SUMMARY
Ninety percent of the 500,000 annual new cases of visceral leishmaniasis occur in India/Bangladesh/Nepal, Sudan and Brazil. Importantly, 80-90% of human infections are sub-clinical or asymptomatic, usually associated with strong cell-mediated immunity. Understanding the environmental and genetic risk factors that determine why two people with the same exposure to infection differ in susceptibility could provide important leads for improved therapies. Recent research using candidate gene association analysis and genome-wide linkage studies (GWLS) in collections of families from Sudan, Brazil and India have identified a number of genes/regions related both to environmental risk factors (e.g. iron), as well as genes that determine type 1 versus type 2 cellular immune responses. However, until now all of the allelic association studies carried out have been underpowered to find genes of small effect sizes (odds ratios or OR<2), and GWLS using multicase pedigrees have only been powered to find single major genes, or at best oligogenic control. The accumulation of large DNA banks from India and Brazil now makes it possible to undertake genome-wide asscociation studies (GWAS), which are ongoing as part of phase two of the Wellcome Trust Case Control Consortium. Data from this analysis should seed research into novel genes and mechanisms that influence susceptibility to visceral leishmaniasis.
Keywords: leishmaniasis, candidate genes, genome scans
INTRODUCTION
The parasitic disease visceral leishmaniasis (VL) caused by protozoa of the Leishmania donovani species complex (L. donovani, L. archibaldi, L. infantum/chagasi) is associated with liver, spleen and lymph gland enlargement, fever, weight loss, anaemia, and is fatal unless treated. Ninety percent of the 500,000 annual new cases occur in India/Bangladesh/Nepal, Sudan and Brazil (http://www.who.int/inf-fs/en/fact116.html). Importantly, 80-90% of human infections are sub-clinical or asymptomatic, usually associated with strong cell-mediated immunity (positive skin-test delayed type hypersenstivity (DTH+); lymphocyte proliferation; interferon-γ T-cell response) to leishmanial antigen [1, 2, 3, 4]. Understanding the environmental and genetic risk factors that determine why two people with the same exposure to infection differ in susceptibility could provide important leads for improved therapies. Indeed, the intersect of studies on human genetic variation with gene expression studies have the power to influence one of the major bottlenecks in drug development, that of choosing the best targets that represent key points of therapeutic intervention [5]. One of the major aims of genetic studies is to identify genes/mechanisms/pathways that contribute to the pathogenesis of disease, for example by influencing trafficking or survival of the parasite in host macrophages, or the development of a protective type 1 immune response. Genetics can also provide concrete evidence for the role of modifiable environmental variables (e.g. iron, as might be indicated by an association between polymorphism at SLC11A1 and susceptibiltiy to intramacrophage pathogens, cf. below) in determining disease outcome [6]. Knowledge gained through both avenues can translate into improved interventions.
Although most genetic studies of VL undertaken to date have been underpowered, some common genes have emerged by studying different populations, while founder effects and population sub-structure in Africa have provided an interesting avenue to identifying new genes that contribute to susceptibility to VL. These studies are highlighted here. For the future, more recent advances in study design and technology promise to provide the power to examine candidate genes with confidence, and to find novel genes influencing the complex phenotypes of VL or DTH response using genome-wide association studies (GWAS).
APPROACHES TO GENETIC STUDIES
Traditional approaches to genetic analysis of complex diseases have included allelic association analysis of candidate genes and linkage analysis using multicase families. The linkage test, usually reported as a LOD score (logarithm of the odds for linkage), is based on genetic recombination events in families and maps disease susceptibility genes into intervals of 10-20 centiMorgans (~10-20 Mb). This approach has generally been used to undertake genome-wide linkage scans (GWLS), i.e. to search for new regions of the genome carrying susceptibility loci, the first such study being to look for genes controlling the complex disease type 1 diabetes [7]. Such studies typically genotype all members of multicase families for 400-500 highly polymorphic microsatellite markers spaced at 10-20 centimorgan (cM) intervals across the genome. Due to the multiple testing problem related to studying large numbers of polymorphic markers, Lander and Kruglyak [8] proposed a classification for reporting the results of genome-wide scan data based on the number of times one would expect to see a result at random in a dense, complete genome scan. The thresholds they propose are: “suggestive linkage” where statistical evidence would be expected to occur one time at random in a genome scan; “significant linkage” 0.05 times; “highly significant linkage” 0.001 times; and “confirmed linkage” where significant linkage from an initial scan has been confirmed with a nominal P value of ≤0.01 in a second independent study. The first three categories correspond to point-wise significance levels of 7×10-4, 2×10-5 and 3×10-7 (LOD scores 2.2, 3.6 and 5.4). Although some authors consider these thresholds to be over conservative [9], they serve as a guide to evaluate the significance associated with the nominal point-wise P values reported by most authors (cf. VL studies below). Allelic association tests determine direct association between alleles at particular loci, e.g. a candidate gene, or haplotypes of closely linked markers (i.e. markers in linkage disequilibrium or LD with each other) and a disease phenotype. Until recently, this approach was used largely to analyse candidate genes. However, with the advent of technologies that allow upwards of 500,000 single nucleotide polymorphisms (SNPs) to be assayed simultaneously, the so-called “SNP-chip” technology, GWAS have become possible (cf. below). Allelic association is measured over smaller intervals, usually <1 Mb depending on the extent of LD in the population under investigation. For example, LD generally extends over larger intervals in Caucasian compared to African populations [10]. Allelic association studies can be undertaken using either population-based sampling (e.g. case-control), or family-based collections of case-parent trios. Logistic regression analysis is usually used to analyse case-control samples, facilitating adjustment for data on environmental variables. Family-based allelic association tests (e.g. FBAT [11]) based on the transmission disequilibrium test (TDT) [12], which looks for a bias in transmission of alleles from heterozygous parents to affected offspring, are used to analyse case-parent trios/families. Robust tests can be applied to data from multicase families for association testing, taking pedigree or family clustering or known linkage to a region into account. Case-control sampling can be a problem in ethnically admixed populations, where mis-matching of cases and controls can lead to type I errors. The TDT approach, which uses family-based controls, is therefore preferable in ethnically admixed populations. A case/pseudo-control strategy [13] and conditional logistic regression analysis can also be used for trios, where the case is the actual genotype transmitted from parents to the affected offspring, and the pseudo-controls are the 1-3 genotypes (depending on phase) that could have been transmitted. This allows for easy adjustment for data on environmental variables, and extension of the analysis to determine whether multiple loci/SNPs within a gene show independent main effects, or whether one SNP carries all of the information for that gene (i.e. is a haplotype tagging SNP or tag-SNP for markers in LD across the region of the gene associated with disease).
COMPLEXITY AND HERITABILITY OF LEISHMANIASIS SUSCEPTIBILITY
Studies in mice (reviewed [14]) provided early support for a strong genetic component to susceptibility to L. donovani infection. In this defined model system, it was possible to demonstrate that different genes control innate versus adaptive immunity, as expected for a complex disease. In humans, studies based on ethnic differences [15, 16], familial aggregation and segregation analysis [15, 17, 18, 19], and a high relative risk (λ2S = 34) of disease in further siblings of affected sibling pairs [18] support a genetic hypothesis, with longitudinal studies showing a strong interplay between environmental and host factors during outbreaks [1, 19, 20]. Segregation analysis undertaken following total population surveys that measured DTH responses in Peru [21] and Brazil [22] support multifactorial genetic control over a sporadic model. We recently undertook a GWLS for the quantitative DTH response in families from Natal, Brazil [23]. Familial correlations were estimated using the FCOR program of the SAGE (Statistical Analysis for Genetic Epidemiology) software package (version 4.8 [1987]). Heritability (h2) was estimated from the sibling correlations r using the equation h2=2r. In these families there were 440 sibling pairs with a correlation of 0.42, 212 grandparent-grandchild pairs with a correlation of 0.265, and 90 cousin pairs with a correlation of 0.13. This is consistent with genetic control of the DTH response, where first-degree relatives have a stronger correlation than second- or third-degree relatives. Estimated heritability of the DTH immune response was 84%, suggesting a substantial genetic component to variation in induration size, as determined by the DTH skin test.
A number of labs, including our respective labs, have undertaken both candidate gene [24, 25, 26, 27, 28, 29, 30, 31, 32, 33] and GWLS [23, 34, 35, 36] for the VL phenotype (cf. below). In Brazil we also analyzed candidate genes/gene regions [27, 32] using the DTH response as a qualitative trait and, as alluded to above, carried out a GWLS analyzing DTH as a quantitative trait (QTL) [23]. It should be noted, however, that all of the allelic association studies carried out to date were underpowered to find multiple genes of small effect sizes (odds ratios or OR<2). Similarly, all GWLS using multicase pedigrees were only sufficiently powered to find single major loci, or at best oligogenic control.
