Abstract
We report the first experimental measurements of Ramachandran Ψ-angle distributions for intrinsically disordered peptides: the N-terminal peptide fragment of tumor suppressor p53 and its P27 mutant form. To provide atomically detailed views of the conformational distributions, we performed classical, explicit-solvent molecular dynamics simulations on the microsecond timescale. Upon binding its partner protein, MDM2, wild-type p53 peptide adopts an α-helical conformation. Mutation of Pro27 to serine results in the highest affinity yet observed for MDM2-binding of the p53 peptide. Both UV resonance Raman spectroscopy (UVRR) and simulations reveal that the P27S mutation decreases the extent of PPII helical content and increases the probability for conformations that are similar to the α-helical MDM2-bound conformation. In addition, UVRR measurements were performed on peptides that were isotopically labeled at the Leu26 residue preceding the Pro27 in order to determine the conformational distributions of Leu26 in the wild-type and mutant peptides. The UVRR and simulation results are in quantitative agreement in terms of the change in the population of non-PPII conformations involving Leu26 upon mutation of Pro27 to serine. Finally, our simulations reveal that the MDM2-bound conformation of the peptide is significantly populated in both the wild-type and mutant isolated peptide ensembles in their unbound states, suggesting that MDM2 binding of the p53 peptides may involve conformational selection.
1. Introduction
Intrinsically disordered proteins, which adopt heterogeneous ensembles of conformations under physiological conditions,1,2 comprise at least a third of some eukaryotic genomes.3 These proteins form a structural continuum that ranges from extended, unstructured states, to molten globules (with secondary structure, but no specific tertiary structure), and finally, to mostly folded proteins with flexible or disordered regions.4 Many of these proteins adopt distinct conformations only upon binding to their partner proteins, suggesting a new paradigm for protein-protein recognition.4 A classic example is the intrinsically disordered N-terminal peptide fragment of tumor suppressor protein p53 (residues 17–29), which adopts an α-helical conformation upon binding to the MDM2 oncoprotein.5 NMR studies show that this peptide is at least partially preorganized in its unbound state for binding to MDM2.6 In particular, mutation of Pro27 to a serine further preorganizes the peptide by increasing its α-helical content; as a result, the mutant peptide has the highest MDM2 affinity observed (Kd = 47 nM).6 Not surprisingly, this P27S mutation results in significant conformational changes for the Leu26 peptide bond that precedes the proline.6
Although numerous NMR studies have provided high-resolution structural information about the N-terminal domain of p53,6–11 a detailed view of its conformational diversity in the unbound state has been lacking. An incisive approach for determining the conformational diversity of the peptide is UV resonance Raman (UVRR) spectroscopy, which can be used to determine the distributions of backbone torsional Ψ angles.12–15 In addition, molecular dynamics (MD) simulations can be used to characterize the conformational diversity of the peptide with atomistic detail. However, due to their large computational expense, only short simulations (≤ 150 ns) have been conducted to explore the dynamics of the unbound p53 peptide.16–20
In this work, we use UVRR spectroscopy and microsecond-timescale classical (i.e. molecular mechanics) molecular dynamics (MD) simulations to examine the p53 N-terminal peptide conformational dependence on the P27S mutation. Both UVRR measurements and MD simulations show a decrease in the PPII content and an increase in the non-PPII content upon P27S mutation. In addition, our simulations reveal that the α-helical conformations that are characteristic of the MDM2-bound state are significantly populated in the unbound state of both the wild-type and mutant p53 peptides, suggesting that MDM2 might bind the p53 peptide through conformational selection.
2. Experimental and Computational Methods
2.1. Experimental Methods
Samples
The wild-type p53 peptide fragment (residues 17–29) with the sequence Acetyl-ETFSDLWKLLPEN-NH2 and the mutant P27S peptide with the sequence Acetyl-ETFSDLWKLLSEN-NH2 were synthesized by CHI SCIENTIFIC (≥95% purity). Isotopically labeled peptides with perdeuterated Leu26 were synthesized by AnaSpec (≥ 95% purity). Peptide samples were prepared at 1 mg/ml concentrations (~ 0.6 mM) at pH 7. Samples of Trp and Phe (≥ 95% purity) were purchased from Sigma and prepared at ~0.6 mM concentrations.
CD spectra
CD spectra were measured for the wild-type and P27S mutant peptides by using a Jasco-715 spectropolarimeter with a 0.02 cm path length cuvette. We co-added ten individual spectra.
UVRR spectra
UVRR spectra were measured for both the wild-type and P27S mutant peptides using an spectrometer that was described in detail by Bykov et al.21 Briefly, 204 nm light was utilized to enhance the peptide bond vibrations by exciting within the π → π* electronic transitions of the peptide bonds.22 The 204 nm UV light (2 mW average power, 100 μm diameter spot, 25–40 ns pulse width) was obtained by mixing the 3rd harmonic with the 816 nm fundamental of a 1 kHz repetition rate tunable Ti:Sapphire laser system (DM20-527 TU-L-FHG) from Photonics Industries. The 229 nm UV light (1 mW average power, 100 μm diameter spot size), which was produced by an intracavity frequency doubled Ar+ laser (Coherent, FReD 400) was utilized to enhance the Trp aromatic ring vibrations.23
The sample was circulated in a free surface, temperature-controlled stream. A 165° sampling backscattering geometry was used. The collected light was dispersed by a double monochromator onto a back thinned CCD camera with a Lumogen E coating (Princeton Instruments-Spec 10 System). We utilized 5 min accumulation times, and four accumulations were co-added. Internal standard concentrations of 0.2 M NaClO4 were used for the Raman studies. Raman spectra were normalized to the peak height of the 932 cm−1 ClO4− band.
