Abstract
We describe here a unique ethanol-inducible process for expression of recombinant proteins in transgenic plants. The process is based on inducible release of viral RNA replicons from stably integrated DNA proreplicons. A simple treatment with ethanol releases the replicon leading to RNA amplification and high-level protein production. To achieve tight control of replicon activation and spread in the uninduced state, the viral vector has been deconstructed, and its two components, the replicon and the cell-to-cell movement protein, have each been placed separately under the control of an inducible promoter. Transgenic Nicotiana benthamiana plants incorporating this double-inducible system demonstrate negligible background expression, high (over 0.5 × 104-fold) induction multiples, and high absolute levels of protein expression upon induction (up to 4.3 mg/g fresh biomass). The process can be easily scaled up, supports expression of practically important recombinant proteins, and thus can be directly used for industrial manufacturing.
Keywords: plant-made pharmaceuticals, TMV
Inducible expression of recombinant proteins and other molecules holds a major promise for yield optimization as it allows to separate the “growth” and the “production” phases in a manufacturing process. Indeed, although living cells can be engineered to express reasonably high levels of recombinant protein while continuing to perform other functions necessary for growth and development, at a certain expression level, energy, material, and other constraints impose limits on such “dual” growth plus production performance. Following the successful development of inducible systems relying on small molecules as chemical inducers for microbial biotechnology purposes (1), similar attempts have been made with eukaryotic cells and organisms (2).
However, when plants are used as expression hosts, recombinant protein expression is still mainly achieved using transgenic plants containing constitutively expressed transgenes, or rely on transient expression using standard nonreplicating vectors or replicating viral vectors (3). A number of inducible expression systems have nevertheless been developed for plants in the past 25 y (4–7). However, all solutions proposed so far are still lacking either one or both of the two performance criteria required for practical usefulness: low background expression in the uninduced state and high absolute yield upon induction. Tight control of expression in the uninduced state has been achieved using a number of inducible systems borrowed from nonplant organisms (8–14); however, all those systems rely on expression driven by a modified constitutive promoter and do not provide high expression levels upon induction. Such levels of expression can in fact easily be achieved without induction (by using the unmodified promoter), because, in case of nontoxic proteins, they are fully compatible with plant growth and impose little or no penalty on general biomass yield. The attention of researchers then turned to systems that have a better recombinant protein yield potential such as systems based on viral replicons (15). Unfortunately, the inducible systems developed so far still provide insufficient protein yields (16, 17), or show high leakiness (18), which may eventually lead to transgene silencing (19).
We have previously developed a transient expression process, magnifection, that is based on expression from viral RNA replicons delivered into plant cells systemically using Agrobacterium. Magnifection allows production of recombinant proteins at yields up to 5 g per kg of fresh leaf biomass (20, 21), which approaches the biological limits for protein expression. Such high yields are possible because of the transient nature of the process, which allows the use of very potent amplicons derived from RNA viruses such as Tobacco mosaic virus (TMV) or Potato virus X, without limiting biomass accumulation, which takes place prior to infection. Magnifection is an efficient and fast process but requires special equipment and is expensive to perform on a large scale, as transfection of entire plants is achieved by vacuum-infiltration in special apparatuses, and because the plants must be grown in trays or pots and have to be inverted for the infiltration process step (21). Furthermore, containment of the genetically engineered agrobacteria used for transfection is an additional constraint.
To bypass these limitations, we propose here an ethanol-inducible system that provides efficient release of viral RNA replicons from proreplicons contained in a stably transformed cassette. To achieve tight control of replicon activation in the uninduced state, the viral vector has been deconstructed, and its two components, the replicon and the movement protein, have been placed separately under control of an inducible promoter. Transgenic Nicotiana benthamiana plants incorporating this double-inducible system demonstrate negligible background expression, high induction multiples, as well as very high absolute levels of protein expression upon induction.
Results
Transgenic Plants Containing a Full Viral Vector Proreplicon Cannot Be Obtained.
