Abstract
Fruiting body lectins have been proposed to act as effector proteins in the defense of fungi against parasites and predators. The Marasmius oreades agglutinin (MOA) is a Galα1,3Gal/GalNAc-specific lectin from the fairy ring mushroom that consists of an N-terminal ricin B-type lectin domain and a C-terminal dimerization domain. The latter domain shows structural similarity to catalytically active proteins, suggesting that, in addition to its carbohydrate-binding activity, MOA has an enzymatic function. Here, we demonstrate toxicity of MOA toward the model nematode Caenorhabditis elegans. This toxicity depends on binding of MOA to glycosphingolipids of the worm via its lectin domain. We show further that MOA has cysteine protease activity and demonstrate a critical role of this catalytic function in MOA-mediated nematotoxicity. The proteolytic activity of MOA was dependent on high Ca2+ concentrations and favored by slightly alkaline pH, suggesting that these conditions trigger activation of the toxin at the target location. Our results suggest that MOA is a fungal toxin with intriguing similarities to bacterial binary toxins and has a protective function against fungivorous soil nematodes.
Keywords: C. elegans, Calcium, Carbohydrate-binding Protein, Fungi, Glycolipids, Lectin, Protease, Toxins
Introduction
A wide variety of lectins with different structures and specificities has been isolated from fungi (1). Recently, cytoplasmic fungal lectins, also referred to as fruiting body lectins, were proposed to be part of a fungal defense system against parasites and predators. This hypothesis is based on the carbohydrate binding-dependent toxicity of several fungal lectins toward nematodes, insect larvae, and amoebae (2). The exclusive and abundant expression of these lectins in fruiting bodies or sclerotia of the host fungus may result in special protection of these reproductive organs. It is proposed that predators such as fungivorous nematodes ingest the cytoplasmic lectin when feeding on the content of the fungal cell and that binding of the lectin to specific glycans in the intestine of the worm induces toxicity by an unknown mechanism (2–5).
The Marasmius oreades agglutinin (MOA)3 is a B-type erythrocyte-agglutinating lectin that was isolated from fruiting bodies of the fairy ring mushroom. The dimeric lectin consists of 293-residue protomers and has been shown to specifically recognize Galα1,3-containing structures, exhibiting the highest affinity for the branched blood group B trisaccharide Galα1,3(Fucα1,2)Gal (6). Crystal structures of MOA in complex with the xenotransplantation epitope and the blood group B trisaccharide revealed an N-terminal ricin B-type lectin domain with three canonical carbohydrate-binding sites and a C-terminal domain that is involved in dimerization. The C-terminal domain shows structural homology to catalytically active proteins such as peptide N-glycanases, N-acetyltransferases, transglutaminases, and cysteine proteases. This structural similarity is limited to a putative catalytic triad consisting of Cys-215, His-257, and Glu-274 (7, 8). However, no catalytic activity of MOA has been reported so far. Moreover, the biological target glycan and the function of the lectin remained unclear. Like other fruiting body lectins, MOA lacks a classical secretion signal and is thus predicted to be localized in the cytoplasm. An endogenous function of the lectin is therefore unlikely, as glycosylated structures are generally secreted or localized in the extracellular space (9). Accordingly, no glycoconjugate recognized by MOA has thus far been identified in fungi.
In this study, we investigated the possible role of MOA as a fungal defense molecule against nematodes using Caenorhabditis elegans as a genetically tractable model system. First, we demonstrate carbohydrate binding-dependent nematotoxicity of MOA and identify glycosphingolipids as the ligands of MOA in C. elegans. Second, we show that MOA-mediated toxicity is dependent on the previously suggested catalytic function that we identified as Ca2+-dependent cysteine protease activity. Our results suggest that MOA is a fungal toxin that protects the fruiting body from predators such as fungivorous nematodes.
EXPERIMENTAL PROCEDURES
Strains, Cultivation Conditions, Cloning, and Site-directed Mutagenesis
Strains, primer sequences, and experimental procedures can be found under supplemental “Materials and Methods.”
C. elegans Toxicity Assays and Statistical Analysis
Biotoxicity assays with C. elegans were performed as described (10). Results were analyzed using a t test for pairwise comparisons.
Protein Expression, Purification, and Biotinylation
MOA and MOA(C215A) were expressed recombinantly in Escherichia coli and purified as described (6, 11). Detailed information can be found under supplemental “Materials and Methods.” Truncated Cry5B (t-Cry5B) was prepared as described previously (12). For TLC overlay analysis, purified MOA and t-Cry5B were labeled with EZ-Link sulfo-NHS-biotin (Pierce) using a 20-fold molar excess of labeling reagent. Labeling proceeded for 2 h on ice, followed by desalting on a PD-10 column.
Screen for MOA-resistant C. elegans Mutants Using Mos1-mediated Insertional Mutagenesis and Light Microscopy of C. elegans
Mos1-mediated insertional mutagenesis, stereomicroscopy, and differential interference contrast microscopy of C. elegans were performed as described (3).
Preparation and Analysis of Glycolipids from C. elegans
Glycolipid extraction from C. elegans N2, bre-2, and bre-4 strains and TLC overlay staining were performed as described by Barrows et al. (13). For overlay analysis, biotinylated MOA and t-Cry5B were used at a concentration of 150 nm.
