Abstract
Retinitis pigmentosa (RP) is a genetically heterogeneous heritable disease characterized by apoptotic death of photoreceptor cells. We used exome sequencing to identify a homozygous Alu insertion in exon 9 of male germ cell-associated kinase (MAK) as the cause of disease in an isolated individual with RP. Screening of 1,798 unrelated RP patients identified 20 additional probands homozygous for this insertion (1.2%). All 21 affected probands are of Jewish ancestry. MAK encodes a kinase involved in the regulation of photoreceptor-connecting cilium length. Immunohistochemistry of human donor tissue revealed that MAK is expressed in the inner segments, cell bodies, and axons of rod and cone photoreceptors. Several isoforms of MAK that result from alternative splicing were identified. Induced pluripotent stem cells were derived from the skin of the proband and a patient with non-MAK–associated RP (RP control). In the RP control individual, we found that a transcript lacking exon 9 was predominant in undifferentiated cells, whereas a transcript bearing exon 9 and a previously unrecognized exon 12 predominated in cells that were differentiated into retinal precursors. However, in the proband with the Alu insertion, the developmental switch to the MAK transcript bearing exons 9 and 12 did not occur. In addition to showing the use of induced pluripotent stem cells to efficiently evaluate the pathogenicity of specific mutations in relatively inaccessible tissues like retina, this study reveals algorithmic and molecular obstacles to the discovery of pathogenic insertions and suggests specific changes in strategy that can be implemented to more fully harness the power of sequencing technologies.
Keywords: disease modeling, next generation sequencing, retinal degeneration, photoreceptor differentiation
Primary cilia are sensory organelles found on the surface of almost all vertebrate cells, and the outer segments of photoreceptor cells are highly modified examples of these organelles (1). Mutations in genes that encode critical components of the outer segment apparatus are common causes of photoreceptor loss (2), and such loss often leads to night blindness, constricted visual fields, intraretinal pigmentation, and electrophysiological abnormalities, which are recognized clinically as retinitis pigmentosa (RP). It is estimated that mutations in more than 100 genes will eventually be shown to cause this phenotype (3, 4). At present, fewer than one-half of these mutations have been identified (4). About 20% of all nonsyndromic RP kindreds display a clearly autosomal recessive mode of inheritance, whereas another 30% consist of isolated cases that are frequently autosomal recessive as well (5).
The majority of sequence variations that are currently known to cause Mendelian disease in humans is within or immediately adjacent to DNA sequences that are ultimately translated into protein (i.e., the exome) (6). For this reason, a number of strategies have been devised for interrogating the exomes of individuals affected with Mendelian disorders in search of disease-causing variations (7, 8). The primary limitations to these approaches include the large amount of background variation in the human genome, the imperfection of current exome capture methods, the relatively high frequency of heterozygous recessive disease-causing variants in outbred populations, and the high cost of validating putative disease-causing mutations in large cohorts of patients and controls. Many of these limitations can be overcome when there are multiple affected individuals in kindred. In such cases, linkage analysis, often coupled with a homozygosity hypothesis, can be used to dramatically reduce the number of variants requiring validation in a disease cohort.
Many physicians envision the day that exomic or even genomic sequence information from individual patients can be used routinely to facilitate their care. However, to achieve this routine use, analytic methods and human polymorphism databases will need to be developed to the degree that disease-causing mutations can be reliably detected without relying on positional information. Identifying additional affected individuals with mutations in the same gene will always strengthen the association of the gene with a given phenotype, but other emerging technologies can also contribute to such associations. For example, skin-derived induced pluripotent stem cells (iPSCs) can be used to investigate the function or dysfunction of a mutant gene product in tissues such as retina that are inaccessible to molecular analysis in living patients (9, 10). In this study, we sequenced the exome of an individual with RP who had no family history of eye disease and only one living sibling, and we identified a likely disease-causing homozygous mutation. We validated this finding in a large cohort of RP patients and then, used skin-derived iPSCs from the proband to investigate the mechanism through which the mutation causes disease.
Results
Next Generation Sequencing Detects an Alu Insertion in Male Germ Cell-Associated Kinase.
Genomic DNA from a patient affected with autosomal recessive RP was fragmented and hybridized to an Agilent exome capture reagent, and the eluted fragments were sequenced on both Illumina and ABI sequencing instruments. The Illumina sequencing experiment yielded 71 million uniquely mapped paired-end sequences, with each end being 50 bp in length, whereas the ABI experiment yielded 157 million uniquely mapped single-end sequences, with each end being 50 bp in length. These sequences were aligned to the reference human genomic sequence (hg19) using BWA (11) and BFAST (12). Departures from the reference were identified with GATK (13). In both experiments, more than 20,000 sequence variations were detected (Table S1). These variants were prioritized using the following criteria: GATK variation quality score greater than 50, presence in coding sequence or within 5 bp of a splice junction, annotated frequency of less than 2% in all available population databases, presence in two or fewer exome sequences from other individuals analyzed by our laboratory, prediction to alter protein structure, and lack of complete genotype sharing with the proband’s unaffected sibling (as determined by genome-wide SNP genotyping) (Table S1). After applying these filters to the data obtained from both sequencing instruments, an average of 417 plausible disease-causing sequence variations, in an average of 405 genes, were identified by each sequencing platform. Approximately 75% of the observed variations were identified by both sequencing instruments. Although there were no instances in which two plausible disease-causing variants were observed in a gene previously associated with RP, there were two genes (ABCA4 and USH2A) that each harbored a single variant previously associated with autosomal recessive RP (Asp1532Asn and Cys759Phe, respectively) (14–16). Sanger