Abstract
Hypoxia stabilizes the tumour suppressor p53, allowing it to function primarily as a transrepressor; however, the function of p53 during hypoxia remains unclear. In this study, we showed that p53 suppressed BNIP3 expression by directly binding to the p53-response element motif and recruiting corepressor mSin3a to the BNIP3 promoter. The DNA-binding site of p53 must remain intact for the protein to suppress the BNIP3 promoter. In addition, taking advantage of zebrafish as an in vivo model, we confirmed that zebrafish nip3a, a homologous gene of mammalian BNIP3, was indeed induced by hypoxia and p53 mutation/knockdown enhanced nip3a expression under hypoxia resulted in cell death enhancement in p53 mutant embryos. Furthermore, p53 protected against hypoxia-induced cell death mediated by p53 suppression of BNIP3 as illustrated by p53 knockdown/loss assays in both human cell lines and zebrafish model, which is in contrast to the traditional pro-apoptotic role of p53. Our results suggest a novel function of p53 in hypoxia-induced cell death, leading to the development of new treatments for ischaemic heart disease and cerebral stoke.
Keywords: apoptosis, BNIP3, hypoxia, p53, zebrafish
Introduction
BNIP3 was first identified using the adenovirus E1B-19K protein as bait in a yeast two-hybrid screen (Boyd et al, 1994; Yasuda et al, 1998). BNIP3 and BNIP3-like (BNIP3L) protein constitute a subfamily of BH3-only proteins with similar functions (Matsushima et al, 1998; Chen et al, 1999; Mellor and Harris, 2007; Chinnadurai et al, 2008; Giam et al, 2008; Zhang and Ney, 2009). Unlike other BH3-only proteins, BNIP3 fails to interact with anti-apoptotic members (Ray et al, 2000; Sowter et al, 2003; Burton and Gibson, 2009). Instead, transcriptional mechanisms that involve the hypoxia-inducible factor (HIF) complex under hypoxic conditions indirectly regulate BNIP3 apoptotic activity by modulating expression levels. The promoter of BNIP3 contains a functional hypoxia-response element (HRE) and its mRNA and protein expression are dramatically increased in multiple cell types in response to hypoxia (Kim et al, 1997b; Bruick, 2000; Guo et al, 2001; Sowter et al, 2001; Kothari et al, 2003; Zhang et al, 2008). In cultured cells, enforced expression of BNIP3 induces cell death characterized by localization of BNIP3 at the mitochondria, opening of the mitochondrial permeability transition pore and loss of mitochondrial membrane potential (Δψm) (Vande Velde et al, 2000). Notably, this BNIP3-mediated apoptosis occurs without cytochrome c release from the mitochondria or caspase activation (Chen et al, 1997; Vande Velde et al, 2000; Regula et al, 2002; Prabhakaran et al, 2007), although other studies have shown that BNIP3 also appears to mediate release of cytochrome c via a novel mechanism (Lomonosova and Chinnadurai, 2008). Enforced expression of a BNIP3 mutant lacking its transmembrane domain (BNIP3ΔTM) partially blocks hypoxia-induced cell death, further supporting a critical role for the TM domain in hypoxia-induced apoptosis (Vande Velde et al, 2000; Kothari et al, 2003; Burton and Gibson, 2009). A truncated BNIP3 protein lacking part of the PEST domain, and all of the TM domain, CD domain and BH3 domain detected in the SkOv3 ovarian cancer cell line is also found to inhibit hypoxia-induced cell death (Bristow et al, 2011). Evidence indicates that BNIP3 may mediate apoptosis of cardiomyocytes under conditions of ischaemic stress based on the findings that hypoxia activates BNIP3 transcription and acidosis induces BNIP3 activity by promoting membrane translocation (Crow, 2002; Kubasiak et al, 2002; Regula et al, 2002; Graham et al, 2004; Webster et al, 2005, 2006; Frazier et al, 2006; Hamacher-Brady et al, 2007; Dorn and Kirshenbaum, 2008; Kubli et al, 2008; Weidman et al, 2008). In addition, in a knockout mouse model, BNIP3 has a role in cellular response to ischaemia/reperfusion injury in the heart, further supporting its importance in ischaemic heart disease (Diwan et al, 2007). Moreover, BNIP3 is also identified to induce autophagic-type cell death, as characterized by the formation of autophagic vacuoles, and localization and processing of LC3 in epithelial-derived cells, glioma cells and fibroblasts (Bristow et al, 2011). Recent studies suggested that BNIP3 may have a dual function in the myocardium, where it regulated both mitochondrial turnover via autophagy and cell death (Gustafsson, 2011).
Abundant evidence indicates that p53 functions as a tumour suppressor through its ability to induce apoptosis in cells under stressful conditions (Symonds et al, 1994; Miyashita and Reed, 1995; Zhu et al, 1998; Attardi and Jacks, 1999; Aurelio et al, 2000; Oda et al, 2000a; Schmitt et al, 2002). Given that hypoxia represents a major stress during tumour progression, the role of p53 in mediating apoptosis under hypoxia has been extensively investigated (Graeber et al, 1996; Giaccia and Kastan, 1998; Stempien-Otero et al, 1999; Denko et al, 2000; Pan et al, 2004; Hammond and Giaccia, 2005). Studies have shown that tumours with wild-type p53 undergo significant hypoxia-induced apoptosis while tumours with mutated p53 do not (Graeber et al, 1996). In addition, p53 levels stabilize under severe hypoxia, with evidence supporting both HIF-1α-dependent or -independent mechanisms (An et al, 1998; Blagosklonny et al, 1998; Alarcon et al, 1999; Koumenis et al, 2001; Suzuki et al, 2001; Chen et al, 2003; Hammond and Giaccia, 2006). As a well-defined transcription factor, p53 transactivates many target genes in response to genotoxic stress, thereby mediating apoptosis in cultured cells and tumour tissues (Miyashita and Reed, 1995; Levine, 1997; Roperch et al, 1998; Venot et al, 1998; Soengas et al, 1999, 2001; Attardi et al, 2000; Zhu et al, 2000; Oda et al, 2000b; Nakano and Vousden, 2001; Wang et al, 2001; Fei et al, 2002, 2004; Sax et al, 2002; Jeffers et al, 2003; Mirza et al, 2003; Shibue et al, 2003; Villunger et al, 2003; Yu et al, 2003; Menendez et al, 2009). However, despite the number of p53-activated apoptosis induced genes identified to date, including Bax, Puma, Noxa and Perp, none has been shown to mediate p53-dependent cell death under hypoxia (Hammond and Giaccia, 2005). In fact, under hypoxic conditions, p53 cannot recruit coactivators such as CBP or p300 to the promoters of target genes, and as a result, p53 cannot up-regulate these genes (Koumenis et al, 2001; Schmid et al, 2004; Hammond and Giaccia, 2005).
p53 also functions as a transrepressor and mounting evidence suggests that, under hypoxia, p53 primarily induces apoptosis by repressing gene expression rather than promoting it (Sabbatini et al, 1995; Murphy et al, 1996, 1999; Ahn et al, 1999). Work by Koumenis et al (2001) demonstrated that p53 acts as a transrepressor during hypoxia by forming a complex with the transcriptional corepressor, mSin3a. The identification of genes repressed by p53 during hypoxia may provide a better understanding of how p53 induces apoptosis in this environment. However, even though the list of p53-repressed genes is growing, relatively few have been shown to have a direct role in apoptosis, and even fewer in hypoxia-induced apoptosis (Wang et al, 2001).
The prevailing view is that p53 promotes apoptosis, but this may not always be the case. Notably, hypoxia-induced apoptosis of cardiac myocytes appears to occur independently of p53; in these cells, hypoxia does not affect expression of p53 target genes Bax and Bak (Graham et al, 2004). In addition, cardiac myocytes from p53 wild-type mice are more resistant to hypoxia-induced apoptosis than myocytes from p53 null mice, as revealed by DNA fragmentation analysis (Graham et al, 2004). Furthermore, Matoba et al (2006) reported that the endurance of p53 null mice is significantly reduced as compared with that of wild-type littermates as measured by a swimming stress test, implying that wild-type p53 status could help mice to survive longer under hypoxic conditions. Interestingly, as a putative downstream gene of p53, Bax has been implicated in hypoxia-induced apoptosis; however, Bax-deficient cells undergo similar amounts of apoptosis under hypoxic conditions (Alarcon et al, 1999). These observations suggest that p53 might have a protective role against the hypoxia-induced apoptotic pathway, contrasting to its traditional role for inducing cell death.
Zebrafish has three copies of BNIP3 homologous genes (bnip3 (ENSDARG00000045033), bnip3b (ENSDARG00000022832) and nip3a (AF493987)) in database. Heart-targeted overexpression of nip3a in zebrafish embryos caused abnormal heart development and cardiac dysfunction, implying an important role of nip3a in heart homeostasis (Wang et al, 2006). However, whether zebrafish BNIP3 homologous genes are induced by hypoxia and have similar roles as that of mammalian BNIP3 remain unclear. P53 mutant zebrafish have been well characterized (Berghmans et al, 2005). Similar to that of p53 null mice, multiple organ tumours were developed in p53 mutant zebrafish, indicating that zebrafish p53 has similar tumour suppressive function as that of mammalian p53 (Berghmans et al, 2005; Patton et al, 2005). To further investigate the relationship between BNIP3 and p53 by in vivo model, we took advantage of both p53 mutant zebrafish embryos and their counterpart wild-type embryos to explore the regulation of nip3a by p53.
In this paper, we demonstrate that p53 directly transrepresses BNIP3 expression to protect cells against hypoxia-induced cell death in vitro and in vivo. The finding that p53 may have a protective role in hypoxia-induced apoptosis may provide a better understanding of p53's function during hypoxia and a new approach for developing treatments for ischaemia heart disease and cerebral strokes.
Results
p53 suppressed BNIP3 promoter activity efficiently under normoxic and hypoxic conditions
Because p53 directly induces expression of BNIP3L under hypoxia (Fei et al, 2004), we were interested in determining if p53 could also regulate expression of the closely related BNIP3. To evaluate the potential relationship between p53 and BNIP3, we performed a series of promoter assays. We transfected a human BNIP3 promoter luciferase reporter (Kothari et al, 2003) and increasing amounts of Myc-tagged human p53 expression vector, along with a Renilla control, into p53-deficient Saos-2 cells. Cells were then incubated under normoxic conditions. Surprisingly, p53 dramatically inhibited activity of the BNIP3 promoter reporter in a dose-dependent manner (Figure 1A). Western blots using anti-myc antibody verified p53 expression (Figure 1B). We obtained similar results in HEK293 cells and Hela cells (data not shown). To test whether p53's suppression of the BNIP3 promoter is evolutionary conserved, we performed the same assays using a rodent BNIP3 promoter reporter (Bruick, 2000) in Saos-2, 293 and Hela cells. p53 inhibited activity of the rodent BNIP3 promoter as well in all cell lines (data not shown).