THE ISSUE OF SAMPLE SIZE AND POWER
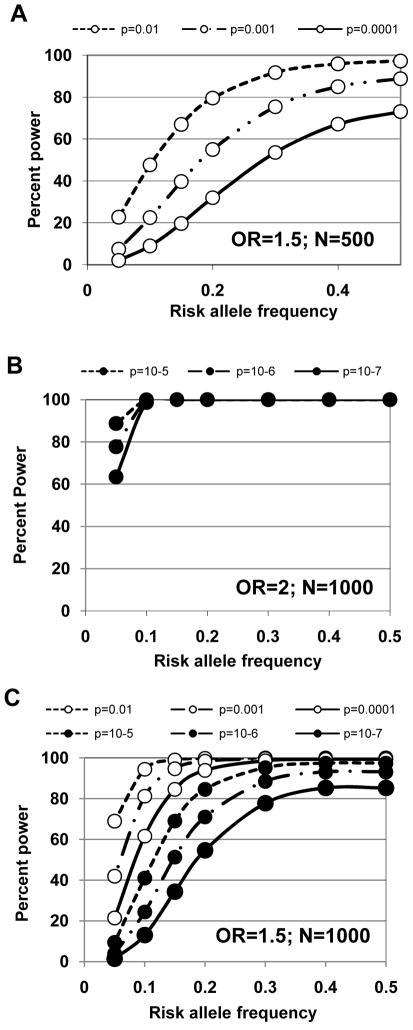
One major problem with all candidate gene studies for infectious diseases reported to date is that they were under-powered (reviewed [37]). Until recently, this was a general problem in genetic analysis of complex disease, along with issues relating to study design and population history (reviewed [38, 39]). Small sample sizes also preclude definitive conclusions being drawn from a number of VL studies (Table 1) reporting no association for candidate genes [24, 25, 29, 30, 40, 41]. Figure 1 compares power to detect association at OR=1.5 or OR=2, given different risk allele frequencies, P values, and sample sizes. This shows that 500 trios or case-control pairs have little power to detect association for small effect sizes (OR=1.5). Even with 1000 trios or case-control pairs, power is limited for risk alleles with frequency <0.2 for an effect size (OR) 1.5, although low frequency (e.g. 0.10) risk alleles with larger effect sizes (OR >2) may be detected. All published VL studies have sample sizes <300 cases, severely limiting power even for hypothesis-driven candidate genes.
Table 1.
Summary of reported linkages or associations between candidate genes and VL or DTH phenotypes*. Adapted and updated from [37].
| A. Papers Reporting Significant Linkage (L) or Association (A) | ||||||
|---|---|---|---|---|---|---|
| Candidate Gene | Population | Phenotype | Sample Size | Reported Results (L, A) | Year | Reference |
| MHC Class I Region: | ||||||
| A25 | Iranian | VL | Ca=52; Co=222 | RR=13.27; P=0.004 (A) | 1995 | [28] |
| MHC Class II Region: | ||||||
| DRB1*15; *16 | Tunisian | VL | Ca=156; Co=154 | RR=0.54; P=0.04 (Pc>0.05) (A) | 2001 | [29] |
| DQB1*0201 | Tunisian | VL | Ca=156; Co=154 | RR=0.46; P=0.03 (Pc>0.05) (A) | 2001 | [29] |
| HLA-DR msat | Brazilian | VL | 183 fam (104 aff) | P=0.02 (A) | 2002 | [27] |
| MHC Class III Region: | ||||||
| TNFA (TNFa/-308bp haplotype) | Brazilian | DTH+ | 183 fam (381 DTH+) | P=0.0265 (A) | 2002 | [27] |
| TNFA -308bp | Brazilian | DTH+ | 183 fam (381 DTH+) | P=0.0006 (A) | 2002 | [27] |
| SLC11A1 (formerly NRAMP1): | ||||||
| SLC11A1 (GT(n)) | Sudanese | VL | 37 fam (74 aff) | LOD=1.32; P=0.007 (L) | 2003 | [24] |
| SLC11A1 (469+14G/C) | Sudanese | VL | 67 fam (168 aff) | χ2=9.03; P=0.0027 (A) | 2004 | [26] |
| Chr 5q23.3–q31.1: | ||||||
| IL4 (IL4RP1; IL4RP2) | Sudanese | VL+PKDL | 67 fam (177 aff) | OR=2.5; OR=1.68; P<0.003 (A) | 2003 | [25] |
| IL4 | Brazilian | DTH- | 102 fam (190 DTH-) | OR 3.14; P=0.006 (A) | 2007 | [32] |
| Between LECT2-TGFBI | Brazilian | DTH- | 102 fam (190 DTH-) | OR 3.00; P=0.042 (A) | 2007 | [32] |
| LECT2 | Brazilian | DTH+ | 102 fam (323 DTH+) | OR 2.25; P=0.005 (A) | 2007 | [32] |
| TGFBI | Brazilian | DTH+ | 102 fam (323 DTH+) | OR 1.94; P=0.003 (A) | 2007 | [32] |
| Chr 17q11.2-q21.3: | ||||||
| CCL1 | Brazilian | VL | 98 fam (176 aff) | Z=2.18; P=0.02; Pc=0.03 (L) | 2007 | [36] |
| CCL16 | Brazilian | VL | 98 fam (176 aff) | Z=2.21; P=0.02; Pc=0.03 (L) | 2007 | [36] |
| Other Candidates: | ||||||
| IFNGR1 (INT6 msat, a10&a11) | Sudanese | PKDL | 26 fam (54 aff) | OR=3.0 & OR=2.18; P<0.03 (A) | 2003 | [25] |
| IFNGR1 (INT6 msat, a12) | Sudanese | PKDL | 26 fam (54 aff) | OR=0.15; P=0.014 (A) | 2003 | [25] |
| IFNGR1 −470ins/delTT, -270T/C, -56T/C, +95T/C | Sudanese | PKDL | 80 trios | χ210df=21.97; P=0.015 (A) | 2007 | [31] |
| B. Papers Reporting No Significant Linkage (L) or Association (A) | ||||||
| Candidate Gene | Population | Phenotype | Sample Size | Reported Results (L, A) | Year | Reference |
| MHC Class I Region: | ||||||
| HLA -A; -B | Indian | VL | Ca=51; Co=46 | P=0.21; P=0.22 (A) | 1997 | [41] |
| MHC Class II Region: | ||||||
| DR | Indian | VL | Ca=51; Co=46 | P=0.40 (A) | 1997 | [41] |
| DQB1; DQA1; DRB1 | Brazilian | VL | 117 fam (214 aff) | ns (L) | 2002 | [30] |
| MHC Class III Region: | ||||||
| TNFA (-308bp) | Tunisian | VL | Ca=156; Co=154 | ns (A) | 2001 | [29] |
| LTA/TNFB | Tunisian | VL | Ca=156; Co=154 | ns (A) | 2001 | [29] |
| HSP70-2 (PstI) | Tunisian | VL | Ca=156; Co=154 | ns (A) | 2001 | [29] |
| HSP70-hom (NcoI) | Tunisian | VL | Ca=156; Co=154 | ns (A) | 2001 | [29] |
| LTA/TNFB | Brazilian | VL | 117 fam (214 aff) | ns (A) | 2002 | [30] |
| TNFA (-308bp; -238bp; TNFa) | Brazilian | VL | 117 fam (214 aff) | ns (A) | 2002 | [30] |
| SLC11A1 (formerly NRAMP1): | ||||||
| SLC11A1 | Brazilian | VL | 46 fam (~92 aff) | ns (L) | 1997 | [40] |
| Chr 5q23.3–q31.1: | ||||||
| IL4 | Sudanese | VL | 37 fam (74 aff) | ns (L) | 2003 | [24] |
| IL9 | Sudanese | VL+PKDL | 67 fam (177 aff) | ns (A) | 2003 | [25] |
| IRF1-IL5-IL13-IL4-IL9-LECT2-TGFBI | Brazilian | VL | 102 fam (123 aff) | ns (A) | 2007 | [32] |
| IRF1-IL5-IL13-IL4--IL9 | Brazilian | DTH+ | 102 fam (323 DTH+) | ns (A) | 2007 | [32] |
| IRF1-IL5-IL13-IL9 | Brazilian | DTH- | 102 fam (190 DTH-) | ns (A) | 2007 | [32] |
| Chr 17q11.2-q21.3: | ||||||
| NOS2A,CRLF3,CCL2,CCL7,CCL11,CCL8, CCL13,CCL5,CCL15,CCL23,CCL18,CCL3, CCL4,PSMD3,CSF3,THRA1,CCR7,STAT5B, STAT5A,STAT3,CCR10,PSME3,FZD2, ADAM11,TBX21,SP2,SCAP1,TOB1 | Brazilian | VL | 98 fam (176 aff) | ns | 2007 | [36] |
| Other Candidates: | ||||||
| IFNG (JAP Intronic msat) | Sudanese | VL | 37 fam (74 aff) | ns (L) | 2003 | [24] |
| IFNG (−179G/A, +874T/A) | Sudanese | PKDL | 80 trios | ns (A) | 2007 | [31] |
| IFNGR1 (INT6 msat) | Sudanese | VL alone | 53 fam (123 aff) | ns (A) | 2003 | [25] |
| IFNGR1 (FA1 Intronic msat) | Sudanese | VL | 37 fam (74 aff) | ns (L) | 2003 | [24] |
Abbreviations: aff= affected; Ca = case; Co = control; DTH = delayed type hypersensitivity; fam = nuclear families; INT = intron; LOD = log10 likelihood for linkage; msat = microsatellite; ns = not significant; OR = odds ratio; P = probability; Pc = probability corrected for multiple testing; PKDL = post Kala-azar dermal leishmaniasis; RR = relative risk; VL = visceral leishmaniasis.
