Abstract
PAPD5 is one of the seven members of the family of noncanonical poly(A) polymerases in human cells. PAPD5 was shown to polyadenylate aberrant pre-ribosomal RNAs in vivo, similar to degradation-mediating polyadenylation by the noncanonical poly(A) polymerase Trf4p in yeast. PAPD5 has been reported to be also involved in the uridylation-dependent degradation of histone mRNAs. To test whether PAPD5 indeed catalyzes adenylation as well as uridylation of RNA substrates, we analyzed the in vitro properties of recombinant PAPD5 expressed in mammalian cells as well as in bacteria. Our results show that PAPD5 catalyzes the polyadenylation of different types of RNA substrates in vitro. Interestingly, PAPD5 is active without a protein cofactor, whereas its yeast homolog Trf4p is the catalytic subunit of a bipartite poly(A) polymerase in which a separate RNA-binding subunit is needed for activity. In contrast to the yeast protein, the C terminus of PAPD5 contains a stretch of basic amino acids that is involved in binding the RNA substrate.
Keywords: polyadenylation, noncanonical poly(A) polymerase, TRAMP4, RNA surveillance
INTRODUCTION
The addition of a poly(A) tail to the 3′ end of mRNAs in the nucleus of eukaryotic cells is catalyzed by the classical or canonical poly(A) polymerases (PAPs) and is an important step during mRNA maturation. The poly(A) tail confers stability to the mRNA and allows transport out of the nucleus and efficient translation in the cytoplasm. In contrast, the functions of the so-called noncanonical PAPs or Cid1-like proteins (after the founding member of the family) are only partially understood. Enzymes of this group are involved in cytoplasmic polyadenylation of mRNAs, but also in polyadenylation-mediated degradation of RNAs, and some members of the family even catalyze the addition of uridyl residues to RNA substrates (Schmidt and Norbury 2010). Like the canonical PAPs, Cid1-like proteins belong to the superfamily of polymerase β-like nucleotidyl transferases. Canonical and noncanonical PAPs have similar catalytic and central domains but differ in their nucleotide-base recognition motif. Interestingly, only a few Cid1-like proteins contain an RNA-binding domain (Martin and Keller 2007).
One well-studied member of the Cid1-like protein family is Trf4p, a PAP involved in nuclear RNA surveillance in Saccharomyces cerevisiae. Polyadenylation of RNAs by Trf4p facilitates their subsequent degradation by the nuclear exosome, a 3′–5′ exonuclease complex (Kadaba et al. 2004, 2006; Vanacova et al. 2005), or in some cases induces the processing of the RNA by exosome-catalyzed trimming of the 3′ end (Egecioglu et al. 2006). A stimulatory effect of polyadenylation on the exosome activity was first shown to allow efficient degradation of a hypomodified, unstable tRNA (LaCava et al. 2005; Vanacova et al. 2005). Later, rRNAs, snRNAs, and snoRNAs, as well as so-called cryptic unstable transcripts (CUTs), spliced-out introns, and different mRNAs were identified as substrates of this polyadenylation-mediated RNA quality-control mechanism (LaCava et al. 2005; Wyers et al. 2005; Davis and Ares 2006; Dez et al. 2006; Egecioglu et al. 2006; Carneiro et al. 2007; Houseley et al. 2007; San Paolo et al. 2009; Callahan and Butler 2010).
Trf4p lacks an RNA-binding domain and is therefore inactive on its own. In vivo it is part of the TRAMP4 complex that contains Trf4p as the catalytic subunit, one of the zinc knuckle proteins Air1p or Air2p, thought to confer the RNA-binding function to the complex, and the RNA helicase Mtr4p (LaCava et al. 2005; Vanacova et al. 2005). A similar complex, SpTRAMP, was identified in fission yeast (Win et al. 2006; Bühler et al. 2008). In Drosophila melanogaster, overexpression of a homolog of Trf4p was shown to lead to the polyadenylation of snRNAs (Nakamura et al. 2008), whereas the human homolog PAPD5 is involved in the polyadenylation-mediated degradation of aberrant pre-rRNAs (Shcherbik et al. 2010).
The human PAPD5 protein was also reported to function in the degradation of replication-dependent histone mRNAs. These mRNAs do not have a poly(A) tail but end in a stem–loop structure. Here, however, the degradation pathway does not start with polyadenylation of the RNA, but with the addition of a short oligo(U) tail, which is recognized by the Lsm1-7 complex and subsequently leads to decapping and degradation of the mRNAs. A knockdown of either PAPD5 or of the mitochondrial poly(A) polymerase results in the stabilization of histone mRNAs (Mullen and Marzluff 2008). However, this effect could not be observed by other investigators (Schmidt et al. 2010). Instead, the knockdown of the uridyl transferase ZCCHC11, a different member of the human Cid1-like proteins, was shown to strongly reduce the oligouridylation of histone mRNAs, leading to their stabilization (Schmidt et al. 2010). It is therefore possible that PAPD5, depending on the RNA substrate or the presence of different protein cofactors, can catalyze the polyadenylation as well as the oligouridylation of RNAs. To better understand the function of PAPD5, we set out to investigate the activity of the protein in vitro and to explore its function in mammalian cells through cross-linking and immunoprecipitation (CLIP) (Hafner et al. 2010).