Figure 1.
p53 suppressed BNIP3 promoter activity. (A) Dose-dependent suppression of human BNIP3 promoter activity by human p53 in Saos-2 cells under normoxia. (B) The expression of Myc–p53 under normoxia was confirmed by western blot using anti-Myc antibody. α-Tubulin was used as a loading control. (C) Human BNIP3 promoter activity was induced under hypoxia (1% O2) in Saos-2 cells. (D) The suppression of human BNIP3 promoter activity by human p53 in Saos-2 cells was further confirmed under normoxia. (E) The suppression of human BNIP3 promoter activity by human p53 in Saos-2 cells was confirmed under hypoxia. Data are reported as mean±s.d. of three independent experiments performed in triplicate; the statistical analysis was performed using GraphPad Prism 5.0 (t-tests).
Sometimes, overexpression construct could produce mutant form protein, which actually functioned as dominant-negative factor. To rule out the possibility that the expression vectors of p53 used in this study produced dominant-negative form of p53, we cotransfected either Myc-tagged p53 or HA-tagged p53 together with promoter luciferase reporters of five well-defined p53 downstream genes (Mdm2-luc., Bax-luc., p21-luc., Puma-luc. and p27-luc.), and observed that the p53 overexpression constructs could indeed dramatically up-regulate the luciferase activity of these five promoter reporters (data not shown).
We next evaluated if p53 could also suppress BNIP3 promoter activity under hypoxic conditions (1% O2) using the p53-deficient Saos-2 cells in similar promoter assays. The first set of assays tested the response of the BNIP3 promoter to hypoxia (Figure 1C); the second set further confirmed p53's suppression of BNIP3 promoter activity under normoxic conditions (Figure 1D) and the third set tested the effect of p53 expression on BNIP3 promoter activity under hypoxic conditions (Figure 1E). As shown in Figure 1C, hypoxia (1% O2) indeed induced BNIP3 promoter activity as previously reported (Kothari et al, 2003). Moreover, p53 suppressed BNIP3 promoter activity significantly (P<0.0001; Figure 1D) when cells were subjected to normoxic condition. p53 also suppressed activation of the BNIP3 promoter under hypoxia (P=0.0052; Figure 1E). Similar results were also obtained in HEK293 cells and Hela cells (data not shown). This observation that p53 effectively suppressed BNIP3 promoter activity under hypoxic conditions is in contrast to its ability to activate the BNIP3L promoter (Fei et al, 2004). Given the similar role of both BNIP3 and BNIP3L in hypoxia-induced apoptosis (Mellor and Harris, 2007), these results suggest that p53 might have a novel function through suppressing BNIP3 expression in this pathway.
The potential p53-binding site in BNIP3 promoter was responsible for p53 inhibiting BNIP3 promoter activity under normoxia and hypoxia
In the human BNIP3 promoter, two HREs (HRE1 and HRE2) have been identified, with HRE2 serving a critical function in the response of the gene to hypoxia (Kothari et al, 2003). Considering that p53 suppresses BNIP3 promoter activity under hypoxic conditions, we next evaluated the importance of HRE1 or HRE2 in this response. Using PCR, we introduced a single mutation into HRE1 or HRE2 or simultaneous mutations into both HRE1 and HRE2 (Figure 2A and B). After transfecting Hela cells with the appropriate constructs, we then conducted a series of promoter assays under either normoxic or hypoxic conditions. The results indicated that p53 still suppressed the activity of either the single or double HRE mutants (Figure 2D). However, as reported previously, the activity of HRE2 mutants' response to hypoxia was dramatically decreased (Figure 2D) (Kothari et al, 2003).
Figure 2.
p53 suppression of BNIP3 required p53RE (RE1 and RE2), but not HREs in the BNIP3 promoter. (A) The schematic depiction of the BNIP3 promoter with mutations in the hypoxia-response elements (HRE1 and HRE2) and p53REs (RE1 and RE2). (B) The HRE mutants of BNIP3 promoter. The nucleotide substitutions and the positions are indicated. (C) The RE mutants of the BNIP3 promoter. The nucleotide substitutions and the positions are indicated. (D) The suppressive activity of human p53 on HRE mutants of human BNIP3 promoter was evaluated in Hela cells under normoxia and hypoxia. (E) The human p53 completely lost its suppressive activity on the BNIP3 promoters mutated in either p53-RE deletions (RE1-del and RE1/2-del) or p53-RE substitutions (RE1-mut and RE2-mut) under normoxia and hypoxia conditions. Data are reported as mean±s.d. of three independent experiments performed in triplicate; the statistical analysis was performed using GraphPad Prism 5.0 (t-tests).
We suspected that a p53-response element (RE) in the BNIP3 promoter may be responsible for p53's suppressive activity; however, the promoter appeared to lack a classic p53RE (Menendez et al, 2009). Recently though, the p53RE responsible for either transactivation or repression has been redefined (Wang et al, 2009). Based on the new characterization of a p53RE, we changed some of the search parameters, which led to the identification of a candidate p53RE that covers the promoter region spanning from –1146 to –1112. This potential p53RE comprises two decamer motifs, RYRCWWGRWW and WWWCWWGRRR, separated by a spacer of 16 bp. We then did deletion and substitution mutations in this candidate p53RE (Figure 2A and C), and performed promoter assays under both normoxia and hypoxia using these promoter mutants. As shown in Figure 2E, either deletion (RE1-del and RE1/2-del) or substitution (RE1-mut and RE2-mut) in the candidate p53RE of the BNIP3 promoter ablated the ability of p53 to repress activity. Interestingly, under hypoxia, the activity of all RE mutants (RE1-del, RE1/2, RE1-mut and RE2-mut) was higher than that of wild-type BNIP3 promoter, which suggests that the endogenous p53 expressed in Hela cells may suppress BNIP3 promoter activity effectively. Taken together, we concluded that the potential p53RE in the BNIP3 promoter is required for p53 suppression under both normoxia and hypoxia.
The full-length p53 and DNA-binding site of p53 were required for p53's suppressive function on BNIP3 promoter activity
p53 has three well-characterized functional domains; a transactivation domain (aa 1–100), a DNA-binding domain (aa 102–292) and an oligomerization/regulation domain (aa 323–393). In order to determine which region of p53 is responsible for its suppression of the BNIP3 promoter, we performed domain mapping, using five deletion mutants that were cloned into the pCGN-HAM vector to generate HA-tagged fusion proteins (Figure 3A). The expression of wild-type p53 and the five deletion mutants was checked by western blots using a monoclonal antibody against HA. Except for HA–p53(1–100), Hela cells expressed all of the constructs efficiently (Figure 3B). As reported previously, p53(1–100) is targeted for ubiquitin proteasomal degradation, which likely accounts for why HA–p53(1–100) was not detected (Scheffner et al, 1993). After performing a series of promoter assays, we found that none of the deletion mutants could suppress BNIP3 promoter activity, under either normoxia or hypoxia (Figure 3C and D), suggesting that the full length of p53 is required for its suppression of the BNIP3 promoter.
Figure 3.
The full-length and DNA-binding site of p53 were required for p53 suppression of BNIP3. (A) The schematic depiction of the p53 mutants. (B) The expression of the p53 mutants was verified by western blot using a monoclonal antibody against HA, GAPDH was used as a loading control. (C) The suppressive activity of p53 mutants on the BNIP3 promoter was measured by promoter assays under normoxia in Hela cells. (D) The suppressive activity of p53 mutants on the BNIP3 promoter was evaluated by promoter assays under hypoxia in Hela cells. (E) The expression of HA-tagged wild-type p53 and DNA-binding site mutated p53 (R175) was verified by western blot using anti-HA antibody under normoxia and hypoxia. (F) The suppressive activity of wild-type p53 and mutated p53 (R175H) on the BNIP3 promoter was checked by promoter assays under normoxia. (G) The suppressive activity of wild-type p53 and mutated p53 (R175H) on BNIP3 promoter was determined by promoter assays under hypoxia. Data of luciferase reporter assays are reported as mean±s.d. of three independent experiments performed in triplicate; the statistical analysis was performed using GraphPad Prism 5.0 (t-tests).
Previous studies have shown that the ability of p53 to serve as a transcription factor or transrepressor requires the protein to bind to the target promoter DNA (Wang et al, 2009). To determine if this holds true for the suppression of BNIP3, we used a p53 DNA-binding site mutant (p53(R175H)) in promoter assays under both normoxia and hypoxia. The expression of HA-tagged p53(R175H) was checked by western blot (Figure 3E). The results show that p53(R175) had no any detectable suppressive effects on BNIP3 promoter activity (Figure 3F and G). Taken together, these observations suggest that the suppressive activity of p53 likely requires direct binding of the protein to the BNIP3 promoter.
P53 directly bound to the BNIP3 promoter
To determine whether p53 can directly bind to the candidate p53RE site in human BNIP3, we performed a chromatin immunoprecipitation (ChIP) assay using lysates from Hela cells. The positions of PCR primers used to detect BNIP3, p21 and β-actin promoter fragments are indicated in Figure 4A. A monoclonal anti-p53 antibody precipitated the 123-bp DNA fragment corresponding to site RE of BNIP3 promoter (Figure 4B3, line 4 from left to right), which was further confirmed by sequencing (data not shown). In contrast, no DNA was recovered from cells immunoprecipitated with a mouse IgG control (Figure 4B3, line 3). In order to confirm the reliability of the ChIP assay, we also used a negative control that detected 252 bp of the β-actin promoter region and a positive control that detected 270 bp of the p21 promoter region (Saramaki et al, 2006). As shown in Figure 4B1, no DNA fragment corresponding to the β-actin promoter was recovered from cells immunoprecipitated with either the anti-p53 antibody or mouse IgG control. However, a 270-bp DNA fragment corresponding to the p53-binding site of the p21 promoter was recovered from cells immunoprecipitated with the anti-p53 antibody, but not with the mouse IgG control (Figure 4B2). Furthermore, to determine the extent of p53 binding to BNIP3 promoter, we performed semi-quantitative real-time PCR analysis. The results showed that anti-p53 antibody could indeed pull down 123 bp of BNIP3 promoter region more significantly compared with mouse IgG (Figure 4C; P=0.0474).
