Abstract
Ultraspiracle protein/retinoid X receptor (USP/RXR) is a nuclear receptor and transcription factor which is an essential component of a heterodimeric receptor complex with the ecdysone receptor (EcR). In insects this complex binds ecdysteroids and plays an important role in the regulation of growth, development, metamorphosis and reproduction. In some holometabolous insects, including Lepidoptera and Diptera, USP/RXR is thought to have experienced several important shifts in function. These include the acquisition of novel ligand-binding properties and an expanded dimerization interface with EcR. In light of these recent hypotheses, we implemented codon-based likelihood methods to investigate if the proposed shifts in function are reflected in changes in site-specific evolutionary rates across functional and structural motifs in insect USP/RXR sequences, and if there is any evidence for positive selection at functionally important sites. Our results reveal evidence of positive selection acting on sites within the loop connecting helices H1 and H3, the ligand-binding pocket, and the dimer interface in the holometabolous lineage leading to the Lepidoptera/Diptera/Trichoptera. Similar analyses conducted using EcR sequences did not indicate positive selection. However, analyses allowing for variation across sites demonstrated elevated non-synonymous/synonymous rate ratios (d N/d S), suggesting relaxed constraint, within the dimerization interface of both USP/RXR and EcR as well as within the coactivator binding groove and helix H12 of USP/RXR. Since the above methods are based on the assumption that d S is constant among sites, we also used more recent models which relax this assumption and obtained results consistent with traditional random-sites models. Overall our findings support the evolution of novel function in USP/RXR of more derived holometabolous insects, and are consistent with shifts in structure and function which may have increased USP/RXR reliance on EcR for cofactor recruitment. Moreover, these findings raise important questions regarding hypotheses which suggest the independent activation of USP/RXR by its own ligand.
Introduction
USP/RXR, a group II nuclear receptor and transcription factor which belongs to the steroid receptor superfamily, is found across a diversity of Metazoan species ranging from sponges to mammals [1]. First characterized in Drosophila melanogaster, USP is the insect homolog of vertebrate RXR [2], [3]. In insects USP/RXR is involved in an array of functions including metamorphosis, reproduction, growth and development (see [4], [5], [6], [7], [8] for examples). USP/RXR is known primarily as the partner of the ecdysone receptor (EcR). In response to the binding of 20-hydroxyecdysone (20E) to EcR, heterodimerization between USP/RXR and EcR occurs and the complex then binds DNA to transactivate genes [9], [10], [11]. However, it is the controversy over whether or not USP/RXR binds juvenile hormone (JH) in insects, and is activated independent of EcR, which has led to the most debate regarding the function and evolution this receptor [12], [13], [14].
In vertebrates, 9-cis retinoic acid (9cRA) was identified as the high affinity ligand of the RXR receptor [15], [16], [17]. However, in insects no natural ligand has been conclusively identified, and USP/RXR remains an orphan receptor. JH has been suggested as a candidate ligand [12], but only experimental evidence from the dipteran D. melanogaster supports this hypothesis. In cell lines expressing Drosophila USP, the application of JH III induces the transcription of a transfected promoter, suggesting that JH binds USP resulting in a functional outcome [13], [18], [19]. Fluorescence-binding assays have shown that Drosophila USP binds not only JH III but also the JH precursors farnesol, farnesoic acid, and methyl farnesoate [20]. However, JH does not appear to directly bind with USP/RXR in less derived insects such as the holometabolous Tribolium castaneum or the hemimetabolous Locusta migratoria [14], [21]. There is also little evidence to suggest a high affinity for retinoids in insects [2], [21]. However, recent displacement binding experiments have shown that USP/RXR may bind 9cRA and all-trans RA at the high nanomolar range in L. migratoria [22]. Given the small quantity of RA found in Locusta, the physiological significance of this in vitro finding is unclear.
Lineage specific variation is also evident in the structure of USP/RXR. A comparative analysis of structural data demonstrates key differences in the ligand-binding pocket (LBP) of USP/RXR in insects. Crystallography data have shown that Dipteran and Lepidopteran USP share a conserved ligand-binding domain (LBD) with a large hydrophobic cavity capable of accepting a natural ligand [23], [24], [25]. In both the Diptera and Lepidoptera, USP copurified from the bacterial expression system with a phospholipid occupying the LBP. However, the identity of the endogenous ligand is unknown. In contrast, crystal structures of USP/RXR from two less derived insects, the red flour beetle T. castaneum and the sweet potato whitefly Bemisia tabaci, reveal a collapsed LBP [21], [26]. Given the taxonomic relationships of insects from which USP/RXR has been crystallized, this data implies a derived open pocket in higher insects. Despite the open LBP, Dipteran and Lepidopteran USP may not function independently as ligand-dependent transcription factors. In these species the ligand-dependent activation function (AF-2) domain in α-helix H12, is locked in an antagonistic conformation [23], [24], [25]. As a consequence of interactions between H3, H11–H12 and a highly conserved insertion in the loop connecting helices H1 and H3 (L1–3), residues in H12 occupy the coactivator groove (H3, loop L3–4 and H4), precluding the agonist conformation and preventing the binding of transcriptional coactivators [23], [24], [27].
These striking lineage-specific differences in both structure and function have sparked an interest in exploring the molecular evolution of insect USP/RXR, in part to understand the evolution of endocrine regulation, but also to aid in the design of selective pesticides which target the USP/RXR-EcR complex. The use of relative-rate tests revealed high divergence rates in the Lepidoptera and Diptera for both USP/RXR and EcR, particularly in the LBD of USP/RXR [21], [27]. These results suggested that the heterodimer may have coevolved to become functionally divergent in the Holometabola. It was later shown that this event was unique to the Mecopterida, a suborder of insects which includes the Diptera, Trichoptera, Mecoptera, Siphonaptera and Lepidoptera [28], [29]. More recently, this work has been extended to the heterodimer interface between USP/RXR and EcR. By reconstructing and modeling the ancestral Mecopterida heterodimer, Iwema et al. [30] demonstrated that an expanded dimerization surface was common to this branch. Furthermore, the data suggested that this enlarged surface was the result of torsion in the structure of USP/RXR caused by the position of loop L1–3 in these insects.
Based on experimental data from Drosophila, crystallography work from Heliothis virescens, and molecular evolutionary studies, a model for the functional evolution of USP/RXR has been proposed. Iwema et al. [21], [30] and Tocchini-Valentini et al. [31] suggest the following three groups of USP/RXR: 1. The RXR type which evolved retinoid-binding early, a function retained in the Cnidaria, Mollusca, and Chordata; 2. The Non-Mecopterida type which lost ligand-binding function along an ancient arthropod lineage, a state found in insect orders such as the Hymenoptera, Coleoptera, Orthoptera and Dictyoptera; 3. Finally, the Mecopterida USP type which may have gained a novel ligand-binding function and an enlarged dimerization interface with EcR during the evolution of some higher insects.
To investigate these hypotheses of USP/RXR evolution, we implemented codon-based maximum likelihood methods to estimate the ratio of non-synonymous (d N) to synonymous (d S) substitutions across an insect phylogeny and independently along the Mecopterida lineage. This ratio serves as an indicator of selective constraint and such methods have been successfully used to detect positive selection, the signature of functional gain, in a variety of insect gene families and vertebrate nuclear receptors [32], [33], [34], [35], [36]. We also examined among-site variation in d N/d S across a dataset of Mecopterida and Non-Mecopterida insects using random-sites models to investigate changes in site-specific evolutionary rates across key structural and functional motifs. Additionally, several of these analyses were repeated with a dataset of EcR sequences to examine evolutionary constraint in the heterodimer. Finally, we employed newly developed methods which allow the independent estimation of d N and d S in order to determine the effect among-site variation in d S on the patterns of site-specific evolutionary rates observed under random-sites analyses.
Methods
Sequence data collection and dataset assembly
USP/RXR and EcR sequences were collected from literature and GenBank using a combination of BLAST and keyword searches (Table S1). Where possible, EcR-A isoform data were used in order to incorporate a larger portion of the protein in subsequent analyses. Both USP/RXR and EcR sequences were not available for all species, and in such cases sequences from the most closely related insects were used instead. Amino acid multiple sequence alignments for USP/RXR and EcR were constructed using ClustalW [37] as implemented in MEGA 4 [38] and adjusted by eye to ensure structural motifs were maintained. Poorly aligned regions and major gaps were deleted (see supporting Figure S1, S2).
To explore selective constraint across the entire phylogeny, both USP/RXR and EcR alignments were truncated such that only the well conserved LBD was included for use in branch and branch-sites analyses (termed USP/RXR LBD and EcR LBD datasets). To examine the variation in evolutionary rates across sites and across functional domains within each group, the USP/RXR and EcR alignments were each split into Mecopterida and Non-Mecopterida datasets (termed USP/RXR A/B-LBD and EcR A/B-LBD datasets for Mecopterida and Non-Mecopterida respectively).
Estimation of evolutionary rates
In order to test for positive selection and examine changes in site-specific evolutionary rates codon-based likelihood methods were used to estimate d N/d S (ω) ratios across the USP/RXR gene. Under no selective pressure, sequences evolve neutrally, and this is indicated by ω = 1, whereas ω<1 indicates purifying selection, and ω>1 positive selection [39], [40]. To estimate ω several branch-specific [41], [42], branch-site [43], [44], and random-sites [45], [46] models were implemented using the codeml program of the PAML software package (version 4.2b; [47]). Random-sites models which allow for variation in d S [48] were also implemented using the HyPhy software package (version 1.0; [49]). In addition, many of these analyses were also carried out for EcR to examine evolutionary constraint in the USP/RXR-EcR heterodimer.