Distributions of backbone torsional Ψ angles were calculated from 204 nm excited UVRR spectra using the methodology of Mikhonin et al.13–15 (see Fig. S5–S7 and text Supporting Information). Prior to these calculations, Raman bands due to Trp and Phe aromatic rings (Fig. S1) were subtracted from the spectra. UVRR measurements excited by 229 nm light were performed to determine the extent of solvent exposure of the Trp residues in the wild-type and mutant peptides. The differences in solvent exposure between the wild-type and mutant peptides were calculated based on differences in the Raman cross sections of the Trp Raman bands.23
2.2. Computational Methods
MD simulations
To obtain extensive sampling of conformations of the wild-type and P27S mutant p53 peptides, ten 1-μs simulations were performed for each peptide in explicit solvent, with each simulation starting from a different conformation. One of the ten simulations was started from the MDM2-bound conformation of the peptide; each of the remaining nine simulations was started from a different random coil conformation. Heavy-atom coordinates of the bound conformations were taken from the crystal structure of the MDM2-p53 peptide complex;5 heavy-atom coordinates of the random coil conformations were generated using the Sosnick group’s unfolded state web server (http://godzilla.uchicago.edu/cgi-bin/unfolded.cgi)24 in conjunction with the SCWRL3.0 side-chain prediction program.25 Consistent with the peptides synthesized for the experiments (see above), each random coil conformation was capped with acetyl and amino groups at the N terminus and C terminus, respectively. Hydrogen atoms were added using ionization states present in neutral solution. Each model was solvated in dodecahedral boxes of TIP3P water26 (total of 3498 molecules) with a minimum solutewall distance of 12 Å, then charge-neutralized by adding two Na+ counterions.
MD simulations were performed using the GROMACS 4.0 software package27 and the OPLS-AA/L force field28 in the NPT ensemble (constant number of atoms, pressure, and temperature). The temperature was maintained at 30 °C using the Nose Hoover thermostat29,30 and the pressure was maintained at 1 atm using the Parrinello-Rahman barostat31 with time constants for coupling set to 0.5 and 4 ps, respectively. Van der Waals interactions were switched off smoothly between 8 and 9 Å; a long-range analytical dispersion correction was applied to the energy and pressure to account for the truncation of these interactions.32 Real-space electrostatic interactions were truncated at 10 Å while the long-range components of these interactions were calculated using particle mesh Ewald (PME)33 and periodic boundary conditions. To enable a 2-fs time step, bonds to hydrogen were constrained to their equilibrium lengths with the LINCS algorithm.34
To relieve unfavorable interactions, each model was subjected to energy minimization followed by a two-stage equilibration with harmonic position restraints on all non-hydrogen atoms of the peptide. During the first stage, the energy-minimized system was equilibrated for 20 ps at constant temperature (30 °C) and volume. During the second stage, the system was equilibrated for 2 ns at constant temperature (30 °C) and pressure (1 atm). After equilibration, fully unrestrained production simulations were carried out for 1 μs at 30 °C and 1 atm. Each 1-μs simulation required a month of calendar time using 16 CPUs in parallel on two quad-core 2.66GHz Xeon nodes of a Linux cluster at the University of Pittsburgh’s Center for Molecular and Materials Simulations. To avoid bias towards the starting conformation, only the latter 900 ns of each simulation was used for subsequent analysis, using conformations sampled every 100 ps. Based on the distribution of alternate conformations, the ensembles of simulations for each peptide are converged (see Ramachandran plots in Fig. S2).
Kinetic clustering of peptide conformations
The conformations of the wild-type peptide ensemble were clustered based on kinetic similarity using the MSMBuilder software package.35 In short, conformations are grouped according to geometric (and assumed kinetic) similarity into a large number of microstates, and these microstates are in turn grouped by kinetic similarity into a small number of macrostates. The matrix of transition probabilities between these macrostates is then capable of describing the long-timescale kinetics of the system, assuming that the transitions between macrostates are Markovian on some sufficiently long timescale (the Markov time of the system).36–38 In our case, all 90,000 conformations from the unbound wild-type peptide ensemble were clustered based on backbone RMSD into 500 microstates (average microstate radius of 1.8 Å). These 500 microstates were then grouped into four macrostates which showed Markovian behavior on timescales longer than 50 ns (see Fig. S3). The conformations from the P27S mutant peptide ensemble were then assigned to these same macrostates by geometric similarity to the microstates determined above. The resulting macrostate populations were then examined to determine the population shift induced by mutation. Populations of each macrostate for the wild-type and mutant peptides were determined by Monte Carlo sampling of the macrostate-to-macrostate transition matrices for the wildtype and mutant peptide ensembles.37
Calculation of NMR chemical shifts and J-coupling constants
Chemical shifts of α- and amide protons were computed for each conformation in the peptide ensemble using the SHIFTX program,39 then averaged for comparison to the experimentally measured values. J-coupling constants between α and amide protons (3JαN) were computed for each conformation in the peptide ensemble, then averaged for comparison to the experimentally measured values. These constants were computed using a version of the Karplus equation that has been used for the analysis of peptides: 3JαN = A cos2 (θ − 60°) + B cos(θ − 60°) + C where with A = 6.51, B = −1.76, C = 1.60,40 and θ is the dihedral angle between the protons that are separated by three bonds. In this case, the dihedral angle is the backbone torsional angle Φ.
