Abstract
Lymphotactin (Ltn) is a unique chemokine that under physiological solution conditions displays large-scale structural heterogeneity, defining a new category of ‘metamorphic proteins’. Previous Ltn studies have indicated that each form is required for proper function, but the mechanism of interconversion remains unknown. Here we have investigated the temperature dependence of kinetic rates associated with interconversion and unfolding by stopped-flow fluorescence to determine transition-state free energies. Comparisons of derived thermodynamic parameters revealed striking similarities between interconversion and protein unfolding. We conclude that Ltn native state rearrangement proceeds by way of a large-scale unfolding process rather than a unique intermediate structure.
Chemokines are small secreted signaling proteins that direct leukocyte migration toward areas of inflammation during an immune response. This task is dependent upon two functional roles associated with chemokines: 1) high affinity binding and activation of specific G-protein-coupled receptors (GPCRs) resulting in cell chemotaxis (1), and 2) binding to glycosaminoglycans (GAGs) found in the extracellular matrix resulting in establishment of a signaling gradient toward the area of injury or infection (1). Chemokines are classified into four subfamilies (CX3C, CXC, CC and C) based on the configuration of disulfide bonds within the N terminus (2). Lymphotactin (Ltn/XCL1) is the defining member of the C-type subfamily, which contains only one of the two disulfide bonds found in all other chemokines. Ltn is unique in the way it bridges these two functional aspects of chemokine physiology, transforming between a monomeric canonical chemokine fold that can stimulate its cognate GPCR XCR1 (3), and a dimeric β-sheet configuration capable of binding extracellular GAGs with high affinity (4) (Figure 1a). Depending on solution conditions Ltn adopts the conserved chemokine fold, termed Ltn10 (10 °C, 200 mM NaCl), or the unique dimer configuration, termed Ltn40 (40 °C, 0mM NaCl) (5). At equilibrium under near-physiological conditions (37 °C, 150 mM NaCl) both conformations are equally populated and each is required for proper biological function (4). The unique structural interconversion of Ltn has led to further classification as a ‘metamorphic protein’. These newly discovered proteins are capable of adopting different folded conformations for the same primary sequence. However unlike other members of this emerging class, Ltn is the only naturally occurring protein known to populate two distinct folds in a native-state equilibrium that involves a complete restructuring of core residues (6).
Figure 1.
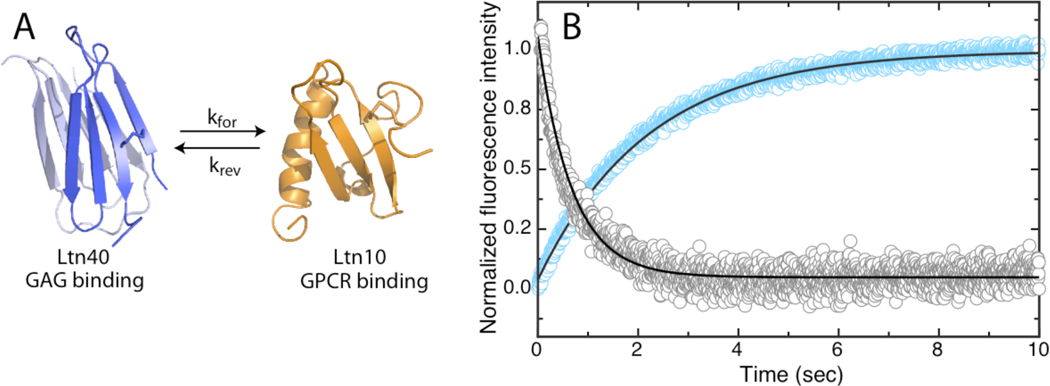
A) Native state structures of Ltn depicting kinetic rate (kforward or kreverse) associated with interconversion. B) Stopped-flow fluorescence detection of Ltn interconversion. Representative kinetic traces of Ltn40→Ltn10 and Ltn10→Ltn40 conversion induced by addition of 200mM NaCl (blue) and 60uM low molecular weight heparin (gray), respectively, are shown with best-fit single exponential curves (black).