The amino acid alignments described above for each of the six datasets were converted into nucleotide data. A tree reflecting current understanding among major insect lineages was used for both USP/RXR and EcR (Figure 1; [50], [51] Insecta; [52] Coleoptera; [53] Lepidoptera; [54] Diptera; [55], [56] Hymenoptera). Gene trees generated with our USP/RXR and EcR alignments, using both maximum-likelihood [57] and neighbor-joining [38], [58] methods, were generally consistent with these known inter-species relationships (see supporting Figure S3). However, low branch support for many of the ordinal relationships resulted in trees with poor resolution of the Non-Mecopterida, therefore all analyses were only performed using the species trees. For analyses carried out on the USP/RXR and EcR A/B-LBD datasets, the trees were modified to include only Mecopterida or Non-Mecopterida taxa. However, this resulted in a tree bifurcated at the root with two clades when only the Mecopterida were considered, so T. castaneum was added as an outgroup to form a tripartition tree. Correspondingly, the T. castaneum sequence was added to both USP/RXR and EcR A/B-LBD Mecopterida datasets.
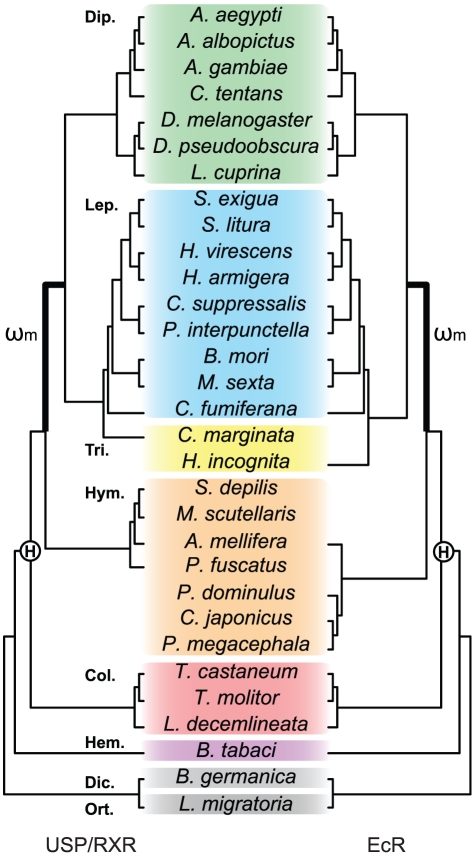
Figure 1. Phylogeny of insect species used in our analysis.
The topology of the trees used for the USP/RXR (left) and EcR (right) datasets are based on known taxonomic relationships. Species contained in the Diptera (Dip.) are shaded green, Lepidoptera (Lep.) blue, Trichoptera (Tri.) yellow, Hymenoptera (Hym.) orange, Coleoptera (Col.) red, Hemiptera (Hem.) purple, Dictyoptera (Dic.) and Orthoptera (Ort.) grey. The origin of holometabolous insects is indicated by an encircled bold H. The highlighted branch is the foreground lineage for detecting positive selection, where a separate d N/d S ratio was estimated for the Mecopterida (ωm).
It is important to note that our dataset was composed of insect taxa which diverged over 140 million years ago [59], and that the saturation of synonymous substitution rates over such large distances may result in the estimation of d N/d S ratios higher than the actual value [60]. However, this would affect all estimations across all datasets, and our primary objective was to examine relative differences and patterns in substitution rates.
Branch and branch-site models
To investigate the gain in function hypothesized to have occurred in Mecopterida USP/RXR, branch models were implemented which allowed for an additional ω parameter along the lineage leading to the Mecopterida (Two-ratios), or alternatively along the lineage leading to the ancestor of Mecopterida and Hymenoptera. Codon frequencies were estimated using the F3x4 matrix and models were run from varying starting ω values ranging above and below 1, or κ (relative transition to transversion rate ratio) values ranging from 0 to 5 in order to test for convergence. Likelihood ratio tests (LRTs; [61]) were then conducted to determine statistically significant differences between nested models. Models with the additional ω parameter were compared against M0 where only one ω value was estimated across the phylogeny. Since coevolution of USP/RXR and EcR has been suggested, the above branch models were also applied to the EcR LBD dataset.
Given the elevation in ω identified by branch models, variation in selective pressure among codon sites in Mecopterida USP/RXR was examined using branch-site models applied to the USP/RXR LBD dataset. Branch-site models allow the use of a Bayes Empirical Bayes (BEB) analysis [62] to identify specific positively selected sites within the gene along a given branch. A model with Mecopterida designated as foreground, Model A alt, was compared against a stringent model for positive selection, Model A null, and a less stringent model, M1a. Positive selection is detected if ‘Model A alt’ is a better fit than ‘Model A null’ where classes of positively selected sites have ω set to 1, whereas relaxed purifying selection (possibly suggestive of positive selection) is indicated if ‘Model A alt’ is a better fit than M1a which does not allow a class of sites with ω>1 (referred to as test 2 and test 1 of positive selection by [44]). Sites identified as under positive selection by BEB analysis with a posterior probability (P)>0.95 were subsequently mapped onto the 1.65 Å crystal structure of H. virescens USP (PBD 1G2N) or the 2.90 Å crystal structure of the H. virescens USP-EcR heterodimer (PBD 1R1K) using the VMD software package [63].
Random-sites models
In order to compare among-site variation in d N/d S across functional domains of USP/RXR and EcR between Mecopterida and Non-Mecopterida insects, random-sites models were implemented using PAML [45], [46]. Codon frequencies and initial starting values were the same as those used in the above branch and branch-site models. Comparison between M3, with three discrete classes of ω, and null M0 tests for variation in selection pressure among sites, but does not explicitly incorporate positive selection. However, comparisons between both M2a and null M1a (discrete distribution), and M8 and null M7 (where ω is drawn from a beta distribution), test for positive selection by allowing for an additional class of sites with ω>1. Posterior probabilities for site classes under sites models can be calculated either by the Naïve Empirical Bayes (NEB) approach [45], [46] or by the BEB approach [62] which considers sampling errors in the estimation of ω. For models such as M8 where BEB data was available, the posterior mean ω at each site was plotted using site class assignments as calculated by BEB, not NEB.
Random-sites models that incorporate variation in d S
To account for the effect of variation of synonymous substitution rates across sites, we implemented several codon selection analyses using the dNdSRateAnalysis program of the HyPhy software package [49]. Similar to M3 in PAML the variable nonsynonymous rates model, or ‘Nonsynonymous’, implemented in HyPhy assumes a constant d S (αS = 1), but samples d N (βS) values from a given rate distribution. However, the dual variable rates model, or ‘Dual’, estimates d S (αS) and d N (βS) independently, sampling both from a given rate distribution [48]. In this study, both models were run using the MG94×REV core rate matrix with the GDD (general discrete distribution) rate distribution method using the ‘Independent Discrete’ setting. For each model three synonymous and nonsynonymous rate classes (3×3) were specified. All models were run several times using the randomized initial value option to find the global optimum. To test for site-to-site variation in d S across both USP/RXR and EcR A/B-LBD datasets for the Mecopterida and Non-Mecopterida, nested Dual and Nonsynonymous models were compared using LRTs.
Results
Branch models
The current theory of USP/RXR functional evolution proposes that the ligand-binding function was lost in an ancient arthropod lineage followed by a subsequent gain in function along the Mecopterida lineage. Additionally, it has been suggested that Mecopterida USP/RXR acquired an expanded dimerization interface with EcR. We tested these hypotheses of functional gain by estimating evolutionary rates across a dataset composed of insect USP/RXR ligand-binding domain sequences (USP/RXR LBD dataset), using codon-based models of substitution (Figure 1). Likelihood scores and ω (d N/d S) values, as calculated by PAML, are shown in Table 1. Branch models implemented in PAML allow ω to be freely estimated along specified foreground branches while all other background branches are constrained to the same ω across the phylogeny (Two-ratios model). A branch model analysis, for which the Mecopterida lineage was set as foreground, demonstrated an elevated value (ωm = 0.166) compared to the background (ω = 0.040), but cannot distinguish between relaxed constraint or positive selection at a subset of sites (Table 1). However, the likelihood ratio test (LRT) indicated that the additional parameter did not yield a significantly better fit for the data than the null model M0 where one ω is estimated across the entire phylogeny (p = 0.347).
Table 1. Parameter estimates for branch and branch-site models (LBD).
| LRT | ||||||||
| Model | np | lnL | κ | Parameter Estimates | Positively Selected Sites | Null | df | p-value |
| USP/RXR | ||||||||
| M0: one-ratio | 53 | −11110.02 | 1.537 | ω = 0.041 | - | |||
| Branch-specific models: | ||||||||
| Two-ratios | 54 | −11109.58 | 1.533 | ω0 = 0.040, ω1 = 0.166 | - | M0 | 1 | 0.347 |
| Branch-site models: | ||||||||
| M1aa | 54 | −11097.30 | 1.544 | ω0 = 0.040, ω1 = 1.000 | - | |||
| p 0 = 0.990, p 1 = 0.010 | ||||||||
| Model A nullb | 55 | −11082.55 | 1.535 | ω0 = 0.039, ω1 = 1.000 | - | |||
| ω2a = 1.000, ω2b = 1.000 | ||||||||
| p 0 = 0.814, p 1 = 0.009 | ||||||||
| p 2a = 0.175, p 2b = 0.002 | ||||||||
| Model A alt | 56 | −11079.65 | 1.536 | ω0 = 0.040, ω1 = 1.000 | 30 sites: 222, 230, 231, | M1a | 2 | 2.174×10−08 * |
| ω2a = 8.122, ω2b = 8.122 | 252, 253,272, 302, 403, | Br-s A nullb | 1 | 0.016* | ||||
| p 0 = 0.796, p 1 = 0.009 | 411 (at P>0.95) | |||||||
| p 2a = 0.194, p 2b = 0.002 | 296, 301, 324, 398 | |||||||
| (at P>0.99) | ||||||||
| EcR | ||||||||
| M0: one-ratio | 53 | −9701.18 | 1.661 | ω = 0.029 | ||||
| Branch-specific models: | ||||||||
| Two-ratios | 54 | −9700.11 | 1.664 | ω0 = 0.029, ω1 = 0.008 | M0 | 1 | 0.143 | |
NOTE – np is the number of parameters for each model, df is the degrees of freedom, while p is the proportion of sites in a given site class. Sites under positive selection are listed according to the site numbering of the H. virescens reference sequence, accession number AX383958.