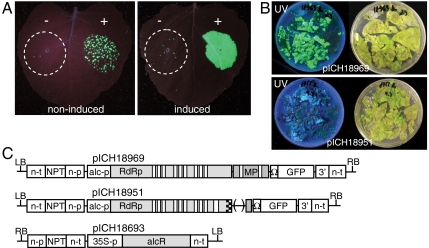
A first construct, pICH18969, was made, based on a previously described TMV-vector containing several introns for optimized nuclear export of the viral RNA (21). Although this vector lacks the gene coding for the coat protein, it is still able to replicate and move from cell-to-cell and is thus referred to as full vector. Expression of the viral replicon was under control of the Aspergillus nidulans alcohol dehydrogenase (alcA) promoter (Fig. 1C). The alcA promoter is active only upon binding of the AlcR transcriptional activator in the presence of ethanol. A second construct, pICH18693, was thus made containing the alcR gene cloned under control of the constitutive CaMV 35S promoter. The functionality of these constructs was tested transiently by agroinfiltration of Nicotiana benthamiana leaves and then treating the plants with ethanol 2 d later (time necessary for transient expression of AlcR). Coinfiltration of the viral vector and the AlcR construct resulted in the formation of some green fluorescent protein (GFP) spots in the absence of ethanol, but to confluent GFP foci in a leaf treated with ethanol (Fig. 1A), demonstrating ethanol inducibility of the viral vector construct. In the absence of AlcR, the number of GFP foci was much smaller, although not null. Despite the low leakiness of the system in transient tests, stable transformation of N. benthamiana with construct pICH18969 could not be achieved, due to background release of movement-competent viral vector in leaf explants during the transformation procedure. This release of viral vectors could be seen as strong GFP fluorescence of the leaf explants under UV light (Fig. 1B, upper box).
Fig. 1.
Test of inducible full viral vector pICH18969. (A) Agroinfiltration for transient expression of vector pICH18969 (OD600 = 2.5 × 10-3) without (−) or with (+) the transcriptional activator AlcR (construct pICH18693; OD600 = 0.25). The leaf shown on the right was treated with ethanol 2 d after infiltration. (B) Leaf disc transformation with vectors pICH18969 and pICH18951. Leaf explants pictured 10 d after transformation. The same petri dish is shown under white light (right) and UV light (left). (C) Maps of the T-DNA regions of constructs pICH18969, pICH18951 and pICH18693. RB and LB, right and left borders; 35S-p, CaMV 35S promoter; alc-p, alcA promoter from Aspergillus nidulans; n-p and n-t, promoter and terminator of the Agrobacterium nopaline synthase gene; NPT, neomycin phosphotransferase II for selection of transgenic plants; RdRp, viral RNA-dependent RNA polymerase; MP, movement protein for cell-to-cell movement; 3′, viral 3′-nontranslated region; Ω, translational enhancer from TMV. Introns in the RdRp and MP are shown as white boxes. The hatched part of RdRp in construct pICH18951 indicates silent mutations.
Deconstructed Virus—Transient Tests.
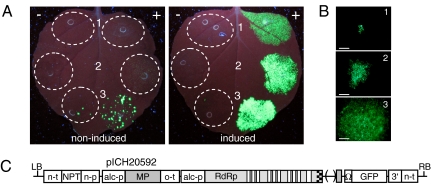
Because transformation of a full vector was not successful, the viral vector was deconstructed, i.e., the viral movement protein (MP) that is responsible for cell-to-cell movement was deleted, resulting in construct pICH18951 (Fig. 1C). Unintended release of viral replicons during agrotransformation, or later in transgenic plants containing such a construct, may sporadically lead to a few cells with actively replicating viral RNA, but because cell-to-cell movement is lacking, this should not lead to whole plant infection and may thus be tolerable. Accordingly, we observed only single GFP expressing cells in N. benthamiana leaf explants during transformation of this construct (Fig. 1B, lower box) followed by successful regeneration of transgenic plants (see below). Although cell-to-cell movement would not be required in a scenario where all cells of a transgenic plant become induced and start viral replication, this is not likely to happen under real conditions. Therefore, in order to obtain high-level recombinant protein expression, the MP gene was added back to the construct, but outside of the viral replicon (in trans) and under control of the alcA promoter, resulting in construct pICH20592 (Fig. 2C). Using this arrangement, viral replicons will be able to move from one cell to the next upon induction, but the double level of control on both viral vector release and movement is expected to provide a tighter control.
Fig. 2.
Test of deconstructed vectors pICH18951 and pICH20592. (A) Agroinfiltration for transient expression of vectors pICH18951 (1), pICH20592 (2), and pICH18969 (3) without (−) or with (+) AlcR (construct pICH18693; OD600 = 0.25). Agrobacteria for viral constructs were used at OD600 = 0.025 (pICH18951, pICH20592) or OD600 = 2.5 × 10-3 (pICH18969). The leaf shown on the right was treated with ethanol 2 d after infiltration. (B) Microscopic view of single transfection events after agroinfiltration at OD600 = 2.5 × 10-6 and induction with ethanol. Bars represent 2 μm. (C) Map of the T-DNA region of construct pICH20592. o-t, terminator of the Agrobacterium octopine synthase gene. All other elements are explained in Fig. 1.