Normal mode calculations
Normal mode calculations of various peptides in the gas phase were carried out using the Gaussian’03 Suite of programs.41 We optimized the geometry and calculated the vibrational frequencies of the peptides using density functional theory (DFT)42–44 with the B3LYP functional45–47 and 6–311+G** basis set. (The same level of theory and basis set was used in a previous study to calculate the dependence of the AmII p band on peptide conformation.48) Normal mode composition analysis was done by calculating the potential energy distribution using the GAR2PED Gaussian output processing utility written by J.M.L. Martin and C. Van Alsenoy.49
3. Results and Discussion
3.1. Temperature dependence of peptide conformations
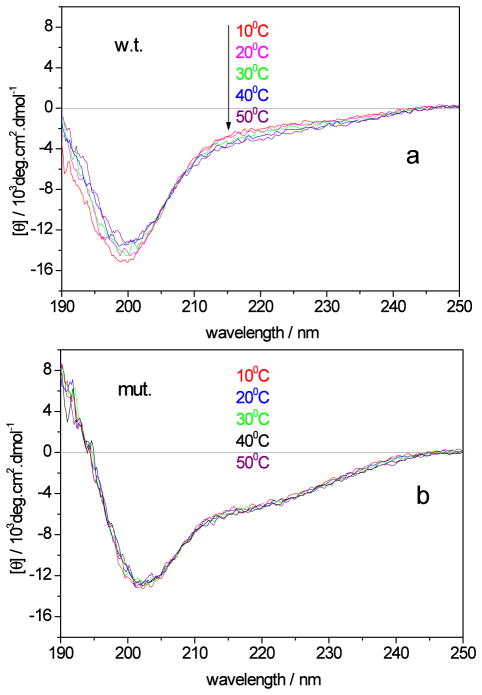
To examine the temperature dependence of secondary structure in the wild-type and mutant p53 peptides, we performed both CD spectroscopy and UVRR measurements at temperatures ranging from 10 to 50 °C. Fig. 1a shows the temperature dependence of the CD spectra of the wild-type peptide. At 10 °C, the CD spectrum (red) shows a strong negative band at 200 nm with a negative shoulder at ~220 nm. As the temperature increases, the 200 nm band slightly decreases in amplitude, while the ~220 nm band becomes slightly more negative. These CD spectra indicate that the wild-type peptide mainly adopts extended conformations.50–52 Fig. 1b shows the temperature dependence of the CD spectra of the P27S mutant peptide. At 10 °C, the CD spectrum (red) shows a trough at 203 nm, and a more negative shoulder at ~220 nm than that of the wild-type peptide. This indicates an increased α-helix content in the mutant peptide, consistent with a previous study.6 As the temperature increases, the mutant peptide CD spectrum does not change, indicating that the increased α-helix conformation does not melt below 50 °C.
Figure 1.
Temperature dependence of the CD spectra of a) the p53 wild-type peptide, and b) the mutant peptide.
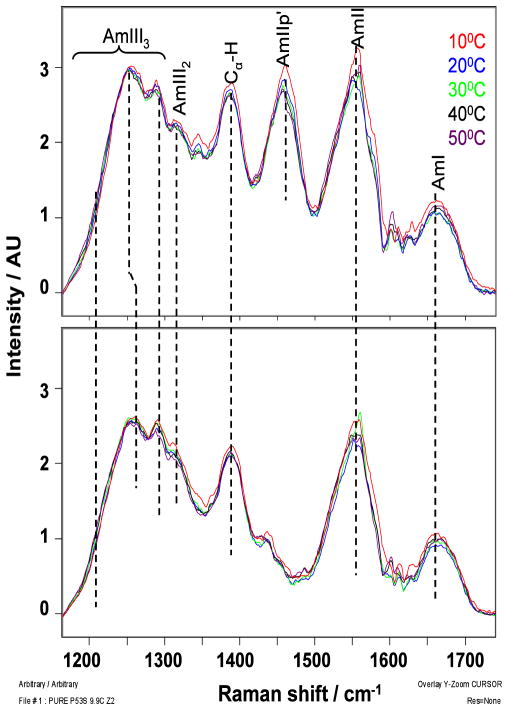
Fig. 2 shows the temperature dependence of the 204 nm excited UVRR spectra of the wild-type and mutant peptides. The AmI band (~1660 cm−1) arises from vibration consisting mainly of C=O stretching. The AmII band (~1550 cm−1) derives from out-of-phase motion involving C-N stretching and N-H bending. The AmII′p band (~1455 cm−1) results from the C-N stretching mode of Pro27. The Cα-H bending band (~1390 cm−1) derives from a Cα-H bending vibration that is resonance enhanced because of its coupling to N-H bending and C-N stretching.12 The intensity of the Cα-H bending band will increase as the population of non-helical conformations increases.14 The bands in the AmIII region arise from vibrations which involve in-phase contributions of C-N stretching and N-H bending. The AmIII3 band frequency depends upon the Ramachandran Ψ angle due to the Ψ-angle dependent coupling between N-H bending and the (C)Cα-H bending motions.12,14
Figure 2.
Temperature dependence of the 204 nm excited UVRR spectra of the p53 wild-type peptide (top) and the mutant peptide (bottom); contributions from Trp and Phe aromatic rings (Fig. S1) have been removed.
The AmIII3 region of the wild-type peptide shows bands at ~1250 cm−1 and ~1290 cm−1, with a shoulder at ~1200 cm−1. Previous studies showed that an AmIII3 band at ~1248 cm−1 indicates a PPII-like conformation, while AmIII3 bands at ~1290 cm−1 and ~1200 cm−1 indicate different turn conformations.25 As the temperature increases, the ~1250 cm−1 AmIII3 band downshifts very slightly while its intensity slightly decreases; the Cα-H bending band intensity also decreases with increasing temperature. Such temperature-induced Raman spectral changes are not due to conformational changes; they result from the temperature dependence of the hydrogen bonding between water and the peptide bond amide nitrogen.13 The lack of additional temperature-induced spectral changes indicates a temperature independent conformational distribution.
The bands in the AmIII3 region of the mutant peptide (~1255 cm−1 and ~1290 cm− 1, with a shoulder at ~1200 cm−1) indicate that its conformational distribution is also temperature independent. The AmIII3 and the Cα-H band intensities of the mutant peptide are slightly decreased relative to that of the wild-type peptide. This Raman intensity hypochromism indicates a greater population of α-helical conformations for the mutant peptide.22 We calculated the non-PPII fractional increase of the mutant peptide by using the methodology of Xiong et al.53,54 (see Supporting Information). The calculated increase in the non-PPII content of the mutant peptide is 0.20 ± 0.02.