Figure 4.
p53 and mSin3a can bind to the BNIP3 promoter directly as revealed by ChIP assays. (A) Schematic diagram depicts the fragments of the BNIP3, p21 and actin genes that were amplified. The positions of PCR primers used to detect BNIP3, p21 and β-actin promoter fragments are indicated with arrows. (B) Hela cells were treated with formaldehyde to create cross-links between p53 and chromatin. The chromatin was isolated, sheared and immunoprecipitated (IP) using a monoclonal antibody against p53 or mouse IgG was used as a control. The presence of chromatin fragments corresponding to p21 (B2), BNIP3 (B3) or β-actin (B1) gene promoter were assessed by PCR using gene-specific primers. The p21 promoter was used as a positive control (B2). (C) Semi-quantitative RT–PCR analysis for ChIP assays. (D) The secondary ChIP analysis for corepressor molecule mSin3a. After the DNA–protein complex pulled down by anti-p53 antibody was eluted by p53 mini-peptide, anti-mSin3A antibody could not pull down β-actin promoter region (negative control) (D1), but could pull down p53-binding region in the BNIP3 promoter (D2). Semi-quantitative RT–PCR analysis confirmed that mSin3A binds to BNIP3 promoter dramatically (D3). Data of RT–PCR analysis are reported as mean±s.d. of three independent experiments performed in triplicate; the statistical analysis was performed using GraphPad Prism 5.0 (t-tests).
If p53 binds to the BNIP3 promoter and represses its expression, a transcriptional repressor complex should be recruited to the BNIP3 promoter by p53. As reported, for acting its suppressive role, p53 associates with corepressor molecules such as mSin3a and HDACs to form a repression complex on the promoter of target genes (Hammond and Giaccia, 2005). To further confirm that p53 represses BNIP3 expression through recruiting corepressor to the BNIP3 promoter, we conducted the secondary ChIP assays. After eluting the protein–DNA complex from anti-p53-conjugated agarose protein A/G beads by p53 mini-peptide, we performed ChIP assays using anti-mSin3a antibody. As shown in Figure 4D, anti-mSin3a antibody precipitated the 123-bp DNA fragment corresponding to RE site of BNIP3 promoter (Figure 4D2, line 4 from left to right), which was further verified by sequencing. No DNA was recovered from the protein–DNA complex immunoprecipitated with a rabbit IgG control (Figure 4D2, line 3). As predicted, β-actin promoter region and p21 promoter region could not be recovered from the protein–DNA complex immunoprecipitated with either rabbit IgG or anti-mSin3a antibody (Figure 4D1 and data not shown). Semi-quantitative RT–PCR further confirmed that anti-mSin3a antibody pulled down 123 bp of the BNIP3 promoter more dramatically than that of rabbit IgG (Figure 4D3; P=0.0007). However, in semi-quantitative RT–PCR assays for the p21 promoter, no signal could be detected from the complex immunoprecipitated with either anti-mSin3a antibody or rabbit IgG (data not shown).
Based on the results, we conclude that p53 directly binds to the candidate p53 site in the human BNIP3 promoter and recruits corepressor mSin3a to the BNIP3 promoter.
Endogenous p53 stimulation suppressed endogenous BNIP3 expression
To confirm that BNIP3 is a p53-suppressed gene, we examined BNIP3 expression in HCT116 cells, which has intact and functional p53 (Polyak et al, 1996), after exposure to the chemotherapeutic agent, 5-flurouracil (5-Fu). As expected, 5-Fu effectively induced endogenous p53 expression in HCT116 cells under both normoxic and hypoxic conditions (Figure 5A and C). In addition, the p53 protein level was elevated under hypoxia, as reported previously (An et al, 1998) (Figure 5A and C). Western blot analysis was performed using an antibody against human BNIP3 (Abcam, cat# ab10433) to check endogenous BNIP3 expression. As previously observed, this antibody could detect multiple endogenous BNIP3 protein bands migrated from 30 kDa monomer to 60 kDa dimmer in several human cell lines (Tracy et al, 2007; Zhang et al, 2008; Bellot et al, 2009). As expected, lower level of BNIP3 protein was expressed in wild-type p53-expressing HCT116 cells after treatment with 5-Fu as compared with HCT116 cells treated with the control vehicle under both normoxia and hypoxia (Figure 5A and C). In addition, semi-quantitative RT–PCR revealed lower levels of BNIP3 mRNA expression in HCT116 cells after treatment with 5-Fu as compared with HCT116 cells treated with the control vehicle under both normoxia and hypoxia (Figure 5B and D), which further refined that p53 suppressed BNIP3 expression in transcriptional level. Similar results were obtained in Hela cells (Supplementary Figure S1).
Figure 5.
Endogenous p53 stimulation by 5′-Fu (fluorouracil) treatment inhibited BNIP3 expression under normoxia and hypoxia in HCT116 cells. (A, C) The expression of endogenous p53 was stimulated by 5-Fu (middle panels in A, C) and the expression of BNIP3 was inhibited by p53 up-regulation (upper panels in A, C) under normoxia (A) and hypoxia (C), as revealed by western blot, using monoclonal antibodies against BNIP3 and p53, respectively. β-Actin was used as the loading control (bottom panels in A, C). BNIP3 antibody can detect multiple endogenous BNIP3 bands between 30 and 60 kDa. (B, D) The expression of endogenous BNIP3 mRNA was suppressed by p53 up-regulation under normoxia (B) and hypoxia (D) as revealed by semi-quantitative RT–PCR (P=0.0014 and P=0.0145, respectively). The primers to amplify human 18S RNA was used as an internal control. Data of RT–PCR analysis are reported as mean±s.d. of three independent experiments performed in triplicate; the statistical analysis was performed using GraphPad Prism 5.0 (t-tests).
Endogenous p53 knockdown resulted in the up-regulation of endogenous BNIP3 expression
To further evaluate the ability of p53 to suppress BNIP3 gene expression endogenously, we examined BNIP3 protein levels by western blot and mRNA levels by semi-quantitative RT–PCR in wild-type p53-expressing HCT116 cells after p53 depletion by p53 siRNA treatment. Endogenous p53 was indeed knocked down by siRNA treatment under both normoxia (about four-fold) and hypoxia (about three-fold) as revealed by western blot (Figure 6A and C). In contrast to p53 stimulation by 5-Fu treatment, p53 knockdown caused a significant increase of endogenous BNIP3 protein levels and mRNA levels (Figure 6A–D). Similar results were obtained in Hela cells (Supplementary Figure S2).
Figure 6.
p53 knockdown by p53 siRNA up-regulated BNIP3 expression under normoxia and hypoxia in HCT116 cells. (A, C) the expression level of endogenous p53 protein was knocked down by p53 siRNA treatment (middle panels in A, C) and the expression level of endogenous BNIP3 protein was increased (upper panels in A, C) after p53 knockdown under normoxia (A) and hypoxia (C), as revealed by western blot using monoclonal antibodies against p53 and BNIP3. β-Actin was used as the loading control (bottom panels in A, C). The relative fold of p53 protein was marked underlined. (B, D) BNIP3 mRNA was up-regulated by p53 knockdown under normoxic (B) and hypoxic (D) conditions as revealed by semi-quantitative RT–PCR (P=0.01 and P=0.0001, respectively). The primers to amplify human 18S RNA was used as an internal control. Data of RT–PCR analysis are reported as mean±s.d. of three independent experiments performed in triplicate; the statistical analysis was performed using GraphPad Prism 5.0 (t-tests).
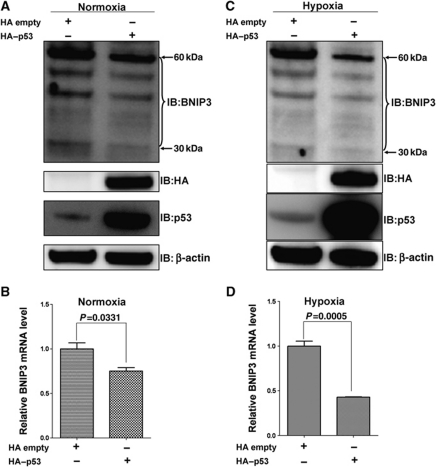
Exogenous p53 overexpression suppressed endogenous BNIP3 expression
To reevaluate the suppressive ability of p53 on BNIP3 expression, we examined BNIP3 protein levels and mRNA in HCT116 cells after transiently transfected p53 expression vector (HA-tagged p53). Exogenous p53 overexpression was confirmed by western blot using either an antibody against HA tag or an antibody against p53 (Figure 7A and C). Consistent with endogenous p53 stimulation by 5-Fu treatment, p53 overexpression caused decrease of endogenous BNIP3 protein levels and mRNA levels (Figure 7A–D). Taken together, p53 appears to suppress BNIP3 expression effectively.
Figure 7.
p53 overexpression suppressed BNIP3 expression under normoxia and hypoxia in HCT116 cells. (A, C) The expression level of endogenous BNIP3 protein was reduced by p53 overexpression under normoxia (A) and hypoxia (C), as revealed by western blot using monoclonal antibodies against BNIP3, HA tag and p53. β-Actin was used as the loading control. (B, D) The expression of endogenous BNIP3 mRNA was suppressed by p53 overexpression under normoxia (B) and hypoxia (D) as revealed by semi-quantitative RT–PCR (P=0.0331 and P=0.0005, respectively). The primers to amplify human 18S RNA was used as an internal control. Data of RT–PCR analysis are reported as mean±s.d. of three independent experiments performed in triplicate; the statistical analysis was performed using GraphPad Prism 5.0 (t-tests).
Endogenous p53 knockdown resulted in the down-regulation of endogenous BNIP3L expression and endogenous p53 stimulation up-regulated endogenous BNIP3 expression
It has been reported that BNIP3L, a BNIP3 homologous gene, is induced by p53 under hypoxia and apoptosis is reduced in hypoxia-exposed cells with functional p53 following BNIP3L knockdown (Fei et al, 2004). To validate the approaches used for determining the suppression of BNIP3 by p53 in this study, we also examined the expression of BNIP3L in HCT116 cells with endogenous p53 knockdown or stimulation under hypoxia. As shown in Supplementary Figure S3, endogenous p53 knockdown mediated by pSuper-p53 RNAi transfection resulted in the down-regulation of endogenous BNIP3L expression (Supplementary Figure S3A) and endogenous p53 stimulation mediated by 5-Fu treatment up-regulated endogenous BNIP3 expression (Supplementary Figure S3B). These results were consistent with the observations reported by Fei et al (2004) previously, which also validated the results about p53 suppression of BNIP3 obtained in this study.
p53 efficiently protected against hypoxia-induced cell death mediated by suppressing BNIP3
Among the known activities of BNIP3, mediating hypoxia-induced apoptosis explains the majority of its effects. Here, we showed that p53 could suppress BNIP3 expression effectively under hypoxic conditions. These data led us to hypothesize that p53 might protect against hypoxia-induced cell death through BNIP3 suppression. Subsequently, we intended to know whether p53 had a protection role in hypoxia-induced cell death. HCT116 cells were chosen for this assay due to its wild-type p53 intact. Because overexpression of p53 in HCT116 induces growth arrest (Polyak et al, 1996), which may disturb further apoptosis assays, we analysed hypoxia-induced apoptosis in p53 knockdown status, but not p53 overexpression status. The results indicate that compared with GFP siRNA transfection, the cell death ratios in p53 siRNA-treated HCT116 cells increased significantly under hypoxia (Supplementary Figure S4; P<0.0001). Similar results were obtained in Hela cells (Supplementary Figure S5; P<0.0001).