The new construct was tested transiently and, like the construct lacking MP, showed negligible background in the absence of AlcR (Fig. 2A). Coinfiltration with AlcR in noninduced leaves resulted in a low number of fluorescent cells, which increased strongly when the leaf was treated with ethanol. Although both constructs didn’t reach the same level of expression as pICH18969, the benefit of providing MP in trans was clear. The extent of cell-to-cell movement was demonstrated in a further infiltration, where the agrobacteria were used at low cell density (OD600 = 2.5 × 10-6), allowing to distinguish single transfection events (Fig. 2B). After ethanol induction, expression from pICH18951 was restricted to single cells, whereas fluorescent cell clusters were found with both constructs containing MP. Because MP was not part of the viral replicon in case of pICH20592, it is only expressed in the initially transfected cell. Movement of the replicon is thus limited to 1–2 cell layers adjacent to the transfected cell resulting in fluorescent foci smaller than those seen with the full viral vector.
Generation of Stable Transgenic Plants.
Constructs pICH18693 (AlcR), pICH18951 (viral vector, no MP), and pICH20592 (viral vector, MP in trans) were transformed independently into N. benthamiana. Successful DNA integration in primary transformants was evaluated by agroinfiltration with the complementing constructs and induction with ethanol (Fig. S1A). Several plants with the expected phenotypes were selected and crossed in order to combine the AlcR activator and the inducible vector in the same plant. Progeny from these crosses were screened by ethanol induction (Fig. S1B) and plants with the strongest expression were selected. Plant lines containing all elements for high-level inducible expression (i.e., plants containing AlcR, viral vector and MP) were propagated for three generations until no segregation was observed any more.
Optimizing Induction Conditions.
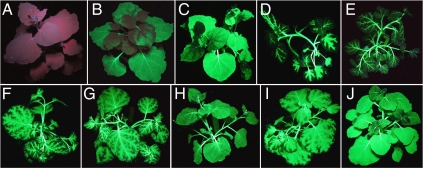
The transgenic plants described above were used to optimize the induction conditions (Fig. 3). The tested parameters include ethanol concentration, application route, and single vs. repeated treatments. A single spray with up to 20% ethanol was inefficient, but repeated spraying (up to 10 times, twice a day) with 4% ethanol resulted in a quite uniform expression in the leaves (Fig. 3 B and C). However, expression seemed to be restricted to the cells in immediate contact with the inducer. Root drenching with up to 4% ethanol was very efficient in inducing expression in the stem and along the major leaf veins (Fig. 3D). Higher concentrations and repeated treatments with 4% ethanol usually were harmful for the plants. Repeated watering with lower concentrations of ethanol gave only minor improvements (Fig. 3 E and F). Good results were obtained by a single combined treatment of root drenching and spraying with 4% ethanol (Fig. 3G). A further improvement could be achieved by incubating the plants for 24 h in ethanol vapor with or without a prior spray- or root-treatment (Fig. 3 H–J). Other chemical compounds known from the literature for their inducing capacity (acetaldehyde, isopropanol, and butanol) were also tested, but found to be less effective and/or more toxic.
Fig. 3.
Test of different ethanol treatments. Transgenic plants containing constructs pICH18693 (alcR) and pICH20592 (viral vector, MP) were treated with ethanol using different application methods: (A) noninduced plant; (B) one spray per day with 4% ethanol on five subsequent days; (C) same as (B) with two sprays per day; (D) root drenching with 4% ethanol; (E) root drenching repeated three times with 1% ethanol; (F) same as (E) with 2% ethanol; (G) root drenching and spraying with 4% ethanol; (H) incubation in ethanol vapor for 24 h without any pretreatment; (I) root drenching with 4% ethanol followed by incubation in ethanol vapor; and (J) spraying with 4% ethanol followed by incubation in ethanol vapor.
Quantification of Recombinant Protein Expression.
