously designated Lnx-like), a factor we studied previously (Supplementary Figures S1 and S2) (Ro and Dawid, 2009, 2010). Besides having transcriptional repressor activity (Fajas et al, 2001), E4f1 has been reported to act as a BMI1 modulator (Chagraoui et al, 2006) and as an atypical ubiquitin ligase (Le Cam et al, 2006), suggesting multiple functions for E4f1 during development. While Lnx2b can act as an E3 ubiquitin ligase (Ro and Dawid, 2009), it did not modulate the stability or transcriptional repressor activity of E4f1 (Supplementary Figure S1A and B; unpublished data). Since early embryonic lethality of E4f1 mutant mice impeded further analysis of its developmental role (Le Cam et al, 2004), we exploited zebrafish for studying the function of E4f1. Zebrafish e4f1 is expressed maternally and zygotically without exhibiting significant spatio-temporal differences (Supplementary Figure S3). Injection of e4f1 translation-blocking morpholino (MO) caused developmental defects beginning by 11 h post-fertilization (h.p.f.), leading to delayed tail elongation and, starting from early somitogenesis, shortened and curled-up tails (Figure 1A and B). Because the e4f1 morphant phenotype was largely rescued by the injection of human E4F1 mRNA that does not contain the MO target site (Figure 1C), we conclude that the MO effects are specific to the function of E4f1 rather than reflecting non-specific effects. Further, these observations suggest that the function of E4f1 is conserved from teleosts to tetrapods. Overexpression of E4f1 by injection of zebrafish e4f1 mRNA at the same level that achieved rescue of morphants did not result in a visible phenotype by 24 h.p.f. (Figure 1D), possibly due to the fact that the embryo already contains substantial levels of E4f1 (Supplementary Figure S3).
Figure 1.
Depletion of E4f1 causes caudal defects. (A–C) The tail defects of e4f1 morphants (e4f1 MO; 5 ng) were largely rescued by injection of 100 pg of human E4F1 mRNA. Injected reagents are shown at bottom left. (B) Tail shortened and kinked in 74% of embryos (n=159). (C) Most embryos showed rescued tail (69%, n=160). (D) Overexpression of E4f1 by injection of a similar level of zebrafish e4f1 RNA as used in the rescue experiment (C; 100 pg) did not cause a visible phenotype. (E) A moderate amount of dkk1 mRNA (12.5 pg) was injected alone or together with e4f1 mRNA (150 pg) or e4f1 MO (5 ng). Tail defects were classified into five levels at 26 h.p.f. Enforced expression of E4f1 mitigated tail defects due to Wnt inhibition, whereas e4f1 MO injection aggravated the effect. Numbers of embryos (N at top) are based on three independent experiments. Lateral views of embryos at 26 h.p.f. Scale bar, 200 μm.
The development of the caudal domain of the embryo is controlled by several signalling pathways among which the Wnt pathway has an important position (Shimizu et al, 2005; Nordström et al, 2006; Iimura et al, 2009; Mallo et al, 2009). The formation of short tails by depleting E4f1 suggests that this factor has a positive influence on tail development, as does the Wnt pathway. An embryo with attenuated Wnt signalling might then represent a sensitized test object in which the effect of E4f1 can be analysed. This expectation is borne out by the data in Figure 1E. Classification of tail defects into five categories allowed us to quantify the results. As expected, injection of a moderate level of RNA encoding the Wnt inhibitor Dkk1 led to intermediate levels of tail shortening. These effects were substantially ameliorated by co-injection of e4f1 mRNA with over 50% of the embryos rescued to normal development; in contrast injection of the e4f1 MO strongly enhanced the tail phenotype, with over 50% of embryos showing complete loss of the tail (Figure 1E). These results strongly support our conclusion that E4f1 is a positive factor in tail development, and further suggest that it carries out this function in conjunction with the Wnt signalling pathway.
E4f1 together with Wnt regulates cdx4 expression
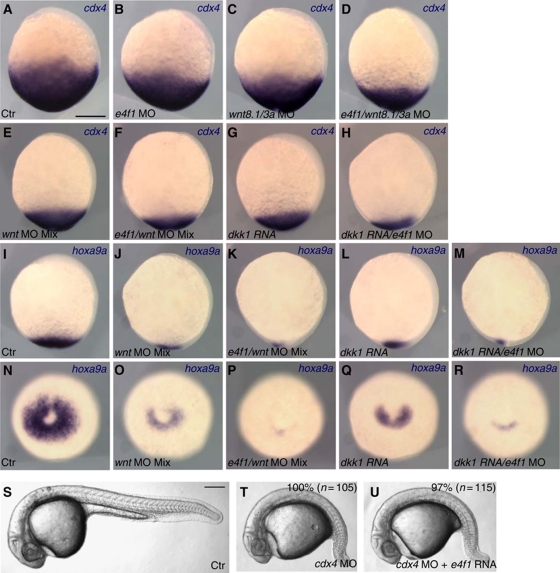
Cdx transcription factors have a major role in caudal body development in all vertebrates (van den Akker et al, 2002; Shimizu et al, 2005; Nordström et al, 2006; Faas and Isaacs, 2009; Young and Deschamps, 2009; Young et al, 2009). Therefore, we asked whether the tail defects in e4f1 morphants correlate with effects on the expression of cdx4, the major caudal determinant in zebrafish. e4f1 morphants exhibited a modest reduction of the cdx4 expression domain at the bud stage (49%, n=37), similar to embryos injected with a wnt3a/wnt8.1 MO mixture (94%, n=32) (Figure 2A–C). The expression domain of cdx4 became further compromised when wnt3a/wnt8.1/e4f1 expression was blocked simultaneously (Figure 2D; 100%, n=33). Similar to a previous report, extensive blocking of Wnt signalling through depletion of wnt3a/wnt8.1/wnt8.2 (100%, n=24; Figure 2E) or by overexpression of a high level of Dkk1 (100%, n=30; Figure 2G) caused strong reduction of cdx4 expression (Shimizu et al, 2005). Expression of cdx4 was reduced further when e4f1 and Wnt function was blocked simultaneously (Figure 2F; 70%, n=30 and Figure 2H; 60%, n=30). The reduction of cdx4 expression in the e4f1 morphants was visible as early as the onset of gastrulation (Supplementary Figure S4A, C and D). In contrast to the effects of the MO, overexpression of E4f1 did not significantly alter the expression level of cdx4 (Supplementary Figure S4B and D), consistent with the lack of a visible phenotype in embryos injected with the same levels of e4f1 mRNA (Figure 1D). The expression levels of other caudal genes that are regulated by posterior Wnt signal were also compromised in the e4f1 morphant embryos (Supplementary Figure S4D) (Dorsky et al, 2003; Li et al, 2011). These results indicate that E4f1 cooperates with Wnt signalling in the regulation of cdx4 expression in the caudal domain of the embryo.
Figure 2.
E4f1 modulates Wnt-dependent regulation of cdx4 and hoxa9a expression. (A–H) cdx4 expression in the caudal region at the end of gastrulation is reduced by inhibiting Wnt signalling and further attenuated by e4f1 depletion. (I–R) Expression of hoxa9a at the end of gastrulation is likewise regulated by Wnt signalling and E4f1. Injected reagents are listed at the bottom, probe for in situ hybridization at top right. wnt MO mix indicates a mixture of wnt3a MO (2 ng), wnt8.1 MO (2 ng) and wnt 8.2 MO (2 ng); 5 ng of e4f1 MO and 25 pg of dkk1 mRNA were injected. (A–M) Lateral view, anterior is up and dorsal to the right. (N–R) Posterior view, dorsal is up. (S–U) E4f1 acts downstream of Cdx4. E4f1 mRNA (150 pg) does not rescue tail defects caused by cdx4 MO (2 ng). Scale bars, 200 μm.
Cdx4 regulates caudal development and erythropoiesis through target genes among which several Hox genes hold an important position. Therefore, we examined the expression of hoxa9a, a direct target gene of Cdx4 (Davidson et al, 2003; Shimizu et al, 2005) after manipulating Wnt and E4f1 function. hoxa9a expression was reduced in wnt3a/wnt8.1/wnt8.2 morphants (86%, n=28) or Dkk1 overexpressing embryos (100%, n=33), and reduced further by co-injection of e4f1 MO (wnt3a/wnt8.1/wnt8.2/e4f1 MO, 45%, n=31; dkk1 RNA+e4f1 MO, 64%, n=25) (Figure 2I–R). These observations confirm that the changes in cdx4 expression noted above have functional consequences for downstream genes. Since Cdx4 is critical for primitive erythropoiesis in zebrafish through the activation of the posterior hox gene cluster (Davidson et al, 2003), we analysed expression of gata1, a marker for primitive erythropoietic progenitors (Davidson and Zon, 2004). gata1 expression in the posterior lateral plate mesoderm was severely reduced in cdx4 morphants, similar to the kgg mutant (Davidson et al, 2003) (Supplementary Figure S5A and B). To visualize synergistic effects, we injected moderate amounts of dkk1 mRNA to dampen rather than eliminate Wnt signalling; in these embryos, gata1 expression was slightly compromised, similar to the reduction seen in e4f1 morphants (Supplementary Figure S5C and D). Notably, gata1 expression was strongly reduced in embryos co-injected with low-level dkk1 mRNA plus e4f1 MO, but could be rescued by co-injection of cdx4 mRNA (Supplementary Figure S5E and F). By contrast, the pax2.1 stripe in the lateral cells of the pronephric primordium was less sensitive to levels of e4f1 and Wnt signalling (Supplementary Figure S5G–L). These data indicate that Wnt and E4f1 coordinately modulate caudal tissue formation and primitive erythropoiesis through the regulation of cdx4 expression. As Wnt signalling regulates caudal development through Cdx4 and we show that E4f1 cooperates with Wnt in regulating cdx4 expression, we predict that E4f1 should act upstream of Cdx4. This is the case as E4f1 overexpression failed to rescue tail development in cdx4 morphants (Figure 2S–U).