To understand whether the enhancement of cell death in p53 knockdown HCT116 cells under hypoxia was mediated by p53 suppression of BNIP3, we further conducted cotransfection experiments by taking advantage of dominant-negative form of BNIP3 (dnBNIP3(1–153 aa)), which has been revealed to block BNIP3-induced cell death specifically and efficiently (Vande Velde et al, 2000; Kothari et al, 2003; Burton and Gibson, 2009). As shown in Figure 8, the overexpression of dnBNIP3 reduced cell death significantly in HCT116 cells transfected with pSuper-GFP siRNA plus HA–dnBNIP3 expression vector compared with cells transfected with pSuper-GFP siRNA plus HA empty vector (Figure 8A1, A2 and B, column 2 compared with column 1 from left to right; P=0.0288), consistent with the observations reported previously (Kothari et al, 2003). p53 knockdown mediated by pSuper-p53 siRNA transfection enhanced cell death (Figure 8A1, A3 and B, column 3 compared with column 1 from left to right; P=0.0432), similar to what was observed in p53 knockdown cells mediated by p53 siRNA treatment (Supplementary Figures S4 and S5). However, the overexpression of dnBNIP3 in p53 knockdown cells reduced cell death significantly (Figure 8A3, A4 and B, column 4 compared with column 3 from left to right; P=0.0015).
Figure 8.
p53 protected against hypoxia-induced cell death mediated by p53 suppression of BNIP3. (A) HCT116 cells were transfected with pSuper-GFP siRNA plus HA empty vector (control), pSuper-GFP siRNA plus HA–dnBNIP3 (HA–dnBNIP3(1–153 aa)), pSuper-p53 siRNA plus HA empty vector or pSuper-p53 siRNA plus HA–dnBNIP3, respectively, under hypoxia. Hoechst staining was used to detect dying cells. The dying cells were indicated with red arrows. (B) The quantitative analysis was used for the dead cell ratio (%). p53 knockdown enhanced hypoxia-induced cell death significantly (column 3 compared with column 1 from left to right, P=0.0432), but overexpression of dnBNIP3(1–153 aa) in p53 knockdown cells significantly reduced hypoxia-induced cell death (column 4 compared with column 3, P=0.0015) and resulted in cell death ratio reaching to a similar level of the control (column 4 compared with column 1, P=0.1979). Data are reported as mean±s.d. representing nine randomly selected fields of three wells in six-well plate; the statistical analysis was performed using GraphPad Prism 5.0 (t-tests). (C) The expression of HA–dnBNIP3 was confirmed by western blot using a monoclonal antibody against HA. α-Tubulin detection was used as a loading control.
Taken together, these observations suggest that p53 may protect against hypoxia-induced cell death mediated by suppressing BNIP3 expression. Due to extensive apoptosis-induction role of p53 in most cell lines when overexpression, we were unable to directly test the protection role of p53 in hypoxia-induced cell death. However, further zebrafish in vivo model might provide additional evidences to refine the conclusion obtained by cell culture system.
p53 inhibited hypoxia-induced autophagy mediated by suppressing BNIP3
Multiple evidences indicate that p53 can modulate autophagy in a dual manner, either induction of autophagy or inhibition of autophagy, leading to both cell survival and cell death (Tasdemir et al, 2008b; Maiuri et al, 2010). Hypoxia and BNIP3 also induce autophagy (Rouschop and Wouters, 2009; Zhang and Ney, 2009). Moreover, hypoxia-induced BNIP3 expression increases autophagic cell death (Tracy et al, 2007). These observations led us to figure out whether p53 also modulates autophagy through suppressing BNIP3 under hypoxia and whether this autophagy modulation contributes to the cell death response.
To monitor cell autophagy, initially, we constructed a vector for expressing LC3–GFP fusion protein by PCR (Tracy et al, 2007; Tasdemir et al, 2008a). When HCT116 cells were transfected with HA–dnBNIP3 without p53 knockdown, the autophagy was reduced significantly under hypoxia (Supplementary Figure S6A1, A2 and B, column 2 compared with column 1 from left to right; P=0.0325), indicating that BNIP3 blocking inhibited cell autophagy, consistent with BNIP3's function revealed previously (Daido et al, 2004; Tracy et al, 2007). However, when p53 was knocked down by pSuper-p53 siRNA transfection in HCT116 cells under hypoxia, the autophagy was increased dramatically (Supplementary Figure S6A1, A3 and B, column 3 compared with column 1 from left to right; P=0.0254), indicating that p53 inhibition could induce autophagy, consistent with what was reported previously (Tasdemir et al, 2008a). Furthermore, when cells were transfected with dnBNIP3 together with pSuper-p53 siRNA under hypoxia, the autophagy was reduced significantly (Supplementary Figure S6A3, A4 and B, column 4 compared with column 3 from left to right; P=0.014), indicating that BNIP3 blocking in p53 knockdown cells could reduce autophagy efficiently.
The behaviours of p53 suppression of BNIP3 in modulating hypoxia-induced autophagy were quite similar to that in modulating hypoxia-induced cell death if not identical. Because of the barrier encountered for tracing LC3–GFP puncta (LC3 vacuoles) in dying cells, it was hard to get a clear picture, showing that autophagic cells would eventually go through dying under hypoxia. However, given the well-defined function of BNIP3 for inducing autophagic cell death in cancer cells under hypoxia (Daido et al, 2004; Tracy et al, 2007), we thought that p53 suppression of BNIP3 change autophagy might contribute to cell death response in p53 knockdown cells under hypoxia, at least in partial. Of course, to what extent of this contribution definitely needs to be further investigated.
Taken together, these observations not only suggest that the p53 can inhibit hypoxia-induced autophagy through suppression of BNIP3, but also imply that the inhibition of autophagy by p53 may contribute to p53's role in protecting against hypoxia-induced cell death.
Zebrafish nip3a is a homologous gene of mammalian BNIP3
As reported, p53 mutant zebrafish exhibited phenotypes similar to that of p53 null mice (Berghmans et al, 2005). To figure out the regulation of BNIP3 by p53 in vivo, we intended to take advantage of zebrafish model.
Initially, we sought to identify zebrafish homologous gene of BNIP3. After searching in the database, three copies of zebrafish BNIP3 were identified (bnip3 (ENSDARG00000045033), bnip3b (ENSDARG00000022832) and nip3a (AF493987)). Phylogenetic analysis indicated that nip3a was much closer to human BNIP3 (Figure 9A), which was 52.58% identical in amino acid (Figure 9B). The dominant-negative form lacking mitochondrial membrane potential (Δψm) of human BNIP3 (153 amino acids in N-terminal) corresponds to 181 amino acids of zebrafish nip3a N-terminal (Figure 9B). Whole mount in situ hybridization analysis revealed that the expression of zebrafish nip3a was ubiquitous during early embryogenesis (Figure 9C1 and C2) and restricted in specific regions, such as retina (r), midbrain (mb), hindbrain (hb) and pectorial fin primordium (p) at 24 h.p.f. (Figure 9C3), implying its maternal expression.
Figure 9.
Zebrafish nip3a is a homologous gene of human BNIP3. (A) Phylogenetic analysis for human BNIP3 and zebrafish nip3a (AF493987), bnip3 (ENSDARG00000045033) and bnip3b (ENSDARG00000022832) by the methods of Maximum Parsimony (MP), Minimum Evolution (ME) and Neighbor-Joining (NJ). The bootstrap value for 1000 replacement was indicated on the phylogenetic tree. (B) The amino-acid alignment of human BNIP3 and zebrafish nip3a. The identical amino acids were marked by dots and the deleted amino acids were marked by dashes. The N-terminal region serving as dominant-negative portion of human BNIP3 separating from its C-terminal region was marked by red colour, which encompassed 153 amino acids of human BNIP3 N-terminus and 181 amino acids of zebrafish nip3a N-terminus. (C) Whole mount in situ hybridization analysis for zebrafish nip3a expression at 40 epiboly, 90% epiboly and 24 h post-fertilization (h.p.f.) during embryogenesis. nip3a was ubiquitously expressed at 40% epiboly and 90% epiboly, but higher expressed in retina (r), midbrain (mb), hindbrain (hb) and pectorial fin primordium (p), lower expressed in pronephric duct (pd) at 24 h.p.f.
Subsequently, to figure out whether nip3a was induced by hypoxia similar to that of human BNIP3, we treated 48 h.p.f. embryos by hypoxia (2% O2) for 2 h (Figure 10A), 4 h (Figure 10B) and 8 h (Figure 10C), respectively, and observed that nip3a was indeed induced by hypoxia dramatically in both wild-type and p53 mutant embryos as revealed by whole mount in situ hybridization assays (Figure 10A–C) and semi-quantitative RT–PCR assays (Figure 10D). However, the expression of bnip3 or bnip3b, other two zebrafish homologous genes of BNIP3 was not induced by hypoxia as revealed by in situ hybridization (data not shown). Both phylogenetic analysis and hypoxia-induction assays clearly suggested that zebrafish nip3a resembled mammalian BNIP3.
Figure 10.
Zebrafish nip3a expression was induced by hypoxia (2% O2) in both wild-type embryos and p53 mutant embryos; the induction of nip3a expression by hypoxia in p53 mutant embryos was higher than that in wild-type embryos. (A) For 50 h.p.f. embryos, the expression of nip3a was induced by hypoxia (2% O2) in wild-type embryos (A1, A2; n=20, respectively) and p53 mutant embryos (A3, A4; n=20, respectively). (B) For 52 h.p.f. embryos, the expression of nip3a was induced by hypoxia (2% O2) in wild-type embryos (B1, B2; n=20, respectively) and p53 mutant embryos (B3, B4; n=20, respectively); the expression of nip3a induced by hypoxia (2% O2) for 4 h in p53 mutant embryos (B4; n=20) was higher than that in wild-type embryos (B2; n=20). (C) For 56 h.p.f. embryos, the expression of nip3a was induced by hypoxia (2% O2) in wild-type embryos (C1, C2; n=20, respectively) and p53 mutant embryos (C3, C4; n=20, respectively); the expression of nip3a induced by hypoxia (2% O2) for 8 h in p53 mutant embryos (C4; n=20) was higher than that in wild-type embryos (C2; n=20). h.p.f., hours post-fertilization; WT, wild-type; p53 mut, p53 mutant. (D) Semi-quantitative RT–PCR analysis confirmed that the expression of nip3a in p53 mutant embryos was higher than that in wild-type embryos significantly (P<0.05; n=20, respectively) under both normoxia and hypoxia; the expression of nip3a in wild-type embryos was induced significantly after 4 h-2% O2 treatments (for 2 h-2% O2 treatment, P=0.0725, n=20; for 4 h-2% O2 treatment, P=0.0094, n=20; for 8 h-2% O2 treatment, P=0.0008, n=20).