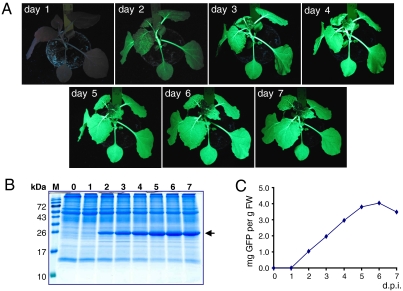
First, we followed the progress of expression after induction. Transgenic plants were induced by a combination of root drenching and vapor. Every 24 h, pictures were taken (Fig. 4A) and plants were harvested. The aerial parts of the plants were ground in liquid nitrogen and extracted for analysis by SDS-PAGE (Fig. 4B) and fluorescence spectrometry (Fig. 4C). GFP fluorescence was first detected 2 d after induction, correlating with the appearance of a strong band at 27 kDa in the protein gel. The highest expression levels were found from day five to seven after induction.
Fig. 4.
Time course of GFP expression after induction. (A) Pictures of the same plant taken under UV light at 1–7 d after ethanol treatment. (B) Total protein extracts of different plants harvested before (day 0) or at different times after induction (day 1–7). The arrow points to GFP (27 kDa). M: molecular mass standard. (C) GFP concentration measured by fluorescence spectrometry on the same samples as in (B).
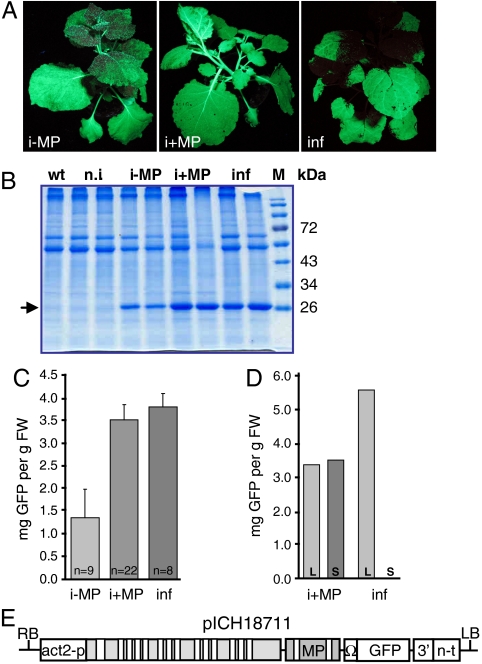
Next, we compared the level of expression obtained after ethanol induction with the levels obtained using the established magnifection process. Several transgenic plants [transformed with pICH18951 (no MP) or pICH20592] were induced by the root drenching-vapor method. In parallel, the constitutive and highly expressing full viral vector pICH18711 (21) was vacuum infiltrated into N. benthamiana plants (Fig. 5A). Entire plants (leaves and stems) were harvested seven days after induction/infiltration, homogenized, and analyzed by SDS-PAGE (Fig. 5B). A strong band corresponding to GFP was seen in induced and infiltrated plants whereas no such band was visible in samples from a wild type plant (WT) and from noninduced transgenic plants (n.i.). Induced plants without MP (i-MP) had lower expression levels than plants containing an inducible MP (i+MP) or the vacuum-infiltrated plants (inf). The amount of expression in different plants was then quantified by fluorescence spectrometry (Fig. 5C; data for individual plants are given in Table S1). GFP was expressed at a level of 2.7–4.3 mg/g fresh weight (FW) from induced plants with MP, which was in the same range as expression from infiltrated plants (3.3–4.1 mg/g FW). In contrast, induced plants without MP gave weaker expression (1.35 ± 0.63 mg/g FW), suggesting that the proportion of cells with replicating viral RNA is lower in plants lacking MP. We suspect that a fraction of cells in these plants was either not accessible to the inducer or that the effective inducer concentration was not high enough in all cells. This view is supported by the expression pattern in plants induced by root drenching where the expression decreases with increasing distance from the main leaf veins indicating a diffusion gradient of the inducer (Fig. 3 D–G).
Fig. 5.
Quantification of expression. (A) Pictures of induced plants and of a vacuum-infiltrated plant. i-MP, induced transgenic plant lacking MP (pICH18951/pICH18693); i+MP, induced transgenic plant containing MP (pICH20592/pICH18693); inf, WT plant infiltrated with pICH18711; (B) SDS-PAGE with crude plant extracts. n.i., noninduced transgenic plants, M, molecular mass standard. The arrow points to GFP. (C, D) Fluorescence spectrometric measurements of GFP concentration. (C) Average GFP concentration in extracts from the entire aerial part of induced or infiltrated plants. Error bars indicate standard deviation. n, number of plants analyzed. (D) GFP amount in leaf (L) and stem (S) tissue extracted separately for individual plants. (E) Map of the T-DNA region of construct pICH18711 used for vacuum infiltration. Act2-p, promoter of the A. thaliana actin2 gene. All other elements are explained in Fig. 1.
