Abstract
Depolarization of nerve terminals stimulates rapid dephosphorylation of two isoforms of dynamin I (dynI), mediated by the calcium-dependent phosphatase calcineurin (CaN). Dephosphorylation at the major phosphorylation sites Ser-774/778 promotes a dynI-syndapin I interaction for a specific mode of synaptic vesicle endocytosis called activity-dependent bulk endocytosis (ADBE). DynI has two main splice variants at its extreme C terminus, long or short (dynIxa and dynIxb) varying only by 20 (xa) or 7 (xb) residues. Recombinant GST fusion proteins of dynIxa and dynIxb proline-rich domains (PRDs) were used to pull down interacting proteins from rat brain nerve terminals. Both bound equally to syndapin, but dynIxb PRD exclusively bound to the catalytic subunit of CaNA, which recruited CaNB. Binding of CaN was increased in the presence of calcium and was accompanied by further recruitment of calmodulin. Point mutations showed that the entire C terminus of dynIxb is a CaN docking site related to a conserved CaN docking motif (PXIXI(T/S)). This sequence is unique to dynIxb among all other dynamin variants or genes. Peptide mimetics of the dynIxb tail blocked CaN binding in vitro and selectively inhibited depolarization-evoked dynI dephosphorylation in nerve terminals but not of other dephosphins. Therefore, docking to dynIxb is required for the regulation of both dynI splice variants, yet it does not regulate the phosphorylation cycle of other dephosphins. The peptide blocked ADBE, but not clathrin-mediated endocytosis of synaptic vesicles. Our results indicate that Ca2+ influx regulates assembly of a fully active CaN-calmodulin complex selectively on the tail of dynIxb and that the complex is recruited to sites of ADBE in nerve terminals.
Keywords: Calcineurin, Endocytosis, Phosphatase, Protein-Protein Interactions, Synaptosomes, Dynamin
Introduction
The dynamins are a superfamily of up to seven cellular GTPase enzymes, which each play a role in membrane fission or fusion (1). There are three “classical dynamins,” dynI,2 -II, and -III, which mediate the internal fission of vesicles from cell membranes. The neuronal isoform of dynamin, dynI, is essential for all forms of synaptic vesicle (SV) endocytosis in central nerve terminals (2, 3). At least three different SV endocytosis modes are proposed to exist in nerve terminals. The kiss-and run mode has no defining biochemical or morphological markers, and thus its existence is still a matter of controversy (4). Clathrin-mediated endocytosis (CME) retrieves single SVs from the plasma membrane and is the dominant endocytosis mode during mild stimulation (5). In contrast, activity-dependent bulk endocytosis (ADBE) invaginates large areas of plasma membrane to form bulk endosomes (BEs) from which SVs can later bud and rejoin the recycling pool (6, 7). ADBE is only triggered during high intensity stimulation and is the dominant SV endocytosis mode under these conditions (8).
CME and ADBE appear to utilize similar or the same endocytic proteins (9, 10), with some exceptions (3, 11). In agreement, dynI GTPase activity is essential for both modes (3). However, ADBE is additionally reliant on the dephosphorylation and rephosphorylation of dynI (3, 12, 13). DynI is the founding member of a group of endocytic proteins called the dephosphins that are dephosphorylated by the Ca2+- and calmodulin-dependent protein phosphatase calcineurin (CaN, or protein phosphatase 2B) upon nerve terminal depolarization (14–17). CaN is the only protein phosphatase directly regulated by Ca2+ and calmodulin (18–20). It is a heterodimer of a 60-kDa catalytic subunit, CaNA, and a 19-kDa regulatory subunit, CaNB. DynI interacts directly with CaN in a strictly Ca2+-dependent manner via its PRD (21, 22); however, no functional role has been identified for this interaction. In contrast, the activity-dependent dephosphorylation of dynI by CaN is essential for the triggering of ADBE. DynI dephosphorylation permits its association with syndapin I, a protein that is essential for ADBE but not CME (3, 23). Thus, CaN activity is required to trigger ADBE in central neurons to increase endocytic capacity under periods of unusually intense activity.
DynI is a modular protein, containing an N-terminal GTPase, middle, GTPase effector, pleckstrin homology domain, and proline-rich domain (PRD). The dynI PRD is located at the C terminus, which is the main site of phosphorylation and an interaction platform for a variety of Src homology 3 (SH3) domains from other proteins. Rat dynI has three sites for alternative splicing that produce up to eight mRNAs (UniProtKB accession number P21575) (24); however, there is limited understanding of the functional roles of these splice variants in neurons (25–27). The dynI third splice site produces three alternative C-terminal tails beginning at residue 845, dynIxa (long), dynIxb (short), or dynIxd (intermediate). The two major spliced tails (xa and xb) have an approximate ratio of 5:4 (xa/ (xb + xd)) in nerve terminals, with dynIxd expression being ∼10% of the others (27). DynIxa has a unique 20-residue tail after Pro-844, 844PSRSGQASPSRPESPRPPFDL864, which includes several potential additional binding sites for SH3 domain-containing proteins. In contrast, the unique 7-residue tail of dynIxb, 844PRITISDP851, does not. Both dynIxa and dynIxb are phosphorylated and dephosphorylated upon electrical or chemical depolarization to approximately equal extents using metabolic 32Pi labeling, suggesting that they are coordinately regulated (28). However, quantitative mass spectrometry (which also detects unlabeled phosphorylation sites) revealed major differences in the distribution of phosphate between these sites in the dynIxa and dynIxb (27). Thus, the presence of alternative binding platforms and differential phosphorylation of key sites supports the hypothesis that these dynI splice variants may have unique functional roles in nerve terminals.
To date, the only reported differences between any of the endogenously expressed dynI isoforms is a differential phosphoregulation of dynIxa in synaptosomes (27, 29). DynI is phosphorylated at seven sites in nerve terminals, with the majority (69%) of metabolic labeling with 32Pi occurring on Ser-774 and Ser-778 in a region of the PRD called the phospho-box (13, 29). However, two additional phosphosites in nerve terminals are unique for dynIxa, Ser-851 and Ser-857 (29). Among these two sites, Ser-857 is phosphorylated by Dyrk 1A/minibrain kinase in vitro, and this appears to affect amphiphysin I binding to this isoform (30, 31).
In an attempt to uncover different roles for different dynI splice variants in nerve terminals, we searched for protein partners whose interaction may be specific for either dynIxa or dynIxb. We report here that CaN uniquely associates with the short (xb) tail and not with the long (xa) tail of dynI. We characterized the dynIxb-CaN interaction, mapped the binding site, and found that the interaction was associated with a specific endocytic mode, ADBE. Our results support the model whereby dynI dephosphorylation by CaN is the primary trigger for ADBE, leading to the formation of a dynI-syndapin I complex as the next step in the process.