Zebrafish nip3a expression was suppressed by p53 in vivo
To figure out whether nip3a was also suppressed by p53 in vivo, we compared the expression level of nip3a mRNA in p53 mutant embryos and their counterpart wild-type embryos under normoxia and hypoxia by whole mount in situ hybridization (Supplementary Figure S7; Figure 10A–D). Obviously, the expression level of nip3a mRNA in p53 mutant embryos is much higher than that in wild-type embryos under normoxia (Supplementary Figure S7; P<0.0001). Furthermore, after hypoxia treatment (2% O2), the induction of nip3a expression in p53 mutant embryos was also much higher than that in wild-type embryos (Figure 10A–D). These observations indicated a significant influence of p53 status in vivo on nip3a expression under both normoxia and hypoxia.
To further confirm the suppressive role of p53 on nip3a expression under hypoxia in vivo, we overexpressed p53 in both wild-type embryos and p53 mutant embryos by microinjection of p53 mRNA (200–500 pg/per embryo). As shown in Figure 11, nip3a expression was sufficiently suppressed by exogenous p53 overexpression (Figure 11A3, A4, A9, A10 and B). The expression of injected myc–p53 mRNA was confirmed by western blot (Figure 11C). On the contrary, p53 knockdown mediated by microinjection of p53 morpholino (p53-MO, 8 ng/per embryo) in wild-type embryos enhanced nip3a expression significantly (Figure 11A1, A2, A5 and A6 and B, column 3 compared with column 1 from left to right; P=0.0008), similar to what was observed in p53 mutant embryos (Figure 10). The efficiency and specificity for p53-MO mediating p53 knockdown were confirmed by western blot (Figure 11D).
Figure 11.
Under hypoxia, the expression of zebrafish nip3a in wild-type embryos was inhibited by p53 overexpression and up-regulated by p53 knockdown. (A) Under hypoxia (2% O2, 8 h), the expression of nip3a in wild-type embryos injected with synthesized zebrafish p53 mRNA (200–500 pg/per embryo) was reduced (A3, A4; n=20) compared with the embryos without injection (A1, A2; n=20); the expression of nip3a in wild-type embryos injected p53 morpholino (8 ng/per embryo) was increased (A5, A6; n=20) compared with the embryos without injection (A1, A2); the expression of nip3a in p53 mutant embryos injected with synthesized p53 mRNA was decreased (A9, A10; n=20) compared with embryos without injection (A7, A8; n=20). (B) Semi-quantitative RT–PCR analysis further confirmed the results shown in (A). Data of RT–PCR analysis are reported as mean±s.d. of three independent experiments performed in triplicate; the statistical analysis was performed using GraphPad Prism 5.0 (t-tests). (C) The expression of injected p53 mRNA was confirmed by western blot. (D) The specificity and efficiency of p53-morpholono (p53-MO)-mediated p53 knockdown was confirmed by western blot.
Together, these observations indicate that p53 could suppress nip3a expression effectively in vivo, consistent with the results obtained by cell culture system.
Zebrafish p53 protected against hypoxia-induced cell death in vivo
To better understand whether p53 could also protect against hypoxia-induced cell death in zebrafish model, we sought to compare dying cell ratio and embryo survival rate between wild-type embryos and p53 mutant embryos. Of note, compared with wild-type embryos, more p53 mutant embryos exhibited serious defect during embryogenesis and higher embryo death rate even under normoxia condition, which might be misleading for the explanation of results from further cell death assays and survival rate assays. To minimize this effect, initially, we did a series of pilot experiments. We noticed that if p53 mutant embryos were still normal in general morphology after 48 h post-fertilization, they would exhibit similar morphogenesis and survival rate as that of wild-type embryos during the following developing process under normoxia condition (data not shown). Therefore, we let both wild-type embryos and p53 mutant embryos to develop under normoxia condition for 48 h, and then picked out embryos with any detectable defects from both wild-type and p53 mutant embryos under dissection microscope. Then, we performed following cell death assays and survival rate assays.
For cell death assays, we separated embryos in two sets and performed parallel treatments. One set was put into normoxia incubator and another set was put into hypoxia incubator (2% O2). At different time points (2, 4, 6 and 8 h), we conducted dying cell staining for embryos (Qi et al, 2010). As shown in Figure 12A, under normoxia, there was no significant difference in dying cell number between wild-type embryos (n=21) and p53 mutant embryos (n=17). However, after hypoxia treatment, the dying cell number was gradually increased in p53 mutant embryos (n=23/23), but not in wild-type embryos (n=0/25) (Figure 12B1–B28). The quantitative analysis by Image J software showed that the dying cell number increased significantly in p53 mutant embryos compared with that in wild-type embryos under hypoxia (Figure 12B7, B14, B21 and B28; P<0.05, respectively), which was also exhibited in a time-cause-dependent manner (Figure 12B7, B14, B21 and B28).
Figure 12.
p53 mutant embryos exhibited more dying cells and higher embryo death ratio compared with wild-type embryos under hypoxia; the overexpression of nip3 dominant-negative mRNA (nip3a DN mRNA (1–181 aa)) could rescue cell death in p53 mutant embryos under hypoxia. (A) Under normoxia, no dying cells were observed in both p53 mutant embryos and wild-type embryos at 48 h.p.f. (A1–A8). (B) Under hypoxic, more dying cells were observed in p53 mutant embryos (B4, B5, B6; B11, B12, B13; B18, B19, B20; B25, B26 and B27) compared with that in wild-type embryos (B1, B2, B3; B8, B9, B10; B15, B16, B17; B22, B23 and B24). The dying cells were marked by red arrows. The quantitative analysis by Image J software showed that the number of dying cell induction by hypoxia in p53 mutant embryos was much greater than that in wild-type embryos (B7, B14, B21 and B28; P<0.05, respectively). The cell death induction by hypoxia in p53 mutant embryos was exhibited in a time-cause-dependent manner (B6, B7, B13, B14, B20, B21, B27 and B28). The dying cells were monitored by Acridine Orange staining. Data of dying cell number are reported as mean±s.d., representing three different fields of each embryo with the same square; the statistical analysis was performed using GraphPad Prism 5.0 (t-tests). (C) The survival curve of p53 mutant embryos and wild-type embryos under hypoxia. Compared with the p53 mutant embryos, more wild-type embryos survived under the same hypoxia condition. (D) Overexpression of nip3a dominant-negative mRNA could rescue cell death in p53 mutant embryos under hypoxia (D5–D8) (n=10/19).
The dead cell induction in embryos will eventually cause embryo death, thus, we further analysed the embryo survival rate between p53 mutant embryos and wild-type embryos under hypoxia. After 48 h post-fertilization, we put the embryos into hypoxia incubator (2% O2) and treated them for additional 24 h. The dead embryos were counted at every 6 h interval. As indicated in Figure 12C, starting from 12 h after hypoxia treatment, p53 mutant embryos (n=226) exhibited higher death rate compared with wild-type embryos (n=243), which might imply that intact p53 presented in wild-type embryos accounted for the protection of hypoxia-induced cell death (Figure 12C).
As reported, human BNIP3 Δψm (153 aa in N-terminus) could block hypoxia-induced cell death mediated by BNIP3 induction efficiently (Vande Velde et al, 2000; Kothari et al, 2003; Burton and Gibson, 2009). To further figure out whether the identical region of nip3a corresponding to human BNIP3 Δψm had similar cell death-blocking function and whether that loss of p53 function enhanced hypoxia-induced cell death in vivo was mediated by loss of p53 suppression on nip3a, we synthesized mRNA encoding 181 amino acids of zebrafish nip3a N-terminus and performed microinjection. As shown in Figure 12D, for wild-type embryos (n=21), hypoxia treatment for 8 h did not cause cell death. As expected, the injection of nip3a DN mRNA did not cause any difference in dying cell staining for wild-type embryos (n=25) (Figure 12D1, D2, D3 and D4). However, for p53 mutant embryos, the injection of nip3a DN mRNA could rescue cell death dramatically (n=10/19) (Figure 12D5, D6, D7 and D8), but the injection of the same amount of GFP mRNA (200–500 ng/individual) (control) could not do so (n=0/23) (data not shown). The expression of injected nip3a DN mRNA was confirmed by western blot (Supplementary Figure S8). Taken together, these results suggest that p53 can protect against hypoxia-induced cell death in vivo, which might be dependent upon p53 suppression of nip3a expression.
Discussion
p53 suppresses BNIP3 expression under normoxic and hypoxic conditions
Under hypoxia, the p53 protein becomes stabilized (An et al, 1998; Blagosklonny et al, 1998; Alarcon et al, 1999; Koumenis et al, 2001; Suzuki et al, 2001; Chen et al, 2003; Hammond and Giaccia, 2005); however, BNIP3 up-regulation under hypoxia occurs independently of p53 (Guo et al, 2001). As reported, p53 induces BNIP3L under hypoxia, leading to the proposal that BNIP3L serves as a unique target of p53 to mediate p53-dependent apoptosis during hypoxia (Fei et al, 2004). In this study, we reevaluated the regulation of BNIP3L by p53 under hypoxia and confirmed the results reported previously (Supplementary Figure S3) (Fei et al, 2004), which also indicates that the results about p53 suppression of BNIP3 revealed by cell culture system shown here is reliable.
Of note, it was reported that BNIP3 induced by p53 overexpression in high metastatic ability lung cancer cell lines (Anip3973 and 95D) resulted in cell apoptosis, but not in low metastatic ability lung cancer cell lines (AGZY83-a and 95C) (Yan et al, 2006). However, this report did not provide convince evidences to show that BNIP3 was a direct downstream target of p53, which might only reflect correlation between p53 and BNIP3 at some extent in special cell types (Yan et al, 2006). Here, we present data showing that p53 suppressed BNIP3 promoter activity, inhibited BNIP3 expression and bound to the BNIP3 promoter directly. Moreover, we demonstrated that the ability of p53 to suppress BNIP3 required p53 to bind to the promoter.