As the next step, we analyzed the different expression patterns of induced and infiltrated plants. Whereas agroinfiltration is restricted to leaves and leaf parts with expanded intercellular space, ethanol induction is effective throughout the plant including roots, stems, petioles, and young leaves, although expression is not as uniform in leaves as with infiltration-based transfection (Fig. 5A). In order to estimate the contribution of the different tissues to overall expression, leaves were extracted separately from stems/petioles and GFP fluorescence was measured (Fig. 5D). As expected, no fluorescence could be detected in the stem of an infiltrated plant, whereas leaf and stem tissue of an induced plant gave approximately the same relative expression levels. Fluorescence in leaves from an induced plant was lower than from an infiltrated plant, but this was compensated by the higher expression level in stems and petioles of the induced plant. These tissues make up 35–40% of the total plant biomass leading to an overall yield in induced plants as high as 92% of the yield in pICH18711-infiltrated plants (Fig. 5C).
Quantification of Induction Level.
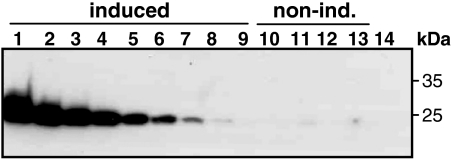
The relative increase of expression upon induction in transgenic plants was determined by Western blot using a GFP-specific antibody. GFP was still detectable in a 1/5,000 dilution of the extract from an induced plant (Fig. 6, lane 8), whereas no signal was detected in the undiluted extracts from three out of four noninduced plants (lanes 10, 12, and 13). In one of the noninduced plants (lane 11) a weak signal could be detected that was as strong as the signal of the 1/5,000 dilution of the induced plant. These results show that there is indeed very low background expression and that induction leads to an at least 0.5 × 104-fold increase in expression.
Fig. 6.
Level of induction determined by Western blot using anti-GFP antibody. Lane 1–9: serial dilution of an extract from an induced transgenic plant. Dilution factors are: 1∶20, 1∶50, 1∶100, 1∶200, 1∶500, 1∶1,000, 1∶2,000, 1∶5,000 and 1∶10,000. Lanes 10–13: extracts from different noninduced transgenic plants. Lane 14: extract from WT plant.
Discussion
High yields of recombinant proteins in plants have already been achieved with a number of expression systems, but all technologies currently available provide partial solutions only. High expression has been reported for nuclear transgenes in seeds (22), with yields of up to 10 g of recombinant protein (lysozyme or lactoferrin) per kg of seed (23). However, because the seed harvest per hectare (5–10 tons/ha) is much lower than the green biomass harvest (100 to 300 tons/ha), the actual yield per hectare is still relatively low. Recombinant proteins can also be expressed in chloroplasts (i.e., in green biomass) at extremely high yield of up to 50% of total soluble protein (24, 25). However, proteins synthesized in plastids are not glycosylated, greatly narrowing the range of pharmaceutical proteins that can be made using this technology. Transient expression systems, such as magnifection, also provide very high absolute and relative yields of recombinant proteins in green biomass, but large scale production is limited to greenhouse production and is therefore relatively expensive. The system described in this paper provides a solution that is inexpensive, indefinitely scalable, does not impose limitations on the class of proteins to be produced, and provides very high expression levels.
The major advantage of viral vectors, i.e., autonomous amplification and spread of genetic information, also presents a major challenge for developing an inducible viral vector system in plants, because background release of a single functional viral replicon might lead to uncontrollable replication and movement of the viral RNA, leading to silencing (19), or even preventing regeneration of a transgenic plant. Because no inducible system can guarantee a zero level of background activation under noninducing conditions, solutions must be found to bypass this problem. At one end of the spectrum of possible solutions, rather than trying to prevent release of viral replicons under noninducing condition, transgenic plants can be regenerated containing a constitutively silenced replicon, allowing the plants to be grown without the detrimental effects of viral replication, and silencing can be released by switching the plants to a lower temperature (18). Such system is however limited by the difficulty to strictly control temperature, even in the relatively controlled setting of a greenhouse. Alternatively, silenced plants can be crossed with plants transgenic for a suppressor of silencing (26). This approach is however not suitable for proteins exhibiting toxicity for plant cells, or if the released viral vectors replicate at levels that inhibit plant growth.