E4f1 regulates expression of a Cdx4 reporter and is antagonized by Lnx2b
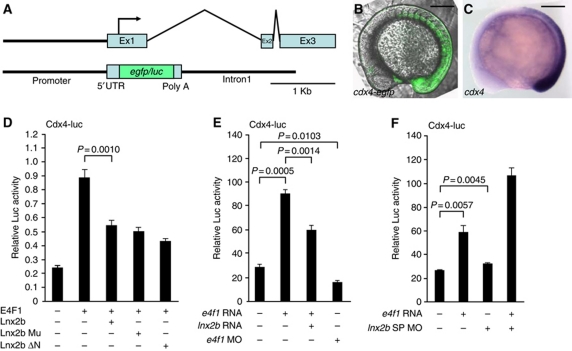
To study cdx4 regulation further, we constructed a cdx4 reporter by inserting the egfp or luciferase (luc) coding region at the translation start site in exon 1 of cdx4, retaining ∼1.6 kb of upstream region and of intron 1 (Figure 3A). The cdx4 genomic structure, composed of three exons separated by a relatively long first intron and short second intron, is well conserved from fish to humans (Figure 3A). The cdx4-egfp reporter was expressed in a similar domain as the resident cdx4 gene in stable transgenic fish (Figure 3B and C), validating the use of this reporter to study cdx4 regulation. A positive role of E4f1 in cdx4 induction suggested above (Figure 2) was supported by transient transfection assays in 293T cells in which E4f1 increased the activity of cdx4-luc (Figure 3D). As we had identified E4f1 as a binding partner of Lnx2b, we tested whether Lnx2b affects E4f1-dependent regulation of the cdx4-luc reporter. We found that Lnx2b counteracted E4f1-dependent cdx4-luc activation. Because E3 ubiquitin ligase-dead mutant forms of Lnx2b (Lnx2b Mu, Lnx2b ΔN) (Ro and Dawid, 2009) also antagonized E4f1-dependent reporter activation, we conclude that the E3 ubiquitin ligase activity is dispensable and that Lnx2b modulates E4f1 activity as a scaffold protein (Figure 3D). In zebrafish embryos as in cultured cells, overexpression of E4f1 enhanced cdx4-luc activity, depletion of E4f1 attenuated the activity, and Lnx2b counteracted reporter activation (Figure 3E). To assess the physiological relevance of the low-level expression of lnx2b in the caudal tissue, we depleted zygotically expressed Lnx2b using splice-blocking MO (SP-MO) in order to avoid defects in dorsoventral axis specification that arise when maternal and zygotic Lnx2b were simultaneously depleted using translation-blocking MOs (Ro and Dawid, 2009; Supplementary Figure S6A). Depletion of zygotically expressed Lnx2b alone was not enough to stimulate cdx4-luc activity, presumably due to the low level of Lnx2b expression, but cdx4-luc reporter activity was synergistically increased by the co-injection of lnx2b SP-MO and e4f1 mRNA (Figure 3F). These data indicate that E4f1 activates cdx4 transcription, modulated by the opposing action of Lnx2b.
Figure 3.
E4f1 and Lnx2b modulate cdx4 expression. (A) The zebrafish cdx4 genomic region and reporter constructs. Three exons (Ex1∼Ex3) and two introns are depicted; the translation initiation site is indicated with an arrow. The egfp or firefly luciferase (luc) coding region was inserted between the 5′ flanking (−1494 to +181) and the coding region of the first exon plus first intron (1637 bp) of cdx4. (B) Stable transgenic embryo carrying the cdx4-egfp transgene, at the 12-somite stage. (C) cdx4 expression at the equivalent stage as (B). (D–F) Luciferase activity under the control of the cdx4 regulatory region. (D) Luciferase activity of a transiently transfected cdx4-luc reporter in 293T cells. Lnx2b Mu, stabilized Lnx2b containing two point mutations (H63A, C66A) in the RING domain; Lnx2b ΔN, N-terminal RING domain deleted mutant (Ro and Dawid, 2009). (E, F) cdx4-luc reporter plasmid (30 pg) was co-injected with the indicated mRNAs or MOs into zebrafish embryos. e4f1 mRNA (100 pg), lnx2b mRNA (100 pg), e4f1 MO (5 ng) and lnx2b SP-MO (10 ng) were injected at the 1–4 cell stage. Luciferase activity was measured in triplicate or more at the 3–5-somite stage. Statistical significance of differences is shown in the figure. (B, C) Lateral views, anterior is left and dorsal is up. Scale bars, 200 μm.
We also asked whether E4f1 regulates cdx4 induction through direct binding to the promoter. The cdx4 5′ upstream region contains a putative E4f1-binding element TGACGTCAG, although its sequence does not conform exactly to the consensus TGACGTAAC (core sequence is underlined) (Fernandes and Rooney, 1997; Hofmayer et al, 2009). Nevertheless, we failed to detect binding of E4f1 to the cdx4 promoter in electrophoretic mobility shift assays (data not shown). In addition, a cdx4-luc reporter in which the putative E4f1-binding element was mutated to TAACGTCCG (mutated nucleotides underlined) could still be activated by E4f1 overexpression as effectively as the wild type (WT) (data not shown). Therefore, we conclude that E4f1-dependent cdx4 upregulation is likely to be mediated by an indirect mechanism.
As lnx2b is expressed in the caudal domain (Ro and Dawid, 2009) where it modulates the regulation of cdx4 by E4f1, we asked whether lnx2b is regulated by Wnt signalling. Wnt depletion abolished lnx2b expression in tailbud embryos except in Kupffer's vesicle, and Wnt activation by LiCl enhanced both lnx2b and cdx4 expression (Supplementary Figure S7A–K). In addition, putative LEF/TCF-binding elements are clustered in the upstream region of the lnx2b gene, and at least one of these sites binds Tcf3 in vivo (Supplementary Figure S6B). To our surprise we found that lnx2b expression was strongly enhanced after MO-dependent depletion of cdx1a/cdx4, and was further elevated by treatment with LiCl (Supplementary Figure S7L and M). These data indicate that Wnt signalling activates both cdx4 and lnx2b expression, but Cdx proteins suppress lnx2b. These relationships suggest a feedback loop that may contribute to robustness of cdx4 regulation, as further discussed below.
Tcf3 regulates cdx4 by repression and Wnt/E4f1-mediated derepression
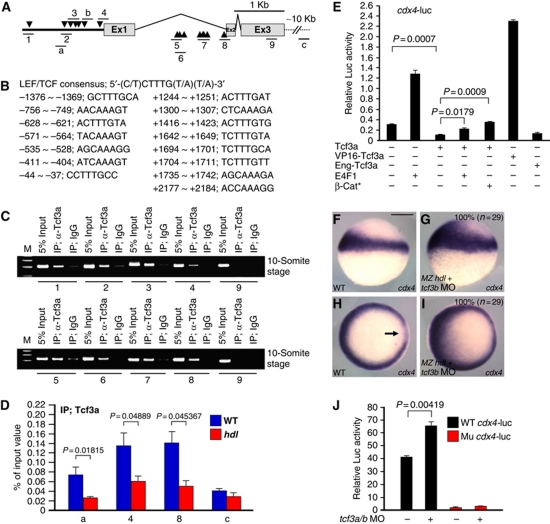
Tcf3 is a major mediator of the Wnt pathway in the early embryo, acting as a repressor in this context, as removal of Tcf3 yields the same phenotype as zygotic Wnt activation (Fredieu et al, 1997; Kim et al, 2000; Dorsky et al, 2003; Merrill et al, 2004; Gribble et al, 2009). We surmised that E4f1 acts by modulating Tcf3 function in cdx4 regulation. Therefore, we analysed the cdx4 5′ flanking region as well as the first intron, and identified several LEF/TCF consensus elements (Figure 4A and B). We then carried out chromatin immunoprecipitation (ChIP) analysis for several regions containing predicted Tcf3-binding elements and found that they are occupied by endogenously expressed Tcf3a in the cdx4-egfp transgenic embryo (Figure 4C; Supplementary Figure S8). The specific binding of Tcf3a to the cdx4 promoter and first intron regions was confirmed by comparing ChIP analysis obtained using WT with Tcf3a mutant zebrafish (tcf3a/hdl; maternal/zygotic MZ hdl animals were used) (Kim et al, 2000) (Figure 4D). The ChIP signal generated by WT fish was robust, but was reduced substantially when mutant fish were used.