EXPERIMENTAL PROCEDURES
Materials
Leupeptin was from Merck. Tissue culture plastics were from Falcon (Franklin Lakes, NJ). Tetramethylrhodamine-dextran, penicillin/streptomycin, phosphate-buffered saline, minimal essential medium, and fetal calf serum were from Invitrogen. All other materials were from Sigma unless otherwise stated. Antibodies to PP2B-A (CaN-A), PP2B-B (CaN-B), amphiphysin I, and syndapin I were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); antibodies to dynI (hudy1) or calmodulin were from Millipore; and secondary antibodies were all from Dako. Three penetratin-based peptides, RRMKWKK-GVPRITISDP (short peptide), RRMKWKK-GVARATASDP (mutant short peptide), and RRMKWKK-GVPSRSGQASPSRPESPRPPFDL (long peptide), were synthesized by Auspep (Parkville, Australia). The pBJ5mCaNB (mouse, plasmid 17872), pBJ5-CaNA (mouse, plasmid 11785), and pGEX-CaNA(2–347) (human, plasmid 13251) plasmids were from Addgene Inc.
Plasmid Construction and Protein Expression
GST-tagged dynIxa PRD plasmid (long tail, rat, amino acids 746–864) was constructed into pGEX-4T-1 by subcloning the coding region from GFP-tagged dynIaa (23). The sequence encoding the dynIxb PRD (short tail, rat sequence, amino acids 746–851) was amplified from dynIab in pCR3.1 expression vector (24) with the oligonucleotides 5′-CGGCGAATTCAACACGACCACCGTCAGCACGCCC-3′ and 5′-CTGCAGAATTGCGGCCGCTTAGGGGTCACTGATAGTG-3′ and then subcloned into a pGEX-4T-1 vector (GE Healthcare). Point mutants in dynIxb PRD were generated using the QuikChange site-directed mutagenesis kit (Stratagene). The plasmid expressing GST-amphiphysin I SH3 domain (human, amino acids 545–695) was as previously reported (23). GST-α-adaptin appendage domain (amino acids 701–938 in pGEX2T expression vector) was previously described (32). GST constructs were transformed into Escherichia coli JM109 by heat shock, and expressed recombinant proteins bound to glutathione (GSH)-Sepharose beads were prepared according to the manufacturer's instructions. Cloning and expression of His6-syndapin 1-FL was as described previously (33).
Pull-down Experiments and Mass Spectrometry
Crude (P2) synaptosomes were prepared from rat brains (25) and lysed in ice-cold buffer containing 20 mm Tris (pH 7.4), 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, 1 mm EGTA, 20 μg/ml leupeptin, 1 mm phenylmethylsulfonyl fluoride (PMSF), and EDTA-free Complete-protease inhibitor tablets (Roche Applied Science). The homogenate was centrifuged at 75,600 × g for 30 min at 4 °C. The extract was precleared with GSH-Sepharose beads for 1 h and then incubated with the GSH-Sepharose beads coated with GST recombinant proteins at 4 °C for 1 h. The beads were isolated and washed extensively with ice-cold lysis buffer and eluted in 2× concentrated SDS sample buffer. Bound proteins were separated by 7.5–15% gradient SDS-PAGE and stained with colloidal Coomassie Blue. Protein bands were then excised from gels and identified by LC-MS/MS (QSTAR XL mass spectrometer, AB SCIEX) and searched against protein databases as described (27). See supplemental Table 1 for more details.
Synaptosomes and 32Pi Labeling
P2 synaptosomes were prepared from adult rat brains and labeled with 0.75 mCi/ml 32Pi for 1 h at 37 °C in a low calcium Krebs-like buffer (118.5 mm NaCl, 4.7 mm KCl, 1.18 mm MgCl2, 0.1 mm K2HPO4, 20 mm Hepes, 10 mm glucose, with 0.1 mm calcium, pH 7.4) and washed (25). In some experiments, the synaptosomes were depolarized by the addition of KCl to 41 mm for 10 s, were immediately lysed in ice cold lysis buffer (25 mm Tris, pH 7.4, containing 1% Triton X-100, 150 mm NaCl, 1 mm EGTA, 2 mm EDTA, 50 mm NaF, 20 μg/ml leupeptin, 1 mm PMSF, and EDTA-free protease inhibitor mixture), and were centrifuged at 20,442 × g for 20 min at 4 °C. Pull-down experiments were performed sequentially. First, the GST-amphiphysin I-SH3 domain on GSH-Sepharose beads was used to pull down dynI and synaptojanin from the lysate. Then GST-α-adaptin ear domain on GSH-Sepharose beads was used in a subsequent pull-down from the same lysate to isolate AP180 and amphiphysin. The washed beads were heated in SDS-PAGE sample buffer, and proteins were resolved on SDS gels and subjected to autoradiography. Densitometry was performed on storage phosphor screens scanned with a Typhoon TRIO imager, and phosphorylation was quantified with ImageQuant TL software (GE Healthcare).
Cell Culture and Electron Microscopy
Primary cultures of cerebellar granule neurons were prepared from the cerebella of 7-day-old Sprague-Dawley rat pups as described previously (34). Cultures were processed for electron microscopy as described previously (3) with minor modifications. Cultures were removed into incubation medium (170 mm NaCl, 3.5 mm KCl, 0.4 mm KH2PO4, 20 mm TES, 5 mm NaHCO3, 5 mm glucose, 1.2 mm Na2SO4, 1.2 mm MgCl2, 1.3 mm CaCl2, pH 7.4) and after 10 min were incubated with short (xb) peptide (30 or 60 μm) for a further 15 min. Cultures were then stimulated with 800 action potentials (delivered at 80 Hz, 1-ms pulse width) in incubation medium supplemented with horseradish peroxidase (HRP; 10 mg/ml) and peptide where indicated. This stimulation protocol activates both CME and ADBE (8). Neurons were then immediately fixed in a 2% solution of glutaraldehyde in PBS for 30 min at 37 °C. After washing in 100 mm Tris, pH 7.4, cultures were exposed to 0.1% diaminobenzidine and 0.2% H2O2 in 100 mm Tris, pH 7.4. Upon development of color, cultures were washed with 100 mm Tris and then postfixed with 1% osmium tetroxide for 30 min. After washing, cultures were poststained with 2% uranyl acetate for 15 min and then dehydrated using an ethanol series and polypropylene oxide and then embedded using Durcupan. Samples were sectioned, mounted on grids, and viewed using an FEI (Hillsboro, OR) Tecnai 12 transmission electron microscope. Intracellular structures that were <100 nm in diameter were arbitrarily designated to be SVs, whereas larger structures were designated to be endosomes. All experiments were performed on cultures between 8 and 10 days in vitro. For transfection of CaNA or CaNB into mammalian cells, COS-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, and the transfection reagent was FuGENE 6 (Roche Applied Science).