RESULTS
PAPD5 is a poly(A) polymerase in vitro
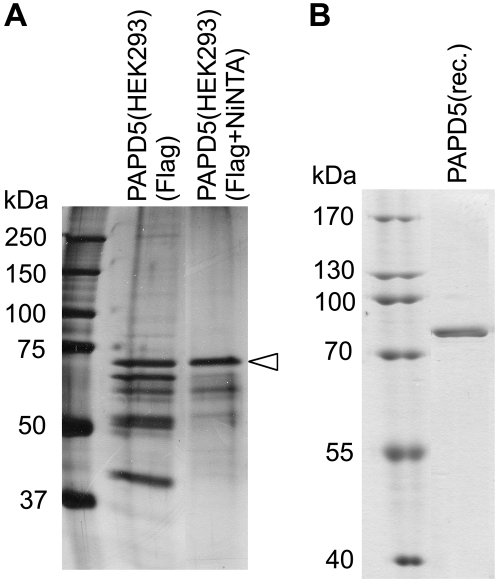
PAPD5 (UniProt accession number Q8NDF8) is involved in the polyadenylation of aberrant ribosomal RNA precursors in mouse (Shcherbik et al. 2010). In addition, it may also be responsible for the uridylation-mediated degradation of histone mRNAs (Mullen and Marzluff 2008). However, this function of PAPD5 is controversial (Schmidt et al. 2010). To test the activity of PAPD5 in vitro, the coding sequence was amplified from cDNA of HeLa cells. The protein was expressed in HEK293 cells as a fusion protein with an N-terminal Flag tag and a His8 tag. After cell lysis, the protein was bound to anti-Flag agarose, washed, and eluted with Flag peptide. The protein was further purified via nickel affinity chromatography (Fig. 1A).
FIGURE 1.
PAPD5 preparations used in the analysis. (A) PAPD5 was expressed as a fusion protein with N-terminal His8 and Flag tag in HEK293 cells and purified via anti-Flag agarose (Flag), followed by metal chelating affinity chromatography (Flag + NiNTA). PAPD5 is marked by an arrowhead. (B) Bacterially expressed PAPD5 after metal chelating affinity chromatography (for details, see Materials and Methods).
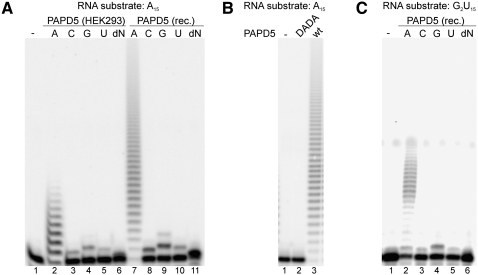
To test the nucleotidyltransferase activity of PAPD5, the oligoribonucleotide A15 was incubated with the protein either in the presence of ATP, CTP, GTP, or UTP, or in a mixture of all four dNTPs (Fig. 2A). When PAPD5 and A15 were incubated in the presence of ATP, the incorporation of several nucleotides into the RNA substrate was observed. In contrast, the incorporation of the other three ribonucleotides was low and limited to single residues, showing a strong preference of PAPD5 for ATP. In addition, PAPD5 is able to discriminate against dNTPs, as these were not used as substrates.
FIGURE 2.
PAPD5 preferentially adds AMP residues to RNA substrates. (A) PAPD5 preparations from HEK293 cells (HEK293) or recombinant protein expressed in E. coli (rec.) were incubated with ribo-oligonucleotide A15 in the presence of ATP (A), CTP (C), GTP (G), UTP (U), or a mixture of all four dNTPs (dN), respectively. (B) PAPD5 wild-type protein (wt) or catalytic site mutant (DADA) expressed in E. coli were incubated with ribo-oligonucleotide A15 in the presence of ATP. (C) PAPD5 expressed in E. coli was incubated with ribo-oligonucleotide G2U15 in the presence of ATP (A), CTP (C), GTP (G), UTP (U), or a mixture of all four dNTPs (dN), respectively. Lane 1 always shows the RNA substrate incubated in the absence of protein.
PAPD5 does not need a cofactor for polyadenylation in vitro
To further analyze the properties of PAPD5, the protein was expressed in Escherichia coli. To prevent contamination with bacterial poly(A) polymerase, an expression strain with a disruption in the gene for poly(A) polymerase was used. Two PAPD5 expression constructs were tested: a His6 fusion and a combination of a C-terminal His6 tag and an N-terminal fusion of a GB1 tag that was shown to increase the solubility of recombinant proteins (Zhou et al. 2001). Because the activity of both wild-type fusion proteins was the same but the yield of the GB1-tagged protein was higher, the latter construct was used for further experiments. The purified protein is shown in Figure 1B.
Trf4p, the poly(A) polymerase of the TRAMP4 complex in baker's yeast, was previously shown to be inactive on its own. Trf4p contains no obvious RNA-binding domain and is believed to rely on the zinc-knuckle proteins Air1p or Air2p as the substrate-binding domain of the complex. Indeed, only the combination of Trf4p with either Air1p or Air2p has poly(A) polymerase activity (LaCava et al. 2005; Vanacova et al. 2005).
Surprisingly, the bacterially expressed PAPD5 (Fig. 1B) showed the same activity as the protein purified from HEK293 cells. When the ribo-oligonucleotide A15 was incubated with PAPD5 in the presence of different nucleotides, efficient incorporation could only be observed with ATP (Fig. 2A). A PAPD5 variant in which two aspartate residues (D177, D179) in the catalytic domain were exchanged to alanine could not catalyze the addition of nucleotides to the RNA substrate (PAPD5 DADA) (Fig. 2B). Therefore, the observed polyadenylation activity is an intrinsic activity of PAPD5, and a protein cofactor is not needed for activity.
The nucleotide incorporation experiment was repeated with other RNA substrates: Ribo-oligonucleotide G2U15 was used to test whether the last nucleotide of the RNA primer influences the specificity for the nucleotide substrate (Fig. 2C). In addition, in vitro–synthesized tRNAiMet (a known substrate for yeast Trf4p) and an in vitro–synthesized fragment of the 3′ UTR of mouse histone H2a mRNA comprising the stem–loop structure were tested as substrates (results not shown). Again, only AMP incorporation could be observed. Therefore, the specificity for ATP incorporation does not depend on the last nucleotide of the RNA primer but results from nucleotide specificity of the protein. The specificity for ATP incorporation was also observed when a reaction buffer containing manganese instead of magnesium was used (results not shown).
As reported for yeast Trf4p (LaCava et al. 2005), the polyadenylation reaction catalyzed by PAPD5 is not processive: In all cases, the reaction products were heterogeneous in size, the smallest product usually corresponding to the incorporation of a single nucleotide. When DNA oligonucleotides were used as primers, none of the nucleotide substrates was incorporated (results not shown), showing that the reaction is RNA-specific.