3.2. Conformational diversity of peptides
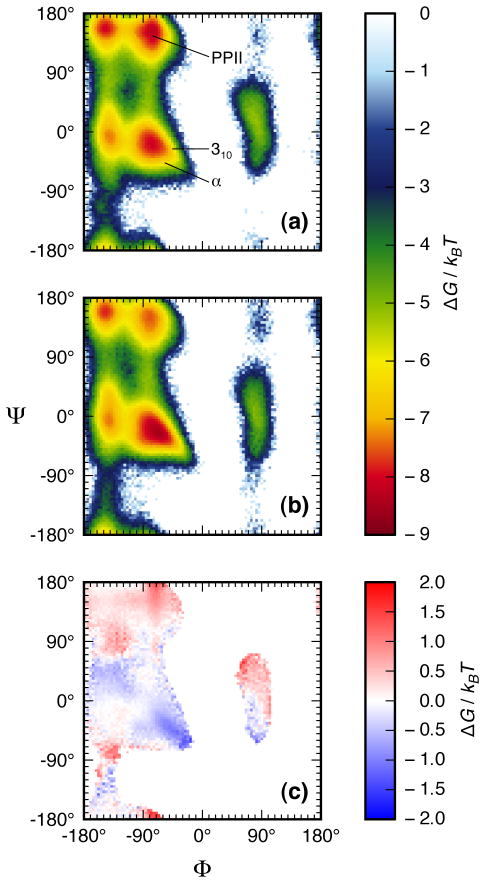
As shown by Ramanchandran plots that were computed from our simulations (Fig. 3), the mutant P27S peptide ensemble has a decreased population of PPII-like conformations and an increased population of non-PPII conformations (which would include the MDM2-bound α-helical peptide conformation) relative to the wild-type peptide ensemble. Ψ-angle distributions were computed from the UVRR spectra (Fig. 2), allowing direct comparison of the UVRR and simulation results.
Figure 3.
Ramachandran plots computed from explicit solvent MD simulations for a) the wild-type p53 peptide ensemble and b) the P27S mutant p53 peptide ensemble. A difference plot of the b) relative to a) is presented in c).
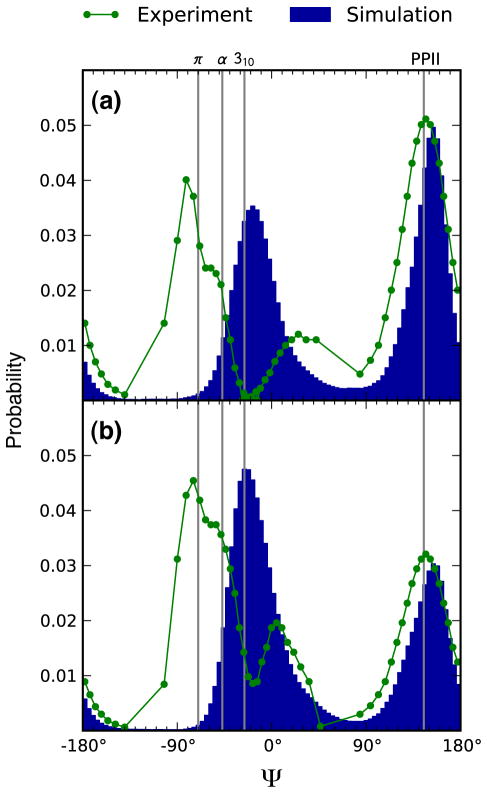
Based on the UVRR spectra, the wild-type peptide Ψ-distribution (Fig. 4a) shows a broad PPII-like Ψ angle region with an average Ψ angle of ~145°. The PPII-like conformations dominate, with a negligible α-helix contribution. The probability distribution also contains Type I′ or Type III′ β turn regions with Ψ angles centered at ~30°, a γ-turn region with Ψ angles centered at ~ −60°, and a Type V β turn region with Ψ angles centered at −80°. The mutant peptide Ψ-angle distribution (Fig. 4b) shows a decreased PPII contribution and a significantly increased α-helix-like conformation contribution. The contribution of Type I/I′ or Type II/II′ β turn regions with Ψ angles centered at ~0° significantly increases. The Type V β turn contribution also increases.
Figure 4.
Distributions of backbone Ψ angles for the a) wild-type and b) P27S mutant peptides at 30 °C determined from explicit solvent MD simulations (blue) and UVRR spectroscopy (green). Source data for these plots are provided in Supporting Information (Data S1–S4).
A comparison of the Ψ-angle distributions from simulations with those calculated from the UVRR spectra reveals that the relative populations of PPII and non-PPII conformations (including α-helix and 310 helix conformations) in the wild-type and mutant peptide ensembles are in excellent agreement (Fig 4). Integration of the Ψ-angle distributions in the non-PPII region (between −140° and 84°) gives non-PPII populations of 0.38 and 0.46 for the wild-type experimental and simulation Ψ-angle distributions, respectively. Conversely, the non-PPII populations of the P27S peptide are 0.60 and 0.61 for Ψ-angle distributions from experiment and simulation, respectively. In contrast to a recent simulation study, which revealed little or no formation of 310 helices for helical peptides,55 the 310 helix was the dominant non-PPII conformation in our simulations involving the p53 peptides. Given that the dominant non-PPII conformation in the UVRR experiments is the α-helix, exhibiting i+4 → i instead of the i+3 → i hydrogen bonding of a 310 helix, our simulation result may be an artifact of the OPLS-AA/L force field.28
We also computed chemical shifts and J-coupling constants using our simulations and compared these values to those measured by NMR experiments.6 The computed chemical shifts were within experimental error. In particular, the relative RMS deviations between computed and experimentally measured chemical shifts (in reference to the experimental value) are 0.084 and 0.069 ppm for the α- and amide protons of the wild-type peptide and 0.086 and 0.070 ppm for the α- and amide protons of the mutant P27S peptide, respectively (Tables S1 and S2). The J-coupling constants that were computed from our simulations are those between the Cα and amide protons (3JαN). We were unable to compare our results for the wild-type peptide to experimental results, because the 3JαN-coupling constants for most of the residues in the wild-type p53 peptide have not been determined due to spectral overlap (Table S3). For the mutant peptide, no significant correlation (R2 = 0.12) was found between the computed and experimental6 J-couplings (Table S4). It is therefore likely that our simulations do not provide sufficient conformational sampling to reproduce the experimental J-couplings, which report on averages over timescales that may extend into the millisecond range.56