In Vitro Protease Assay
Briefly, 20 μg of denatured RNase A was incubated with 0.1 ng to 1 μg of purified recombinant MOA or MOA(C215A) in assay buffer (40 mm Tris-HCl (pH 8.0), 1 mm CaCl2, and 1% Nonidet P-40) in a final reaction volume of 30 μl for 1 h at 37 °C. Samples were analyzed by SDS-PAGE and Coomassie Blue staining. Detailed information is provided under supplemental “Materials and Methods.”
Cleavage Site Specificity
Bovine asialofetuin, bovine casein, and human hemoglobin (40 μg each) were digested with 100 ng of MOA in 40 mm Tris-HCl (pH 8.0) and 1 mm CaCl2 for 1 h at 37 °C. The reaction was stopped by the addition of 25 mm EDTA. Samples were desalted using C18 ZipTips (Millipore) and analyzed on an LTQ Orbitrap XL mass spectrometer (Thermo Fischer) coupled to a nano-HPLC system (Eksigent Technologies, Dublin, CA). Detailed information on MS analysis and data evaluation is provided under supplemental “Materials and Methods.”
RESULTS
MOA Inhibits C. elegans Development and Amoebal Growth Dependent on Its Carbohydrate-binding Ability
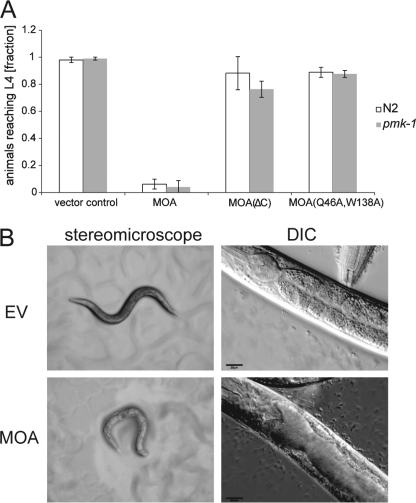
On the basis of results with other fruiting body lectins (2), we tested MOA for toxicity toward the nematode C. elegans and the amoeba Acanthamoeba castellanii. These biotoxicity assays involved feeding the organisms MOA-expressing E. coli as described previously (10) and revealed a clear toxicity of MOA toward C. elegans and A. castellanii (Fig. 1 and supplemental Fig. S1). Both C. elegans and A. castellanii were fed E. coli cells expressing authentic MOA, the C-terminal deletion mutant MOA(ΔC), or the carbohydrate binding-deficient variant MOA(Q46A,W138A) (7). In a control experiment, test organisms were grown on vector-containing E. coli transformants. In the case of C. elegans, severe inhibition of larval development was observed for N2 wild-type and pmk-1(km25) mutant animals feeding on MOA-expressing bacteria compared with the vector control (p < 0.05) (Fig. 1A). The pmk-1(km25) mutation in the MAPK pathway was shown previously to cause hypersensitivity to different kinds of stress, including exposure to fungal lectins (3). In contrast, MOA(Q46A,W138A) had no significant effect on the development of the worms (p > 0.05). Similar results were obtained with A. castellanii (p < 0.05) (supplemental Fig. S1). Previous studies showed that Gln-46 and Trp-138 are involved in glycan coordination (7, 8) and that the binding of the double mutant to thyroglobulin is reduced compared with wild-type MOA (14). The lack of toxicity of MOA(Q46A,W138A) suggests that the nematotoxicity of MOA is dependent on the carbohydrate-binding activity of the protein. This is also supported by the decreased nematotoxicity of MOA(ΔC) (p < 0.05), a mutant that showed reduced carbohydrate-binding activity possibly due to loss of dimerization (Fig. 1A) (14).
FIGURE 1.
Recombinant MOA displays carbohydrate binding-dependent toxicity toward C. elegans. E. coli BL21(DE3) cells expressing MOA, the C-terminal deletion mutant MOA(ΔC), or the carbohydrate binding-deficient variant MOA(Q46A,W138A) or containing an empty vector were fed to C. elegans N2 wild-type and pmk-1(km25) mutant worms. A, MOA inhibits C. elegans development. The fraction of C. elegans L1 larvae having developed to the L4 stage was scored after 72 h of feeding on a lawn of respective E. coli BL21 cells. Bars represent the average of nine biological replicates. Error bars indicate S.D. B, MOA damages the C. elegans intestine. C. elegans N2 wild-type L4 larvae were fed MOA-expressing (lower panels) and control, i.e., empty-vector (EV) containing (upper panels) E. coli BL21(DE3) cells and examined after 24 h under a stereomicroscope (left panels) and by differential interference contrast (DIC) microscopy (right panels). Scales bars = 20 μm.
The physiological effects of intoxication were studied by comparative light and differential interference contrast microscopy of C. elegans L4 larvae fed MOA-expressing or vector-containing E. coli cells. MOA-intoxicated animals showed an expanded lumen of the anterior intestine compared with control animals (Fig. 1B). A similar phenotype was observed with other nematotoxic fungal lectins (3) and bacterial toxins (15).