Figure 1.
Percent power (Y-axis) of N=500 (A) or N=1000 (B,C) trios of case-control pairs to detect allelic association for a risk allele with effect size (OR) 1.5 (A,C) or 2 (B), given different risk allele frequencies (X-axis) and P-values (10-2 to 10-7 as indicated on key for different lines on each graph). Greater robustness to type one error, i.e. lower thresholds for P-values, is required for GWAS (cf. in text).
CANDIDATE GENE STUDIES OF VL AND DTH PHENOTYPES
A number of candidate gene studies have been reported for VL (Table 1) [24, 25, 26, 27, 28, 29, 30, 31]. Important amongst these are ones arising from murine studies, where both innate immunity under the control of the Slc11a1 (formerly Lsh/Ity/Bcg/Nramp1) gene [42, 43] and acquired immunity directed by the major histocompatibility complex (H-2 in mice, HLA in man) [44] were shown to be important. The latter form part of a broader analysis of genes that control T helper 1 (Th1) versus T helper 2 (Th2) immune responses.
The innate resistance gene SLC11A1
Recent interest has focused on the role of innate immunity in driving the adaptive immune response, particularly in relation to intra-macrophage pathogens. In mice, the archetypal innate resistance gene was first identified as a gene controlling VL caused by Leishmania donovani sensu strictu (reviewed in Ref. [45]). This gene, originally designated Lsh, Ity or Bcg, was also shown to influence innate resistance to Salmonella typhimurium, Mycobacterium bovis BCG, M lepraemurium and M. intracellulare. Following its identification by positional cloning [46], it was renamed the natural resistance associated macrophage protein (Nramp1). This is now superseded by the functional designation solute carrier family 11a (proton-coupled divalent metal ion transporters) member 1 or Slc11a1, consistent with formal demonstration that the proteins encoded by murine Slc11a1 and human SLC11A1 function as proton/divalent cation (Fe2+, Zn2+ and Mn2+) antiporters [47, 48]. The protein localises to the late endosomal/lysosomal compartment of macrophages [49] and has many pleiotropic effects on macrophage function (reviewed in Ref. [45]). In humans, SLC11A1 has been linked to genetic susceptibility to leprosy in Vietnam and to tuberculosis in Brazil and Aboriginal Canadians (reviewed in Ref. [50]). SLC11A1 is globally associated with TB, with both 5’ and 3’ polymorphisms contributing independently to disease risk [51, 52, 53]. SLC11A1 is associated with HIV [54] and a wide range of autoimmune diseases in humans (reviewed in Ref. [50]).
Polymorphism at SLC11A1 has been linked [24, 26] and associated [26] with VL in Sudan, the latter study by us demonstrating allelic association with 5’ (GTn, 274C/T, 469+14G/C) but not 3’ (D543N, 3’UTR TGTG, 3’UTR CAAA) markers within SLC11A1. To date, only the promoter GTn is known to be functional in regulating expression of SLC11A1 [55], modulated by SNPs at −237 bp [56] and −86 bp (H.S. Mohamed & J.M. Blackwell, unpublished) in the promoter region. The activity of the GTn functions by binding Hypoxia-Inducible Factor 1 alpha (HIF1α) to a sequence element within the repeat [57]. Preliminary data from a subset of the Brazilian cohort also shows association of VL with the 274C/T (χ2=5; p=0.03) and 469+14G/C (χ2=4.28; p=0.04) polymorphisms (S.E. Jamieson, J.M. Blackwell, M.E. Wilson, S.M. Jeronimo, unpublished). These data require robust analysis in a larger sample. So far the region of the genome that includes SLC11A1 has also failed to register suggestive or significant linkage on any GWLS [23, 34, 35, 36], but this could also be due to lack of power. It will be of interest to see whether there are positive results for SLC11A1 in ongoing GWAS where much larger samples sizes are being examined for Brazil and India (cf. below).
Genes associated with Th1 versus Th2 responses
Clinical VL is a complex disease phenotype so we expect multiple genes to influence susceptibility to disease. In particular, genes that regulate induction of an adaptive T cell response will be important. In mice [44] we know, for example, that the right H-2 haplotype controlling adaptive immunity can overcome innate susceptibility caused by mutation at Slc11a1. Preliminary reports (Table 1) of failure to link class II (DR/DQ) and/or class III (TNFA, LTA, TNFa) HLA genes to VL in Brazil [30] or Sudan [24] were underpowered. Case-control studies in Iran [28] (52 cases; 222 controls) and Tunisia [29] (156 cases; 154 controls) reporting association with class I (A25, OR=13.27, P=0.004) and class II (DRB1*15*16, OR=0.54, P=0.04; DQB1*0201, OR=0.46, P=0.03) genes were also underpowered and not robust to multiple testing correction. Preliminary studies in Brazil show associations at TNFA when VL cases are compared to DTH+ (i.e. >5mm induration) indviduals, and a bias in transmission of alleles at TNFA (59 haplotype transmissions, P=0.0265; 36 TNFA -308 bp transmissions, P=0.0006) from heterozygous parents to DTH+ individuals in families [27]. Again, the HLA region has failed to provide positive linakges on GWLS carried out to date, suggesting that this is not a major locus controlling susceptibiltihy to VL [23, 34, 35, 36]. The HLA region requires robust analysis in a larger sample.
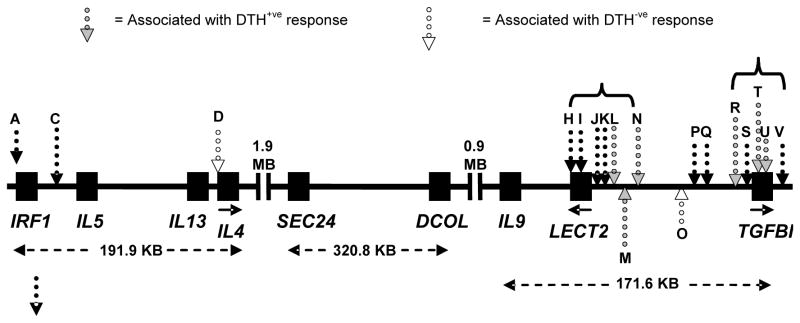
In murine leishmaniasis polarisation of the adaptive immune response down Th1 versus Th2 pathways is associated with resistance and susceptibility in different mouse strains [58]. The region on murine chromosome 11 that has conserved synteny with human Chromosome 5q23-q33 and carries the genes encoding interleukin 4 (IL-4) and other type 2 cytokines is linked to visceralisation of L. major in susceptible BALB/c mice [59]. In humans IL-4 is detected in all Brazilian VL patient sera [60], and is at a 13-fold higher level in Indian VL patients compared with controls [61]. Interestingly, non-exposed individuals also show differences in patterns of IFN-γ and/or IL-4 production upon stimulation with L. donovani antigens [62], suggesting an inherent bias in response. Candidate gene analysis of IL4 and IL9 within this cluster (Figure 2), whose cytokine products IL-4 and IL-9 mediate Th2 responses, provided evidence for association at IL4 but not IL9 in Sudan [25]. We recently reported [32] multiple independent associations across this gene cluster for the DTH phenotype in Brazil (Figure 2). DTH was analyzed as a qualitative trait, either as DTH+ (>5mm) or DTH- (<5mm; resident ≥3 years in an areas with >40% infection rate). No associations were observed for VL, which may reflect low power (107 VL trios). No association was observed DTH+ (176 trios) and SNPs at IL4, but two independent associations were observed in separate LD blocks at LECT2 (OR 2.25; P=0.005; 95% CI 1.28-3.97; Figure 2: markers L, M, N) and TGFBI (OR 1.94; P=0.003; 95% CI 1.24-3.03; Figure 2: markers R, T, U). Independent associations were observed for DTH- (118 trios) and SNP rs2070874 (Figure 2: marker D) at IL4 (OR 3.14; P=0.006; 95% CI 1.38-7.14) and SNP rs30740 (Figure 2: marker O) between LECT2 and TGFBI (OR 3.00; P=0.042; 95% CI 1.04-8.65). The former is interesting in relation to the innate role that early IL-4 production plays in determining outcome of L. major infection in BALB/c mice. In relation to other genes in the region, LECT2 encodes the leukocyte cell-derived chemotaxin 2 that has neutrophil chemotactic activity. This could be relevant to the DTH+ phenotype, although its principal expression in human hepatocytes and endothelial cells of hepatic arteries, portal veins and central veins make it an unlikely candidate. TGFBI encodes a protein whose transcript is upregulated by TGF-β in adenocarcinoma cells. The gene product keratoepithelin contains an N-terminal signal peptide and a C-terminal RGD motif similar to other adhesion proteins, and is expressed in many tissues. Expression in skin epithelial cells could contribute to DTH, but further candidates, SMAD5 and its antisense transcript DAMS, located ~100 kb distal to TGFBI have not been studied. SMAD5 plays a critical role in the signalling by which TGFβ inhibits cell proliferation. As for SLC11A1 and HLA loci, the 5q23-q33 region has not been positive on GWLS carried out to date [23, 34, 35, 36]. Lack of power precludes speculation as to whether this represents a biological divergence between mouse and man, or simply the fact that we are comparing inbred mouse data with outbred human populations and underpowered human genetic studies. The 5q23-q33 results require robust validation and replication in larger samples, using a more complete set of tag-SNPs across the region.