Figure 4.
Tcf3 acts as a repressor in regulating cdx4. (A) Schematic diagram of cdx4 genomic region. Arrowheads indicate putative LEF/TCF-binding elements. Numbers 1 to 9 and a to c indicate amplicons tested in ChIP experiments. (B) LEF/TCF consensus sequence and sequences of the putative binding elements in the cdx4 gene. (C) ChIP of embryo chromatin with anti-Tcf3a antibody. Amplicons 1–8 encompassing LEF/TCF-binding elements were precipitated, but exonic amplicon 9 and amplicon c located ∼10 Kb downstream of amplicon 8 were not. IgG was used as negative control. (D) Comparison of the q-PCR yield, based on triplicate analysis, of indicated amplicons after ChIP with anti-Tcf3a antibody between WT and MZ hdl/tcf3a−/− mutant embryos. Statistical significance is indicated in the figure. (E) cdx4-luc reporter activity in 293T cells. Combined DNA mixtures were used for transfection as indicated. Tcf3a and En-Tcf3a suppressed cdx4-luc transgene activity, while E4f1 and stabilized β-catenin relieved repression. VP16–Tcf3a acts as a strong transcriptional activator. (F–I) At the shield stage, cdx4 is expressed in the margin in a ventral-to-dorsal gradient in uninjected embryos, with a gap in the shield region (arrow in H). Depletion of Tcf3 by injection of tcf3b MO (2 ng) into MZ hdl/tcf3a−/− mutant embryos, expanded cdx4 expression into the organizer and ventral-animal domain. (J) Zebrafish embryos were co-injected with tcf3a/b MO and WT cdx4-luc or LEF/TCF-binding site null mutant construct (Mu cdx4-luc; see Supplementary Figure S9). The embryos were harvested at 6.5 h.p.f. for measurement of reporter activity. Luciferase activity was assayed at least in triplicate. (F) Scale bar, 200 μm.
Since Tcf3 primarily acts as transcriptional repressor, we reasoned that cdx4 induction should be suppressed by overexpression of Tcf3. Using the cdx-luc reporter we found that Tcf3a suppressed basal activity, whereas E4f1 stimulated it, as shown above (Figures 3D–F and 4E). Notably, E4f1 and the classical Wnt pathway activator β-catenin (β-cat* is a stabilized form) similarly increased reporter activity to about basal level, suggesting that derepression rather than activation takes place under these conditions. This view is supported by the fact that an artificial activator, a VP16–Tcf3a fusion protein, increased reporter activity strongly whereas the artificial repressor, Eng-Tcf3a, repressed at a similar level as WT Tcf3a (Figure 4E).
If Tcf3 does suppress cdx4 induction in vivo, cdx4 levels should be increased after depletion of Tcf3. In contrast, if Tcf3 conversion to an activator by Wnt stimuli is a prerequisite for cdx4 induction, Tcf3 depletion would preclude cdx4 induction. In gastrula embryos, cdx4 is expressed around the margin in a ventral-to-dorsal gradient that results in a gap in the organizer region in which relatively low levels of Wnt signalling are maintained (Erter et al, 2001; Lekven et al, 2001) (Figure 4F and H). To suppress both Tcf3 isoforms, we introduced tcf3b MO (Dorsky et al, 2003) into MZ hdl mutant embryos. These embryos showed increased cdx4 expression without any gap in the organizer (Figure 4G and I), indicating that Tcf3 functions as a transcriptional repressor to restrict cdx4 expression to tissues with high levels of Wnt activity. Derepression of cdx4 by the depletion of Tcf3a/b was further validated by measuring cdx4-luc activity. WT cdx-luc reporter was stimulated by injecting a tcf3a/b MO mixture into the embryos, while a mutant reporter construct lacking LEF/TCF-binding sites (Supplementary Figure S9A) lost basal activity and failed to respond to the MO injection (Figure 4J). In contrast, overexpression of Tcf3a significantly reduced WT cdx4-luc reporter activity (Supplementary Figure S9B). Furthermore, whereas both LEF/TCF consensus sites in the upstream region and the first intron contribute to the basal expression of cdx4, only the consensus sites in the 5′ flanking region mediate the suppressive action of Tcf3a (Supplementary Figure S9B). Taken together, these data agree with the conclusions of Tcf3 loss-of-function studies (Kim et al, 2000; Dorsky et al, 2003) and support our view that cdx4 activation is the result of derepression, in agreement with a recent report (Hikasa et al, 2010).
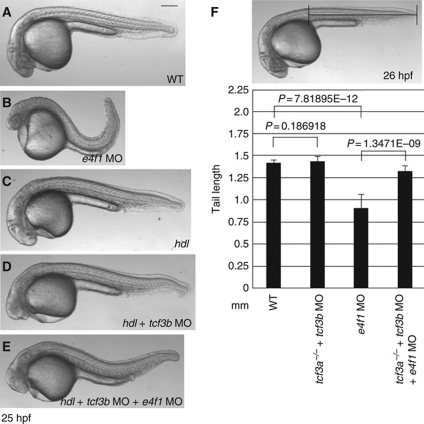
If E4f1 mediates relief of cdx4 from repression by Tcf3, embryos without functional Tcf3 should be refractory to e4f1 MO. We tested this prediction of an epistatic relationship between Tcf3 and E4f1 in the sensitive assay of tail formation. As shown above (Figure 1B), E4f1 depletion leads to reduced tail length (Figure 5B and F). Removal of Tcf3 affects head development but has almost no effect on the tail, as previously reported (Kim et al, 2000; Dorsky et al, 2003) (Figure 5C, D and F). Importantly, embryos lacking Tcf3 (MZ hdl embryos injected with tcf3b MO) did not show tail defects after depletion of E4f1, whereas WT embryos exhibited shortened tails. In other words, the reduced tail length of e4f1 morphants was largely rescued by concurrent depletion of Tcf3a and b (Figure 5E and F). At the same time the head phenotype resulting from Tcf3a/b depletion was unaffected by the e4f1 MO (Figure 5D and E). These results strongly support a physiological function of E4f1 in caudal body formation: if e4f1 MO effects on tail length were due to off-target effects, it should not be possible to rescue tail length by co-depletion of Tcf3. Consistent with the lack of an effect on tail length, alteration of E4f1 levels did not substantially affect the elevated cdx4 expression in embryos depleted of Tcf3 (Supplementary Figure S10).
Figure 5.
Tcf3 depleted embryos resist e4f1 MO injection. (A–E) Morphology of embryos injected as indicated in lower right. (A) Control embryo; (B) e4f1 MO (5 ng)-injected embryo; (C) MZ hdl/tcf3a−/− mutant embryo; (D) tcf3b MO (2 ng)-injected MZ hdl/tcf3a−/− embryo; (E) e4f1 MO (5 ng) was co-injected with tcf3 MO (2 ng) into MZ hdl/tcf3a−/− embryos. Note that the tail defect in (B) is largely rescued in (E), but the head defect in (D) is not. (F) The tail length of 20 randomly selected embryos from each group was measured using ImageJ, as indicated in the photograph. Statistical significance is shown in the histogram. Scale bar, 200 μm.