Fluorescence Imaging of Dextran Uptake
The uptake of tetramethylrhodamine-dextran (40 kDa, 50 μm) was monitored as described previously (3) in cells stimulated with a train of 800 action potentials (80 Hz, 10 s). Where indicated, cultures were incubated with 60 μm xb peptide for 15 min before and during stimulation. Dextran loading was determined by the number of fluorescent puncta in a defined field of view (130 × 130 μm) using a ×20 air objective at 550-nm excitation and >575-nm emission. Thresholding analysis was performed to discount regions too large to represent individual nerve terminals (diameter greater than 2 μm). The average number of dextran puncta/field for each experiment (usually 10 fields of view/experiment) were averaged for the same conditions. Experiments were performed across multiple culture preparations; however, to ensure that the density of nerve terminals was consistent between fields and experimental conditions, for each set of cultures, every experimental condition was performed (between 8–10 days in vitro).
RESULTS
Calcineurin Selectively Binds the Short DynIxb Splice Variant
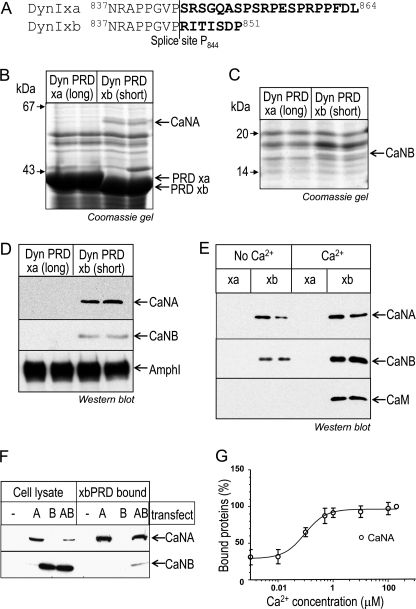
DynIxa and dynIxb differ in the amino acid sequences of their alternatively spliced tails (Fig. 1A) (24, 25). To search for potential isoform-selective binding partners for the two tails, both PRDs were expressed as GST fusion proteins bound to GSH-Sepharose beads and incubated with rat brain synaptosomal lysate for pull-down experiments. Proteins at about 58 and 19 kDa selectively associated with one form, dynIxb PRD (Fig. 1, B and C). The 58-kDa protein was unequivocally identified by LC-MS/MS sequencing as the catalytic subunit of the serine/threonine-protein phosphatase 2B (CaN), also known as CaNA, and the 19-kDa protein was identified as the regulatory subunit of calcineurin, CaNB. The 58-kDa protein had 19 matching peptides with 38% sequence coverage of CaNA α isoform 1 and 10 matching peptides with 36% sequence coverage of CaNA α isoform 2 (supplemental Table 1). It also had eight matching peptides with 20% sequence coverage of CaNA β isoform, three peptides being unique to the β not α isoform (supplemental Table 1). Therefore, both CaNA α and β forms were unequivocally detected. The 19-kDa protein revealed seven matching peptides with 53% sequence coverage of CaNB, matching both known isoforms 1 and 2 (supplemental Table 2). The identity of these proteins as CaNA and CaNB was independently confirmed by Western blotting with specific antibodies (Fig. 1D). The blot was stripped and reprobed with anti-amphiphysin I antibodies, and amphiphysin I was found to bind to xa and xb to about the same extent (Fig. 1D). The results show that both CaNA and CaNB specifically interact with the short dynIxb splice variant but do not yet reveal which is the primary contact.
FIGURE 1.
CaN specifically associates with short tailed dynIxb in a Ca2+-regulated manner. A, amino acid sequence alignment of the C terminus of dynIxa (long) and dynIxb (short) splice variants. The amino acids in boldface type are the alternatively spliced sequences. B and C, CaN specifically binds to dynIxb PRD. Synaptosomal lysates were incubated with GST-dynI PRD (either xa or xb) coupled to GSH-Sepharose beads. Bound proteins were separated by SDS-PAGE and stained with Coomassie Blue. The proteins bound to dynIxb PRD at about 58 kDa (B) and about 19 kDa (C) were identified by LC-MSMS as the A and B subunits of protein phosphatase 2B (calcineurin, CaNA, and CaNB). D, bound proteins from pull-down experiments with dynI PRD (xa or xb) were subjected to Western blot analysis with antibodies against CaNA, CaNB, and amphiphysin I. E, binding of CaNA and CaNB subunits to GST-dynIxb PRD is increased in the presence of Ca2+, and CaM binds to dynIxb PRD only in the presence of Ca2+. Proteins from synaptosomal lysate were pulled down by GST-dynI PRD (xa or xb) in the absence or presence of 200 μm Ca2+. CaN or CaM binding was revealed by Western blot. F, CaNB binds dynIxb PRD via CaNA. COS7 cells were transiently transfected with either CaNA or CaNB or cotransfected with both. Cell lysates were incubated with GST-dynIxb PRD coupled to GSH-Sepharose beads. Bound proteins were subjected to Western blot analysis with antibodies against CaNA and CaNB. Left, protein expression in cell lysates; right, proteins bound to dynIxb PRD. G, synaptosomal lysates were used for pull-down experiments with GST-dynIxb PRD in the presence of EGTA (first data point) or various concentrations of Ca2+. Bound proteins were subjected to Western blot with antibodies against CaNA, and the amount of proteins bound was quantified by densitometric analysis of the blots. Data are normalized to 100% against maximum binding in 200 μm Ca2+. All gels and blots are representative of at least three (B–E) or two (G) independent experiments, with some data for gels and blots being shown in duplicate (B–E). Error bars, S.E.
We next explored the potential Ca2+ dependence of this interaction. In the presence of 200 μm Ca2+, both CaNA and CaNB increased binding to the dynIxb PRD (Fig. 1E and supplemental Fig. S1, A and B). Interestingly, an additional band at 21 kDa was also detected in the Coomassie-stained gel, binding specifically to dynIxb PRD (supplemental Fig. S1B). LC-MS/MS clearly identified six tryptic peptides with 50% sequence coverage of calmodulin (CaM) (supplemental Table 3). This was confirmed by immunoblot analysis with specific anti-CaM antibodies, which also revealed that CaM bound to the dynIxb-CaN complex only in the presence of Ca2+ (Fig. 1E, bottom). Because CaNA and CaNB are known to be a complex, we asked which is the primary contact with dynIxb. We transfected CaNA or CaNB, or we cotransfected both, into COS7 cells. The cell lysates were incubated with GST-dynIxb PRD on GSH-Sepharose beads. Western blots showed that CaNA directly associated with dynIxb PRD (Fig. 1F, top), whereas CaNB did not bind alone, but bound dynIxb through CaNA (Fig. 1F, bottom). The amount of CaNA bound to the dynIxb PRD in the presence of various concentrations of Ca2+ was further explored by Western blot, and the level of bound CaNA was quantified by densitometry. Maximal binding of CaNA to dynIxb PRD occurred at about 1 μm free Ca2+, with an estimated EC50 for Ca2+ of 0.1 μm (Fig. 1G). This result is consistent with the previously reported binding of dynI to CaN for calcium (EC50 in the range of 0.1–0.4 μm (21)). This affinity was similar to the in vitro affinity of CaN for calcium (Kd value is less than 1 μm (35)). The results show that it is the catalytic subunit that interacts with dynIxb and that it has a high degree of constitutive binding.