Null model for test 1 of positive selection from Zhang et al. [44].
Null model for test 2 from Zhang et al. [44], a more stringent test of positive selection.
*Significant (p-value<5%).
Since coevolution of USP/RXR and EcR has been suggested, a branch model for which the Mecopterida lineage was set as foreground was also fit to a dataset composed of EcR ligand-binding domain sequences (EcR LBD dataset). Unlike USP/RXR, the foreground lineage demonstrated a decreased evolutionary rate (ωm = 0.008) suggestive of stronger purifying selection (Table 1). However, the LRT again indicated that the additional parameter did not yield a significantly better fit for the data than M0 (p = 0.143).
In a separate analysis, the lineage leading to the ancestor of Hymenoptera and Mecopterida was also freely estimated across both USP/RXR and EcR LBD sequences to determine if ω was elevated in the branch preceding the Mecopterida. However, in both datasets, ω was found to be below background (ω = 0.002 and ω = 0.0001, respectively; data not shown). The added parameter along that branch also did not yield a statistically better fit than M0 in either dataset (p = 0.072 and p = 0.084, respectively; data not shown). Overall, the branch model results, which only allow for a constant ω parameter across sites, suggest a weak trend where evolutionary rates might be elevated in USP/RXR along the Mecopterida lineage.
Branch-sites models
The insignificance of the branch models tests may have been a consequence of a lack of positive selection, or positive selection acting only on a few sites within the gene along a given branch. Therefore branch-site models were implemented to estimate ω for each site in the USP/RXR LBD dataset along the specified foreground lineage. Branch-site Model A alt demonstrated that a proportion of sites in the USP/RXR gene have ω>1 in the Mecopterida lineage (Table 1). When compared to both M1a and branch-site Model A null, a more stringent test for positive selection, the LRT showed that the result was statistically significant (p = 2.174×10−8 and p = 0.016, respectively; Table 1).
Using a Bayes Empirical Bayes (BEB) analysis in Model A alt, a class of sites with ω>1 was identified (Table S2). Nine sites showed ω>1 at a posterior probability (P) 0.99>P>0.95, and four at P>0.99 (Table 1). These sites mapped onto important structural and functional regions of the H. virescens USP [24], [25] crystal structure (Figure 2A). Of the 13 sites, one directly interacted with the ligand, and three others lay near ligand-binding sites (Figure 2B). Several sites lay within loop L1–3, and were involved in a hydrogen bond network with loop L11–12 and H3 which stabilizes the position of structural elements in Mecopterida USP/RXR (Figure 2C). Although only one site is known to form direct contact with EcR in the crystal structure of the heterodimer, several others fell within H9, a component of the dimerization core (Figure 2D). One site was located in the coactivator groove in loop L3–4, and two others lay in the region immediately adjacent to loop L5-S1. In addition, several of the sites with P<0.95 also participated in interactions with EcR or were located beside sites which do so (supporting Table S2). Overall, these results indicate significantly elevated ω values along the Mecopterida lineage, indicating positive selection in regions of the protein important for ligand-binding, structural stability and dimerization.
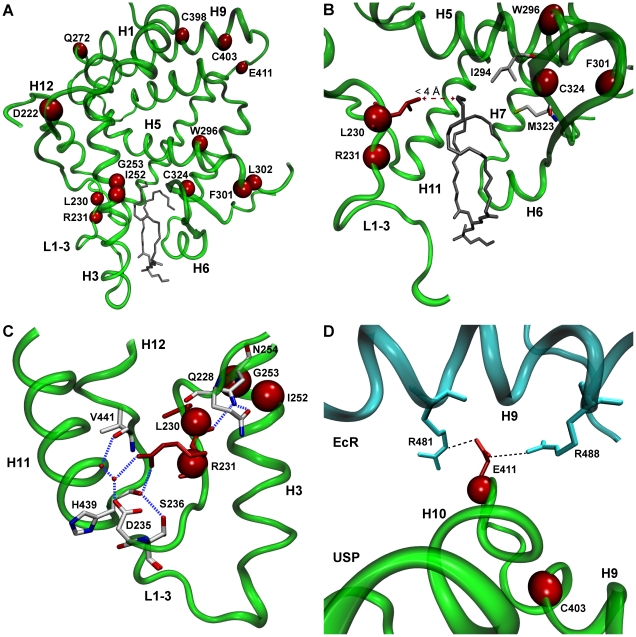
Figure 2. Location of putative positively selected sites in Mecopterida USP/RXR.
(A) The distribution of sites inferred by PAML to be under positive selection (P>0.95) along the Mecopterida branch (red spheres) across the crystal structure of H. virescens USP (green) with the ligand shown in grey. (B) Positively selected sites (red spheres) located near ligand binding sites (white sidechains). Only L230 directly interacts with the phospholipid ligand (grey) via van der Waals (red dashes). (C) The involvement of sites L203 and R231 in a hydrogen bond network between H3, loop L1–3 and H12. Hydrogen bonds are indicated by dotted blue lines and waters by small red dots (adapted from [24]). The backbone and sidechains of residues not under positive selection are shown in white, with blue and red indicating nitrogen and oxygen respectively. (D) Polar interactions (dashed black lines) between the positively selected site E411 in USP loop L9–10 (green) and two arginine residues in H9 of the EcR (cyan). The images were created using PDB 1G2N and 1R1K, site numbering according to H. virescens USP (AX383958) and EcR (Y09009).
Random-sites models
In order to compare among-site variation in selection pressure across USP/RXR functional domains between Mecopterida and Non-Mecopterida insects, random-sites models were conducted using PAML with datasets for each group which spanned all domains (USP/RXR A/B-LBD datasets) (Table 2). M3 was the best fit for the Mecopterida and Non-Mecopterida datasets (p = 2.231×10−127 and p = 5.063×10−47, respectively), suggestive of significant among-site variation in ω. However, M3 is generally a better fit than the null M0 for most proteins as a single ω value across all sites is unlikely. For both the Mecopterida and Non-Mecopterida USP/RXR A/B-LBD datasets, tests of positive selection were not significant. Neither M2a nor M8 were a better fit than the respective null models M1a and M7 (p = 1.0 in both cases). Although no positively selected sites were identified in either Mecopterida or Non-Mecopterida USP/RXR, ω was in fact, elevated at particular regions of the gene (Figure 3A, B). Site class assignments were generally consistent across models, with peaks in ω occurring at the same codon sites, so only the results for M8 are presented (for M3 results see supporting Figure S4). Overall, more sites with elevated ω values were observed in the Mecopterida compared to the Non-Mecopterida.
Table 2. Parameter estimates for USP/RXR gene (A/B-LBD).
| LRT | |||||||
| Model | np | lnL | κ | Parameter Estimates | Null | df | p-value |
| Mecopterida: | |||||||
| M0: one-ratio | 35 | −10594.82 | 1.532 | ω = 0.035 | |||
| M1a: neutral | 36 | −10531.81 | 1.622 | ω0 = 0.029, ω1 = 1.000 | |||
| p 0 = 0.950, p 1 = 0.050 | |||||||
| M2a: selection | 38 | −10531.81 | 1.622 | ω0 = 0.029, ω1 = 1.000, ω2 = 1.000 | M1a | 2 | 1.000 |
| p 0 = 0.950, p 1 = 0.002, p 2 = 0.048 | |||||||
| M3: discrete (K = 3) | 39 | −10297.50 | 1.564 | ω0 = 0.002, ω1 = 0.046, ω2 = 0.194 | M0 | 4 | 2.231×10−127 * |
| p 0 = 0.457, p 1 = 0. 450, p 2 = 0.092 | |||||||
| M7: beta | 36 | −10299.37 | 1.557 | p = 0.417, q = 9.184 | |||
| M8: beta& ω | 38 | −10299.37 | 1.557 | p = 0.417, q = 9.185 | |||
| p 0 = 0.99999, (p 1 = 0.00001) | M7 | 2 | 1.000 | ||||
| ω = 1.000 | |||||||
| Non-Mecopterida: | |||||||
| M0: one-ratio | 19 | −5537.53 | 1.625 | ω = 0.016 | |||
| M1a: neutral | 20 | −5496.97 | 1.789 | ω0 = 0.013, ω1 = 1.000 | |||
| p 0 = 0.967, p 1 = 0.033 | |||||||
| M2a: selection | 22 | −5496.97 | 1.789 | ω0 = 0.013, ω1 = 1.000, ω2 = 1.000 | M1a | 2 | 1.000 |
| p 0 = 0.967, p 1 = 0.019, p 2 = 0.014 | |||||||
| M3: discrete (K = 3) | 23 | −5426.21 | 1.735 | ω0 = 0.001, ω1 = 0.020, ω2 = 0.119 | M0 | 4 | 5.063×10−47 * |
| p 0 = 0.521, p 1 = 0.394, p 2 = 0.085 | |||||||
| M7: beta | 20 | −5429.44 | 1.728 | p = 0.300, q = 13.893 | |||
| M8: beta& ω | 22 | −5429.44 | 1.728 | p = 0.300, q = 13.895 | |||
| p 0 = 0. 99999, (p 1 = 0. 00001) | M7 | 2 | 1.000 | ||||
| ω = 1.000 | |||||||
NOTE – np is the number of parameters for each model, df is degrees of freedom, while p 0–p 2 is the proportion of sites in a given site class, and p, q describe the beta distribution.