While many of the distinctive structural and functional properties of each Ltn conformation have been characterized (3–5, 7, 8), the exact mechanism of structural interconversion has remained elusive. Because native-state interconversion disrupts every hydrogen bond defining the secondary structure and replaces it with a different pairing (4) we speculated that the transition proceeds through a chiefly disordered state. As an alternative to global unfolding, a structured intermediate could lower the free energy barrier separating Ltn10 and Ltn40 on the conformational energy landscape. Recently, a specific transition-state intermediate resembling a Ltn40 monomer with some Ltn10 tertiary contacts was proposed on the basis of coarse-grained molecular dynamic simulations of the Ltn10-Ltn40 equilibrium (9). As a first step toward a mechanistic description of this process, we studied the temperature dependence of Ltn interconversion and unfolding by stopped-flow fluorescence to determine if the Ltn structural transformation is thermodynamically similar to protein unfolding, or employs a low energy transition-state structure.
Ltn contains one tryptophan (W55) that can report on the conformational state of the protein (8). For instance in the Ltn10 conformation W55 is buried within a hydrophobic pocket formed by residues A49, A53, and V59, and displays maximum fluorescence intensity at 330nm. In contrast W55 in Ltn40 is solvent exposed resulting in reduced, red-shifted fluorescence emission relative to the Ltn10 profile. It has been shown that conformational equilibrium can be shifted to favor formation of Ltn10 or Ltn40 by changing solution conditions through addition of NaCl or low molecular weight heparin, respectively (8). By measuring the change in fluorescence intensity in Ltn samples subjected to rapid mixing with either salt or heparin solutions we were able to monitor conversion to each Ltn state (Figure 1b).
The resulting time-dependent fluorescence intensities were fit to single exponential curves to yield kinetic rates. Analysis revealed a Ltn40 → Ltn10 conversion rate (kfor) of 0.4 s−1 at 308 K, and a Ltn10 → Ltn40 conversion rate (krev) of 1.4 s−1 at 308 K, in excellent agreement with previously determined values of kfor = 0.4 s−1 and krev = 1.2 s−1 derived from equilibrium measurements of interconversion by 2D exchange NMR (8). Based on the large difference in protein concentration employed in fluorescence and NMR experiments (30µM vs. 1mM, respectively) and the absence of heparin in 2D exchange measurements, the results indicated that conformational change is likely a rate limiting process with rate constants independent of total protein or heparin concentrations. This analysis is further supported by estimates of the Ltn dimer Kd in the low micromolar range (5) that would predict a koff rate > 10s−1 (assuming diffusion controlled association ~107 M−1s−1), which is considerably faster than the measured conversion rate of 0.4s−1.
The temperature dependence of both conversion rates was then studied to probe the kinetic barrier between the two Ltn conformers. A complete description of experimental conditions is presented in the supplemental material. The rate constants of interconversion were related to the free energy difference between the corresponding state (Ltn10 or Ltn40) and the transition-state by the Eyring equation:
| (equation 1) |
that can be rewritten as,
| (equation 2) |
where kB is Boltzmann’s constant, h is Planck’s constant, R is the gas constant, and T is temperature. While we acknowledge the limited validity of Eyring analysis as it applies to protein thermodynamics as previously discussed by d’Avignon et al (10), this approach has been used extensively in the context of protein interconversion (11), as well as protein folding and unfolding (12–15). Plots of ln(k/T) vs. 1/T (Figure 2) for the forward and reverse reactions were separately fit to equation 2 allowing estimation of the enthalpic and entropic contributions to the interconversion transition-state.
Figure 2.