E4F1 dissociates HDAC1–Gro/TLE corepressor proteins from Tcf3
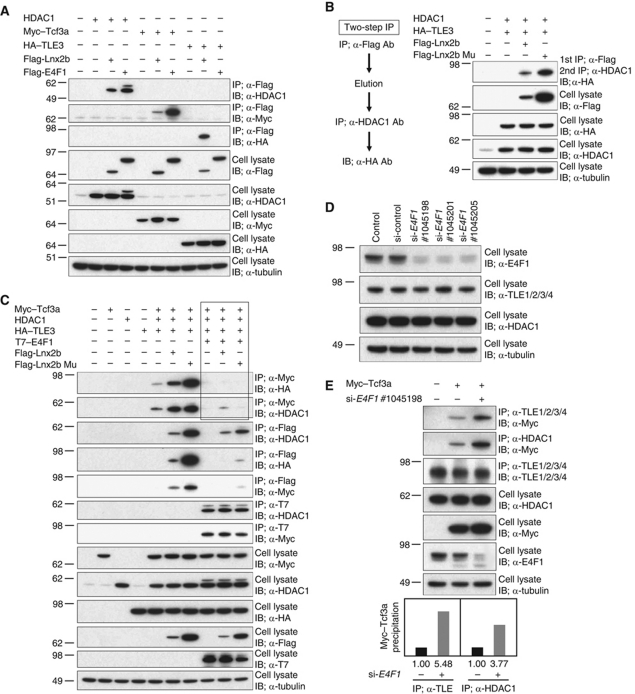
The observations described above, in agreement with earlier data, indicate that Wnt signalling governs cdx4 expression in zebrafish as it does in the mouse (Shimizu et al, 2005; Pilon et al, 2006). As E4f1 has been shown to be a transcriptional repressor (Fajas et al, 2001) and we found that E4f1 does not bind to the cdx4 regulatory region (see above), we tested whether the stimulation of cdx4 expression by E4f1 and its attenuation by Lnx2b might be achieved by protein–protein interactions. There is ample evidence that LEF/TCF transcriptional repressor activity depends upon association with Gro/TLE and HDAC1 (Cavallo et al, 1998; Billin et al, 2000; Brantjes et al, 2001; Yamaguchi et al, 2005; Ye et al, 2009). Initially, we tested whether E4f1 and Lnx2b can associate with Tcf3 and corepressor proteins. E4f1 co-precipitated with HDAC1 as previously reported (Colombo et al, 2003) and also with Tcf3, but not with TLE3 (Figure 6A). Lnx2b could be co-precipitated with all proteins tested, albeit with different yields (Figure 6A). To investigate whether Lnx2b, HDAC1 and Gro/TLE form a complex, we performed sequential co-IP (Figure 6B). Cells transfected with HDAC1, Flag-Lnx2b and HA–TLE3 were immunoprecipitated with anti-Flag antibody, eluted using Flag peptide, and subjected to a second IP with anti-HDAC1 antibody; the immune complex contained HA–TLE3 (Figure 6B). Thus, Lnx2b, HDAC1 and Gro/TLE can form a triple complex, independently of the E3 ubiquitin ligase activity of Lnx2b. Next, we tested whether the yield of repressor complex is modified by E4f1, and found that E4f1 destabilized Tcf3/corepressor association, even in the presence of Lnx2b (Figure 6C; key results are outlined). Thus, we conclude that E4f1 acts to dissociate corepressors from Tcf3, which is expected to abolish its repressor function. To check the role of E4f1 at endogenous levels of expression, we tested these protein–protein interactions after depletion of E4F1 with specific siRNAs in 293T cells; because the levels of TCF7L1 (the human Tcf3 orthologue) are low, we co-transfected Myc-tagged Tcf3a (Figure 6D and E). We observed that treatment of the cells with E4F1 siRNAs resulted in increased association of the endogenously expressed Gro/TLE and HDAC1 with Myc–Tcf3a (Figure 6E), indicating that E4F1 negatively influences Tcf3 repressor complex formation or stability at physiologically relevant levels.
Figure 6.
E4f1 and Lnx2b modulate binding of Tcf3 to corepressor molecules. (A–E) 293T cells were transfected with the indicated plasmids. (A) Co-IP at 48 h post-transfection and blotting was carried out with the antibodies indicated. Lnx2b co-precipitated with Tcf3, TLE3 and HDAC1, while E4f1 co-precipitated with HDAC1 and Tcf3 but not with TLE3. Note that Lnx2b mutant accumulated to a higher level than WT due to self-ubiquitylation and destabilization of the latter (Ro and Dawid, 2009). (B) Two-Step IP. Lysates of cells transfected with plasmids encoding HDAC1 and HA-tagged TLE3, plus Flag-Lnx2b or Flag-Lnx2b Mu, were first precipitated with anti-Flag antibody. Proteins eluted by 3xFlag peptide were subjected to a second IP with anti-HDAC1 antibody; co-precipitated TLE3 was detected by anti-HA blotting. (C) Lnx2b, WT or mutant, enhanced co-precipitation yields of Tcf3a with HDAC1 or TLE3. By contrast, E4f1 overexpression dissociated the corepressors and WT or Mu Lnx2b from Tcf3a; the lanes showing this critical observation are outlined in the figure. Association between E4f1 and Tcf3a and between E4f1 and HDAC1 was not altered by the presence of Lnx2b, and the association between Lnx2b/Lnx2b Mu and HDAC1 was not greatly altered by E4f1. (D) All three E4F1 siRNAs tested knocked downed E4F1 levels. (E) Depletion of E4F1 facilitated binding of endogenous HDAC1 and TLE family proteins to Tcf3a. The histogram at the bottom shows quantification of this experiment using ImageJ.
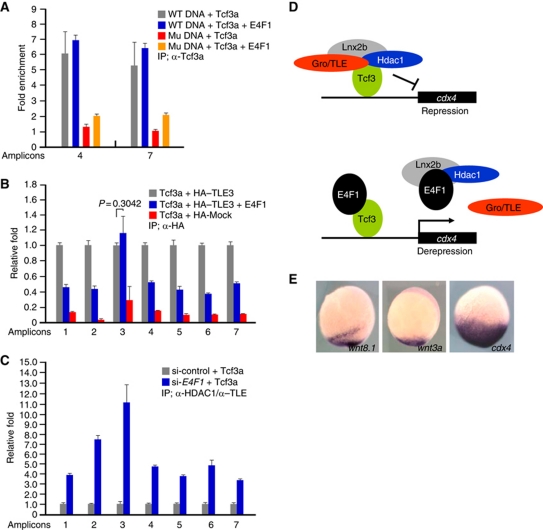
A possible mechanism for E4f1 action in cdx4 derepression might be interference with Tcf3 binding to cis-regulatory DNA elements, similar to the effects of Kaiso, I-mfa and HIPK2 (Snider et al, 2001; Ruzov et al, 2009; Hikasa et al, 2010). To map the critical domains of Tcf3 required for association with E4f1, we constructed several deletion mutants of Tcf3. Even though both the Gro/TLE interaction domain and the HMG DNA-binding domain of Tcf3 are required for its binding to E4f1 (Supplementary Figure S11), ChIP assays revealed that E4f1 does not interfere with the binding of Tcf3 to the regions containing LEF/TCF-binding elements in the cdx4 gene (Figure 7A). In this respect, E4f1 behaves similarly to β-catenin, extensively studied as modulator of LEF/TCF activity (Hurlstone and Clevers, 2002).
Figure 7.
Tcf3 remains bound to the cdx4 promoter in the presence of E4f1 while the corepressors are removed. (A–C) ChIP analyses in 293T cells transfected with the indicated DNAs. Amplicons are identified in Figure 4A. (A) PCR amplified DNA fragments including WT or mutant cdx4 regulatory elements (Supplementary Figure S9Aa and d, respectively) were co-transfected with Tcf3a with or without E4F1, as indicated. Samples were immunoprecipitated with anti-Tcf3a antibody, or IgG as control. Amplicons 4 and 7 were used for q-PCR. Fold enrichment is calculated setting the IgG control as one. Note that E4F1 did not interfere with Tcf3a binding to the DNA, while mutant DNA was recovered at low yield. (B) ChIP q-PCR assay showing that E4F1 dissociates TLE3 from the Tcf3-binding sites in the cdx4 promoter; the aberrant behaviour of amplicon 3 is not understood. (C) ChIP q-PCR analysis of cdx4 regulatory region after IP with anti-HDAC1 and anti-TLE antibodies after depleting endogenously expressed E4F1. cdx4-luc plasmid and Tcf3a were co-transfected with or without E4F1 siRNA. Note that ChIP yields increased significantly in E4F1-siRNA-treated cells. q-PCR was carried out at least in triplicate, and standard error of the mean is shown in the panels. A proposed mechanism is shown in (D). In this model, a complex composed of Tcf3–Gro/TLE–HDAC1–Lnx2b represses the cdx4 gene; E4f1 dissociates the complex, preserving Tcf3 binding to the DNA. (E) Expression of wnt3a and wnt8.1 is much more restricted in the caudal domain at the bud stage than cdx4. A proposed role for E4f1 derepression in achieving this pattern is discussed in the text.
A different mechanism that could explain E4f1-mediated derepression of the cdx4 gene is based on the protein–protein interaction studies shown in Figure 6. These experiments suggest that derepression could be achieved by dissociation of corepressor proteins from Tcf3, even though Tcf3 remains attached to the promoter. This hypothesis is supported by ChIP experiments that showed removal of the Groucho (Gro) family corepressor TLE3 from the Tcf3-binding sites in the cdx4 promoter/first intron after the addition of E4f1; this was true for six genomic regions containing Tcf3-binding sites (Figure 7B). Consistently, ChIP yields of LEF/TCF-binding sites in the cdx4 gene after co-precipitation with endogenously expressed HDAC1 and TLE increased substantially after depletion of E4F1 (Figure 7C). These data support the view that E4f1-dependent cdx4 derepression is due to the dissociation of corepressor proteins from Tcf3 (Figure 7D). We suggest that E4f1 can act on the cdx4 promoter in a similar way and with comparable efficiency (Figure 4E) as β-catenin to relieve Tcf3-mediated repression of the cdx4 gene.