A C-terminal stretch of basic amino acids is involved in binding of the RNA primer
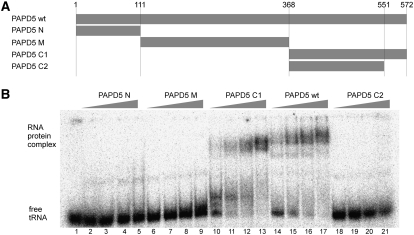
The amino acid sequence of PAPD5 does not contain any motif known to bind nucleic acids. However, the polyadenylation activity of PAPD5 observed in vitro and in the absence of other protein cofactors argues for the presence of a domain in the protein that is involved in binding of the RNA substrate. To analyze the RNA binding of PAPD5, electrophoretic mobility shift experiments were performed. PAPD5 was incubated with in vitro–synthesized tRNA, and the reaction mixture was resolved on a nondenaturing polyacrylamide gel. Indeed, a shift of the RNA band to a band of lower mobility could be observed, showing that PAPD5 is able to bind RNA. To determine which part of the protein is responsible for RNA binding, protein variants comprising only the N terminus including the catalytic domain (amino acids 1–111, PAPD5 N), the central domain (amino acids 112–368, PAPD5 M), or the C terminus of PAPD5 (amino acids 369–572, PAPD5 C1) were analyzed separately. Of these, only the C-terminal part, PAPD5 C1, could shift the tRNA (Fig. 3), indicating the presence of an RNA-binding function in the C terminus of PAPD5.
FIGURE 3.
The C terminus of PAPD5 is involved in RNA binding. (A) Schematic representation of the truncation variants of PAPD5 tested for RNA binding. (B) Electrophoretic mobility shift assay of PAPD5 variants. Increasing amounts of PAPD5 full-length protein or fragments comprising the N terminus, central part, or C terminus of the protein were incubated with radioactively labeled in vitro–synthesized human tRNAiMet, and the reactions were resolved on a native polyacrylamide gel. Protein concentrations between 0.5 and 5 nM were tested for the full-length protein and the protein PAPD5 C2, 1–20 nM for the other variants. Lane 1 shows the tRNA after incubation in the absence of protein.
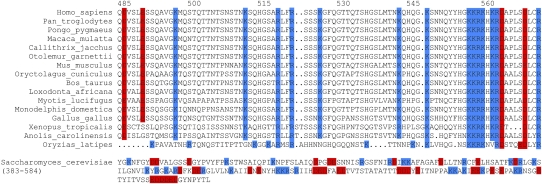
Inspection of the amino acid sequence showed the C terminus of human PAPD5 to contain a stretch of basic amino acids (amino acids 557–563), a motif that is conserved among higher eukaryotes, but not in the yeast protein Trf4p (Fig. 4). To test the influence of the basic motif on the RNA-binding ability of PAPD5, a deletion variant of the C-terminal part lacking this motif was used (amino acids 369–551, PAPD5 C2). Whereas the C terminus binds to RNA (Fig. 3, PAPD5 C1), the deletion variant showed no shift of the RNA in EMSA experiments, even when used in higher concentrations than the original C-terminal fragment (Fig. 3, PAPD5 C2).
FIGURE 4.
The basic motif in the C terminus of PAPD5 is conserved among higher eukaryotes. Amino acid sequences of PAPD5 homologs (Ensembl) were aligned with ClustalW (Chenna et al. 2003) and refined manually (upper panel). The amino acid sequence of the yeast protein Trf4p is shown for comparison (lower panel). Basic amino acids are labeled in blue, acidic amino acids in red.
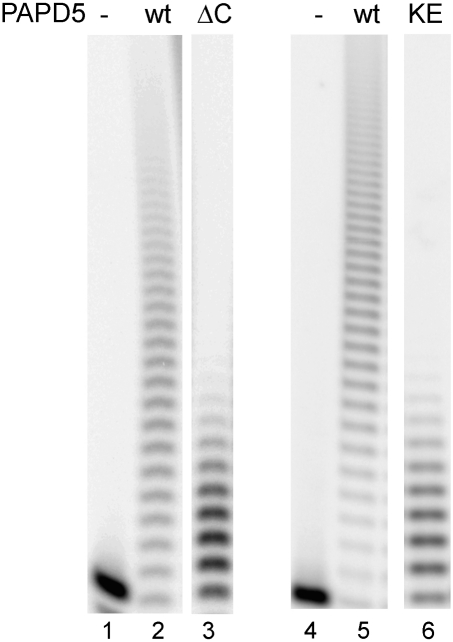
Next, we wanted to analyze the influence of this basic amino acid stretch on the catalytic activity of PAPD5. Therefore, a protein variant PAPD5ΔC, comprising amino acids 1–551, i.e., lacking only the basic stretch, was constructed and tested for polyadenylation activity. In addition, a point mutant of PAPD5 was constructed in which a lysine residue in the basic domain was exchanged to glutamate (PAPD5 K560E). The variants were expressed in E. coli and purified as the wild-type protein. For both variants, the yield of soluble protein was comparable to the wild type. In comparison to the wild-type protein, both variants showed a reduced activity when incubated with the A15 substrate in the presence of ATP, resulting in fewer incorporated nucleotides and therefore shorter polyadenylation products (Fig. 5).
FIGURE 5.
Mutations in the basic motif of PAPD5 lead to loss of activity. PAPD5 wild-type protein (wt), C-terminal deletion mutant (ΔC, amino acids 1–551), or a point mutant in the basic motif (KE, position 560) expressed in E. coli were incubated with ribo-oligonucleotide A15 in the presence of ATP.
To further examine the influence of the basic stretch of amino acids on the polyadenylation activity of PAPD5, the wild-type protein and the variants were incubated with different concentrations of either ATP or A15 substrate to determine the kinetic parameters of the reaction. To this end, radioactive ATP was used as nucleotide substrate, and the radioactively labeled reaction products were separated from nonincorporated nucleotides with DE81 filters. The polyadenylation activity was measured as total incorporation of nucleotides into the A15 substrate, independent of the length of the resulting product.
