Abstract
Background
Cortinarius species in section Calochroi display local, clinal and circumboreal patterns of distribution across the Northern Hemisphere where these ectomycorrhizal fungi occur with host trees throughout their geographical range within a continent, or have disjunct intercontinental distributions, the origins of which are not understood. We inferred evolutionary histories of four species, 1) C. arcuatorum, 2) C. aureofulvus, 3) C. elegantior and 4) C. napus, from populations distributed throughout the Old World, and portions of the New World (Central- and North America) based on genetic variation of 154 haplotype internal transcribed spacer (ITS) sequences from 83 population samples. By describing the population structure of these species across their geographical distribution, we attempt to identify their historical migration and patterns of diversification.
Results
Models of population structure from nested clade, demographic and coalescent-based analyses revealed genetically differentiated and geographically structured haplotypes in C. arcuatorum and C. elegantior, while C. aureofulvus showed considerably less population structure and C. napus lacked sufficient genetic differentiation to resolve any population structure. Disjunct populations within C. arcuatorum, C. aureofulvus and C. elegantior show little or no morphological differentiation, whereas in C. napus there is a high level of homoplasy and phenotypic plasticity for veil and lamellae colour. The ITS sequences of the type specimens of C. albobrunnoides and C. albobrunnoides var. violaceovelatus were identical to one another and are treated as one species with a wider range of geographic distribution under C. napus.
Conclusions
Our results indicate that each of the Calochroi species has undergone a relatively independent evolutionary history, hypothesised as follows: 1) a widely distributed ancestral population of C. arcuatorum diverged into distinctive sympatric populations in the New World; 2) two divergent lineages in C. elegantior gave rise to the New World and Old World haplotypes, respectively; and 3) the low levels of genetic divergence within C. aureofulvus and C. napus may be the result of more recent demographic population expansions. The scenario of migration via the Bering Land Bridge provides the most probable explanation for contemporaneous disjunct geographic distributions of these species, but it does not offer an explanation for the low degree of genetic divergence between populations of C. aureofulvus and C. napus. Our findings are mostly consistent with the designation of New World allopatric populations as separate species from the European counterpart species C. arcuatorum and C. elegantior. We propose the synonymy of C. albobrunnoides, C. albobrunnoides var. violaceovelatus and C. subpurpureophyllus var. sulphureovelatus with C. napus. The results also reinforce previous observations that linked C. arcuatorum and C. aureofulvus displaying distributions in parts of North America and Europe. Interpretations of the population structure of these fungi suggest that host tree history has heavily influenced their modern distributions; however, the complex issues related to co-migration of these fungi with their tree hosts remain unclear at this time.
Background
Several investigators [1-13] have explored intercontinental patterns of ectomycorrhizal fungus species distributions in a phylogenetic context, but there is little information on the origin and patterns of speciation in these fungi. Mushrooms in the genus Cortinarius, section Calochroi (calochroid clade), with over 100 species, represent one of the most conspicuous ectomycorrhizal members of contemporary Northern Hemisphere forest ecosystems [14]. Previous studies of species in Calochroi show various patterns of geographic distributions, including local (C. cisticola), clinal (C. arcuatorum) or circumboreal (C. aureofulvus, C. aureopulverulentus, C. cupreorufus, C. elegantior) distributions. In most instances fungus species distributions correspond to that of their host trees within continents and often disjunct distributions across the Northern Hemisphere show a similar pattern [15-18]. Overall, the patterns of species distribution in the Calochroi roughly matches the range of genera of the host tree families Fagaceae (Castanea, Castanopsis, Chrysolepis, Fagus, Notholithocarpus, Quercus) and Pinaceae (Abies, Larix, Picea, Pinus, Pseudotsuga, Tsuga) [17-21], however, a few Calochroi species form ectotrophic associations with members of the Betulaceae (Alnus, Corylus, Carpinus) [22,23], Cistaceae (Cistus, Helianthenum) [24,25] and Malvaceae (Tilia) [24,25]. Host specificity of Calochroi species may be restricted to a single host species, with host-switching events being less common and often restricted to closely related plant species [17,18,20]. Interestingly, allopatric populations/species on an intercontinental scale display relatively high host fidelities at the host genus level, suggesting potential co-evolutionary/co-migratory trends [14].
Species with a broad distribution provide excellent models for examining their present patterns of spatial variation, especially where there is an obligate association as with an ectotrophic fungus. An evaluation of contemporary geographic patterns of these species can provide a better understanding of speciation and habitat requirement as well as determine host preference and host-switching events that have occurred over time. To date a major difficulty in many mushroom genera, including species of Cortinarius, section Calochroi is the difficulty of identifying species [14,26], which historically has been a major impediment to understanding their patterns of geographical distribution, ecology, diversity and evolutionary relationships. There are a limited number of unique morphological features [14] that can be used to construct robust evolutionary hypotheses within and between closely related species in section Calochroi. Based on similarities in basidiomata morphology and colour some widespread species with largely allopatric distributions in the Old World (Europe) and the New World (North America) (e.g. C. arcuatorum, C. aureofulvus, C. aureopulverulentus, C. cupreorufus (as C. orichalceus), and C. olivellus) were treated as conspecific [17,18,20]. On the other hand, the discovery of certain phenotypic differences between European populations and their North American counterparts has led to the recognition of infraspecific taxa, for example C. elegantior var. americanus [17]. Similarly, detailed observations of morphological variation between European collections of C. elegantior have resulted in a number of forms, subspecies and varieties [19]. In our recent phylogenetic study [14], we confirmed close phylogenetic affinities between some disjunct populations/species, but we also found that several European species and their disjunct North American counterparts showed significant divergence from one another.
Recently the use of DNA sequence data have provided an independent tool for delineating species boundaries, and for revealing patterns of evolution history, identifying convergent morphological features and assessing/estimating species diversity. An evaluation of ITS rDNA sequences published by us [14] revealed distinct intra- and/or inter-continental phylogeographic structuring within some species, sometimes accompanied by relatively little morphological divergence. In contrast, we also found relatively low levels of genetic divergence within and between species with allopatric distributions in North America and Europe that display substantial colour variation in major components of the basidiomata. While, considerable progress has been made in reconstructing interspecific relationships within the section Calochroi through analyses of morphological features and DNA sequences [14,26], however, further studies are needed to examine how sequence divergence relates to the separation or divergence of species over time at continental and inter-continental levels.
Differences in genetic structure among extant species can be used to infer their evolutionary histories. As previously stated, rapidly evolving ITS regions of rDNA from disjunct species of section Calochroi found in regions of Europe and North America show different levels of sequence divergence. However, to date there has not been sufficient data to draw consistent conclusions concerning the genetic variation and divergence of these species. For example, maximum likelihood (ML) analyses [14] suggest a complex evolutionary history within C. arcuatorum, including divergence into four distinctive monophyletic groups with similar morphological features. In contrast, a close phylogenetic relationship was found between species considered to be morphologically separate from one another; European C. napus and North American C. albobrunnoides (vars. albobrunnoides and violaceovelatus and C. subpurpureophyllus var. sulphureovelatus). To gain a more complete understanding of the evolutionary histories of Calochroi species (including phylogeographic structure, character variation, and gene flow), we selected four species for study: 1) C. arcuatorum, 2) C. aureofulvus, 3) C. elegantior and 4) C. napus. Our species concept is anchored in the morphology (macroscopic, chemical, microscopic characteristics) of the basidiomata or mushrooms, which is the structure associated with sexual reproduction of basidiospores. Morphological and molecular evidences show that the four species treated here are each clearly distinct from other known Cortinarius species. The four selected species are somewhat rare to uncommon (especially C. aureofulvus and C. napus) across their known geographical range. They are mainly restricted to more calcareous soils, exhibit intercontinental distributions within boreo-nemoral areas, or in the case of C. arcuatorum extend into in the meridional zones [14]. Across their known distributions, C. arcuatorum is associated with oak (Quercus), beech (Fagus), hornbeam (Carpinus) and tan oak (Notholithocarpus) species, and with spruce (Picea) and Douglas fir (Pseudotsuga menziesii) in Wyoming. The tree hosts grow from lowland regions to high elevations, up to 3000 m in Costa Rica, and up to 2200 m in Wyoming. Three species, C. aureofulvus, C. elegantior and C. napus are associated with conifers including spruce (Picea), fir (Abies), pine (Pinus), Douglas fir (Pseudotsuga menziesii) and hemlock (Tsuga) species in subalpine regions or, less commonly, in mid- to low elevation forests (western elegantior clade and C. subpurpureophyllus var. sulphureovelatus (= C. napus)). Although these host tree genera also occur in Asia, so far there is no survey of these Cortinarius species for this region for inclusion in our study.
Here to infer historical events associated with speciation, we collected population samples within four species over geographical distributions spanning Europe, North America and Central America (Figure 1), used nested clade, demographic and coalescent-based approaches to analyse the fast-evolving ITS region of the nuclear ribosomal DNA. Coalescent-based analyses incorporate random fluctuations in mutation, genetic drift and sampling errors into the calculation of demographic parameters, whereas these variations are underestimated by phylogenetic approaches [27]. We used these analyses to address the following specific questions. What are the mechanisms responsible for the contemporary patterns of geographic distribution within the four species, including centres of origin and potential migration routes? And, are species with allopatric distributions sufficiently distinct from one another within and between continents to merit a specific designation? To address these questions, we compared DNA sequence variation of species at continental and intercontinental scales. We also evaluated the phenotypical and ecological features of the Calochroi species. Special emphasis was placed on detecting highly labile traits and their patterns of geographic variation among population samples represented by individual collection containing one to several basidiomata from one group or cluster found growing in a relatively small patch (genet).
