Abstract
The risk of distant recurrence in breast cancer patients is difficult to assess with current clinical and histopathological parameters, and no validated serum biomarkers currently exist. Using a recently developed recombinant antibody microarray platform containing 135 antibodies against 65 mainly immunoregulatory proteins, we screened 240 sera from 64 patients with primary breast cancer. This unique longitudinal sample material was collected from each patient between 0 and 36 mo after the primary operation. The velocity for each serum protein was determined by comparing the samples collected at the primary operation and then 3–6 mo later. A 21-protein signature was identified, using leave-one-out cross-validation together with a backward elimination strategy in a training cohort. This signature was tested and evaluated subsequently in an independent test cohort (prevalidation). The risk of developing distant recurrence after primary operation could be assessed for each patient, using her molecular portraits. The results from this prevalidation study showed that patients could be classified into high- versus low-risk groups for developing metastatic breast cancer with a receiver operating characteristic area under the curve of 0.85. This risk assessment was not dependent on the type of adjuvant therapy received by the patients. Even more importantly, we demonstrated that this protein signature provided an added value compared with conventional clinical parameters. Consequently, we present here a candidate serum biomarker signature able to classify patients with primary breast cancer according to their risk of developing distant recurrence, with an accuracy outperforming current procedures.
Keywords: affinity proteomics, monitoring disease, prognosis, tumour relaps, malignancy
Survival from breast cancer has improved during the past decades in the Western world. In Sweden, there has been a steady decrease in age-standardized breast cancer mortality in women up to 70 y of age during the past 5 decades, and the decrease in countries such as the United States, United Kingdom, Holland, Germany, and France has been observed since the late 1980s (1, 2). Major reasons for this substantial decrease are earlier surgery of primary breast cancer because of the introduction of mammography screening and adjuvant systemic therapy in the form of both cytotoxic and antihormonal therapy. In particular, the use of the antiestrogen tamoxifen in postmenopausal women and, in more recent years, of the aromatase inhibitors has prolonged the survival of many women. However, the number needed to treat is around 10 even for the best cytotoxic combinations and tamoxifen (3) and is around 20–40 for the third-generation aromatase inhibitors (4) because of the lack of optimal predictors of these therapies. Furthermore, ∼30% of all patients with primary breast cancer will develop distant recurrence (5), and then the possibility of a cure is very limited. To improve survival further, we need to offer better treatments with increased efficacy and lower toxicity, with the therapy for the individual patient based on the clinical and molecular characteristics of the tumor. Traditional prognostic indices, such as the Nottingham Prognostic Index (6, 7), have proven valuable in identifying patients with poor prognosis. More recently, genomic studies have opened the possibility of prognosticating recurrence-free survival, using expression analysis of a multitude of different genes (8, 9). However, a simple way to predict the likelihood of a later recurrence, i.e., an indicator that allows risk assessment for breast cancer metastasis, would be highly desirable. In this respect, in contrast to predictors based upon tumor characteristics at the time of surgery, serum is a particular valuable source, because it is useful not only for the initial screening of the disease but also for continuous monitoring of the therapeutic effect.
The serum biomarkers proposed to date have not yet demonstrated enough prognostic accuracy in breast cancer (10). However, recent developments in affinity proteomics have advanced the field of cancer biomarkers (11–13), and that approach was adopted here to define predictive serum biomarkers associated with tumor relapse in breast cancer patients. We hypothesized that decoding patterns of immunoregulatory serum proteins could reveal important information about the risk of recurrence. Consequently, using minute amounts of nonfractionated serum (14) and a recombinant antibody microarray technology capable of analyzing large numbers of low- and high-abundance protein analytes, we screened samples from breast cancer patients collected over a 3-y period. The samples were collected from breast cancer patients before resection of the primary tumor and then postoperatively every 6–12 mo, resulting in three to five samples per patient. By analyzing the velocity (i.e., the change over time) of the markers, we could stipulate for each patient the risk of developing metastasis after the end of adjuvant systemic therapy. Changes in biomarker serum levels have been used previously, e.g., in calculating prostate-specific antigen (PSA) velocity for the diagnosis of prostate cancer (15). This longitudinal study demonstrates, that a simple blood sample harbors information of tumor relapse in breast cancer patients, brings added value to existing clinical predictors, and outperforms previous attempts to risk classify breast cancer patients. This information would allow refinements in the planning of future clinical studies, improving the current state-of-the-art for adjuvant therapy of primary breast cancer patients.
Results
Serum Biomarker Signature Associated with Tumor Relapse.