MOA Ligand in C. elegans Is a Glycosphingolipid
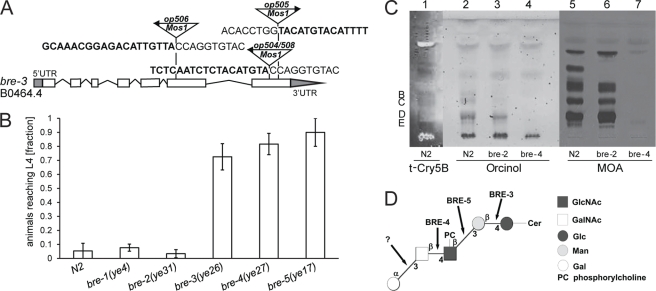
To identify the target glycoconjugate of MOA in C. elegans, a forward genetic screen for mutations conferring MOA resistance was performed. Strain pmk-1(km25) was used to increase the sensitivity of the screen. Four MOA-resistant mutants were isolated in the screen, all of which carried mutations in exons of the bre-3 gene. bre-3(op504) and bre-3(op505) mutants had a Mos1 insertion in the seventh exon of bre-3. bre-3(op506) mutants carried a Mos1 insertion in the fifth exon of the same gene. In an independent screen, one mutant (op508) with a Mos1 insertion site identical to the one in bre-3(op504) was identified (Fig. 2A). bre-3 encodes a glycosyltransferase that acts in a biosynthetic pathway building up an invertebrate-specific glycosphingolipid that is recognized by the Bacillus thuringiensis crystal toxin Cry5B (16). The oligosaccharide core of the ceramide-linked glycan is synthesized by the sequential action of BRE-3 (β1,4-mannosyltransferase), BRE-5 (β1,3-GlcNAc-transferase), BRE-4 (β1,4-GalNAc-transferase), and BRE-2 (putative β1,3-galactosyltransferase). bre-1 encodes an enzyme involved in the conversion of GDP-mannose into GDP-fucose (17). Given the resistance of bre-3 mutant animals, we tested worms with mutations in the other bre genes for sensitivity toward MOA. We observed that also mutations in the bre-4(ye27) and bre-5(ye17) genes conferred resistance to MOA (p < 0.05), whereas bre-1(ye4) and bre-2(ye31) mutant worms were sensitive to intoxication with MOA (p > 0.05) (Fig. 2B). These results suggest that, like bacterial Cry5B, fungal MOA targets a glycosphingolipid but that the species is different from the one recognized by Cry5B.
FIGURE 2.
Toxicity of MOA toward C. elegans is mediated by binding to glycosphingolipids. A, identified insertion sites of the Mos1 transposon in the bre-3 gene of MOA-resistant C. elegans mutants. Arrows above Mos1 elements indicate the orientation of the Mos1 primer oJL115 used for sequencing inverse PCR products of mutant lysates. Boldface letters indicate C. elegans genomic sequences, followed by Mos1 sequence. The gene model is taken from WormBase Release WS213. White boxes indicate exons, and grey boxes at the ends of the gene indicate 5′- and 3′-untranslated regions. B, resistance of C. elegans bre-3(ye26), bre-4(ye27), and bre-5(ye17) mutants to MOA-mediated toxicity. C. elegans mutants of the indicated genotypes were analyzed for development from L1 to L4 as described in the legend to Fig. 1. Bars represent the average of nine biological replicates. Error bars indicate S.D. C, MOA binds glycolipids of N2 and bre-2(ye31) animals, but not bre-4(ye27) animals. Lipids were extracted from N2, bre-2(ye31), and bre-4(ye27) worms; resolved by TLC; and stained with orcinol (lanes 2-4) or overlaid with biotinylated t-Cry5B (lane 1) or MOA (lanes 5–7). The origin is always at the bottom. Letters refer to the glycolipid classification by Griffitts et al. (16). D, tentative structure of the main glycosphingolipid species (component D) recognized by MOA. The core of the ceramide (Cer)-linked oligosaccharide is synthesized by BRE-3, BRE-5, and BRE-4. The terminal α1,3Gal is added by a yet unknown galactosyltransferase.
To confirm the genetic data, direct binding of MOA to glycolipids isolated from N2, bre-2(ye31), and bre-4(ye27) worms was assessed in a TLC overlay. For this purpose, lipids were extracted from C. elegans, partitioned into lower phase lipids and upper phase glycolipids, and subsequently purified as described by Barrows et al. (13). Purified glycolipids were separated by TLC using chloroform/methanol/water (4:4:1) as a solvent and visualized using orcinol sulfate (Fig. 2C, lanes 2–4). The main glycolipid bands were observed in positions similar to those reported previously (12, 16). TLC overlay staining of upper phase glycolipids with MOA revealed binding to N2 and bre-2(ye31) but not bre-4(ye27) glycolipids (Fig. 2C, lanes 5–7). This is consistent with MOA susceptibility of N2 and bre-2(ye31) worms and resistance of bre-4(ye27) animals. To identify the glycolipid species that is recognized by MOA, comparative overlay staining with t-Cry5B as described by Ideo et al. (12) was performed. The pattern of the orcinol staining and the t-Cry5B overlay of N2-derived glycolipids (Fig. 2C, lane 1) indicated that the main glycolipid species recognized by MOA corresponds to component D, according to the classification of Griffitts et al. (16), that has been characterized previously (18). Binding to the glycan part of this glycosphingolipid of the arthroseries (Galα1,3GalNAcβ1,4GlcNAcβ1,3Manβ1,4Glc) (Fig. 2D) (16, 18) is in agreement with the Galα1,3Gal specificity of MOA. Finally, MS and MS/MS analysis of glycolipids extracted from the main TLC band recognized by MOA further supported the presence of component D, including the ceramide moiety composed of a saturated hydroxy fatty acid with 22 and 24 carbon atoms, respectively (supplemental Fig. S2). Besides component D, MOA was able to bind to a number of unidentified glycolipid species from both N2 and bre-2(ye31) animals (Fig. 2C, lanes 5 and 6). These glycolipid species were absent in the bre-4(ye27) animals.