sequencing of the entire coding sequence of both of these genes confirmed the presence of the heterozygous variants identified by exome sequencing but failed to detect a second disease-causing variation in either gene. We then turned our attention to the list of genes that harbored two plausible disease-causing variations. The filtered exome sequencing data predicted four genes to have homozygous truncating mutations (i.e., nonsense, frameshift, or splice variants), but none of these genes were confirmed to be homozygous by Sanger sequencing. Similarly, the exome sequencing data predicted four genes to each harbor two different truncating mutations in the compound heterozygous state; however, only one of these genes was confirmed by Sanger sequencing to harbor two different variants, and of these variants, one variant was frequently seen in the normal population. There were six genes predicted to have one truncating and one missense variant, and of these 12 variants, only 5 were present in the data from both sequencers. The ABI dataset contained two mutations in male germ cell-associated kinase (MAK) that were not present in the Illumina dataset: a 2-bp insertion, Lys433 ins2aaaTT, and a missense mutation, Asp435Tyr, both in exon 9. When we amplified MAK exon 9 to confirm these two variants, we were surprised to observe a larger than expected fragment (Fig. 1). Sanger sequencing of this fragment revealed a homozygous insertion of a 353-bp Alu repeat between codons 428 and 429. If translated, this insertion would result in the insertion of 31 incorrect amino acids followed by a premature termination. Careful inspection of the raw sequence data revealed the reason for the different MAK findings from the two sequencers (Figs. S1 and S2). Briefly, the blocking reagents used to remove repetitive sequences during exome fragment capture differ slightly between sequencing platforms. Genomic fragments bearing the Alu–MAK junction were completely removed during the exome capture portion of the ABI sequencing experiment, whereas these fragments were not completely removed during the exome capture portion of the Illumina experiment. Paradoxically, it was the removal of the junction fragments in the ABI experiment that led to the detection of the mutation. The absence of the normal exon 9 MAK sequence from the proband’s exome fragment library led to the capture of a chimeric DNA molecule during the exome capture step. This molecule was interpreted by the sequence analysis algorithm as a compound heterozygous mutation. In contrast, although the Alu–MAK junction fragment was present in the raw data from the Illumina sequencer, the GATK analysis algorithm trimmed all of the Alu sequence from the ends of these junction fragment reads, because the sequence did not match the reference genome, resulting in an artifactually normal MAK sequence.
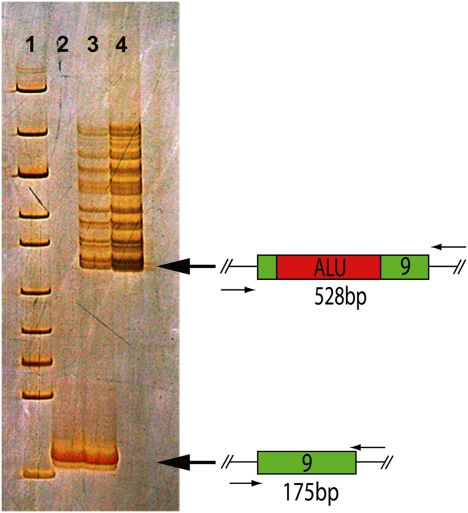
Fig. 1.
Identification of an Alu repeat insertion in exon 9 of MAK. Exon 9 amplification products are resolved on a silver-stained polyacrylamide gel. Lane 1, size standards; lane 2, normal control; lane 3, a heterozygous carrier of the Alu insertion; lane 4, the homozygous proband affected with MAK-associated RP.
Alu Insertion in MAK Causes RP in Individuals of Jewish Ancestry.
The 353-bp Alu insertion is easily detectable on silver-stained 6% polyacrylamide gels (Fig. 1, lane 4), and we used this method to screen 277 unrelated individuals without eye disease (554 chromosomes) and failed to find a single instance of the mutation. Next, we screened 1,798 unrelated individuals with RP and a family history compatible with autosomal recessive inheritance, and we identified 21 homozygous individuals, 1 individual heterozygous for the Alu insertion, and 2 individuals with heterozygous missense variants (Arg394Thr and Asp435Tyr). Sequencing of the entire coding sequence of MAK in the three heterozygous RP patients failed to identify a second disease-causing mutation in any of them. These patients may have mutations in noncoding portions of MAK or mutations in another RP-causing gene. In two families, the proband had an affected relative (a sibling in one family and a first cousin in the other family). In both cases, the affected relative was also homozygous for the Alu insertion. A total of five unaffected first degree relatives had samples in the collection available for study. None of these individuals were homozygous for the Alu insertion. There was no known consanguinity among any of the kindreds harboring the Alu insertion, but all 21 families reported Jewish ancestry. In one family, the unaffected mother had emigrated from Turkey, whereas the unaffected father had emigrated from Russia, suggesting that the founder of this mutation may have lived before the separation of the Sephardic and Ashkenazi groups in the Middle Ages.
Alu Insertion in MAK Is Not Associated with Other Photoreceptor Degenerations.
Many genes that cause nonsyndromic RP have also been associated with other retinal diseases such as Leber congenital amaurosis (LCA), Bardet Biedl syndrome (BBS), Usher syndrome, cone rod dystrophy, and Stargardt disease (3, 16–19). To investigate the possibility that the Alu insertion in MAK is also involved in any of these phenotypes, we screened 454 probands with LCA, 125 probands with BBS, 109 probands with Usher syndrome, 175 probands with cone rod dystrophy, and 202 probands with Stargardt disease. Each of these subjects had been previously screened for mutations in genes that commonly cause these phenotypes and had no mutations found. No instances of the Alu insertion in MAK were observed in any of these 1,065 individuals. To estimate the carrier frequency of the Alu insertion in MAK in the North American population, we screened an additional 2,675 individuals without photoreceptor disease and failed to find any additional instances of this variant. The number of unrelated homozygotes observed among RP patients (21/1,798) was significantly higher (P < 10−8, Fisher’s exact test) than the number observed among unrelated people without photoreceptor disease (0/2,952).
MAK Is Expressed in both Rod and Cone Photoreceptors in the Human Retina.
MAK was localized to human retina using monoclonal and polyclonal antibodies. In photoreceptor cells, labeling was predominately observed in the outer nuclear layer, axons, and inner segments (Fig. 2 A and C). Outer segments that contained abundant rhodopsin (Fig. 2A) did not show strong MAK expression, which was generally not observed distal to the connecting cilium. Immunolabeling with anti-MAK antibodies was identical with both monoclonal and polyclonal antibodies, and this pattern was not observed when primary antibodies were omitted (Fig. 2B). The localization of MAK to photoreceptor cells was also observed in foveal cones (Fig. 2C), in which inner segments as well as the Henle’s fiber layer of axons were strongly positive.