*Significant (p-value<5%).
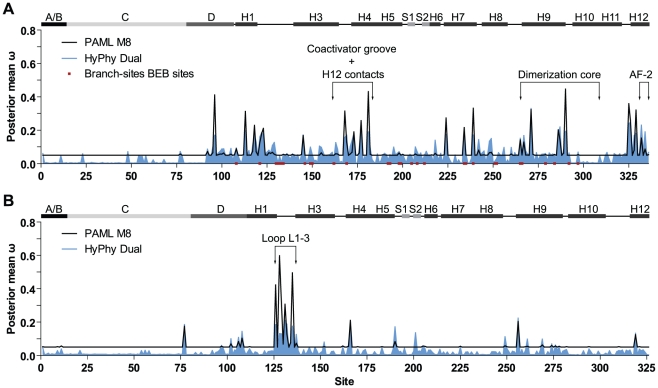
Figure 3. Posterior mean ω at each amino acid site across the USP/RXR gene.
The values of ω as estimated by M8 in PAML (black line) and the Dual model of HyPhy (shaded blue) are shown for each codon site across the gene for the Mecopterida (A) and Non-Mecopterida (B). All sites inferred to be under positive selection by branch-site analysis the along the Mecopterida branch are shown as red boxes on the x-axis in of the Mecopterida plot. A schematic of USP/RXR secondary structure is shown above both plots to illustrate the position of each functional domain (A/B, C, and D) as well as the helices (H1–H12) and β sheets (S1–S2) of the ligand-binding domain. The schematic for the Mecopterida and Non-Mecopterida genes are based of the crystal structures of H. virescens and B. tabaci USP/RXR, respectively. Site numbering is based on the USP/RXR alignment given in supporting figure S1.
In the Mecopterida, there were a dramatic number of sites with increased ω in the LBD and D domain compared to the A/B and C domains (Figure 3A). These sites clustered within H1, the amino-terminal of loop L1–3, loop L3–4, H4, H7, H9 and H12 of the LBD (see supporting Table S3 for details). Two sites were located beside ligand-binding sites and six other sites lay near residues which make dimerization contacts with EcR. Three sites in H12 lay near residues that interact with loop L1–3, H3 and the coactivator binding groove. Four sites lay in the coactivator binding groove, a region blocked in the Mecopterida as a consequence of contacts with H12 and loop L1–3 [23], [24]. Several of the above sites also overlapped with those identified to be under positive selection along the Mecopterida lineage by branch-site analysis (Figure 3A, red squares). However, many sites differed between these types of analyses as branch-sites models compare the specified foreground branch to the background lineages, whereas random-sites models detect among-site rate variation within a clade. Unlike the Mecopterida, only a handful of sites with an elevated ω were observed in the Non-Mecopterida USP/RXR dataset (Figure 3B; supporting Table S3). These were primarily located in loop L1–3, a region with extensive sequence variation among Non-Mecopterida species [14], [64], [65]. There were also a few sites with elevated ω in H4, H9, H12 and the carboxy-terminal DNA-binding domain (C domain). Overall, among-site variation in the USP/RXR suggests that regions involved in dimerization and the blockage of the coactivator binding groove may be under relaxed constraint within the Mecopterida clade.
We also sought to examine among-site variation across EcR functional domains (EcR A/B-LBD datasets), as shifts in USP/RXR structure and function may also have affected the molecular evolution of EcR (Table 3). As with USP/RXR, M3 also yielded the best fit for both Mecopterida and Non-Mecopterida datasets (p = 3.320×10−97 and 9.273×10−32, respectively). No positively selected sites were identified and neither M2a nor M8 were a better fit than the null models M1a and M7 for either dataset (p = 1.0 in both cases). As in the USP/RXR analyses, d N/d S site-profiles were plotted for EcR based on the M8 result (Figure 4A, B; see supporting Figure S4 for M3 results). Again, more sites with elevated ω values were observed in Mecopterida compared to the Non-Mecopterida.
Table 3. Parameter estimates for EcR gene (A/B-LBD).
| LRT | |||||||
| Model | np | lnL | κ | Parameter Estimates | Null | df | p-value |
| Mecopterida: | |||||||
| M0: one-ratio | 35 | −10215.15 | 1.690 | ω = 0.029 | |||
| M1a: neutral | 36 | −10198.25 | 1.737 | ω0 = 0.028, ω1 = 1.000 | |||
| p 0 = 0.988, p 1 = 0.012 | |||||||
| M2a: selection | 38 | −10198.25 | 1.737 | ω0 = 0.028, ω1 = 1.000, ω2 = 1.000 | M1a | 2 | 1.000 |
| p 0 = 0.988, p 1 = 0.008, p 2 = 0.003 | |||||||
| M3: discrete (K = 3) | 39 | −9987.57 | 1.721 | ω0 = 0.000, ω1 = 0.023, ω2 = 0.107 | M0 | 4 | 3.320×10−97 * |
| p 0 = 0.404, p 1 = 0.393, p 2 = 0.203 | |||||||
| M7: beta | 36 | −9988.17 | 1.725 | p = 0.357, q = 10.575 | |||
| M8: beta& ω | 38 | −9988.17 | 1.719 | p = 0.357, q = 10.575 | |||
| p 0 = 0. 99999, (p 1 = 0. 00001) | M7 | 2 | 1.000 | ||||
| ω = 1.000 | |||||||
| Non-Mecopterida: | |||||||
| M0: one-ratio | 19 | −6357.75 | 2.022 | ω = 0.016 | |||
| M1a: neutral | 20 | −6357.74 | 2.029 | ω0 = 0.016, ω1 = 1.000 | |||
| p 0 = 0.999, p 1 = 0.001 | |||||||
| M2a: selection | 22 | −6357.74 | 2.029 | ω0 = 0.016, ω1 = 1.000, ω2 = 1.000 | M1a | 2 | 1.000 |
| p 0 = 0.999, p 1 = 0.001, p 2 = 0.000 | |||||||
| M3: discrete (K = 3) | 23 | −6281.96 | 2.085 | ω0 = 0.000, ω1 = 0.013, ω2 = 0.0737 | M0 | 4 | 9.273×10−32 * |
| p 0 = 0.412, p 1 = 0.435, p 2 = 0.153 | |||||||
| M7: beta | 20 | −6282.43 | 2.085 | p = 0.326, q = 17.365 | |||
| M8: beta& ω | 22 | −6282.43 | 1.906 | p = 0.326, q = 17.365 | |||
| p 0 = 0.99999, (p 1 = 0.00001) | M7 | 2 | 1.000 | ||||
| ω = 1.000 | |||||||
NOTE – np is the number of parameters for each model, df is the degrees of freedom, while p 0–p 2 is the proportion of sites in a given site class, and p, q describe the beta distribution.
*Significant (p-value<5%).
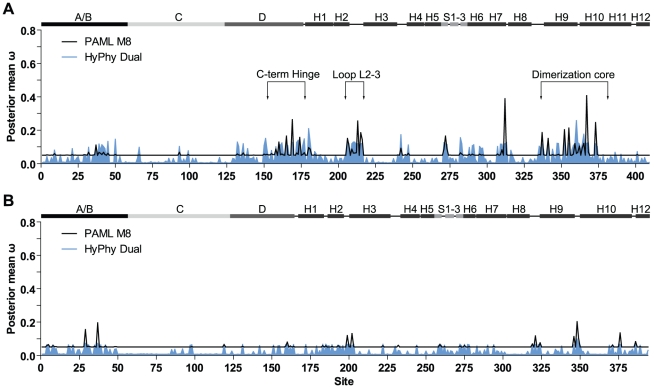
Figure 4. Posterior mean ω at each amino acid site across the EcR gene.
The values of ω as estimated by M8 in PAML (black line) and the Dual model of HyPhy (shaded blue) are shown for each codon site across the gene for the Mecopterida (A) and Non-Mecopterida (B). A schematic of EcR secondary structure is shown above both plots to illustrate the position of each functional domain (A/B, C, and D) as well as the helices (H1–H12) and β sheets (S1–S3) of the ligand-binding domain. The schematic for the Mecopterida and Non-Mecopterida genes are based of the crystal structures of H. virescens and B. tabaci EcR, respectively. Site numbering is based on the EcR alignment given in supporting figure S2.
In the Mecopterida there was a concentration of sites with elevated ω in carboxy-terminal D domain and the dimerization core (Figure 4A; supporting Table S4). Several sites were located in loop L2–3, a region which varies in sequence and is not well modeled in insect crystal structures [21], [25], [26]. One site in the β sheet region was located between several ligand-binding sites. Six sites in H7, H9 and H10 were located near residues which make contact with USP/RXR. Three sites at the amino-terminus of H10 fell into a region associated with dimerization and ligand-binding specificity. Substitutions in this region confer Aedes aegypti-like sensitivity to ecdysone to D. melanogaster EcR, which is normally only responsive to 20E [66]. As in USP/RXR, there was far less among-site variation in ω across Non-Mecopterida EcR (Figure 4B; supporting Table S4). Three sites in loop L9–10 and H10 align with the aforementioned region in A. aegypti which affects sensitivity to ecdysone. In addition, three other sites were located near ligand-binding sites. Overall, these results indicate that the dimerization core of EcR, like that of USP/RXR, may also be under relaxed constraint in the Mecopterida clade.
