Abstract
Histone H3 lysine 4 trimethylation (H3K4me3) is a major hallmark of promoter-proximal histones at transcribed genes. Here, we report that a previously uncharacterized Drosophila H3K4 methyltransferase, dSet1, and not the other putative histone H3K4 methyltransferases (Trithorax; Trithorax-related protein), is predominantly responsible for histone H3K4 trimethylation. Functional and proteomics studies reveal that dSet1 is a component of a conserved H3K4 trimethyltransferase complex and polytene staining and live cell imaging assays show widespread association of dSet1 with transcriptionally active genes. dSet1 is present at the promoter region of all tested genes, including activated Hsp70 and Hsp26 heat shock genes and is required for optimal mRNA accumulation from the tested genes. In the case of Hsp70, the mRNA production defect in dSet1 RNAi-treated cells is accompanied by retention of Pol II at promoters. Our data suggest that dSet1-dependent H3K4me3 is responsible for the generation of a chromatin structure at active promoters that ensures optimal Pol II release into productive elongation.
Keywords: Drosophila , dSet1, heat shock, H3 lysine 4 methylation, transcription elongation
Introduction
Genomic DNA of eukaryotic cells is organized into a nucleoprotein structure called chromatin. The fundamental building block of chromatin is the nucleosome core particle, which consists of 147 base pairs of DNA wrapped around an octamer of the histones H2A, H2B, H3, and H4. Nucleosomes can form an obstacle to processes requiring access to the DNA such as transcription. Covalent modifications of histones at particular residues can alter the properties of nucleosomes by both changing chromatin's compactness and accessibility and by specifying new interactions of histones with transcription factors. One of these modifications, histone H3 lysine 4 trimethylation (H3K4me3), has been shown to be a major conserved mark of chromatin at nucleosomes immediately downstream of promoters of transcribed genes in yeast, Drosophila, and mammals (Pokholok et al, 2005; Barski et al, 2007; Guenther et al, 2007; Schuettengruber et al, 2009). Although nucleosomes carrying this modification are targeted by a number of transcription and chromatin regulators (Martin et al, 2006; Shi et al, 2006; Taverna et al, 2006; Sims et al, 2007; Vermeulen et al, 2007, 2010), its contribution to transcription remains mysterious. In baker's yeast, the mutation in the gene coding for the H3K4 methyltransferase, SET1, does not cause obvious transcription defects during vegetative growth, while in human cells, the functional study of this mark is hampered by the existence of several genes encoding H3K4 trimethyltransferases (Eissenberg and Shilatifard, 2010).
H3K4 methylation is introduced into nucleosomes by the type-2 histone lysine methyltransferases (KMT2s; Allis et al, 2007). The prototypic KMTs from Drosophila are Suppressor of variegation 3–9, Enhancer of zeste, and Trithorax (Trx), and they share similarity in their catalytic SET domain. Of these, only Trx is H3K4-specific, and has been proposed to be the major KMT2 in flies (Shilatifard, 2008; Eissenberg and Shilatifard, 2010). Flies possess a second, Trithorax-related protein (Trr), which functions in the regulation of hormone-response gene expression (Sedkov et al, 1999, 2003). The two Drosophila Trx relatives are related to the mammalian Mixed lineage leukaemia (Mll)1–4 KMT2s (Eissenberg and Shilatifard, 2010). While some Mll complexes contain subunits found in the yeast Set1 complex (COMPASS; Miller et al, 2001; Roguev et al, 2001; Nagy et al, 2002), Trx/Mll relatives have also been shown to assemble into complexes that are unrelated to COMPASS and contain histone acetyltransferases (Ernst et al, 2001; Petruk et al, 2001; Smith et al, 2004; Dou et al, 2005). These findings suggest a functional diversification of Trx/Mll complexes. Surprisingly, genome-wide studies in flies and mammals revealed that Trx/Mll relatives do not localize to over 97% of H3K4me3 domains, including those adjacent to transcription start sites (TSSs) of most expressed genes (Schuettengruber et al, 2009; Wang et al, 2009; Eissenberg and Shilatifard, 2010; Schwartz et al, 2010). Recent studies led to the identification of two Set1 orthologues in humans and showed that they assemble into complexes similar to yCOMPASS (Lee and Skalnik, 2005; Lee et al, 2007a); however, their contribution to overall H3K4me3 is still obscure. Since a Set1 homologue was not found in Drosophila, the role of Set1 versus Trx/Mll relatives in global, transcription-linked H3K4 trimethylation at promoters is still unclear in multicellular organisms.
Here, we report the identification and characterization of a Set1 homologue from Drosophila, CG40351/dSet1. Proteomics and functional studies revealed that the dSet1 complex is identical in its composition to its human counterpart and has strong H3K4 trimethyltransferase activity towards recombinant nucleosomes. RNAi-mediated knockdown (KD) studies demonstrated that dSet1 is responsible for bulk H3K4 di- and trimethylation, while the KD of Trx or Trr had less pronounced effects on H3K4me2/3. dSet1 co-localizes with H3K4me3 and transcribing Pol II on polytene chromosome, and the loss of the dSet1-complex subunit, dCfp1, diminishes dSet1 and H3K4me3 at transcription puffs. The KD of dSet1 causes reduced mRNA levels at all tested genes, including heat shock (HS) genes. Live cell imaging studies also revealed that EGFP–dSet1 is rapidly recruited to the Hsp70 HS loci upon activation. Photobleaching/recovery assays demonstrated that EGFP–dSet1 is continuously exchanged at the activated hsp70 loci. Moreover, time course HS experiments showed that KD of dSet1 caused increased Pol II levels at the hsp70 promoter during extended HS periods. Our data supports a model in which dSet1-dependent H3K4me3 regulates chromatin changes at promoter-proximal nucleosomes that positively influence the release of Pol II into productive elongation, thereby contributing to optimal mRNA levels.
Results
Characterization of the Drosophila dSet1 complex
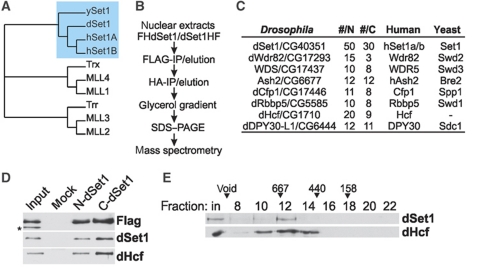
The structural conservation of yeast and human Set1 complexes suggested that Drosophila also might possess a Set1-type protein. Database searches identified CG40351 as the closest match of yeast SET1 and human Set1B. Phylogenetic comparisons of Set1- and Trx/Mll-KMT2s from humans, flies, and yeast indicated that these factors form three distinct subfamilies (Figure 1A). CG40351 falls into the same group with the human and yeast Set1 relatives, while Trx has the highest similarity to MLL1 and MLL4. A third subgroup consists of Trr, MLL2, and MLL3. A comparison of the domain structure of these proteins indicated that the CG40351 protein contains an RNA recognition motif (Tresaugues et al, 2006; Lee et al, 2007a) in the N-terminus (Supplementary Figure S1). This domain is characteristic for Set1-type KMT2s and missing in Trx/Mll-type proteins. These comparative studies also revealed that a variety of domains present in distinct Trx/Mll relatives are absent in CG40351. In summary, the CG40351 protein is structurally very similar to the yeast or human Set1 relatives, but does not have features typical for Trx/Mll-type KMT2s.
Figure 1.
Characterization of the Drosophila dSet1 complex. (A) Phylogenetic comparison of the Set1 and Mll1-4 homologues from yeast, flies, and humans generated with Phylip (bootstrap value 1000, 15 repeats). (B) Purification scheme for FH-dSet1 or dSet1-HF complexes. (C) Identified polypeptides from purified dSet1 complexes compared with human and yeast complexes. #N, #C: unique peptides from N- or C-terminally tagged dSet1. (D) Immunoblot analysis of purified dSet1 complexes. The asterisk marks a non-specific band. (E) Western blot analysis of nuclear extract fractionated by gel filtration. Arrowheads, calibration marker sizes in kDa.