Figure 2.
Diagrammatic representation of the 5q23-q31 region showing results of allelic association studies for VL, DTH+ and DTH- phenotypes in Natal, Brazil. Dotted black arrows with grey filling indicate markers with significant DTH+ associations; dotted black arrows with white filling indicate markers with significant DTH- associations. Markers D and O show independent main effects, indicating that two genes influence DTH-. For DTH+, markers L, M and N are in strong LD and do not show independent effects from each other, but do show independent effects compared to the markers R, T and U. Similarly, markers R, T and U are in LD with each other but not with L, M and N and show independent effects from L, M and N. Within this cluster, R and U show independent effects from each other. Figure adapted from [32].
GWLS FOR VL AND DTH PHENOTYPES
Despite limited power in GWLS for VL, evidence for strong genetic effects in local populations is found in Sudan that relate to recent migration, marital systems, and population substructure. One study [34] reported genome-wide significance for a gene on chromosome 22q12 (LOD score 3.5, nominal P=3×10-5 , λS=1.83 for all families; LOD score 3.9, nominal P=1×10-5 if affected towards the beginning of an outbreak) using multicase families of VL from eastern Sudan. This could indicate important differences in innate immune mechanisms that might operate in a naïve population at the beginning of an outbreak, compared to acquired immune mechanisms important as the outbreak progresses. Follow up studies in this population indicate that polymorphisms at IL2RB likely contribute to this peak of linkage [33]. This is of interest in relation to GWAS data showing that polymorphism at IL2RB contributes to susceptibility to rheumatoid arthritis [63], another disease where regulatory T cells play an important role. Nevertheless, variation at IL2RB is likely to contribute only a minor component of linkage at 22q12 [33]. Other important candidate genes in the region include NCF4 that encodes the p40 subunit of NADPH oxidase, CSF2RB encoding the receptor for GMCSF, and LIF encoding leukemia inhibitory factor. Further studies are required to identify the major genes contributing to the 22q12 peak of linkage in this population in Sudan.
We also reported [35] genome-wide significance for major susceptibility loci at D1S1568 on 1p22 (LOD score 5.65; nominal P =1.72×10-7; empirical P<1×10-5; λS=5.1) and D6S281 on 6q27 (LOD score 3.74; nominal P =1.68×10-5; empirical P <1×10-4; λS=2.3)) using multicase families from villages in eastern Sudan. In this case, the linkages were Y-chromosome-lineage and village-specific. The results suggested strong lineage-specific genes within villages due to founder effect and consanguinity in recent immigrant populations. Fine mapping and identification of aetiological genes within these regions at chromosomes 1p22 and 6q27 has been carried out (M. Fakiola, M. Raju, J.M. Blackwell and colleagues, unpublished data) using a combination of dense tag-SNP genotyping and allelic association analysis across multiple biological candidate genes in these regions, re-sequencing, haplotype and LD analysis, in silico bioinformatics analysis, qualitative RT/PCR analysis of tissue and cellular (cell lines, including control versus classically activated macrophages), and quantitative RT/PCR analysis of gene expression in splenic aspirates from VL patients in India compared to commercially available normal spleen. Allelic association data from India and Brazil (M. Fakiola, JM Blackwell, et al., unpublished) identify the gene (DLL1) encoding delta notch ligand 1 as the aetiological gene under the 6q27 linkage peak. DLL1 is expressed in stromal cells and antigen presenting cells, in particular dendritic cells [64, 65]. Bone marrow stromal cells expressing DLL1 induce the emergence of T/NK cell precursors from human haematopoietic progenitors and are required for T cell lineage specification during early thymocyte development [66, 67]. Bone marrow stromal cells are targeted by L. donovani, particularly those with macrophage characteristics which support long-term growth of parasites and exhibit increased capacity for hematopoiesis [68]. Antigen presenting cells also use Notch signaling to promote Th cell differentiation [69] with DLL1 on dendritic cells interacting with Notch3 of CD4+ T cells to induce the Th1 phenotype [69, 70]. Dll1 in mice can alter susceptibility to L. major in BALB/c mice by promoting a Th1 response [70]. We found that DLL1 was strongly down-regulated in RNA from splenic aspirates of all VL patients compared to control spleen, consistent with a depressed Th1 response in visceral leishmaniasis patients. Re-sequencing failed to identify novel coding variants at DLL1 that could alter function. Bioinformatic analysis, similar to that undertaken previously (32), pinpoint putative regulatory variants in conserved non-coding sequence as the aetiological variants associated with disease.
At least nine biological candidate genes were identified under the 1p22 linkage peak (M. Raju, J.M. Blackwell, et al., unpublished), including BCL10, DDAH1, TGFBR3, GLMN, GFI1, MTF2, DR1, GCLM and VCAM1 which we have investigated in detail using the strategy outlined above. TGFBR3, GLMN, GFI1, MTF2, DR1, and GCLM lie immediately beneath the peak of linkage, whereas BCL10/DDAH1 and VCAM1 lie ~7.5 Mb distal and proximal to the peak of linkage, respectively. The results show independent genetic and functional evidence for association between DDAH1 and VL, and between extended haplotypes across GFI1/MTF2/DR1/GCLM and VL. DDAH1 encoding dimethylarginine dimethylaminohydrolase 1 is of particular interest as an inhibitor of nitric oxide synthase activity [71]. Although previous studies had identified the DDAH2 isoform encoded in the class II region of HLA at 6p21.3 as the isoform expressed in macrophages and inhibiting inducible nitric oxide synthase [71, 72], we found that DDAH1 was expressed in mature macrophages and was strongly downregulated in splenic aspirates from all VL patients compared to normal control spleen. DDAH1 lies between the markers D1S207 and D1S2766 for which the nominal P values for linkage were in the range 5.8×10-4<P<3.04×10-5. Although not at the peak of linkage, these nominal P values still fall within the Lander and Kruglyak [73] criteria for significant linkage on a genome-wide scan. Although a disease locus found in a family based linkage study might normally be found immediately under the peak of linkage [74], the true susceptibility gene can be displaced by up to 10cM (=10Mb) from the linkage peak, particularly in smaller samples [75, 76]. Hence, DDAH1 cannot be discounted as the potential aetiological gene under the 1p22 linkage peak. For the biological candidates directly under the peak of linkage, the extended haplotype associations across GFI1/MTF2/DR1/GCLM are accompanied by some evidence for co-regulation at the mRNA level in splenic aspirates from patients. These genes are also of specific interest. GFI1 encoding growth factor independent 1 transcription factor, has recently emerged as a major determinant driving Th2 differentiation [77], providing another potential gene in the important immunological pathway of Th1 versus Th2 differentiation that may be associated with VL disease. MTF2 encoding metal-response element-binding transcription factor 2 is involved in the activation of metallothionein genes in response to heavy metal ions [78]. MTFs coordinate the expression of genes such as Glycine Cleavage System H Protein, metallothionein, the zinc-transporter-1, which are involved in zinc homeostasis and protection against metal toxicity and oxidative stresses [79]. Recent microarray data shows that metallothionein genes are amongst the most highly upregulated genes in monocyte-derived macrophages infected with L. chagasi [80]. Together with evidence for the role of divalent cation homeostasis in regulating intra-macrophage pathogens like L. donovani provided by the innate resistance gene SLC11A1 (reviewed [45]), this means that candidacy for MTF2 as a VL susceptibility gene is also strong. DR1 encodes the downregulator of transcription 1 (also called negative cofactor 2β/NC2β) that inhibits transcription by binding to the TATA box binding protein [81] (TBP). CIITA, the transactivator of major histocompatibility complex class II molecules in antigen presenting cells, requires participation of, and is extremely sensitive to mutations in, TBP [82]. GCLM encodes the regulatory light subunit of gamma-glutamylcysteine synthetase, also known as glutamate-cysteine ligase, that is the first rate-limiting enzyme in the de novo synthesis of tripeptide glutathione. Glutatione redox status plan an important role in induction of nitiric oxide synthase [83] and hence production of antimicrobial nitric oxide [84], and GSH modulators have been shown to influence parasite loads in lesions and tissues during the course of L. major infection in susceptible BALB/c mice [85]. Work is in progress to further validate these genes as genetic and functional candidates for VL susceptibility at the chromosome 1p22 peak of linkage.