MOA Shows in Vitro Protease Activity
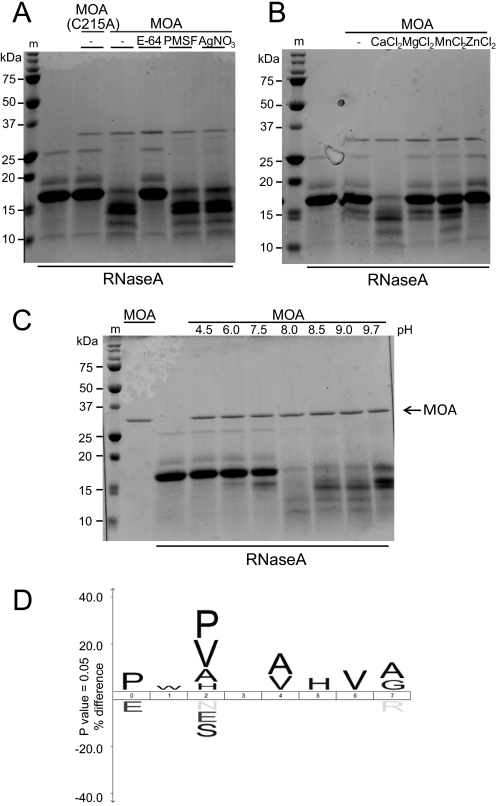
X-ray crystallographic studies of MOA in complex with the xenotransplantation epitope and the blood group B trisaccharide revealed that the C-terminal part of MOA adopts a fold that is observed in catalytically active proteins such as peptide N-glycanases, N-acetyltransferases, transglutaminases, and cysteine proteases (7, 8). As the highest homology was observed with a peptide N-glycanase, we tested purified recombinant MOA in the in vitro peptide N-glycanase assay described by Suzuki (19). However, instead of deglycosylation of the glycoprotein RNase B, degradation of the substrate was observed, indicating that MOA may have protease rather than peptide N-glycanase activity. To confirm this hypothesis, we performed an in vitro protease assay with non-glycosylated RNase A as a substrate (Fig. 3). MOA efficiently degraded denatured RNase A within 1 h at 37 °C. In contrast, MOA(C215A), in which the cysteine residue of the putative catalytic triad was replaced with alanine, had no effect on RNase A, confirming that the observed protease activity was MOA-dependent. In accordance with these results, degradation of the substrate protein by MOA was abolished in the presence of 50 μm E-64, an irreversible, highly selective cysteine protease inhibitor, but not in the presence of 2 mm PMSF, an irreversible serine protease inhibitor, or 2 mm AgNO3, a potent trypsin and chymotrypsin inhibitor (Fig. 3A). On the basis of these results, we conclude that MOA has, in addition to its glycan-binding properties, cysteine protease activity.
FIGURE 3.
Purified recombinant MOA shows in vitro protease activity. Shown is the in vitro protease activity of MOA using denatured RNase A as a substrate. 0.5 (A and B) or 1 (C) μg of purified recombinant MOA or MOA(C215A) was incubated with 20 μg of denatured RNase A in assay buffer for 1 h at 37 °C. The reaction was stopped by the addition of SDS sample buffer and heating at 95 °C for 10 min. Samples were analyzed by SDS-PAGE and Coomassie Blue staining. m, molecular mass standard. A, the catalytic activity of MOA is dependent on Cys-215 and abolished by cysteine protease inhibitor E-64. MOA was incubated with 50 μm E-64, 2 mm PMSF, and 2 mm AgNO3 for 20 min at room temperature before the substrate was added. B, Ca2+ is required for the protease activity of MOA. The reaction was performed in assay buffer without CaCl2 as well as in buffer containing 1 mm CaCl2, MgCl2, MnCl2, and ZnCl2, respectively. C, MOA protease activity has an alkaline pH optimum. Buffers at different pH values ranging from 4.5 to 9.7 were supplemented with 1 mm CaCl2 and 1% Nonidet P-40 and used as assay buffers. D, cleavage site specificity of MOA. The iceLogo (20) was generated from 83 MOA-induced cleavage sites in native model substrates (bovine asialofetuin, bovine casein, and human hemoglobin). MOA-mediated cleavage occurred between positions 4 and 5 of the sequence logo.