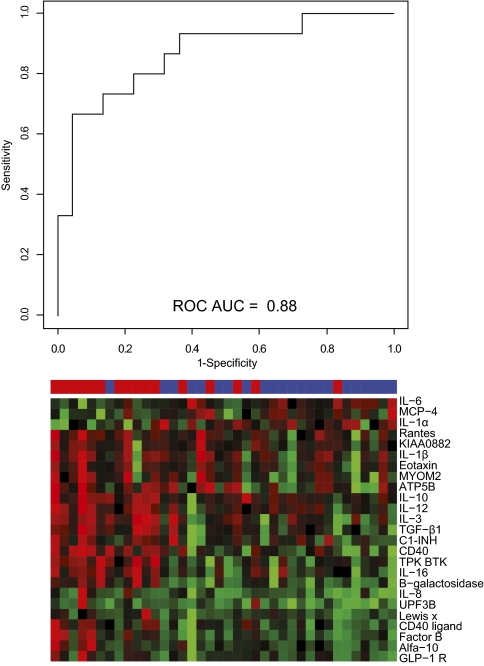
In an attempt to identify a serum biomarker signature predictive of distant recurrence in breast cancer, we analyzed samples previously collected over a 3-yperiod from patients with primary breast cancer (Fig. 1). Finding a classifier for metastatic disease by comparing the absolute serum protein (analyte) levels from samples collected at the time of primary surgery was not possible, as was evident by a receiver operating characteristic (ROC) area under the curve (AUC) of 0.54. Instead, we used more information in the samples by analyzing the analyte velocity. Using the velocity calculated for samples collected 3–6 mo after surgery asthe classifier allowed stratification of patients as having high risk versus low risk for tumor recurrence with an AUC of 0.88 (P = 1.7 × 10−5) (Fig. 2). Hence, this approach demonstrated that there was enough information in the dataset to identify a velocity-driven candidate biomarker signature that allowed identification of a patient group with high risk for developing a distant tumor relapse within a follow-up time of 5 y.
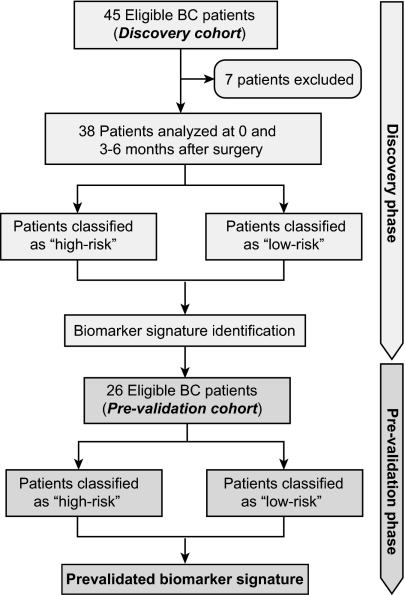
Fig. 1.
Flowchart describing the retrospective study design. Seven patients were excluded because of lack of samples collected at the relevant time points (preoperative and 3–6 mo later). BC, breast cancer.
Fig. 2.
SVM analysis for prediction of metastatic breast cancer. Analyte levels in samples collected 3–6 mo after surgery were compared with those in samples taken at the time of surgery. The differences in all analytes as identified on the arrays were fed to an SVM, which, using a leave-one-out cross-validation procedure, was calibrated to classify the patient as one who will develop metastatic cancer or one who will not. The analysis resulted in a decision value for each patient, significantly separating the groups (Wilcoxon P value of 1.7 × 10−5) and yielding an ROC AUC of 0.88. The heat map shows all analytes displaying a Wilcoxon P value <0.05, with red indicating an increase and green indicating a decrease in biomarker velocity.
Adopting the same strategy for de novo biomarker discovery but using instead the samples collected at 12 mo after surgery, we found a similar qualitative result but with less prediction power, as illustrated by an ROC AUC of 0.75 (Fig. S1). This trend could not be followed further, using samples collected at 24 and 36 mo, because the cohorts of patients not diagnosed with metastatic disease at these time points were too small for statistically relevant de novo biomarker discovery.
Analysis of all analytes (Fig. S2A) showed that increasing analyte velocities were clearly overrepresented in metastatic patients during this period, and the opposite was true for nonmetastatic patients. When the predictive biomarker signature was analyzed in more detail, Lewis X (P = 0.0005), IL-16 (P = 0.002), and CD40 (P = 0.0003) were shown to differ the most in velocity between patients with recurring versus nonrecurring breast cancer during the first 6 mo after surgery, as determined by Wilcoxon signed-rank test. In Fig. S2 B–D, which shows the dynamics for these three analytes, it is evident that a significant increase in the velocity of these analytes during these first months after surgery was more frequent in patients who developed a distant tumor relapse at a later time point.
Prevalidation of the Biomarker Signature Derived from the Discovery Cohort.
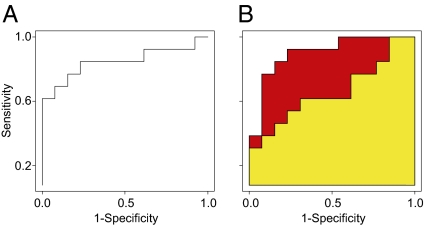
For implementation as risk assessment in clinical use, a signature comprising a small number of biomarkers is desirable. To define the smallest biomarker signature that retains predictive accuracy, we used a backward-elimination strategy to condense the total number of analytes down to the 21 nonredundant biomarkers making the greatest contribution to the classification (Fig. S3). Then, to test the strength of the classification derived from the discovery cohort, two serum samples were analyzed from each of the 26 new patients in the prevalidation cohort. The first sample was collected at the time of the primary operation, and the second was collected 3–6 mo later, following the same procedure used for the discovery cohort. Consequently, 52 independent samples were processed with our antibody microarray platform, as described above, and the velocity of each biomarker was determined. The classifier, now consisting of 21 biomarkers, allowed a stratification of patients into high versus low risk for tumor recurrence with an AUC of 0.85 in the independent prevalidation cohort (Fig. 3A). Of note, our previously reported cancer-associated biomarker signatures, defined using affinity proteomics, overlapped by <30% with the present 21 biomarkers (11, 13, 16). Importantly, our predictive signature did not reflect a general inflammatory response (17).