Figure 1.
A map showing the geographic location of the sampling sites of calochroid population samples across Europe, North America and Central America. Population samples of C. arcuatorum are represented in blue, C. aureofulvus in orange, C. elegantior in red and C. napus in green. Collection numbers correspond to collections in Additional File 4. Distribution of host plant families: green dashed lines indicate distribution of Fagaceae and red dashed lines indicate distribution of Pinaceae, respectively.
Our findings show that the comparison of population samples using phenotypical, ecological and genetic characteristics help to clarify questions concerning the historical events involved in the modern distributions of Calochroi species as well as aspects related to host preferences and host-switching, and the use of a combined phylogenetic, morphological and ecological species definition. We also discuss the utility and limitations of ITS sequences and phenotypic characteristics as identification tools for Cortinarius species with wide geographical distributions.
Results
DNA amplification, phylogenetic placement and relationship
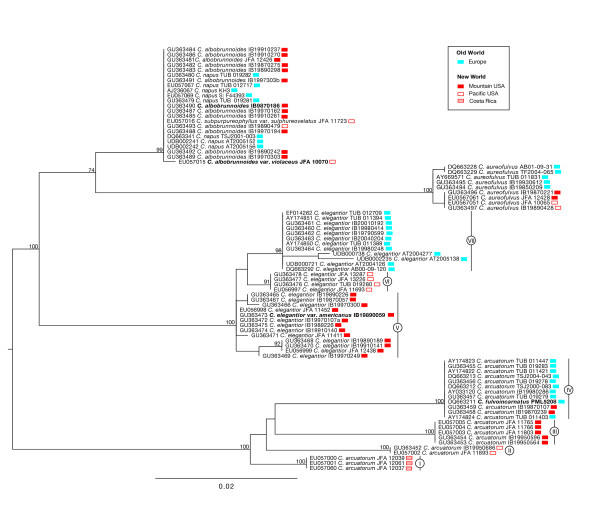
More than 100 fungus population samples, including our own collections and those from other herbaria, were analysed in this study (see the Methods section for more details). We were unable to amplify the ITS region for many collections because they were either poorly conserved or too old, and the sequence quality was too low, or we amplified contaminant fungi that probably colonised the dried basidiomata. Analysis of ITS sequences supported the phylogenetic placement of a total of 83 population samples identified as belonging to C. arcuatorum, C. aureofulvus, C. elegantior and C. napus lineages (data not shown). Each species was recovered as a monophyletic clade with high bootstrap support (Figure 2). The internal phylogeny of C. arcuatorum is composed of four major lineages: a clade including population samples from Costa Rica (I); a clade comprising population samples from Mendocino (Pacific USA) (II); a clade composed of samples from Del Norte Co (Pacific USA) (III); and a large clade containing mostly European and two sample populations from Wyoming (IV), respectively. Within C. elegantior, several population samples had unresolved positions (V), a portion of North American population samples are distributed into one well-supported subclade (VI) and European population samples formed a well-supported subclade (VII). The North American taxa C. albobrunnoides, C. albobrunnoides var. violaceus, and C. subpurpureophyllus var. sulphureovelatus and the European taxon C. napus formed a monophyletic group with not internal subclades. New World and Old World population samples of C. aureofulvus clustered together with no significant genetic differentiation between them.
Figure 2.
Phylogenetic relationships of New World and Old World population samples of Calochroi species based on ML analysis of ITS rDNA sequences. The represented tree was computed from 1000 runs and was mid-point rooted. Numbers above branches are bootstrap values (values < 70 not shown) from 1000 replicates. Assignations I to VII indicate major groups of C. arcuatorum and C. elegantior discussed in the text. Type specimens are printed in bold.
Patterns of nucleotide variation in the ITS region
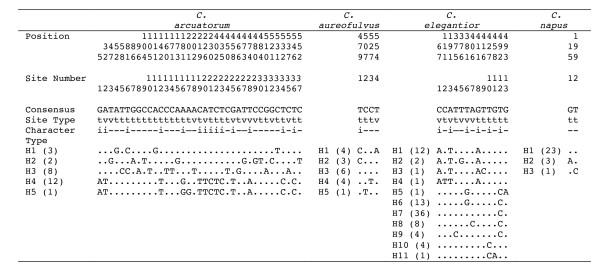
Sequence divergence of the complete ITS region between intercontinental disjunct populations ranged from identical to 4.63% in C. arcuatorum, from 0.16% to 0.35% in C. aureofulvus, from 0.68% to 1.46% in C. elegantior and from identical to 0.17% in C. napus. After removing indels and recombination blocks, twenty-four distinct ITS haplotypes were identified among the 83 population samples from the four species analysed: 5 haplotypes in C. arcuatorum and C. aureofulvus, 11 in C. elegantior, and 3 in C. napus (Figure 3). Nucleotide diversity (π) estimates for the whole population sample were 0.024 ± 1.9 × 10-3 in C. arcuatorum, 0.002 ± 3.0 × 10-4 in C. aureofulvus, 4.2 × 10-3 ± 4.2 × 10-4 in C. elegantior and 5.0 × 10-4 ± 2.0 × 10-4 in C. napus (Table 1). When species were partitioned into sampled areas, population samples showed similar or lower estimates of nucleotide diversity. The average per-nucleotide expected heterogeneity, θw, for the whole sample of each species was identical or higher than partitioned sampled areas.
Figure 3.
Distribution of polymorphic sites after removing indels and recombination blocks in the ITS haplotypes of calochroid species. t, transitions; v, transversions; i, phylogenetically informative sites; -, uninformative sites.
Table 1.
Population statistics, diversity estimates and neutrality tests based on ITS region of calochroid taxa studied
| Population statistics | Tests of neutrality | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Species/ population |
n | s | h | hd | k |
π (SD) |
θw |
Tajima's D |
Fu and Li's D* Statistic |
Fu and Li's F* Statistic |
Fu's Fs Statistic |
| C. arcuatorum | |||||||||||
| Old World | 11 | 1 | 2 | 0.182 | 0.181 | 3.3 × 10-4 (2.6 × 10-4) |
6.1 × 10-4 | -1.128 (NS) |
-1.289 (NS) |
-1.399 (NS) |
-0.410 (NS) |
| New World | 15 | 36 | 4 | 0.685 | 12.229 | 0.021 | 0.019 | 1.635 (NS) |
1.502** | 1.502 (NS) |
9.684 (NS) |
| Mountain USA | 2 | 0 | 1 | 0.000 | 0.000 | 0.000 | 0.000 | ND | ND | ND | ND |
| Pacific USA | 10 | 20 | 2 | 0.356 | 7.111 | 0.013 (5.6 × 10-3) | 0.013 | 0.027 (NS) |
1.567** | 1.326 (NS) |
9.432 (NS) |
| Costa Rica | 3 | 0 | 1 | 0.000 | 0.000 | 0.000 | 0.000 | ND | ND | ND | ND |
| All (l = 559) |
26 | 37 | 5 | 0.698 | 13.741 | 0.024 (1.9 × 10-3) |
0.017 | 1.569 (NS) |
1.568** | 1.843** | 13.357 (NS) |
| C. aureofulvus | |||||||||||
| Old World | 7 | 1 | 2 | 0.571 | 0.571 | 0.001 (2.1 × 10-4) |
7.3 × 10-4 | 1.341 (NS) |
0.953 (NS) |
1.101 (NS) |
0.856 (NS) |
| New World | 11 | 2 | 3 | 0.618 | 0.690 | 1.23 × 10-3 (2.9 × 10-4) |
1.22 × 10-3 | 0.036 (NS) |
-0.033 (NS) |
-0.269 (NS) |
-0.113 (NS) |
| Mountain USA | 2 | 0 | 1 | 0.000 | 0.000 | 0.000 | 0.000 | ND | ND | ND | ND |
| Pacific USA | 9 | 2 | 3 | 0.667 | 0.777 | 0.001 (3.2 × 10-4) |
0.001 | 0.195 (NS) |
-0.221 (NS) |
-0.135 (NS) |
-0.108 (NS) |
| All (l = 560) |
18 | 4 | 5 | 0.804 | 1.346 | 0.002 (3.0 × 10-4) |
0.002 | 0.473 (NS) |
0.211 (NS) |
0.324 (NS) |
-0.505 (NS) |
| C. elegantior | |||||||||||
| Old World | 16 | 3 | 4 | 0.442 | 0.4833 | 9 × 10-4 (3.6 × 10-4) |
0.002 | -1.316 (NS) |
-1.122 (NS) |
-1.351 (NS) |
-1.867 (NS) |
| New World | 67 | 7 | 7 | 0.662 | 1.003 | 0.002 (2.7 × 10-4) |
0.003 | -0.791 (NS) |
-0.420 (NS) |
-0.638 (NS) |
-1.614 (NS) |
| Mountain USA | 62 | 4 | 5 | 0.608 | 0.739 | 0.001 (1.8 × 10-4) |
0.002 | -0.288 (NS) |
-0.153 (NS) |
-0.228 (NS) |
-0.753 (NS) |
| Pacific | 5 | 1 | 2 | 0.400 | 0.400 | 7.8 × 10-4 (4.6 × 10-4) |
9.3 × 10-4 | -0.816 (NS) |
-0.816 (NS) |
-0.771 (NS) |
0.090 (NS) |
| All (l = 516) |
83 | 13 | 11 | 0.760 | 2.139 | 4.2 × 10-3 (4.2 × 10-4) |
0.005 | -0.495 (NS) |
-0.782 (NS) |
-0.808 (NS) |
-1.468 (NS) |
| C. napus | |||||||||||
| Old World | 8 | 0 | 1 | 0.000 | 0.000 | 0.000 | 0.000 | ND | ND | ND | ND |
| New World | 19 | 2 | 3 | 0.368 | 0.385 | 6.8 × 10-4 (2.5 × 10-4) |
1.0 × 10-3 | -0.777 | -0.573 | -0.720 | -0.725 (NS) |
| Mountain USA | 16 | 1 | 2 | 0.325 | 0.325 | 5.8 × 10-4 (2.2 × 10-4) |
5.3 × 10-4 | 0.155 (NS) |
0.688 (NS) |
0.627 (NS) |
0.551 (NS) |
| Pacific USA | 3 | 1 | 2 | 0.667 | 0.666 | 1.1 × 10-3 (5.6 × 10-4) |
0.002 | ND | ND | ND | 0.201 (NS) |
| All (l = 564) |
27 | 2 | 3 | 0.271 | 0.279 | 5.0 × 10-4 (2.0 × 10-4) |
9.2 × 10-4 | -0.977 (NS) |
-0.702 (NS) |
-0.898 (NS) |
-1.088 (NS) |
l, nucleotide sequence length; n, sample size; s, number of segregating sites; h, haplotypes; hd, haplotype diversity; k, average number of nucleotide pair wise differences; π, number of nucleotide differences per site (SD = standard deviation of π); θw, Watterson's estimate of θ per site; ND, not determined because no polymorphism was found; NS, non-significant; **, P < 0.02.