Discussion
E4f1 modulates regulation of cdx4 by Tcf3
Wnt signalling has a major role controlling cell proliferation, differentiation and body patterning and is an important factor in the causation of various tumours (MacDonald et al, 2009). Wnt-dependent Cdx-Hox gene expression is critical for caudal tissue specification including haematopoiesis (Isaacs et al, 1998; Davidson et al, 2003; Shimizu et al, 2005; Pilon et al, 2006, 2007; Lengerke et al, 2008; Young and Deschamps, 2009). Here, we describe novel components and interactions that modulate Wnt signalling during caudal body formation in the zebrafish embryo. We identified a mechanism for caudal region development based on the relief of repression of the cdx4 gene by Tcf3. In agreement with the expected role of a transcriptional repressor (Brannon et al, 1997; Kim et al, 2000; Dorsky et al, 2003; Gribble et al, 2009), Tcf3 suppresses cdx4 induction through direct binding to multiple cis-regulatory elements. Removal of Tcf3 by mutation of the tcf3a gene and using an MO against tcf3b, led to an increase in cdx4 expression (Figure 4F–I), consistent with a repressor function of Tcf3 in cdx4 expression. In this respect, Tcf3 acts similarly in caudal body formation as previously reported for head formation where depletion of Tcf3 gives equivalent phenotypes to activation of the Wnt pathway (Fredieu et al, 1997; Kim et al, 2000; Dorsky et al, 2003). Corepressor proteins such as Gro/TLE and HDAC1 confer repressor activity upon Tcf3 (Brannon et al, 1997; Kim et al, 2000; Dorsky et al, 2003; Gribble et al, 2009), and we show that Tcf3 associates with corepressors in a complex that can be stabilized by Lnx2b (Figure 6B and C). This Tcf3–HDAC1–Gro/TLE repressor complex, even if stabilized by Lnx2b, can be dissociated by E4f1 to mediate derepression of cdx4 (Figure 6C). Results of ChIP experiments indicate that the corepressor complex is bound to LEF/TCF-binding sites in the cdx4 promoter (Figure 7B), and that Tcf3 remains associated with the DNA when E4f1 replaces the corepressors in the complex (Figure 7A–C). Thus, E4f1 exerts its effect on cdx4 expression by protein–protein interaction rather than by binding to the promoter. It should be emphasized that overexpression of stabilized β-catenin did not activate the cdx4-luc transgene when Tcf3 was co-expressed, but instead restored the basal level of expression (Figure 4E). These data are consistent with the view that β-catenin and E4f1 act in a similar manner on the cdx4 promoter, leading to a relief of Tcf3 repression. Recently, Hikasa et al (2010) reported studies on Tcf3-mediated regulation of the vent2 and also the cdx4 gene in Xenopus, showing that Tcf3 phosphorylation leads to its removal from a cognate binding site in the vent2 gene. As E4f1-mediated derepression of cdx4 in zebrafish occurs while Tcf3 remains bound to the promoter, these two mechanisms are distinct, and their possible interaction remains to be analysed.
Four Lef/Tcf factors are expressed during zebrafish embryonic development (Dorsky et al, 1999; Kim et al, 2000; Young et al, 2002; Veien et al, 2005). Since some functional redundancy has been reported among these factors (Galceran et al, 1999; Nagayoshi et al, 2008; Nguyen et al, 2009), it seemed possible that Lef/Tcf factors in addition to Tcf3 are involved in translating the Wnt signal to caudal tissue specification by regulating cdx4 expression. Among these factors, Tcf4 seems to be ruled out as it is not expressed in the caudal domain (Young et al, 2002). In addition, even though lef1 and tcf7 transcripts can be detected in the caudal region, combined inhibition of Lef1/Tcf7 function caused pectoral fin defects but not tail agenesis (Nagayoshi et al, 2008). Thus, our evidence combined with previous reports strongly supports the view that the transcriptional repressor activity of Tcf3 is the key activity to mediate Wnt signalling in caudal development through the regulation of cdx and hox gene expression (Kim et al, 2000; Dorsky et al, 2003; Merrill et al, 2004). Several different signals control the Cdx-Hox pathway through crosstalk and collaboration (Isaacs et al, 1998; Shimizu et al, 2005; Lengerke et al, 2008; Iimura et al, 2009; Mallo et al, 2009; Young and Deschamps, 2009). In our proposed mechanism, E4f1 and Lnx2b have a role in fine-tuning Tcf3-mediated repression of cdx4 by affecting the composition of the repressor complex, with Lnx2b enhancing and E4f1 disrupting the complex (Figure 7D).
Biological role of E4f1 antagonism of Tcf3 repression
The biological significance of the proposed mechanism can be illustrated with Figure 7E. Wnt 3a and Wnt 8.1 are expressed in a tight domain at the caudal tip of the embryo, as reported previously (Lekven et al, 2001; Thorpe et al, 2005). While Wnt factors can diffuse (Yan and Lin, 2009), the effective ligand concentration will become low at a distance from the source, and fine differences in levels might lead to imprecise boundaries of cdx4 activation at the animal edge of the gradient. The presence of a constitutive derepressing factor, E4f1, may contribute to the robustness of the induction of cdx4 in this region of the embryo. This interpretation is consistent with our observation that inhibition of E4f1 expression combined with attenuation of Wnt signalling leads to a gradual diminution of the domain of cdx4 expression (Figure 2).
The E3 ubiquitin ligase Lnx2b appears to have a modest refining role in the interactions that lead to Tcf3-mediated repression and derepression. Lnx2b stabilizes the repressor complex, but does not strongly resist dissociation of the complex by E4f1 (Figure 6C). This is reflected in a measurable but weak inhibition of E4f1-dependent cdx4-luc reporter activity by Lnx2b (Figure 3D–F). These experiments did, however, uncover the interesting fact that Lnx2b acts as a scaffold protein in this situation, independently of its E3 ubiquitin ligase activity. The ability to serve as a binding platform is not a unique property of Lnx2b. For instance, ectodermin/TIF1γ, a ligase involved in Smad4 ubiquitylation, acts as a scaffold protein for recruiting positive elongation factors to erythroid genes to promote transcription (Bai et al, 2010). Recently, Honda et al (2010) reported that PDZRN3 (LNX3) negatively regulates Wnt/β-catenin signalling by reducing LRP6 phosphorylation through an unknown mechanism. It will be intriguing to test whether LNX3 and Lnx2b regulation of the Wnt pathway exhibit overlapping modalities.
The role of Tcf3 is widespread in different cells and tissues. It will be of particular interest to test whether the expression of pathogenic Wnt target genes is modulated by E4F1 and LNX proteins in cells or tissues in which Tcf3 expression is tightly regulated, for instance in skin epithelia, hair follicles and embryonic stem cells (Jiang et al, 2008; Nguyen et al, 2009; Abu-Remaileh et al, 2010). Most recently, Lacroix et al (2010) verified a novel role for E4f1 in skin homoeostasis and epidermal stem cell maintenance by generating skin-specific E4f1 conditional knockout mice. The possible involvement of E4F1 in pathogenesis by dysregulation of Wnt signalling during skin agenesis through the transient hyperplasia of epidermal stem cell could be addressed in further studies.
Materials and methods
Fish embryos
Embryos were obtained from natural spawning of WT (AB*) or MZ hdl mutant lines (Kim et al, 2000).
Cell culture and transfection
293T cells were grown in Dulbecco's Modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics. Cells were transfected using FuGeneHD (Roche). After 24–48 h, cells were harvested and assayed. We used zebrafish lnx2b, tcf3 and β-catenin, and human E4F1, HDAC1 and TLE3 DNAs for transfection.
Transgenic zebrafish
The PCR amplified 5′ flanking region (−1494 to +217) of cdx4 was subcloned into a pSCAC-40 vector (Kim et al, 2008) between the Tol2 Rt arm and the egfp reporter gene. The PCR amplified first intron (1637 bp) of cdx4 was subcloned into the above construct. To generate transgenic animals, 10 pg of plasmid was co-injected with 50 pg of in vitro transcribed mRNA encoding transposase into one-cell stage zebrafish embryos (Kawakami et al, 2004).
Luciferase assay
The cdx4 genomic DNA used for transgenesis above was subcloned into pGL3-Basic Vector (Promega). The 5′ flanking region was inserted between the NheI and BglII sites, and the first intron between the BamHI and SalI sites. 293T cells that were 50% confluent were transfected with 5 ng of pRL-SV40, 100 ng cdx4-luc and 100 ng of other indicated plasmids. Luciferase assays were performed using Dual-Luciferase Reporter Assay System (Promega). Injected embryos (6.5 or 11 h.p.f.) or transfected 293T cells (24 h after transfection) were lysed in Passive Lysis Buffer (Promega). Each luciferase activity was measured at least three times, and significance was assessed by the Student's t-test.