In zebrafish model, we further provide evidence to show that nip3a is a homologous gene of mammalian BNIP3. In addition, we not only show that the expression of nip3a in p53 mutant embryos is much higher than that in their counterpart wild-type embryos under normoxia, but also show that the induction of nip3a in p53 mutant embryos by hypoxia is much higher than that in wild-type embryos. These observations indicate that the p53 status in embryos significantly affect nip3a expression, further supporting the suppressive role of p53 on BNIP3 expression in vivo. Taken together, these observations lead us to propose that p53 directly targets BNIP3 for suppression.
P53 binds to BNIP3 promoter directly and has a repressing function
The literature provides abundant evidence that p53 binds to a large number of p53-RE variants and subsequently activates gene expression (Menendez et al, 2009; Wang et al, 2009). Because of the difficulty in identifying p53RE motifs in promoters of repressed targets, the repressive function of p53 remains poorly characterized. Recently, Wang et al (2009) redefined the p53RE based on whether p53 is acting as a transactivator or transrepressor.
Based on the ChIP assays, we found that p53 could directly bind to a candidate p53RE in the BNIP3 promoter, repressing its activity. Notably, the p53-binding site in BNIP3 promoter is an atypical p53RE that comprises two decamer motifs, RYRCWWGRWW and WWWCWWGRRR, separated by a spacer of 16 bp, where the dinucleotide cores are AA and TT, similar to the dinucleotide core of p202, a p53-repressed target (Datta et al, 1996; D'Souza et al, 2001). Further mutation assays indicate that either AA mutated to GG or AA mutated to CC resulted in abolishing the repressive function of p53 on BNIP3 promoter activity, inconsistent with the results obtained by Wang et al (2009). Our observations extend the definition of p53RE motifs responsible for p53-mediated repression, indicating the need for further studies to identify the unique consensus binding sites responsible for this function of p53.
Furthermore, we provide evidences to show that p53 recruits corepressor mSin3a to the promoter of BNIP3, refining the suppressive role of p53 on BNIP3 expression.
p53 protects against hypoxia-induced cell death
As a classic tumour suppressor, the induction of apoptosis represents an essential role of p53. Hypoxia is a major stress, not only for tumour cells during tumour growth, but also for myocytes and neuron cells during ischaemic heart disease and cerebral stroke, respectively. Thus, the role of p53 in hypoxia-induced cell death has garnered much attention (Graeber et al, 1996; Kim et al, 1997a; Giaccia and Kastan, 1998; Stempien-Otero et al, 1999; Denko et al, 2000; Pan et al, 2004; Hammond and Giaccia, 2005). To date, the literature describes a number of p53 targets that mediate p53-dependent cell death. However, with the exception of BNIP3L, none of the identified p53 targets appears to contribute to the p53-dependent apoptosis induced by hypoxia (Hammond and Giaccia, 2005). In fact, studies have shown that, with hypoxia, p53 fails to recruit coactivators to its target promoters and thereby mainly acts as transrepressor instead of transactivator (Koumenis et al, 2001; Schmid et al, 2004). The number of p53-repressed genes is growing, but again, these targets do not appear to have a direct role in hypoxia-induced apoptosis (Hammond and Giaccia, 2005). Although, hypoxia-induced p53 acts almost entirely as a transrepressor, the underlying mechanism remains unclear (Hammond and Giaccia, 2005).
We demonstrate that p53 protected against hypoxia-induced cell death through directly suppressing BNIP3 expression, revealing a novel function of p53 in hypoxia-induced cell death. Consistent with our results, previous studies have shown that the myocytes from p53 wild-type mice are more resistant to hypoxia-induced apoptosis as compared with the myocytes from p53 null mice (Graham et al, 2004). In addition, p53 null mice had significantly reduced endurance in a swimming test as compared with their wild-type littermates (Matoba et al, 2006).
As reported previously, BNIP3L knockdown in cells with functional p53 reduces hypoxia-induced apoptosis, similar to what we and others observed for BNIP3 (Vande Velde et al, 2000; Prabhakaran et al, 2007), which refines the well-characterized role of BNIP3L and BNIP3 in hypoxia-induced apoptosis pathway. In this study, we demonstrate that p53 knockdown/mutant can enhance hypoxia-induced cell death, in which BNIP3 blocking can reduce cell death as revealed by cell culture system and zebrafish model. However, as a downstream target of p53 under hypoxia, BNIP3L is supposed to go down when p53 is knocked down. Thus, the cell death enhancement by p53 knockdown under hypoxia is not correlated to the p53 up-regulation of BNIP3L, but well correlated to the p53 suppression of BNIP3. These facts imply that the BNIP3 may act as a major effector for mediating p53's protection function in hypoxia-induced cell death pathway, but not BNIP3L.
In addition, we also noticed that the dead cell ratio in cells transfected with pSuper-GFP siRNA plus HA empty vector (average 17%) (Figure 8B) was higher than that in cells transfected with GFP siRNA (average 8.5%) (Supplementary Figure S4B). This phenomena might result from the different transfection reagent used (Lipofectamine 2000 was used for siRNA transfection; VigoFect was used for pSuper-siRNA vector transfection; we noticed that VigoFect had higher transfection efficiency, but was more toxic for cells than that of Lipofectamine 2000). However, due to strict control designed in each batch of experiments, it would not affect the major conclusion obtained in this study.
In this report, we provide several lines of evidence showing that p53 directly bound to a p53RE motif in the BNIP3 promoter and recruited corepressor mSin3a to the BNIP3 promoter, suppressed BNIP3 expression and protected against hypoxia-induced cell death. Moreover, we demonstrate that the intact endogenous p53 could indeed suppress nip3a (BNIP3) expression and protect against hypoxia-induced cell death by zebrafish model in vivo. The novel role for p53 revealed in this study may lay the groundwork for the development of new treatments for ischaemic heart disease and cerebral stoke.
Materials and methods
Detailed Materials and methods are described in the Supplementary data.
Chromatin immunoprecipitation
ChIP assays were performed using a protocol modified from the user manual of an acetyl-histone H3 immunoprecipitation kit (Upstate) and Hela cells were used for assays. Mouse IgG, rabbit IgG and protein A/G agarose beads were purchased from Santa Cruz. Anti-mSin3a antibody (ab3479) was purchased from Abcam. A p53 mini-peptide with the sequence: EPPLSQETFSDLWKL corresponding to aa 11–25 of human p53 protein (which is the same peptide used for developing monoclonal antibody against p53 (DO-1) by Santa Cruz) was synthesized from ChinaPeptides Co., Ltd. (Shanghai, China). The primers used for amplifying the BNIP3 promoter region corresponding to –1169 to –1047 (the transcription initial site was defined as +1) (which contains potential p53-binding sites) were 5′-AGCGTTTCTGGGGCGCACCTTG-3′ and 5′-GGGACTGGGAGGCACTTTTCAGAGGA-3′. p21 promoter region was amplified as a positive control, using primers specific for the p21 promoter region corresponding to –2209 to –2478: 5′-CACCACTGAGCCTTCCTCAC-3′ and 5′-CTGACTCCCAGCACACACTC-3′ (Saramaki et al, 2006). β-Actin promoter region was used as a negative control and amplified using the following primers: 5′-CAGGGCGTGATGGTGGGCA-3′ and 5′-CAAACATGATCTGGGTCATCTTCTC-3′. The PCR products were also cut out from agarose gel and did sequence verification. Semi-quantitative RT–PCR was conducted to determine the extent of p53 binding to BNIP3 promoter.
To determine whether mSin3A, a corepressor molecule, could be recruited to BNIP3 promoter by p53, we further performed the secondary ChIP assays. First, the p53 antibody–protein–DNA complex was obtained by immunoprecipitation using anti-p53 monoclonal antibody; then we eluted the complex by p53 mini-peptide (1 μg/μl; dissolved in Triton buffer) and obtained the protein–DNA complex solution. Subsequently, we used anti-mSin3A antibody to conduct immunoprecipitation. PCR was performed for examining whether mSin3A binds to the same region of BNIP3 promoter as that of p53 and PCR products were cut from agarose gel for sequencing. Semi-quantitative RT–PCR was conducted to determine the extent of mSin3A binding to BNIP3 promoter.
Zebrafish caring, gene cloning, whole mount in situ hybridization, dying cell detection and dead embryo counting
Tp53 mutant zebrafish obtained from Jun Chen (Zhejiang University, Hanzhou, China), originally obtained from A Thomas Look and their counterpart wild-type zebrafish, were bred and maintained under the standard library conditions. The identification of tp53 mutant was conducted by digesting DNA fragment amplified from genomic DNA by PCR using the primers described by Berghmans et al (2005).
Zebrafish wild-type tp53 was amplified from zebrafish cDNA pool using the primers: zp53-SalI-F: 5′-GTCGACATGGCGCAAAACGACAG-3′ and zp53-BamHI-R: 5′-GGATCCATCAGAGTCGCTTCTTCC-3′, then cloned into Psp64 poly(A) vector (Promega) for synthesizing mRNA. To confirm the expression of injected p53 mRNA in embryos by western blot, we also made myc-tagged zebrafish wild-type p53 construct. The dominant-negative form of zebrafish nip3a (nip3a DN) (encoding 181 amino acids in nip3a N-terminus, which is identical to dominant-negative form of human BNIP3 (153 aa of N-terminus)) was amplified from zebrafish cDNA pool using the primers: zNip3-64-Sal1-F: 5′-ACGCGTCGACCTAGTAGAAGCCACCATG-3′, and zNip3-BamH1-181-R: 5′-GGATCCGCTCATTTGAGAAACTCAGC-3′, and also cloned into Psp64 poly(A) vector (Promega). We also made myc-tagged nip3a DN construct for expression verification. For whole mount in situ hybridization probe, the cDNA fragment of zebrafish nip3a was amplified from zebrafish cDNA pool using the primers: zNip3-F: 5′-AAGCCGCCCTACACAGAGCCACC-3′ and zNip3-R 5′-TAGCAGGGGGAGTTGTCAGTC-3′, and cloned into pTA2 vector (Toyobo). mRNA synthesis was performed following the protocol described previously (Liu et al, 2009). For mRNA injection, each embryo was injected newly synthesized mRNA at 200–500 pg/per embryo.
For whole mount in situ hybridization, probe preparation and embryo staining was conducted following the protocol described previously (Liu et al, 2009). The hypoxia treatment for embryos was conducted in the incubator with O2 control (Forma Series II) at 2% O2 (1% O2 treatment causes most of the embryo death at early embryogenesis).
For confirming the expression of injected zebrafish p53 mRNA and injected zebrafish nip3a dominant-negative form mRNA (myc nip3a DN mRNA) or examining the efficiency and specificity of p53-MO-mediated p53 knockdown, we conducted western blot analysis using a monoclonal antibody against myc tag (9E10, Santa Cruz) after injection of the indicated mRNA, expression vectors or p53-MO into embryos for 50–60 h (the number of embryos used for protein preparation is about 40–50).