Familial aggregation is also a feature of VL caused by L. chagasi in northeastern Brazil [17], providing a high relative risk (λ2S=34) of disease in further siblings of affected sibling pairs [18]. We undertook [36] a GWLS for susceptibility genes in this ethnically admixed population using 91 families including 215 affected relatives from 4 ethnically admixed peri-urban populations in northern Brazil. Not surprisingly (because of the ethnic admixture), the primary scan identified multiple regions at low significance level, with weak evidence for linkage retained at chromosomes 6q27 (LOD score 0.99, p=0.016) and 17q21.3 (LOD score 1.67, p=0.003) with refined mapping. The peak at 6q27 was coincident with the peak observed in Sudan, suggesting a common susceptibility gene now mapped in an extended sample of affected child-parent trios from the Brazilian study (M. Fakiola, J.M. Blackwell, et al., unpublished data). The peak at 17q21.3 was within a cluster of immune response genes, multiple members of which had previously been shown by us to contribute to leprosy and tuberculosis in this region of Brazil [86]. Initial analysis of SNPs in genes across the cluster [36] identified the chemokines CCL1 and CCL16 as genes associated with VL in Brazil, but the picture is likely to be more complex. Further analysis using a much larger sample now available in Brazil should provide the power to dissect out the genes contributing to VL susceptibility at 17q21.3. Output from the ongoing GWAS of VL and DTH phenotypes (cf. below) should contribute to this analysis.
As expected, GWLS for the VL phenotype in the ethnically admixed population of Brazil have not achieved genome-wide significance for the numbers of families used in these studies [23, 36], nor have our recent studies of 59 Indian multicase pedigrees (77 nuclear families; 372 individuals) with 156 affected relatives (M. Fakiola, A. Mishra, J.M. Blackwell, S. Sundar and colleagues, unpublished data). The 59 families, comprised of 32 Hindu and 27 Muslim pedigrees, underwent a 10cM GWLS of 515 polymorphic microsatellite markers. Peaks of linkage at p<0.01 were observed for both religious groups on chromosomes 2q12.2 and 11q14.2, with Hindu-specific peaks on 6p25.3 and 8p23 and Muslim-specific peaks on 1p13.1 and Xq23. These regions were further investigated by genotyping 65 additional Indian VL multicase families and 19 additional polymorphic microsatellites to provide a denser 2-5cM refined map. Refined analysis that corrects for overrelatedness in the population retained evidence for linkage at 2q14.1 (D2S363; singlepoint LOD=2.33; P=5×10-4), at 6p25.1 (D6S1617; singlepoint LOD=1.00; P=0.016), at 8p23.1 (D8S516; singlepoint LOD=1.52; P=0.004), at 11q14.2 (D11S1780; singlepoint LOD=2.53; P=3×10-4), and at Xq23 (DXS8055; multipoint LOD=1.58; P=0.004). These results contribute novel population-specific candidate regions for ongoing studies into VL susceptibility in India. The GWAS underway using a much larger sample of VL cases and controls from India (cf. below) should validate (or not) these peaks of linkage and provide data for allelic association mapping and gene identification.
As mentioned above, we also undertook a GWLS using Brazilian families that segregate for both VL and DTH phenotypes [23]. A total of 405 (385 autosomal, 20 sex chromosomal) microsatellite markers were genotyped for 1290 individuals from 191 pedigrees, including 188 VL cases. The VL phenotype was analysed as a qualitative trait, DTH as a continuous quantitative trait based on the actual induration of the skin test (classically a 5 mm induration is considered a positive response). It should be noted that the study was much better power to analyse the quantitative trait, so it cannot be concluded that because a region showed linkage to this trait it was not involved in the qualitative VL trait. Data were analysed using MERLIN (v1.0-alpha) which allowed for inclusion of discordant pairs and variance components (DTH only) [87]. The strongest evidence for linkage for VL occurred at D9S1118 on chromosome 9q (LOD=1.60; P=0.003). This region was not positive for the DTH phenotype, indicating that a gene here might contribute to susceptibility to disease by a mechanism independent of the kind of cell mediated immune response that the DTH reflects. Variance components analysis, also performed in MERLIN [88], included age and gender as covariates in the model. Evidence for linkage for DTH was found on chromosomes 2, 13, 15 and 19, the highest at D15S657 (LOD=2.50; P=0.0003) and D19S246 (LOD=1.93; P=0.0014). Refined mapping by genotyping of additional markers across the peaks of linkage for both the VL and DTH phenotypes is underway. Again, validation of these peaks of linkage and data for allelic association mapping and gene identification should be provided by the GWAS underway (cf. below) for VL and DTH phenotypes for Brazil.
Overall, the results of candidate gene and GWLS point to potential geographic and population differences in the genes controlling susceptibility to VL. This could reflect local adaptation of the parasite to host genetic background, differences in selective pressure leading to different functional variants. However, until the issue of study power is resolved, we cannot speculate further on population-related differences in the genes so far identified in controlling susceptibility to VL.
GWAS FOR VL AND DTH PHENOTYPES
The application of SNP-chip based GWAS has been highly successful in rapidly increasing the number of loci that have been positively associated with complex diseases. For example, the WTCCC study published in 2007 [63] of 14,000 cases of seven common diseases and 3,000 shared controls has itself identified 24 independent association signals at P<5×10-7, 9 of which were in Crohn’s disease, 3 in rheumatoid arthritis, 7 in type 1 diabetes, and 3 in type 2 diabetes. The increased problem of multiple testing associated with genotyping in excess of 500,000 SNPs in large numbers of cases and controls necessitated this stringent threshold P value <5×10-7 to achieve genome-wide significance. However, across all diseases, a large number of further signals, including 58 loci with single point P values between 10-5 and 5×10-7, were identified which are likely to yield additional susceptibility loci. A number of papers providing validation of the original WTCCC data have already been published [89, 90, 91, 92, 93, 94, 95, 96]. The WTCCC study is but one of an increasing number of published GWAS for complex diseases (e.g. [97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113]) now catalogued at http://www.genome.gov/gwastudies/. As pointed out in the recent News Feature on “Genetics by Numbers” in Nature [114], identification of the initial SNP association on a primary GWAS is only the first step on the path to validating and identifying the etiological genes and associated mechanisms of disease susceptibility. Some papers have demonstrated at least two independent etiological variants at one locus (6q23) associated with rheumatoid arthritis, while regions like the HLA complex require intensive fine mapping. For example, following the WTCCC type 1 diabetes study, a combined total of 1,729 polymorphisms across HLA were genotyped in >6000 cases and controls across cohorts, and statistical methods of recursive partitioning and regression applied to pinpoint disease susceptibility to the MHC class I genes HLA-B and HLA-A (risk ratios >1.5; P(combined) = 2.01×10-19 and 2.35×10-13, respectively) in addition to the established associations of the MHC class II HLA-DQB1 and HLA-DRB1 genes [115]. This demonstrates the intensive mapping that must follow any primary GWAS to validate the SNP associations and identify the genes associated with disease, and the statistical power that can be achieved by genotyping large numbers of cases across multiple cohorts. Interestingly, the only GWAS performed in Africa so far (WTCCC, Nature, in press) has found evidence for population substructure between geographically adjacent ethnic groups, making it impossible to impute the HbS causal SNP for malaria using current HapMap reference data because it has independently arisen on different haplotypic backgrounds in different parts of Africa.