Mapping the CaNA Binding Region in the DynIxb Short Tail
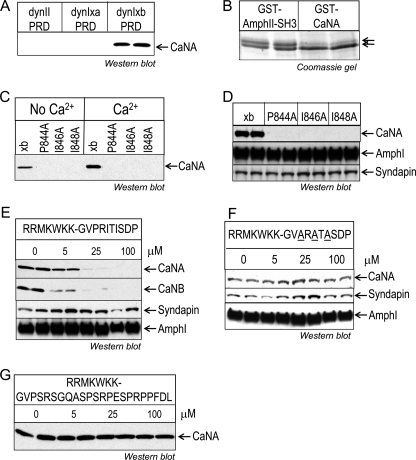
To characterize dynIxb-CaNA binding, we used dynII PRD to pull down proteins from synaptosomal lysates. Note that there is only one known C-terminal tail for dynII, and its amino acid sequence is more similar to that of dynIxa. No CaNA was detected by immunoblot on dynII PRD (Fig. 2A). We also used GST-CaNA(2–347)-coated GSH-Sepharose beads to pull down its binding partners from synaptosomal lysate. The main Coomassie protein band found to bind GST-CaNA(2–347) comigrated on gels with the short form of dynI (dynIxb) (Fig. 2B, note the lower band of the doublet), whereas in contrast, GST-AmpI-SH3 is known to associate with dynI splice variants, including dynIxa, dynIxb, and dynIxd (27). Mass spectrometry analysis of the main band confirmed that CaNA binds dynIxb with considerable specificity (data not shown). Due to the sensitivity provided by mass spectrometry analysis, both dynIxa (in the upper band) and dynIxd (in both upper and lower bands) were detected, but not dynIxc. Considerably less Coomassie protein stain was detected for dynIxa (Fig. 2B, lanes 1 and 2 versus lanes 3 and 4, upper band), despite the fact that dynamin family members are well known to dimerize with each other (36). It was not possible to quantify changes in dynIxd binding, but like dynIxa, it has no specific sequence motif to indicate that it might interact with CaNA directly rather than indirectly via dynIxb. These results suggest that CaNA specifically interacts with the 7-amino acid tail that is unique to dynIxb.
FIGURE 2.
Mapping the CaN binding site within the dynIxb PRD. A, CaN specifically binds to dynIxb PRD but not dynIIxa. Synaptosomal lysates were incubated with GST-dynI PRD (either xa or xb) or GST-dynII PRD coupled to GSH-Sepharose beads, and CaNA binding was revealed by Western blot. B, dynIxb specifically binds to GST-CaNA. Synaptosomal lysates were incubated with either GST-CaNA(2–347) or GST-AmpII-SH3 coupled to GSH-Sepharose beads. Bound proteins were stained with Coomassie Blue after separation by an acrylamide gel with low bis-acrylamide and elevated pH, which is known to resolve the dynI splice variants (27). C, CaN binds to the alternatively spliced C-terminal tail of dynIxb(844–851). Single point mutations were made in the PRITIS motif of dynIxb PRD. The bacterially expressed mutant GST-dynIxb PRDs were used for pull-down experiments from synaptosomal lysates. Bound proteins were subjected to Western blotting with anti-CaNA antibodies. D, mutations in the PRITIS motif do not affect amphiphysin I or syndapin binding to dynIxb PRD. E, peptide competition revealed binding of CaN to the C terminus of dynIxb PRD. The penetratin-tagged peptide mimetic of the short variant, dynIxb(842–851), containing the entire spliced insert was synthesized and used for competition studies. Synaptosomal lysates were incubated with GST-dynIxb PRD immobilized on GSH-Sepharose beads in the absence or presence of 5–100 μm peptide. Binding of proteins was detected by Western blot analysis with antibodies against CaNA, CaNB, syndapin, and amphiphysin I. F, mutant peptide competition determined the specificity of the binding of CaN to the C terminus of dynIxb PRD. The penetratin-tagged mutant peptide containing ARATA was synthesized and used for competition studies. Synaptosomal lysates were incubated with GST-dynIxb PRD immobilized on GSH-Sepharose beads in the absence or presence of 5–100 μm mutant peptide. Binding of proteins was detected by Western blot analysis with antibodies against CaNA, syndapin, and amphiphysin I. G, CaN binding to GST-dynIxb PRD is unaffected by the alternatively spliced C-terminal tail of dynIxa(842–864). The penetratin-tagged peptide dynIxa(842–864) was synthesized and used for a pull-down experiment as in E. CaNA binding to the dynIxb PRD was analyzed by Western blot with anti-CaNA antibodies. All results are representative of two independent experiments. AmphI and AmphII, amphiphysin I and II, respectively.
Our next aim was to identify the binding site for CaN in the short dynIxb variant. Inspection of the dynIxb tail sequence (PRITISDP) revealed a strong similarity to the conserved CaN docking motif PXIXI(T/S) (37, 38). To evaluate whether the “PRITIS” region of dynIxb might be responsible for the CaN interaction, we performed single point mutations on each of the three amino acids conserved in the motif. GST-dynIxb PRD mutated at any of the three sites (P844A, I846A, or I848A) failed to bind any CaN in pull-downs from nerve terminal lysates (Fig. 2, C and D). Binding to amphiphysin I and syndapin were not affected by these mutations (Fig. 2D). To confirm that CaN specifically binds to the short dynIxb tail, a synthetic PRITIS peptide containing dynIxb(842–851) (GVPRITISDP) was fused with a short penetratin heptapeptide RRMKWKK (“short peptide”) and incubated with nerve terminal lysates prior to pull-downs with GST-dynIxb PRD. The peptide reduced binding of CaNA, CaNB, and CaM to the dynIxb PRD in a concentration-dependent manner, both in the absence (Fig. 2E) and presence of Ca2+ (supplemental Fig. S2A), whereas the peptide did not affect syndapin and amphiphysin I binding to dynIxb PRD (Fig. 2E). In contrast, a mutant PRITIS peptide, ARATASDP, fused with RRMKWKK, did not disrupt any of the CaNA-dynIxb, syndapin-dynI, and amphiphysin I-dynI interactions (Fig. 2F). Another peptide containing the long dynIxa tail, dynIxa(842–864) (RRMKWKK-GVPSRSGQASPSRPESPRPPFDL) (long peptide), also did not disrupt the CaN-dynI interaction (Fig. 2G). Thus, we conclude that the PRITIS motif in the dynIxb C-terminal splice site is the docking site for CaN, via the parallel use of both mutagenesis and competitive peptides.
To determine whether CaN binding might be regulated by dynI phosphorylation, we performed pull-down experiments with GST-dynIxb PRD containing single or double point mutations that introduce pseudophosphorylation at Ser-774 and/or Ser-778, which are phosphosites known to regulate the interaction of dynI with syndapin I (23). These were without effect on CaNA binding (supplemental Fig. S2B). The results suggest that dynamin phosphorylation in the phospho-box region does not regulate CaN binding to dynIxb. To determine whether other protein-protein interactions might regulate CaN binding, we prebound the GST-dynIxb PRD with His-tagged full-length syndapin (which is its main binding partner for ADBE) and then performed the pull-down in synaptosomal lysates with this complex. Prebinding of syndapin abolished the subsequent CaNA-dynI interaction (supplemental Fig. S2C).