With the exception of a study where CG40351 was knocked down by RNAi to test its involvement in dosage compensation in flies (Yokoyama et al, 2007), this factor has remained uncharacterized. To further confirm that CG40351 codes for a bona fide Set1 homologue, we characterized its interaction partners by mass spectrometry. For this purpose, we generated cell lines expressing an N- or C-terminally FLAG/HA-tagged CG40351. Matching data from both cell lines would ensure that intact complexes were isolated. CG40351 and associated factors were affinity purified from nuclear extracts followed by glycerol gradient centrifugation and compared by Silverstaining (Supplementary Figure S2). CG40351-peak fractions were then analysed by mass spectrometry (Figure 1B). The data from both purifications revealed that CG40351 is a component of a complex similar to human and yeast COMPASS (Figure 1C; Miller et al, 2001; Lee et al, 2007a), and that the tagging of dSet1 in the N- or C-terminus had no major effect on complex assembly and stability. Antisera against CG40351 recognized the same band that was labelled by anti-Flag antibodies confirming that these antisera were specific in immunoblotting experiments (Figure 1D; Supplementary Figure S3). Western blots on fractions from size exclusion chromatography on nuclear extracts showed that dSet1 complexes eluted in a single peak near a 670-kDa marker protein (Figure 1E). The combined theoretical mass of all dSet1-complex subunits in single copy number is about 632.8 kDa. Immunoblotting experiments confirmed that an identified subunit, Host cell factor (dHcf), was present in Drosophila COMPASS (Figure 1D and E). Its human homologue is a component of hCOMPASS (Lee et al, 2007a). Since dHcf associates with other chromatin complexes (Kusch et al, 2003; Wysocka et al, 2003; Guelman et al, 2006), it eluted in a broader profile from sizing columns. In summary, our data indicates that CG40531 is a component of a conserved dCOMPASS complex and the bona fide Set1 homologue in Drosophila. We, therefore, will refer to CG40351 in the following as dSet1.
dSet1 is responsible for bulk H3K4 trimethylation
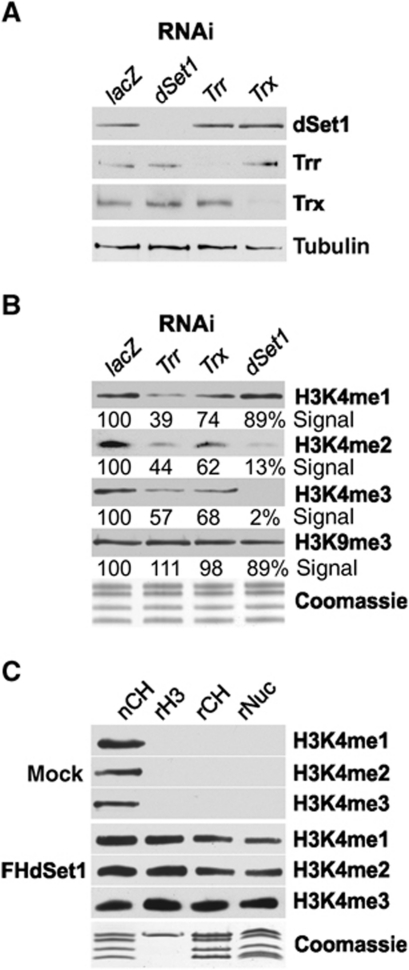
We next conducted RNAi-mediated KD studies to determine the contribution of Trx, Trr, and dSet1 to genome-wide H3K4me1, -me2, and -me3 (Figure 2A and B; Supplementary Figure S4). As determined by signal intensity measurements, the KD efficiency in all cases appears to be higher than 85%. The KD of each KMT2 had no discernable effect on the protein levels of the other factors, and each had a different impact on H3K4 methylation. The loss of Trr most strongly affected H3K4me1, and to a lesser extent H3K4me2 and H3K4me3, relative to lacZ-KD samples (Figure 2B; Supplementary Figure S4), while the KD of Trx had the least influence on H3K4me1, -2, and -3. In dSet1-depleted cells, H3K4me1 was nearly unaffected, while H3K4me2 and -3 showed the most prominent decrease (Figure 2B; Supplementary Figure S4). Note that changes in H3K4me1 or -2 levels in these experiments need to be interpreted cautiously, because flies possess two H3K4-specific demethylases, Su(var)3-3 and Lid, which, respectively, use H3K4me2 or -3 as substrate (Gildea et al, 2000; Eissenberg et al, 2007; Lee et al, 2007b; Rudolph et al, 2007; Secombe et al, 2007; Di Stefano et al, 2011). The results suggested that these KMT2s might in part functionally interact in H3K4 methylation. Combinatorial KDs of Trx/Trr, Trx/dSet1, Trr/dSet1, and triple KDs only showed additive effects, suggesting that these KMT2s act rather independently in H3K4 methylation (not shown).
Figure 2.
dSet1 is required for bulk H3K4 di- and trimethylation. (A) Western blot analysis of nuclear extracts from RNAi-treated cells (top). lacZ: control from cells treated with dsRNA for E. coli lacZ. Tubulin served as loading control. (B) H3K4 methylation changes upon KD of dSet1, Trr, or Trx. Immunoblots of histone extracts from RNAi-treated cells probed with antibodies against methylated H3K4 (me1, me2, me3) or H3K9me3. Values below each lane represent the relative intensity of each band in comparison with the respective lacZ lane as quantified by ImageJ. (C) Purified dSet1 complexes methylate H3K4 in vitro. Immunoblots of KMT assays with the purified dSet1 complex (FHdSet1). Mock, purifications from mock-transfected cells. nCH, native fly histones served as positive control; rH3, recombinant H3; rCH, core histones; rNuc, nucleosomes. Coomassie, loading control.
Using recombinant histone H3, core histones, and nucleosomes as substrates, we next tested if the purified dSet1 complex was capable of trimethylating H3K4 in vitro (Figure 2C). The relative levels of H3K4me1–3 were determined by immunoblots using specific antibodies. To reduce the frequently observed cross-reactivity of anti-methyl-lysine antibodies, competitor H3K4 peptides were added to the reactions (see Materials and methods for details). While mock-purified samples had no discernable H3K4 methylation activity, the purified dSet1 complex was capable of mono-, di-, and trimethylating H3K4 of all three substrates in a processive manner. In vitro studies on other KMT2 complexes revealed that these complexes usually do not fully trimethylate their substrates, but generate substantial quantities of mono- or dimethylated H3K4.
The partial methylation of H3K4 in these reactions is not necessarily indicative of a limited KMT activity of the dSet1 complex in vivo, because H2B ubiquitination is known to enhance H3K4 trimethylation by Set1 complexes (Kim et al, 2009; Takahashi et al, 2009; Chandrasekharan et al, 2010). Moreover, other KMT2 enzymes have been found to be rather inefficient in their methylation activity in vitro (Takahashi and Shilatifard, 2009; Cosgrove and Patel, 2010).
GST fusions of the SET domains of dSet1 (or Trx) showed no KMT activity on H3 and core histones, in contrast to GST-Trr, which had H3K4 monomethyltransferase activity towards H3 and core histones (Supplementary Figure S5). This indicates the catalytic SET domain of dSet, unlike the complete complex, is ineffective as KMT. This was expected, since most SET domain fusions were incapable of trimethylating nucleosomes in vitro (with few exceptions; e.g. HypB; Supplementary Figure S5; Sun et al, 2005).