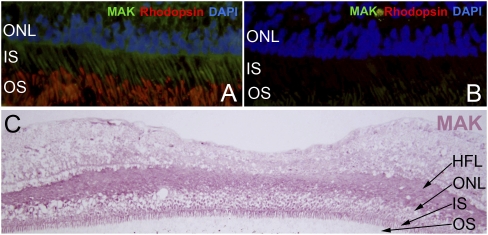
Fig. 2.
MAK is expressed in human photoreceptors. (A) Immunofluorescence labeling of MAK (green) and rhodopsin (red) in human retina. Note the labeling of MAK in photoreceptor cell inner segments (IS) and the restricted labeling of rhodopsin to outer segments (OS). (B) A control section exposed to secondary antibodies only. (C) Colorimetric immunohiostochemical detection of MAK in the human fovea centralis shows strong labeling in Henle’s fiber layer (HFL) as well as the inner segments of foveal cones.
iPSCs Exhibit Retinal-Specific Markers After in Vitro Differentiation.
To enable the investigation of the Alu insertion’s effect at the RNA and protein levels, iPSCs were created from dermal fibroblasts of the proband homozygous for the Alu insertion and a non-MAK–associated RP patient (RP control) by lentiviral transduction of four transcription factors: Oct4, Sox2, KLF4, and c-Myc (Fig. 3A). Similarly generated iPSCs from a normal individual (normal control) were obtained as a gift from George Q. Daley (Children’s Hospital Boston, Harvard Medical School, Boston, MA). Three to four weeks after transduction, iPSC colonies were manually isolated and expanded. After three rounds of expansion, iPSCs were harvested, RNA was extracted, and RT-PCR was used to assess pluripotency marker gene expression. As shown in Fig. 3, iPSCs from both RP patients expressed the pluripotent/iPSC genes NANOG, Sox2, c-Myc, and KLF4 (Fig. 3 B and C). iPSC lines were differentiated to a retinal cell lineage using our previously published stepwise differentiation protocol (20) (Fig. 3D). This protocol takes advantage of current knowledge of retinal cell development and uses extrinsic chemical signaling to manipulate cell fate. At 33 d postdifferentiation, pluripotent iPSCs generated from both RP control and MAK-associated RP human fibroblasts adopted a postmitotic retinal cell morphology (Fig. 3 E and F) and expressed the retinal differentiation markers Pax6, Chx10, Otx2, and recoverin (Fig. 3 G and H).
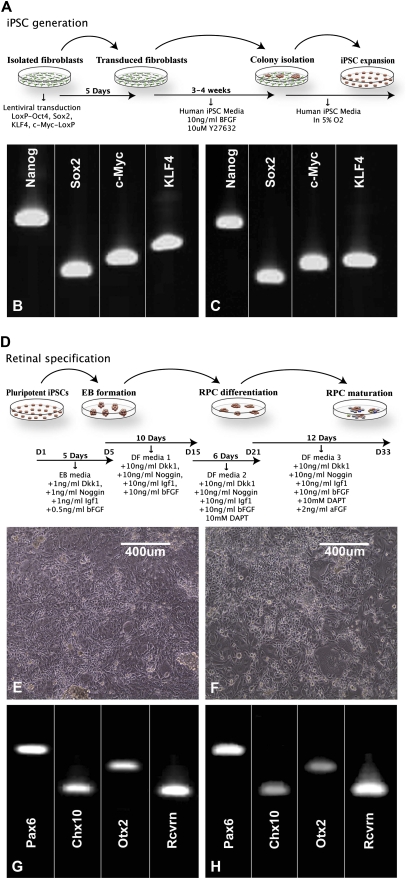
Fig. 3.
Production of retinal neurons from RP-control and RP-MAK iPSCs. (A) Schematic diagram illustrating the iPSC generation paradigm used in this study. (B and C) RT-PCR analysis of undifferentiated RP-control (B) and RP-MAK (C) iPSCs for expression of the iPSC/pluripotency markers Nanog, Sox2, c-Myc, and KLF4. (D) Schematic diagram illustrating the retinal differentiation paradigm used in this study. (E and F) Microscopic analysis of RP-control (E) and RP-MAK (F) iPSCs at 33 d postdifferentiation. (G and H) RT-PCR analysis of differentiated RP-control (G) and RP-MAK (H) iPSCs for the expression of the retinal specification/photoreceptor genes Pax6, Chx10, Otx2, and recoverin. At 33 d postdifferentiation, pluripotent iPSCs generated from both RP-control and RP-MAK human fibroblasts adopted a postmitotic retinal cell morphology and expressed the retinal specification/photoreceptor genes Pax6, Chx10, Otx2, and recoverin. (Scale bar: 400 μm.)
Alu Insertion in Exon 9 Results in Loss of the Retina-Specific Isoform of MAK.
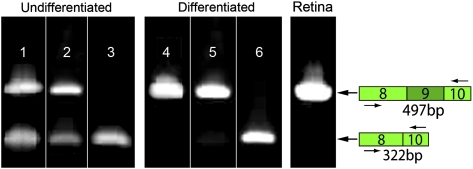
Thirty-three days after the initiation of the differentiation protocol described above, the RP control sample exhibited only the MAK transcript bearing exon 9, whereas the sample from the RP patient with the Alu insertion in MAK exhibited only the transcript lacking exon 9 (Fig. 4A). Similar analysis of RNA extracted from the neural retina of a normal adult human eye donor revealed only the exon 9-containing MAK transcript (Fig. 4A). Thus, the Alu insertion seems to disrupt the correct splicing of exon 9, thereby preventing mature retinal cells from expressing the correct MAK isoform. The existence of multiple mRNA transcripts in stem cells and a single transcript in adult tissues has been previously observed with other genes (21). Western blotting reveals very little MAK protein in undifferentiated IPSCs (Fig. 4B). As the cells differentiate into retinal precursors, the line derived from the RP control patient begins to express a MAK protein very similar in size to the protein seen in adult human retina (98.4 kDa) (Fig. 4B). In contrast, the iPSCs derived from the RP patient with the MAK Alu insertion express a protein that is about 40 kDa smaller (Fig. 4B).