Structural homology to catalytically active proteins was first observed in a crystal structure of MOA in complex with the xenotransplantation epitope (7). A second crystal structure of MOA complexed with the blood group B trisaccharide revealed that binding of two Ca2+ ions close to the hypothetical active site in the C-terminal domain causes a conformational change, opening up a large cleft containing the potential active site (8). To investigate the influence of divalent cations on MOA protease activity, reactions in assay buffers supplemented with 1 mm CaCl2, MgCl2, MnCl2, and ZnCl2, respectively, were performed. In the presence of Ca2+ ions, proteolytic degradation of RNase A by MOA was observed. In contrast, MOA showed hardly any activity in buffer supplemented with MgCl2, MnCl2, or ZnCl2 or in the absence of divalent cations (Fig. 3B). This observation is in accordance with the structural data and suggests that Ca2+ binding activates the catalytic activity of MOA. We found that a minimal concentration of 50 μm was required for MOA to exhibit protease activity and that this activity further increased upon raising the Ca2+ concentration to 5 mm (supplemental Fig. S3).
To test the influence of pH on the reaction, protease assays in buffers of different pH values were performed. Complete degradation of RNase A by MOA was observed at pH 8.0. At higher pH, MOA only partially degraded the substrate, whereas under acidic conditions, the enzyme was not active (Fig. 3C). Therefore, MOA protease activity is favored by a slightly alkaline pH. The presence or absence of the carbohydrate ligand Galα1,3Galβ1,4GlcNAc did not affect the proteolytic activity of MOA (supplemental Fig. S4).
To characterize the substrate specificity of MOA, we tested its activity toward different denatured and native protein substrates in the in vitro assay. BSA, FITC-labeled casein, asialofetuin, and hemoglobin were degraded when denatured before incubation with MOA (supplemental Fig. S5A). In the native state, only FITC-labeled casein, asialofetuin, and hemoglobin were susceptible to MOA protease activity, whereas BSA, RNase A, and RNase B were not degraded (supplemental Fig. S5B). Proteolytic degradation of FITC-labeled casein by MOA could also be detected in a fluorometric assay, where it showed a 3.6-fold higher activity/mol at pH 8.0 than the cysteine protease papain at pH 7.5 (supplemental Fig. S6). As MOA did not act on every tested native protein substrate, we conclude that the protease has a certain substrate specificity.
To investigate the cleavage site specificity of MOA, native asialofetuin, casein, or hemoglobin was digested with purified MOA, and the resulting peptides were analyzed by LC-electrospray ionization-MS/MS. This procedure yielded 83 cleavage sites that were used to create an iceLogo (20) of the preferred sequence specificity, where the cleavage occurs between positions 4 and 5 (Fig. 3D). The results of this analysis suggest that MOA prefers proline and proline or valine at positions 0 and 2, respectively. A digest of the C. elegans proteome with MOA yielded similar results (data not shown), confirming the specificity observed for the model substrates.
Protease Activity of MOA Is Required for Nematotoxicity
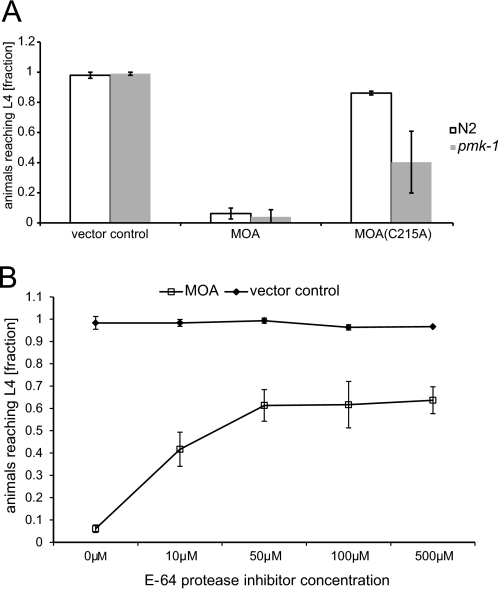
To test whether the protease activity of MOA plays a role in toxicity toward C. elegans, worms were fed E. coli expressing the catalytic site mutant MOA(C215A). This single-site mutant formed dimers like wild-type MOA as confirmed by analytical gel filtration (supplemental Fig. S7). The toxicity of MOA(C215A) was significantly reduced in N2 animals and pmk-1(km25) mutants compared with wild-type MOA (p < 0.05) (Fig. 4A). Similar results were obtained for A. castellanii (p < 0.05) (supplemental Fig. S1). On the basis of these results, we conclude that, in addition to carbohydrate binding, the cysteine protease activity of MOA is necessary for full toxicity. In C. elegans, the contribution of the catalytic domain to the toxicity of MOA is also made apparent by the fact that pmk-1(km25) mutant and N2 wild-type worms were equally susceptible to wild-type MOA (Figs. 1A and 4A). This is in contrast to nematotoxic fungal lectins lacking a catalytic domain, which display a higher toxicity to pmk-1(km25) mutant worms (2, 3). Accordingly, abolishment of the catalytic activity, as in the case of MOA(C215A) and MOA(ΔC), resulted in higher susceptibility of pmk-1(km25) worms compared with N2 worms (Figs. 1A and 4A). The higher toxicity of MOA(C215A) in comparison with MOA(ΔC) can be explained by the lack of dimerization in the latter case. These results suggest that the toxicity mechanism of MOA differs from that of nematotoxic lectins without a catalytic domain.
FIGURE 4.