CaN Dephosphorylation of both DynIxa and DynIxb Splice Variants Requires CaN Binding to the Short Form
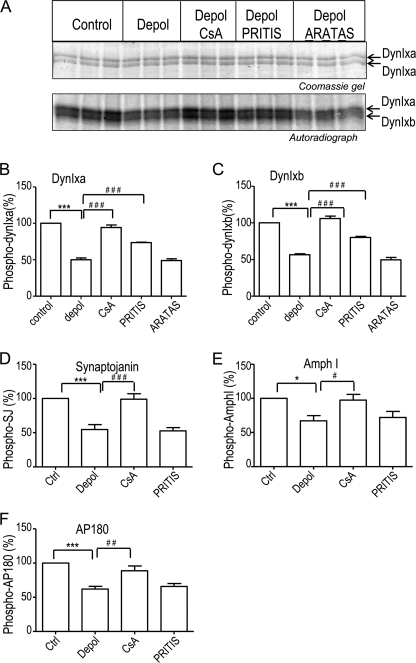
CaN dephosphorylates both dynIxa and dynIxb to an approximately equal extent during nerve terminal depolarization (28). To determine whether dynI dephosphorylation by CaN requires CaN binding to dynIxb, we utilized 32Pi-labeled synaptosomes that were depolarized with KCl either in the absence or presence of the dynIxb peptide or the CaN inhibitor cyclosporin A. The dynIxb peptide and its inactive mutant control peptide were tagged with a penetratin heptapeptide to facilitate entry into synaptosomes (3, 39). Labeled synaptosomal lysates were subjected to a pull-down with GST-amphiphysin I-SH3 domains on GSH beads, which extracted dynI and synaptojanin. The lysates were then subjected to a second sequential pull-down with GST-α-adaptin ear domain to further isolate two other dephosphins, AP180 and amphiphysin I. Autoradiography revealed that cyclosporin A prevented CaN-mediated dephosphorylation of all four dephosphins in response to the KCl depolarization (Fig. 3A and supplemental Fig. S3, A and B; data shown in triplicate), but it did not affect the phosphorylation of the four proteins under the control conditions (supplemental Fig. S3, C and D). At a single concentration (100 μm), the dynIxb peptide greatly reduced dynI dephosphorylation. Surprisingly, it affected the upper and lower bands of the dynI doublet to the same extent (Fig. 3A), despite the fact that CaN docking could be demonstrated only to the shorter xb form. When data from multiple independent experiments were pooled and quantified, it was revealed that both dynIxa and -xb dephosphorylation were significantly blocked by the xb peptide (Fig. 3, B and C). The effect was specific because the peptide did not prevent the dephosphorylation of the three other dephosphins examined (Fig. 3, D–F, and supplemental Fig. S3, A and B). In contrast, the inactive mutant peptide (100 μm) did not have any effects on any of the four proteins under any conditions (Fig. 3, A–C, and supplemental Fig. S3, A–D). Therefore, CaN binding to a single dynamin splice variant regulates KCl-evoked dephosphorylation of two splice variants but not dephosphorylation of at least three other dephosphins.
FIGURE 3.
The CaN-dynIxb interaction regulates KCl-evoked dynI dephosphorylation but not that of three other dephosphins. A, synaptosomes were metabolically labeled with 32Pi, washed, and incubated with or without either 100 μm PRITIS or 100 μm ARATAS peptide or with 30 μm cyclosporin A for 15 min. Synaptosomes were then stimulated with 41 mm KCl for 10 s. After lysis, dynI and synaptojanin were isolated by pull-down with GST-amphiphysin I-SH3 bound to GSH-Sepharose beads, and dynIxa and -xb were separated by 12% acrylamide gel with 0.08% bisacrylamide and pH 9.2 (27). Proteins in the 100 kDa region containing the dynamins were visualized by Coomassie Blue staining to demonstrate even sample loading (top). Phosphorylated dynIxa and -xb were visualized by autoradiography of triplicate samples (bottom). Synaptojanin was visualized by a 7.5–15% gradient SDS-PAGE (supplemental Fig. S3). Following this pull-down, two additional dephosphins, AP180 and amphiphysin I, were extracted by a second sequential pull-down with GST-α-adaptin ear domain bound to GSH-Sepharose beads of the same lysates (supplemental Fig. S3). B–F, quantitative analysis of synaptosomal phosphorylation from several experiments, such as those shown in A and supplemental Fig. 3, A and B for dynIxa (B), dynIxb (C), synaptojanin (D), amphiphysin I (E), and AP180 (F). Quantified data are presented as a ratio of 32P levels (from phosphorimager data) normalized to the total protein from densitometry of the gel, and results were then normalized to levels in the unstimulated controls. Results are the mean ± S.E. for n = 8 (from three biologically independent experiments). One-way analysis of variance was applied (***, p < 0.001; **, p < 0.01; *, p < 0.05; all pairs were compared).
The CaN-DynIxb Interaction Regulates ADBE but Not CME
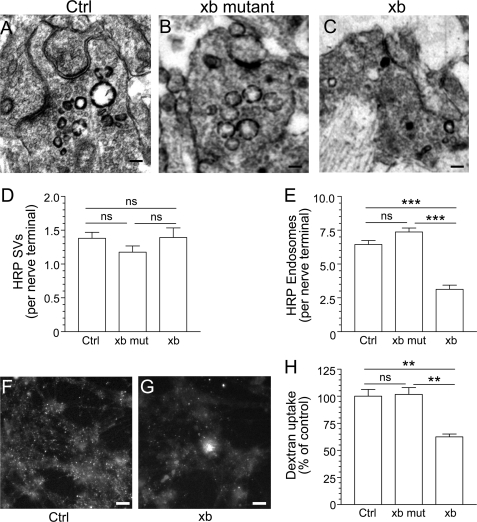
The activity-dependent dephosphorylation of dynI by CaN and the subsequent interaction with syndapin I is specifically required for the ADBE mode of SV endocytosis, whereas dynI GTPase activity is required for both ADBE and CME (2, 3, 23). To determine whether the CaN-dynIxb interaction is required for either SV endocytosis mode, the dynIxb peptide (dynIxb(842–851), GVPRITISDP) was introduced to primary cultures of cerebellar granule neurons, in which expression of dynIxa, -xb, and -xd was confirmed by pull-downs, Western blots, and mass spectrometry (supplemental Fig. S4, A and B). HRP was used as a fluid phase marker for parallel monitoring of both ADBE and CME because it has been extensively characterized in this neuronal system (3, 12, 40). ADBE is detected as the very rapid appearance of large BE structures (>100 nm in diameter) filled with electron-dense reaction product in nerve terminals, whereas CME is detected as electron-dense small SVs (<100 nm in diameter). Both ADBE and SVs were activated in the same nerve terminal after stimulation with a high intensity train of 800 action potentials (80 Hz), as revealed by a mix of small and large electron-dense vesicles (Fig. 4A). Both the PRITIS and inactive mutant peptides were without effect on the number of HRP-labeled small diameter SVs at 60 μm, but only the PRITIS peptide reduced the number of large diameter HRP-labeled endosomes (Fig. 4, B and C). Quantification of the number of HRP-loaded SVs or BEs per nerve terminal from multiple independent experiments shows that the PRITIS peptide inhibits ADBE without a significant effect on CME (Fig. 4, D and E).