Another possibility is the use of cell suspension cultures rather than intact plants (17, 27, 28). In this case, background release of a viral vector in individual cells does not affect other cells in the culture. However, although reported expression levels were quite high and reproducible, the specific requirements of cell suspension cultures like tailored growth media and sterile working conditions seriously limit the scalability of these systems.
Potentially more versatile and powerful are systems that control activation and release of the viral replicons. Two systems based on geminivirus (17) or the tripartite Brome mosaic virus (16) control release and amplification of the replicon by placing the replicase under control of an inducer. Both systems require continuous or repeated induction in order to maintain replication and expression. Additionally, both viral replicons do not have cell-to-cell movement ability, limiting expression to the cells that have been reached by the inducer. In contrast, the system that we have developed allows release of viral RNA that is autonomously replicating and is capable of cell-to-cell movement. The double level of control on both the release of viral RNA and on cell-to-cell movement of the released replicons allows regeneration of transgenic plants that have a WT phenotype. Under noninducing conditions, rare individual cells expressing GFP can be found. Fortunately, this very low number of cells containing replicating vector does not lead to silencing of the plant, as evidenced by the high level of expression obtained upon induction with ethanol. This design allows to reach extremely high level of recombinant protein expression, comparable to levels obtained transiently using magnifection, but without the need for plant infiltration.
Many of the inducible viral systems that have been previously reported are based on mammalian hormones and their corresponding receptors (estrogen, glucocorticoids) (16, 27–29) and, although these systems are well characterized and show good inducibility, the use of hormones or hormone analogs is restricted to closed environments and small scale applications. We chose ethanol for our inducible viral system because ethanol is relatively inexpensive and nontoxic and the system exhibits a low background in the uninduced state. It should be however noted that our double-inducible system is not limited to ethanol as the inducer, but could easily be adapted to other inducible systems that might function well in the field such as those based on ecdyson-receptor agonists (like the insecticides tebufenozide or methoxyfenozide) (12, 13, 30).
In conclusion, this report describes the successful implementation of a highly expressing viral vector into transgenic plants without the limitations previously reported for such systems. Preliminary results in our lab indicate that this technology is well suited for the expression of different valuable proteins and thus expands the toolbox for recombinant protein production in plants.
Methods
Constructs.
All plasmids used for cloning were described earlier (21). pICH18969 is a fully assembled viral vector derived from pICH17272. First, a SpeI site was introduced by PCR at the transcriptional start site of pICH17272. Then, the alcA promoter was amplified by PCR from A. nidulans genomic DNA and fused with a minimal 35S promoter sequence (10) added as primer extension. The PCR product was cloned as KpnI-SpeI fragment into the modified viral vector. To make pICH18951, the 3′-region of the viral polymerase (RdRp) and the MP in pICH18969 were replaced by an XhoI-HindIII fragment from pICH16141 containing silent mutations in the RdRp and a 575 bp deletion in the MP. An alcA promoter-MP fusion construct (pICH19940) was made by replacing the 35S promoter in construct pICH10745 with an EcoRI-XhoI digested PCR-product of the alcA promoter. pICH20592 was made by cloning a BsaI-PstI fragment from pICH19940 into BsaI-NsiI digested pICH18951.
The AlcR coding sequence was also amplified by PCR and cloned as NcoI-EcoRI fragment into a small, pBIN19 derived binary vector containing the 35S promoter and the nopaline synthase terminator, giving construct pICH18693. The identity and correctness of all PCR-products was confirmed by sequencing.
Agroinfiltration.
Infiltrations were done as described (20). In brief, agrobacterium overnight cultures were grown in LB medium to high cell density (OD600 = 2.0–4.0) and were diluted between 1∶5 and 1∶106 directly into infiltration buffer (10 mM MES, pH 5.5; 10 mM MgSO4) to reach the desired concentration. Bacterial suspensions were infiltrated into N. benthamiana leaves using a syringe without needle. The magnifection process (vacuum infiltration of whole plants) was carried out as described (21).
Plant Transformation and Regeneration.
N. benthamiana was transformed by leaf disk transformation and selected on kanamycin-containing medium using a slightly modified standard protocol (31). Regenerated plants were transferred to the greenhouse and tested by agroinfiltration with the complementing construct for the presence and expression of the transgene. Plants transformed with pICH18951 or pICH20592 were infiltrated with pICH18693. Plants transformed with pICH18693 were infiltrated with pICH20592. Plants were induced with ethanol two days after infiltration.
Ethanol Induction.