Protease activity of MOA is required for full toxicity toward C. elegans. A, MOA(C215A) exhibits reduced toxicity toward C. elegans. C. elegans N2 wild-type and pmk-1(km25) mutant worms were seeded onto lawns of E. coli BL21(DE3) cells expressing MOA or MOA(C215A) or containing an empty vector and analyzed for development from L1 to L4 as described in the legend to Fig. 1. Bars represent the average of nine biological replicates. Error bars indicate S.D. B, cysteine protease inhibitor E-64 diminishes MOA-mediated toxicity toward C. elegans. L1 stage C. elegans pmk-1(km25) worms were seeded onto a lawn of E. coli BL21(DE3) cells expressing MOA mixed with E-64 diluted in M9 buffer at the indicated concentrations. Development of the animals from L1 to L4 was analyzed as described in the legend to Fig. 1. As a control, the experiment was performed with E. coli BL21(DE3) cells containing an empty vector. The data shown represent the average of nine biological replicates. Error bars indicate S.D.
As the protease activity of MOA could be inhibited by E-64 in vitro, pmk-1(km25) animals were grown on MOA-expressing bacteria in the presence of this highly selective cysteine protease inhibitor at 10–500 μm. The cell-permeable inhibitor significantly reduced MOA-mediated toxicity in a dose-dependent manner. On the other hand, E-64 did not impair development of worms feeding on vector-containing E. coli (Fig. 4B). These results confirm the role of MOA cysteine protease activity in nematotoxicity. As observed for the catalytic site mutant in the case of pmk-1(km25) worms, toxicity was not completely abolished by inhibiting protease activity even at the highest concentration of E-64. The remaining toxicity at inhibitor concentrations higher than 50 μm can be referred to the toxicity mediated by the lectin domains of the dimer, which are not affected by E-64.
DISCUSSION
Fruiting body lectins have been proposed to be part of the fungal defense system against predators and parasites (2). In agreement with such a function, we have demonstrated that the fruiting body lectin MOA is toxic toward the nematode C. elegans and the amoeba A. castellanii. As MOA mutants deficient in carbohydrate binding exhibited no or significantly reduced toxicity for both organisms, we conclude that the lectin activity of the protein is essential for its toxicity. Morphological changes in MOA-intoxicated worms included an enlargement of the intestinal lumen, an effect that has been reported previously for the fungal lectin CGL2 from Coprinopsis cinerea (3) and the B. thuringiensis crystal toxin Cry5B (15). These results suggest that binding of the lectin to a target glycan in the worm intestinal epithelium is essential for nematotoxicity. We hypothesize that the lectin is stored in the cytoplasm of the fungal cell and is released and transferred to the intestines of fungivorous nematodes upon predation.
Using C. elegans genetics, we could identify the target glycoconjugate of MOA in this organism that is responsible for its toxicity. Similar to the nematotoxic Cry5B toxin from B. thuringiensis, MOA binds to different species of the invertebrate-specific arthroseries of glycosphingolipids in C. elegans. Based on TLC overlays and MS analysis, the main species recognized by MOA is component D, according to the annotation of Griffitts et al. (16). This finding is consistent with the genetic results, as bre-3, bre-5, and bre-4 (but not bre-2) encode glycosyltransferases involved in the biosynthesis of this specific glycosphingolipid. In addition to this previously identified glycosphingolipid species, MOA bound to several unknown glycolipids present in N2 and bre-2(ye31) worms, but not in bre-4(ye27) worms. These results indicate the presence of multiple additional glycosphingolipid species carrying terminal Galα1,3Gal in C. elegans. The full sensitivity of bre-1(ye4) worms suggests that, in contrast to glycosphingolipid-binding Cry5B, fucosylation of the lipid is not required for MOA binding.
The putative catalytic activity of MOA was investigated in in vitro assays with purified recombinant protein. In these assays, MOA showed proteolytic activity toward several native and denatured model substrates. The lack of activity toward some native protein substrates suggests a certain substrate specificity. The proteolytic activity of MOA was dependent on a cysteine residue in the predicted catalytic site and on the presence of Ca2+ and had a pH optimum of 8.0. The C-terminal domain of MOA does not show homology to any existing proteinase family currently classified in the MEROPS Database. Interestingly, the peptidase domain of MOA is the smallest in terms of residues that has ever been described and is even smaller than the HIV-1 retropepsin. The crystal structure suggests that the active-site cysteine is very close to the calcium-binding sites, and the glutamine that would be an ideal candidate for forming the oxyanion hole is also a calcium ligand. Thus, the chemistry around the active site of this protease domain appears to be very “busy.”4
Intriguingly, we could show that, in addition to carbohydrate binding, the proteolytic activity of MOA is required for full toxicity toward C. elegans, as toxicity of a catalytic site mutant was decreased and the toxic effects of MOA were reduced in the presence of the highly specific cysteine protease inhibitor E-64. The equal susceptibility of N2 and pmk-1(km25) worms to wild-type MOA as opposed to catalytically inactive MOA variants suggests that the catalytic activity adds an additional twist to this lectin with regard to its mechanism of toxicity.
As determined by in vitro digests of different model substrates, MOA preferentially cleaves sequences with proline or valine at position P2. Interestingly, a similar sequence specificity has been described for calpains, a family of calcium-dependent, non-lysosomal cysteine proteases that are expressed ubiquitously in mammals and many other organisms (21) and are involved in regulation of signal transduction pathways, cell motility, and apoptosis (22, 23). In contrast to calpain, which proteolyzes its substrates in a limited manner, exhibiting modulatory functions by cutting interdomain regions, our analysis using model substrates suggests that MOA digests protein substrates to small peptides. The specificity for sequences containing proline may allow MOA to act on a wide range of proteins, as proline residues are generally located in turns at the protein surface. However, further studies with more physiological substrate proteins are required to strengthen this hypothesis.