In summary, our studies indicate that the dSet1 complex has a major role in H3K4me3 in Drosophila and suggest that it might be responsible for deposition of this mark at active promoters. The KD of the two Trx relatives also had some impact on H3K4me3 levels, but their overall contribution is substantially less compared with dSet1.
dSet1 extensively overlaps with H3K4 trimethylation on polytene chromosomes
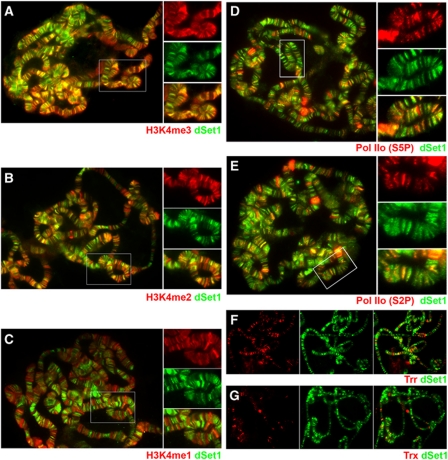
The immunological analyses from RNAi-treated cells indicated that dSet1 has a major role in H3K4 trimethylation in Drosophila S2 cells in immunoblotting experiments (Figure 2B). After confirming that the antibodies against dSet1 were also specific in immunofluorescence staining experiments (Supplementary Figure S6), we next assessed the extent to which dSet1 regulates H3K4 methylation in distinct chromosomal regions during later developmental stages. For this purpose, we co-labelled polytene chromosomes from third-instar larvae with antibodies against dSet1 and H3K4me1, -me2, or -me3 in the presence of competitor peptides (Figure 3A–C). As shown in Figure 3A, the signals of dSet1 and H3K4me3 nearly fully overlapped on polytene chromosomes. H3K4me2 and dSet1 also co-localized at a large number of transcription puffs; however, many H3K4me2 signals were also found in dSet1-negative and DAPI-positive chromatin regions lacking dSet1 (Figure 3B; Supplementary Figure S7). H3K4me1 and dSet1 only overlapped in a few transcription puffs, while the majority of H3K4me1 was found in compacted chromosomal regions (Figure 3C; Supplementary Figure S7).
Figure 3.
dSet1 co-localizes with and is required for H3K4 trimethylation at transcription sites. (A) Chromosomes co-stained with antibodies against H3K4me3 (red) and dSet1 (green). (B) Chromosomes labelled with anti-H3K4me2 (red) and anti-dSet1 (green) antibodies. (C) Chromosomes stained for H3K4me1 (red) and dSet1 (green). (D) Chromosomes co-stained with antibodies against Pol IIo (S5P; red) and dSet1 (green). (E) Chromosomes double labelled with antibodies against Pol IIo (S2P; red) and dSet1 (green). (F) Chromosomes co-labelled for Trr (red) and dSet1 (green). (G) Chromosome spread stained for Trx (red) and dSet1 (green). Yellow/orange signals in the channel merges indicate co-localization. Boxed areas in (A–E) are shown in magnification to the right. Magnifications in (D) and (E) were enhanced for the red channels for better visualization of Pol IIo.
Since H3K4me3 is universally found at the promoters of transcriptionally active genes, we hypothesized that dSet1 may be involved in global transcription regulation. To examine the extent of dSet1 involvement in H3K4 methylation at sites of transcription, we next compared the distributions of dSet1 and transcriptionally engaged Pol II on polytene chromosomes by indirect immunofluorescence microscopy. We examined the co-localization of dSet1 with Pol II phosphorylated at serine 5 of its CTD (a mark of paused and productively elongating Pol II in metazoans; Boehm et al, 2003; Gomes et al, 2006) and elongating serine 2-phosphorylated Pol II (Figure 3D and E), respectively. Our results revealed considerable co-localization between dSet1 and Pol IIo. At most puffs, the dSet1 signals were narrower than Pol IIo bands, and usually concentrated at the edge of transcription puffs (Supplementary Figure S8). While many dSet1-positive regions did not appear to contain easily discernable levels of Pol IIo, a careful inspection of these sites revealed that most contain low levels of Pol IIo (Figure 3D and E, magnifications). Although we cannot exclude that some regions are only positive for dSet1, adjustments of the Pol IIo channel intensity suggested that the vast majority of dSet1-positive bands also labelled for Pol IIo.
To assess the relative abundance and potential contribution of the Trx relatives to H3K4 methylation on polytene chromosomes, we next compared their distributions to that of dSet1. These studies revealed that dSet1-positive regions were far more abundant than Trr or Trx signals (Figure 3F and G). We also noticed that Trr-containing bands are more abundant than Trx-positive ones, which is consistent with previous reports (Kuzin et al, 1994; Chinwalla et al, 1995; Sedkov et al, 2003). In summary, dSet1 appears to be the most abundant of the three KMT2s on polytene chromosomes, and nearly completely overlaps with H3K4me3. This provides further support for the notion that dSet1 is the principal histone H3K4 trimethyltransferase with possible involvement in transcription at the global level.
dCfp1 is required for chromatin association of dSet1 and global H3K4 trimethylation
Recent studies showed that Cfp1, a subunit of hCOMPASS, is required for the restriction of Set1 to euchromatin and directs it to unmethylated CpG islands near active genes (Tate et al, 2010; Thomson et al, 2010). Our mass-spectrometric data indicated that the dSet1 complex contains the fly homologue of this factor, dCfp1 (Figure 1C); however, the lack of CpG islands in flies suggested that the protein might have a different role in the regulation of dSet1-dependent H3K4 methylation.
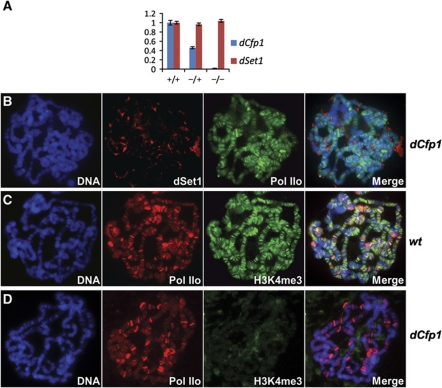
To get a first insight into the role of dCfp1, we decided to analyse polytene chromosomes from flies carrying a P element insertion immediately upstream of the TSS of the dCfp1 gene, which leads to about a 98% reduction of dCfp1 mRNA levels in salivary glands of third-instar larvae as assessed by reverse transcription/quantitative PCR (RT/qPCR; Figure 4A). In these animals, dSet1 expression is not reduced; however, the protein is not detectable on polytene chromosomes, while Pol IIo distribution appeared essentially normal (Figure 4B). Furthermore, H3K4me3, which normally co-localizes extensively with Pol IIo (Figure 4C), was not detectable at transcription puffs in these mutants (Figure 4D). We conclude that dCfp1 is crucial for dSet1 association with chromatin and H3K4 trimethylation at transcription puffs. Additionally, and in contrast to human Cpf1, which prevented the mislocalization of Set1A to heterochromatin, dCpf1 was critical for general chromosomal association of dSet1.
Figure 4.
Loss of dCfp1 abolishes chromosomal association of dSet1 and H3K4me3. (A) RT/qPCR data for dCfp1 (blue) or dSet1 (red) using total RNA from salivary glands of wild-type (+/+), dCfp1 heterozygotes (+/−), or dCfp1 homozygous mutant (−/−) larvae. All values were normalized against rp49. Error bars represent the s.e.m. of three independent RNA preparations. (B) Polytene chromosomes from dCfp1 homozygous larvae co-stained with antibodies against dSet1 (red) and Pol IIo (green). (C) Polytene chromosomes of wild-type larvae co-labelled with antibodies against Pol IIo (red) and H3K4me3 (green). (D) Polytene chromosomes of dCfp1 homozygous mutants co-stained with the same antibodies as in (C). (B–D) DNA was counterstained with DAPI. Yellow-orange signals in the merged channels (right panels) indicate co-localization.