These recent successes of GWAS for complex diseases using sufficiently powered large sample sizes (reference Figure 1) stimulated us to collect larger DNA banks that would permit this approach to be applied to the study of VL susceptibility. The accumulation of DNA from a total of 1217 VL cases in multicase families or trios/sibships from India (total sample 3630 individuals), together with 1000 separately ascertained unrelated age-, village- and sex-matched controls, and 626 VL cases and 1160 DTH+/900 DTH- individuals in families (total 2882 individuals with parents) from Brazil, underpinned a successful bid to phase two of the WTCCC. The strategy is to undertake SNP-chip analysis of 1000 genetically unrelated VL cases and 1000 unrelated controls from India, and a family-based GWAS using all the DNAs in Brazilian families, i.e. a total of 4880 SNP-chips. Positive hits in the Indian study will be validated using FBAT analysis of dense tagging SNPs in the 1217 VL cases in families to control for possible sub-structure in this population. Across both Indian and Brazilian studies, identification of novel susceptibility and resistance genes, and associated functional etiological variants, based on these GWAS will involve validation of positive allelic association signals using dense tag-SNPs, re-sequencing, in silico bioinformatic analysis, and mRNA and protein expression analysis in clinical and experimental samples. The results of these GWAS should be in the public domain by late 2009, and will provide a wealth of new data that will seed many novel functional studies on mechanisms of disease that can be translated into better interventions for the future.
Acknowledgments
We acknowledge the many members of our laboratories who have contributed to research reviewed here. We thank the communities in Sudan, Brazil and India who have participated in our studies. Our research is funded by The Wellcome Trust and the NIH.
Footnotes
Disclosures The authors declare no conflicts of interest
References
- 1.Badaro R, Jones TC, Lorenco R, et al. A prospective study of visceral leishmaniasis in an endemic area of Brazil. J Infect Dis. 1986;154:639–49. doi: 10.1093/infdis/154.4.639. [DOI] [PubMed] [Google Scholar]
- 2.Badaro R, Jones TC, Carvalho EM, et al. New perspectives on a subclinical form of visceral leishmaniasis. J Infect Dis. 1986;154:1003–9. doi: 10.1093/infdis/154.6.1003. [DOI] [PubMed] [Google Scholar]
- 3.Ho M, Siongok TK, Lyerly WH, et al. Prevalence and disease spectrum in a new focus of visceral leishmaniasis in Kenya. Trans R Soc Trop Med Hyg. 1982;76:741–6. doi: 10.1016/0035-9203(82)90095-5. [DOI] [PubMed] [Google Scholar]
- 4.Sacks DL, Lal SL, Shrivastava SN, et al. An analysis of T cell responsiveness in Indian Kala-azar. J Immunol. 1987;138:908–13. [PubMed] [Google Scholar]
- 5.Vallance P, Levick M. Drug discovery and development in the age of molecular medicine. Clin Pharmacol Ther. 2007;82(4):363–6. doi: 10.1038/sj.clpt.6100333. [DOI] [PubMed] [Google Scholar]
- 6.Ebrahim S, Davey Smith G. Mendelian randomization: can genetic epidemiology help redress the failures of observational epidemiology? Hum Genet. 2008;123(1):15–33. doi: 10.1007/s00439-007-0448-6. [DOI] [PubMed] [Google Scholar]
- 7.Davies JL, Kawaguchi Y, Bennett ST, et al. A genome-wide search for human type 1 diabetes susceptibility genes. Nature. 1994;371(6493):130–6. doi: 10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- 8.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–7. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 9.Sawcer S, Jones HB, Feakes R, et al. A genome screen in multiple sclerosis reveals susceptibility loci on chromosome 6p21 and 17q22. Nat Genet. 1996;13(4):464–8. doi: 10.1038/ng0896-464. [DOI] [PubMed] [Google Scholar]
- 10.Reich DE, Cargill M, Bolk S, et al. Linkage disequilibrium in the human genome. Nature. 2001;411(6834):199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 11.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet. 2001;9(4):301–6. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 12.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–16. [PMC free article] [PubMed] [Google Scholar]
- 13.Cordell HJ, Clayton DG. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am J Hum Genet. 2002;70(1):124–41. doi: 10.1086/338007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blackwell JM. Genetic susceptibility to leishmanial infections: studies in mice and man. Parasitology. 1996;112:S67–S74. [PubMed] [Google Scholar]
- 15.Zijlstra EE, El Hassan AM, Ismael A, et al. Endemic kala-azar in Eastern Sudan, a longitudinal study on the incidence of clinical and subclinical infection and post-kala-azar dermal leishmaniasis. Am J Trop Med Hyg. 1994;51:826–36. doi: 10.4269/ajtmh.1994.51.826. [DOI] [PubMed] [Google Scholar]
- 16.Ibrahim ME, Lambson B, Yousif AO, et al. Kala-azar in a high transmission focus: an ethnic and geographic dimension. Am J Trop Med Hyg. 1999;61(6):941–4. doi: 10.4269/ajtmh.1999.61.941. [DOI] [PubMed] [Google Scholar]
- 17.Cabello PH, Lima AM, Azevedo ES, et al. Familial aggregation of Leishmnaia chagasi infection in northeastern Brazil. Am J Trop Med Hyg. 1995;52:364–5. doi: 10.4269/ajtmh.1995.52.364. [DOI] [PubMed] [Google Scholar]
- 18.Peacock CS, Collins A, Shaw MA, et al. Genetic epidemiology of visceral leishmaniasis in northeastern Brazil. Genet Epidemiol. 2001;20(3):383–96. doi: 10.1002/gepi.8. [DOI] [PubMed] [Google Scholar]
- 19.Bucheton B, Kheir MM, El-Safi SH, et al. The interplay between environmental and host factors during an outbreak of visceral leishmaniasis in eastern Sudan. Microbes Infect. 2002;4(14):1449–57. doi: 10.1016/s1286-4579(02)00027-8. [DOI] [PubMed] [Google Scholar]
- 20.Jeronimo SM, Duggal P, Braz RF, et al. An emerging peri-urban pattern of infection with Leishmania chagasi, the protozoan causing visceral leishmaniasis in northeast Brazil. Scand J Infect Dis. 2004;36(6-7):443–9. doi: 10.1080/00365540410020451. [DOI] [PubMed] [Google Scholar]
- 21.Shaw MA, Davies CR, Llanos-Cuentas EA, et al. Human genetic susceptibility and infection with Leishmania peruviana. Am J Hum Genet. 1995;57:1159–68. [PMC free article] [PubMed] [Google Scholar]
- 22.Feitosa MF, Axevedo E, Lima AM, et al. Genetic causes involved in Leishmania chagasi infection in northeastern Brazil. Genet Mol Biol. 1999;22:1–5. [Google Scholar]
- 23.Jeronimo SM, Duggal P, Ettinger N, et al. Genetic predisposition to symptomatic versus self-curing infection with the protozoan Leishmania chagasi: A genome wide scan. J Infect Dis. 2007;196:1261–9. doi: 10.1086/521682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bucheton B, Abel L, Kheir MM, et al. Genetic control of visceral leishmaniasis in a Sudanese population: candidate gene testing indicates a linkage to the NRAMP1 region. Genes Immun. 2003;4(2):104–9. doi: 10.1038/sj.gene.6363927. [DOI] [PubMed] [Google Scholar]
- 25.Mohamed HS, Ibrahim ME, Miller EN, et al. Genetic susceptibility to visceral leishmaniasis in The Sudan: linkage and association with IL4 and IFNGR1. Genes Immun. 2003;4:351–5. doi: 10.1038/sj.gene.6363977. [DOI] [PubMed] [Google Scholar]
- 26.Mohamed HS, Ibrahim ME, Miller EN, et al. SLC11A1 (formerly NRAMP1) and susceptibility to visceral leishmaniasis in The Sudan. Eur J Hum Genet. 2004;12:66–74. doi: 10.1038/sj.ejhg.5201089. [DOI] [PubMed] [Google Scholar]
- 27.Karplus TM, Jeronimo SM, Chang H, et al. Association between the tumor necrosis factor locus and the clinical outcome of Leishmania chagasi infection. Infect Immun. 2002;70(12):6919–25. doi: 10.1128/IAI.70.12.6919-6925.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faghiri Z, Tabei SZ, Taheri F. Study of the association of HLA class I antigens with kala-azar. Hum Hered. 1995;45:258–61. doi: 10.1159/000154309. [DOI] [PubMed] [Google Scholar]
- 29.Meddeb-Garnaoui A, Gritli S, Garbouj S, et al. Association analysis of HLA-class II and class III gene polymorphisms in the susceptibility to mediterranean visceral leishmaniasis. Hum Immunol. 2001;62(5):509–17. doi: 10.1016/s0198-8859(01)00237-3. [DOI] [PubMed] [Google Scholar]
- 30.Peacock CS, Sanjeevi CB, Shaw MA, et al. Genetic analysis of multicase families of visceral leishmaniasis in northeastern Brazil: no major role for class II or class III regions of HLA. Genes Immun. 2002;3(6):350–8. doi: 10.1038/sj.gene.6363852. [DOI] [PubMed] [Google Scholar]