Chimeric lectins homologous to MOA are present in the basidiomycetes Polyporus squamosus (24) and Schizophyllum commune (25). In P. squamosus lectin, the catalytic triad and the four Asp residues taking part in Ca2+ binding are conserved. However, the lectin domain of P. squamosus lectin has been shown to be specific for Neu5Acα2,6Galβ1,4Glc/GlcNAc groups of N-glycans (24, 26, 27). In contrast, S. commune agglutinin has a similar carbohydrate-binding specificity as MOA (to be published elsewhere), but the catalytic residues Cys, His, and Glu of MOA are replaced with Ser, Ser and Asp, respectively, and only one of the four Asp residues involved in Ca2+ binding is conserved, suggesting that the cysteine protease activity is not conserved in this homolog.
As one possible toxicity mechanism, binding of MOA to glycosphingolipids in the nematode intestinal epithelium may result in digestion of surrounding proteins and ultimately lead to disintegration of the intestinal epithelium. A similar mechanism has been proposed for the maize cysteine protease Mir1-CP, which has been shown to impair the growth of fall armyworm (Spodoptera frugiperda) larvae by disrupting the insect's peritrophic matrix in the intestine (28–30). Such proteases are used by plants, besides lectins and protease inhibitors, as defense molecules against herbivores (31), in line with our hypothesis of MOA being involved in fungal defense. Interestingly, Mir1-CP was shown recently to synergize the insecticidal activity of B. thuringiensis toxins (32).
On the other hand, the carbohydrate binding-deficient but protease-proficient variant MOA(Q46A,W138A) was absolutely nontoxic even at the high lectin concentrations in the biotoxicity assay with recombinant bacteria (Fig. 1), and the lumen of the C. elegans intestine is thought to be slightly acidic based on the pH optima of digestive hydrolases (33), arguing against such a mechanism. In addition, the nature of its ligand (receptor) and the structural and functional organization of MOA are strikingly similar to bacterial AB5 toxins such as the cholera and Shiga toxins. These proteins consist of a nontoxic B-subunit that binds to specific glycolipids and an enzymatically active A-subunit that induces toxicity by blocking the GTPase activity of Gα and activating adenylate cyclase (cholera toxin) or by inactivating ribosomal protein biosynthesis (Shiga toxin). The intoxication process by these proteins is initiated by attachment of the toxin via the B-subunit, followed by internalization of the AB5-receptor complex by receptor-mediated endocytosis and retrograde transport to the endoplasmic reticulum (34). Finally, the A-subunit of these toxins is translocated to the cytoplasm by the sec translocon. This final step is omitted in the case of the subtilase cytotoxin SubAB, which induces cell death by exhibiting highly specific protease activity toward the endoplasmic reticulum-resident chaperone BiP (35). It is remarkable that SubAB produces pathological features in mice that strongly resemble the hemolytic uremic syndrome in humans (35), a characteristic that has also been observed for MOA (36). These results suggest that SubAB and MOA may have a similar toxicity mechanism. In accordance with such a hypothesis, the combination of neutral pH (37) and a Ca2+ concentration of >100 μm in the endoplasmic reticulum (38) meets the requirements of MOA to exhibit protease activity. On the other hand, MOA is not expected to be catalytically active at the neutral pH but low Ca2+ concentration (∼100 nm) of the cytosol (37, 39), which allows storage of this toxic protein in an inactive form in the cytoplasm of the fungal cell. Similarly, the slightly acidic pH in the intestinal lumen of the nematode should inhibit the proteolytic activity of MOA despite the presumably high Ca2+ concentrations in this compartment (33). Such regulation of its proteolytic activity via pH and Ca2+ concentration could explain the apparent lack of autoproteolytic activation or of pronounced substrate specificity of MOA. We therefore hypothesize that, upon binding to the glycosphingolipid receptor present on intestinal epithelial cells in the nematode, MOA is internalized to the early endosomal compartment by endocytosis and transported via the Golgi to the endoplasmic reticulum, where its catalytic domain is activated and may induce toxicity by degrading proteins that are in the folding process (supplemental Fig. S8). Further experiments are required to corroborate this hypothetical toxicity mechanism.
Supplementary Material
Acknowledgments
We thank I. J. Goldstein (Department of Biological Chemistry, University of Michigan Medical School, Ann Arbor, MI) and H. Tateno (National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan) for providing plasmids pT7LO-MOA and pT7LOH-MOA(Q46A,W138A), R. Aroian (University of California, San Diego, La Jolla, CA) for providing plasmid pQE9(Cry5B), and Dr. P. Nanni (Functional Genomics Center, University and ETH Zürich) for support in mass spectrometry.
Note Added in Proof
A study (Cordara, G., Egge-Jacobsen, W., Johansen, H. T., Winter, H.C., Goldstein, I. J., Sandvig, K., and Krengel, U. (2011) Biochem. Biophys. Res. Commun. 408, 405–410) describing the protease activity of MOA was published while this paper was under review.