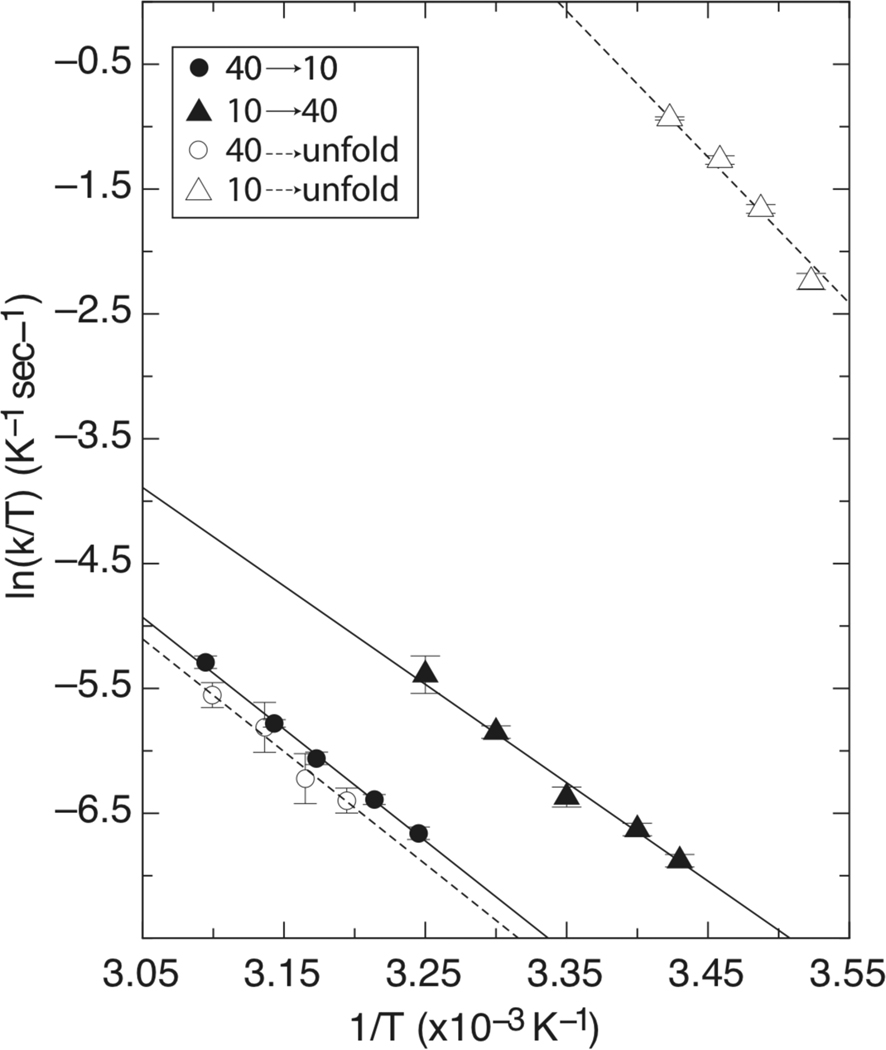
Eyring plots generated from Ltn interconversion (solid lines) and urea induced unfolding (dashed lines) experiments.
Between 308 K and 323 K, kfor exhibited a linear Eyring relationship with values ranging from 0.4 s−1 to 1.6 s−1, respectively (Figure 2). Eyring analysis revealed that a large enthalpic barrier (17.7 kcal/mol) and a modest entropic barrier (−0.8 kcal/mol at 298 K) must be overcome during Ltn40 to Ltn10 interconversion. A list of all thermodynamic parameters determined is displayed in Table 1.
Table 1.
Thermodynamic parameters for Ltn interconversion and unfolding determined by stopped-flow fluorescence.
| Parameter | Value | |
|---|---|---|
| Ltn40 →Ltn10 | ΔG‡ | 18.5 kcal/mol |
| ΔH‡ | 17.7 kcal/mol | |
| ΔS‡ | −2.7 cal/mol·K (−0.8 kcal/mol @ 298K) |
|
| Ltn10→Ltn40 | ΔG‡ | 17.7 kcal/mol |
| ΔH‡ | 15.6 kcal/mol | |
| ΔS‡ | −7.0 cal/mol·K (−2.1 kcal/mol @ 298K) |
|
| Ltn40→unfold | ΔG‡ | 18.6 kcal/mol |
| ΔH‡ | 17.9 kcal/mol | |
| ΔS‡ | −2.4 cal/mol·K (−0.7 kcal/mol @ 298K) |
|
| Ltn10→unfold | ΔG‡ | 14.1 kcal/mol |
| ΔH‡ | 23.2 kcal/mol | |
| ΔS‡ | 30.5 cal/mol·K (9.1 kcal/mol @ 298K) |
Similar analysis of the krev rates measured between 291 K and 308K also resulted in a linear Eyring dependence with rates between 0.3 s−1 and 1.4 s−1, respectively (Figure 2). Eyring analysis again indicated large enthalpic (15.6 kcal/mol) and modest entropic (−2.1 kcal/mol at 298 K) barriers associated with the Ltn10 → Ltn40 interconversion transition state at 298K. The temperature dependence of interconversion rates therefore indicates that similar enthalpic and entropic contributions are required for both the Ltn10→Ltn40 and Ltn40→Ltn10 transformations. The large enthalpic barrier (ΔH‡) clearly reflected disruption of non-covalent bonding interactions. However it was unclear if these values represented partial loss of secondary and tertiary structures or a global unfolding event. Entropy changes of the transition-state (ΔS‡) arise from two effects that are typically counterbalanced in globular proteins: conformational freedom of the polypeptide chain, and ordering of solvent molecules around nonpolar residues (the ‘hydrophobic effect’). Because Ltn interconversion is a global structural change with a large ΔH‡, it is unlikely that the individual entropic factors would be unchanged in the transition state. Thus, we reasoned that the small ΔS‡ might correspond to large opposing changes in both solvent exposure and chain entropy, which appears to be most consistent with a global unfolding event. Therefore we investigated the temperature dependence of Ltn10 and Ltn40 unfolding to determine if the resulting transition states were thermodynamically similar to the interconversion process.