3.3. Does binding occur by conformational selection?
A potential mechanism for the binding of the p53 peptide to the MDM2 oncoprotein is conformational selection57, where the MDM2-bound conformation of the p53 peptide is already significantly populated in the unbound state, interconverting with alternate conformations; the presence of MDM2 then selects for the bound conformation by stabilizing this conformation over all other conformations through binding.
To explore the potential for conformational selection, we determined the population of the bound conformation in the unbound state of the p53 peptide from the kinetic clustering model described above. In the wild-type peptide ensemble, the bound state has the highest population (48 ± 15%) among the four kinetically distinct states (macrostates); as expected, this population is even greater in the mutant peptide ensemble (89 ± 6%). This supports the notion that the P27S mutation improves binding of p53 to MDM2 by allowing the unbound peptide to access its bound conformation more readily. We note that the p53 peptide may also bind MDM2 through the “fly-casting” mechanism, contacting its MDM2 partner at a distance, then folding as it reels in its MDM2 partner; this mechanism has been proposed as a general explanation for a kinetic advantage to being intrinsically disordered vs. folded.58 Our results do not rule out the possibility of fly casting; they simply provide support for conformational selection as a viable mechanism of MDM2-p53 binding.
3.4. Conformational diversity of a selected peptide residue: Leu26
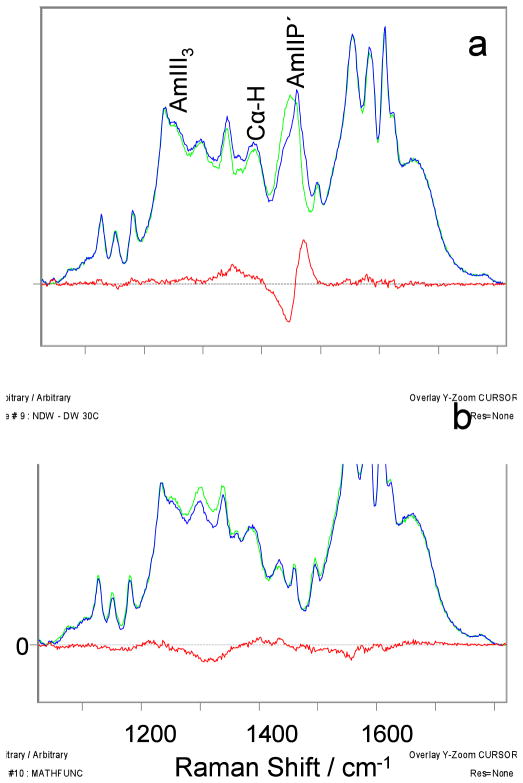
As mentioned above, mutation of Pro27 to serine results in significant conformational changes in the preceding residue, Leu26.6 The conformation of the Leu26 peptide bond can be highlighted by perdeuterating the Leu26 of the p53 wild-type and mutant peptides and then measuring UVRR spectra of the resulting isotope labeled peptides.59,60 Fig. 5 shows the 204 nm excited UVRR spectra of the perdeuterated and non-deuterated Leu26 of the wild-type and mutant p53 peptides and their difference spectra. The difference spectrum (Fig. 5a) shows that perdeuterating the Leu residue preceding Pro27 significantly downshifts the AmII′p band as shown by the ~1445 cm−1 trough and the ~1472 cm−1 peak. This 27 cm−1 AmII′p downshift must result from an AmII′p normal mode compositional change since no conformational change will occur upon isotopic substitution.
Figure 5.
204 nm excited UVRR spectra of Leu26 perdeuterated and non-deuterated p53 a) wild-type peptide, and b) P27S mutant peptide, and their difference spectra (red) at 30 °C. All spectra were normalized to the intensity of the AmI band.
Quantum mechanical calculations were therefore performed to calculate the normal modes of Acetyl-LP-NH2 and its perdeuterated leucine derivative. In the non-deuterated peptide the AmII′p vibration arises mainly from C-N stretching (28%) coupled out-of-phase to Leu Cα-H in-plane bending (22%) (Table 1); Cα-C stretching (10%) and C=O in-plane bending (7%) contribute less. Perdeuteration of Leu26 eliminates the Cα-H bending contribution to the AmII′p normal mode and increases the C-N stretching contribution to 46%. Additionally, the perdeuterated Leu-Pro AmII′p vibration acquires small contributions from C-N stretching (8%) and deformation (6%) of the proline ring. The Cα-C and C=O stretching contributions are preserved. These AmII′p normal mode composition changes give rise to a calculated 19 cm−1 downshift compared to that of the non-perdeuterated peptide. A similar calculation for Acetyl-AP-NH2 and its perdeuterated Ala derivative shows similar normal mode composition changes (Table 1) that result in a 17 cm−1 AmII′p frequency downshift.
Table 1.