Tests of neutrality and population subdivision
The neutrality tests performed for the whole-species data sampling had non-significant values; therefore, the equilibrium model of neutral evolution could not be rejected. Only in C. arcuatorum were significant values detected for Fu and Li's D*, and Fu and Li's F* tests, suggesting background selection for the whole sample (Table 1). The geographical distributions and frequency of the haplotypes are given in Additional File 1. With regard to C. arcuatorum, haplotype H1 (I) was only found in those samples obtained from Costa Rica (Jardín de Dota and Parque Prusia), while haplotypes H2 (II) and H3 (III) are specific to California (Mendocino and Del Norte Counties, respectively). The most frequent haplotype (H4, IV; 12 population samples) is distributed throughout the Old World (Germany, France, Italy, Sweden) and also occurs in the New World (Wyoming), see population samples IB19870107 and IB19870239. The single haplotype H5 (IV) is restricted to the Old World. In C. aureofulvus, haplotypes H1 and H2 are restricted to the Old World (Austria, France, Germany, Sweden), whereas haplotypes H4 and H5 were found in the New World (Washington State) population samples. Haplotype H3 was found in population samples from the Pacific Northwest (Washington State) and Mountain USA (Colorado and Wyoming). In C. elegantior, H7 (V) and haplotypes H1 (VII) were predominant in the New World (Wyoming) and the Old World (Austria, France, Germany and Switzerland), respectively. Haplotypes H2, H3 and H4 are restricted to Old World and haplotypes H5, H6 (VI), H8, H9, H10 and H11 were observed only in those population samples obtained from Old World. With regard to C. napus, the predominant haplotype H1 was shared by different taxa in the New World (Pacific: Oregon and Washington State, and Mountain USA: Wyoming and Colorado) and the Old World (Germany, Norway, Sweden). Haplotype H2 was found in the Wyoming populations and haplotype H3 is represented by a single population sample (JFA 10070) from the Pacific (Washington State).
Hudson's tests were carried out to estimate population genetic structures within and among population samples from each area (Table 2). Analyses of populations showed significant differentiation between Costa Rica/Pacific USA (California) and Mountain USA (Wyoming)/Europe population samples of C. arcuatorum (P < 0.001, KST = 0.6540, KS = 5.0769, KT = 14.6769); between Europe/Pacific USA (Washington State, Oregon) and Mountain USA (Wyoming, Colorado) population samples of C. elegantior (P < 0.001, KST = 0.0833, KS = 3.1645, KT = 3.4522) and between Europe/Pacific USA (Washington State) and Mountain USA (Wyoming, Colorado) population samples of C. aureofulvus (P = 0.001, KST = 0.3183, KS = 1.4166, KT = 2.0784). Non-significant genetic differentiation between Europe/Pacific (Washington State, Oregon) and Mountain USA (Wyoming, Colorado) was found for C. napus population samples (P = 0.2240, KST = 0.0518, KS = 0.3998, KT = 0.4216). Alternatively, significant P values (P < 0.001) for Hudson's tests indicated that New World and Old Wold population samples of C. arcuatorum (KST = 0.4619, KS = 7.8968, KT = 14.6769), C. aureofulvus (KST = 0.3731, KS = 1.3027, KT = 2.0784) and C. elegantior (KST = 0.3927, KS = 2.0963, KT = 3.4522) were genetically differentiated, while New World and Old World macro-populations of C. napus (P = 0.6840, KST = -0.0251, KS = 0.4322, KT = 0.4216) were not significantly differentiated from each other.
Table 2.
Population subdivision in calochroid taxa inferred from Hudson's test statistics Ks (upper right matrix) and Kst (lower left matrix)
| Old World | Mountain USA | Pacific USA | Costa Rica | |
|---|---|---|---|---|
| C. arcuatorum | ||||
| Old World | 0.1818 (NS) | 3.7041*** | 0.1636** | |
| Mountain USA | -0.1818 (NS) | 7.6666** | 0.0000 (NS) | |
| Pacific USA | 0.7511*** | 0.4025** | 6.8148* | |
| Costa Rica | 0.9754** | 1.0000 (NS) | 0.3185* | |
| C. aureofulvus | ||||
| Old World | 0.5714* | 1.3399*** | ||
| Mountain USA | 0.3949* | 1.8888 (NS) | ||
| Pacific USA | 0.4044*** | -0.1052 (NS) | ||
| C. elegantior | ||||
| Old World | 1.7739*** | 0.6607*** | ||
| Mountain USA | 0.4581*** | 1.9434*** | ||
| Pacific USA | 0.7411*** | 0.1880*** | ||
| C. napus | ||||
| Old World | 0.3908 (NS) | 0.0952 (NS) | ||
| Mountain USA | -0.0081 (NS) | 0.5655 (NS) | ||
| Pacific USA | 0.4761 (NS) | 0.0329 (NS) | ||
Significance, including incompatible sites and recombination blocks, was evaluated by performing 1000 permutations using SNAP Map. NS, non-significant; *0.01 ≤ P < 0.05; **0.001 ≤ P < 0.01; ***P < 0.001.
Network analyses
To clarify the phylogenetic relationship among haplotypes within each species, networks were constructed using haplotypes from ITS sequences with non-recombining blocks (Figure 4a-d). Nested clade analyses for C. arcuatorum resulted in one network with two connected Old World haplotypes (H4 and H5) and three single unconnected New World haplotypes (H1, H2 and H3) (Figure 4a). Analyses of C. aureofulvus (Figure 4b) yielded a network comprising five haplotypes: haplotypes H3, H4 and H5 were scattered throughout the New World, whereas haplotypes H1 and H2 were restricted to the Old World. The haplotypes H1, H2, H4 and H5 were derived from haplotype H3. Similarly, in C. elegantior, haplotypes were contained in a single network where haplotype H7 has an ancestral position to the other haplotypes and the Old World haplotypes H1, H2, H3 and H4 grouped together (Figure 4c). The Old World haplotypes H1, H2, H3 and H4 appear to be more related to the Pacific haplotypes H10 (Oregon and Washington State) and H11 (Washington State). In C. napus, haplotype H1 is ancestral to the other haplotypes and was shared by both New World and Old World populations, whereas the haplotypes H2 and H3 were restricted to Mountain USA (Wyoming) and Pacific (Washington State) in the New World, respectively (Figure 4d).
Figure 4.
TCS haplotype networks based on non-recombining ITS sequences. a. C. arcuatorum. b. C. aureofulvus. c. C. elegantior. d. C. napus. Parsimony probabilities were set at 95%. Sizes of circular and rectangular areas are proportional to the number of individuals with that haplotype. Haplotypes with a blue background are from the Old World and those with red backgrounds are from the New World. Haplotypes comprising New and Old World populations are indicated by lilac backgrounds. Distributions of ecological and morphological features are abbreviated as follows: F, Fagaceae; P*, Pinaceae (pure stand in Wyoming); P, Pinaceae; B, Betulaceae, M1, populations of C. arcuatorum with larger spores; M2, some populations of C. elegantior with sulphur yellow veil and lacking of veil patches on the pileus surface.
Migration analyses
The four species showed different histories of recombination: two distinct recombination blocks were detected in C. arcuatorum, four in C. aureofulvus, nine in C. elegantior and one in C. napus, respectively. There was no evidence of recombination within each of these blocks. Migration parameter estimates, including population mutation rate, time of divergence and direction of migration estimates between New World and Old World macro-population samples, are shown in Additional File 2. These analyses showed that migrations between New World and Old World macro-population samples within C. arcuatorum and C. aureofulvus were approximately equal in both directions and that these have occurred at low rates. While gene flow estimates in C. elegantior indicate that migration appeared to be slightly higher (with overlapping confidence intervals) from the New World to the Old World (m1 = 0.34) rather than vice versa (m2 = 0.16).