Unfolding rates were determined by stopped-flow fluorescence for both Ltn forms using urea as the chemical denaturant. Final kinetic rates for each unfolding transition (ku10 or ku40) used in the analysis were extrapolated to 0 M denaturant based on a two-state unfolding model (Supplemental material, Figure S1). Selection of each Ltn conformer present at the start of denaturant mixing experiments was accomplished by adjustment of temperature and solution conditions. Ltn10 unfolding was measured by buffering solutions in 20 mM NaH2PO4, with 200 mM NaCl, at temperatures below 298K, while the Ltn40 unfolding was measured by buffering solutions in 20mM NaH2PO4 with no added salt at temperatures above 313 K. The Ltn10 unfolding rates were measured by single exponential fitting of decay curves. Eyring analysis revealed a linear dependence with extrapolated unfolding rates of ku10 = 3.4 to 4.7 sec-−1 between 283 K and 291 K, yielding an enthalpic barrier of 23.2 kcal/mol and an entropic contribution of 9.1 kcal/mol calculated at 298 K (Equation 2 and Figure 2). Analysis of Ltn40 unfolding rates were conducted by single exponential fitting of unfolding curves collected between 313 K and 323 K. Extrapolated unfolding rates of ku40 = 0.5 to 1.3 sec-−1 were determined between 313 K and 323 K, resulting in an enthalpic barrier of Ltn40unfold of 17.9 kcal/mol and entropic contribution of −0.7 kcal/mol calculated at 298 K (Equation2 and Figure 2).
Comparison of Ltn interconversion parameters (Table 1) reveals very similar barrier heights that correspond to a ΔGLtn40-Ltn10 of 0.8 kcal/mol separating the two conformational states. This small free energy difference is consistent with NMR measurements of the Ltn conformational equilibrium indicating that Keq ~ 1 at 298 K (8). Both conversion events reveal that a large enthalpic barrier must be crossed, consistent with the disruption of hydrogen bonding networks that must accompany Ltn structural rearrangement (5). Furthermore the slightly negative entropy change likely describes a reduction in water entropy from solvent exposure of hydrophobic core residues that exceeds the increase in chain entropy associated with the loss of tertiary structure, based on the virtually identical thermodynamic signature that characterize both Ltn40→Ltn10 conversion and Ltn40 unfolding. Thermodynamic parameters derived from Ltn10 unfolding also a show a large enthalpic signature, but reveal a favorable entropic contributions leading to a free energy barrier height that is less than structural interconversion. We attribute this disparity in barrier height to the most obvious difference in reaction pathways for the two processes: Ltn10 ↔ Ltn40 interconversion requires formation and disassembly of both tertiary and quaternary contacts, while Ltn10 unfolding reports only on the disruption of tertiary structure. Hence the difference in barrier heights would be expected given that additional free energy is required to bring two monomer subunits together during the Ltn10 → Ltn40 interconversion reaction.