FIGURE 4.
The CaN-dynIxb interaction is essential for ADBE. Cerebellar granule neuron cultures were incubated with 10 mg/ml HRP, and loading was stimulated by a train of 800 action potentials delivered at 80 Hz. The cultures were incubated with either wild-type (xb) or mutant containing ARATA (xb mut) penetratin-tagged peptide dynIxb(842–851) (60 μm) for 15 min before and during stimulation. A–C, HRP-labeled structures in typical fields of view either in the absence of peptide (A, Ctrl) or in the presence of mutant (B) or wild-type peptide (C). Scale bar, 150 nm in all images. D, mean number of HRP-labeled SVs generated per nerve terminal in the presence of the peptide. E, mean number of HRP-labeled endosomes generated per nerve terminal in the presence the peptide. Data were pooled from either four (Ctrl) or three (xb mut and xb) independent experiments (Ctrl, n = 173 nerve terminals; xb mut, n = 119; xb, n = 87; all mean ± S.E.; ***, p < 0.001 compared with control, one-way analysis of variance; ns, not significantly different). F and G, dextran loading in typical fields of view in cerebellar granule neurons either in the absence of xb peptide (F) or in its presence (G). Scale bars (F and G), 15 μm. H, granule neurons were incubated with 50 μm tetramethyrhodamine-dextran and loading was stimulated by a train of 800 action potentials (80 Hz) followed by immediate dextran washout. Where indicated, cultures were incubated with 60 μm either wild-type or mutant xb peptide 15 min before and during stimulation. The data shown are mean number of dextran puncta per field of view as a percentage of control (n = 4 ± S.E.; **, p < 0.01, one-way analysis of variance).
To independently confirm a block of ADBE, we used a complementary assay based on uptake of the large (40-kDa) fluorescent tetramethylrhodamine-dextran. This dextran selectively labels ADBE when applied during intense stimulation because it is too large to enter single SVs. In agreement with the HRP experiment, dextran uptake into nerve terminals does not occur during mild neuronal activity when only CME is active (8). The addition of the dynIxb peptide (60 μm) significantly reduced the uptake of dextran evoked by a train of 800 action potentials (80 Hz) when compared with control conditions, whereas the inactive mutant peptide did not have any effect (Fig. 4, F–H). These results suggest that the interaction between dynIxb and CaN is required for ADBE but not for CME.
DISCUSSION
We have shown that a single neuronal splice variant of the classical GTPase dynI, dynIxb, possesses a selective docking site at its extreme C terminus for the catalytic subunit of the Ca2+-dependent protein phosphatase CaN. This docking site is unique to dynIxb and conforms to the consensus PXIXI(T/S) sequence observed in other CaN interaction domains. Inhibition of the CaN-dynIxb interaction resulted in a selective block of dynI dephosphorylation over other CaN substrates and a selective inhibition of ADBE over other SV endocytosis modes.
Our finding that CaN regulates dynI function through binding to the unique 7-amino acid sequence of the dynIxb variant at PRITISDP, encompassing the entire spliced tail, provides a surprising functional specificity for a single splice variant. This amino acid sequence or motif is unique across all classical dynamins and reported splice variants. We confirmed that dynII did not bind CaN. Our study extends a previous report that dynI binds the CaN complex (21) but reveals that the binding is specific to a single splice variant and is not a broader dynamin property. We also find that the CaN-dynIxb interaction occurs in the absence of Ca2+, which differs from the previous study showing that the association was strictly Ca2+-dependent (21). We also observed a large increase in the binding of the complex of CaNA and CaNB to dynIxb in the presence of Ca2+. However, CaNB interacted with dynIxb indirectly through CaNA. CaNA associates with the CaNB subunit in the presence or absence of Ca2+ (35). Moreover, CaM also binds CaNA in a Ca2+-dependent manner (35). Analogously, the transcription factor “nuclear factor of activated T cells” (NFAT) interacts more strongly with CaM-activated CaNA than inactive CaNA (41), and CaN binding to TRESK is increased in the presence of Ca2+ and CaM (42). Our results suggest that an active CaN complex is likely to be assembled on the tail of dynIxb, allowing CaM and CaNB to synergistically activate CaN phosphatase activity in the presence of increased intracellular Ca2+ (18, 43, 44). This suggests that Ca2+ influx regulates assembly of fully active CaN locally onto the tail of dynIxb in an ordered manner.
Mammalian CaN does not bind to the vast majority of its substrates. However, it does bind to a selective small group of four, NFAT (45), the two-pore domain K+ channel TRESK (42), AKAP79 (46), and neutral sphingomyelinase 2 (47). They share in common a short consensus docking sequence motif, PXIXI(T/S). This was first found in the sequence SPRIEIT in the NFAT family of proteins (37, 38) and subsequently in the others (42, 46–48). We found that GST-CaNA(2–347) pulled down mainly the short form of dynI from rat synaptosomal lysate despite the fact that it has been reported that different dynI isoforms can assemble with each other to form either dimmers or tetramers (36). In that study, more than 30–40% of one dynamin isoform could be co-immunoprecipitated by the antibodies against another isoform (36). However, our results show a surprisingly specific interaction of dynIxb with CaNA, and this raises the possibility that in nerve terminals, the short form might not necessarily occur as a multimer with the long form. This raises the possibility that there might be different pools of dynamin in neurons, some of which may contain just one single isoform, and is consistent with markedly different stoichiometry of phosphorylation of dynIxa and -xb in verve terminals (29).
We defined the CaN binding site in dynIxb by site-directed mutagenesis and peptide competition as being PRITIS, a sequence that conforms to the original CaN docking sequence SPRIEIT in NFAT. Docking of CaN to a select group of its substrates appears to be a conserved model to provide strong specificity to CaN signaling, despite the apparent availability of a much larger substrate selection. For example, the transcriptional activity of NFAT is dependent on its dephosphorylation by CaN (49–51). Disruption of the CaN-NFAT1 interaction with a SPRIEIT peptide inhibited NFAT dephosphorylation both in vitro and in vivo and also inhibited its nuclear translocation and gene expression in T-cells, without inhibiting the ability of CaN to dephosphorylate other endogenous proteins (37). Similarly, we discovered that the peptide containing the dynIxb(842–851) PXIXIS motif prevented CaN interaction with dynIxb in vitro and selectively inhibited KCl-evoked dynI dephosphorylation in nerve terminals but not that of three other well characterized dephosphins, amphiphysin I, AP180, and synaptojanin. This demonstrates that the peptide did not block CaN activity but only its recruitment to dynIxb. Thus, the specific anchoring of CaN to dynIxb may be employed in the nerve terminal for ADBE in a similar manner to that seen with NFAT1 in the cell body for transcriptional regulation.