Fig. 3.
Prevalidation of the classification of patients into high- or low-risk groups for breast cancer recurrence. (A) An SVM was calibrated on the discovery dataset and tested on the prevalidation dataset using antibody array data derived from the 21-biomarker signature generated using backward elimination, as described in SI Materials and Methods. The ROC AUC for this classification was 0.85. (B) An SVM was calibrated using conventional clinical data (yellow) giving an ROC AUC = 0.66. The combination of the 21-biomarker signature and conventional clinical parameters (red) resulted in an ROC AUC = 0.90, demonstrating the added value in the array data.
To investigate if the signal identified at 3–6 mo was still detectable after 12 mo, the support vector machine (SVM) model based on the 21 biomarkers was applied on the 12-mo samples available in the discovery cohort. The classification resulted in an AUC of 0.86, showing that the identified signal still was present after 1 y.
The 21-biomarker signature was developed following the relevant REMARK guidelines for tumor biomarker studies (16). Furthermore, the effectiveness of the backward-elimination strategy also was evaluated by comparing the performance of the identified 21-biomarker signature with that of 1,000 randomly selected signatures of the same size. Each random signature was used to calibrate an SVM model in the same way described for the original 21-candidate signature. The candidate signature was shown to outperform 99.6% of the random signatures, based on AUC values (Fig. S4), demonstrating that the backward-elimination strategy indeed had produced a high-performing signature. Finally, a Kaplan–Meier plot was constructed to quantify further how the velocity-driven biomarker signature classified the patients into high- and low-risk groups. A log rank test based on the decision values from the SVM indicated a highly significant (P = 0.00037) difference between the groups (Fig. S5).
Furthermore, functional annotation of the signature proteins provided some insight into the underlying mechanisms, because several proteins [CCL2, CD40, factor B, IL-5, IL-6, IL-9, IL-13, IL-18, IL-12α, Lewis X/sialyl Lewis X, TNF-β, and serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 (SERPING1)] that displayed increasing velocities, indicating tumor recurrence, are known to be involved in cell migration and infiltration. In addition, several of the proteins in our biomarker signature, such as IL-1β, IL-8, regulated on activation, normal t expressed and secreted. (RANTES), and CD40L, are involved in the NF-κB pathway, which has been implicated in metastatic breast cancer.
Effect of Adjuvant Therapy on Classification into High- or Low-Risk Groups.
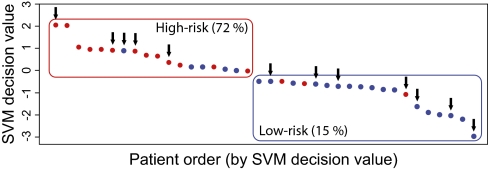
To test if the classification, as displayed in Fig. 2, was dependent on the therapy received by the patients in the discovery cohort, we analyzed the effect of adjuvant chemotherapy on the classification into high or low risk for breast cancer recurrence. In Fig. 4, the SVM decision values for all patients, as well as patients receiving chemotherapy during the first 3–6 postoperative months, are indicated by arrows. No stratification of the individual patients receiving chemotherapy could be detected. Similar results were obtained with patients receiving adjuvant endocrine treatment and patients treated with anti-inflammatory drugs; previously these treatments had been identified as confounding factors when absolute levels of analytes were measured (18). Hence, the classification into high- and low-risk groups for distant tumor relapse was not biased by a particular adjuvant therapy. Interestingly, because a few patients who received chemotherapy still were classified after 6 mo as belonging to the high-risk group, any beneficial effect of that particular therapy was not evident. Consequently, based on the molecular portrait derived from our microarray analysis, these patients could be selected for another treatment regime.
Fig. 4.
Chemotherapy was found not to be a confounding factor for our candidate biomarker signature. The SVM decision values for all of the patients are plotted, and patients receiving chemotherapy during the first 3–6 mo are indicated by arrows. When patients were categorized in risk groups based on the SVM decision value, 72% of patients in the high-risk group (red) but only 15% in the low-risk group (blue) later developed metastasis. When results were correlated to the patients who received adjuvant treatment (black arrows; 5 of 18 patents in the high-risk group and 7 of 20 patients in the low-risk group), it was evident that this factor did not bias the analysis.
Molecular Biomarker Signature Versus Conventional Diagnostic Parameters.
The power of molecular diagnostics sometimes has been questioned, in particular in relation to the conventional clinical parameters (19), such as lymph node status, tumor size, histological grade, and estrogen receptor (ER) and progesterone receptor (PgR) status. Therefore it is essential to compare the performance of our serum predictor with the performance of predictors based upon such clinical markers. We compared the combination of conventional parameters with our serum biomarker signature, which displayed an ROC AUC of 0.85 (Fig. 3A). Two additional SVM models were calibrated in the discovery dataset, using the clinical data and a combination of the clinical and microarray data, respectively. The models were tested using the patients in the prevalidation cohort, and the result was displayed as ROC curves. The AUC for the predictor using the conventional clinical parameters was 0.66. Consequently, the molecular signature based on analyte velocities had improved predictive power. Even more importantly, combining the conventional clinical parameters with our 21-biomarker signature as an approach for risk classification resulted in an AUC of 0.90 (Fig. 3B). Consequently, the two different sets of variables did not contain overlapping information with respect to the clinical outcome, as evident by a Pearson correlation analysis, which showed that the analyte velocities had weak correlations (≤0.35) with the conventional clinical markers. This analysis supports the fact that our protein serum approach identified unique information not present in conventional markers, in contrast to correlations observed between clinical markers and gene microarray profiling (19). For further comparison of our data and available decision-making tools, we investigated Adjuvant! Online (www.adjuvantonline.com), which estimates the risk for tumor relapse in breast cancer. The patient age, ER status, tumor grade, tumor size, and the number of positive nodes were entered online, and the calculated risk for relapse within 10 y was recorded for each patient. The patients then were sorted according to their estimated risk, and, using the true outcome for the patients in relation to distant recurrence, an ROC displaying an AUC of 0.60 was generated. This result based on conventional clinical parameters is in agreement with our analysis, which gave an AUC of 0.66. The disease-free follow-up time was 5 y in our prevalidation cohort.