Calculated amide II′p frequencies and potential energy distributions for Acetyl-L-P-NH2 and its perdeuterated lecucine isotopomer; Acetyl-A-P-NH2 and its perdeuterated alanine isotopomer.
| Freq. (cm−1)* | Potential energy distributions (>5%) | |
|---|---|---|
| Acetyl-L-P-NH2 | 1444 | C-N s (28) -Cα-H inp b (22) -Cα-C s (10) -C=O inp b (7) |
| Acetyl-L(D)-P-NH2 | 1425 | C-N s (46) -Cα-C s (9) -C=O inp b (9) –C-N s (in Pro ring) (8) Pro ring def (6) |
| Acetyl-A-P-NH2 | 1445 | C-N s (31) -Cα-H inp b (18) –Cα-C s (11) -C=O inp b (7) - C=O inp b (6) |
| Acetyl-A(D)-P-NH2 | 1428 | C-N s (41) -Cα-C s (10) -C=O inp b (8) C-H inp b (Pro) (7) -C-N s (Pro ring) (7) Pro ring def (5) |
Our calculated AmII′p frequencies are about 15 cm−1 lower than the Fig. 1 measured values because we did not consider the effect of hydrogen bonding or the impact of the solvent dielectric constant; we previously showed that a 25 cm−1 upshift in the AmII′p frequency will occur due to hydrogen bonding.48
The Cα-H bending band intensity of the perdeuterated Leu26 p53 mutant peptide slightly decreases relative to that of the natural abundance mutant (Fig. 5b), indicating that the Leu26-Ser27 mutant peptide bond possesses a weak Cα-H bending band intensity. By using the same methodology for calculating the non-PPII fractional increases of the mutant peptide above, we calculated the non-PPII content of this bond. The calculated non-PPII content of Leu26 is 0.72 ± 0.11.
Based on our MD simulations, the calculated non-PPII content (including α-helical conformations) of Leu26 is 0.63, which is in quantitative agreement with experiment. These results are consistent with the expectation that the Leu26-Ser27 peptide bond becomes more α-helical upon introducing the P27S mutation due to the fact that Pro is known to disrupt formation of α helices.
The P27S difference spectrum also shows a negative band centered at 1320 cm−1 and only small difference features between 1200 cm−1 and 1270 cm−1. Previous studies indicate that the difference spectrum for the PPII conformation between hydrogenated (Cα-H) versus deuterated (Cα-D) of a mainly polyalanine peptide shows a strong positive band at 1248 cm−1 and a strong negative band at 1326 cm−1. In contrast, the difference spectrum for the α-helical conformation shows only a weak positive band at 1258 cm−1 and a weak negative band at 1287 cm−1 (Fig. S2). The lack of a strong positive feature at 1248 cm−1 in the P27S difference spectrum indicates little, if any PPII content. The relatively flat region between 1200 cm−1 and 1270 cm−1 in Fig. 5b might result from a broad distribution of conformations with Ψ angles around the α-helix and turn peptide bond conformation that would not show AmIII3 intensities between 1200–1250 cm−1. The 1320 cm−1 trough results from the Cα-D deuterated species bands that show up in this region.12,61 It is important to note that the Leu26-Ser27 mutant peptide bond, even if 100% α-helix will not show the Fig. S4 difference spectrum (that derives from a long α-helical peptide with its intramolecular hydrogen bonding).
3.5. Solvent exposure of the Trp residue
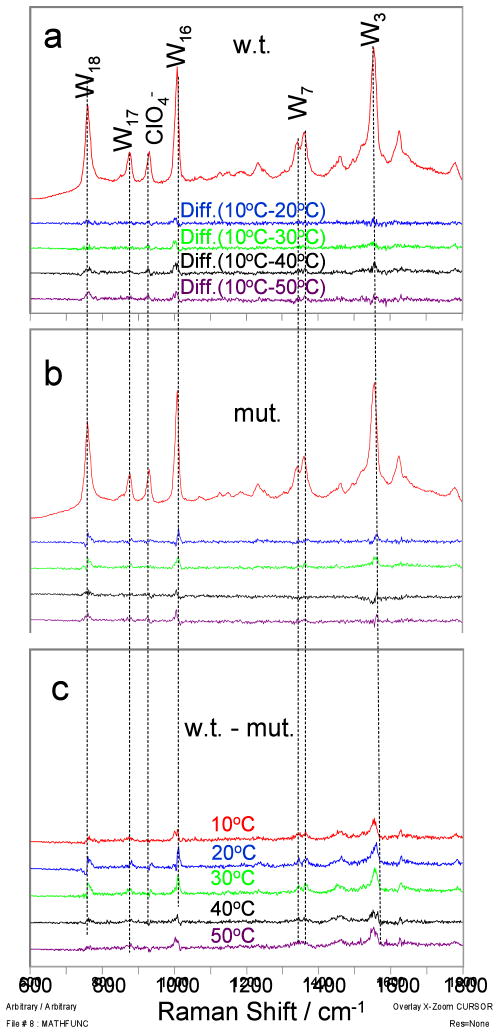
We measured UVRR excited by 229 nm light to determine the difference in solvent exposure of the Trp residue (Trp23) in the wild-type and mutant p53 peptides. The resulting UVRR spectra (Fig. 6) are dominated by the in-plane Trp aromatic ring vibrations at 759 cm−1 (W18), 876 cm−1 (W17), 1008 cm−1 (W16), 1341 cm−1 (W7 2), 1361 cm−1 (W7 1) and 1555 cm−1 (W3). Previous studies showed that the 229 nm Trp band intensities increase as the Trp residue becomes less exposed to solvent.23 The Trp band intensities of the wild-type and mutant peptides are essentially temperature independent, indicating temperature independent solvent exposure of the Trp in both peptides. The difference spectra (Fig. 6c) between the wild-type and mutant peptides show that the peptides have similar Trp band intensities [wild-type average intensities were only 10 ± 6% (deviation between intensities) greater than the mutant intensities], suggesting that the Trp residue is similarly exposed to solvent in both peptides. Consistent with experiment, our simulations resulted in an insignificant difference (29 ± 64 Å2; uncertainty is one standard deviation) in the average solvent accessible surface areas of the Trp residue in the wild-type and mutant peptides.