Inductions were performed in different ways. Root drenching was done with ethanol concentrations up to 10% (v/v) using 40 mL for a single plant grown in a pot with 9 cm diameter. Spraying with up to 20% (v/v) ethanol was done using a hand-hold spray bottle until run-off. For vapor induction, plants were transferred to a tightly closed plastic container (126 L) containing a tray with 1 l of 4% (v/v) ethanol and incubated for 24 h.
Protein Extraction and Analysis.
For quantification of GFP, whole plants or plant parts were extracted in 4–6 volumes (i.e., 4–6 mL per gram FW) of buffer (200 mM Tris/HCl, pH 8.0; 5 mM EDTA; 0.1% Tween 20) for 45 s at high speed in a Waring blender. Small leaf samples (30–100 mg) were frozen in liquid nitrogen, ground to fine powder, and suspended in extraction buffer. GFP fluorescence was measured by fluorescence spectrometry as described (20). rGFP from Roche Diagnostics was used as a standard. Denaturing protein electrophoresis and blotting were done according to standard procedures using equipment from Bio-Rad. Coomassie-staining was done with PageBlue™ staining solution from Fermentas. Western blots were probed with an anti-GFP antibody (dilution 1∶1,000; Roche Diagnostics) and detected with a goat anti-mouse-HRP conjugate (1∶10,000; Sigma) and the ECL chemiluminescence reagent (GE Healthcare).
Supplementary Material
Footnotes
Conflict of interest statement: All authors are current or former employees of Icon Genetics GmbH.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102928108/-/DCSupplemental.
References
- 1.Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2006;72:211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 2.Weber W, Fussenegger M. Inducible product gene expression technology tailored to bioprocess engineering. Curr Opin Biotechnol. 2007;18:399–410. doi: 10.1016/j.copbio.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Klimyuk V, Marillonnet S, Knäblein J, McCaman M, Gleba Y. In: Modern Biopharmaceuticals. Knäblein J, editor. Weinheim: Wiley-VCH; 2005. pp. 893–917. [Google Scholar]
- 4.Corrado G, Karali M. Inducible gene expression systems and plant biotechnology. Biotechnol Adv. 2009;27:733–743. doi: 10.1016/j.biotechadv.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Moore I, Samalova M, Kurup S. Transactivated and chemically inducible gene expression in plants. Plant J. 2006;45:651–683. doi: 10.1111/j.1365-313X.2006.02660.x. [DOI] [PubMed] [Google Scholar]
- 6.Padidam M. Chemically regulated gene expression in plants. Curr Opin Plant Biol. 2003;6:169–177. doi: 10.1016/s1369-5266(03)00005-0. [DOI] [PubMed] [Google Scholar]
- 7.Wang R, Zhou X, Wang X. Chemically regulated expression systems and their applications in transgenic plants. Transgenic Res. 2003;12:529–540. doi: 10.1023/a:1025852307127. [DOI] [PubMed] [Google Scholar]
- 8.Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- 9.Bohner S, Lenk II, Rieping M, Herold M, Gatz C. Technical advance: transcriptional activator TGV mediates dexamethasone-inducible and tetracycline-inactivatable gene expression. Plant J. 1999;19:87–95. doi: 10.1046/j.1365-313x.1999.00503.x. [DOI] [PubMed] [Google Scholar]
- 10.Caddick MX, et al. An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nat Biotechnol. 1998;16:177–180. doi: 10.1038/nbt0298-177. [DOI] [PubMed] [Google Scholar]
- 11.Gatz C, Frohberg C, Wendenburg R. Stringent repression and homogeneous de-repression by tetracycline of a modified CaMV 35S promoter in intact transgenic tobacco plants. Plant J. 1992;2:397–404. doi: 10.1111/j.1365-313x.1992.00397.x. [DOI] [PubMed] [Google Scholar]
- 12.Martinez A, Sparks C, Hart CA, Thompson J, Jepson I. Ecdysone agonist inducible transcription in transgenic tobacco plants. Plant J. 1999;19:97–106. doi: 10.1046/j.1365-313x.1999.00504.x. [DOI] [PubMed] [Google Scholar]
- 13.Tavva VS, Dinkins RD, Palli SR, Collins GB. Development of a tightly regulated and highly inducible ecdysone receptor gene switch for plants through the use of retinoid X receptor chimeras. Transgenic Res. 2007;16:599–612. doi: 10.1007/s11248-006-9054-y. [DOI] [PubMed] [Google Scholar]