Discussion
Breast cancer patients are treated either with local-regional therapy alone or by the addition of systemic therapy, resulting in improved clinical outcome. However, a considerable number of patients are overtreated, resulting in side effects for the patients and increasing costs for the health care providers. Consequently, prognostic parameters that would aid in making rational treatment decisions by stratifying patients into different risk groups are in high demand. Furthermore, predictive parameters for monitoring disease progression and therapy efficacy would be highly desirable, allowing evidence-based selection of adjuvant therapies and avoidance of overtreatment. Patients with the same stage of disease can respond very differently to therapy, showing that traditional parameters fail to classify breast cancer patients accurately according to their clinical need. However, recent progress in gene-expression profiling has shown promise in predicting the clinical outcome of breast cancer (8, 9) and in predicting the risk of developing local recurrence after breast-conserving surgery (20), as well as in earlier diagnosis (21). Because prediction of clinical outcome of breast cancer based on gene-expression profiles requires tumor tissue, gene-based approaches do not allow continuous disease monitoring and assessment of treatment efficacy after the initial tumor resection. For serial sampling, the preferred choice would be serum, but serum analysis using traditional proteomic approaches has not been possible because of technical limitations. (22). Furthermore, single biomarkers have not demonstrated added value clinically and consequently have not been accepted as routine tests in the breast cancer clinic (10). However, with the introduction of affinity proteomics in the microarray format (23, 24), the complexity of the proteome (25) has become much less of an issue (26).
In addition to the more classical plasma proteins, serum also consists of tissue leakage proteins (25), and much evidence suggests today that among these tissue leakage proteins are proteins indicative of a variety of human diseases. For example, because the human immune system is a very early indicator of disturbances in homeostasis caused by disease (27), a proteomic screen of nonimmunoglobulin low- and high-abundance immunoregulatory serum proteins might reveal a growing tumor. Based on this rationale, we designed a recombinant antibody microarray platform (28–30) displaying high sensitivity and reproducibility and focused on serum proteins associated with the immune system. This affinity proteomics approach recently has allowed the identification of several serum biomarker signatures distinguishing between different cancer indications and healthy individuals (11, 12, 18, 31), thereby demonstrating the power of the platform.
To investigate whether information transiently stored in serum could be deciphered and aid in the management of breast cancer, we screened 240 sera from a total of 64 patients with primary breast cancer for molecular portraits associated with the risk of developing distant relapse. Comparing samples collected longitudinally over a period of 3 y, we could not decipher molecular patterns in the serum proteome using static analyte levels; this result indicated that the signals either were too weak or were composed of parallel rather than orthogonal variability vectors. However, when instead we analyzed the change in analyte intensity over time, we could classify the patients successfully into groups at high and low risk for developing metastatic breast cancer. It was clear that the discriminatory biomarkers increased in intensity during the first 3–6 mo if a patient belonged to the high-risk group, and vice versa for the low-risk patients. De novo biomarker discovery at 12 mo resulted in patterns with less accuracy for risk assessment, as illustrated in Fig. S1. This reduced accuracy might be explained by diminishing signal integrity or by a decrease in the signal-to-noise ratio. Most previous proteomic studies are limited by the use of only a single cohort of patients to discover potential biomarkers. The present study used longitudinally collected samples, which are scarce and consequently were limited in number, from breast cancer patients. Clinical studies in which the number of samples is limited and a large number of marker candidates are analyzed are inherently susceptible to overfitting. To minimize confounding factors and to determine if our velocity-driven biomarker signature truly was associated with a risk for tumor relapse, it was necessary first to fix the classifier signature and then to apply it to samples from a completely independent patient cohort. Consequently, we first condensed the number of analytes to a 21-candidate biomarker signature, using our unbiased backward-elimination strategy (SI Materials and Methods). This strategy resulted in the identification of the smallest discriminatory signature, which was composed of members recognizing orthogonal patterns in the dataset, thus minimizing the risk of classifier overfitting. This biomarker signature is unlike a list that includes biomarkers only on the basis of low P values. An SVM classifier then was calibrated, using the identified biomarkers in the discovery cohort, and was used to test blindly the patients in the independent prevalidation cohort (with a sample size estimated to yield sufficient statistical power). It should be noted that the new samples were analyzed using a new batches of arrays and antibodies, thereby minimizing any systematic biases. This blind classification of the independent cohort yielded an AUC of 0.85. This small decrease of predictive power, compared with that observed in the discovery cohort, was expected, because classifiers always perform better in the dataset from which they originally were derived. Still, the AUC of 0.85 obtained in the secondary, independent cohort demonstrates is that the information harbored in a serum sample can be decoded, paving the way for developing a more personalized approach to the treatment of breast cancer patients. Of note, the signature did not seem to be affected by the adjuvant therapy received by the patients. This observation by itself demonstrates the need for an accurate prediction of metastatic breast cancer, because several of the patients in the high-risk group might have benefitted from a consecutive and different adjuvant treatment, just as some of the patients in the low-risk group may have been overtreated. It also should be noted that the identified candidate-biomarker signature was shown to be predictive only during the 5-y follow-up time, although breast cancer patients can have recurrences more than10 y after primary operation. Consequently, the predictive power of this proteomic signature still is limited in this regard.