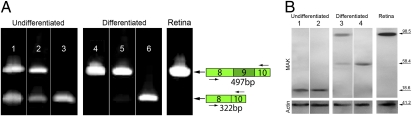
Fig. 4.
Insertion of an Alu repeat into MAK exon 9 results in improper splicing and loss of the mature MAK protein. (A) RT-PCR analysis of MAK exon 9 expression using a primer pair spanning exons 8–10 in undifferentiated (lanes 1–3) and differentiated (lanes 4–6) iPSCs and adult human retina (lane 7). Undifferentiated iPSCs from the normal control (lane 1) and RP control (lane 2) individuals exhibit two species, one with and one without exon 9. In contrast, undifferentiated iPSCs from the patient with the Alu insertion in exon 9 (lane 3) only express the transcript lacking exon 9. Because the iPSCs are differentiated into retinal precursors, the transcript lacking exon 9 is lost from the cells derived from the normal control (lane 4) and the RP control (lane 5), whereas it remains the only MAK RNA species in the cells derived from the proband with the Alu insertion (lane 6). When analyzed with these primers, adult human retina exhibits only transcripts bearing exon 9 (lane 7). (B) Western blot analysis of MAK expression in undifferentiated (lanes 1 and 2) and differentiated (lanes 3 and 4) iPSCs and adult human retina (lane 5). Undifferentiated RP-control (lane 1) and RP-MAK (lane 2) iPSCs express an 18-kDa MAK protein. Because these cells are differentiated into retinal precursors, the RP-control cells (lane 3) express two versions of MAK, one of 58 kDa and another of ∼98 kDa. The differentiated RP-MAK iPSCs (lane 4) also express the 58-kDa protein but fail to express the 98-kDa isoform. Adult human retina (lane 5) expresses only the 98-kDa MAK isoform.
Retinal Isoform of MAK Contains a Retina Specific Exon.
A Northern blot of RNA extracted from the retina of a normal human eye donor revealed the largest and most abundant transcript to be slightly larger than 2,000 nt in length (Fig. S3). To determine the sequence composition of this transcript, RT-PCR was performed using primers corresponding to sequences in the first and last exons of the largest known human MAK transcript (NM_005906.3). DNA sequencing of this amplification product revealed a 75-bp exon that was not previously known to exist in humans (Fig. S4). This exon lies between exons 11 and 12 of the longest previously published human MAK transcripts, and it is highly homologous to a similar exon recently observed in mice (22). This 14-exon retinal transcript (the newly recognized exon is now exon 12) contains 648 codons and encodes a protein with a predicted size of 73.3 kDa. The difference between the predicted and observed (Fig. 4B) sizes is likely caused by posttranslational modifications such as glycosylation. We then performed a phylogenetic survey to determine the evolutionary origin of exon 12. Although orthologs to MAK can be found in Arabidopsis and Oryza, exon 12 seems to have arisen after the separation of reptiles and amphibians (Fig. 5). An ORF homologous to human exon 12 exists in the genomic sequences of lizard, chicken, and six mammals, but it is absent from zebrafish and frog. MAK transcripts amplified from the retina of adult zebrafish and frogs lack exon 12 (Fig. S4). Comparison of the exon 12 sequences of eight species reveals 10 invariant amino acids (Fig. 6). Given the amount of scrutiny that the human retinal transcriptome has received, it is surprising that exon 12 has not been previously detected. Fig. 7 shows a likely explanation for how this oversight occurred. Of the 73 independent human MAK expressed sequence tags (ESTs) in public databases, 22 are confined to the 3′ UTR, and another 18 are limited to the sequence encoded by the first four exons. Only three published ESTs contain sequences from exons 10 and 11, and none of these ESTs were derived from retina.
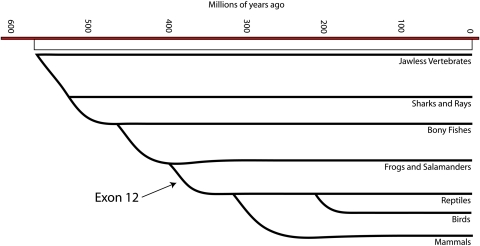
Fig. 5.
The retina-specific MAK exon 12 in phylogeny. Schematic diagram illustrating the time at which MAK exon 12 appeared during evolution. Modified from Reece et al. (34). Modern fish and amphibians lack exon 12, but modern amniotes (reptiles, birds, and mammals) all exhibit it in their genomic sequences.
Fig. 6.
Exon 12 amino acid homology among eight amniotes. Blue shading indicates amino acids that are shared among all eight species. Yellow shading indicates a departure from the consensus sequence.
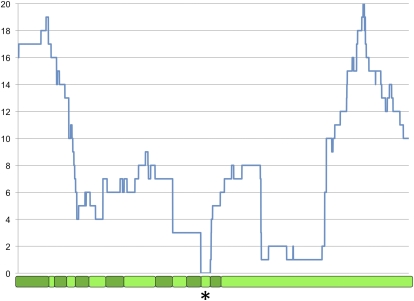
Fig. 7.
Relative representation of MAK coding sequences in publicly available databases of ESTs. The coding sequence of the 14-exon retinal transcript of MAK is depicted along the x axis. Odd-numbered exons are dark green, and even-numbered exons are light green. Exon 12 is marked with an asterisk. The number of times a specific nucleotide is found among the 73 available MAK ESTs is shown on the y axis. The 75 nt of the retina-specific exon 12 were not present in any of these ESTs.
Inclusion of Exon 12 Requires Inclusion of Exon 9.
We used RT-PCR to investigate the tissue distribution of MAK exon 12 in humans. Fig. 8A shows that the exon 12-containing transcript is only expressed in retina. In contrast, all adult tissues studied except for smooth muscle express exon 9 (Fig. 8B). When iPSC-derived retinal progenitor cells begin to express exon 9 at 33 d postdifferentiation, they also begin to express exon 12 (Fig. 9, lanes 4 and 5). Disruption of the developmental inclusion of exon 9 by the Alu insertion also results in the loss of exon 12 (Fig. 9, lane 6).