IP and western blotting
IP was performed by mixing lysates of transfected cells with 20 μl of Protein G Sepharose 4 Fast Flow (GE Healthcare) in M-PER Mammalian Protein Extraction Reagent (Pierce) or in CompLysis Protein Extraction Reagent (SignaGen) with Complete protease inhibitor cocktail (Roche; hereafter CPIC). The total volume of lysate mixture was adjusted to 1 ml with NP-40 IP buffer (150 mM NaCl, 1 mM DTT, 10 mM Tris–Cl pH 7.5, 0.2% NP-40, CPIC). Precipitates were washed four times with 1 ml ice-cold NP-40 IP buffer, and were analysed by western blotting. After separation in Nu-PAGE 4–12% Bis-Tris Gel (Invitrogen) and transfer onto PVDF membrane, the membrane was immersed in blocking solution (4% skim milk or 2% BSA in TBS with 0.05% Tween-20) at room temperature for 30 min, incubated with primary antibodies diluted in blocking solution for 1 h, followed by incubation with HRP-conjugated secondary antibody (Jackson, 1:5000) for 1 h, all at room temperature. The detection was performed using the ECL detection system (Pierce). The antibodies were supplied from Roche (rat HA), Sigma (mouse Flag M2), Clontech (mouse Myc), Millipore (rabbit Myc; rabbit HDAC1 used for Two-Step IP in Figure 6B and ChIP in Figure 7C), Vector (rabbit IgG), Santa Cruz Biotechnology (goat HDAC1 N-19, used for IP in Figure 6E), Cell Signaling Technology (rabbit TLE1/2/3/4 for ChIP in Figure 7C), Proteintech Group, Inc. (rabbit HDAC1, used for immunoblotting), Abcam (rabbit HA used for ChIP in Figure 7B), Bethyl Laboratories Inc. (rabbit E4f1) and Calbiochem (mouse T7, mouse α-tubulin). Anti-rabbit Tcf3a antibody that was used for ChIP was a gift of Dr Dorsky (Gribble et al, 2009).
Two-Step IP
Plasmid mixtures of HDAC1, HA–TLE3, Flag-Lnx2b, Flag-Lnx2b Mu (Ro and Dawid, 2009) and Flag-Mock vector were transfected into 293T cells. After 48 h, the cell lysates were incubated with Protein G Sepharose beads and anti-Flag M2 antibody for 6 h at 4°C. The beads were washed with ice-cold NP-40 IP buffer three times, and proteins were eluted with 300 μl modified NP-40 IP buffer (250 mM NaCl, 1 mM DTT, 10 mM Tris–Cl pH 7.5, 0.2% NP-40, CPIC and 3xFLAG peptide (300 μg/ml, Sigma)) for 2 h at 4°C. The eluted products were adjusted to 1 ml with salt diluted NP-40 IP buffer (final NaCl concentration 150 mM). After adding anti-HDAC1 antibody and Protein G Sepharose beads, the samples were incubated for 6 h at 4°C with gentle agitation, and then washed with ice-cold NP-40 IP buffer three times. Immunoblotting was performed as above.
siRNA
Cultured 293T cells in 60 mm dish were used for siRNA transfection. In all, 1 μg of plasmid encoding Tcf3a was co-transfected with 5 μg of siRNA. si-E4F1 #1045198, 5′-GGCAAGCGCUACAAGACUA-3′; si-E4F1 #1045201, 5′-CACAGUGUUGGUGGAGUUC-3′; si-E4F1 #1045205, 5′-CUAUAGAGACUUCACCCGU-3′. Negative control siRNA (5′-CCUACGCCACCAAUUUCGU-3′, Bioneer Inc.) was used for normalization of transfection amount. X-tremeGENE siRNA Transfection Reagent (Roche) was used according to the manufacturer's instructions.
Chromatin immunoprecipitation
ChIP was performed as described previously with small modification (Nelson et al, 2006), using polyclonal Tcf3a antibody, epitope tag antibodies or control IgG. Two hundred dechorionated embryos (10-somite stage) were fixed with 2.2% paraformaldehyde (PFA) dissolved in PBS for 15 min at RT. After quenching with 125 mM Glycine, the cross-linked chromatin was sheered to ∼300 bp length using the Bioruptor Next Gen (Diagenode). Following IP, PCR (26–40 cycles) using AccuPower PCR PreMix (Bioneer Inc.) or q-PCR using LightCycler 480 (Roche) was performed. The following primer sets were used for amplifying the indicated amplicons (∼200 bp):
lnx2b amplicon #1 forward primer, 5′-CATTGTTTGGTGCATGCCCCAACTCTG-3′;
lnx2b amplicon #1 reverse primer, 5′-ATAACAATGGGAACCTGCTGTAGTGTAC-3′;
lnx2b amplicon #2 forward primer, 5′-GAGTTGCACACGTTCTCATGTTGGTGAGC-3′;
lnx2b amplicon #2 reverse primer, 5′-CAGGCAATTAAGGCAGGTGTGGATCACCG-3′;
tcf3a amplicon #1 forward primer, 5′-GAGAGCCCGCATTTCCATAAACCGG-3′;
tcf3a amplicon #1 reverse primer, 5′-GTCCCATCTGCATAGTACCCATGCC-3′;
tcf3a amplicon #2 forward primer, 5′-CTTGTAAAATGTTATCACGTGTGCC-3′;
tcf3a amplicon #2 reverse primer, 5′-CTATAATTACAAAGTAATCTTTATAGC-3′;
tcf3a amplicon #3 forward primer, 5′-GTGATTATTAATTTTACATTGAC-3′;
tcf3a amplicon #3 reverse primer, 5′-GCACAATATAACAGGAAATATTATC-3′;
tcf3a amplicon #4 forward primer, 5′-GAATGCAAATCTAAAGGTTTTGAAG-3′;
tcf3a amplicon #4 reverse primer, 5′-CACTGGGATCCAAGGCCCGACTC-3′;
tcf3a amplicon #5 forward primer, 5′-CTCAGGTTTTGAGGGTTATCTGTC-3′;
tcf3a amplicon #5 reverse primer, 5′-TTTACAGTTAGGACGTGTGACTCTG-3′;
tcf3a amplicon #6 forward primer, 5′-TCAGGCGGCGACTTCTTTCAACACC-3′;
tcf3a amplicon #6 reverse primer, 5′-AAAAGCACAACTGAACACACAAAG-3′;
tcf3a amplicon #7 forward primer, 5′-GGATGCATATCAAACAAGTGCAGC-3′;
tcf3a amplicon #7 reverse primer, 5′-GCCTACTATAAACCAAGATCACATC-3′;
tcf3a amplicon #8 forward primer, 5′-TGGACTTATGTATGCCTTTCGCAAC-3′;
tcf3a amplicon #8 reverse primer, 5′-TTACCTGTCTCTCTGAAAGCCCGAG-3′;
tcf3a amplicon #9 forward primer, 5′-CAGGTACGGAGTTTCATGGGACGTG-3′;
tcf3a amplicon #9 reverse primer, 5′-TTGCTGAAATCAAACGTTATACCCG-3′.
tcf3a amplicon #a forward primer, 5′-CAATTGAATTTTTGGTAATAGCC-3′;
tcf3a amplicon #a reverse primer, 5′-GTTGTGTTAATCATAGTCAATGTA-3′;
tcf3a amplicon #b forward primer, 5′-GATAATATTTCCTGTTATATTGTG-3′;
tcf3a amplicon #b reverse primer, 5′-AAGCATTCGATTTGAAAGGTGTAG-3′;
tcf3a amplicon #c forward primer, 5′-TGCTGTAGTGAAGACTGCATTAGTCC-3′;
tcf3a amplicon #c reverse primer, 5′-GTATATATACAGATCTGTATAAAAAGAC-3′.
For ChIP experiments in cultured cells, 293T cells were transfected with the indicated combination of DNAs and siRNAs: cdx4-luc plasmid, cdx4 promoter and first intron DNA fragments amplified by PCR, pCS2+/Tcf3a, pCS2+/MT-Tcf3a (gift of Dr Chitnis), pCS2+/HA–TLE3, pCS2+/E4F1, control siRNA and E4F1 siRNA. Empty vector was used as a negative control (Mock). After 24 h transfection, the cells were fixed in 1.42% PFA in PBS for 12 min and quenched with 125 mM glycine. Sheared cross-linked chromatin was immunoprecipitated with 2–5 μg of the indicated antibodies. After extensive washing with ChIP-IP buffer (150 mM NaCl, 50 mM Tris–HCl (pH 7.5), 5 mM EDTA, 0.5% NP-40, 1% Triton X-100), the precipitated DNA fragments were eluted and then analysed using the LightCycler 480 (Roche).
Whole mount in situ hybridization
The embryos were collected at the appropriated stage and fixed in 4% PFA in PBS. Antisense riboprobes were generated with linearized template DNA using appropriate RNA polymerase following the manufacture's instructions (Roche). Proteinase K treatment (10 μg/ml) was performed for 1 to 10 min depending on the stages of embryos. The hybridized probes were detected using pre-absorbed anti-digoxigenin-AP Fab fragments (Roche) diluted (1:2000) in blocking solution (PBS, 0.1% Tween-20, 5% sheep serum, 0.2% Blocking reagent (Roche)). After staining, the embryos were fixed and examined under the microscope.