Random-sites models that incorporate variation in d S
Traditional d N/d S methods estimate d N and d S as a ratio, and assume a constant d S [45], [46]. However, this may be an unrealistic assumption for some datasets, and synonymous substitutions are thought to be under selection in flies, nematodes and yeast [67]. Recently, models have been developed to allow for the independent estimation of d N and d S. To examine the effect of among-site variation in d S on the estimation of ω in our datasets, we implemented several such models in the HyPhy software package (see supporting Tables S5, S6, S7 for parameter estimates). Overall, the results of HyPhy are similar to those of PAML (Figure 3 and 4, shaded blue). For all datasets the regions with elevated ω were consistent between both methods, with some variation in peak height. The baseline of the d N/d S site-profile plots appears elevated for the PAML M8 results compared to the HyPhy results because the former employs BEB methods, which account for sampling error in the maximum likelihood d N/d S (and proportion) estimates, whereas the latter does not.
For both USP/RXR and EcR A/B-LBD datasets in the Mecopterida site-to-site variation in d S was significant when the Dual model compared to the Nonsynonymous null model where d S is fixed at all sites (p = 0.037 and 0.025 respectively; Table 4). However, this was not the case with either dataset for the Non-Mecopterida in which significant variation in d S among sites was not detected. Given the small size of the Non-Mecopterida datasets and the limited taxonomic sampling, we may lack the statistical power to estimate d N and d S accurately in this group. Overall the HyPhy results indicate that despite significant among-site variation in d S within some datasets, the patterns in substitution rates across both the USP/RXR and EcR A/B-LBD datasets are the same when d N and d S are estimated independently.
Table 4. Synonymous rate variation among sites (HyPhy).
| MG94×REV Nonsynonymous GDD 3 | MG94×REV Dual GDD 3×3 | LRT | ||||
| Dataset | np | Log L | np | Log L | df | p-value |
| Mecopterida USP/RXR (A/B-LBD) | 43 | −10237.10 | 47 | −10232.01 | 4 | 0.037* |
| Non-Mecopterida USP/RXR (A/B-LBD) | 27 | −5415.51 | 31 | −5410.81 | 4 | 0.052 |
| Mecopterida EcR (A/B-LBD) | 43 | −9946.36 | 47 | −9940.77 | 4 | 0.025* |
| Non-Mecopterida EcR (A/B-LBD) | 27 | −6269.43 | 31 | −6269.40 | 4 | 0.999 |
NOTE – np is the number of parameters for each model, Log L is the logarithm of the maximum likelihood value, and df is the degrees of freedom used in the LRT calculation when comparing Dual versus Nonsynonymous models.
*Significant (p-value<5%).
Discussion
Our results indicate that USP/RXR is under positive selection along the branch leading to the Mecopterida, with positively selected sites tending to be located in regions involved in ligand-binding, interactions with loop L1–3 and dimerization (Figure 2). Furthermore, random-sites analyses showed that ω was elevated across the Mecopterida clade compared to the Non-Mecopterida for both USP/RXR and EcR (Figure 3, 4). Sites with elevated ω tended to be concentrated in components of the dimerization interface, suggesting relaxed constraint in this region among the Mecopterida (Figure 3A, 4A). There were also several sites with elevated ω in both proteins located near sites important for ligand-binding. Overall, these results are consistent with the acquisition of functional gains with respect to ligand-binding and dimerization in USP/RXR in the Mecopterida.
Although higher relative substitution rates have been shown in the Mecopterida, our study represents the first instance of codon-based likelihood phylogenetic methods being used to estimate d N/d S ratios across the USP/RXR gene [21], [27], [28]. The elevated d N/d S values we observed along the Mecopterida lineage are consistent the with high amino acid substitution rates reported previously [28]. Strikingly, the regions identified as under either positive selection or relaxed constraint play a major role in defining the structure of USP/RXR in the Mecopterida. In order to function as ligand-dependent transcription factors, nuclear receptors must undergo a shift in the position of H12 upon ligand binding to generate an interface for coactivators [68]. Residues in the AF-2 domain and the coactivator binding groove become accessible, allowing the binding of transcriptional coactivators [68], [69], [70]. However, the position of loop L1–3 in Mecopterida USP/RXR prevents shifts in the H12 position, regardless of ligand binding, thereby locking the receptor in an antagonist conformation [23], [24].
These conformational changes are evident in the evolutionary history of USP/RXR. Studies using reconstruction and homology modeling of the ancestral Mecopterida USP/RXR have investigated the origin of an expanded interface with EcR; these novel contacts were a result of intramolecular epistasis whereby the position of loop L1–3 created torsion in the protein, shifting the position of loop L8–9 closer to EcR [30]. We identified sites under positive selection along the Mecopterida lineage in the loop L1–3 region which interact with the ligand and contribute to a hydrogen bond network with H3 and loop L11–12 (Figure 2). Site R385 (according to H. virescens) in H9, a Mecopterida-specific contact with EcR, was also shown to be under positive selection along the Mecopterida lineage by branch-site analysis. Furthermore, sites surrounding R385 appear to be under relaxed constraint within the Mecopterida in our random-sites analysis (Figure 3A). This was also the case at sites near S447 (according to H. virescens) in EcR H7, which establishes contact with USP/RXR R385 (Figure 4A). This is consistent with the acquisition of an expanded dimer interface between USP/RXR and EcR in the Mecopterida.
It has been postulated that the expansion of this interface may reflect a strengthened inter-dependency between USP/RXR and EcR for activation [30]. Based on structural studies, Diptera and Lepidoptera USP/RXR cannot bind the canonical NR-box LXXLL motif of coactivators because H12 sits within the binding site [23], [24], [71], [72]. Our random-sites analyses revealed sites with elevated ω within both H12 and the coactivator binding groove (loop L3–4, H4) of Mecopterida USP/RXR (Figure 3A). These results are consistent with a change in the function of these regions, leading to a relaxation of evolutionary constraint at these sites. Without a novel interaction interface, such shifts may render USP/RXR more dependent on partner proteins for coactivator recruitment [23], [73]. Interestingly, similar shifts in cofactor interfaces may also have occurred in EcR. An elevation in ω was observed in the carboxy-terminal D domain of Mecopterida EcR, a region implicated in corepressor binding in the mammalian thyroid hormone receptor [74]. The biological significance of an increased dependence on partner proteins remains unclear, but there are some possibilities.
One mechanism which may have led to increased evolutionary rates in the Mecopterida dimer interface is the increased recruitment of USP/RXR as a protein hub. In D. melanogaster and A. aegypti, USP/RXR is known to interact with many other proteins such as Seven-up (Svp), hormone receptors 38 and 78 (HR38, 78), as well as the Methoprene-tolerant gene product (MET) [75], [76], [77], [78]. Hubs at major branch points in protein-protein interaction networks tend to display evidence of elevated ω rates and positive selection [79]. The strong positive selection we observed could be congruent with such a role for USP/RXR. Indeed network level analyses suggest that several proteins in the ecdysone cascade, including USP/RXR and EcR, experienced an increased evolutionary rate at the base of the Mecopterida [80]. However, it is unclear if this is due to functional divergence or simply parallel accelerations across the network in order to maintain interactions. Similarly, the elevated evolutionary rates we detected within the dimerization interface of the Mecopterida clade may be the result of a combination of strong positive selection and purifying selection, not simply relaxed constraint. Substitutions in this region may affect both the affinity and specificity of protein-protein interaction, but might not necessarily improve both of these attributes. In fact, there may be some trade-offs, whereby some substitutions confer a tighter affinity for one protein, but reduce the specificity of interactions with other partner proteins. However, at this point the residues involved in the interface between USP/RXR and alternate partner proteins remain unknown. Furthermore, the network of USP/RXR protein interactions is unknown in lower insects, and a lack of sequence data from outside of the Holometabola prevented the analysis of alternate USP/RXR partners in our study. Therefore, further work is needed to clarify the protein-protein interactions of USP/RXR among insect lineages.
The link between shifts in USP/RXR molecular evolution and shifts in physiology at the organismal level has been difficult to establish. Studies have shown that the role of USP/RXR in molting and metamorphosis seems conserved across insect lineages [6], [7]. Differences in the reproductive biology of insect lineages have been largely unexplored, but may hold more promise [28]. Although there are some exceptions, JH is the major hormone regulating reproductive events such as vitellogenesis in most insect groups [81]. In the Lepidoptera, there is a decreased dependence on JH for vitellogenesis, whereas in the Diptera, vitellogenesis is largely regulated by ecdysteroids. In many Diptera, JH is instead required during previtellogenesis in order for tissues to acquire competence to 20E [82]. The site of vitellogenin production also varies between insect lineages. In many basal lineages, the fat body appears to be the sole site of synthesis, in others, both the ovary and fat body contribute, whereas in some Diptera, the ovary may be the major source [83], [84]. Thus, a stronger relationship between USP/RXR and EcR for heterodimerization is also consistent with the increased dependence on ecdysteroids for the regulation of reproductive events in some higher insects.