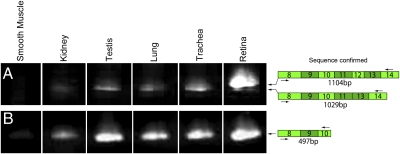
Fig. 8.
MAK exon 12 is retina-specific, but MAK exon 9 is not. (A) In adult tissues, RT-PCR analysis of MAK exon 12 expression using a primer pair spanning exons 8–14 reveals two splice variants, a retina-specific transcript containing exon 12 and a transcript lacking exon 12 that is also expressed in testis, lung, and trachea. (B) RT-PCR analysis of MAK exon 9 expression using a primer pair spanning exons 8–10 reveals a single transcript containing exon 9 in all adult tissues that express MAK. This finding is in contrast to the pattern seen in undifferentiated iPSCs, in which a subset of MAK transcripts lack exon 9 (Fig. 4).
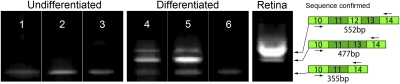
Fig. 9.
Insertion of an Alu repeat into MAK exon 9 prevents the expression of retina-specific MAK exon 12. RT-PCR analysis of MAK exon 12 in undifferentiated (lanes 1–3) and differentiated (lanes 4–6) iPSCs and adult human retina (lane 7). All iPSCs (lanes 1–6) express a small fraction of MAK transcripts that lack exons 12 and 13. This transcript is not seen in adult human retina (lane 7), and its biological significance is unknown. Adult human retina exhibits two transcripts, one with and one without exon 12, and the latter is significantly more abundant than the former. Differentiated iPSCs from the normal control (lane 4) and RP control (lane 5) individuals also exhibit these two species, but their relative abundance is reversed. Differentiated iPSCs derived from the patient with the Alu insertion in MAK exon 9 do not express any RNA bearing exon 12, suggesting that proper splicing of exon 9 is required for inclusion of exon 12.
Point Mutations in Exons 9 and 12 Are Not a Common Cause of RP.
The frequency of homozygosity of the exon 9 Alu insertion among RP patients (21 of 22 probands) suggests that individuals within the population from which these patients were drawn are not randomly mating and/or that there are few, if any, other MAK alleles in the population that are capable of causing the RP phenotype. The fact that all of the affected homozygotes have known Jewish ancestry indicates that the first of these possibilities is true. To investigate the second possibility, we used single-strand conformational polymorphism (SSCP) to screen 1,798 unrelated autosomal recessive RP patients for mutations in two alternatively spliced MAK exons, 9 and 12. Because SSCP is less sensitive than automated DNA sequencing, we also screened 242 of these patients for mutations in these two exons using automated DNA sequencing. Although we observed a few rare heterozygous sequence variants in RP patients that were not seen in control individuals, when we sequenced the entire MAK coding region in these patients, we did not identify a second disease-causing mutation in any of them.
Discussion
MAK was initially identified by screening a human genomic library for sequences that cross-hybridize with the v-ros oncogene, and it was described as a testis-enriched gene with two transcripts that result in a complex pattern on Western blot (23, 24). Expression of Mak in rat was found to be strongest in meiotic spermatocytes, with down-regulation in postmeiotic spermatids (24). Despite its expression during the cell cycle in developing germ cells, targeted deletion of Mak in mice does not result in impaired fertility (25). Similarly, the majority of individuals with MAK-associated RP in our cohort each had two or more children (range = 0–4). Thus, human fertility also seems to be unaffected by the loss of MAK function. In contrast, studies of the retina in Mak−/− mice reveal a dramatic effect of Mak deficiency on photoreceptor cell structure, function, and viability (22). Mice homozygous for Mak deletion show elongated connecting cilia, reduced electrical response to light stimulation, protein mislocalization, and ultrastructural defects in outer segment morphogenesis (22).
The evidence that the Alu insertion into exon 9 of MAK causes autosomal recessive RP is compelling. This variant was observed 43 times among 3,596 alleles of unrelated RP patients and was not observed among 5,904 alleles of unrelated individuals without photoreceptor disease (P < 10−18, Fisher’s exact test); 42 of these alleles were observed among 21 homozygotes. Unaffected parents of these homozygotes are heterozygous for the insertion, whereas other unaffected first-degree relatives are either heterozygous or lack the insertion entirely. Analysis of iPSCs shows that the Alu insertion in MAK results in the loss of a retinal isoform that has been conserved since the divergence of amphibians and reptiles. MAK is known to be involved in regulation of cilia length in photoreceptors, and its loss is known to cause photoreceptor death in mice (22).
RP is a very genetically heterogeneous disorder, and it is estimated that mutations in more than 100 different genes are capable of causing the autosomal recessive form of this disease (4). As a result, most RP genes will be associated with less than 1% of all cases of this disease. One might reasonably ask whether it is important to identify disease genes that cause less than 1% of a given phenotype, and if it is important, whether it can be accomplished in an efficient and comprehensive fashion. Finding genes that cause small fractions of rare diseases is important for several reasons. First, it contributes to our knowledge of the normal function of human genes and the structures and biological pathways dependent on them. Second, it improves the ability to provide an accurate and mechanistically detailed diagnosis to patients. Third, it lays the groundwork for the development and delivery of mechanism-specific therapies, including viral-mediated gene replacement. Fourth, it hastens the discovery of additional disease genes by enriching patient cohorts for patients whose causative genes remain to be identified. Fifth, it allows clinically relevant genotype–phenotype and ethnicity–phenotype relationships to be observed. For example, the MAK-Alu insertion mutation seems to be a common cause of RP among people of Jewish ancestry, a population that has had a successful and ongoing program of carrier testing aimed at reducing the incidence of genetic disorders.