CaN dephosphorylates dynI only during periods of high activity. This event triggers an interaction with syndapin I for ADBE (3, 52). Our finding that the dynI tail PRITIS peptide dynIxb(842–851) selectively inhibits both ADBE and dynI phosphorylation adds considerable support to the model in which the action of CaN in ADBE is primarily mediated by dynI dephosphorylation. It may initially appear contradictory that dynI is selectively involved with only one mode of SV endocytosis when the use of the dynamin GTPase inhibitor dynasore has shown that dynI is essential for all modes (2, 3). However, there is a large pool of non-phosphorylated dynI in nerve terminals representing 80% of the total (27) that is not regulated by the signaling cascades that regulate ADBE (6, 13). Therefore, we propose that multiple pools of dynI reside in nerve terminals and that CaN binding and phosphorylation mechanistically separate a phospho-pool for ADBE, whereas the bulk of residual dynI is potentially available for CME or other functions. The ADBE-triggering action could therefore occur in two steps: first, the Ca2+-dependent recruitment of CaN to dynIxb (this study) and, second, the dephosphorylation of dynI, thereby triggering its binding to syndapin I (3, 13). Our results show that prior binding of syndapin to dynIxb prevents CaN recruitment. Because phosphorylation of dynI regulates its interaction with syndapin but not CaN, we hypothesize that the wild type dynI dissociates from CaN to associate with syndapin to play a key role in ADBE. Interestingly, both splice variants are dephosphorylated by CaN, suggesting that ADBE might utilize either variant.
Our results provide further evidence that CaN is required for ADBE via its interaction with dynIxb and dephosphorylation of dynI. In agreement, previous studies in chromaffin cells demonstrate a CaN-sensitive form of endocytosis that was only observed during high activity (53, 54). However, there are multiple CaN substrates in nerve terminals (e.g. the dephosphins) that are dephosphorylated independently of the CaN-dynIxb interaction and are linked to other SV endocytosis or trafficking modes (17, 55). This suggests that CaN activity may play other yet unidentified roles in the SV life cycle. Recent studies have raised the possibility of new functions for CaN in SV turnover, both during SV retrieval and during SV trafficking between intracellular pools (56, 57). It will be interesting to determine whether the targeted dephosphorylation of specific dephosphins may be responsible for these potential additional functions. How such targeting occurs remains to be determined, but possibilities include differential dephosphorylation by specific trains of neuronal activity, anchoring in different nerve terminal subdomains, or differential association with CaN-inhibitory proteins (58).
In summary, we found that a short alternatively spliced insert in dynI confers the ability of a single splice variant to bind to an additional distinct partner, in addition to shared common binding partners like syndapin, endophilin, and amphiphysin. The CaN-dynIxb interaction, like the dynI-syndapin I interaction, is required for ADBE but not for CME in neurons. Our results suggest that CaN is specifically recruited to sites of ADBE by a selective docking to the tail of a single dynI splice variant. To the best of our knowledge, this is the first report of a selective cellular function for one of the dynI splice variants. Our results support a model whereby dynI dephosphorylation by CaN is the primary trigger for ADBE during intense neuronal stimulation, leading to the formation of a dynI-syndapin I complex as the next step in the process of bulk endocytosis.
Supplementary Material
Acknowledgments
We thank Alan Prescott and John James for help with the electron microscopy analysis and the NHMRC for financial support.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. S1–S4.
- dyn
- dynamin
- SV
- synaptic vesicle
- CaN
- calcineurin
- CaM
- calmodulin
- PRD
- proline-rich domain
- SH3
- Src homology 3
- ADBE
- activity-dependent bulk endocytosis
- CME
- clathrin-mediated endocytosis
- TES
- N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid
- BE
- bulk endosome
- NFAT
- nuclear factor of activated T cells.
REFERENCES
- 1. Anggono V., Robinson P. J. (2009) in Encyclopedia of Neuroscience (Squire L. R. ed) Academic Press, Inc., Oxford [Google Scholar]
- 2. Newton A. J., Kirchhausen T., Murthy V. N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17955–17960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clayton E. L., Anggono V., Smillie K. J., Chau N., Robinson P. J., Cousin M. A. (2009) J. Neurosci. 29, 7706–7717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang Q., Li Y., Tsien R. W. (2009) Science 323, 1448–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Granseth B., Odermatt B., Royle S. J., Lagnado L. (2006) Neuron 51, 773–786 [DOI] [PubMed] [Google Scholar]
- 6. Clayton E. L., Cousin M. A. (2009) J. Neurochem. 111, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheung G., Jupp O. J., Cousin M. A. (2010) J. Neurosci. 30, 8151–8161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clayton E. L., Evans G. J., Cousin M. A. (2008) J. Neurosci. 28, 6627–6632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jockusch W. J., Praefcke G. J., McMahon H. T., Lagnado L. (2005) Neuron 46, 869–878 [DOI] [PubMed] [Google Scholar]
- 10. Wu L. G., Ryan T. A., Lagnado L. (2007) J. Neurosci. 27, 11793–11802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kasprowicz J., Kuenen S., Miskiewicz K., Habets R. L., Smitz L., Verstreken P. (2008) J. Cell Biol. 182, 1007–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Evans G. J., Cousin M. A. (2007) J. Neurosci. 27, 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clayton E. L., Sue N., Smillie K. J., O'Leary T., Bache N., Cheung G., Cole A. R., Wyllie D. J., Sutherland C., Robinson P. J., Cousin M. A. (2010) Nat. Neurosci. 13, 845–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. LaPointe M. C., Deschepper C. F., Wu J. P., Gardner D. G. (1990) Hypertension 15, 20–28 [DOI] [PubMed] [Google Scholar]