dSet1 is required for optimal transcription from tested genes
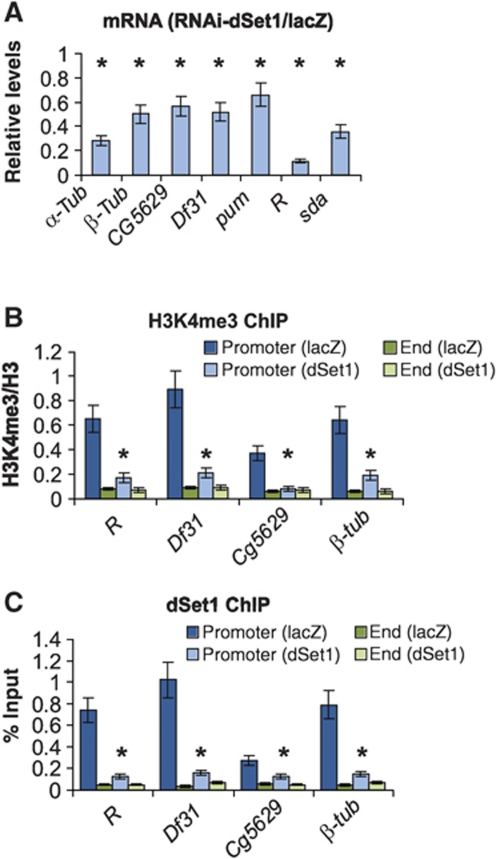
To test whether dSet1-dependent H3K4me3 has a role in transcription, we studied the expression changes on seven genes upon the KD of dSet1 by RT/qPCR. These genes were chosen by their relative expression levels as previously published (Muse et al, 2007). As shown in Figure 5A, the expression of all genes showed a significant drop (40–90% reduction) compared with cells treated with lacZ dsRNA (P<0.005, t-test). Notably, highly expressed genes (R, sda, α-tubulin) showed the strongest reduction, while lower level expressed genes (e.g. CG5629, pum, β-tubulin) were less dependent on dSet1.
Figure 5.
dSet1 is required for transcription and promoter-proximal H3K4 methylation. (A) RT/qPCR values from seven transcribed genes, expressed as ratio of mRNA levels from dSet1 KD versus lacZ-KD samples (=1). All values were normalized against levels of rp49. (B) ChIP/qPCR values of H3K4me3 at promoters versus ends of transcribed genes in dSet1 KD and lacZ-KD cells. The values are plotted as relative enrichment of IP chromatin compared with inputs after normalization using antibodies against H3. (C) ChIP assays evaluating the levels of dSet1 at promoters and ends of transcribed genes, plotted as percent IP versus input chromatin. Error bars represent the s.e.m. of three independent RNAi treatments (*P<0.01, t-test).
We next tested whether the KD of Set1 would affect H3K4me3 at the promoters versus the ends of one strongly (R), two moderately (β-tubulin, Df31), as well as one low-level expressed gene (CG5629) by competitive chromatin immunoprecipitation/qPCR assays (cChIP/qPCR; Figure 5B). In order to account for differences in nucleosome density, the data was normalized using antibodies against H3. While the TSSs of all four genes were enriched for H3K4me3 in controls, the KD of dSet1 significantly diminished H3K4me3 here (P<0.005, t-test). At the 3′ ends, H3K4 methylation levels were low and unchanged in the KD samples. ChIP/qPCR experiments confirmed that dSet1 is mainly enriched at the promoters of these genes (Figure 5C). We also looked at a subset of transcriptionally active genes that are modulated by diverse sets of regulatory elements and tested the RNAi KD effect of all putative histone H3K4 methyltransferases on the levels of histone H3K4me3 at the 5′-end region of these genes. In almost all cases, the KD of dSet1, not Trr or Trx, resulted in a notable decrease in the H3K4me3 levels (Supplementary Figure S9).
dSet1 is rapidly recruited to activated hsp70 loci
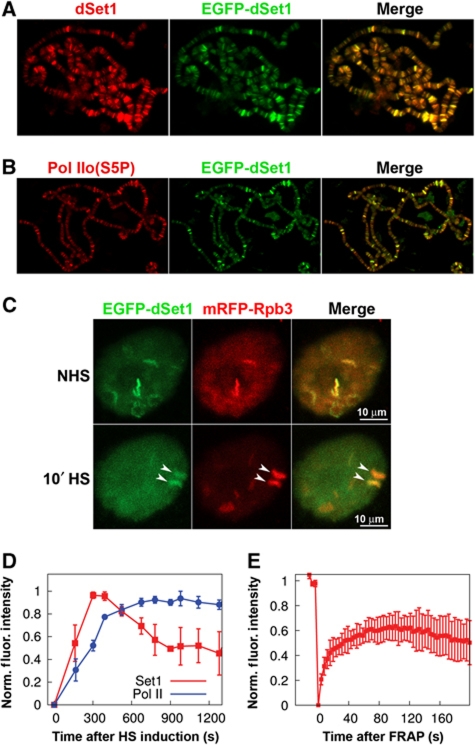
Previous studies showed that hsp loci become trimethylated at H3K4 as early as 2.5 min after HS (Adelman et al, 2006). Therefore, we sought to more closely examine the association and recruitment kinetics of dSet1 to the Hsp70 genes at the 87A and 87C loci. We generated fly lines carrying a transgene allowing for the GAL4-dependent expression (Rorth, 1996) of an EGFP–dSet1 fusion protein and confirmed by immunoblotting experiments that EGFP–dSet1 is detectable in the salivary gland extracts of third-instar larvae (Supplementary Figure S10). Confocal reflection microscopy in combination with laser-scanning confocal microscopy (LSCM) of EGFP confirmed that EGFP–dSet1 predominantly accumulated in the nuclei of the salivary glands (Supplementary Figure S11). Polytene immunostainings using antibodies against GFP showed that EGFP–dSet1 does not localize to DAPI-intense heterochromatic regions (Supplementary Figure S12), and a co-staining using antibodies against dSet1 confirmed that EGFP–dSet1 protein is targeted to the same chromosomal regions as the endogenous protein (Figure 6A). The two showed essentially a complete overlap, indicating that the N-terminal tagging of dSet1 did not negatively affect its chromosomal targeting. Staining with anti-GFP antibodies on polytene chromosomes from wild-type flies further confirmed that the anti-GFP antibodies did not exhibit any unspecific cross-reactivity to chromosomal proteins, including Pol IIo (Supplementary Figure S13). Like untagged dSet1, the EGFP–dSet1 extensively co-localizes with transcriptionally active Pol II (Figures 3D and 6B).
Figure 6.
EGFP–dSet1 is rapidly recruited to the activated Hsp70 loci. Polytene chromosomes from transgenic third-instar larvae expressing EGFP–dSet1 double labelled with antibodies against (A) GFP (green) and dSet1 (red); (B) GFP (green) and Pol IIo (red); (C) laser-scanning microscopy (maximum intensity projections) shows co-localization of EGFP–dSet1 and mRFP–Rpb3 prior and after a 10-min HS in living salivary gland cells. Arrows denote the 87A and 87C HS loci that harbour multiple copies of Hsp70. (D) Recruitment of EGFP–dSet1 and mRFP–Rpb3 (Pol II) to Hsp70 loci as a function of time after HS induction (n=6, error bars represent s.e.m). (E) FRAP recovery plot showing recovery of EGFP–dSet1 signals at the Hsp70 loci. Error bars represent s.e.m. (n=8).
Having established that the N-terminal tagging of dSet1 did not detectably affect its chromosomal targeting, we next sought to closely examine the association and recruitment kinetics of EGFP–dSet1 to the major hsp70 puffs upon HS by LSCM in living salivary gland cells (Yao et al, 2006, 2007, 2008). As observed on polytene chromosomes (Figure 6B), EGFP–dSet1 showed strong co-localization with mRFP–Rpb3 (Pol II subunit) at many loci prior to HS induction in living cells (Figure 6C). Similar to previous reports, Pol II was rapidly recruited to the hsp70 loci and reached a plateau around 8 min after HS (Yao et al, 2007). EGFP–dSet1 also appeared quickly at activated Hsp70 loci with its signal reaching peak intensity at about 6 min after HS induction and slowly declining between 8 and 15 min to about 50% of the maximal levels (Figure 6D; Supplementary Figure S14).