When the concentration of the RNA substrate A15 was varied, the maximal velocities for the wild-type protein and the variants were similar (Table 1). However, the KM of the proteins with changes in the C-terminal basic motif was increased by a factor of ∼3 to 5. In contrast, there was no significant difference in the apparent affinity for ATP. The concentration of ATP during A15 titration was in much higher excess than the concentration of the RNA primer when ATP was titrated; therefore, the kcat values determined by titration of ATP are lower than the values observed for the titration of A15. This result confirms the participation of the stretch of basic amino acids in the binding of the RNA substrate by PAPD5.
TABLE 1.
Kinetic parameters of PAPD5 variants
Specificity for RNA structure
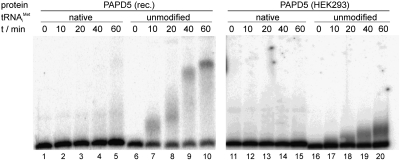
The yeast TRAMP complex was shown to specifically polyadenylate unmodified initiator tRNA, but not the native and therefore fully modified tRNA (Vanacova et al. 2005). This is thought to be a result of a more compact tertiary structure of the native tRNA. It was shown recently that a core complex consisting of the central part of Trf4p and two out of the five zinc knuckles of Air2p is sufficient to discriminate between the two tRNA substrates (Hamill et al. 2010). To test whether PAPD5 shows a similar ability, PAPD5 purified from HEK293 cells as well as bacterially expressed PAPD5 were incubated with either in vitro–synthesized yeast tRNAiMet or the native tRNA purified from yeast cells in the presence of ATP. Interestingly, both enzyme preparations were able to polyadenylate the in vitro transcript but not the native tRNA, whereas at the same time E. coli poly(A) polymerase polyadenylated both tRNA substrates (Fig. 6; results not shown). Therefore, similarly to the yeast Trf4p, PAPD5 seems to be sensitive to the structure of the RNA substrate.
FIGURE 6.
PAPD5 adds poly(A) tails to the unmodified in vitro transcript of S. cerevisiae tRNAiMet, but not to the native tRNA isolated from yeast. PAPD5 protein expressed in E. coli (PAPD5 rec.) or in HEK293 cells was incubated with the different RNA substrates in the presence of ATP, and the reactions were stopped after the reaction times indicated (0, 10, 20, 40, 60 min).
Subcellular localization of PAPD5
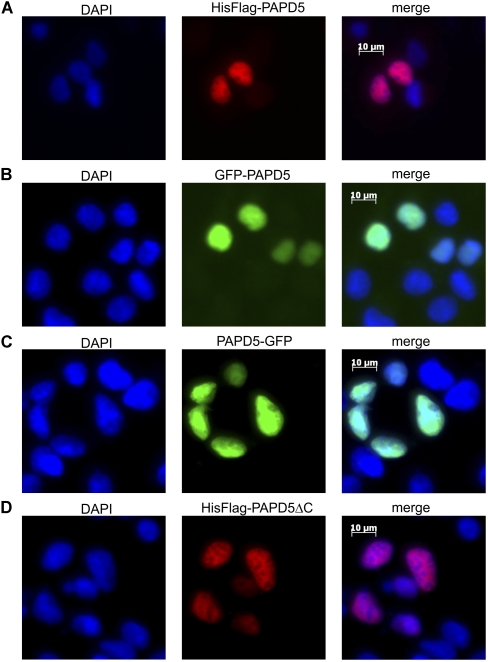
In yeast, Trf4p is part of a RNA surveillance machinery residing in the nucleus. To determine the localization of PAPD5, the protein was detected in cells expressing Flag-tagged PAPD5 with anti-Flag antibody followed by a secondary antibody conjugated to a fluorescent dye (Fig. 7A). In addition, PAPD5 fusion proteins with green fluorescent protein (GFP) fused to the N or C terminus were expressed transiently (Fig. 7B,C). All fusion proteins were detected in the nucleus as shown by colocalization of the fluorescence signals with the DAPI stain, no staining of the cytoplasm could be observed. The basic stretch of amino acids at the C terminus of PAPD5 that is involved in RNA binding does not play a role for localization of PAPD5, because a Flag-tagged deletion variant corresponding to PAPD5ΔC, i.e., lacking the basic stretch, is still exclusively located in the nucleus (Fig. 7D).
FIGURE 7.
PAPD5 is located in the nucleus. (A) PAPD5 was expressed as a fusion protein with N-terminal His8-Flag tag in HEK293 cells. The protein was detected with anti-Flag antibody and Cy3-conjugated secondary antibody. Nuclei stained with DAPI (left panel); the tagged protein signal (middle); the merged picture (right panel). (B,C) PAPD5 was expressed as a fusion protein with green fluorescent protein fused to the N terminus (B) or the C terminus (C). Panels as in A. (D) PAPD5ΔC (amino acids 1–551) was expressed as a fusion protein with N-terminal His8-Flag tag in HEK293 cells. The protein was detected with anti-Flag antibody and Cy3-conjugated secondary antibody. Panels as in A. DAPI stain of the nucleus (blue), Cy3 staining (red), and GFP signal (green).