Genealogical analyses
Coalescent genealogies for C. arcuatorum and C. elegantior showed that two ancestral lineages gave rise to the extant New and Old World haplotypes (Figures 5 and 6). Genealogies inferred for C. aureofulvus (Figure 7) and C. napus (data not shown) did not have enough genetic polymorphisms to resolve transoceanic disjunction events. In C. arcuatorum, a widely distributed (New World (Wyoming) - Old World (Europe)) ancestral macro-population could be the origin of extant haplotypes. In C. elegantior, an ancestral macro-population diverged at 0.7 into two distinct lineages in the New World, whereas Old World haplotypes diverged at 0.4, respectively. The oldest mutations of both New World and Old World macro-populations of C. arcuatorum have a mean age of 0.9, while the C. elegantior macro-populations have a mean age of approximately 0.7. In C. arcuatorum, extant Old World haplotypes are younger than New World haplotypes, T < 0.2 and T > 0.5, respectively. In C. elegantior, New World haplotype deep divergence was older (T > 0.6) than New World haplotypes (T = 0.4), but extant New World haplotypes are descendents from more recent lineages.
Figure 5.
Coalescent-based genealogy of the internal transcribed spacers (ITS1 and ITS2), showing the distribution mutations for New World (CR = Costa Rica; Pacific, CA = California; Mountain USA, WY = Wyoming) and Old World (EU = Europe) populations of C. arcuatorum performed using Genetree. The inferred genealogy is a rooted genealogy based on five million simulations of the coalescent. Simulations were carried out assuming a constant population size. Maximum likelihood estimates of the tree with the highest root probability (likelihood = 1.16 × 10-18) and standard deviation (SD = 9.53 × 10-16) showing the distribution of mutations. The direction of divergence is from the top of the genealogy (oldest = past) to bottom (youngest = present); coalescence is from the bottom (present) to the top (past). The numbers below each branch indicate the corresponding haplotype and its frequency in each macro-population and sampling area. The time scale is in coalescent units of effective population size.
Figure 6.
Coalescent-based genealogy of the internal transcribed spacers (ITS1 and ITS2), showing the distribution mutations for New World (Pacific, WA = Washington State, OR = Oregon; Mountain USA, WY = Wyoming) and Old World (EU = Europe) populations of C. elegantior performed using Genetree. The inferred genealogy is a rooted genealogy based on five million simulations of the coalescent. Simulations were carried out assuming a constant population size. Maximum likelihood estimates of the tree with the highest root probability (likelihood = 2.55 × 10-15) and standard deviation (SD = 4.68 × 10-13) showing the distribution of mutations. The direction of divergence is from the top of the genealogy (oldest = past) to bottom (youngest = present); coalescence is from the bottom (present) to the top (past). The numbers below each branch indicate the corresponding haplotype and its frequency in each macro-population and sampling area. The time scale is in coalescent units of effective population size.
Figure 7.
Coalescent-based genealogy of the internal transcribed spacers (ITS1 and ITS2), showing the distribution mutations for New World (Pacific, WA = Washington State; Mountain USA, WY = Wyoming, CO = Colorado) and Old World (EU = Europe) populations of C. aureofulvus performed using Genetree. The inferred genealogy is based on five million simulations of the coalescent. Simulations were carried out assuming a constant population size. Maximum likelihood estimates of the tree with the highest root probability (likelihood = 1.75 × 10-8) and standard deviation (SD = 1.61 × 10-6) showing the distribution of mutations. The direction of divergence is from the top of the genealogy (oldest = past) to bottom (youngest = present); coalescence is from the bottom (present) to the top (past). The numbers below each branch indicate the corresponding haplotype and its frequency in each macro-population and sampling area. The time scale is in coalescent units of effective population size.
Basidioma phenotypical features
Detailed descriptions of the macro- and microscopical structures for each species are given in Additional File 3. The most relevant phenotypical features are summarised below.
(i) Basidioma colour
There was little or no detectable variation of basidiomata colour within C. arcuatorum, C. aureofulvus and C. elegantior (see Figures 8, 9 and 10). However, population samples of C. napus, showed considerable variation in colour of both lamellae and veil (Figure 11).
Figure 8.
Basidioma morphology and colouration within C. arcuatorum. a. Costa Rican population JFA 12039. b. Costa Rican population JFA 12061. c. Californian (Mendocino) population IB19950686, d. Californian (Patricks Creek Campground) population JFA 11765. e. Californian (Patricks Creek Campground) population IB19950564. f. European population TUB 011403. Photos c and e courtesy of M. Moser.
Figure 9.
Basidioma morphology and colouration of C. aureofulvus. a. Washington State population IB19890428. b. Colorado population JFA 12428. c. Swedish population IB19850209. Photos a and c courtesy of M. Moser.
Figure 10.
Basidioma morphology and colouration within C. elegantior. a. Washington State (Table Mountain) population JFA 13287. b. Washington State (Table Mountain) population JFA 13287, a basidioma showing KOH reaction. c. Washington (Snohomish County) population TUB 019280. d. Wyoming (Teton National Forest) population of C. elegantior var. americanus IB19890059 (holotype). e. Wyoming (Teton National Forest) population IB19890189; NH3 is showed on stipe context. f. Austrian population IB19790599 (one basidioma shows a red wine KOH reaction). Photos d, e and f courtesy of M. Moser.
Figure 11.
Basidioma morphology and colouration within C. napus. a. Washington State (Rainy Pass) population IB19890479. b. Washington (Chelan County) population sample of C. albobrunnoides var. violaceovelatus IB19890186 (typus). c. Oregon (Lincoln County) population sample of C. subpurpureophyllus var. sulphureovelatus JFA 11723. d. Wyoming population (Shoshone National Forest) IB19970303. e. Wyoming (Shoshone National Forest) population IB19910237. f. European population TUB 019282. Photos a, b, d and e courtesy of M. Moser.
(ii) Basidioma size
Some variation in basidioma size was observed within C. arcuatorum, where populations from Costa Rica and some from Europe presented somewhat slender basidiomata in comparison with other population samples (Figure 8).
(iii) Colour reaction of dried specimens
A pink to pinkish orange colour reaction with 40% KOH on both pileus surface and mycelia at the stipe base characterised the basidiomata of C. arcuatorum populations. Population samples of C. napus yielded a greyish to dark red brown colour reaction on the pileus surface, whereas the mycelia showed a pale pink to intense pink colour reaction. All population samples in C. aureofulvus had a black reaction on both pileus surface and mycelia on the stipe bulb, while in C. elegantior, both the pileus surface and mycelia turned wine red (Figure 12).
Figure 12.
Colour change reactions with KOH 40% mycelia at the stipe bulb and on pileus surface of dried specimens. a - c: C. arcuatorum: a. Blood colour on mycelia in IB19870239. b. Blood colour on pileus surface in IB19870239. c. Pink on pileus surface in TUB 019283. d - f: C. aureofulvus: d. Black on mycelia in IB19890428. e. Purple on pileus surface in IB19930612. f. Reaction in (e), turning black after some minutes. g - i: C. elegantior var. americanus IB198959 holotype: g. Wine red on mycelia. h. Intense wine red on pileus surface. i. Reaction in (g), after a while becoming dark wine red. j. Pink with orange tinge on mycelia in C. napus TUB 019282. k - l: C. albobrunnoides. k. Pink with orange tinge on mycelia in C. albobrunnoides IB19890298. l. Dark red-brown on pileus surface in C. albobrunnoides IB19970303. Note: photographs were taken in natural daylight.
Discussion
The markedly high species diversity within the genus Cortinarius together with their obligate ectomycorrhizal status makes the factors that have driven their speciation of particular interest to a broad spectrum of biologists. In addition, the occurrence of widespread taxa within Cortinarius raises intriguing questions about the origins of transoceanic disjunct species and the influence of the historical events that have created contemporary geographical distributions.
Phylogeographical structure and historical demography of Calochroi species
While it is difficult to determine the origin and migration routes of ectomycorrhizal fungi between North America, Europe and Asia, one can gain some insight into the question by looking at host migration patterns and assuming co-migration of plant and fungus. Genetic data reveal considerable divergence between populations across the core range of distribution of C. arcuatorum and C. elegantior in the New World (Figure 4a, c; Table 2). Since the divergent ancestral lineages were found in the New World, we postulate this area as the centre of origin for C. arcuatorum and C. elegantior, and that Old World populations are the result of relatively recent demographic expansion (Figures 4a, c, 5 and 6). Migration with host plants [28] through Beringia is one of the most plausible hypotheses to explain the disjunct population distributions between the New World and Old World, but migrations through the North Atlantic land bridge [29] or via long-distance dispersal [30], though a less likely explanation cannot be completely ruled out. Genetic breaks between C. arcuatorum populations within the New World have probably been triggered by paleogeographic and paleoclimatic events, e.g. the rise of the Sierra Nevada and other mountains followed by desertification of the Great Basin, or other events such as Pliestocene glaciation [30,31]. The European populations of C. arcuatorum and most of the North American populations occur mainly with trees of the family Fagaceae, with the exception of the population samples in Wyoming, where this species occurs with conifers. These known population locations are currently geographically isolated from members of the Fagaceae; however, it is likely that Picea and/or Tsuga were sympatric with Quercus in the past [32], allowing for the possibility of a host switch from Quercus to a conifer host.