We also utilized fluorescence recovery after photobleaching (FRAP) to study the local exchange dynamics of dSet1 at the 87A and 87C HS puffs (Figure 6E) and other developmental loci associated with dSet1 (Supplementary Figure S15). In the case of the hsp70 puffs, we observed a fast rate of recovery, indicating a rapid exchange of dSet1 at these genes. The reduced extent of recovery of dSet1 levels to about 60% of the initial levels could be explained by the fact that the photobleaching was performed at T=8 min after HS induction, when the dSet1 intensity already had begun to decrease (Figure 6D). We also carried out FRAP analysis at various developmental loci and observed variable recovery profiles for EGFP–dSet1 (Supplementary Figure S15). At some loci, EGFP–dSet1 recovery was rapid and nearly complete, suggesting that dSet1-exchange rates are high in these regions. In other domains, dSet1 signal recovery rates and levels were substantially slower. These results support the dynamic association of dSet1 with active transcription loci, which most likely correlates with transcription levels of these genes as suggested by the RNAi expression studies (Figure 5A). In summary, our live cell imaging studies reveal both a rapid recruitment of dSet1 to newly activated genes as well as a dynamic association between the recruited dSet1 and activated HS and developmental genes.
dSet1 is required for optimal Pol II release from HS promoters
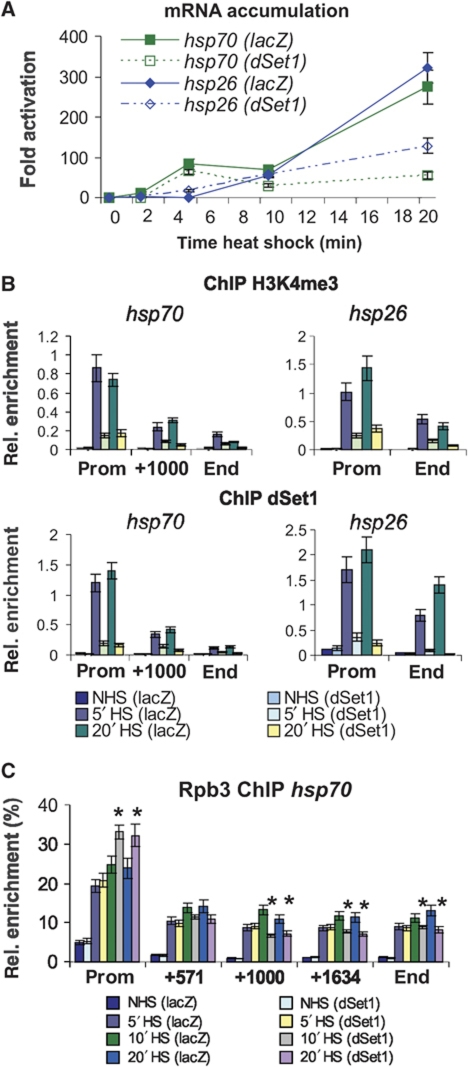
dSet1 and H3K4 methylation become detectable at the hsp loci upon their activation (Figure 6; Adelman et al, 2006). To gain insight into the role of dSet1 in transcription control at these genes, we performed time course/HS experiments and compared the levels of the hsp70 and hsp26 mRNAs by RT/qPCR at 0, 2, 5, 10, and 20 min after HS induction between control and dSet1-depleted samples (Figure 7A). The KD of dSet1 had no major impact on the expression of these genes within the first 10 min as evidenced by the similar mRNA accumulation kinetics in both samples. The strongest increase of hsp mRNA accumulation was observed between 10 and 20 min in the controls. Interestingly, only a slight increase in mRNA levels was found in dSet1-depleted cells during this period. Similar observations were made on the hsp83 gene, which, unlike Hsp70, has relatively high basal transcription levels (Supplementary Figure S16). In summary, these data indicate that dSet1-dependent H3K4me3 is most important for maximal transcription up-regulation during later stages of HS induction.
Figure 7.
dSet1 is required for the maintenance of transcription from HS genes. (A) RT/qPCR data using hsp70- and hsp26-specific amplicons from dSet1 KD (dotted lines) or lacZ-KD cells (solid lines) at 0′, 2′, 5′, 10′, and 20′ at 37°C. Fold activation, signals were plotted relative to values from cells at 25°C. (B) ChIP/qPCR data from dSet1 KD cells using antibodies against H3K4me3 (middle) or dSet1 (bottom) at different HS times (0′, 5′, 20′; see legend in bottom). Amplicons at promoters (prom), about 1000 bp downstream of the TSS (+1000), and at the 3′ end (end) of the hsp70 locus were used. The hsp26 gene only spans 1010 bp. NHS, non-heat shock; HS, heat shock. (C) Pol II levels along the axis of the hsp70 gene significantly change upon the loss of dSet1 after 10′ HS (*P<0.01, t-test). Plotted are qPCR values from anti-Rpb3-ChIP using hsp70-specific amplicons normalized to signals from anti-H3CT ChIP with an amplicon targeting a non-transcribed, intergenic region. Samples from NHS, 5′, 10′, or 20′ min HS are compared. All error bars represent the s.e.m. of at least three independent RNAi treatments.
To determine the kinetics of H3K4 trimethylation along the axis of the hsp loci, we performed cChIP/qPCR experiments. As shown in Figure 7B, H3K4me3 levels were low and constant along the body of both hsp genes prior to HS. After 5 min of transcription activation, its levels had substantially increased at the TSSs of both genes, with some minor elevation of H3K4me3 levels within the first 1000 bp downstream of the TSSs. No major increase of H3K4me3 was observed after 5 min, showing that its maximal levels are reached earlier than changes in mRNA levels. The KD of dSet1 led to a strong reduction of promoter-proximal H3K4me3 during HS. ChIP experiments confirmed that both hsp genes are direct targets of dSet1 and that dSet1 is recruited with the same rapid kinetics as observed for H3K4 trimethylation (Figure 7B). We observed the same kinetics of dSet1 recruitment and H3K4 trimethylation at the hsp83 locus; however, the effects were less dramatic since hsp83 is already expressed at high levels in S2 cells (Supplementary Figure S16). In summary, dSet1-dependent H3K4me3 at the TSSs of the hsp loci reaches peak levels as early as 5 min after activation.
Recent studies have shown that the rate-limiting step of transcription at many genes is the release of ‘paused’ Pol II into productive elongation (Core and Lis, 2008). Since H3K4me3 at the hsp promoters is strongly reduced in the dSet1-KD samples, we tested whether the distribution of Pol II along the axis of the activated hsp70 gene changes upon the loss of dSet1. As shown in Figure 7C, HS time course/ChIP experiments with antibodies against the Pol II subunit, Rpb3, revealed that the KD of dSet1 led to a modest but reproducible increase in Pol II levels in the hsp70 promoter/5′-end region after 10 min of HS. This was also accompanied with a reduction of Pol II levels in downstream regions (P<0.01; t-test) and suggested that the KD of dSet1 may be negatively affecting the rate of Pol II release into productive elongation. This effect on the entry into productive elongation could be responsible for the reduction of mRNA levels in dSet1 KD, which correlated with transcription levels of the affected genes (Figure 5A) and occurred during phases of maximal transcription up-regulation (Figure 7A); however, it could also partly be explained by defects in RNA processing, which would lead to increased RNA degradation.
Discussion
Here, we have characterized the Drosophila dSet1 complex and demonstrated that it is a component of a conserved complex responsible for bulk H3K4me3. dSet1 is required for H3K4me3 at all transcribed genes tested and its loss leads to a reduction of transcription levels accompanied by the accumulation of Pol II at promoters, which occur during phases of maximal transcription up-regulation. Our findings support a model in which dSet1-dependent H3K4me3 generates a chromatin architecture facilitating the passage of Pol II through promoter-proximal nucleosomes during multiple rounds of transcription.
dSet1 complex is the main H3K4 trimethyltransferase in Drosophila
To this date, the role of Trx/Mll versus Set1 orthologues in transcription-linked H3K4 methylation is not fully understood. Our studies support that dSet1 and Trx/Mll relatives form distinct subfamilies that most likely have non-overlapping functions in H3K4 methylation (Figures 1A, 2B, 3F and G; Supplementary Figure S1). While it has been proposed that Trx functions in complexes similar to Set1 in flies (Eissenberg and Shilatifard, 2010), our study along with other reports support that Set1 homologues have a broader role in transcription-linked H3K4 methylation (Petruk et al, 2001; Schwartz et al, 2010; Figures 2B, 3F and G).