The Acridine Orange staining was used for dying cell staining in embryos (Qi et al, 2010). Image J software was used to quantify the number of dying cells based on the photos taken under a Leica M205 FA fluorescent dissection microscope and GraphPad Prism 5 was used for statistical analysis (t-tests). The dying cell numbers were counted from the field with the same square and three different fields of each embryo were chosen for counting. The results were presented with error bars (mean±s.d.) and P-values.
For comparing the survival rate between p53 mutant embryos and wild-type embryos under hypoxia, we let the embryos to develop for 48 h (48 h.p.f.) under normoxia condition, and then put embryos without any detectable defects into the incubator with O2 control at 2% O2. The number of dead embryos was counted after 6 h interval. Three independent experiments were conducted.
Supplementary Material
Acknowledgments
We are grateful to Drs Spencer Gibson, Richard Bruick, Bert Vogelstein, William Tansey, A Thomas Look, Jun Chen, R Agami, H Shu and Yonghua Sun for the generous gift of reagents. WX is supported by ‘973’ Grant 2010CB126306, 2007CB815705, NSFC Grant 31071212, 91019008 and 20890113, National Transgene Project 2009ZX08010-021B, CAS Grant KSCX2-EW-N-004-3 and NSF of Hubei Province 2008CDA012.
Author contributions: XF performed the experiments and wrote the paper; XL performed the experiments; WZ did data analysis; WX designed the experiments and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahn J, Murphy M, Kratowicz S, Wang A, Levine AJ, George DL (1999) Down-regulation of the stathmin/Op18 and FKBP25 genes following p53 induction. Oncogene 18: 5954–5958 [DOI] [PubMed] [Google Scholar]
- Alarcon R, Koumenis C, Geyer RK, Maki CG, Giaccia AJ (1999) Hypoxia induces p53 accumulation through MDM2 down-regulation and inhibition of E6-mediated degradation. Cancer Res 59: 6046–6051 [PubMed] [Google Scholar]
- An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM (1998) Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature 392: 405–408 [DOI] [PubMed] [Google Scholar]
- Attardi LD, Jacks T (1999) The role of p53 in tumour suppression: lessons from mouse models. Cell Mol Life Sci 55: 48–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi LD, Reczek EE, Cosmas C, Demicco EG, McCurrach ME, Lowe SW, Jacks T (2000) PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev 14: 704–718 [PMC free article] [PubMed] [Google Scholar]
- Aurelio ON, Kong XT, Gupta S, Stanbridge EJ (2000) p53 mutants have selective dominant-negative effects on apoptosis but not growth arrest in human cancer cell lines. Mol Cell Biol 20: 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM (2009) Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 29: 2570–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, Morris JP, Liu TX, Schulte-Merker S, Kanki JP, Plasterk R, Zon LI, Look AT (2005) tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci USA 102: 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV, An WG, Romanova LY, Trepel J, Fojo T, Neckers L (1998) p53 inhibits hypoxia-inducible factor-stimulated transcription. J Biol Chem 273: 11995–11998 [DOI] [PubMed] [Google Scholar]
- Boyd JM, Malstrom S, Subramanian T, Venkatesh LK, Schaeper U, Elangovan B, D′Sa-Eipper C, Chinnadurai G (1994) Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell 79: 341–351 [DOI] [PubMed] [Google Scholar]
- Bristow N, Burton TR, Henson ES, Ong-Justiniano C, Brown M, Gibson SB (2011) Truncated forms of BNIP3 act as dominant negatives inhibiting hypoxia-induced cell death. Biochim Biophys Acta 1812: 302–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick RK (2000) Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci USA 97: 9082–9087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton TR, Gibson SB (2009) The role of Bcl-2 family member BNIP3 in cell death and disease: NIPping at the heels of cell death. Cell Death Differ 16: 515–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Li M, Luo J, Gu W (2003) Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function. J Biol Chem 278: 13595–13598 [DOI] [PubMed] [Google Scholar]
- Chen G, Cizeau J, Vande Velde C, Park JH, Bozek G, Bolton J, Shi L, Dubik D, Greenberg A (1999) Nix and Nip3 form a subfamily of pro-apoptotic mitochondrial proteins. J Biol Chem 274: 7–10 [DOI] [PubMed] [Google Scholar]
- Chen G, Ray R, Dubik D, Shi L, Cizeau J, Bleackley RC, Saxena S, Gietz RD, Greenberg AH (1997) The E1B 19K/Bcl-2-binding protein Nip3 is a dimeric mitochondrial protein that activates apoptosis. J Exp Med 186: 1975–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G, Vijayalingam S, Gibson SB (2008) BNIP3 subfamily BH3-only proteins: mitochondrial stress sensors in normal and pathological functions. Oncogene 27(Suppl 1): S114–S127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow MT (2002) Hypoxia, BNip3 proteins, and the mitochondrial death pathway in cardiomyocytes. Circ Res 91: 183–185 [DOI] [PubMed] [Google Scholar]
- D'Souza S, Xin H, Walter S, Choubey D (2001) The gene encoding p202, an interferon-inducible negative regulator of the p53 tumor suppressor, is a target of p53-mediated transcriptional repression. J Biol Chem 276: 298–305 [DOI] [PubMed] [Google Scholar]
- Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S (2004) Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res 64: 4286–4293 [DOI] [PubMed] [Google Scholar]
- Datta B, Li B, Choubey D, Nallur G, Lengyel P (1996) p202, an interferon-inducible modulator of transcription, inhibits transcriptional activation by the p53 tumor suppressor protein, and a segment from the p53-binding protein 1 that binds to p202 overcomes this inhibition. J Biol Chem 271: 27544–27555 [DOI] [PubMed] [Google Scholar]
- Denko NC, Green SL, Edwards D, Giaccia AJ (2000) p53 checkpoint-defective cells are sensitive to X rays, but not hypoxia. Exp Cell Res 258: 82–91 [DOI] [PubMed] [Google Scholar]
- Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, Li H, Kirshenbaum LA, Hahn HS, Robbins J, Jones WK, Dorn GW (2007) Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest 117: 2825–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn GW II, Kirshenbaum LA (2008) Cardiac reanimation: targeting cardiomyocyte death by BNIP3 and NIX/BNIP3L. Oncogene 27(Suppl 1): S158–S167 [DOI] [PubMed] [Google Scholar]
- Fei P, Bernhard EJ, El-Deiry WS (2002) Tissue-specific induction of p53 targets in vivo. Cancer Res 62: 7316–7327 [PubMed] [Google Scholar]
- Fei P, Wang W, Kim SH, Wang S, Burns TF, Sax JK, Buzzai M, Dicker DT, McKenna WG, Bernhard EJ, El-Deiry WS (2004) Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell 6: 597–609 [DOI] [PubMed] [Google Scholar]
- Frazier DP, Wilson A, Graham RM, Thompson JW, Bishopric NH, Webster KA (2006) Acidosis regulates the stability, hydrophobicity, and activity of the BH3-only protein Bnip3. Antioxid Redox Signal 8: 1625–1634 [DOI] [PubMed] [Google Scholar]
- Giaccia AJ, Kastan MB (1998) The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev 12: 2973–2983 [DOI] [PubMed] [Google Scholar]
- Giam M, Huang DC, Bouillet P (2008) BH3-only proteins and their roles in programmed cell death. Oncogene 27(Suppl 1): S128–S136 [DOI] [PubMed] [Google Scholar]
- Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, Giaccia AJ (1996) Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature 379: 88–91 [DOI] [PubMed] [Google Scholar]
- Graham RM, Frazier DP, Thompson JW, Haliko S, Li H, Wasserlauf BJ, Spiga MG, Bishopric NH, Webster KA (2004) A unique pathway of cardiac myocyte death caused by hypoxia-acidosis. J Exp Biol 207: 3189–3200 [DOI] [PubMed] [Google Scholar]
- Guo K, Searfoss G, Krolikowski D, Pagnoni M, Franks C, Clark K, Yu KT, Jaye M, Ivashchenko Y (2001) Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Differ 8: 367–376 [DOI] [PubMed] [Google Scholar]
- Gustafsson AB (2011) Bnip3 as a dual regulator of mitochondrial turnover and cell death in the myocardium. Pediatr Cardiol 32: 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, Gottlieb RA, Gustafsson AB (2007) Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ 14: 146–157 [DOI] [PubMed] [Google Scholar]
- Hammond EM, Giaccia AJ (2005) The role of p53 in hypoxia-induced apoptosis. Biochem Biophys Res Commun 331: 718–725 [DOI] [PubMed] [Google Scholar]
- Hammond EM, Giaccia AJ (2006) Hypoxia-inducible factor-1 and p53: friends, acquaintances, or strangers? Clin Cancer Res 12: 5007–5009 [DOI] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, McKinnon PJ, Cleveland JL, Zambetti GP (2003) Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4: 321–328 [DOI] [PubMed] [Google Scholar]
- Kim CN, Wang X, Huang Y, Ibrado AM, Liu L, Fang G, Bhalla K (1997a) Overexpression of Bcl-X(L) inhibits Ara-C-induced mitochondrial loss of cytochrome c and other perturbations that activate the molecular cascade of apoptosis. Cancer Res 57: 3115–3120 [PubMed] [Google Scholar]
- Kim CY, Tsai MH, Osmanian C, Graeber TG, Lee JE, Giffard RG, DiPaolo JA, Peehl DM, Giaccia AJ (1997b) Selection of human cervical epithelial cells that possess reduced apoptotic potential to low-oxygen conditions. Cancer Res 57: 4200–4204 [PubMed] [Google Scholar]
- Kothari S, Cizeau J, McMillan-Ward E, Israels SJ, Bailes M, Ens K, Kirshenbaum LA, Gibson SB (2003) BNIP3 plays a role in hypoxic cell death in human epithelial cells that is inhibited by growth factors EGF and IGF. Oncogene 22: 4734–4744 [DOI] [PubMed] [Google Scholar]
- Koumenis C, Alarcon R, Hammond E, Sutphin P, Hoffman W, Murphy M, Derr J, Taya Y, Lowe SW, Kastan M, Giaccia A (2001) Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol Cell Biol 21: 1297–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubasiak LA, Hernandez OM, Bishopric NH, Webster KA (2002) Hypoxia and acidosis activate cardiac myocyte death through the Bcl-2 family protein BNIP3. Proc Natl Acad Sci USA 99: 12825–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubli DA, Quinsay MN, Huang C, Lee Y, Gustafsson AB (2008) Bnip3 functions as a mitochondrial sensor of oxidative stress during myocardial ischemia and reperfusion. Am J Physiol Heart Circ Physiol 295: H2025–H2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ (1997) p53, the cellular gatekeeper for growth and division. Cell 88: 323–331 [DOI] [PubMed] [Google Scholar]