Given the similar thermodynamic profiles of the transition states investigated in this study, we conclude that Ltn interconversion likely requires a global unfolding transition and does not pass through a structured low energy intermediate. This scenario is consistent with a previous H/D exchange study conducted by 2-D 1H-15N- HSQC analysis (8), that showed regardless of temperature or salt concentration Ltn protein samples lost all detectable amide signals within the experimental dead time (5 min.), reflecting an absence of stable hydrogen bonding networks within the timescale of interconversion (kex ~1.5 sec−1). Interestingly, a recent report on the structural rearrangement between two β-strand registers of the ARNT PAS-B mutant Y456T (11) also concluded that the protein interconverts by unfolding to a chiefly disordered state, and refolds into either conformation. An analogous process is likely at work in Ltn interconversion, however a complete mechanistic understanding is still being elucidated. Given that chemokine-mediated recruitment of cells in vivo depends on distinct functions (GAG binding and GPCR activation) (16) that are supplied independently by each of the folded Ltn native state structures, our results suggest that the ability to access the unfolded state is an essential feature of its biological activity. Considering the close structural similarity between the two ARNT PAS-B mutant forms (11) and the disparate structural states of Ltn, these results also suggest that perhaps global unfolding is a common mechanism when structural rearrangement requires disruption of a large number of hydrogen bonds.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Jacek Zielonka of the Free Radical Research Center at the Medical College of Wisconsin for access to the stopped-flow fluorescence instrument. This research was supported by NIH grants AI063325 and AI058072 to B.F.V.
Footnotes
SUPPORTING INFORMATION. Material and Methods for experimental protocols and Figures S1 are provided in Supporting Information. This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Thelen M, Stein JV. How chemokines invite leukocytes to dance. Nat Immunol. 2008;9:953–959. doi: 10.1038/ni.f.207. [DOI] [PubMed] [Google Scholar]
- 2.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 3.Tuinstra RL, Peterson FC, Elgin ES, Pelzek AJ, Volkman BF. An engineered second disulfide bond restricts lymphotactin/XCL1 to a chemokine-like conformation with XCR1 agonist activity. Biochemistry. 2007;46:2564–2573. doi: 10.1021/bi602365d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc Natl Acad Sci U S A. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuloglu ES, McCaslin DR, Markley JL, Volkman BF. Structural rearrangement of human lymphotactin, a C chemokine, under physiological solution conditions. J Biol Chem. 2002;277:17863–17870. doi: 10.1074/jbc.M200402200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murzin AG. Biochemistry. Metamorphic proteins. Science. 2008;320:1725–1726. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- 7.Peterson FC, Elgin ES, Nelson TJ, Zhang F, Hoeger TJ, Linhardt RJ, Volkman BF. Identification and characterization of a glycosaminoglycan recognition element of the C chemokine lymphotactin. J Biol Chem. 2004;279:12598–12604. doi: 10.1074/jbc.M311633200. [DOI] [PubMed] [Google Scholar]
- 8.Volkman BF, Liu TY, Peterson FC. Chapter 3. Lymphotactin structural dynamics. Methods Enzymol. 2009;461:51–70. doi: 10.1016/S0076-6879(09)05403-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camilloni C, Sutto L. Lymphotactin: how a protein can adopt two folds. J Chem Phys. 2009;131:245105. doi: 10.1063/1.3276284. [DOI] [PubMed] [Google Scholar]
- 10.d’Avignon DA, Bretthorst GL, Holtzer ME, Holtzer A. Thermodynamics and kinetics of a folded-folded' transition at valine-9 of a GCN4-like leucine zipper. Biophysical Journal. 1999;76:2752–2759. doi: 10.1016/s0006-3495(99)77428-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans MR, Gardner KH. Slow transition between two beta-strand registers is dictated by protein unfolding. J Am Chem Soc. 2009;131:11306–11307. doi: 10.1021/ja9048338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen BL, Baase WA, Schellman JA. Low-temperature unfolding of a mutant of phage T4 lysozyme. 2. Kinetic investigations. Biochemistry. 1989;28:691–699. doi: 10.1021/bi00428a042. [DOI] [PubMed] [Google Scholar]
- 13.Manyusa S, Whitford D. Defining folding and unfolding reactions of apocytochrome b5 using equilibrium and kinetic fluorescence measurements. Biochemistry. 1999;38:9533–9540. doi: 10.1021/bi990550d. [DOI] [PubMed] [Google Scholar]
- 14.Tan YJ, Oliveberg M, Fersht AR. Titration properties and thermodynamics of the transition state for folding: comparison of two-state and multi-state folding pathways. J Mol Biol. 1996;264:377–389. doi: 10.1006/jmbi.1996.0647. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Matthews CR. Thermodynamic properties of the transition state for the rate-limiting step in the folding of the alpha subunit of tryptophan synthase. Biochemistry. 1994;33:6356–6362. doi: 10.1021/bi00186a040. [DOI] [PubMed] [Google Scholar]
- 16.Proudfoot AE, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, Borlat F, Wells TN, Kosco-Vilbois MH. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc Natl Acad Sci U S A. 2003;100:1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.