- 31.Salih MA, Ibrahim ME, Blackwell JM, et al. IFNG and IFNGR1 gene polymorphisms and susceptibility to post-kala-azar dermal leishmaniasis in Sudan. Genes Immun. 2007;8(1):75–8. doi: 10.1038/sj.gene.6364353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeronimo SM, Holst AK, Jamieson SE, et al. Genes at human chromosome 5q31.1 regulate delayed-type hypersensitivity responses associated with Leishmania chagasi infection. Genes Immun. 2007;8(7):539–51. doi: 10.1038/sj.gene.6364422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bucheton B, Argiro L, Chevillard C, et al. Identification of a novel G245R polymorphism in the IL-2 receptor beta membrane proximal domain associated with human visceral leishmaniasis. Genes Immun. 2007;8(1):79–83. doi: 10.1038/sj.gene.6364355. [DOI] [PubMed] [Google Scholar]
- 34.Bucheton B, Abel L, El-Safi S, et al. A major susceptibility locus on chromosome 22q12 plays a critical role in the control of kala-azar. Am J Hum Genet. 2003;73(6):1052–60. doi: 10.1086/379084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller EN, Fadl M, Mohamed HS, et al. Y chromosome lineage- and village-specific genes on chromosomes 1p22 and 6q27 that control visceral leishmaniasis in The Sudan. PLoS Genet. 2007;3:679–88. doi: 10.1371/journal.pgen.0030071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jamieson SE, Miller EN, Peacock CS, et al. Genome-wide scan for visceral leishmaniasis susceptibility genes in Brazil. Genes Immun. 2007;8(1):84–90. doi: 10.1038/sj.gene.6364357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burgner D, Jamieson SE, Blackwell JM. Genetic susceptibility to infectious diseases: big is beautiful, but will bigger be even better? Lancet Infect Dis. 2006;6(10):653–63. doi: 10.1016/S1473-3099(06)70601-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cordell HJ, Clayton DG. Genetic association studies. Lancet. 2005;366(9491):1121–31. doi: 10.1016/S0140-6736(05)67424-7. [DOI] [PubMed] [Google Scholar]
- 39.Palmer LJ, Cardon LR. Shaking the tree: mapping complex disease genes with linkage disequilibrium. Lancet. 2005;366(9492):1223–34. doi: 10.1016/S0140-6736(05)67485-5. [DOI] [PubMed] [Google Scholar]
- 40.Blackwell JM, Black GF, Peacock CS, et al. Immunogenetics of leishmanial and mycobacterial infections: The Belem Family Study. Phil Trans Roy Soc B. 1997;352:1331–45. doi: 10.1098/rstb.1997.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh N, Sundar S, Williams F, et al. Molecular typing of HLA class I and class II antigens in Indian kala-azar patients. Trop Med Int Health. 1997;2:468–71. [PubMed] [Google Scholar]
- 42.Bradley DJ. Genetic control of natural resistance to Leishmania donovani. Nature. 1974;250:353–4. doi: 10.1038/250353a0. [DOI] [PubMed] [Google Scholar]
- 43.Bradley DJ. Regulation of Leishmania populations within the host. II Genetic control of acute susceptibility of mice to Leishmania donovani infection. Clin Exp Immunol. 1977;30:130–40. [PMC free article] [PubMed] [Google Scholar]
- 44.Blackwell J, Freeman J, Bradley D. Influence of H-2 complex on acquired resistance to Leishmania donovani infection in mice. Nature. 1980;283:72–4. doi: 10.1038/283072a0. [DOI] [PubMed] [Google Scholar]
- 45.Blackwell JM, Goswami T, Evans CAW, et al. SLC11A1 (formerly NRAMP1) and disease. Cell Microbiol. 2001;3:773–84. doi: 10.1046/j.1462-5822.2001.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vidal SM, Malo D, Vogan K, et al. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73:469–85. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 47.Goswami T, Bhattacharjee A, Babal P, et al. Natural-resistance-associated macrophage protein 1 is an H+/bivalent cation antiporter. Biochem J. 2001;354(Pt 3):511–9. doi: 10.1042/0264-6021:3540511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Techau M, Valdez-Taubas J, Popoff J-F, et al. Evolution of differences in transport function in Slc11a family members. J Biol Chem. 2006;282:35646–56. doi: 10.1074/jbc.M707057200. [DOI] [PubMed] [Google Scholar]
- 49.Searle S, Bright NA, Roach TIA, et al. Localisation of Nramp1 in macrophages: modulation with activation and infection. J Cell Sci. 1998;111:2855–66. doi: 10.1242/jcs.111.19.2855. [DOI] [PubMed] [Google Scholar]
- 50.Blackwell JM, Searle S, Mohamed H, et al. Divalent cation transport and susceptibility to infectious and autoimmune disease: continuation of the Ity/Lsh/Bcg/Nramp1/Slc11a1 gene story. Immunol Lett. 2003;85(2):197–203. doi: 10.1016/s0165-2478(02)00231-6. [DOI] [PubMed] [Google Scholar]
- 51.Li HT, Zhang TT, Huang QH, et al. Meta-analysis on NRAMP1 gene polymorphisms and tuberculosis susceptibility in east-asia population. Zhonghua Liu Xing Bing Xue Za Zhi. 2006;27(5):428–32. [PubMed] [Google Scholar]
- 52.Li HT, Zhang TT, Zhou YQ, et al. SLC11A1 (formerly NRAMP1) gene polymorphisms and tuberculosis susceptibility: a meta-analysis. Int J Tuberc Lung Dis. 2006;10(1):3–12. [PubMed] [Google Scholar]
- 53.Bellamy R, Ruwende C, Corrah T, et al. Variation in the NRAMP1 gene is associated with susceptibility to tuberculosis in West Africans. N Engl J Med. 1998;338:640–4. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 54.Marquet S, Sanchez FO, Arias M, et al. Variants of the human NRAMP1 gene and altered human immunodeficiency virus infection susceptibility. J Infect Dis. 1999;180(5):1521–5. doi: 10.1086/315091. [DOI] [PubMed] [Google Scholar]
- 55.Searle S, Blackwell JM. Evidence for a functional repeat polymorphism in the promoter of the human NRAMP1 gene that correlates with autoimmune versus infectious disease susceptibility. J Med Genet. 1999;36(4):295–9. [PMC free article] [PubMed] [Google Scholar]
- 56.Zaahl MG, Robson KJ, Warnich L, et al. Expression of the SLC11A1 (NRAMP1) 5’-(GT)n repeat: opposite effect in the presence of -237C-->T. Blood Cells Mol Dis. 2004;33(1):45–50. doi: 10.1016/j.bcmd.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Bayele HK, Peyssonnaux C, Giatromanolaki A, et al. HIF-1 regulates heritable variation and allele expression phenotypes of the macrophage immune response gene SLC11A1 from a Z-DNA forming microsatellite. Blood. 2007;110(8):3039–48. doi: 10.1182/blood-2006-12-063289. [DOI] [PubMed] [Google Scholar]
- 58.Locksley RM, Scott P. Helper T-cell subsets in mouse leishmaniasis induction, expansion and effector function. Immunol Today. 1991;12:A58–A61. doi: 10.1016/S0167-5699(05)80017-9. [DOI] [PubMed] [Google Scholar]
- 59.Roberts M, Mock BA, Blackwell JM. Mapping of genes controlling Leishmania major infection in CXS recombinant inbred mice. Eur J Immunogenet. 1993;20:349–62. doi: 10.1111/j.1744-313x.1993.tb00154.x. [DOI] [PubMed] [Google Scholar]
- 60.Zwingenberger K, Harms G, Pedrosa C, et al. Determinants of the immune response in visceral leishmaniasis: evidence for predominance of endogenous interleukin 4 over interferon-gamma production. Clin Immunol Immunopathol. 1990;57(2):42–9. doi: 10.1016/0090-1229(90)90038-r. [DOI] [PubMed] [Google Scholar]
- 61.Sundar S, Reed SG, Sharma S, et al. Circulating T helper 1 (Th1) cell- and Th2 cell-associated cytokines in Indian patients with visceral leishmaniasis. Am J Trop Med Hyg. 1997;56(5):522–5. doi: 10.4269/ajtmh.1997.56.522. [DOI] [PubMed] [Google Scholar]
- 62.Kurtzhals JA, Kemp M, Poulsen LK, et al. Interleukin-4 and interferon-gamma production by Leishmania stimulated peripheral blood mononuclear cells from nonexposed individuals. Scand J Immunol. 1995;41(4):343–9. doi: 10.1111/j.1365-3083.1995.tb03577.x. [DOI] [PubMed] [Google Scholar]
- 63.Consortium WTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson AC, Robey EA, Huang YH. Notch signaling in lymphocyte development. Curr Opin Genet Dev. 2001;11(5):554–60. doi: 10.1016/s0959-437x(00)00232-x. [DOI] [PubMed] [Google Scholar]
- 65.Yamaguchi E, Chiba S, Kumano K, et al. Expression of Notch ligands, Jagged1, 2 and Delta1 in antigen presenting cells in mice. Immunol Lett. 2002;81(1):59–64. doi: 10.1016/s0165-2478(01)00326-1. [DOI] [PubMed] [Google Scholar]
- 66.Jaleco AC, Neves H, Hooijberg E, et al. Differential effects of Notch ligands Delta-1 and Jagged-1 in human lymphoid differentiation. J Exp Med. 2001;194(7):991–1002. doi: 10.1084/jem.194.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmitt TM, Ciofani M, Petrie HT, et al. Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions. J Exp Med. 2004;200(4):469–79. doi: 10.1084/jem.20040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cotterell SE, Engwerda CR, Kaye PM. Leishmania donovani infection of bone marrow stromal macrophages selectively enhances myelopoiesis, by a mechanism involving GM-CSF and TNF-alpha. Blood. 2000;95(5):1642–51. [PubMed] [Google Scholar]
- 69.Amsen D, Blander JM, Lee GR, et al. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117(4):515–26. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 70.Maekawa Y, Tsukumo S, Chiba S, et al. Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity. 2003;19(4):549–59. doi: 10.1016/s1074-7613(03)00270-x. [DOI] [PubMed] [Google Scholar]