Recent studies showed that Trx might even have a role in development independent of its KMT activity (Schwartz et al, 2010). A C-terminal cleavage product of Trx (Trx-Cter) containing the SET domain was found at sites lacking H3K4me3. Trx-Nter distributed over large domains of active PcG-target genes, which also lacked H3K4me3, but were acetylated at H3K27 (H3K27ac). H3K27ac is introduced by CBP, and its KAT activity depends on its association with Trx-Nter (Tie et al, 2009). This acetylation protects H3K27 from methylation by PcG complexes, suggesting that Trx can function in positive gene regulation independent of its KMT activity. Consistent with this model, the loss of the elongation factor, Kismet, abolished the association of Trx with chromatin, while global H3K4me3 levels were unaffected (Srinivasan et al, 2008). In these studies, chromosomal H3K4me3 was also unchanged in trx mutants. These findings are consistent with our observations that H3K4me3 nearly fully overlapped with dSet1 and depended at most sites on the dSet1 complex (Figures 2B, 3A, 4B and D).
Our studies and previous reports support that Trr has a more widespread role in H3K4 methylation compared with Trx (Sedkov et al, 2003; Figures 2B, 3F and G). The SET domain of Trr had robust monomethyltransferase activity in vitro, and we observed that the KD of Trr affected H3K4me1 (Figure 2B; Supplementary Figure S5). Since isolated SET domains of KMT2s are only capable of monomethylating H3K4 (Cosgrove and Patel, 2010), further functional studies on purified Trr complexes will be necessary to address whether this factor has limited H3K4 KMT activity. In summary, dSet1 complexes are likely to regulate H3K4me3 in most chromatin regions, including the promoters of the majority of active genes, while Trx and Trr might function in diverse complexes with primary roles in developmental gene regulation.
Conserved function of Set1 complexes in H3K4 trimethylation
The characterization of the yeast and human Set1 complexes supported a functional and structural conservation between these complexes (Lee and Skalnik, 2005; Eissenberg and Shilatifard, 2010). Our proteomics studies indicate that this is also the case for the Drosophila Set1 complex, which is identical in its composition to the human complex (Figure 1C). We further observed functional similarities between the Cfp1 subunit of Set1 complexes in metazoans. In dCfp1 mutants, dSet1 dissociated from chromatin and H3K4me3 levels on polytene chromosomes were severely reduced (Figure 4B and D). In humans, Cfp1 is essential for the targeting of hCOMPASS to euchromatin (Tate et al, 2010), while yeast yCfp1p/Spp1p is required for the trimethyltransferase activity of yCOMPASS (Takahashi et al, 2009; Chandrasekharan et al, 2010; Murton et al, 2010).
Our in vitro methyltransferase assays revealed that dSet1 complexes trimethylate H3K4 in recombinant nucleosomes (Figure 2C). In vivo, H2B ubiquitination stimulates nucleosomal H3K4 trimethylation by Set1 (Takahashi and Shilatifard, 2009; Eissenberg and Shilatifard, 2010). Our data suggests that H2Bub has no direct regulatory role on the dSet1 complex, which is consistent with studies reporting that H2Bub prevents the loss of H2A/H2B heterodimers by Pol II-dependent transcription in vivo (Takahashi et al, 2009; Chandrasekharan et al, 2010). This might explain why the stable association of Set1 complexes with nucleosomes is less dependent on H2Bub in a transcription-independent context. In conclusion, our data supports that dCOMPASS and its counterparts in other eukaryotes are highly conserved at the structural and functional level.
Role of dSet1-dependent H3K4me3 in transcription
The role of Set1 in transcription initiation versus elongation is still under debate (Eissenberg and Shilatifard, 2010). It is well established that the 5′ end of the hsp genes are occupied by paused Pol II (Saunders et al, 2006; Core and Lis, 2008). Our data indicates that promoter occupancy by Pol II at the HS and many other loci does not depend on dSet1. Pol II levels were also mostly unchanged at transcription puffs in dCfp1 mutants, and the KD of dSet1 did not cause a drop in Pol II levels at the activated Hsp70 gene (Figures 4B, D and 7C). Furthermore, dSet1 and H3K4me3 levels increased at hsp loci after their activation (Figures 6 and 7), further supporting that dSet1 accumulation positively correlates with transcriptional activity. In fact, dSet1-dependent H3K4me3 appears to directly correlate with gene expression levels, and the KD of dSet1 has stronger impact on highly expressed genes such as α-tubulin (84B), sda, R, or the hsp loci (Figures 5A and 7). Interestingly, the hsp26, hsp70, and hsp83 mRNA accumulation defects occurred in dSet1-depleted cells during phases of maximal up-regulation between 10 and 20 min post-HS (Figure 7; Supplementary Figure S16). Recent genome-wide studies on the role of Ash2, a subunit of the dSet1 complex (Figure 1C), revealed that Ash2-dependent H3K4me3 was most critical for the expression of strongly expressed genes (Perez-Lluch et al, 2011), which is fully consistent with our studies.
The retention of Pol II at the hsp70 promoter in dSet1-depleted cells is accompanied with a decrease in Pol II density in the body of the gene (Figure 7C). This suggests that dSet1 might have a role in the release of Pol II into productive elongation, which has been shown to be the rate-limiting step in transcription (Core and Lis, 2008). This appears to be different from yeast, in which the promoter occupancy of Pol II was reduced at the MET16 locus in cells lacking SET1 (Santos-Rosa et al, 2003). Given the relatively modest effect of dSet1 KD on the Pol II density across the body of Hsp70 gene, it is likely that dSet1-dependent H3K4me3 has other functions in transcription; however, these roles must be restricted to promoter-proximal nucleosomes due to the accumulation of dSet1 in the 5′ regions of active genes (Figures 5 and 7). Considering that H3K4me3 is required for the recruitment of pre-mRNA processing and elongation factors to the 5′ regions of genes (Sims et al, 2007), it cannot be excluded that the observed hsp mRNA accumulation defects are due in part to the degradation of improperly processed pre-mRNAs. We did not observe major changes in the phosphorylation states of Pol IIo along the axis of the activated hsp70 gene (not shown). This was not unexpected, since serine 5 phosphorylation and Pol II pausing at the hsp70 promoters precedes H3K4me3 (Adelman et al, 2006), supporting that this process is independent of H3K4me3.
Our data support a model in which dSet1-dependent H3K4me3 successively leads to the generation of a chromatin structure facilitating the repeated passage of Pol II through promoter-proximal chromatin. Since the release of Pol II into productive elongation is rate limiting (Core et al, 2008; Seila et al, 2008), H3K4me3 might form a docking platform for other chromatin modifiers, which generate nucleosomes that are less refractive to Pol II passage. Felsenfeld and co-workers showed that the nucleosomes at promoters of highly expressed genes are enriched for H3.3 and H2A.Z in hyperacetylated form (Jin et al, 2009). The candidate H3.3-exchange factor, CHD1, was shown to bind to H3K4-trimethylated nucleosomes, and H3.3 is the main target for H3K4 trimethylation at active genes (Schwartz and Ahmad, 2005; Konev et al, 2007; Hodl and Basler, 2009; Jin et al, 2009). It is tempting to speculate that H3K4me3 might contribute to the generation of unstable nucleosomes containing H3.3 and H2A.Z, which would be consistent with the observed continuous exchange of dSet1 at the activated hsp70 puffs (Figures 6D and E). On the other hand, we cannot rule out the possibility that histone H3K4me3-independent functions of the dSet1 complex are critical for its role in transcription.