Figure 6.
229 nm excited UVRR spectra of the p53 wild-type and mutant peptides. a) 10 °C spectra of the wild-type peptide and difference spectra between the 10 °C spectrum and that at higher temperature, b) temperature difference spectra of the mutant peptide; c) difference spectra between wild-type and mutant peptide. All spectra were normalized to the peak height of the 932 cm−1 ClO4− band.
4. Conclusions
To our knowledge, we have reported the first experimentally measured distributions of Ramachandran Ψ angles for intrinsically disordered peptides. These measurements were performed on an N-terminal p53 peptide and its P27S mutant form using UVRR. To provide atomically detailed views of these conformational distributions, we also performed explicit solvent MD simulations on the microsecond timescale. Based on the Ψ-angle distributions determined from both UVRR and simulations, PPII-like conformations were found to dominate the wild-type peptide ensemble and to significantly decrease in population in the mutant peptide ensemble. For each peptide, the relative populations of PPII-like conformations to non-PPII conformations from experiment and simulations are in excellent agreement. We also determined the Ψ-angle distributions for the residue preceding the proline residue in both peptides (Leu26) using isotopically labeled UVRR spectroscopy and simulations. Results from experiment and simulation are in quantitative agreement in terms of the non-PPII content of the Leu26- serine peptide bond. Even though the mutant peptide is more preorganized for MDM2 binding, no significant differences in solvent exposure of the Trp residue was found between the wild-type and mutant peptides using either 229 nm excited UVRR measurements or simulations. Finally, our simulations reveal that the MDM2-bound conformation of the peptide is significantly populated in both the wild-type and mutant P27S peptide ensembles and that this population is significantly greater in the mutant peptide ensemble. Thus, mutation of Pro27 to serine preorganizes the peptide for binding, supporting the notion that MDM2 binding of the peptide might involve conformational selection.
Supplementary Material
Acknowledgments
This work was supported by NIH grant 1RO1EB009089 and NSF CAREER Award MCB-0845216. Computational resources were provided by the Center for Simulation and Modeling at the University of Pittsburgh.
Footnotes
Supporting Information Available: Methods for determination of Ψ-angle distributions from UVRR spectra; Figures S1–S7; Table S1–S4; Data S1–S4.
References
- 1.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Biochemistry. 2002;41:6573. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 2.Wright PE, Dyson HJ. J Mol Biol. 1999;293:321. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 3.Dunker AK, Obradovic Z. Nat Biotechnol. 2001;19:805. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- 4.Dyson HJ, Wright PE. Nat Rev Mol Cell Biol. 2005;6:197. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 5.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Science. 1996;274:948. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 6.Zondlo SC, Lee AE, Zondlo NJ. Biochemistry. 2006;45:11945. doi: 10.1021/bi060309g. [DOI] [PubMed] [Google Scholar]
- 7.Botuyan MV, Momand J, Chen Y. Fold Des. 1997;2:331. doi: 10.1016/S1359-0278(97)00047-3. [DOI] [PubMed] [Google Scholar]
- 8.Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, Chang J, Sung YC, Choi KY, Han KH. Journal of Biological Chemistry. 2000;275:29426. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- 9.Bell S, Klein C, Muller L, Hansen S, Buchner J. Journal of Molecular Biology. 2002;322:917. doi: 10.1016/s0022-2836(02)00848-3. [DOI] [PubMed] [Google Scholar]
- 10.Dawson R, Muller L, Dehner A, Klein C, Kessler H, Buchner J. Journal of Molecular Biology. 2003;332:1131. doi: 10.1016/j.jmb.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Vise PD, Baral B, Stancik A, Lowry DF, Daughdrill GW. Proteins. 2007;67:526. doi: 10.1002/prot.21364. [DOI] [PubMed] [Google Scholar]
- 12.Asher SA, Ianoul A, Mix G, Boyden MN, Karnoup A, Diem M, Schweitzer-Stenner R. J Am Chem Soc. 2001;123:11775. doi: 10.1021/ja0039738. [DOI] [PubMed] [Google Scholar]
- 13.Asher SA, Mikhonin AV, Bykov S. J Am Chem Soc. 2004;126:8433. doi: 10.1021/ja049518j. [DOI] [PubMed] [Google Scholar]
- 14.Mikhonin AV, Bykov SV, Myshakina NS, Asher SA. J Phys Chem B. 2006;110:1928. doi: 10.1021/jp054593h. [DOI] [PubMed] [Google Scholar]
- 15.Mikhonin AV, Myshakina NS, Bykov S, Asher SA. J Am Chem Soc. 2005;127:7712. doi: 10.1021/ja044636s. [DOI] [PubMed] [Google Scholar]
- 16.Espinoza-Fonseca LM, Trujillo-Ferrara JG. Biochemical and Biophysical Research Communications. 2006;343:110. doi: 10.1016/j.bbrc.2006.02.129. [DOI] [PubMed] [Google Scholar]
- 17.Chen HF, Luo R. J Am Chem Soc. 2007;129:2930. doi: 10.1021/ja0678774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dastidar SG, Lane DP, Verma CS. J Am Chem Soc. 2008;130:13514. doi: 10.1021/ja804289g. [DOI] [PubMed] [Google Scholar]