The coalescent-based analysis suggests that extant populations diverged from a quite recent common ancestor early in the evolution of C. elegantior with little gene flow among disjunt populations (Figure 6; Additional File 2). Despite this, very little morphological change has evolved between New World and Old World lineages. Hence, the close relatedness of Pacific and European haplotypes of C. elegantior could indicate an expansion northward along the western side of the mountains through the Bering Land Bridge, which resulted in the extant populations in New World (Figure 4c). This evolutionary scenario is compatible with the migration of Tricholoma (matsutake) between western North America and eastern Asia, as postulated by Chapela & Garbelotto [8]. The phylogeographical patterns observed in C. elegantior support the hypothesis that the current distributions of populations are closely associated with the historical events of their host plants. Similarly, conifers are the primary hosts for populations of C. aureofulvus and for C. napus. In most parts of their ranges, these Cortinarius species are sympatric or allopatric, although their frequency of occurrence varies, with C. aureofulvus being the least commonly encountered, C. napus being moderately frequent, and C. elegantior populations being the most frequently encountered, especially at higher elevations with Picea. Populations of C. elegantior and C. napus are most frequently encountered in subalpine conifer forests; however, they also occur in more mesic mid-elevations and coastal sites. Picea is currently a main host plant genus for C. aureofulvus, C. elegantior and C. napus in western North America; this has probably been the case for an extended period of time, with a shift in abundance of Picea from eastern to western North America several times over the last 21,000 years [32]; however, Abies, Pinus and Tsuga are also potential hosts for these species. Douglas fir, another possible host for these species, appeared in western North America in the mid-Pleistocene [33]. Recent phylogenetic studies of Picea indicate that it originated in North America (P. breweriana and P. sitchensis as basal taxa) and that the present distribution of Picea could be a result of two dispersal events, one from North America to Asia by the Bering Land Bridge [34], and a second from Asia to Europe. Most of the northeastern Asian species and the European P. abies could have arisen from a recent radiation [35]. The literature also notes that the earliest (Paleocene) fossil of Picea is from Montana and that it is well represented in the Eocene in western North America, but was not common in Asia until the Oligocene and not present in Europe until the Pliocene. Therefore, it is fully possible that populations within C. aureofulvus, C. elegantior and C. napus have long been associated with Picea and could have migrated with this and possible other host plants during glacial and interglacial periods for millions of years. The findings presented here, in conjunction with those of Dentinger et al. [4] and Wu et al. [7] strongly support the key role of Beringia in the phylogeographic processes leading to speciation and intraspecific population structures in the Northern Hemisphere. Subsequently, the comigration with their associated phanerogams [28,35] through e.g. short-distance spore dispersal may be an important means explaining the expansion of these fungi from Asia to Europe.
We cannot estimate times for the divergence of disjunct populations of Calochroi with certainty, owing to a lack of fossil records. However, it is plausible to hypothesise that they co-migrated with their host plants, and that the lineages may have subsequently diverged as a result of fragmentation and geographic isolation of ancestral populations due to geologic and subsequent climatic changes. However, each species has a somewhat different biogeographical history that likely has been influenced by different biotic and abiotic factors, such as dispersal potential and host (phanerogam) history. The occurrence of these taxa in Asia has not been documented to date. Consequently further sampling across this region is necessary in order to better understand their historical distributions across the Northern Hemisphere. The recent work on porcini mushrooms [4] supports the idea of a potential link between European, Asian and North American species in the section Calochroi.
ITS sequences and phenotype as identification tools for Cortinarius species with wider ranges of geographical distribution; utility and limitations
Nested clade, demographic and coalescent-based analyses based on the ITS region of C. arcuatorum, C. aureofulvus, C. elegantior and C. napus allowed us to infer their evolutionary histories. Overall, C. arcuatorum and C. elegantior exhibited geographically structured haplotypes, with evidence for a more ancient population expansion. These findings agree with those of Geml et al. [9,11], who observed a similar population structure in the widespread fly agaric Amanita muscaria. Interestingly, geographically structured populations within C. arcuatorum and C. elegantior were often accompanied by some divergence in basidioma colouration and size in the absence of any described species (see Additional File 3). By contrast, some populations within C. arcuatorum and C. aureofulvus with disjunct distributions in North America and Europe exhibited little morphological differentiation and had identical to relatively low levels of ITS sequence divergence (identical to 0.3% (2 bp) divergence). Two hypotheses might explain this low degree of nucleotide variation. First, these taxa might be relatively young, having recently spread across their ranges, and, accordingly, having had insufficient time to accumulate ITS sequence divergence. An alternative hypothesis is that these results could be a consequence of low mutation rates at the locus studied. Although some differences in colouration of the basidiomata were found between western populations of C. albobrunnoides var. albobrunnoides, C. albobrunnoides var. violaceovelatus and C. subpurpureophyllus var. sulphureovelatus and European populations of C. napus, these differences were not sufficient to consider them as separate taxa. This suggests that variation in colouration alone might not necessarily reflect different species or subspecies. For example, violet pigments in the basidiomata are highly labile because they are sensitive to solar radiation and the age of the basidiomata. Therefore, it is evident that it is important to do careful fieldwork and to consider the variation of macroscopical and ecological features before they are used for taxonomic purposes, especially characters related to colouration. According to our analyses, colour reactions with KOH are relatively stable among populations, whereas ITS sequences, in certain instances, provide several genetic markers to distinguish species as well as population structure within the section Calochroi.
Conclusions
Based on variations in ITS DNA polymorphism, this study revealed different evolutionary histories for New World and Old World populations within Cortinarius arcuatorum, C. aureofulvus, C. elegantior and C. napus. Nested clade, demographic and coalescent-based analyses provide powerful tools for assessing the effect of historical and contemporary events of the geographic distributions of these species, proposed as follows: (i) an ancestral population of C. arcuatorum putatively with a Wyoming-Europe distribution evolved into at least three distinct lineages in association with a variety of fagaceous and coniferous trees in the New World; (ii) two divergent lineages in C. elegantior gave rise to New World and Old World haplotypes that occur almost exclusively in association with Picea (apart from one Washington State population in Tsuga/Abies forest); (iii) low genetic diversity among New World and Old World populations of C. aureofulvus; iv) low genetic diversity among New World and Old World populations of C. napus. The results for C. aureofulvus and C. napus could be due to recent demographic expansion, but the origin of such transoceanic disjunct distributions remains unclear. The scenario of a spatial expansion through the Bering Land Bridge appears to be the most probable explanation for modern transoceanic disjunct distributions; however, it does not represent a complete explanation for the remarkably low degree of genetic differentiation between populations of these two species. Our study reveals patterns of species diversification restricted to the New World within C. acuatorum and C. elegantior, however, the relatively few morphological innovations found among allopatric populations suggest that diversification events may have been driven by ecological opportunities afforded by the shift to new host species, edaphic and climate conditions. The morphological and genetic data described here support a single species C. napus (to include C. albobrunnoides var. albobrunnoides, C. albobrunnoides var. violaceovelatus and C. subpurpureophyllus var. sulphureovelatus) with transoceanic distribution. Similarly, C. aureofulvus shows disjunct distributions in New World and Old World. Colour reactions with KOH represent a powerful tool for recognising these Calochroi species, whereas features related mainly to veil and lamellae colouration displayed high levels of homoplasy and phenotypic plasticity. Interpretations of the population structures suggest that host tree history as influenced by past events (glaciation, mountain building, and associated factors), was one of the driving forces that shaped the modern distributions of Calochroi species. Finally, our work shows the need for further studies that focus on careful fieldwork in the context of the variation of both morphology and ecology, as well as the use of more genetic information, to draw more complete models of population histories in these and other species of Cortinarius.
Taxonomic implications
See Additional File 3 for information of the following taxonomic novelties:
Cortinarius elegantio-montanus Garnica & Ammirati (= Cortinarius elegantior (Fr.) Fr. var. americanus M.M. Moser & McKnight, 1995), new synonym and new status.
Cortinarius elegantio-occidentalis Garnica & Ammirati, new species - allied to Cortinarius elegantior (Fr.) Fr., 1838.
Cortinarius fulvo-arcuatorum Garnica & Ammirati, new species - allied to Cortinarius arcuatorum Rob. Henry, 1939.
Cortinarius jardinensis Garnica, Ammirati & Halling, new species - allied to Cortinarius arcuatorum Rob. Henry, 1939.
Cortinarius lilaciotinctus Garnica & Ammirati, new species - allied to Cortinarius arcuatorum Rob. Henry, 1939.
Cortinarius napus Fr. 1838 (= Cortinarius albobrunnoides var. albobrunnoides M.M. Moser & McKnight, 1995; Cortinarius albobrunnoides M.M. Moser & K. McKnight var. violaceovelatus M.M. Moser & J. Ammirati, 1996; and Cortinarius subpurpureophyllus A.H. Smith var. sulphureovelatus M.M. Moser, 2000), new synonym.
The authors declare that they have no competing interests.
Methods
Population sampling
We collected population samples within C. arcuatorum Rob. Henry, C. aureofulvus M.M. Moser, C. elegantior (Fr.) Fr. var. elegantior (including C. elegantior (Fr.) Fr. var. americanus M.M. Moser & McKnight) and C. napus Fr. (including C. albobrunnoides M.M. Moser & McKnight, C. albobrunnoides var. violaceovelatus M.M. Moser & Ammirati, and C. subpurpureophyllus A.H. Sm. var. sulphureovelatus M.M. Moser) groups, following their distribution ranges in Europe, Central America and western North America. The following specimens were initially used for DNA sequencing: 1 collection of C. albobrunnoides from the University of Washington Herbarium (WTU) and 13 from Innsbruck Herbarium (IB) and 8 collections of C. arcuatorum (including two named C. cf. sodagnitus IB19870239 and IB19870107); 36 collections of C. elegantior (including the type material of C. elegantior var. americanus IB19890059) from Europe and North America deposited at (IB) and 4 from WTU; 14 collections of C. aureofulvus and 7 collections of C. napus from IB. An attempt was made to sample from throughout the distribution range of these species, although sampling was dictated by the availability of recently collected and accurately documented herbarium specimens. These four taxa were selected because they represent a diverse set of species/populations, each with a monophyletic lineage displaying allopatric distributions in Europe and North America, as previously revealed in research by the authors [14]. In order to incorporate as much genetic information as possible from a wide range of geographical locations for these species, sequences were obtained from GenBank http://www.ncbi.nlm.nih.gov/ and UNITE http://unite.ut.ee/ databases. Details of the population samples used for this study and the geographic location of the sampling sites are provided in Additional File 4.