- Liu JX, Hu B, Wang Y, Gui JF, Xiao W (2009) Zebrafish eaf1 and eaf2/u19 mediate effective convergence and extension movements through the maintenance of wnt11 and wnt5 expression. J Biol Chem 284: 16679–16692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonosova E, Chinnadurai G (2008) BH3-only proteins in apoptosis and beyond: an overview. Oncogene 27(Suppl 1): S2–S19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G (2010) Autophagy regulation by p53. Curr Opin Cell Biol 22: 181–185 [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM (2006) p53 regulates mitochondrial respiration. Science 312: 1650–1653 [DOI] [PubMed] [Google Scholar]
- Matsushima M, Fujiwara T, Takahashi E, Minaguchi T, Eguchi Y, Tsujimoto Y, Suzumori K, Nakamura Y (1998) Isolation, mapping, and functional analysis of a novel human cDNA (BNIP3L) encoding a protein homologous to human NIP3. Genes Chromosomes Cancer 21: 230–235 [PubMed] [Google Scholar]
- Mellor HR, Harris AL (2007) The role of the hypoxia-inducible BH3-only proteins BNIP3 and BNIP3L in cancer. Cancer Metastasis Rev 26: 553–566 [DOI] [PubMed] [Google Scholar]
- Menendez D, Inga A, Resnick MA (2009) The expanding universe of p53 targets. Nat Rev Cancer 9: 724–737 [DOI] [PubMed] [Google Scholar]
- Mirza A, Wu Q, Wang L, McClanahan T, Bishop WR, Gheyas F, Ding W, Hutchins B, Hockenberry T, Kirschmeier P, Greene JR, Liu S (2003) Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene 22: 3645–3654 [DOI] [PubMed] [Google Scholar]
- Miyashita T, Reed JC (1995) Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80: 293–299 [DOI] [PubMed] [Google Scholar]
- Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, George DL (1999) Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev 13: 2490–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M, Hinman A, Levine AJ (1996) Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes Dev 10: 2971–2980 [DOI] [PubMed] [Google Scholar]
- Nakano K, Vousden KH (2001) PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 7: 683–694 [DOI] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N (2000a) Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288: 1053–1058 [DOI] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, Taya Y (2000b) p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102: 849–862 [DOI] [PubMed] [Google Scholar]
- Pan Y, Oprysko PR, Asham AM, Koch CJ, Simon MC (2004) p53 cannot be induced by hypoxia alone but responds to the hypoxic microenvironment. Oncogene 23: 4975–4983 [DOI] [PubMed] [Google Scholar]
- Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, Berghmans S, Mayhall EA, Traver D, Fletcher CD, Aster JC, Granter SR, Look AT, Lee C, Fisher DE, Zon LI (2005) BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol 15: 249–254 [DOI] [PubMed] [Google Scholar]
- Polyak K, Waldman T, He TC, Kinzler KW, Vogelstein B (1996) Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev 10: 1945–1952 [DOI] [PubMed] [Google Scholar]
- Prabhakaran K, Li L, Zhang L, Borowitz JL, Isom GE (2007) Upregulation of BNIP3 and translocation to mitochondria mediates cyanide-induced apoptosis in cortical cells. Neuroscience 150: 159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, Yaghi NK, Lim H, Garcia BA, Brizuela L, Zhao K, Roberts TM, Shi Y (2010) Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466: 503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R, Chen G, Vande Velde C, Cizeau J, Park JH, Reed JC, Gietz RD, Greenberg AH (2000) BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J Biol Chem 275: 1439–1448 [DOI] [PubMed] [Google Scholar]
- Regula KM, Ens K, Kirshenbaum LA (2002) Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ Res 91: 226–231 [DOI] [PubMed] [Google Scholar]
- Roperch JP, Alvaro V, Prieur S, Tuynder M, Nemani M, Lethrosne F, Piouffre L, Gendron MC, Israeli D, Dausset J, Oren M, Amson R, Telerman A (1998) Inhibition of presenilin 1 expression is promoted by p53 and p21WAF-1 and results in apoptosis and tumor suppression. Nat Med 4: 835–838 [DOI] [PubMed] [Google Scholar]
- Rouschop KM, Wouters BG (2009) Regulation of autophagy through multiple independent hypoxic signaling pathways. Curr Mol Med 9: 417–424 [DOI] [PubMed] [Google Scholar]
- Sabbatini P, Chiou SK, Rao L, White E (1995) Modulation of p53-mediated transcriptional repression and apoptosis by the adenovirus E1B 19K protein. Mol Cell Biol 15: 1060–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saramaki A, Banwell CM, Campbell MJ, Carlberg C (2006) Regulation of the human p21(waf1/cip1) gene promoter via multiple binding sites for p53 and the vitamin D3 receptor. Nucleic Acids Res 34: 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sax JK, Fei P, Murphy ME, Bernhard E, Korsmeyer SJ, El-Deiry WS (2002) BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol 4: 842–849 [DOI] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM (1993) The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75: 495–505 [DOI] [PubMed] [Google Scholar]
- Schmid T, Zhou J, Kohl R, Brune B (2004) p300 relieves p53-evoked transcriptional repression of hypoxia-inducible factor-1 (HIF-1). Biochem J 380: 289–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW (2002) Dissecting p53 tumor suppressor functions in vivo. Cancer Cell 1: 289–298 [DOI] [PubMed] [Google Scholar]
- Shibue T, Takeda K, Oda E, Tanaka H, Murasawa H, Takaoka A, Morishita Y, Akira S, Taniguchi T, Tanaka N (2003) Integral role of Noxa in p53-mediated apoptotic response. Genes Dev 17: 2233–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soengas MS, Alarcon RM, Yoshida H, Giaccia AJ, Hakem R, Mak TW, Lowe SW (1999) Apaf-1 and caspase-9 in p53-dependent apoptosis and tumor inhibition. Science 284: 156–159 [DOI] [PubMed] [Google Scholar]
- Soengas MS, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, McCombie R, Herman JG, Gerald WL, Lazebnik YA, Cordon-Cardo C, Lowe SW (2001) Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature 409: 207–211 [DOI] [PubMed] [Google Scholar]
- Sowter HM, Ferguson M, Pym C, Watson P, Fox SB, Han C, Harris AL (2003) Expression of the cell death genes BNip3 and NIX in ductal carcinoma in situ of the breast; correlation of BNip3 levels with necrosis and grade. J Pathol 201: 573–580 [DOI] [PubMed] [Google Scholar]
- Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL (2001) HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res 61: 6669–6673 [PubMed] [Google Scholar]
- Stempien-Otero A, Karsan A, Cornejo CJ, Xiang H, Eunson T, Morrison RS, Kay M, Winn R, Harlan J (1999) Mechanisms of hypoxia-induced endothelial cell death. Role of p53 in apoptosis. J Biol Chem 274: 8039–8045 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Tomida A, Tsuruo T (2001) Dephosphorylated hypoxia-inducible factor 1alpha as a mediator of p53-dependent apoptosis during hypoxia. Oncogene 20: 5779–5788 [DOI] [PubMed] [Google Scholar]
- Symonds H, Krall L, Remington L, Saenz-Robles M, Lowe S, Jacks T, Van Dyke T (1994) p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell 78: 703–711 [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D′Amelio M, Criollo A, Morselli E, Zhu C, Harper F, Nannmark U, Samara C, Pinton P, Vicencio JM, Carnuccio R, Moll UM, Madeo F, Paterlini-Brechot P, Rizzuto R, Szabadkai G et al. (2008a) Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 10: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Orhon I, Kepp O, Morselli E, Criollo A, Kroemer G (2008b) p53 represses autophagy in a cell cycle-dependent fashion. Cell Cycle 7: 3006–3011 [DOI] [PubMed] [Google Scholar]
- Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF (2007) BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol 27: 6229–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Velde C, Cizeau J, Dubik D, Alimonti J, Brown T, Israels S, Hakem R, Greenberg AH (2000) BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol Cell Biol 20: 5454–5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L (1998) The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J 17: 4668–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, Adams JM, Strasser A (2003) p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302: 1036–1038 [DOI] [PubMed] [Google Scholar]
- Wang B, Xiao Z, Ren EC (2009) Redefining the p53 response element. Proc Natl Acad Sci USA 106: 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wu Q, Qiu P, Mirza A, McGuirk M, Kirschmeier P, Greene JR, Wang Y, Pickett CB, Liu S (2001) Analyses of p53 target genes in the human genome by bioinformatic and microarray approaches. J Biol Chem 276: 43604–43610 [DOI] [PubMed] [Google Scholar]
- Wang WD, Huang CJ, Lu YF, Hsin JP, Prabhakar VR, Cheng CF, Hwang SP (2006) Heart-targeted overexpression of Nip3a in zebrafish embryos causes abnormal heart development and cardiac dysfunction. Biochem Biophys Res Commun 347: 979–987 [DOI] [PubMed] [Google Scholar]
- Webster KA, Graham RM, Bishopric NH (2005) BNip3 and signal-specific programmed death in the heart. J Mol Cell Cardiol 38: 35–45 [DOI] [PubMed] [Google Scholar]
- Webster KA, Graham RM, Thompson JW, Spiga MG, Frazier DP, Wilson A, Bishopric NH (2006) Redox stress and the contributions of BH3-only proteins to infarction. Antioxid Redox Signal 8: 1667–1676 [DOI] [PubMed] [Google Scholar]
- Weidman D, Shaw J, Bednarczyk J, Regula KM, Yurkova N, Zhang T, Aguilar F, Kirshenbaum LA (2008) Dissecting apoptosis and intrinsic death pathways in the heart. Methods Enzymol 446: 277–285 [DOI] [PubMed] [Google Scholar]
- Yan J, Yun H, Yang Y, Jing B, Feng C, Song-bin F (2006) Upregulation of BNIP3 promotes apoptosis of lung cancer cells that were induced by p53. Biochem Biophys Res Commun 346: 501–507 [DOI] [PubMed] [Google Scholar]
- Yasuda M, Theodorakis P, Subramanian T, Chinnadurai G (1998) Adenovirus E1B-19K/BCL-2 interacting protein BNIP3 contains a BH3 domain and a mitochondrial targeting sequence. J Biol Chem 273: 12415–12421 [DOI] [PubMed] [Google Scholar]
- Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L (2003) PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci USA 100: 1931–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL (2008) Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 283: 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang J, Ney PA (2009) Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ 16: 939–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhang S, Jiang J, Chen X (2000) Definition of the p53 functional domains necessary for inducing apoptosis. J Biol Chem 275: 39927–39934 [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhou W, Jiang J, Chen X (1998) Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J Biol Chem 273: 13030–13036 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.