Finally, dSet1-dependent H3K4me3 is likely to recruit factors that generate a chromatin architecture at promoters, which is critical for optimal transcription levels. In fact, a number of transcription and chromatin regulators were confirmed to interact with nucleosomes trimethylated at H3K4 (Eissenberg and Shilatifard, 2010); however, repressive complexes like the HDAC1 complexes were also found to be recruited by this mark (Shi et al, 2006). Future studies are necessary to identify other key regulators that interact with dSet1 complexes in the regulation of maximal transcription levels.
Materials and methods
Tissue culture, complex purification, and mass spectrometry
The full-length open reading frame of dSet1 (clone CG40351, Flybase) was amplified from 1 μg of total RNA, and error-free clones were introduced into FLAGHA or HAFLAG expression vectors that allow for expression from the inducible metallothionein promoter (Kusch et al, 2004). Stable transgenic Drosophila S2 cell lines were generated and the tandem-affinity purifications were performed as described before (Kusch et al, 2004). In brief, S2 cells were transfected with the expression vectors that carry a hygromycin resistance cassette. After selection, test, and expansion of stable transfectants, the cells were expanded in suspension culture, and expression of the recombinant dSet1 proteins was induced by the addition of copper sulphate. After an induction of 2 days, nuclear extract from 2 × 109 cells was prepared for the tandem-affinity purifications. The affinity-purified proteins were concentrated in a 14% Tris–Tricine gel and excised as a single gel slice for mass-spectrometric analyses (UMDNJ/Rutgers Proteomics core facilities). A detailed description can be found in the supplement. The size exclusion chromatography was performed as previously described (Kusch et al, 2003). Either pooled dSet1-peak fractions or 0.5 mg of nuclear extracts were separated on a Superose 6 1/24 column (GE Healthcare).
Recombinant proteins and antibody production
The following protein fragments were used to generate His6 or GST fusions as previously described (Kusch et al, 2003, 2004): dSet1: aa 263–599 (antigen), aa 1324–1641 (SET domain); Trr: aa 2199–2410; Trx: aa 3412–3759 (cDNA was a gift of A Mazo). The HypB expression construct was a gift of B Li. Anti-dSet1 antibodies were generated by Covance, Inc.
Recombinant histones and KMT assays
Histone purifications, generation of histone octamers, and nucleosomal arrays have been described previously (Kusch et al, 2004). For KMT assays, 125 ng of H3 or 500 ng of octamers were incubated with 50 fmol of purified FHdSet1 complexes or controls in KMT buffer (40 mM Tris–Cl (pH 8.5), 40 mM NaCl, 0.5 mM EDTA, 1 mM MgCl2, 1 mM DTT, 5% glycerol, 50 μM S-adenosyl methionine) for 1 h and 30 min at 37°C. The samples were separated in 14% Tris–Tricine SDS gels prior to western analyses.
Immunological methods
Co-immunoprecipitation assays were performed as previously described (Kusch et al, 2004). For western blots, the following antibodies were used: anti-dSet1 (guinea pig: 1:3000); anti-dHcf (rabbit, 1:2000); anti-Trr (Sedkov et al, 2003); rabbit, 1:1500 (a gift of A Mazo); anti-Trx (Smith et al, 2004); rabbit, 1:1000 (a gift of A Mazo); anti-H3K4me3 (rabbit, 1:6000), anti-H3K4me2 (rabbit, 1:6000), anti-H3K4me1 (rabbit, 1:6000), anti-H3 (1:25000, all from Active Motif); anti-tubulin (mouse, 1:2000, Sigma); anti-Flag M2 (1:1000, Agilent); anti-GST (rabbit, 1:1000), and anti-GFP (Goat, 1:2000, Rockland). Band intensities were determined using ImageJ (NIH).
Polytene chromosomes were dissected from third-instar Oregon R or y1w76c23P[w+mCy+mDint2=EPgy2]EY23011 larvae, which carry a P element insertion 84 bp upstream of the TSS of dCfp1. RT/qPCR confirmed that dCfp1 expression is reduced by >98% in homozygous animals. The fixation and staining protocols are published elsewhere (Kusch et al, 2003; Srinivasan et al, 2008). The following antibody concentrations were used: anti-dSet1 (1:40; guinea pig), anti-H3K4me1, -me2, -me3 (1:15; rabbit), anti-GFP (1:50, chicken, Abcam), anti-phosphorylated Pol II (H5, H14; both 1:40; Covance; anti-PIIS5P, anti-PIIS2P, both 1:25, gift of D. Eick; Chapman et al, 2007), anti-Trr, and anti-Trx (both 1:40, rabbit; a gift of A Mazo); 5 fmol of antibody-unspecific competitor H3 (aa1-17; K4me0-3) peptides (a gift of M Vermeulen) was added in stainings with methyl-H3K4-specific antibodies. Microscopy was performed on a Deltavision II Deconvolution system (Applied Scientific). Pictures were processed using ImageJ (NIH) and Photoshop (Adobe). For the GFP-dSet1 experiments, image acquisition and data analysis was carried out using Zeiss Axioplan 2 microscope and velocity imaging software.
Laser-scanning confocal imaging
Drosophila salivary glands were dissected from third-instar larva into 5:6 Grace's Media, as previously described (Yao et al, 2008) and were used immediately for imaging. Dissected salivary glands were transferred with medium to a Bioptechs FCS3 Closed Chamber System with a 0.2-mm spacer. A Carl Zeiss LSM510 META microscope was used to obtain confocal images using identical C-Apochromat 63x, 1.2 NA, water immersion objectives to image during NHS, and HS. For HS, the heated objective was swapped in for unheated objective, and HS times were started at the moment the objective contacted the immersion medium.
Kinetic analysis of Set1 recruitment
A scan zoom of 3 × was used to image an individual nucleus from each salivary gland. We obtained a 35-μm pre-activation z-stack, imaging every 1 μm with both channels alternating every slice. Then the pre-heated objective was moved into position. Time intervals were around 150 s and the time series lasted 20 min. Images were taken at a resolution of 512 × 512 pixels, using 12-bit colour depth.
Fluorescence recovery after photobleaching
A circular region of interest limited to the dimensions of the Hsp70 loci was selected for photobleaching GFP, using 100% laser power (488 nm laser). Under these conditions, the samples were photobleached to about 40–60% of the initial intensity. Post-acquisition, the images were corrected for photobleaching occurring over the course of imaging by measuring the fluorescence at a small nuclear region and the sections containing the Hsp70 loci were identified and mean fluorescence intensity was measured. FRAP curves were double normalized so that pre-bleach images equal one, and first images after the bleach equal 0.
Confocal reflection microscopy
Drosophila salivary glands were dissected and prepared for imaging as above. Images were taken using a Ph2 Plan-NEOFLUAR 25 × 0.8 NA oil immersion objective on the Carl ZeissLSM510 META. Glands were illuminated with 488 nm laser light. The reflected light was detected using a band pass 390–465 filter and the GFP fluorescence was detected using a band pass 500–550 filter.
HS, RNAi, RT/qPCR, and ChIP/qPCR
RNAi was performed as previously described (Kusch et al, 2004). For RT, total RNA from S2 cells or 20 salivary glands was isolated using Trizol following standard protocols (Invitrogen). A total of 5 μg of DNase-treated RNA, 200 U of MMLV RT, and a mix of reverse primers were used for the first strand synthesis. Reactions with 5′ primers served as negative controls. The gene rp49 was used as calibrator. qPCR was performed with 0.25% of the first strand reaction using a Realplex4 system (Eppendorf).
HS experiments were performed as described (Ardehali et al, 2009). ChIP was performed using chromatin from 5 × 107 S2 cells. The preparation of chromatin and ChIP were performed as previously published (Schwartz et al, 2010). The following antibody amounts were used: anti-dSet1 (2 μl); anti-H3K4me3 (1.5 μl, 5 fmol of competitor peptides), and anti-Rpb3 (4 μl). Significance of the data was calculated using Student's paired two-tailed t-test assuming equal variance. Primer sequences are listed in the supplement.