DNA extraction, amplification, sequencing and sequence editing
Total genomic DNA was extracted from dried lamella fragments using the DNAeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the standard protocol. The internal transcribed spacers (ITS1, ITS2), the 5.8 S ribosomal subunit and the D1/D2 regions of the nucLSU were amplified with the primer combination ITS1F (5'-CTTGGTCATTTAGAGGAAGTAA-3') [36]/NL4 (5'-GGTCCGTGTTTCAAGACGG-3') [37]. Subsequently, those specimens that did not give any amplification products were amplified with the primer combinations ITS1F/ITS4 (5'-TCCTCCGCTTATTGATATGC-3') [38] or ITS1 (5'-TCCGTAGGTGAACCTGCGG -3') [38]/ITS4 and 5.8SR (5'-TCGATGAAGAACGCAGCG-3')/LR3 (5'-CCGTGTTTCAAGACGGG-3') [39] or 5.8SR/NL4. Amplifications of ITS1, ITS2, the 5.8S ribosomal subunit and the D1/D2 regions of the nucLSU were carried out using a touch-down program with the following conditions: initial denaturation at 94°C for 3 min; 10 cycles with temperatures ranging from 60°C in the first cycle to 51°C, each cycle decreasing by 1°C; 25 cycles with an annealing temperature of 50°C, each cycle consisted of an annealing step of 0.5 min; an elongation step of 72°C for 1 min and a denaturation step of 94°C for 0.5 min and a final elongation phase at 72°C for 7 min. Alternatively, amplifications of the ITS region for those specimens with negative results using Taq DNA polymerase (Invitrogen Corporation, Carlsbad, CA, USA) were carried out using 25 μL Phusion™ High-Fidelity DNA polymerase-mediated reactions (Finnzymes Oy, Keilaranta, Finland) with an initial heating of 95°C for 60 seconds, followed by 35 cycles of denaturation at 94°C; 30 seconds, annealing at 50°C-55°C; 30 seconds and extension at 72°C; and 45-90 seconds, followed by a final extension of 72°C for 10 minutes. Amplification products were electrophoresed in a 0.7% agarose gel and stained with ethidium bromide for visualization of the bands. PCR products were cleaned using ExoSAP-IT® (USB Corporation, Cleveland, OH, USA) reagent diluted 1: 20 according to the manufacturer's instructions. Both DNA strands were cycle-sequenced with the amplification primers and, in some cases, internal primers as indicated in Garnica et al. [40]. Clean PCR products were sequenced in both directions with a 1: 6 diluted dye terminator sequencing kit (Big Dye 3.1; Applied Biosystems, Foster City, CA, USA) on an ABI Prism 3130 × l Genetic Analyzer (Applied Biosystems). Unpublished molecular data from our lab indicate non-sequence divergence among individuals within the same collection; therefore, we sequenced only one individual per collection. Forward and reverse sequence fragments were assembled and edited using Sequencher version 4.1 (Gene Codes Corporation, Ann Arbor, MI, USA).
Sequence identity, alignments, phylogenetic placement and variability
To detect potential contaminant sequences, we first screened our sequences against those available in the GenBank database http://www.ncbi.nlm.nih.govhttp:///[41]) using the Basic Local Alignment Search Tool (BLAST). To establish the phylogenetic placement of the newly generated ITS sequences and those from Garnica et al. [14], we automatically aligned them in MAFFT v5.7 [42] with the E-INS-i option. Our data matrix, which included 637 sequences and 739 nucleotide positions, was analysed by ML inference as implemented in RAxML version 7.0.3 [43]. The best-known likelihood tree under the GTRMIX model of nucleotide substitution was computed from 100 runs starting from distinct randomised maximum parsimony starting trees. A total of 1000 non-parametric bootstrap replicates [44] were run on the original alignment. Graphical processing of the trees with best likelihood and bootstrapping were generated using TreeViewPPC version 1.6.6 [45] and PAUP* 4.0b10 [46]. Subsequently, sequences for each taxon were aligned separately in MAFFT. Within each taxon set, all polymorphic sites were rechecked from the chromatograms. Isolates from fungal specimens are n + n and therefore provide ambiguous haplotype data for heterozygotes. Heterozygous sites appear as two coincident peaks at the same site in the forward and reverse sequence chromatograms. To resolve the haplotype structure within the heterozygote, we used Clark's haplotype subtraction algorithm [47]. The DNA sequences newly generated in this study were submitted to GenBank (accession numbers GU363452-GU363497).
All sequences within one species were collapsed into haplotypes removing indels and excluding infinite-site violations using Map as implemented in SNAP Workbench [48,49]. A site-incompatibility matrix was calculated to assess compatibility among all variable characters, with incompatible characters being subsequently removed. Sites are called compatible if there is a phylogeny which both can evolve on without homoplasy. Two or more variable sites showing an identical pattern of compatibility are considered to form a recombination block [50]. When more than one recombination blocks were found, we used RecMin [51] as implemented in SNAP Workbench to estimate the minimum number of recombination events. Conversely, all sites being compatible with each other were considered as evidence for no recombination. Sequence polymorphism (haplotype h and nucleotide π diversities) was estimated using DnaSP version 5 [52]. Estimates were calculated for the entire dataset comprising all sequences of one species, as well as for each subpopulation sampled.
Neutrality tests and population subdivision
Identical ITS sequences were collapsed into haplotypes using SNAP Map [49] after excluding indels and infinite-site violations. Site compatibility matrices were generated from each haplotype datasets using SNAP Clade and Matrix [53]. Sequence variations were tested for deviations from neutrality by using Tajima's D [54], Fu and Li's D* and F* [55], and Fu's Fs [56] statistics with DNASP v 5.00.07 [52]. Tests of neutrality assume a constant population size, no recombination and no migration. The D* test by Fu and Li is based on assessing selection by counting external (recent) and internal (ancient) mutations in the genealogy and comparing the values to their expectations under the assumption of neutral evolution. Significant Fu and Li's tests suggest background selection, while a significant Fu's Fs value indicates population growth and genetic hitchhiking. As small sample sizes and population subdivisions are known to limit the power of the neutrality tests [57,58], we subsequently tested our data for population subdivision. To test for population genetic differentiation, we used SNAP Map [49] to generate the appropriate sequence file, Septomatrix to convert the sequence file to a distance matrix, and Permtest [57] to test for geographic intraspecific subdivision among the different areas sampled, as implemented in SNAP Workbench [48]. Population genetic differentiation analyses were calculated according to the sampled areas in Central America (CR = Costa Rica), Pacific USA (WA = Washington State, OR = Oregon, CA = California), Mountain USA (WY = Wyoming, CO = Colorado) and Europe (EU). In order to increase sample sizes, populations were grouped into macro-populations with regard to the directions of migration hypothesized as follows: for C. arcuatorum, these were Costa Rica (CR) + Pacific (CA) + Mountain USA (WY); for C. aureofulvus, these were Pacific USA (WA) + Mountain USA (WY, CO); for C. elegantior, Pacific (WA, OR) + Mountain USA (WY); for C. napus, Pacific USA (WA, OR) + Mountain USA (WY, CO). Each macro-population was compared against the EU macro-population. The main criteria used to define the macro-populations were: geographical location of the sampling sites, geological events and host tree histories. Alternatively, population samples were analysed as follows: for C. arcuatorum in Central America (CR) + Pacific (CA) macro-populations was compared against Mountain USA (WY, CO) + Europe (EU) macro-populations, whereas in C. aureofulvus and C. elegantior Pacific USA (WA, OR) macro-populations were compared against Mountain USA (WY) + Europe (EU) macro-populations. Hudson's test statistics were evaluated under the null hypothesis of no genetic differentiation applying, 1,000 permutations per species sample. For this purpose only, we used the non-reduced datasets containing the recombination blocks (see above), as recombination has been shown to improve the power of Hudson's test [57].
Haplotype network analyses
We constructed haplotype networks by using the program TCS version 1.21 [59]. Parsimony probability was set at 95%; therefore, haplotypes with a probability of parsimony higher than 95% would be connected and those with a probability lower than 95% would be unlinked.
Migration analysis
For those species showing significant population subdivision according to Hudson's test, we conducted migration analysis using the IM program [60]. As the program model assumes neutrality and no recombination, the reduced dataset that contained non-recombinant characters only was submitted to analysis. This program simultaneously infers multiple genealogical parameters like effective population sizes for ancestral (θA) and modern (θ1 and θ2) populations, time since divergence between populations (t) and migration rates (m1 and m2) between two subpopulations using a Markov Chain Monte Carlo (MCMC) approach. We used chain lengths of 10,000,000 steps for C. elegantior dataset and 20,000,000 steps for C. arcuatorum and C. aureofulvus to guarantee a sufficiently large effective sample size (ESS), as well as a flat or presumably complete appearance of the posterior distribution curve. For each dataset, we executed a total of 15 independent simulations of the chain (starting from different random number seeds for each) and evaluated the estimated means of migration rates m1 and m2. Subsequently, a one-sided t-test was used to statistically assess the hypothesis of directional migration between the two subpopulations.