USP/RXR is involved in many reproductive events in higher insects such as egg chamber formation and chorion gene expression in Drosophila and regulation of the cyclicity of vitellogenesis in A. aegypti [75], [85], [86]. Ovarian morphology and structure also varies among insects groups, particularly in lineages between the basal Non-Mecopterida and Mecopterida, such as the Paraneoptera, Coleoptera and Neuropterida [87], [88]. The genes which control reproductive events differ among ovary types as well. For example, the set of genes responsible for chorion formation in meroistic ovaries differs from those in the panoistic type ovary, found in the basal Non-Mecopterida [87], [89]. In addition, USP/RXR and EcR are highly expressed in nurse cells, a cell type unique to the meroistic ovary [85], [90]. Thus, shifts in ovarian morphology, development and the endocrine control of reproduction may have also required plasticity in nuclear receptors. However, the role of USP/RXR in the reproductive events of basal insects is not known, as is its role in highly derived insects contained within the Mecopterida.
Finally, it is important to consider caveats of d N/d S based methods. In recent years a debate has emerged regarding the use of these methods to reliably detect positive selection because of potential false positives identified in some genes [91], [92], [93]. However, in our dataset the positively selected sites identified along the Mecopterida lineage by branch-site analysis were consistent with the structural and experimental work of other researchers. Furthermore, we used patterns of d N/d S variation across sites, not merely positive selection, to draw conclusion regarding USP/RXR evolution. Finally, we utilised new models which allow for among-site variation in d S and observed strikingly similar patterns in site-specific evolutionary rates even across these different methods of estimating d N/d S.
While computational methods, such as those used here, are useful in identifying patterns of selective constraint acting on USP/RXR across insect lineages, the functional changes generated by the substitutions identified here remain speculative. For example, we cannot clarify whether or not sites under positive selection near the ligand-binding pocket are related to the potential acquisition of JH binding. Further experimental evidence is required to understand the physiological implications of these evolutionary shifts. In particular, examining the role of USP/RXR in the reproductive system of more basal insects, for example, with the use of techniques such as RNAi, may shed light on these issues. Additionally, the apparent increase in the rigidity of Mecopterida USP/RXR makes this an excellent system for the use molecular dynamics studies to investigate subtle shifts in structure. Future work in this area may yield more insights into the role of the positively selected sites identified in this study which impact the conformation of the receptor complex. Overall, our results highlight the need for more comparative physiological work in the basal Non-Mecopterida and provide critical site-specific information for the design of future site directed mutagenesis studies on USP/RXR.
Supporting Information
Alignment of insect USP/RXR sequences. The green and orange bars indicate the Mecopterida and Non-Mecopterida taxa, respectively, and the schematic below the alignment shows the USP/RXR domain structure. For ease of viewing one alignment is shown. However, the complete alignment was not used for analysis as some regions do not align (e.g. D domain). Only the carboxy-terminal E/F (*), or ligand-binding domain, was used for the branch and branch-sites analyses reported in table 1. Arrows indicate where poorly aligned regions and major gaps were deleted. Full length sequences were used for the random-sites and HyPhy analyses where the larger dataset was split into Mecopterida and Non-Mecopterida only datasets in order to compare evolutionary rates between the two groups. For clarity, site numbering for the full length Mecopterida (green) and Non-Mecopterida (orange) datasets is shown above and below the alignment, respectively. Note that species names have been abbreviated to six characters, complete names can be found in supporting table S1.
(PDF)
Alignment of insect EcR sequences. The green and orange bars indicate the Mecopterida and Non-Mecopterida taxa, respectively, and the schematic below the alignment shows the EcR domain structure. For ease of viewing one alignment is shown. However, the complete alignment was not used for analysis as some regions do not align (e.g. D domain). Only the carboxy-terminal E (*), or ligand-binding domain, was used for the branch analyses reported in table 1. Arrows indicate where poorly aligned regions and major gaps were deleted. Full length sequences were used for the random-sites and HyPhy analyses where the larger dataset was split into Mecopterida and Non-Mecopterida only datasets in order to compare evolutionary rates between the two groups. For clarity, site numbering for the full length Mecopterida (green) and Non-Mecopterida (orange) datasets is shown above and below the alignment, respectively. Note that species names have been abbreviated to six characters, complete names can be found in supporting table S1.
(PDF)
USP/RXR and EcR gene trees. Gene trees for USP/RXR and EcR were generated using the alignment of LBD sequences given in supporting figures S1 and S2. Maximum-likelihood trees for USP/RXR (A) and EcR (C) were constructed in PhyML [57] using the WAG substitution model, with four rate categories to estimate the gamma parameter shape. Neighbor-joining [58] trees for USP/RXR (B) and EcR (D) were constructed in MEGA 4 [38] using the Poisson correction model, with the pair-wise deletion of gaps. For all analyses 100 bootstrap replicates were performed, and nodes with values less than 60 were later collapsed. Each tree was then rooted along the branch leading to B. germanica and L. migratoria. Note that species names have been abbreviated, see supporting table S1.
(PDF)
d N/ d S site-profile plots for PAML model M3. The values of ω as estimated by M3 in PAML using the NEB method are shown for each codon site across Mecopterida USP/RXR (A), Non-Mecopterida USP/RXR (B), Mecopterida EcR (C) and Non-Mecopterida EcR (D). A schematic of USP/RXR and EcR secondary structure is shown above each plot to illustrate the position of each functional domain (A/B, C, and D) as well as the helices (H1–H12) and β sheets of the ligand-binding domain. These schematics are based on the crystal structure of each gene in H. virescens and B. tabaci. Site numbering is the same as figures 3 and 4.
(PDF)
Sequence data information.
(DOC)
Putative positively selected sites.
(DOC)
Sites with elevated ω in the USP/RXR gene.
(DOC)
Sites with elevated ω in the EcR gene.
(DOC)
Statistics for rate distributions inferred by HyPhy.
(DOC)
Rate class assignment for MG94×REV Nonsynonymous GDD 3.
(DOC)
Rate class assignment for MG94×REV Dual GDD 3×3.
(DOC)
Acknowledgments
We thank Jingjing Du and Cameron J. Weadick of the department of Ecology and Evolutionary Biology at the University of Toronto for their insightful comments, technical expertise and critiques of earlier drafts.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by Natural Sciences and Engineering Council of Canada (NSERC) Discovery grants (SST and BSWC) and an Early Researcher Award to BSWC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wiens M, Batel R, Korzhev M, Müller WEG. Retinoid X receptor and retinoic acid response in the marine sponge Suberites domuncula. J Exp Biol. 2003;206:3261–3271. doi: 10.1242/jeb.00541. [DOI] [PubMed] [Google Scholar]
- 2.Oro AE, McKeown M, Evans RM. Relationship between the product of the Drosophila ultraspiracle locus and the vertebrate retinoid X receptor. Nature. 1990;347:298–301. doi: 10.1038/347298a0. [DOI] [PubMed] [Google Scholar]
- 3.Mangelsdorf DJ, Ong ES, Dyck JA, Evans RM. Nuclear receptor that identifies a novel retinoic acid response pathway. Nature. 1990;345:224–229. doi: 10.1038/345224a0. [DOI] [PubMed] [Google Scholar]
- 4.Hall BL, Thummel CS. The RXR homolog Ultraspiracle is an essential component of the Drosophila ecdysone receptor. Development. 1998;125:4709–4717. doi: 10.1242/dev.125.23.4709. [DOI] [PubMed] [Google Scholar]
- 5.Barchuk AR, Figueiredo VLC, Simões ZLP. Downregulation of ultraspiracle gene expression delays pupal development in honeybees. J Insect Physiol. 2008;54:1035–1040. doi: 10.1016/j.jinsphys.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Martín D, Maestro O, Cruz J, Mané-Padrós D, Bellés X. RNAi studies reveal a conserved role for RXR in molting in the cockroach Blattella germanica. J Insect Physiol. 2006;52:410–416. doi: 10.1016/j.jinsphys.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Tan A, Palli SR. Edysone receptor isoforms play distinct roles in controlling molting and metamorphosis in the red flour beetle, Tribolium castaneum. Mol Cell Endocrinol. 2008;291:42–49. doi: 10.1016/j.mce.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu JJ, Tan AJ, Palli SR. The function of nuclear receptors in regulation of female reproduction and embryogenesis in the red flour beetle, Tribolium castaneum. J Insect Physiol. 2010;56:1471–1480. doi: 10.1016/j.jinsphys.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas HE, Stunnenberg HG, Stewart AF. Heterodimerization of the Drosophila ecdysone receptor with retinoid X receptor and ultraspiracle. Nature. 1993;362:471–475. doi: 10.1038/362471a0. [DOI] [PubMed] [Google Scholar]
- 10.Yao TP, Forman BM, Jiang ZY, Cherbas L, Chen JD, et al. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature. 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 11.Yao TP, Segraves WA, Oro AE, McKeown M, Evans RM. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]