Importantly, this study also reveals some gaps in our current methods of disease gene discovery. The closure of these gaps represents an opportunity to accelerate progress to the goal of identifying all genes responsible for inherited disease. Before the introduction of the PCR, it was difficult to investigate the structure of genes at the nucleotide level in individual patients. As a result, most disease-causing mutations that were discovered in the late 1970s and early 1980s were relatively large insertions or deletions that were detectable with Southern blotting. The introduction of PCR and automated DNA sequencing in the mid-1980s made it possible to efficiently screen large cohorts of patients for variations in specific sequences, but these methods are relatively insensitive to large insertions and deletions. As a result, most disease-causing mutations discovered in the past 20 y have been variations that affect fewer than 10 nt. Like its predecessors, next generation sequencing has its own strengths and weaknesses as a mutation finding tool, and these aspects will shape the observations that can be made with this approach until the weaknesses are fully understood and overcome. All current exome capture methods are imperfect, and some will undersample as much as 20% of the human exome—perhaps even more if one considers currently unrecognized exons such as MAK exon 12. When one uses exome capture methods to study autosomal recessive diseases in an outbred population, where compound heterozygotes may be more common than homozygotes among affected individuals, both mutant alleles will not always be identified. Thus, one does not have the luxury of discounting plausible disease-causing heterozygous variants such as those variants observed in ABCA4 and USH2A in the proband of this study. Such variants often number in the hundreds in a typical exome sequencing experiment. Thus, improved methods are needed to evaluate their functional effects in cells and tissues as well as to determine their relative frequencies in cohorts of patients and control individuals.
This study illustrates that current methods used to analyze exome sequencing data are relatively insensitive to large insertions and deletions, despite the wide recognition of the importance of these classes of mutation to human disease. We were fortunate in this experiment that one of two sequencing approaches that we used detected the presence of the disease-causing mutation, albeit by a somewhat indirect mechanism. Many groups are working to overcome these analytical blind spots (26, 27), and as these methods are perfected, many more Short INterspersed Elements- and Long INterspersed Elements-associated mutations will undoubtedly be identified by exome sequencing.
One of the challenges of studying human diseases of the retina and brain is the lack of access to the affected tissue for molecular and cell biological studies. In the past, investigators have had to infer the pathogenicity of mutations that they detected by showing a statistically significant association between these variations and the disease phenotype. Another strategy has been to use various types of animal models to show that the variations observed in patients are likely to be responsible for disease. In this study, we show the use of iPSCs derived from the skin of affected individuals for the investigation of the pathophysiologic mechanism. In a matter of weeks, these cells can be induced to mimic the development of the retina in a human embryo, and in this case, they revealed that the insertion of an Alu element into exon 9 of MAK prevented the normal switch to a retinal-specific isoform bearing exons 9 and 12. Such cells can also be used to test the efficacy of various aspects of gene replacement vectors (e.g., the promoter, vector serotype, artificial introns, etc.) before testing them in animal models.
In summary, we have identified a cause of RP using a combination of next generation sequencing, SSCP screening, and Sanger sequencing of a large validation cohort, molecular analysis of patient-derived iPSCs, and molecular and histochemical analysis of human donor tissue. The discovery of a pathogenic Alu insertion, despite algorithmic and molecular obstacles, and the presence of a tissue-specific unannotated exon in the disease-causing gene suggest some specific changes in strategy that can be implemented to more fully harness the mutation identification power of sequencing technologies.
Methods
Human Subjects.
All subjects provided written informed consent for this research study, which was approved by the Institutional Review Boards of the participating centers and adhered to the tenets set forth in the Declaration of Helsinki.
DNA Extraction.
Blood samples were obtained from all subjects. DNA was extracted by using the manufacturer’s specifications for whole-blood DNA extraction using Gentra Systems’s Autopure LS instrument.
Exon Capture.
Targeted enrichment of exons was performed using the Agilent SureSelect All Exon Capture platform per the manufacturer’s instructions. This capture platform includes 38 Mb targeted features.
Next Generation DNA Sequencing.
Sequencing of the captured genomic DNA was performed using the manufacturer’s instructions on an ABI SOLiD 4hq at the University of Iowa’s DNA Core Facility and an Illumina HiSeq sequencer at the Hudson Alpha Institute in Huntsville, AL.
SSCP Analysis PCR.
Twelve and one-half nanograms each patient's DNA were used as template in a 8.35-μL PCR containing 1.25 μL 10× buffer (100 mM Tris⋅HCL, pH 8.3, 500 mM KCl, 15 mM MgCl2), 300 μm each dCTP, dATP, dGTP, and dTTP, 1 pmole each primer, and 0.25 units Biolase polymerase (Biolase). Samples were denatured for 5 min at 94 °C and incubated for 35 cycles under the following conditions: 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s in a DNA thermocycler.
SSCP Analysis.
After amplification, 5 μL stop solution (95% formamide, 10 mM NaOH, 0.05% Bromophenol Blue, 0.05% Xylene Cyanol) were added to each sample. Amplification products were denatured for 3 min at 94 °C and electrophoresed on 6% polyacrylamide and 5% glycerol gels at 25 W for ∼3 h at room temperature. After electrophoresis, gels were stained with silver nitrate.
Automated DNA Sequencing.
Abnormal PCR products identified by SSCP analysis were confirmed using automated DNA sequencing with dye termination chemistry on an ABI 3730 sequencer. All sequencing was bidirectional.
Immunohistochemistry.