Genealogical analyses
Ancestral intraspecific genealogies were inferred using Genetree Version 9 [61] in SNAP Workbench [48]. The genealogies with the highest root probability, ages of mutations, the times to the most recent common ancestor (TMRCA), and geographic distribution of the mutations were assessed from coalescent simulations. Coalescent simulations were carried out assuming an infinite-site model, constant size and population subdivision. As for the previous analyses, a reduced dataset was used that lacked incompatible and recombinant sites to meet the model assumptions. The backward migration matrix was set up by averaging the backward migration rate estimates obtained from the preceding IM simulations. Genealogical analyses for each species were estimated using 4,000,000 steps per coalescent simulation. For the most likely tree topology two independent runs using different starting seeds were executed, both resulting in the same topology.
Distribution of phenotypic features among populations
To evaluate the distribution of morphological and macrochemical features of the basidiomata of the populations used in this study, we analysed the macro- and microscopical morphological structures in detail, as well as analysing the macrochemical reactions with KOH following standard procedures. Shape, measurement and colour of the microscopic features were obtained from dried specimens by mounting them in 3% KOH. Measurements of basidiospores (n = 51) are given as length and breadth, where the values in brackets represent the uncommon extremes. Basidiospore length/width quotient (Q), mean and standard deviation values were calculated.
In accordance with article 29 and 30A.2 of the International Code of Botanical Nomenclature, copies of this article are deposited at the following ten botanical and/or publicly accessible libraries: 1) Departamento de Botánica, Instituto de Biología U.N.A.M., Circuito exterior s/n, Cuidad Universitaria, Copilco, Coyoacán, Distrito Federal. C.P. 04510, A.P. 70-233 Mexico; 2) Marian Koshland Bioscience and Natural Resources Library, 2101 Valley Life Sciences Bldg. #6500, University of California, Berkeley, CA 94720-6500, USA; 3) Natural Sciences Library, Box 352900, University of Washington, Seattle, WA 98195, USA; 4) Serial and Electronic Resources, Washington State University Libraries, PC Box 645610, 100 Dairy Rd, Pullman, WA 99164-5610, USA; 5) Acquisitions, The LuEsther T. Mertz Library, The New York Botanical Garden, 2900 Southern Blvd, Bronx, NY 10458, USA; 6) Librarian, Canadian Forest Service, Natural Resources Canada, Pacific Forestry Centre Victoria, 506 West Burnside Road, BC Canada V8Z 1M5; 7) UBC Herbarium, Dept. of Botany, #3529-6270, University Blvd. Vancouver, B. C. Canada V6T 1Z4; 8) Plant Research Library, Wm. Saunders Bldg. #49, C.E.F., Otawa, ON, Canada; 9) Main Library, Herbarium, Royal Botanic Gardens, Kew, Surrey, TW9 3AB, UK; 10) Herbarium MSB, Ludwig-Maximilians-Universität München, Menzinger Straße 67, 80638 München, Germany; and 11) Herbarium Tubingense (TUB), University of Tübingen, Auf der Morgenstelle 1, D-74076 Tübingen, Germany.
Authors' contributions
SG, JA and FO conceived the study design; SG generated the DNA sequences and wrote the manuscript; PS and SG performed population genetics analyses; macro- and microscopical descriptions of North American populations were made by JA; macroscopical descriptions of European populations were carried out by BO; SG studied the European collections microscopically. All authors read and approved the final manuscript.
Supplementary Material
Identity of haplotypes of calochroid taxa inferred from ITS rDNA sequences. Haplotype frequencies are shown in parentheses. In some cases, a single collection can carry two or more different haplotypes.
Population mutation rate, effective sample size (ESS), time of divergence and direction of migration estimates within C. arcuatorum, C. aureofulvus and C. elegantior population samples. The parameters are as follows: θ1, θ2 and θA, are the mean population rates for the Old World, the New World and the ancestral population, respectively; t is the mean time of divergence of populations from a common ancestor; m1 represents the mean number of migrations into the Old World and m2 is the mean number of migrations into the New World. Numbers in parenthesis are standard deviations. Reliable estimation of the highest probability density (HPD) intervals could not be achieved, as the posterior distribution turned out to be incomplete in several cases. All values represent arithmetic means taken from 15 independent runs per species.
Taxonomy. Macroscopical descriptions are based on fresh material, whereas microscopical structures were analysed from dried specimens.
Population samples used in this study and their respective voucher numbers, GenBank numbers, collection date, collection site, host tree(s), collector and determinator. Holotype specimens are marked in bold. Herbarium abbreviations: IB = Herbarium Innsbruck, Austria; JFA = J. F. Ammirati material deposited at the Burke Museum, Herbarium, University of Washington Herbarium, Seattle, USA (WTU), Costa Rican National Biodiversity Institute (INB), and University of Costa Rica, San José (USJ); TUB = Herbarium Tübingense, University of Tübingen; S = Herbarium Stockholm, Sweden.
Contributor Information
Sigisfredo Garnica, Email: sigisfredo.garnica@uni-tuebingen.de.
Philipp Spahn, Email: philipp.spahn@uni-tuebingen.de.
Bernhard Oertel, Email: b.oertel@uni-bonn.de.
Joseph Ammirati, Email: cort@u.washington.edu.
Franz Oberwinkler, Email: franz.oberwinkler@uni-tuebingen.de.
Acknowledgements
The authors are grateful to Sabine Silberhorn for technical assistance with lab work and Gunther Saar for providing dried specimens. Special thanks to Regine Kuhnert-Finkernagel and Ursula Peintner from IB herbarium (Innsbruck Herbarium, Austria) for kindly facilitating use of herbarium specimens, colour slides and Meinhard Moser's original macroscopical descriptions. We thank Estella Leopold and Caroline Stromberg (University of Washington), and Cathy Whitlock (Montana State University) for discussions of plant geography. Editor and two anonymous reviewers contributed important suggestions and guidance. This work was supported by grants from the Deutsche Forschungsgemeinschaft (OB 24/27-1, 29), and funding from the Universidad de Costa Rica, Instituto Nacional de Biodiversidad (INBio), Costa Rica; the New York Botanical Garden (Roy Halling) and the D. E. Stuntz Memorial Foundation, Seattle, Washington.
References
- Kretzer A, Li Y, Szaro T, Bruns TD. Internal transcribed spacer sequences from 38 recognized species of Suillus sensu lato: Phylogenetic and taxonomic implications. Mycologia. 1996;88:776–785. doi: 10.2307/3760972. [DOI] [Google Scholar]
- Martin F, Díez J, Dell B, Delaruelle C. Phylogeography of the ectomycorrhizal Pisolithus species as inferred from nuclear ribosomal DNA ITS sequences. New Phytol. 2002;153:345–357. doi: 10.1046/j.0028-646X.2001.00313.x. [DOI] [Google Scholar]
- den Bakker HC, Zuccarello GC, Kuyper TW, Noordellos ME. Phylogeographic patterns in Leccinum sect. Scabra and the status of the arctic-alpine species L. rotundifoliae. Mycol Res. 2007;111:663–72. doi: 10.1016/j.mycres.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Dentinger BTM, Ammirati JF, Both EE, Desjardin DE, Halling RE, Henkel TW, Moreau P-A, Nagasaw E, Soytong K, Taylor AF, Watling R, Moncalvo JM, McLaughlin DJ. Molecular phylogenetics of porcini mushrooms (Boletus section Boletus) Mol Phylogenet Evol. 2010;57:1276–92. doi: 10.1016/j.ympev.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Rochet J, Moreau P-A, Manzi S, Gardes M. Comparative phylogenies and host specialization in the alder ectomycorrhizal fungi Alnicola, Alpova and Lactarius. BMC Evol Biol. 2011;11:40–53. doi: 10.1186/1471-2148-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling R. Ectomycorrhizae: Co-evolution, significance, and biogeography. Ann Missouri Bot Gard. 2001;88:5–13. doi: 10.2307/2666128. [DOI] [Google Scholar]
- Wu QX, Mueller GM, Lutzoni FM, Huang YQ, Guo SY. Phylogenetic and biogeographic relationships of Eastern Asian and Eastern North American disjunct Suillus species (Fungi) as inferred from nuclear ribosomal RNA ITS sequences. Mol Phylogenet Evol. 2000;17:27–37. doi: 10.1006/mpev.2000.0812. [DOI] [PubMed] [Google Scholar]
- Chapela IH, Garbelotto M. Phylogeography and evolution in matsutake and close allies inferred by analyses of ITS sequences and AFLPs. Mycologia. 2004;96:730–741. doi: 10.2307/3762107. [DOI] [PubMed] [Google Scholar]
- Geml J, Laursen GA, O'Neill K, Nusbaum HC, Taylor DL. Beringian origins and cryptic speciation events in the fly agaric (Amanita muscaria) Mol Ecol. 2006;15:225–239. doi: 10.1111/j.1365-294X.2005.02799.x. [DOI] [PubMed] [Google Scholar]
- Hosaka K, Castellano MA, Spatafora JW. Biogeography of Hysterangiales (Phallomycetidae, Basidiomycota) Mycol Res. 2008;112:449–462. doi: 10.1016/j.mycres.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Geml J, Tulloss RE, Laursen GA, Sazanova NA, Taylor DL. Evidence for strong inter- and intracontinental phylogeographic structure in Amanita muscaria, a wind-dispersed ectomycorrhizal basidiomycete. Mol Phylogenet Evol. 2008;48:694–701. doi: 10.1016/j.ympev.2008.04.029. [DOI] [PubMed] [Google Scholar]