Finally, the backward-elimination strategy also was interrogated by sampling 1,000 random 21-marker signatures, calibrating SVM models in the discovery cohort based on each of these signatures and by comparing their predictive power in the prevalidation cohort with that of the original 21-biomarker signature (AUC of 0.85). The latter was found to outperform almost all randomly sampled biomarker combinations, demonstrating that the rationale for our strategy was sound.
Interestingly, by applying the identified 21-biomarker signature from the SVM calibrated using the 3- to 6-mo velocities to samples collected at 12 mo, an accurate classification still could be achieved (AUC 0.86). This result supports the notion that the signal-to-noise ratio had decreased but that the information was still present a year later when a predefined biomarker signature was applied. At later time points the accuracy (as measured by AUC) decreased significantly.
To have a clinical impact, the serum-based biomarkers must outperform the traditional prognostic parameters of clinical status (age, menopausal status), histopathological status (histological grade, lymph node status, tumor size), and hormone receptor (ER, PgR) status (14). Existing precalibrated predictors based upon such parameters cannot be used straightforwardly in this case, because they typically assume relapse times longer than the 5 y used in our study and would be disfavored in a comparison. Therefore, we assessed the predictive power of the traditional parameters by calibration and blind testing of an SVM, with these markers as input values. We demonstrated that even their combined power, represented by a ROC AUC 0.66, was significantly lower than our prevalidated serum biomarker signature. Importantly, when we combined the traditional clinical parameters with our 21-biomarker signature, the predictive power increased even further, demonstrating that the serum markers provided clinically added value. Of note, this combined predictive power was independent of classical patient- and tumor-related prognostic factors, making it potentially useful for clinicians.
Finally, we performed a functional annotation using signal pathway analysis focusing on the condensed biomarker signature. For example, CCL2, CD40, factor B, IL-5, IL-6, IL-9, IL-13, IL-18, IL-12α, Lewis X/sialyl Lewis X, TNF-β, and SERPING1 have been implicated previously in cell migration and infiltration. Briefly, IL-6 is a multifunctional cytokine involved in angiogenesis, a process crucial for tumor growth and progression (32). Serum levels of IL-6 also have been associated previously with different cancers, including metastatic breast cancer (33). IL-8 and IL-18 are autocrine cytokines of the tumor also involved in angiogenic processes and have been investigated previously as diagnostic and prognostic factors for breast cancer (34, 35). Furthermore, RANTES (CCL5), Monocyte chemotactic protein-1 (MCP-1), and IL-1β in our signature have been reported to support invasiveness in human breast carcinoma cells and to play important roles in disease progression (36, 37). TGF-β stimulates cell invasion, and activation of TGF-β signaling has been identified as supporting breast cancer metastasis (38, 39). Of note, IL-1β, IL-8, RANTES, and CD40 ligand are all involved in the NF-κB pathway. Finally, overexpression of the carbohydrate sialyl Lewis X has been identified on invasive breast cancer cells and could be correlated to malignancy and a poor prognosis (40). Taken together, the biological functions of several of the defined biomarkers have been associated with metastatic breast cancer, lending indirect support to our findings. However, the rationale for using a network of the biomarkers for risk assessment needs further investigation.
In conclusion, we have addressed a clinically defined problem and demonstrated that serum biomarkers can deliver improved clinical value, with high accuracy, outperforming other attempts to classify breast cancer patients by risk of recurrence after primary operation. Furthermore, the defined biomarker signature was not a result of general inflammation but was associated specifically with the risk of breast cancer relapse. Finally, the data indicated that risk classification based on our serum signature provided a better means of guiding adjuvant therapy, reducing the rate of both over- and undertreatment.
Materials and Methods
Samples and Array Analysis.
Serum samples were collected from two independent cohorts of patients, denoted as the “discovery cohort” and the “prevalidation cohort” (Table 1). In the discovery cohort, 188 samples were collected from 38 patients diagnosed with primary breast cancer. Written informed consents were collected during the preoperative visit at the Department of Surgery (Lund University Hospital, Lund, Sweden), where the serum samples were collected. Time and date were recorded when the blood samples were drawn. Serum were stored at −80 °C for later analysis and labeled with serial codes to enable blinded analyses. For the majority of the patients, the preoperative visit took place less than a week before the surgery. Blood was drawn a second time at the first follow-up (3–6 mo later) and then approximately every 12 mo for 3 y. This study was approved by the Regional Ethical Committee (Lund, Sweden). In the prevalidation cohort, 52 serum samples were drawn, as described for the discovery cohort, from 26 different patients with newly diagnosed breast cancer. These samples were collected at a later stage than the samples in the discovery cohort. In total 240 serum samples were collected from the two cohorts. Patients with different tumor-related prognostic factors as well as patients who received different adjuvant therapies were selected (Table 1). Patients who did not develop distant recurrence were followed up for 5 y (Table 1).