- 12.Jones G, Sharp PA. Ultraspiracle: An invertebrate nuclear receptor for juvenile hormones. Proc Natl Acad Sci U S A. 1997;94:13499–13503. doi: 10.1073/pnas.94.25.13499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Fang F, Chu YX, Jones D, Jones G. Activation of transcription through the ligand-binding pocket of the orphan nuclear receptor ultraspiracle. Eur J Biochem. 2002;269:6026–6036. doi: 10.1046/j.1432-1033.2002.03293.x. [DOI] [PubMed] [Google Scholar]
- 14.Hayward DC, Dhadialla TS, Zong ST, Kuiper MJ, Ball EE, et al. Ligand specificity and developmental expression of RXR and ecdysone receptor in the migratory locust. J Insect Physiol. 2003;49:1135–1144. doi: 10.1016/j.jinsphys.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Levin AA, Sturzenbecker LJ, Kazmer S, Bosakowski T, Huselton C, et al. 9-Cis retinoic acid stereoisomer binds and activates the nuclear receptor RXRα. Nature. 1992;355:359–361. doi: 10.1038/355359a0. [DOI] [PubMed] [Google Scholar]
- 16.Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, et al. 9-Cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 17.Mangelsdorf DJ, Borgmeyer U, Heyman RA, Zhou JY, Ong ES, et al. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 18.Wozniak M, Chu YX, Fang F, Xu Y, Riddiford L, et al. Alternative farnesoid structures induce different conformational outcomes upon the Drosophila ortholog of the retinoid X receptor, ultraspiracle. Insect Biochem Mol Biol. 2004;34:1147–1162. doi: 10.1016/j.ibmb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Fang F, Xu Y, Jones D, Jones G. Interactions of ultraspiracle with ecdysone receptor in the transduction of ecdysone- and juvenile hormone-signaling. FEBS J. 2005;272:1577–1589. doi: 10.1111/j.1742-4658.2005.04578.x. [DOI] [PubMed] [Google Scholar]
- 20.Jones G, Jones D, Teal P, Sapa A, Wozniak M. The retinoid-X receptor ortholog, ultraspiracle, binds with nanomolar affinity to an endogenous morphogenetic ligand. FEBS J. 2006;273:4983–4996. doi: 10.1111/j.1742-4658.2006.05498.x. [DOI] [PubMed] [Google Scholar]
- 21.Iwema T, Billas IML, Beck Y, Bonneton F, Nierengarten H, et al. Structural and functional characterization of a novel type of ligand-independent RXR-USP receptor. EMBO J. 2007;26:3770–3782. doi: 10.1038/sj.emboj.7601810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nowickyj SM, Chithalen JV, Cameron D, Tyshenko MG, Petkovich M, et al. Locust retinoid X receptors: 9-Cis-retinoic acid in embryos from a primitive insect. Proc Natl Acad Sci U S A. 2008;105:9540–9545. doi: 10.1073/pnas.0712132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clayton GM, Peak-Chew SY, Evans RM, Schwabe JWR. The structure of the ultraspiracle ligand-binding domain reveals a nuclear receptor locked in an inactive conformation. Proc Natl Acad Sci U S A. 2001;98:1549–1554. doi: 10.1073/pnas.041611298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Billas IML, Moulinier L, Rochel N, Moras D. Crystal structure of the ligand-binding domain of the ultraspiracle protein USP, the ortholog of retinoid X receptors in insects. J Biol Chem. 2001;276:7465–7474. doi: 10.1074/jbc.M008926200. [DOI] [PubMed] [Google Scholar]
- 25.Billas IML, Iwema T, Garnier JM, Mitschler A, Rochel N, et al. Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor. Nature. 2003;426:91–96. doi: 10.1038/nature02112. [DOI] [PubMed] [Google Scholar]
- 26.Carmichael JA, Lawrence MC, Graham LD, Pilling PA, Epa VC, et al. The X-ray structure of a hemipteran ecdysone receptor ligand-binding domain: comparison with a lepidopteran ecdysone receptor ligand-binding domain and implications for insecticide design. J Biol Chem. 2005;280:22258–22269. doi: 10.1074/jbc.M500661200. [DOI] [PubMed] [Google Scholar]
- 27.Bonneton F, Zelus D, Iwema T, Robinson-Rechavi M, Laudet V. Rapid divergence of the ecdysone receptor in Diptera and Lepidoptera suggests coevolution between ECR and USP-RXR. Mol Biol Evol. 2003;20:541–553. doi: 10.1093/molbev/msg054. [DOI] [PubMed] [Google Scholar]
- 28.Bonneton F, Brunet FG, Kathirithamby J, Laudet V. The rapid divergence of the ecdysone receptor is a synapomorphy for Mecopterida that clarifies the Strepsiptera problem. Insect Mol Biol. 2006;15:351–362. doi: 10.1111/j.1365-2583.2006.00654.x. [DOI] [PubMed] [Google Scholar]
- 29.Kristensen NP. Phylogeny of endopterygote insects, the most successful lineage of living organisms. Eur J Entomol. 1999;96:237–253. [Google Scholar]
- 30.Iwema T, Chaumot A, Studer RA, Robinson-Rechavi M, Billas IML, et al. Structural and evolutionary innovation of the heterodimerization interface between USP and the ecdysone receptor ECR in insects. Mol Biol Evol. 2009;26:753–768. doi: 10.1093/molbev/msn302. [DOI] [PubMed] [Google Scholar]
- 31.Tocchini-Valentini GD, Rochel N, Escriva H, Germain P, Peluso-Iltis C, et al. Structural and functional insights into the ligand-binding domain of a nonduplicated retinoid X nuclear receptor from the invertebrate chordate amphioxus. J Biol Chem. 2009;284:1938–1948. doi: 10.1074/jbc.M805692200. [DOI] [PubMed] [Google Scholar]
- 32.Kelleher ES, Markow TA. Duplication, selection and gene conversion in a Drosophila mojavensis female reproductive protein family. Genetics. 2009;181:1451–1465. doi: 10.1534/genetics.108.099044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu DD, Wang GD, Irwin DM, Zhang YP. A profound role for the expansion of trypsin-like serine protease family in the evolution of hematophagy in mosquito. Mol Biol Evol. 2009;26:2333–2341. doi: 10.1093/molbev/msp139. [DOI] [PubMed] [Google Scholar]
- 34.Smadja C, Shi P, Butlin RK, Robertson HM. Large gene family expansions and adaptive evolution for odorant and gustatory receptors in the pea aphid, Acyrthosiphon pisum. Mol Biol Evol. 2009;26:2073–2086. doi: 10.1093/molbev/msp116. [DOI] [PubMed] [Google Scholar]
- 35.Bulmer MS, Crozier RH. Variation in positive selection in termite GNBPs and Relish. Mol Biol Evol. 2006;23:317–326. doi: 10.1093/molbev/msj037. [DOI] [PubMed] [Google Scholar]
- 36.Chen CY, Opazo JC, Erez O, Uddin M, Santolaya-Forgas J, et al. The human progesterone receptor shows evidence of adaptive evolution associated with its ability to act as a transcription factor. Mol Phylogenet Evol. 2008;47:637–649. doi: 10.1016/j.ympev.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson JD, Higgins DG, Gibson TJ. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 39.Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977;267:275–276. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- 40.Yang ZH, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang ZH. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 42.Yang ZH, Nielsen R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol. 1998;46:409–418. doi: 10.1007/pl00006320. [DOI] [PubMed] [Google Scholar]
- 43.Yang ZH, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 44.Zhang JZ, Nielsen R, Yang ZH. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 45.Nielsen R, Yang ZH. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang ZH, Nielsen R, Goldman N, Pedersen AMK. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang ZH. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 48.Pond SK, Muse SV. Site-to-site variation of synonymous substitution rates. Mol Biol Evol. 2005;22:2375–2385. doi: 10.1093/molbev/msi232. [DOI] [PubMed] [Google Scholar]
- 49.Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 50.Whiting MF, Carpenter JC, Wheeler QD, Wheeler WC. The Strepsiptera problem: phylogeny of the holometabolous insect orders inferred from 18S and 28S ribosomal DNA sequences and morphology. Syst Biol. 1997;46:1–68. doi: 10.1093/sysbio/46.1.1. [DOI] [PubMed] [Google Scholar]
- 51.Wheeler WC, Whiting M, Wheeler QD, Carpenter JM. The phylogeny of the extant hexapod orders. Cladistics. 2001;17:113–169. doi: 10.1111/j.1096-0031.2001.tb00115.x. [DOI] [PubMed] [Google Scholar]
- 52.Hunt T, Bergsten J, Levkanicova Z, Papadopoulou A, John OS, et al. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science. 2007;318:1913–1916. doi: 10.1126/science.1146954. [DOI] [PubMed] [Google Scholar]
- 53.Weller SJ, Friedlander TP, Martin JA, Pashley DP. Phylogenetic studies of ribosomal RNA variation in higher moths and butterflies (Lepidoptera: Ditrysia). Mol Phylogenet Evol. 1992;1:312–337. doi: 10.1016/1055-7903(92)90007-4. [DOI] [PubMed] [Google Scholar]
- 54.Yeates DK, Wiegmann BM. Congruence and controversy: toward a higher-level phylogeny of Diptera. Annu Rev Entomol. 1999;44:397–428. doi: 10.1146/annurev.ento.44.1.397. [DOI] [PubMed] [Google Scholar]
- 55.Brothers DJ. Phylogeny and evolution of wasps, ants and bees (Hymenoptera, Chrysidoidea, Vespoidea and Apoidea). Zool Scr. 1999;28:233–249. [Google Scholar]
- 56.Castro LR, Dowton M. The position of the Hymenoptera within the Holometabola as inferred from the mitochondrial genome of Perga condei (Hymenoptera: Symphyta: Pergidae). Mol Phylogenet Evol. 2005;34:469–479. doi: 10.1016/j.ympev.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 57.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 58.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 59.Gao TP, Ren D, Shih CK. The first Xyelotomidae (Hymenoptera) from the Middle Jurassic in China. Ann Entomol Soc Am. 2009;102:588–596. [Google Scholar]
- 60.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 61.Felsenstein J. Evolutionary trees from DNA sequences: a maximum-likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 62.Yang ZH, Wong WSW, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 63.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 64.Hayward DC, Bastiani MJ, Trueman JWH, Turman JW, Riddiford LM, et al. The sequence of Locusta RXR, homologous to Drosophila Ultraspiracle, and its evolutionary implications. Dev Genes Evol. 1999;209:564–571. doi: 10.1007/s004270050290. [DOI] [PubMed] [Google Scholar]