Human donor eyes were obtained from the Iowa Lions Eye Bank after informed consent of the donors’ families. Some samples were frozen for RNA analysis or protein biochemistry. Extramacular retina was separated from the underlying RPE-choroid layer and flash-frozen in liquid nitrogen within 6 h of death. In addition, for histological analysis, tissues were fixed in 4% paraformaldehyde within 6 h of death. After 2–4 h fixation, wedges of macular and extramacular retina were cryopreserved in sucrose and embedded in optimal cutting temperature solution for cryostat sectioning. Immunohistochemistry using monoclonal antibodies directed against either MAK (Santa Cruz) or rhodopsin (Santa Cruz) or using polyclonal antibodies directed against MAK (Chemicon) using either immunofluorescence or colorimetry was performed as described previously (28). Control and experimental sections were photographed and adjusted using Adobe Photoshop using identical exposures and processing.
iPSC Generation.
iPSCs were generated from adult human dermal fibroblasts (RP MAK and RP control) by lentiviral transduction of the four transcription factors Oct4, Sox2, Klf4, and c-MYC as previously described (20). At 5 d postinfection, cells were passaged onto a monolayer of inactive mouse embryonic fibroblasts and fed daily with pluripotency media [DMEM F-12 media (Gibco), 15% heat-inactivated FBS (Lifeblood Medical Inc.), 0.0008% β-mercaptoethanol (Sigma-Aldrich), 1% 100× nonessential amino acids (NEAA) (Gibco), 1 × 106 units/L leukemia inhibitory factor (LIF/ESGRO; Millipore), 1% penicillin/streptomycin (Gibco), 0.2% Fungizone (Gibco)]. At 3–5 wk postinfection, iPSC colonies were picked, passaged, and clonally expanded for additional experimentation.
iPS Cell Differentiation.
To maintain pluripotency, adult human iPSCs were cultured on Matrigel-coated cell culture surfaces in bFGF containing pluripotency media. To begin differentiation, iPS cells were removed from the culture substrate by incubation in a 1 mg/mL type 4 collagenase (Sigma-Aldrich) solution, resuspended in embryoid body media [DMEM F-12 media (Gibco) containing 10% knockout serum replacement (Gibco), 2% B27 supplement (Gibco), 1% N2 supplement (Gibco), 1% l-glutamine (Gibco), 1% 100× NEAA (Gibco), 1% penicillin/streptomycin (Gibco), 0.2% Fungizone (Gibco), 1 ng/mL noggin (R&D Systems), 1 ng/mL Dkk-1 (R&D Systems), 1 ng/mL IGF1 (R&D Systems), 0.5 ng/mL bFGF (R&D Systems)], and plated at a density of ∼50 cell clumps/cm2 in ultra low cluster plates (Corning). Cell clumps were cultured for 5 d as indicated above, after which the embryoid bodies were removed, washed, and plated at a density of 25–30/cm2 in fresh differentiation media 1 [DMEM F-12 media (Gibco), 2% B27 supplement (Gibco), 1% N2 supplement (Gibco), 1% l-glutamine (Gibco), 1% 100× NEAA (Gibco), 10 ng/mL noggin (R&D Systems), 10 ng/mL Dkk-1 (R&D Systems), 10 ng/mL IGF1 (R&D Systems), 1 ng/mL bFGF (R&D Systems)] in six-well culture plates coated with poly-d-lysine (10 mg/mL; BD Bioscience), collagen (25 μg/mL; BD Bioscience), laminin (100μg/mL; Gibco), and fibronectin (100 μg/mL; Sigma-Aldrich). Cultures were fed every other day for 10 d with differentiation media 1 and then every other day for an additional 6 d with differentiation media 2 [differentiation media 1 + 10 uM Notch signaling inhibitor DAPT (Calbiochem)] followed every other day for an additional 12 d with differentiation media 3 [differentiation media 2 + 2 ng/mL aFGF (R&D Systems)].
Immunoblotting.
For Western blot analysis, undifferentiated and differentiated iPSCs were homogenized in lysis buffer [50 mM Tris⋅HCl, pH 7.6, 150 mM NaCl, 10 mM CaCl2, 1% triton X-100, 0.02% NaN3 (Sigma-Aldrich)] and centrifuged, supernatants were isolated, and protein concentrations were determined using a BCA protein assay (Pierce). Equivalent amounts of protein (50 μg) were subjected to SDS/PAGE (8–10% acrylamide), transferred to PVDF, and probed with primary antibodies targeted against MAK (Abgent) and tubulin (used as a loading control; Abcam). Blots were visualized with ECL reagents (GE Healthcare) and exposed to X-ray film (Fisher).
RNA Isolation and RT-PCR.
Total RNA was extracted from primary tissues/cultures using the RNeasy Minikit (Qiagen) using the provided instructions. Briefly, human tissues/cultures (human retina, human iPSCs, zebrafish tissues, mouse tissues, and Xenopus laevis tissues) were lysed, homogenized, and diluted in 70% ethanol to adjust binding conditions. Samples were spun using RNeasy spin columns and washed, and RNA was eluted with RNase-free water. Human RNA samples isolated from the trachea, lung, testis, kidney, and smooth muscle were obtained commercially (Clontech); 1 μg RNA was reverse-transcribed into cDNA using the random hexamer (Invitrogen) priming method. All PCR reactions were performed in a 50-μL reaction containing 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 100 ng DNA, 1.0 U Platnium Taq (Invitrogen), and 20 pmol each gene-specific primer (Integrated DNA Technologies). All cycling profiles incorporated an initial denaturation temperature of 94 °C for 10 min through 35 amplification cycles (30 s at 94 °C, 30 s at the annealing temperature of each primer, and 1 min at 68 °C) and a final extension at 68 °C for 5 min. PCR products were separated by gel electrophoresis using 2% agarose e-gels (Invitrogen). The sequences of the primers used in this experiment are given in Table S2.
Northern Blotting.
To identify MAK expression in human tissue, 20 μg total RNA, obtained as indicated in Methods, RNA Isolation and RT-PCR, were separated by gel electrophoresis, transferred to a nitroceluose membrane, and hybridized with a full-length human MAK cDNA (OriGene). The probe was labeled with 32P-dCTP using GE Ready-To-Go DNA labeling beads and purified using GE ProbeQuant G-50 Micro Columns (GE). Hybridization and autoradiography were performed as described previously (29).
Identification of MAK Orthologs.
Entrez Gene (30) was used to identify the previously computed orthologs of MAK. Those sequences were imported into MEGA5 (31) for automated alignment, and additional orthologs were identified using BLAST against the Reference Sequence database of mRNAs. The consensus amino acid sequence of the portion of MAK encoded by exon 12 was determined by identifying the amino acid that was present in the greatest number of species for each of the exon’s 25 codons.
Analysis of ESTs.
The human EST track from the UCSC genome browser (32) was used to assess the alignment of ESTs to the full-length MAK mRNA. Each EST was also evaluated in dbEST (33) by accession number to determine the tissue of origin.