- Halling RE, Osmundson TW, Neves M-A. Pacific boletes: Implications for biogeographic relationships. Mycol Res. 2008;112:437–447. doi: 10.1016/j.mycres.2007.11.021. [DOI] [PubMed] [Google Scholar]
- Matheny PB, Aime MC, Bougher NL, Buyck B, Desjardin DE, Horak E, Kropp BR, Lodge DJ, Trappe JM, Hibbett DS. Out of the palaeotropics? Historical biogeography and diversification of the cosmopolitan mushroom family Inocybaceae. J Biogeogr. 2009;36:577592. [Google Scholar]
- Garnica S, Weiß M, Oertel B, Ammirati J, Oberwinkler F. Phylogenetic relationships in Cortinarius, section Calochroi, inferred from nuclear DNA sequences. BMC Evol Biol. 2009;9:1. doi: 10.1186/1471-2148-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman CH. Cortinarius Fries. North American Flora. 1932;10:282–348. [Google Scholar]
- Smith AH. Studies in the genus Cortinarius I. Contributions from the University of Michigan Herbarium. 1939;2:1–42. [Google Scholar]
- Moser M, McKnight KH, Ammirati JF. Studies on North American cortinarii I. New and interesting taxa from the Greater Yellowstone area. Mycotaxon. 1995;55:301–346. [Google Scholar]
- Moser M, Ammirati JF. Studies on North American cortinarii IV. New and interesting Cortinarius species (subgenus Phlegmacium) from oak forests in Northern California. Sydowia. 1997;49:25–48. [Google Scholar]
- Moser M. Die Gattung Phlegmacium (Schleimköpfe) Bad Heilbrunn, Germany: Julius Klinkhardt; 1960. [Google Scholar]
- Moser M, McKnight KH, Sigl M. In: Plant and their Environments. Proceedings of the First Biennial Scientific Conference on the Greater Yellowstone Ecosystem. Despain DG, editor. 1994. The genus Cortinarius (Agaricales) in the Greater Yellowstone area. Mycorrhizal host associations and taxonomic considerations; pp. 239–246. [Google Scholar]
- Moser M, Ammirati JF. Studies in North American cortinarii VI. New and interesting taxa in subgenus Phlegmacium from the pacific states of North America. Mycotaxon. 2000;74:1–36. [Google Scholar]
- Moser MM. Some aspects of Cortinarius associated with Alnus. Journal des JEC. 2001;4:47–76. [Google Scholar]
- Soop K, Knuntsson T. Un nuveau cortinaire de la section Calochroi. Journal des JEC. 2005;7:51–55. [Google Scholar]
- Frøslev TG, Jeppesen TS, Laessøe T. Seven new calochroid and fulvoid species of Cortinarius. Mycol Res. 2006;110:1046–1058. doi: 10.1016/j.mycres.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Frøslev TG. PhD thesis. University of Copenhagen, Faculty of Science; 2007. Systematics of Cortinarius with emphasis on section Calochroi. [Google Scholar]
- Frøslev TG, Jeppesen TS, Laessøe T, Kjøller R. Molecular phylogenetics and delimitation of species in Cortinarius section Calochroi (Basidiomycota, Agaricales) in Europe. Mol Phylogenet Evol. 2007;44:217–227. doi: 10.1016/j.ympev.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Nordborg M. Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nat Rev Genet. 2002;3:380–390. doi: 10.1038/nrg795. [DOI] [PubMed] [Google Scholar]
- Manos PS, Stanford AM. The historical biogeography of Fagaceae: tracking the tertiary history of temperate and subtropical forests of the Northern Hemisphere. Intl J Plant Sci. 2001;162:S77–S93. doi: 10.1086/323280. [DOI] [Google Scholar]
- Denk T, Grimsson F, Zetter R. Episodic migration of oaks to Iceland: Evidence for a North Atlantic "land bridge" in the latest Miocene. Am J Bot. 2010;97:276–287. doi: 10.3732/ajb.0900195. [DOI] [PubMed] [Google Scholar]
- Milne RI. Northern hemisphere plant disjunctions: a window on tertiary land bridges and climatic change? Ann Bot - London. 2006;98:465–472. doi: 10.1093/aob/mcl148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitzendanner MA, Strenge DD, Soltis PS. Chloroplast DNA intraspecific phylogeography of plants from the Pacific Northwest of America. Plant Syst Evol. 1997;206:353–373. doi: 10.1007/BF00987957. [DOI] [Google Scholar]
- Williams JW, Shuman BN, Webb T III, Bartlein PJ, Leduc PL. Late-quaternary vegetation dynamics in North America: scaling from taxa to biomes. Ecol Monogr. 2004;74:309–334. doi: 10.1890/02-4045. [DOI] [Google Scholar]
- Hermann RK. In: Forest Research Laboratory Special Publication 2b. Corvallis, OR, editor. Oregon State University, College of Forestry; 1985. The genus Pseudotsuga: Ancestral History and Past Distribution. [Google Scholar]
- Brubaker LB, Anderson PM, Edwards ME, Lozhkin AV. Beringia as a glacial refugium for boreal trees and shrubs: new perspectives from mapped pollen data. J Biogeogr. 2005;32:833–848. doi: 10.1111/j.1365-2699.2004.01203.x. [DOI] [Google Scholar]
- Ran J-H, Wie X-X, Wang X-Q. Molecular phylogeny and biogeography of Picea (Pinaceae): Implications for phylogeographical studies using cytoplasmic haplotypes. Mol Phylogenet Evol. 2006;41:405–419. doi: 10.1016/j.ympev.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns D. ITS primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- O'Donnell K. In: The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics. Reynolds DR, Taylor JW, editor. Wallingford, UK: CAB International; 1993. Fusarium and its near relatives; pp. 225–233. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. In: PCR Protocols: A Guide to Methods and Amplifications. Innis MA, Gelfand H, Sninsky JS, White TJ, editor. New York: Academic Press; 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics; pp. 315–322. [Google Scholar]
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnica S, Weiß M, Oertel B, Oberwinkler F. Phylogenetic relationships of European Phlegmacium species (Cortinarius, Agaricales) Mycologia. 2003;95:1155–1170. doi: 10.2307/3761917. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Page RDM. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4.0b10. Sunderland, Massachusetts: Sinauer Associates; 2002. [Google Scholar]
- Clark AG. Inference of haplotypes from PCR-amplified samples of diploid populations. Mol Biol Evol. 1990;7:111–122. doi: 10.1093/oxfordjournals.molbev.a040591. [DOI] [PubMed] [Google Scholar]
- Prince EW, Carbone I. SNAP: workbench management tool for evolutionary population genetic analysis. Bioinformatics. 2005;21:402–404. doi: 10.1093/bioinformatics/bti003. [DOI] [PubMed] [Google Scholar]
- Aylor DL, Price EW, Carbone I. SNAP: combine and map modules for multilocus population genetic analysis. Bioinformatics. 2006;22:1399–1401. doi: 10.1093/bioinformatics/btl136. [DOI] [PubMed] [Google Scholar]
- Carbone I, Liu Y-C, Hillman BI, Milgroom MG. Recombination and migration of Cryphonectria hypovirus 1 as inferred from gene genealogies and the coalescent. Genetics. 2004;166:1611–1629. doi: 10.1534/genetics.166.4.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SR, Griffiths RC. Bounds on the minimum number of recombination events in a sample history. Genetics. 2003;163:375–394. doi: 10.1093/genetics/163.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- SNAP Clade and Matrix, Version 2. http://www.cals.ncsu.edu/plantpath/faculty/carbone/home.html
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li W-H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX. Statistical test of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Mol Biol Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- Simonsen KL, Churchill GA, Aquadro CF. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics. 1995;141:413–429. doi: 10.1093/genetics/141.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths RC, Tavaré S. Ancestral inference in population genetics. Stat Sci. 1994;9:307–319. doi: 10.1214/ss/1177010378. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identity of haplotypes of calochroid taxa inferred from ITS rDNA sequences. Haplotype frequencies are shown in parentheses. In some cases, a single collection can carry two or more different haplotypes.
Population mutation rate, effective sample size (ESS), time of divergence and direction of migration estimates within C. arcuatorum, C. aureofulvus and C. elegantior population samples. The parameters are as follows: θ1, θ2 and θA, are the mean population rates for the Old World, the New World and the ancestral population, respectively; t is the mean time of divergence of populations from a common ancestor; m1 represents the mean number of migrations into the Old World and m2 is the mean number of migrations into the New World. Numbers in parenthesis are standard deviations. Reliable estimation of the highest probability density (HPD) intervals could not be achieved, as the posterior distribution turned out to be incomplete in several cases. All values represent arithmetic means taken from 15 independent runs per species.
Taxonomy. Macroscopical descriptions are based on fresh material, whereas microscopical structures were analysed from dried specimens.
Population samples used in this study and their respective voucher numbers, GenBank numbers, collection date, collection site, host tree(s), collector and determinator. Holotype specimens are marked in bold. Herbarium abbreviations: IB = Herbarium Innsbruck, Austria; JFA = J. F. Ammirati material deposited at the Burke Museum, Herbarium, University of Washington Herbarium, Seattle, USA (WTU), Costa Rican National Biodiversity Institute (INB), and University of Costa Rica, San José (USJ); TUB = Herbarium Tübingense, University of Tübingen; S = Herbarium Stockholm, Sweden.