Ribosomal RNAs are in vivo targets of PAPD5
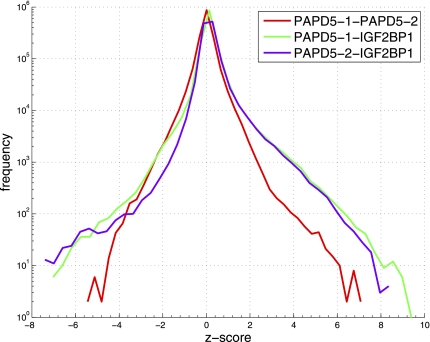
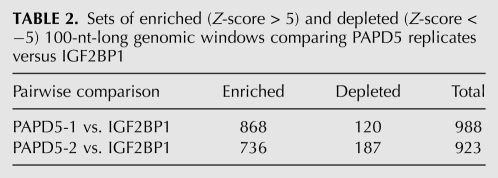
PAPD5 was able to polyadenylate a variety of RNA substrates in vitro, ranging from oligo(A) and oligo(U) to different tRNAs as well as the 3′ UTR of histone mRNAs. To determine the potential substrate spectrum of PAPD5 in vivo, RNAs bound to the protein in HEK293 cells were probed by the PAR-CLIP technique (Hafner et al. 2010). Briefly, HEK293 cells expressing Flag-tagged PAPD5 were grown in medium containing 4-thio-uridine and RNA–protein complexes were UV-cross-linked and immunoprecipitated. The covalently bound RNAs were extracted, adapter oligonucleotides were ligated to the 5′ and 3′ ends, and the RNA was reverse-transcribed. The cDNA was amplified and the sequences determined by deep sequencing. We then compared the results obtained in two separate PAPD5 PAR-CLIP experiments and those previously obtained for the insulin growth factor 2 binding protein 1 (IGF2BP1) (Hafner et al. 2010) to identify genomic regions from which PAPD5 targets derive. The IGF2BP1 PAR-CLIP sample contained 24,914,594 reads and the PAPD5 PAR-CLIP samples 4,949,000 and 5,563,677 reads, respectively. The proportion of reads mapping with at most one error to the human genome was 45.64% in the case of IGF2BP1 and 32.77% and 27.13% for PAPD5 replicates. We normalized the count of mapped reads across samples and calculated the number of reads originating in each 100-nt-long window of the genome, and the variance of these counts in the PAPD5 PAR-CLIP replicates with the method described by Balwierz et al. (2009). We then computed the Z-score indicating the significance of the enrichment or depletion in reads in a PAPD5 sample compared to the IGF2BP1 sample. The distributions of the Z-scores across all genomic regions (Fig. 8) show a much higher similarity of PADP5 replicates relative to the IGF2BP1 sample. With cut-off values of −5 and 5, at which only a very small number of genomic regions will be considered differentially represented between PAPD5 replicates, we obtained 988 regions for the first PAPD5 replicate and 923 for the second (Table 2) that were differentially expressed relative to IGF2BP1. Four hundred thirty-four regions were differentially expressed between both PAPD5 replicates and the IGF2BP1 sample. Of these, 358 regions were enriched, whereas 76 were depleted in PAPD5 samples (Supplemental Table S1). One hundred ninety-two out of the 358 enriched windows corresponded to repeat elements, 61 of which were rRNA. These latter windows were located in genomic regions annotated as rRNA-like in the Genome Browser of the University of California Santa Cruz. They contained reads that we also annotated as rRNA-derived, based on the fact that they mapped with at most one error to the rRNA subunits. In contrast, only two of the 76 regions depleted in PAPD5 binding had rRNA annotations (Fisher's exact test, p < 0.001), indicating that rRNAs could be the primary targets of PAPD5 in human cells.
FIGURE 8.
Distribution of pairwise Z-scores computed for 100-nt-long windows in pairs of samples. The reads obtained in two separate PAPD5 PAR-CLIP experiments as well as those previously obtained for the insulin growth factor 2 binding protein 1 (IGF2BP1) (Hafner et al. 2010) were mapped to the genome. After normalization, the Z-scores, giving a measure of the differential expression of individual 100-nt-long windows in each pair of experiments, were calculated.
TABLE 2.
Sets of enriched (Z-score > 5) and depleted (Z-score < −5) 100-nt-long genomic windows comparing PAPD5 replicates versus IGF2BP1
DISCUSSION
PAPD5 belongs to the family of Cid1-like proteins or noncanonical PAPs. In this family of nucleotidyltransferases, several members do not contain a recognizable RNA-binding domain. For the noncanonical PAP Trf4p from yeast, it was shown that nucleotidyltransferase activity can only be observed for the complex of Trf4p and one of the two zinc-knuckle proteins Air1p or Air2p, but not for Trf4p alone (LaCava et al. 2005; Vanacova et al. 2005). Therefore, it is thought that Trf4p is the catalytic subunit of this bipartite PAP, whereas the zinc-knuckle proteins confer RNA-binding specificity to the complex. Such a modular organization of a catalytic subunit that could even be combined with different RNA-binding subunits to result in an active nucleotidyltransferase may allow the targeting of different classes of substrates, depending on the RNA-binding subunit used. For the TRAMP4 complex in yeast, it was shown that two out of the five zinc knuckles of Air2p are necessary and sufficient for the polyadenylation of a short RNA oligonucleotide. However, efficient polyadenylation of a tRNA required the most N-terminal zinc knuckle in addition. Although the other two zinc knuckles seem to be dispensable for the tRNA substrate, they may be important for the recognition of other TRAMP substrates (Hamill et al. 2010).