- 19.Massova I, Kollman PA. J Am Chem Soc. 1999;121:8133. [Google Scholar]
- 20.Zhong H, Carlson HA. Proteins: Structure, Function, and Bioinformatics. 2005;58:222. doi: 10.1002/prot.20275. [DOI] [PubMed] [Google Scholar]
- 21.Bykov S, Lednev I, Ianoul A, Mikhonin A, Munro C, Asher SA. Applied Spectroscopy. 2005;59:1541. doi: 10.1366/000370205775142511. [DOI] [PubMed] [Google Scholar]
- 22.Sharma B, Bykov SV, Asher SA. Journal of Physical Chemistry B. 2008;112:11762. doi: 10.1021/jp801110q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi ZH, Chen XG, Holtz JSW, Asher SA. Biochemistry. 1998;37:2854. doi: 10.1021/bi971160z. [DOI] [PubMed] [Google Scholar]
- 24.Jha AK, Colubri A, Freed KF, Sosnick TR. Proc Natl Acad Sci USA. 2005;102:13099. doi: 10.1073/pnas.0506078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canutescu AA, Shelenkov AA, Dunbrack JRL. Protein Science. 2003;12:2001. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jorgensen W, Chandrasekhar J, Madura J, Impey R, Klein M. J Chem Phys. 1983;79:926. [Google Scholar]
- 27.Hess B, Kutzner C, van der Spoel D, Lindahl E. J Chem Theory Comput. 2008;4:435. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 28.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. J Phys Chem B. 2001;105:6474. [Google Scholar]
- 29.Nose SJ. J Chem Phys. 1984;81:511. [Google Scholar]
- 30.Hoover WG. Phys Rev A. 1985;31:1695. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 31.Parrinello M, Rahman A. J Appl Phys. 1981;52:7182. [Google Scholar]
- 32.Shirts MR, Pitera JW, Swope WC, Pande VS. J Chem Phys. 2003;119:5740. [Google Scholar]
- 33.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J Chem Phys. 1995;103:8577. [Google Scholar]
- 34.Hess B, Bekker H, Berendsen H, Fraaije J. J Comput Chem. 1997;18:1463. [Google Scholar]
- 35.Bowman GR, Huang X, Pande VS. Methods. 2009;49:197. doi: 10.1016/j.ymeth.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chodera JD, Swope WC, Pitera JW, Dill KA. Multiscale Model Simul. 2006;5:1214. [Google Scholar]
- 37.Noe F. J Chem Phys. 2008;128:244103. doi: 10.1063/1.2916718. [DOI] [PubMed] [Google Scholar]
- 38.Chodera JD, Singhal N, Pande VS, Dill KA, Swope WC. J Chem Phys. 2007;126:155101. doi: 10.1063/1.2714538. [DOI] [PubMed] [Google Scholar]
- 39.Neal S, Nip AM, Zhang HY, Wishart DS. J Biomol NMR. 2003;26:215. doi: 10.1023/a:1023812930288. [DOI] [PubMed] [Google Scholar]
- 40.Vuister GW, Wang AC, Bax A. J Am Chem Soc. 1993;115:5334. [Google Scholar]
- 41.Frisch MJT, GW, Schlegel HB, Scuseria GE, Robb MA, et al. Gaussian 03; Revision C.01 ed. ed. Gaussian, Inc; Wallingford CT: 2004. [Google Scholar]
- 42.Kohn W, Sham LJ. Phys Rev. 1965;140:A1133. [Google Scholar]
- 43.Parr RG, Yang W. Density-functional theory of atoms and molecules. Oxford Univ. Press; Oxford: 1989. [Google Scholar]
- 44.Hohenberg P, Kohn W. Phys Rev. 1964;136:B864. [Google Scholar]
- 45.Becke AD. Journal of Chemical Physics. 1993;98:5648. [Google Scholar]
- 46.Lee C, Yang W, Parr RG. Phys Rev B. 1988;37:785. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 47.Miehlich B, Savin A, Stoll H, Preuss H. Chem Phys Lett. 1989;157:200. [Google Scholar]
- 48.Ahmed Z, Myshakina NS, Asher SA. Journal of Physical Chemistry B. 2009;113:11252. doi: 10.1021/jp809857y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin JML, Van Alsenoy C. GAR2PED. University of Antwerpen; Antwerpen: 1995. [Google Scholar]
- 50.Sreerama N, Woody RW. Biochemistry. 1994;33:10022. doi: 10.1021/bi00199a028. [DOI] [PubMed] [Google Scholar]
- 51.Shi Z, Olson C, Rose G, Baldwin R, Kallenbach N. Proc Natl Acad Sci USA. 2002;99:9190. doi: 10.1073/pnas.112193999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi Z, Chen K, Liu Z, Kallenbach N. CHEMICAL REVIEWS. 2006;106:1877. doi: 10.1021/cr040433a. [DOI] [PubMed] [Google Scholar]
- 53.Xiong K, Asciutto EK, Madura JD, Asher SA. Biochemistry. 2009;48:10818. doi: 10.1021/bi9014709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiong K, Asher SA. Biochemistry. 2010;49:3336. doi: 10.1021/bi100176a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matthes D, de Groot BL. Biophys J. 2009;97:599. doi: 10.1016/j.bpj.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Markwick PR, Showalter SA, Bouvignies G, Bruschweiler R, Blackledge M. J Biolmol NMR. 2009;45:17. doi: 10.1007/s10858-009-9341-z. [DOI] [PubMed] [Google Scholar]
- 57.Pauling L. J Am Chem Soc. 1940;62:2643. [Google Scholar]
- 58.Shoemaker BA, Portman JJ, Wolynes PG. Proc Natl Acad Sci U S A. 2000;97:8868. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ianoul A, Mikhonin A, Lednev IK, Asher SA. Journal of Physical Chemistry A. 2002;106:3621. [Google Scholar]
- 60.Mikhonin AV, Asher SA, Bykov SV, Murza A. Journal of Physical Chemistry B. 2007;111:3280. doi: 10.1021/jp0654009. [DOI] [PubMed] [Google Scholar]
- 61.Oboodi MR, Alva C, Diem M. J Phys Chem. 1984;88:501. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.