- 65.Maestro O, Cruz J, Pascual N, Martín D, Bellés X. Differential expression of two RXR/ultraspiracle isoforms during the life cycle of the hemimetabolous insect Blattella germanica (Dictyoptera, Blattellidae). Mol Cell Endocrinol. 2005;238:27–37. doi: 10.1016/j.mce.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 66.Wang SF, Ayer S, Segraves WA, Williams DR, Raikhel AS. Molecular determinants of differential ligand sensitivities of insect ecdysteroid receptors. Mol Cell Biol. 2000;20:3870–3879. doi: 10.1128/mcb.20.11.3870-3879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharp PM, Averof M, Lloyd AT, Matassi G, Peden JF. DNA sequence evolution: the sounds of silence. Philos Trans R Soc Lond B Biol Sci. 1995;349:241–247. doi: 10.1098/rstb.1995.0108. [DOI] [PubMed] [Google Scholar]
- 68.Wurtz JM, Bourguet W, Renaud JP, Vivat V, Chambon P, et al. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 69.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 70.Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, et al. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell. 2000;5:289–298. doi: 10.1016/s1097-2765(00)80424-4. [DOI] [PubMed] [Google Scholar]
- 71.Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 73.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 74.Hörlein AJ, Näär AM, Heinzel T, Torchia J, Gloss B, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 75.Zhu JS, Miura K, Chen L, Raikhel AS. Cyclicity of mosquito vitellogenic ecdysteroid-mediated signaling is modulated by alternative dimerization of the RXR homologue Ultraspiracle. Proc Natl Acad Sci U S A. 2003;100:544–549. doi: 10.1073/pnas.0235695100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hirai M, Shinoda T, Kamimura M, Tomita S, Shiotsuki T. Bombyx mori orphan receptor, BmHR78: cDNA cloning, testis abundant expression and putative dirnerization partner for Bombyx ultraspiracle. Mol Cell Endocrinol. 2002;189:201–211. doi: 10.1016/s0303-7207(01)00604-9. [DOI] [PubMed] [Google Scholar]
- 77.Baker KD, Shewchuk LM, Kozlova T, Makishima M, Hassell A, et al. The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell. 2003;113:731–742. doi: 10.1016/s0092-8674(03)00420-3. [DOI] [PubMed] [Google Scholar]
- 78.Li YP, Zhang ZL, Robinson GE, Palli SR. Identification and characterization of a juvenile hormone response element and its binding proteins. J Biol Chem. 2007;282:37605–37617. doi: 10.1074/jbc.M704595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alvarez-Ponce D, Aguadé M, Rozas J. Network-level molecular evolutionary analysis of the insulin/TOR signal transduction pathway across 12 Drosophila genomes. Genome Res. 2009;19:234–242. doi: 10.1101/gr.084038.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bonneton F, Chaumot A, Laudet V. Annotation of Tribolium nuclear receptors reveals an increase in evolutionary rate of a network controlling the ecdysone cascade. Insect Biochem Mol Biol. 2008;38:416–429. doi: 10.1016/j.ibmb.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 81.Raikhel AS, Brown MR, Bellés X. Hormonal Control of Reproductive Processes. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Oxford: Pergamon; 2005. pp. 433–491. [Google Scholar]
- 82.Zhu JS, Chen L, Raikhel AS. Posttrapscriptional control of the competence factor βFTZ-F1 by juvenile hormone in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A. 2003b;100:13338–13343. doi: 10.1073/pnas.2234416100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Valle D. Vitellogenesis in insects and other groups - a review. Mem Inst Oswaldo Cruz. 1993;88:1–26. doi: 10.1590/s0074-02761993000100005. [DOI] [PubMed] [Google Scholar]
- 84.Huebner E, Tobe SS, Davey KG. Structural and functional dynamics of oogenesis in Glossina austeni: vitellogenesis with special reference to the follicular epithelium. Tissue Cell. 1975;7:535–558. doi: 10.1016/0040-8166(75)90025-7. [DOI] [PubMed] [Google Scholar]
- 85.Carney GE, Bender M. The Drosophila ecdysone receptor (EcR) gene is required maternally for normal oogenesis. Genetics. 2000;154:1203–1211. doi: 10.1093/genetics/154.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bernardi F, Romani P, Tzertzinis G, Gargiulo G, Cavaliere V. EcR-B1 and Usp nuclear hormone receptors regulate expression of the VM32E eggshell gene during Drosophila oogenesis. Dev Biol. 2009;328:541–551. doi: 10.1016/j.ydbio.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 87.Büning J. Germ cell cluster formation in insect ovaries. Int J Insect Morphol Embryol. 1993;22:237–253. [Google Scholar]
- 88.Trauner J, Büning J. Germ-cell cluster formation in the telotrophic meroistic ovary of Tribolium castaneum (Coleoptera, Polyphaga, Tenebrionidae) and its implication on insect phylogeny. Dev Genes Evol. 2007;217:13–27. doi: 10.1007/s00427-006-0114-3. [DOI] [PubMed] [Google Scholar]
- 89.Irles P, Bellés X, Piulachs MD. Identifying genes related to choriogenesis in insect panoistic ovaries by Suppression Subtractive Hybridization. BMC Genomics. 2009;10:206. doi: 10.1186/1471-2164-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oro AE, McKeown M, Evans RM. The Drosophila retinoid X receptor homolog ultraspiracle functions in both female reproduction and eye morphogenesis. Development. 1992;115:449–462. doi: 10.1242/dev.115.2.449. [DOI] [PubMed] [Google Scholar]
- 91.Nozawa M, Suzuki Y, Nei M. Reliabilities of identifying positive selection by the branch-site and the site-prediction methods. Proc Natl Acad Sci U S A. 2009;106:6700–6705. doi: 10.1073/pnas.0901855106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang Z, Nielsen R, Goldman N. In defense of statistical methods for detecting positive selection. Proc Natl Acad Sci U S A. 2009;106:E95. doi: 10.1073/pnas.0904550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang Z, dos Reis M. Statistical properties of the branch-site test of positive selection. Mol Biol Evol. 2011;28:1217–1228. doi: 10.1093/molbev/msq303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of insect USP/RXR sequences. The green and orange bars indicate the Mecopterida and Non-Mecopterida taxa, respectively, and the schematic below the alignment shows the USP/RXR domain structure. For ease of viewing one alignment is shown. However, the complete alignment was not used for analysis as some regions do not align (e.g. D domain). Only the carboxy-terminal E/F (*), or ligand-binding domain, was used for the branch and branch-sites analyses reported in table 1. Arrows indicate where poorly aligned regions and major gaps were deleted. Full length sequences were used for the random-sites and HyPhy analyses where the larger dataset was split into Mecopterida and Non-Mecopterida only datasets in order to compare evolutionary rates between the two groups. For clarity, site numbering for the full length Mecopterida (green) and Non-Mecopterida (orange) datasets is shown above and below the alignment, respectively. Note that species names have been abbreviated to six characters, complete names can be found in supporting table S1.
(PDF)
Alignment of insect EcR sequences. The green and orange bars indicate the Mecopterida and Non-Mecopterida taxa, respectively, and the schematic below the alignment shows the EcR domain structure. For ease of viewing one alignment is shown. However, the complete alignment was not used for analysis as some regions do not align (e.g. D domain). Only the carboxy-terminal E (*), or ligand-binding domain, was used for the branch analyses reported in table 1. Arrows indicate where poorly aligned regions and major gaps were deleted. Full length sequences were used for the random-sites and HyPhy analyses where the larger dataset was split into Mecopterida and Non-Mecopterida only datasets in order to compare evolutionary rates between the two groups. For clarity, site numbering for the full length Mecopterida (green) and Non-Mecopterida (orange) datasets is shown above and below the alignment, respectively. Note that species names have been abbreviated to six characters, complete names can be found in supporting table S1.
(PDF)
USP/RXR and EcR gene trees. Gene trees for USP/RXR and EcR were generated using the alignment of LBD sequences given in supporting figures S1 and S2. Maximum-likelihood trees for USP/RXR (A) and EcR (C) were constructed in PhyML [57] using the WAG substitution model, with four rate categories to estimate the gamma parameter shape. Neighbor-joining [58] trees for USP/RXR (B) and EcR (D) were constructed in MEGA 4 [38] using the Poisson correction model, with the pair-wise deletion of gaps. For all analyses 100 bootstrap replicates were performed, and nodes with values less than 60 were later collapsed. Each tree was then rooted along the branch leading to B. germanica and L. migratoria. Note that species names have been abbreviated, see supporting table S1.
(PDF)
d N/ d S site-profile plots for PAML model M3. The values of ω as estimated by M3 in PAML using the NEB method are shown for each codon site across Mecopterida USP/RXR (A), Non-Mecopterida USP/RXR (B), Mecopterida EcR (C) and Non-Mecopterida EcR (D). A schematic of USP/RXR and EcR secondary structure is shown above each plot to illustrate the position of each functional domain (A/B, C, and D) as well as the helices (H1–H12) and β sheets of the ligand-binding domain. These schematics are based on the crystal structure of each gene in H. virescens and B. tabaci. Site numbering is the same as figures 3 and 4.
(PDF)
Sequence data information.
(DOC)
Putative positively selected sites.
(DOC)
Sites with elevated ω in the USP/RXR gene.
(DOC)
Sites with elevated ω in the EcR gene.
(DOC)
Statistics for rate distributions inferred by HyPhy.
(DOC)
Rate class assignment for MG94×REV Nonsynonymous GDD 3.
(DOC)
Rate class assignment for MG94×REV Dual GDD 3×3.
(DOC)