Supplementary Material
Acknowledgments
Funding was provided by the Howard Hughes Medical Institute, National Institutes of Health New Innovator Award Program Award DP2 OD007483-01, National Eye Institute Grants EY016822 and EY017451, and the Foundation Fighting Blindness.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 13895.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108918108/-/DCSupplemental.
References
- 1.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 2.Cardenas-Rodriguez M, Badano JL. Ciliary biology: Understanding the cellular and genetic basis of human ciliopathies. Am J Med Genet C Semin Med Genet. 2009;151C:263–280. doi: 10.1002/ajmg.c.30227. [DOI] [PubMed] [Google Scholar]
- 3.Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS. Photoreceptor degeneration: Genetic and mechanistic dissection of a complex trait. Nat Rev Genet. 2010;11:273–284. doi: 10.1038/nrg2717. [DOI] [PubMed] [Google Scholar]
- 4.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 5.Daiger SP, Bowne SJ, Sullivan LS. Perspective on genes and mutations causing retinitis pigmentosa. Arch Ophthalmol. 2007;125:151–158. doi: 10.1001/archopht.125.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng SB, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson P, Krawitz P, Mundlos S. Strategies for exome and genome sequence data analysis in disease-gene discovery projects. Clin Genet. 2011;80:127–132. doi: 10.1111/j.1399-0004.2011.01713.x. [DOI] [PubMed] [Google Scholar]
- 8.Züchner S, et al. Whole-exome sequencing links a variant in DHDDS to retinitis pigmentosa. Am J Hum Genet. 2011;88:201–206. doi: 10.1016/j.ajhg.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin ZB, et al. Modeling retinal degeneration using patient-specific induced pluripotent stem cells. PLoS One. 2011;6:e17084. doi: 10.1371/journal.pone.0017084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashid ST, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homer N, Merriman B, Nelson SF. BFAST: An alignment tool for large scale genome resequencing. PLoS One. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briggs CE, et al. Mutations in ABCR (ABCA4) in patients with Stargardt macular degeneration or cone-rod degeneration. Invest Ophthalmol Vis Sci. 2001;42:2229–2236. [PubMed] [Google Scholar]
- 15.Lewis RA, et al. Genotype/phenotype analysis of a photoreceptor-specific ATP-binding cassette transporter gene, ABCR, in Stargardt disease. Am J Hum Genet. 1999;64:422–434. doi: 10.1086/302251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivolta C, Sweklo EA, Berson EL, Dryja TP. Missense mutation in the USH2A gene: Association with recessive retinitis pigmentosa without hearing loss. Am J Hum Genet. 2000;66:1975–1978. doi: 10.1086/302926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez-Mir A, et al. Retinitis pigmentosa caused by a homozygous mutation in the Stargardt disease gene ABCR. Nat Genet. 1998;18:11–12. doi: 10.1038/ng0198-11. [DOI] [PubMed] [Google Scholar]
- 18.Cremers FP, et al. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt’s disease gene ABCR. Hum Mol Genet. 1998;7:355–362. doi: 10.1093/hmg/7.3.355. [DOI] [PubMed] [Google Scholar]
- 19.Morimura H, et al. Mutations in the RPE65 gene in patients with autosomal recessive retinitis pigmentosa or leber congenital amaurosis. Proc Natl Acad Sci USA. 1998;95:3088–3093. doi: 10.1073/pnas.95.6.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tucker BA, et al. Transplantation of adult mouse iPS cell-derived photoreceptor precursors restores retinal structure and function in degenerative mice. PLoS One. 2011;6:e18992. doi: 10.1371/journal.pone.0018992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vu TH, et al. Developmentally regulated use of alternative promoters creates a novel platelet-derived growth factor receptor transcript in mouse teratocarcinoma and embryonic stem cells. Mol Cell Biol. 1989;9:4563–4567. doi: 10.1128/mcb.9.10.4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Omori Y, et al. Negative regulation of ciliary length by ciliary male germ cell-associated kinase (Mak) is required for retinal photoreceptor survival. Proc Natl Acad Sci USA. 2010;107:22671–22676. doi: 10.1073/pnas.1009437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsushime H, Jinno A, Takagi N, Shibuya M. A novel mammalian protein kinase gene (mak) is highly expressed in testicular germ cells at and after meiosis. Mol Cell Biol. 1990;10:2261–2268. doi: 10.1128/mcb.10.5.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jinno A, Tanaka K, Matsushime H, Haneji T, Shibuya M. Testis-specific mak protein kinase is expressed specifically in the meiotic phase in spermatogenesis and is associated with a 210-kilodalton cellular phosphoprotein. Mol Cell Biol. 1993;13:4146–4156. doi: 10.1128/mcb.13.7.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinkai Y, et al. A testicular germ cell-associated serine-threonine kinase, MAK, is dispensable for sperm formation. Mol Cell Biol. 2002;22:3276–3280. doi: 10.1128/MCB.22.10.3276-3280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hajirasouliha I, et al. Detection and characterization of novel sequence insertions using paired-end next-generation sequencing. Bioinformatics. 2010;26:1277–1283. doi: 10.1093/bioinformatics/btq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alkan C, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullins RF, Skeie JM, Malone EA, Kuehn MH. Macular and peripheral distribution of ICAM-1 in the human choriocapillaris and retina. Mol Vis. 2006;12:224–235. [PubMed] [Google Scholar]
- 29.Swiderski RE, et al. Expression pattern and in situ localization of the mouse homologue of the human MYOC (GLC1A) gene in adult brain. Brain Res Mol Brain Res. 1999;68:64–72. doi: 10.1016/s0169-328x(99)00085-6. [DOI] [PubMed] [Google Scholar]
- 30.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr121. 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita PA, et al. The UCSC Genome Browser database: Update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boguski MS, Lowe TM, Tolstoshev CM. dbEST—database for “expressed sequence tags.”. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 34.Reece JB, et al. Campbell Biology. 9th Ed. San Francisco: Benjamin Cummings; 2010. [Google Scholar]