Microinjection
cDNAs were subcloned into the pCS2+. mRNAs were synthesized using the mMESSAGE mMACHINE kit (Ambion Inc.). RNAs, plasmids or MOs were injected into the yolk of 1–4 cell stage of embryos. MOs were supplied from Gene Tools, LLC. In all, 1 ng of p53 MO was co-injected with other MOs to inhibit possible off-target effects when we analysed the embryos after 24 h.p.f. (Robu et al, 2007). The following represents the sequence of MOs used in our assay; e4f1 ATG MO, 5′-CTTCAGTCATGTCGTTCCAAGCCTC-3′; lnx2b splicing MO (Ro and Dawid, 2009), 5′-GTAAGTGATGCAATACCATCTTCGC-3′; wnt3a MO, 5′-GTTAGGCTTAAACTGACACGCACAC-3′; wnt8.1 MO (Lekven et al, 2001), 5′-ACGCAAAAATCTGGCAAGGGTTCAT-3′; wnt8.2 MO (Lekven et al, 2001); 5′-GCCCAACGGAAGAAGTAAGCCATTA-3′; cdx1a MO (Shimizu et al, 2005), 5′-GTCCAGCAGGTAGCTCACGGACATT-3′; cdx4 MO (Davidson et al, 2003), 5′-CGTACATGATTTGGAAGAAACCCCT-3′; tcf3b MO (Dorsky et al, 2003), 5′-CGCCTCCGTTAAGCTGCGGCATGTT-3′; p53 MO, 5′-GCGCCATTGCTTTGCAAGAATTG-3′.
LiCl treatment
Mid-gastrula (60% epiboly) embryos were raised in fish water containing 0.2 M LiCl for 40 min at 28.5°C. After rinsing several times with fresh fish water, embryos were incubated to the desired stage.
Supplementary Material
Acknowledgments
We thank M Rath and J Gonzales for help with fish maintenance. We especially thank Dr RI Dorsky for the Tcf3a antibody and Drs S-Y Choi, DE Ayer and A Chitnis for other reagents. This work was supported by the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health.
Author contributions: HR and IBD conceived the project, designed the experiments, analysed the data and wrote the manuscript. HR carried out all experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abu-Remaileh M, Gerson A, Farago M, Nathan G, Alkalay I, Zins Rousso S, Gur M, Fainsod A, Bergman Y (2010) Oct-3/4 regulates stem cell identity and cell fate decisions by modulating Wnt/β-catenin signalling. EMBO J 29: 3236–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Kim J, Yang Z, Jurynec MJ, Akie TE, Lee J, LeBlanc J, Sessa A, Jiang H, DiBiase A, Zhou Y, Grunwald DJ, Lin S, Cantor AB, Orkin SH, Zon LI (2010) TIF1γ controls erythroid cell fate by regulating transcription elongation. Cell 142: 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal D, Scholl C, Fröhling S, McDowell E, Lee BH, Döhner K, Ernst P, Davidson AJ, Daley GQ, Zon LI, Gilliland DG, Huntly BJ (2006) Cdx4 dysregulates Hox gene expression and generates acute myeloid leukemia alone and in cooperation with Meis1a in a murine model. Proc Natl Acad Sci USA 103: 16924–16929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billin AN, Thirlwell H, Ayer DE (2000) β-catenin-histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol Cell Biol 20: 6882–6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkowsky JL, Wang X, Fujimoto E, Lee JE, Chien CB, Dorsky RI (2008) Domain-specific regulation of foxP2 CNS expression by lef1. BMC Dev Biol 8: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon M, Gomperts M, Sumoy L, Moon RT, Kimelman D (1997) A β-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev 11: 2359–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantjes H, Roose J, van De Wetering M, Clevers H (2001) All Tcf HMG box transcription factors interact with Groucho-related corepressors. Nucleic Acids Res 29: 1410–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A (1998) Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395: 604–608 [DOI] [PubMed] [Google Scholar]
- Chagraoui J, Niessen SL, Lessard J, Girard S, Coulombe P, Sauvageau M, Meloche S, Sauvageau G (2006) E4f1: a novel candidate factor for mediating BMI1 function in primitive hematopoietic cells. Genes Dev 20: 2110–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Lu YF, Ko TY, Tsai MY, Lin CY, Lin CC, Hwang SP (2009) Zebrafish cdx1b regulates differentiation of various intestinal cell lineages. Dev Dyn 238: 1021–1032 [DOI] [PubMed] [Google Scholar]
- Cheng PY, Lin CC, Wu CS, Lu YF, Lin CY, Chung CC, Chu CY, Huang CJ, Tsai CY, Korzh S, Wu JL, Hwang SP (2008) Zebrafish cdx1b regulates expression of downstream factors of Nodal signaling during early endoderm formation. Development 135: 941–952 [DOI] [PubMed] [Google Scholar]
- Colombo R, Draetta GF, Chiocca S (2003) Modulation of p120E4f1 transcriptional activity by the Gam1 adenoviral early protein. Oncogene 22: 2541–2547 [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Ernst P, Wang Y, Dekens MP, Kingsley PD, Palis J, Korsmeyer SJ, Daley GQ, Zon LI (2003) cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature 425: 300–306 [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Zon LI (2004) The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene 23: 7233–7246 [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Itoh M, Moon RT, Chitnis A (2003) Two tcf3 genes cooperate to pattern the zebrafish brain. Development 130: 1937–1947 [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Snyder A, Cretekos CJ, Grunwald DJ, Geisler R, Haffter P, Moon RT, Raible DW (1999) Maternal and embryonic expression of zebrafish lef1. Mech Dev 86: 147–150 [DOI] [PubMed] [Google Scholar]
- Erter CE, Wilm TP, Basler N, Wright CV, Solnica-Krezel L (2001) Wnt8 is required in lateral mesendodermal precursors for neural posteriorization in vivo. Development 128: 3571–3583 [DOI] [PubMed] [Google Scholar]
- Faas L, Isaacs HV (2009) Overlapping functions of Cdx1, Cdx2, and Cdx4 in the development of the amphibian Xenopus tropicalis. Dev Dyn 238: 835–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajas L, Paul C, Vié A, Estrach S, Medema R, Blanchard JM, Sardet C, Vignais ML (2001) Cyclin A is a mediator of p120E4F-dependent cell cycle arrest in G1. Mol Cell Biol 21: 2956–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes ER, Rooney RJ (1997) The adenovirus E1A-regulated transcription factor E4F is generated from the human homolog of nuclear factor phiAP3. Mol Cell Biol 17: 1890–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores MV, Hall CJ, Davidson AJ, Singh PP, Mahagaonkar AA, Zon LI, Crosier KE, Crosier PS (2008) Intestinal differentiation in zebrafish requires Cdx1b, a functional equivalent of mammalian Cdx2. Gastroenterology 135: 1665–1675 [DOI] [PubMed] [Google Scholar]
- Fredieu JR, Cui Y, Maier D, Danilchik MV, Christian JL (1997) Xwnt-8 and lithium can act upon either dorsal mesodermal or neurectodermal cells to cause a loss of forebrain in Xenopus embryos. Dev Biol 186: 100–114 [DOI] [PubMed] [Google Scholar]
- Galceran J, Fariñas I, Depew MJ, Clevers H, Grosschedl R (1999) Wnt3a-/--like phenotype and limb deficiency in Lef1(-/-)Tcf1(-/-) mice. Genes Dev 13: 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Kaestner KH (2010) Cdx2 regulates endo-lysosomal function and epithelial cell polarity. Genes Dev 24: 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gat U, DasGupta R, Degenstein L, Fuchs E (1998) De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated β-catenin in skin. Cell 95: 605–614 [DOI] [PubMed] [Google Scholar]
- Gribble SL, Kim HS, Bonner J, Wang Xu, Dorsky RI (2009) Tcf3 inhibits spinal cord neurogenesis by regulating sox4a expression. Development 136: 781–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikasa H, Ezan J, Itoh K, Li X, Klymkowsky MW, Sokol SY (2010) Regulation of TCF3 by Wnt-dependent phosphorylation during vertebrate axis specification. Dev Cell 19: 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmayer S, Madisch I, Darr S, Rehren F, Heim A (2009) Unique sequence features of the human adenovirus 31 complete genomic sequence are conserved in clinical isolates. BMC Genomics 10: 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda T, Yamamoto H, Ishii A, Inui M (2010) PDZRN3 negatively regulates BMP-2-induced osteoblast differentiation through inhibition of Wnt signaling. Mol Biol Cell 21: 3269–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanes K, Li TW, Munguia JE, Truong T, Milovanovic T, Lawrence Marsh J, Holcombe RF, Waterman ML (2001) β-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet 28: 53–57 [DOI] [PubMed] [Google Scholar]
- Hurlstone A, Clevers H (2002) T-cell factors: turn-ons and turn-offs. EMBO J 21: 2303–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iimura T, Denans N, Pourquié O (2009) Establishment of Hox vertebral identities in the embryonic spine precursors. Curr Top Dev Biol 88: 201–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs HV, Pownall ME, Slack JM (1998) Regulation of Hox gene expression and posterior development by the Xenopus caudal homologue Xcad3. EMBO J 17: 3413–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH (2008) A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol 10: 353–360 [DOI] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M (2004) A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell 7: 133–144 [DOI] [PubMed] [Google Scholar]