Surprisingly, PAPD5, a human Trf4p homolog lacking a recognizable RNA-binding domain, is able to catalyze the addition of nucleotides to RNA substrates in the absence of a protein cofactor. Analysis of the amino acid sequence revealed a stretch of basic residues in the C terminus of PAPD5 that is highly conserved among higher eukaryotes. When this motif was deleted, the nucleotidyltransferase activity was strongly reduced, and both electrophoretic mobility shift experiments and the analysis of the activity of a point mutant in this part of the protein showed the stretch of basic amino acids to take part in binding of the RNA substrate. PAPD5 was able to incorporate nucleotides to different RNA substrates [oligo(A), oligo(U), tRNA, histone mRNA 3′ end], therefore the RNA-binding function is very likely not sequence-specific, resulting in polyadenylation of all RNAs with a free 3′-hydroxyl end in vitro. However, like the yeast TRAMP complex, PAPD5 discriminates between in vitro–synthesized, unmodified and native, fully modified tRNAs. Very likely, this is due to a difference in the stability of the RNA structure, resulting in the 3′ end of the in vitro transcript being more accessible to the poly(A) polymerase. PAPD5 has been shown to be involved in the polyadenylation of aberrant ribosomal RNAs: Aberrant pre-rRNAs were found to be partially polyadenylated in a murine cell line by reverse transcription with an oligo(dT)-adapter primer followed by PCR with a forward primer specific for the 5′-ETS region of pre-rRNA. After knockdown of PAPD5, the amount of polyadenylated pre-rRNAs was decreased (Shcherbik et al. 2010). In agreement with this finding, the analysis of RNAs cross-linked to PAPD5 in vivo revealed rRNAs to be potential substrates of PAPD5. Ribosomal RNAs were represented in the potential RNA substrates in a high percentage, and they were specifically enriched in the PAPD5 samples compared to a control IGF2BP1 sample. The same was shown for Trf4p-associated RNAs in yeast: 50% of the sequences identified by an in vivo cross-linking approach were mapped to ribosomal sequences (Wlotzka et al. 2011). In addition, other classes of noncoding RNAs as well as pre-mRNAs and mRNAs were found to be associated with PAPD5. Although in Trf4-deletion yeast strains as well as in the cross-linking approach the same classes of RNAs were found to be enriched, the relative proportions of these transcript types were different from that found here for PAPD5 (San Paolo et al. 2009; Wlotzka et al. 2011), and in the PAPD5 samples such targets were not robustly enriched. Whereas in yeast only two noncanonical poly(A) polymerases are present, human cells contain seven members of this protein family. Therefore, other noncanonical poly(A) polymerases may be involved in the polyadenylation of other aberrant RNAs.
The catalytic parameters observed for the wild-type PAPD5 protein (Table 1) are similar to the values determined for bacterial poly(A) polymerase, an enzyme also involved in RNA degradation (Li et al. 1998, 2002). In contrast, canonical poly(A) polymerases show higher maximal velocity even under distributive reaction conditions, reflecting the function of the two different types of polyadenylation. Whereas the canonical poly(A) polymerases add long tails that are immediately bound by poly(A)-binding proteins and allow protection of the mRNA from degradation, polyadenylation-mediated degradation is characterized by the addition of short tails as well as distributive polyadenylation to allow access of the degradation machinery.
In the cell, noncoding RNAs as well as mRNAs are bound by proteins during processing and reside in RNPs to perform their function. In contrast, aberrant RNAs or processing intermediates that—due to misfolding or misprocessing—cannot form the correct interactions to protein partners would be accessible for polyadenylation and degradation. Therefore, PAPD5 could act as a kind of scavenger enzyme, similar to the TRAMP4 complex in yeast, to polyadenylate RNA 3′ ends and thereby tag the corresponding RNA for degradation. Like yeast Trf4p, PAPD5 is located in the nucleus of the cell and may therefore be part of a nuclear surveillance machinery similar to the TRAMP4–exosome system of yeast. In contrast to Trf4p, PAPD5 is able to bind different RNAs, and when His-Flag-tagged PAPD5 was purified from mammalian cells, no copurifying RNA-binding proteins could be identified by mass spectrometry. However, it still remains to be determined whether protein cofactors are necessary for the efficient binding of PAPD5 to substrates, as has been shown for TRAMP4.
When the levels of several of the putative substrates identified by the CLIP experiments were analyzed in cells treated with siRNAs directed against PAPD5 or control siRNA, no significant change could be observed (results not shown). However, mammalian cells contain genes for seven proteins of the Cid1-like family. PAPD7, one of these homologs, is very similar to PAPD5. If more than one of these noncanonical PAPs are able to target the same RNAs, depletion of the cell of a single PAP may not lead to a detectable change in the RNA level. It has been observed for poly(A) and A-rich tails on human rRNA that the knockdown of a single noncanonical PAP does not abolish tail addition (Slomovic et al. 2010). Therefore, the RNA surveillance system is most likely redundant, allowing potentially deleterious aberrant RNAs to be degraded with high efficiency.
MATERIALS AND METHODS
Expression and purification of PAPD5
The coding sequence of PAPD5 was amplified from HeLa cell cDNA. The ORF was cloned into pcDNA5/FRT for expression as a fusion protein with N-terminal His8 and Flag tags in mammalian cells. For expression, an HEK293 cell line, Flp-In-293 (Invitrogen), was used. Cells were cultivated in DMEM medium containing 10% fetal calf serum, and cell lines expressing PAPD5 were selected according to the manufacturer's instructions. For analysis of the subcellular localization, PAPD5 was transiently expressed as an N- or C-terminal GFP fusion protein or with a Flag tag. For detection of Flag-tagged protein, cells were permeabilized, and the fusion protein was stained with mouse anti-Flag antibody (Sigma) and Cy3-conjugated donkey anti-mouse antibody (Dianova).
For expression in E. coli, the ORF of PAPD5 was cloned into pET30 for expression as a fusion protein with an N-terminal GB1 tag and a C-terminal His6 tag (Zhou et al. 2001) or into pETDuet (Novagen) for expression as a C-terminal His6-tag fusion protein. A BL21 (DE3) strain deficient for E. coli poly(A) polymerase was constructed with the TargeTron Gene Knockout System (Sigma-Aldrich). The strain was transformed with the corresponding expression plasmids, grown in LB medium at 37°C to an OD600 of 0.8, and expression of PAPD5 was induced with 1 mM IPTG for 3 h.
For purification of the protein from mammalian cells, the cells were lysed by sonication in buffer containing 50 mM Tris (pH 7.5), 100 mM KCl, 10% glycerol, 0,05% NP-40, 1 mM PMSF, 1 mg/L Pepstatin, and 1 mg/L Leupeptin. The cleared lysate was incubated with M2 anti-Flag agarose (Sigma-Aldrich). The matrix was washed with lysis buffer containing 300 mM KCl, and the protein was eluted with lysis buffer containing 5 mM Flag peptide. The eluate was adjusted to 5 mM imidazole, bound to Ni-NTA agarose (Sigma-Aldrich), washed with buffer containing 25 mM imidazole, and the protein was eluted with buffer containing 250 mM imidazole.