Table 1.
Patient demographics and clinical parameters
| Discovery cohort |
Prevalidation cohort |
|||
| Group | Metastasis | No metastasis | Metastasis | No metastasis |
| Number of patients | 16 | 22 | 13 | 13 |
| Age in y | 54 (14)* | 52 (12) | 59 (13) | 60 (9) |
| Sample 2, collection time after primary operation (in mo) | 4.4 (1.1) | 4.5 (1.1) | 4.9 (2.1) | 4.0 (1.0) |
| Sample 3, collection time after primary operation (in mo) | 13 (1.0) | 13 (1.2) | — | — |
| Time to relapse (months) | 21 (10) | — | 19 (13) | — |
| Tumor size (mm) | 22 (10) | 21 (11) | 38 (23) | 16 (11) |
| Pre-/postmenopausal | 5/11 | 10/12 | 9/4 | 11/2 |
| ER+/ER− | 13/2† | 18/4 | 9/4 | 11/1 |
| PgR+/ PgR− | 9/6 | 13/9 | 7/6 | 7/5 |
| Lymph node+/lymph node− | 12/4 | 10/11 | 6/5 | 3/9 |
| Grade I/II/III | 1/9/5 | 3/13/5 | 0/9/4 | 5/6/1 |
| Ductal/lobular | 16/‡ | 20/2 | 11/2 | 11/2 |
| Radiation/no radiation | 11/5 | 14/8 | 11/2 | 9/4 |
| Adjuvant therapy: hormonal (tamoxifen) | 12 | 16 | 6 | 7 |
| Adjuvant therapy: aromatase inhibitor | 6 | 3 | 4 | 4 |
*Values in parenthesis are SDs.
†In cases where the sum is less than the number in the group, patient data are missing.
‡In the case of ductal/lobular tumor type, patients may have both, resulting in a sum larger than the number of patients in the group.
The recombinant antibody microarray platform contained 135 antibodies against 65 different antigens; i.e., for quality assurance we used two to five different antibody clones against most antigens (Table 2). Twenty-two of these antibodies were newly selected and directed against proteins previously shown to be involved in breast cancer and therefore had not been used in our earlier antibody array applications (11–13). The microarray analysis, including sample preparation, antibody production, array fabrication, and normalization, is described in detail in SI Material and Methods (11, 18).
Table 2.
Summary of biomarkers analyzed by microarray
| Antigen (no. of clones) | Antigen (no. of clones) |
| Alfa-10 (1) | IL-8 (3) |
| Alfa-11 (1) | IL-9 (3)* |
| Angiomotin (2) | IL-10 (3)† |
| APOA4 (3)* | IL-11 (3) |
| ATP5B (3)* | IL-12 (4)* |
| β-Galactosidase (1) | IL-13 (2)*,† |
| BTK (1)* | IL-16 (2) |
| C1q (1)† | IL-18 (3)* |
| C1s (1) | INF-γ (2) |
| C1 esterase inhibitor (1)* | JAK3 (1) |
| C3 (2) | KIAA0882 (3)* |
| C4 (1) | LDL (2) |
| C5 (2) | Leptin (1) |
| CD40 (4)* | Lewisx (2)* |
| CD40 ligand (1) | Lewisy (1) |
| CHX10 (3)* | Lumican (1) |
| Digoxin (1) | OSBPL3 (2)* |
| DUSP9 (1) | MCP-1 (3)*,† |
| Eotaxin (3) | MCP-3 (1) |
| Factor B (2)*,† | MCP-4 (2) |
| GLP-1 (1) | Myomesin (M-protein) 2 (2) |
| GLP-1 R (1) | Procathepsin (1) |
| GM-CSF (3) | Properdin (1) |
| IL-1α (3)* | PSA (1) |
| IL-1β (3) | Rantes (1) |
| IL-1ra (3) | Sialyl Lewis X (1)* |
| IL-2 (3) | TGF-β1 (2) |
| IL-3 (3) | TM peptide (1) |
| IL-4 (3)† | TNF-α (2) |
| IL-5 (3)*,† | TNF-β (4)* |
| IL-6 (4)*,† | UPF3 regulator of nonsense transcripts homolog B (yeast) (3) |
| IL-7 (2)* | VEGF (4) |
*Included in the 21 biomarker signature.
†Antibody specificity against these antigens was validated further by mass spectrometry, protein arrays, or ELISA.
To be able to compare the prediction for distant recurrence using conventional clinical parameters, we assigned numeric values to each of the available binary parameters: In each patient premenopausal/postmenopausal status; expression/absence of ER and/or PgR; ductal/lobular type; and tumor-free/-involved lymph nodes were assigned a value of 1 or −1, respectively, and histological grades I, II, and III were assigned value of −1, 0, and 1, respectively. Tumor size was graded continuously from −1 to 1. An SVM then was calibrated in the discovery cohort, using either the conventional clinical data or a combination of the clinical and microarray data.
Study Design.
In this retrospective study, we used patient groups of similar size in the discovery and prevalidation phase. The entire study design is outlined in Fig. 1. Furthermore, a power analysis based on the observed SVM decision values derived from the discovery cohort was performed to estimate the number of patients required to give a significant classification in the second, independent patient cohort (the prevalidation cohort) (SI Materials and Methods). A cohort size of at least 18 patients was estimated to be needed to reach a statistical power above 80%. The prevalidation cohort contained 26 patients, 50% of whom were identified as having distant breast cancer metastasis within the 5-y period. This sample size corresponded to an estimated statistical power of 93%. The total follow-up time for recurrence-free patients was 5 y.