- 71.Leiper JM, Santa Maria J, Chubb A, et al. Identification of two human dimethylarginine dimethylaminohydrolases with distinct tissue distributions and homology with microbial arginine deiminases. Biochem J. 1999;343(Pt 1):209–14. [PMC free article] [PubMed] [Google Scholar]
- 72.Tran CT, Fox MF, Vallance P, et al. Chromosomal localization, gene structure, and expression pattern of DDAH1: comparison with DDAH2 and implications for evolutionary origins. Genomics. 2000;68(1):101–5. doi: 10.1006/geno.2000.6262. [DOI] [PubMed] [Google Scholar]
- 73.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11(3):241–7. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 74.Weeks DE, Lathrop GM. Polygenic disease: methods for mapping complex disease traits. Trends Genet. 1995;11(12):513–9. doi: 10.1016/s0168-9525(00)89163-5. [DOI] [PubMed] [Google Scholar]
- 75.Hovatta I, Lichtermann D, Juvonen H, et al. Linkage analysis of putative schizophrenia gene candidate regions on chromosomes 3p, 5q, 6p, 8p, 20p and 22q in a population-based sampled Finnish family set. Mol Psychiatry. 1998;3(5):452–7. doi: 10.1038/sj.mp.4000443. [DOI] [PubMed] [Google Scholar]
- 76.Roberts SB, MacLean CJ, Neale MC, et al. Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet. 1999;65(3):876–84. doi: 10.1086/302528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu J, Jankovic D, Grinberg A, et al. Gfi-1 plays an important role in IL-2-mediated Th2 cell expansion. Proc Natl Acad Sci U S A. 2006;103(48):18214–9. doi: 10.1073/pnas.0608981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Inouye C, Remondelli P, Karin M, et al. Isolation of a cDNA encoding a metal response element binding protein using a novel expression cloning procedure: the one hybrid system. DNA Cell Biol. 1994;13(7):731–42. doi: 10.1089/dna.1994.13.731. [DOI] [PubMed] [Google Scholar]
- 79.Andrews GK. Cellular zinc sensors: MTF-1 regulation of gene expression. Biometals. 2001;14:3–4. 223–37. doi: 10.1023/a:1012932712483. [DOI] [PubMed] [Google Scholar]
- 80.Ettinger NA, Wilson ME. Macrophage and T-cell gene expression in a model of early infection with the protozoan, Leishmania chagasi. PLoS Neglect Dis. 2008 doi: 10.1371/journal.pntd.0000252. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goppelt A, Stelzer G, Lottspeich F, et al. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. Embo J. 1996;15(12):3105–16. [PMC free article] [PubMed] [Google Scholar]
- 82.Mahanta SK, Scholl T, Yang FC, et al. Transactivation by CIITA, the type II bare lymphocyte syndrome-associated factor, requires participation of multiple regions of the TATA box binding protein. Proc Natl Acad Sci U S A. 1997;94(12):6324–9. doi: 10.1073/pnas.94.12.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stuehr DJ, Kwon NS, Nathan CF. FAD and GSH participate in macrophage synthesis of nitric oxide. Biochem Biophys Res Commun. 1990;168:558–56. doi: 10.1016/0006-291x(90)92357-6. [DOI] [PubMed] [Google Scholar]
- 84.Hothersall JS, Cunha FQ, Neild GH, et al. Induction of nitric oxide synthesis in J774 cells lowers intracellular glutathione: effect of modulated glutathione redox status on nitric oxide synthase induction. Biochem J. 1997;322:477–81. doi: 10.1042/bj3220477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cruz KK, Fonseca SG, Monteiro MC, et al. The influence of glutathione modulators on the course of Leishmania major infection in susceptible and resistant mice. Parasite Immunol. 2008;30:171–4. doi: 10.1111/j.1365-3024.2007.01014.x. [DOI] [PubMed] [Google Scholar]
- 86.Jamieson SE, Miller EN, Black GF, et al. Evidence for a cluster of genes on chromosome 17q11-q21 controlling susceptibility to tuberculosis and leprosy in Brazilians. Genes Immun. 2004;5:46–57. doi: 10.1038/sj.gene.6364029. [DOI] [PubMed] [Google Scholar]
- 87.Amos CI. Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet. 1994;54(3):535–43. [PMC free article] [PubMed] [Google Scholar]
- 88.Abecasis GR, Cherny SS, Cookson WO, et al. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 89.Beckly JB, Hancock L, Geremia A, et al. Two-stage candidate gene study of chromosome 3p demonstrates an association between nonsynonymous variants in the MST1R gene and Crohn’s disease. Inflamm Bowel Dis. 2008 doi: 10.1002/ibd.20365. [DOI] [PubMed] [Google Scholar]
- 90.Cooper JD, Smyth DJ, Bailey R, et al. The candidate genes TAF5L, TCF7, PDCD1, IL6 and ICAM1 cannot be excluded from having effects in type 1 diabetes. BMC Med Genet. 2007;8:71. doi: 10.1186/1471-2350-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Plenge RM, Cotsapas C, Davies L, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39(12):1477–82. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thomson W, Barton A, Ke X, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39(12):1431–3. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357(5):443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39(7):830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Todd JA, Walker NM, Cooper JD, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39(7):857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–41. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Raelson JV, Little RD, Ruether A, et al. Genome-wide association study for Crohn’s disease in the Quebec Founder Population identifies multiple validated disease loci. Proc Natl Acad Sci U S A. 2007;104(37):14747–52. doi: 10.1073/pnas.0706645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Duggan D, Zheng SL, Knowlton M, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99(24):1836–44. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 99.Burton PR, Clayton DG, Cardon LR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39(11):1329–37. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cronin S, Berger S, Ding J, et al. A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum Mol Genet. 2007 doi: 10.1093/hmg/ddm361. Epub Dec15. [DOI] [PubMed] [Google Scholar]
- 101.Roberts R, Stewart AF, Wells GA, et al. Identifying genes for coronary artery disease: An idea whose time has come. Can J Cardiol. 2007;23(Suppl A):7A–15A. doi: 10.1016/s0828-282x(07)71000-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sulem P, Gudbjartsson DF, Stacey SN, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39(12):1443–52. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 103.Kathiresan S, Manning AK, Demissie S, et al. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cupples LA, Arruda HT, Benjamin EJ, et al. The Framingham Heart Study 100K SNP genome-wide association study resource: overview of 17 phenotype working group reports. BMC Med Genet. 2007;8(Suppl 1):S1. doi: 10.1186/1471-2350-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Florez JC, Manning AK, Dupuis J, et al. A 100K genome-wide association scan for diabetes and related traits in the Framingham Heart Study: replication and integration with other genome-wide datasets. Diabetes. 2007;56(12):3063–74. doi: 10.2337/db07-0451. [DOI] [PubMed] [Google Scholar]
- 106.Scuteri A, Sanna S, Chen WM, et al. Genome-Wide Association Scan Shows Genetic Variants in the FTO Gene Are Associated with Obesity-Related Traits. PLoS Genet. 2007;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Buch S, Schafmayer C, Volzke H, et al. A genome-wide association scan identifies the hepatic cholesterol transporter ABCG8 as a susceptibility factor for human gallstone disease. Nat Genet. 2007;39(8):995–9. doi: 10.1038/ng2101. [DOI] [PubMed] [Google Scholar]
- 108.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39(8):984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 109.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39(8):989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 110.Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39(5):631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 111.Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39(2):207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 112.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–5. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 113.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baker M. Genome studies: genetics by numbers. Nature. 2008;451(7178):516–8. doi: 10.1038/451516a. [DOI] [PubMed] [Google Scholar]
- 115.Nejentsev S, Howson JM, Walker NM, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450(7171):887–92. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]