Supplementary Material
Acknowledgments
We thank D Eick, B Li, A Mazo, and M Vermeulen for reagents and antibodies, H Fernandez for technical help during early phases of the project, H Zheng for assistance with mass spectrometry, and J Werner for the preparation of the polytene chromosome stainings in Figure 6, and Supplementary Figures S12–S14. We also thank W Belden for insightful comments on the manuscript. This work was supported by GM25232 from the National Institutes of Health (to JTL) and a grant from the Busch Biomedical Research Foundation (to TK).
Author contributions: AM, MC, and TK generated the data shown in Figures 1–5, 7, S1–S3, S5–S8, and S16. MBA generated data for Figure 6 and Supplementary Figures S4, S9–S11. KLZ generated the live imaging data for Figure 6 and Supplementary Figure S15. MBA, JTL, and TK wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adelman K, Wei W, Ardehali MB, Werner J, Zhu B, Reinberg D, Lis JT (2006) Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol 26: 250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwein T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y (2007) New nomenclature for chromatin-modifying enzymes. Cell 131: 633–636 [DOI] [PubMed] [Google Scholar]
- Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, Lis JT (2009) Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J 28: 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT (2003) Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol 23: 7628–7637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Chen YC, Sun ZW (2010) Histone H2B C-terminal helix mediates trans-histone H3K4 methylation independent of H2B ubiquitination. Mol Cell Biol 30: 3216–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D (2007) Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318: 1780–1782 [DOI] [PubMed] [Google Scholar]
- Chinwalla V, Jane EP, Harte PJ (1995) The Drosophila trithorax protein binds to specific chromosomal sites and is co-localized with polycomb at many sites. EMBO J 14: 2056–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Lis JT (2008) Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319: 1791–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove MS, Patel A (2010) Mixed lineage leukemia: a structure-function perspective of the MLL1 protein. FEBS J 277: 1832–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano L, Walker JA, Burgio G, Corona DF, Mulligan P, Naar AM, Dyson NJ (2011) Functional antagonism between histone H3K4 demethylases in vivo. Genes Dev 25: 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG (2005) Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121: 873–885 [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Lee MG, Schneider J, Ilvarsonn A, Shiekhattar R, Shilatifard A (2007) The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat Struct Mol Biol 14: 344–346 [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A (2010) Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol 339: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ (2001) MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol 21: 2249–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildea JJ, Lopez R, Shearn A (2000) A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics 156: 645–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM (2006) Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev 20: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, Anderson S, Yates JR III, Washburn MP, Abmayr SM, Workman JL (2006) Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol 26: 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodl M, Basler K (2009) Transcription in the absence of histone H3.3. Curr Biol 19: 1221–1226 [DOI] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G (2009) H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG (2009) RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137: 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, Vershilova E, Pirrotta V, Kadonaga JT, Lusser A, Fyodorov DV (2007) CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science 317: 1087–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR III, Abmayr SM, Washburn MP, Workman JL (2004) Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306: 2084–2087 [DOI] [PubMed] [Google Scholar]
- Kusch T, Guelman S, Abmayr SM, Workman JL (2003) Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol Cell Biol 23: 3305–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzin B, Tillib S, Sedkov Y, Mizrokhi L, Mazo A (1994) The Drosophila trithorax gene encodes a chromosomal protein and directly regulates the region-specific homeotic gene fork head. Genes Dev 8: 2478–2490 [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG (2005) CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem 280: 41725–41731 [DOI] [PubMed] [Google Scholar]
- Lee JH, Tate CM, You JS, Skalnik DG (2007a) Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem 282: 13419–13428 [DOI] [PubMed] [Google Scholar]
- Lee N, Zhang J, Klose RJ, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2007b) The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat Struct Mol Biol 14: 341–343 [DOI] [PubMed] [Google Scholar]
- Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L (2006) The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol 26: 7871–7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A (2001) COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murton BL, Chin WL, Ponting CP, Itzhaki LS (2010) Characterising the binding specificities of the subunits associated with the KMT2/Set1 histone lysine methyltransferase. J Mol Biol 398: 481–488 [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K (2007) RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML (2002) A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci USA 99: 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Lluch S, Blanco E, Carbonell A, Raha D, Snyder M, Serras F, Corominas M (2011) Genome-wide chromatin occupancy analysis reveals a role for ASH2 in transcriptional pausing. Nucleic Acids Res (advance online publication, 9 February 2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A (2001) Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science 294: 1331–1334 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF (2001) The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 20: 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P (1996) A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci USA 93: 12418–12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt A, Jenuwein T, Reuter G (2007) Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell 26: 103–115 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bernstein BE, Karabetsou N, Morillon A, Weise C, Schreiber SL, Mellor J, Kouzarides T (2003) Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol Cell 12: 1325–1332 [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT (2006) Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol 7: 557–567 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G (2009) Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz BE, Ahmad K (2005) Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev 19: 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V (2010) Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet 6: e1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J, Li L, Carlos L, Eisenman RN (2007) The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev 21: 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedkov Y, Benes JJ, Berger JR, Riker KM, Tillib S, Jones RS, Mazo A (1999) Molecular genetic analysis of the Drosophila trithorax-related gene which encodes a novel SET domain protein. Mech Dev 82: 171–179 [DOI] [PubMed] [Google Scholar]
- Sedkov Y, Cho E, Petruk S, Cherbas L, Smith ST, Jones RS, Cherbas P, Canaani E, Jaynes JB, Mazo A (2003) Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature 426: 78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA (2008) Divergent transcription from active promoters. Science 322: 1849–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF et al. (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442: 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A (2008) Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol 20: 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ III, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D (2007) Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ST, Petruk S, Sedkov Y, Cho E, Tillib S, Canaani E, Mazo A (2004) Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol 6: 162–167 [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Dorighi KM, Tamkun JW (2008) Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet 4: e1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XJ, Wei J, Wu XY, Hu M, Wang L, Wang HH, Zhang QH, Chen SJ, Huang QH, Chen Z (2005) Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem 280: 35261–35271 [DOI] [PubMed] [Google Scholar]
- Takahashi YH, Lee JS, Swanson SK, Saraf A, Florens L, Washburn MP, Trievel RC, Shilatifard A (2009) Regulation of H3K4 trimethylation via Cps40 (Spp1) of COMPASS is monoubiquitination independent: implication for a Phe/Tyr switch by the catalytic domain of Set1. Mol Cell Biol 29: 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi YH, Shilatifard A (2009) Structural basis for H3K4 trimethylation by yeast Set1/COMPASS. Adv Enzyme Regul 50: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate CM, Lee JH, Skalnik DG (2010) CXXC finger protein 1 restricts the Setd1A histone H3K4 methyltransferase complex to euchromatin. FEBS J 277: 210–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD (2006) Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell 24: 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, Turner DJ, Illingworth R, Bird A (2010) CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ (2009) CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila polycomb silencing. Development 136: 3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresaugues L, Dehe PM, Guerois R, Rodriguez-Gil A, Varlet I, Salah P, Pamblanco M, Luciano P, Quevillon-Cheruel S, Sollier J, Leulliot N, Couprie J, Tordera V, Zinn-Justin S, Chavez S, van Tilbeurgh H, Geli V (2006) Structural characterization of Set1 RNA recognition motifs and their role in histone H3 lysine 4 methylation. J Mol Biol 359: 1170–1181 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M (2010) Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, Ge K, Krumlauf R, Shilatifard A (2009) Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol 29: 6074–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W (2003) Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev 17: 896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT (2007) Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell 28: 978–990 [DOI] [PubMed] [Google Scholar]
- Yao J, Munson KM, Webb WW, Lis JT (2006) Dynamics of heat shock factor association with native gene loci in living cells. Nature 442: 1050–1053 [DOI] [PubMed] [Google Scholar]
- Yao J, Zobeck KL, Lis JT, Webb WW (2008) Imaging transcription dynamics at endogenous genes in living Drosophila tissues. Methods 45: 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Pannuti A, Ling H, Smith ER, Lucchesi JC (2007) A plasmid model system shows that Drosophila dosage compensation depends on the global acetylation of histone H4 at lysine 16 and is not affected by depletion of common transcription elongation chromatin marks. Mol Cell Biol 27: 7865–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.