Data Analysis Using the SVM.
The analyte velocity was defined as the change (log ratio) in signal for each analyte between the first serum sample, drawn preoperatively, and the postoperative sample drawn at a later time point. An SVM was used to classify the samples as belonging to one of two defined groups (SI Materials and Methods). The analyte velocity was used to calibrate (or “train”) and test the SVM classifier with leave-one-out cross-validation. Because the number of samples is less than the number of analytes, one needs to prune the latter to establish an SVM that is not overfitted and generalizes well in new patient cohorts. To this end, we created an analyte subpanel for the training cohort by selecting analytes that, in the training set, displayed the highest combined discriminatory power. This selection of analytes was made using a cross-validated backward-elimination strategy (SI Materials and Methods). Using this approach, we compiled a list of 21 analytes with the highest scores (Table 2) and calibrated a final SVM model, now termed “frozen.”
During the prevalidation, serum samples from 26 different patients with newly diagnosed breast cancer (the prevalidation cohort) were analyzed and tested with the previously frozen SVM classifier, using the analyte velocities.
Supplementary Material
Acknowledgments
We thank F. Pauly for performing array experiments; Dr. A. S. Albrekt for interactome analysis; and the research nurses M. B. Hedenblad, A. Casslén, K. Henriksson, A. Möller, L. Ågren, A. Weddig, U. Midelund, A. Nilsson, K. Sandström, and S. B. Olsson for sample collection and processing. This study was supported by grants from the Swedish Foundation for Strategic Research (CREATE Health), a Senior Individual Grant (to C.P.), Vetenskapsrådet section NT/M, the Swedish Cancer Society, the Åke Wiberg Foundation, the Mrs, Berta Kamprad Foundation, the Gunnar Nilsson Cancer Foundation, Lund University Hospital Foundation, Skåne County Council's Research and Development Foundation, Governmental Funding of Clinical Research within the National Health Service, and the ProteomeBinder Program.
Footnotes
Conflict of interest statement: Patent application on the biomarker signature (C.A.K.B. and C.W.).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103125108/-/DCSupplemental.
References
- 1.Jemal A, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.La Vecchia C, et al. Cancer mortality in Europe, 2000-2004, and an overview of trends since 1975. Ann Oncol. 2010;21:1323–1360. doi: 10.1093/annonc/mdp530. [DOI] [PubMed] [Google Scholar]
- 3.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 4.Dowsett M, et al. Meta-analysis of breast cancer outcomes in adjuvant trials of aromatase inhibitors versus tamoxifen. J Clin Oncol. 2010;28:509–518. doi: 10.1200/JCO.2009.23.1274. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN. Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol. 2007;608:1–22. doi: 10.1007/978-0-387-74039-3_1. [DOI] [PubMed] [Google Scholar]
- 6.Eifel P, et al. National Institutes of Health Consensus Development Conference Statement: Adjuvant therapy for breast cancer, November 1-3, 2000. J Natl Cancer Inst. 2001;93:979–989. doi: 10.1093/jnci/93.13.979. [DOI] [PubMed] [Google Scholar]
- 7.Goldhirsch A, et al. Panel members. Thresholds for therapies: Highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann Oncol. 2009;20:1319–1329. doi: 10.1093/annonc/mdp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van 't Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 9.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 10.Duffy MJ, Evoy D, McDermott EW. CA 15-3: Uses and limitation as a biomarker for breast cancer. Clin Chim Acta. 2010;411:1869–1874. doi: 10.1016/j.cca.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 11.Ingvarsson J, et al. Detection of pancreatic cancer using antibody microarray-based serum protein profiling. Proteomics. 2008;8:2211–2219. doi: 10.1002/pmic.200701167. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez-Carbayo M, Socci ND, Lozano JJ, Haab BB, Cordon-Cardo C. Profiling bladder cancer using targeted antibody arrays. Am J Pathol. 2006;168:93–103. doi: 10.2353/ajpath.2006.050601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellmark P, et al. Identification of protein expression signatures associated with Helicobacter pylori infection and gastric adenocarcinoma using recombinant antibody microarrays. Mol Cell Proteomics. 2006;5:1638–1646. doi: 10.1074/mcp.M600170-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Wingren C, Ingvarsson J, Dexlin L, Szul D, Borrebaeck CAK. Design of recombinant antibody microarrays for complex proteome analysis: Choice of sample labeling-tag and solid support. Proteomics. 2007;7:3055–3065. doi: 10.1002/pmic.200700025. [DOI] [PubMed] [Google Scholar]
- 15.Carter HB, Pearson JD. PSA velocity for the diagnosis of early prostate cancer. A new concept. Urol Clin North Am. 1993;20:665–670. [PubMed] [Google Scholar]
- 16.McShane LM, et al. Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics. REporting recommendations for tumor MARKer prognostic studies (REMARK) Nat Clin Pract Oncol. 2005;2:416–422. [PubMed] [Google Scholar]