- 15. Bauerfeind R., Takei K., De Camilli P. (1997) J. Biol. Chem. 272, 30984–30992 [DOI] [PubMed] [Google Scholar]
- 16. Marks B., McMahon H. T. (1998) Curr. Biol. 8, 740–749 [DOI] [PubMed] [Google Scholar]
- 17. Cousin M. A., Robinson P. J. (2001) Trends Neurosci. 24, 659–665 [DOI] [PubMed] [Google Scholar]
- 18. Klee C. B., Ren H., Wang X. T. (1998) J. Biol. Chem. 273, 13367–13370 [DOI] [PubMed] [Google Scholar]
- 19. Rusnak F., Mertz P. (2000) Physiol. Rev. 80, 1483–1521 [DOI] [PubMed] [Google Scholar]
- 20. Ye Q., Wang H., Zheng J., Wei Q., Jia Z. (2008) Proteins 73, 19–27 [DOI] [PubMed] [Google Scholar]
- 21. Lai M. M., Hong J. J., Ruggiero A. M., Burnett P. E., Slepnev V. I., De Camilli P., Snyder S. H. (1999) J. Biol. Chem. 274, 25963–25966 [DOI] [PubMed] [Google Scholar]
- 22. Song H. O., Lee J., Ji Y. J., Dwivedi M., Cho J. H., Park B. J., Ahnn J. (2010) Mol. Cells 30, 255–562 [DOI] [PubMed] [Google Scholar]
- 23. Anggono V., Smillie K. J., Graham M. E., Valova V. A., Cousin M. A., Robinson P. J. (2006) Nat. Neurosci. 9, 752–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cao H., Garcia F., McNiven M. A. (1998) Mol. Biol. Cell 9, 2595–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robinson P. J., Sontag J. M., Liu J. P., Fykse E. M., Slaughter C., McMahon H., Südhof T. C. (1993) Nature 365, 163–166 [DOI] [PubMed] [Google Scholar]
- 26. Ruiz-Velasco V., Zhong J., Hume J. R., Keef K. D. (1998) Circ. Res. 82, 557–565 [DOI] [PubMed] [Google Scholar]
- 27. Chan L. S., Hansra G., Robinson P. J., Graham M. E. (2010) J. Proteome Res. 9, 4028–4037 [DOI] [PubMed] [Google Scholar]
- 28. Robinson P. J., Dunkley P. R. (1985) J. Neurochem. 44, 338–348 [DOI] [PubMed] [Google Scholar]
- 29. Graham M. E., Anggono V., Bache N., Larsen M. R., Craft G. E., Robinson P. J. (2007) J. Biol. Chem. 282, 14695–14707 [DOI] [PubMed] [Google Scholar]
- 30. Chen-Hwang M. C., Chen H. R., Elzinga M., Hwang Y. W. (2002) J. Biol. Chem. 277, 17597–17604 [DOI] [PubMed] [Google Scholar]
- 31. Huang Y., Chen-Hwang M. C., Dolios G., Murakami N., Padovan J. C., Wang R., Hwang Y. W. (2004) Biochemistry 43, 10173–10185 [DOI] [PubMed] [Google Scholar]
- 32. Wang L. H., Südhof T. C., Anderson R. G. (1995) J. Biol. Chem. 270, 10079–10083 [DOI] [PubMed] [Google Scholar]
- 33. Rao Y., Ma Q., Vahedi-Faridi A., Sundborger A., Pechstein A., Puchkov D., Luo L., Shupliakov O., Saenger W., Haucke V. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 8213–8218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan T. C., Valova V. A., Malladi C. S., Graham M. E., Berven L. A., Jupp O. J., Hansra G., McClure S. J., Sarcevic B., Boadle R. A., Larsen M. R., Cousin M. A., Robinson P. J. (2003) Nat. Cell Biol. 5, 701–710 [DOI] [PubMed] [Google Scholar]
- 35. Klee C. B., Crouch T. H., Krinks M. H. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 6270–6273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Okamoto P. M., Tripet B., Litowski J., Hodges R. S., Vallee R. B. (1999) J. Biol. Chem. 274, 10277–10286 [DOI] [PubMed] [Google Scholar]
- 37. Aramburu J., Garcia-Cózar F., Raghavan A., Okamura H., Rao A., Hogan P. G. (1998) Mol. Cell 1, 627–637 [DOI] [PubMed] [Google Scholar]
- 38. Aramburu J., Yaffe M. B., López-Rodríguez C., Cantley L. C., Hogan P. G., Rao A. (1999) Science 285, 2129–2133 [DOI] [PubMed] [Google Scholar]
- 39. Cousin M. A., Malladi C. S., Tan T. C., Raymond C. R., Smillie K. J., Robinson P. J. (2003) J. Biol. Chem. 278, 29065–29071 [DOI] [PubMed] [Google Scholar]
- 40. Clayton E. L., Cousin M. A. (2008) Neurochem. Int. 53, 51–55 [DOI] [PubMed] [Google Scholar]
- 41. Garcia-Cozar F. J., Okamura H., Aramburu J. F., Shaw K. T., Pelletier L., Showalter R., Villafranca E., Rao A. (1998) J. Biol. Chem. 273, 23877–23883 [DOI] [PubMed] [Google Scholar]
- 42. Czirják G., Enyedi P. (2006) J. Biol. Chem. 281, 14677–14682 [DOI] [PubMed] [Google Scholar]
- 43. Stemmer P., Klee C. B. (1991) Curr. Opin. Neurobiol. 1, 53–64 [DOI] [PubMed] [Google Scholar]
- 44. Perrino B. A., Ng L. Y., Soderling T. R. (1995) J. Biol. Chem. 270, 340–346 [DOI] [PubMed] [Google Scholar]
- 45. Yu H., van Berkel T. J., Biessen E. A. (2007) Cardiovasc. Drug Rev. 25, 175–187 [DOI] [PubMed] [Google Scholar]
- 46. Dell'Acqua M. L., Dodge K. L., Tavalin S. J., Scott J. D. (2002) J. Biol. Chem. 277, 48796–48802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Filosto S., Fry W., Knowlton A. A., Goldkorn T. (2010) J. Biol. Chem. 285, 10213–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Czirják G., Tóth Z. E., Enyedi P. (2004) J. Biol. Chem. 279, 18550–18558 [DOI] [PubMed] [Google Scholar]
- 49. Jain J., McCaffrey P. G., Miner Z., Kerppola T. K., Lambert J. N., Verdine G. L., Curran T., Rao A. (1993) Nature 365, 352–355 [DOI] [PubMed] [Google Scholar]
- 50. Beals C. R., Clipstone N. A., Ho S. N., Crabtree G. R. (1997) Genes Dev. 11, 824–834 [DOI] [PubMed] [Google Scholar]
- 51. Okamura H., Aramburu J., García-Rodríguez C., Viola J. P., Raghavan A., Tahiliani M., Zhang X., Qin J., Hogan P. G., Rao A. (2000) Mol. Cell 6, 539–550 [DOI] [PubMed] [Google Scholar]
- 52. Andersson F., Jakobsson J., Löw P., Shupliakov O., Brodin L. (2008) J. Neurosci. 28, 3925–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Engisch K. L., Nowycky M. C. (1998) J. Physiol. 506, 591–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chan S. A., Chow R., Smith C. B. (2003) Pflugers Arch. 445, 540–546 [DOI] [PubMed] [Google Scholar]
- 55. Jovanovic J. N., Sihra T. S., Nairn A. C., Hemmings H. C., Jr., Greengard P., Czernik A. J. (2001) J. Neurosci. 21, 7944–7953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun T., Wu X. S., Xu J., McNeil B. D., Pang Z. P., Yang W., Bai L., Qadri S., Molkentin J. D., Yue D. T., Wu L. G. (2010) J. Neurosci. 30, 11838–11847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim S. H., Ryan T. A. (2010) Neuron 67, 797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lai M. M., Luo H. R., Burnett P. E., Hong J. J., Snyder S. H. (2000) J. Biol. Chem. 275, 34017–34020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.