- 14.Zuo J, Niu QW, Chua NH. Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
- 15.Gleba YY, Giritch A. In: Recent Advances in Plant Virology. Caranta C, Aranda MA, Tepfer M, Lopez-Moya JJ, editors. Norfolk, United Kingdom: Caister Academic Press; 2011. pp. 387–412. [Google Scholar]
- 16.Mori M, Fujihara N, Mise K, Furusawa I. Inducible high-level mRNA amplification system by viral replicase in transgenic plants. Plant J. 2001;27:79–86. doi: 10.1046/j.1365-313x.2001.01079.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Mason H. Bean Yellow Dwarf Virus replicons for high-level transgene expression in transgenic plants and cell cultures. Biotechnol Bioeng. 2006;93:271–279. doi: 10.1002/bit.20695. [DOI] [PubMed] [Google Scholar]
- 18.Dujovny G, Valli A, Calvo M, Garcia JA. A temperature-controlled amplicon system derived from Plum pox potyvirus. Plant Biotechnol J. 2009;7:49–58. doi: 10.1111/j.1467-7652.2008.00373.x. [DOI] [PubMed] [Google Scholar]
- 19.Angell SM, Baulcombe DC. Consistent gene silencing in transgenic plants expressing a replicating potato virus X RNA. Embo J. 1997;16:3675–3684. doi: 10.1093/emboj/16.12.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marillonnet S, et al. In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc Natl Acad Sci USA. 2004;101:6852–6857. doi: 10.1073/pnas.0400149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marillonnet S, Thoeringer C, Kandzia R, Klimyuk V, Gleba Y. Systemic Agrobacterium tumefaciens-mediated transfection of viral replicons for efficient transient expression in plants. Nat Biotechnol. 2005;23:718–723. doi: 10.1038/nbt1094. [DOI] [PubMed] [Google Scholar]
- 22.Boothe J, et al. Seed-based expression systems for plant molecular farming. Plant Biotechnol J. 2010;8:588–606. doi: 10.1111/j.1467-7652.2010.00511.x. [DOI] [PubMed] [Google Scholar]
- 23.Hennegan K, et al. Improvement of human lysozyme expression in transgenic rice grain by combining wheat (Triticum aestivum) puroindoline b and rice (Oryza sativa) Gt1 promoters and signal peptides. Transgenic Res. 2005;14:583–592. doi: 10.1007/s11248-004-6702-y. [DOI] [PubMed] [Google Scholar]
- 24.Cardi T, Lenzi P, Maliga P. Chloroplasts as expression platforms for plant-produced vaccines. Expert Rev Vaccines. 2010;9:893–911. doi: 10.1586/erv.10.78. [DOI] [PubMed] [Google Scholar]
- 25.Chebolu S, Daniell H. Chloroplast-derived vaccine antigens and biopharmaceuticals: expression, folding, assembly and functionality. Curr Top Microbiol Immunol. 2009;332:33–54. doi: 10.1007/978-3-540-70868-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallory AC, et al. The amplicon-plus system for high-level expression of transgenes in plants. Nat Biotechnol. 2002;20:622–625. doi: 10.1038/nbt0602-622. [DOI] [PubMed] [Google Scholar]
- 27.Dohi K, et al. Inducible virus-mediated expression of a foreign protein in suspension-cultured plant cells. Arch Virol. 2006;151:1075–1084. doi: 10.1007/s00705-005-0705-8. [DOI] [PubMed] [Google Scholar]
- 28.Huang TK, Plesha MA, Falk BW, Dandekar AM, McDonald KA. Bioreactor strategies for improving production yield and functionality of a recombinant human protein in transgenic tobacco cell cultures. Biotechnol Bioeng. 2009;102:508–520. doi: 10.1002/bit.22061. [DOI] [PubMed] [Google Scholar]
- 29.Tremblay A, Beauchemin C, Seguin A, Laliberte JF. Reactivation of an integrated disabled viral vector using a Cre-loxP recombination system in Arabidopsis thaliana. Transgenic Res. 2007;16:213–222. doi: 10.1007/s11248-006-9038-y. [DOI] [PubMed] [Google Scholar]
- 30.Tavva VS, Dinkins RD, Palli SR, Collins GB. Development of a methoxyfenozide-responsive gene switch for applications in plants. Plant J. 2006;45:457–469. doi: 10.1111/j.1365-313X.2005.02628.x. [DOI] [PubMed] [Google Scholar]
- 31.Horsch RB, Fraley RT, Rogers SG, Sanders PR, Lloyd A. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.