- 17.Chechlinska M, Kowalewska M, Nowak R. Systemic inflammation as a confounding factor in cancer biomarker discovery and validation. Nat Rev Cancer. 2010;10:2–3. doi: 10.1038/nrc2782. [DOI] [PubMed] [Google Scholar]
- 18.Carlsson A, et al. Serum proteome profiling of metastatic breast cancer using recombinant antibody microarrays. Eur J Cancer. 2008;44:472–480. doi: 10.1016/j.ejca.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 19.Edén P, Ritz C, Rose C, Fernö M, Peterson C. “Good Old” clinical markers have similar power in breast cancer prognosis as microarray gene expression profilers. Eur J Cancer. 2004;40:1837–1841. doi: 10.1016/j.ejca.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 20.Niméus-Malmström E, et al. Gene expression profiling in primary breast cancer distinguishes patients developing local recurrence after breast-conservation surgery, with or without postoperative radiotherapy. Breast Cancer Res. 2008;10:R34. doi: 10.1186/bcr1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma P, et al. Early detection of breast cancer based on gene-expression patterns in peripheral blood cells. Breast Cancer Res. 2005;7:R634–R644. doi: 10.1186/bcr1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 23.Ekins RP. Ligand assays: From electrophoresis to miniaturized microarrays. Clin Chem. 1998;44:2015–2030. [PubMed] [Google Scholar]
- 24.Zhu H, Snyder M. Protein chip technology. Curr Opin Chem Biol. 2003;7:55–63. doi: 10.1016/s1367-5931(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 25.Anderson NL, Anderson NG. The human plasma proteome: History, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 26.Borrebaeck CAK, Wingren C. Design of high-density antibody microarrays for disease proteomics: Key technological issues. J Proteomics. 2009;72:928–935. doi: 10.1016/j.jprot.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 27.Storr SJ, et al. Use of autoantibodies in breast cancer screening and diagnosis. Expert Rev Anticancer Ther. 2006;6:1215–1223. doi: 10.1586/14737140.6.8.1215. [DOI] [PubMed] [Google Scholar]
- 28.Steinhauer C, Wingren C, Hager AC, Borrebaeck CAK. Single framework recombinant antibody fragments designed for protein chip applications. Biotechniques. 2002;(Suppl):38–45. [PubMed] [Google Scholar]
- 29.Wingren C, et al. Microarrays based on affinity-tagged single-chain Fv antibodies: Sensitive detection of analyte in complex proteomes. Proteomics. 2005;5:1281–1291. doi: 10.1002/pmic.200401009. [DOI] [PubMed] [Google Scholar]
- 30.Borrebaeck CAK, Wingren C. High-throughput proteomics using antibody microarrays: An update. Expert Rev Mol Diagn. 2007;7:673–686. doi: 10.1586/14737159.7.5.673. [DOI] [PubMed] [Google Scholar]
- 31.Carlsson A, et al. Plasma proteome profiling reveals biomarker patterns associated with prognosis and therapy selection in glioblastoma multiforme patients. Proteomics Clin Appl. 2010;4:591–602. doi: 10.1002/prca.200900173. [DOI] [PubMed] [Google Scholar]
- 32.Cohen T, Nahari D, Cerem LW, Neufeld G, Levi BZ. Interleukin 6 induces the expression of vascular endothelial growth factor. J Biol Chem. 1996;271:736–741. doi: 10.1074/jbc.271.2.736. [DOI] [PubMed] [Google Scholar]
- 33.Ravishankaran P, Karunanithi R. Clinical significance of preoperative serum interleukin-6 and C-reactive protein level in breast cancer patients. World J Surg Oncol. 2011;9:18. doi: 10.1186/1477-7819-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie K. Interleukin-8 and human cancer biology. Cytokine Growth Factor Rev. 2001;12:375–391. doi: 10.1016/s1359-6101(01)00016-8. [DOI] [PubMed] [Google Scholar]
- 35.Metwally FM, El-Mezayen HA, Ahmed HH. Significance of vascular endothelial growth factor, interleukin-18 and nitric oxide in patients with breast cancer: Correlation with carbohydrate antigen 15.3. Med Oncol. 2010 doi: 10.1007/s12032-010-9657-2. in press. [DOI] [PubMed] [Google Scholar]
- 36.Li J, et al. Role for ezrin in breast cancer cell chemotaxis to CCL5. Oncol Rep. 2010;24:965–971. doi: 10.3892/or.2010.965. [DOI] [PubMed] [Google Scholar]
- 37.Soria G, et al. Inflammatory mediators in breast cancer: Coordinated expression of TNFα & IL-1β with CCL2 & CCL5 and effects on epithelial-to-mesenchymal transition. BMC Cancer. 2011;11:130. doi: 10.1186/1471-2407-11-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drabsch Y, Ten Dijke P. TGF-beta signaling in breast cancer cell invasion and bone metastasis. J Mammary Gland Biol Neoplasia. 2011;16:97–108. doi: 10.1007/s10911-011-9217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Zhu H, Chen T, Dai G, Zou L. TGF-beta1 and BRCA2 expression are associated with clinical factors in breast cancer. Cell Biochem Biophys. 2011;60:245–248. doi: 10.1007/s12013-010-9146-4. [DOI] [PubMed] [Google Scholar]
- 40.Jeschke U, et al. Expression of sialyl Lewis X, sialyl Lewis A, E-cadherin and cathepsin-D in human breast cancer: Immunohistochemical analysis in mammary carcinoma in situ, invasive carcinomas and their lymph node metastasis. Anticancer Res. 2005;25(3A):1615–1622. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.