The protein expressed in E. coli was purified via metal chelating chromatography like the protein expressed in mammalian cells, but the cell lysate was adjusted to 25 mM imidazole, and the washing buffer contained 500 mM KCl and 50 mM imidazole.
RNA substrates
A plasmid ptRNAMet for the in vitro transcription of yeast tRNAiMet was obtained from Dr. Bruno Senger (Institut de Biologie Moléculaire et Cellulaire Strasbourg). The plasmid was linearized with BstNI and transcribed with T7 RNA polymerase. The native yeast tRNAMet was a gift of Dr. Gérard Keith (Institut de Biologie Moléculaire et Cellulaire Strasbourg). The plasmid pUC19-H2a for in vitro transcription of the mouse histone H2a mRNA 3′ end with T7 RNA polymerase was obtained from Dr. Sophie Jaeger (Biozentrum, University of Basel). In vitro–transcribed RNAs were purified on 8.3 M urea/10% polyacrylamide gels. For 5′-end labeling, RNAs were treated with alkaline phosphatase and labeled with [γ-32P]ATP (GE Healthcare) and T4 polynucleotide kinase. Labeled RNAs were gel-purified. To allow folding of the structured RNAs, pellets were dissolved in RNase-free water and incubated for 1 min at 65°C, followed by incubation for 10 min on ice after the addition of an equal volume of twice-concentrated assay buffer containing 10 mM MgCl2.
Polyadenylation assays
Polyadenylation assays were carried out in 20-μL reaction mixtures containing 0.07–0.7 pmol of affinity-purified protein, 1 pmol of 5′-end-labeled RNA, 1 mM ATP, 5 mM MgCl2, 25 mM Tris-HCl (pH 7.5), 50 mM KCl, 0.01 mM EDTA, 0.1 mg/mL BSA, 1 mM DTT, and 0.02% NP-40. Reactions were incubated for 30 min at 37°C or the times indicated and stopped by the addition of 25 mM EDTA. The RNA was precipitated by addition of 0.1 volume of 3 M ammonium acetate and three volumes of ethanol. Pellets were resuspended in formamide loading buffer and separated on denaturing polyacrylamide gels. Radioactivity was scanned with a PhosphorImager, and results were analyzed with ImageQuant software (Molecular Dynamics).
For determination of the kinetic parameters, the activities of PAPD5 variants were measured in 20-μL reaction mixtures containing buffer as above with 0.2 μCi of [α-32P]ATP (3000 Ci/mmol) and 0.7–3.5 pmol (0.035–0.17 μM) recombinant protein. The concentration of ATP was varied between 0.01 mM and 1 mM (the concentration of A15 was kept constant at 2.5 μM), and the concentration of unlabeled RNA A15 was varied between 50 nM and 2.5 μM (ATP was kept constant at 1 mM). The reactions were incubated for 20 min at 37°C and stopped by spotting onto DE-81 paper. The filters were washed three times for 10 min in 0.3 M ammonium formate/10 mM Na-pyrophosphate, and the incorporated radioactivity was measured in a scintillation counter.
EMSA
Electrophoretic mobility shift assays (EMSAs) were carried out in 20-μL reaction mixtures containing 0.5–20 nM of affinity-purified protein, 1 nM 5′-end-labeled RNA, 10% glycerol, and 1 mg/L total yeast RNA in buffer as above. Reactions were incubated for 10 min at room temperature and separated on a native 6% polyacrylamide gel in 0.5× TBE. Radioactivity was scanned with a PhosphorImager, and results were analyzed with ImageQuant software (Molecular Dynamics).
PAR-CLIP
RNAs bound to PAPD5 in vivo were analyzed by the PAR-CLIP technique essentially as described in Hafner et al. (2010). Flp-In-293 cells expressing His8-Flag-PAPD5 were grown in medium containing 100 μM 4-thiouridine, RNA–protein complexes were cross-linked by exposure of the cells to UV light (366 nm, 150 mJ/cm2), and lysed in buffer containing 50 mM HEPES (pH 7.4), 150 mM KCl, 2 mM EDTA, 1 mM NaF, 0.5% NP-40, 0.5 mM DTT, 1 mM PMSF, 1 mg/L Pepstatin, and 1 mg/L Leupeptin. After removal of cell debris by centrifugation, the cleared lysate was incubated with RNase T1 to allow bound RNAs to be partially digested to an average size of 40 to 60 nt.
RNA–protein complexes were isolated with M2 anti-Flag antibody (Sigma-Aldrich) coupled to protein G Dynabeads (Invitrogen). The RNA–protein complexes were gel-purified, the RNA was isolated, and linkers were ligated to the 5′ and 3′ ends of the RNA (Hafner et al. 2008). After reverse transcription and amplification, the resulting DNA library was sequenced by Solexa sequencing. Mapping of short reads to the genome (hg18 assembly version from the University of California, Santa Cruz obtained from hgdownload.cse.ucsc.edu) following adaptor removal was performed as described (Hafner et al. 2010).
Normalization and multiplicative error model in PAR-CLIP
To compare the expression of genomic regions across samples, the number of reads needs to be normalized across samples. With several types of high-throughput expression profiling approaches such as SAGE (serial analysis of gene expression), CAGE (cap analysis of gene expression), and chromatin immunoprecipitation (ChIP), it has been reported that the number of reads originating from a genomic position or region has a power-law distribution (Ueda et al. 2004; Zhang et al. 2008; Balwierz et al. 2009). We found that the same holds for our PAR-CLIP data, which we then analyzed as follows. We split the genome into 100-nt-long windows and calculated the number of reads originating in each of these regions in the IGF2BP1 CLIP samples obtained by Hafner et al. (2010) and in our two PAPD5 replicates. The reverse cumulative distribution of the read count in a window (showing the number of windows with a read count at least as high as a given value, which is indicated on the x-axis) was power-law-distributed, as expected (Supplemental Fig. S1A). We standardized these distributions by transforming them to a reference power law distribution with slope −1.25 and a total copy of 1 million reads, as described (Balwierz et al. 2009). The resulting normalized reverse cumulative distributions are shown in Supplemental Figure S1B. Based on the normalized read counts, we estimated the variance in read count per window between the PAPD5 replicates to be 0.41. In contrast, the variances between first and second PAPD5 replicates and IGF2BP1 were estimated to be 0.91 and 1.03, respectively.