- Kim CH, Oda T, Itoh M, Jiang D, Artinger KB, Chandrasekharappa SC, Driever W, Chitnis AB (2000) Repressor activity of headless/Tcf3 is essential for vertebrate head formation. Nature 407: 913–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Kang KH, Kim CH, Choi SY (2008) Real-time imaging of mitochondria in transgenic zebrafish expressing mitochondrially targeting GFP. Biotechniques 45: 331–334 [DOI] [PubMed] [Google Scholar]
- Lacroix M, Caramel J, Goguet-Rubio P, Linares LK, Estrach S, Hatchi E, Rodier G, Lledo G, de Bettignies C, Thépot A, Deraison C, Chébli K, Hovnanian A, Hainaut P, Dubus P, Sardet C, Le Cam L (2010) Transcription factor E4F1 is essential for epidermal stem cell maintenance and skin homeostasis. Proc Natl Acad Sci USA 107: 21076–21081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cam L, Lacroix M, Ciemerych MA, Sardet C, Sicinski P (2004) The E4F protein is required for mitotic progression during embryonic cell cycles. Mol Cell Biol 24: 6467–6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cam L, Linares LK, Paul C, Julien E, Lacroix M, Hatchi E, Triboulet R, Bossis G, Shmueli A, Rodriguez MS, Coux O, Sardet C (2006) E4f1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell 127: 775–788 [DOI] [PubMed] [Google Scholar]
- Lee JE, Wu SF, Goering LM, Dorsky RI (2006) Canonical Wnt signaling through Lef1 is required for hypothalamic neurogenesis. Development 133: 4451–4461 [DOI] [PubMed] [Google Scholar]
- Lekven AC, Thorpe CJ, Waxman JS, Moon RT (2001) Zebrafish wnt8 encodes two Wnt8 proteins on a bicistronic transcript and is required for mesoderm and neurectoderm patterning. Dev Cell 1: 103–114 [DOI] [PubMed] [Google Scholar]
- Lengerke C, Schmitt S, Bowman TV, Jang IH, Maouche-Chretien L, McKinney-Freeman S, Davidson AJ, Hammerschmidt M, Rentzsch F, Green JB, Zon LI, Daley GQ (2008) BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell 2: 72–82 [DOI] [PubMed] [Google Scholar]
- Li Z, Nie F, Wang S, Li L (2011) Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci USA 108: 3116–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X (2009) Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell 17: 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo M, Vinagre T, Carapuço M (2009) The road to the vertebral formula. Int J Dev Biol 53: 1469–1481 [DOI] [PubMed] [Google Scholar]
- Merrill BJ, Pasolli HA, Polak L, Rendl M, García-García MJ, Anderson KV, Fuchs E (2004) Tcf3: a transcriptional regulator of axis induction in the early embryo. Development 131: 263–274 [DOI] [PubMed] [Google Scholar]
- Nagayoshi S, Hayashi E, Abe G, Osato N, Asakawa K, Urasaki A, Horikawa K, Ikeo K, Takeda H, Kawakami K (2008) Insertional mutagenesis by the Tol2 transposon-mediated enhancer trap approach generated mutations in two developmental genes: tcf7 and synembryn-like. Development 135: 159–169 [DOI] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K (2006) Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 1: 179–185 [DOI] [PubMed] [Google Scholar]
- Nguyen H, Merrill BJ, Polak L, Nikolova M, Rendl M, Shaver TM, Pasolli HA, Fuchs E (2009) Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat Genet 41: 1068–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J, McGill MA, Dermer M, Dho SE, Wolting CD, McGlade CJ (2002) LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J 21: 93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström U, Maier E, Jessell TM, Edlund T (2006) An early role for WNT signaling in specifying neural patterns of Cdx and Hox gene expression and motor neuron subtype identity. PLoS Biol 4: e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon N, Oh K, Sylvestre JR, Bouchard N, Savory J, Lohnes D (2006) Cdx4 is a direct target of the canonical Wnt pathway. Dev Biol 289: 55–63 [DOI] [PubMed] [Google Scholar]
- Pilon N, Oh K, Sylvestre JR, Savory JGA, Lohnes D (2007) Wnt signalling is a key mediator of cdx1 expression in vivo. Development 134: 2315–2323 [DOI] [PubMed] [Google Scholar]
- Riedt T, Ebinger M, Salih HR, Tomiuk J, Handgretinger R, Kanz L, Grünebach F, Lengerke C (2009) Aberrant expression of the homeobox gene CDX2 in pediatric acute lymphoblastic leukemia. Blood 113: 4049–4051 [DOI] [PubMed] [Google Scholar]
- Ro H, Dawid IB (2009) Organizer restriction through modulation of Bozozok stability by the E3 ubiquitin ligase Lnx-like. Nat Cell Biol 11: 1121–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro H, Dawid IB (2010) Lnx-2b restricts gsc expression to the dorsal mesoderm by limiting Nodal and Bozozok activity. Biochem Biophys Res Commun 402: 626–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC (2007) p53 activation by knockdown technologies. PLoS Genet 3: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roël G, Hamilton FS, Gent Y, Bain AA, Destrée O, Hoppler S (2002) Lef-1 and Tcf-3 transcription factors mediate tissue-specific Wnt signaling during Xenopus development. Curr Biol 12: 1941–1945 [DOI] [PubMed] [Google Scholar]
- Ruzov A, Hackett JA, Prokhortchouk A, Reddington JP, Madej MJ, Dunican DS, Prokhortchouk E, Pennings S, Meehan RR (2009) The interaction of xKaiso with xTcf3: a revised model for integration of epigenetic and Wnt signaling pathways. Development 136: 723–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl C, Bansal D, Döhner K, Eiwen K, Huntly BJ, Lee BH, Rücker FG, Schlenk RF, Bullinger L, Döhner H, Gilliland DG, Fröhling S (2007) The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. J Clin Invest 117: 1037–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Bae YK, Muraoka O, Hibi M (2005) Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev Biol 279: 125–141 [DOI] [PubMed] [Google Scholar]
- Snider L, Thirlwell H, Miller JR, Moon RT, Groudine M, Tapscott SJ (2001) Inhibition of Tcf3 binding by I-mfa domain proteins. Mol Cell Biol 21: 1866–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoene S, Rawat VP, Heilmeier B, Hoster E, Metzeler KH, Herold T, Hiddemann W, Gökbuget N, Hoelzer D, Bohlander SK, Feuring-Buske M, Buske C (2009) The homeobox gene CDX2 is aberrantly expressed and associated with an inferior prognosis in patients with acute lymphoblastic leukemia. Leukemia 23: 649–655 [DOI] [PubMed] [Google Scholar]
- Thorpe CJ, Weidinger G, Moon RT (2005) Wnt/beta-catenin regulation of the Sp1-related transcription factor sp5l promotes tail development in zebrafish. Development 132: 1763–1772 [DOI] [PubMed] [Google Scholar]
- van den Akker E, Forlani S, Chawengsaksophak K, de Graaff W, Beck F, Meyer BI, Deschamps J (2002) Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development 129: 2181–2193 [DOI] [PubMed] [Google Scholar]
- Veien ES, Grierson MJ, Saund RS, Dorsky RI (2005) Expression pattern of zebrafish tcf7 suggests unexplored domains of Wnt/β-catenin activity. Dev Dyn 233: 233–239 [DOI] [PubMed] [Google Scholar]
- Wang Y, Yabuuchi A, McKinney-Freeman S, Ducharme DM, Ray MK, Chawengsaksophak K, Archer TK, Daley GQ (2008) Cdx gene deficiency compromises embryonic hematopoiesis in the mouse. Proc Natl Acad Sci USA 105: 7756–7761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Tonou-Fujimori N, Komori A, Maeda R, Nojima Y, Li H, Okamoto H, Masai I (2005) Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and notch signaling pathways. Development 132: 3027–3043 [DOI] [PubMed] [Google Scholar]
- Yan D, Lin X (2009) Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol 1: a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, Hsieh J, Bassel-Duby R, Olson EN, Lu QR (2009) HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the β-catenin-TCF interaction. Nat Neurosci 12: 829–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RM, Reyes AE, Allende ML (2002) Expression and splice variant analysis of the zebrafish tcf4 transcription factor. Mech Dev 117: 269–273 [DOI] [PubMed] [Google Scholar]
- Young T, Deschamps J (2009) Hox, Cdx, and anteroposterior patterning in the mouse embryo. Curr Top Dev Biol 88: 235–255 [DOI] [PubMed] [Google Scholar]
- Young T, Rowland JE, van de Ven C, Bialecka M, Novoa A, Carapuco M, van Nes J, de Graaff W, Duluc I, Freund JN, Beck F, Mallo M, Deschamps J (2009) Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev Cell 17: 516–526 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.