Enrichment and depletion of reads in genomic regions in PAR-CLIP
Using the estimated variance above, we calculated the Z-scores for the difference in expression between PAPD5 reads and IGF2BP1 reads in 100-nt-long overlapping genomic windows as described in Balwierz et al. (2009). Inspecting the pairwise Z-score distributions, we chose a cut-off value of 5 and −5 for regions that are enriched and depleted, respectively, in the PAPD5 relative to the IGF2BP1 samples.
Annotation of the sequences
To determine whether the enriched or depleted regions overlapped known elements of the genome, we intersected the genome coordinates of the regions with those of annotated functional elements (UCSC RepeatMasker, mRNA and noncoding RNA tracks for hg18 assembly version) (Smit et al. 1996-2010; Pruitt et al. 2005; Lestrade and Weber 2006; Griffiths-Jones et al. 2008). If the overlap was at least 20 nt, we transferred the annotation of the genomic element to the region that was enriched/depleted in CLIP (Supplemental Table S1).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We are grateful to Markus Landthaler and Thomas Tuschl for their generous help with the CLIP experiments and to Georges Martin and Elmar Wahle for discussions and critical reading of the manuscript. We also thank Bruno Senger and Gérard Keith for clones and native tRNA samples. The work was supported by the University of Basel, the Louis-Jeantet Foundation for Medicine, the Swiss National Science Foundation (grant Nr. 3100A0-102132/2 to W.K.), and the Deutsche Forschungsgemeinschaft (grant Nr. RA 1892/1-1 to C.R.). B.B. is supported by a PhD fellowship from the Werner-Siemens-Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2787011.
REFERENCES
- Balwierz P, Carninci P, Daub C, Kawai J, Hayashizaki Y, Van Belle W, Beisel C, van Nimwegen E 2009. Methods for analyzing deep sequencing expression data: constructing the human and mouse promoterome with deepCAGE data. Genome Biol 10: R79 doi: 10.1186/gb-2009-10-7-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betat H, Rammelt C, Martin G, Mörl M 2004. Exchange of regions between bacterial poly(A) polymerase and CCA adding enzyme generates altered specificities. Mol Cell 15: 389–398 [DOI] [PubMed] [Google Scholar]
- Bühler M, Spies N, Bartel DP, Moazed D 2008. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol 15: 1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan KP, Butler JS 2010. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem 285: 3540–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro T, Carvalho C, Braga J, Rino J, Milligan L, Tollervey D, Carmo-Fonseca M 2007. Depletion of the yeast nuclear exosome subunit Rrp6 results in accumulation of polyadenylated RNAs in a discrete domain within the nucleolus. Mol Cell Biol 27: 4157–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Ares M Jr 2006. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci 103: 3262–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Houseley J, Tollervey D 2006. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J 25: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu DE, Henras AK, Chanfreau GF 2006. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA 12: 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T 2008. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods 44: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp A-C, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill S, Wolin SL, Reinisch KM 2010. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc Natl Acad Sci 107: 15045–15050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Kotovic K, El Hage A, Tollervey D 2007. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J 26: 4996–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT 2006. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ 2006. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 34: D158–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP 1998. Polyadenylation of stable RNA precursors in vivo. Proc Natl Acad Sci 95: 12158–12162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Reimers S, Pandit S, Deutscher MP 2002. RNA quality control: degradation of defective transfer RNA. EMBO J 21: 1132–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W 2007. RNA-specific ribonucleotidyl transferases. RNA 13: 1834–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Möglich A, Keller W, Doublié S 2004. Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase. J Mol Biol 341: 911–925 [DOI] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura R, Takeuchi R, Takata K, Shimanouchi K, Abe Y, Kanai Y, Ruike T, Ihara A, Sakaguchi K 2008. TRF4 is involved in polyadenylation of snRNAs in Drosophila melanogaster. Mol Cell Biol 28: 6620–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR 2005. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 33: D501–D504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Paolo S, Vanacova S, Schenk L, Scherrer T, Blank D, Keller W, Gerber AP 2009. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet 5: e1000555 doi: 10.1371/journal.pgen.1000555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M-J, Norbury CJ 2010. Polyadenylation and beyond: emerging roles for noncanonical poly(A) polymerases. WIREs RNA 1: 142–151 [DOI] [PubMed] [Google Scholar]
- Schmidt M-J, West S, Norbury CJ 2010. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA 17: 39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG 2010. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep 11: 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Fremder E, Staals RHG, Pruijn GJM, Schuster G 2010. Addition of poly(A) and poly(A)-rich tails during RNA degradation in the cytoplasm of human cells. Proc Natl Acad Sci 107: 7407–7412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P 1996. –2010. RepeatMasker Open-3.0. http://www.repeatmasker.org
- Ueda HR, Hayashi S, Matsuyama S, Yomo T, Hashimoto S, Kay SA, Hogenesch JB, Iino M 2004. Universality and flexibility in gene expression from bacteria to human. Proc Natl Acad Sci 101: 3765–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189 doi: 10.1371/journal.pbio.0030189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win TZ, Draper S, Read RL, Pearce J, Norbury CJ, Wang S-W 2006. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol Cell Biol 26: 1710–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlotzka W, Kudla G, Granneman S, Tollervey D 2011. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J 30: 1790–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle J-C, Dufour M-E, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Zhang ZD, Rozowsky J, Snyder M, Chang J, Gerstein M 2008. Modeling ChIP sequencing in silico with applications. PLoS Comput Biol 4: e1000158 doi: 10.1371/journal.pcbi.1000158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Lugovskoy AA, Wagner G 2001. A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. J Biol NMR 20: 11–14 [DOI] [PubMed] [Google Scholar]