This work was supported by the European Commission Marie Curie Program (EuroGlycoArrays ITN), Swiss National Science Foundation Grant 31003A-130671 (to M. K., M. O. H., and M. A.), and ETH Zürich.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” Figs. S1–S9, Tables S1 and S2, and additional references.
N. D. Rawlings, personal communication.
- MOA
- M. oreades agglutinin
- t-Cry5B
- truncated Cry5B.
REFERENCES
- 1. Goldstein I. J., Winter H. C. (2007) Mushroom Lectins, Elsevier, Oxford [Google Scholar]
- 2. Bleuler-Martinez S., Butschi A., Garbani M., Walti M. A., Wohlschlager T., Potthoff E., Sabotic J., Pohleven J., Luthy P., Hengartner M. O., Aebi M., Kunzler M. (2011) Mol. Ecol. 20, 3056–3070 [DOI] [PubMed] [Google Scholar]
- 3. Butschi A., Titz A., Wälti M. A., Olieric V., Paschinger K., Nöbauer K., Guo X., Seeberger P. H., Wilson I. B., Aebi M., Hengartner M. O., Künzler M. (2010) PLoS Pathog. 6, e1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Trigueros V., Lougarre A., Ali-Ahmed D., Rahbé Y., Guillot J., Chavant L., Fournier D., Paquereau L. (2003) Biochim. Biophys. Acta 1621, 292–298 [DOI] [PubMed] [Google Scholar]
- 5. Hamshou M., Smagghe G., Shahidi-Noghabi S., De Geyter E., Lannoo N., Van Damme E. J. (2010) Insect Biochem. Mol. Biol. 40, 883–890 [DOI] [PubMed] [Google Scholar]
- 6. Winter H. C., Mostafapour K., Goldstein I. J. (2002) J. Biol. Chem. 277, 14996–15001 [DOI] [PubMed] [Google Scholar]
- 7. Grahn E., Askarieh G., Holmner A., Tateno H., Winter H. C., Goldstein I. J., Krengel U. (2007) J. Mol. Biol. 369, 710–721 [DOI] [PubMed] [Google Scholar]
- 8. Grahn E. M., Winter H. C., Tateno H., Goldstein I. J., Krengel U. (2009) J. Mol. Biol. 390, 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Varki A., Esko J. D., Colley K. J. (2009) in Essentials of Glycobiology (Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. eds) pp. 37–47, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 10. Künzler M., Bleuler-Martinez S., Butschi A., Garbani M., Lüthy P., Hengartner M. O., Aebi M. (2010) Methods Enzymol. 480, 141–150 [DOI] [PubMed] [Google Scholar]
- 11. Kruger R. P., Winter H. C., Simonson-Leff N., Stuckey J. A., Goldstein I. J., Dixon J. E. (2002) J. Biol. Chem. 277, 15002–15005 [DOI] [PubMed] [Google Scholar]
- 12. Ideo H., Fukushima K., Gengyo-Ando K., Mitani S., Dejima K., Nomura K., Yamashita K. (2009) J. Biol. Chem. 284, 26493–26501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barrows B. D., Griffitts J. S., Aroian R. V. (2006) Methods Enzymol. 417, 340–358 [DOI] [PubMed] [Google Scholar]
- 14. Tateno H., Goldstein I. J. (2004) Arch. Biochem. Biophys. 427, 101–109 [DOI] [PubMed] [Google Scholar]
- 15. Marroquin L. D., Elyassnia D., Griffitts J. S., Feitelson J. S., Aroian R. V. (2000) Genetics 155, 1693–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Griffitts J. S., Haslam S. M., Yang T., Garczynski S. F., Mulloy B., Morris H., Cremer P. S., Dell A., Adang M. J., Aroian R. V. (2005) Science 307, 922–925 [DOI] [PubMed] [Google Scholar]
- 17. Barrows B. D., Griffitts J. S., Aroian R. V. (2007) J. Invertebr. Pathol. 95, 198–200 [DOI] [PubMed] [Google Scholar]
- 18. Gerdt S., Dennis R. D., Borgonie G., Schnabel R., Geyer R. (1999) Eur. J. Biochem. 266, 952–963 [DOI] [PubMed] [Google Scholar]
- 19. Suzuki T. (2005) Methods 35, 360–365 [DOI] [PubMed] [Google Scholar]
- 20. Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2009) Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 21. Tompa P., Buzder-Lantos P., Tantos A., Farkas A., Szilágyi A., Bánóczi Z., Hudecz F., Friedrich P. (2004) J. Biol. Chem. 279, 20775–20785 [DOI] [PubMed] [Google Scholar]
- 22. Saido T. C., Sorimachi H., Suzuki K. (1994) FASEB J. 8, 814–822 [PubMed] [Google Scholar]
- 23. Sorimachi H., Ishiura S., Suzuki K. (1997) Biochem. J. 328, 721–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mo H., Winter H. C., Goldstein I. J. (2000) J. Biol. Chem. 275, 10623–10629 [DOI] [PubMed] [Google Scholar]
- 25. Ohm R. A., de Jong J. F., Lugones L. G., Aerts A., Kothe E., Stajich J. E., de Vries R. P., Record E., Levasseur A., Baker S. E., Bartholomew K. A., Coutinho P. M., Erdmann S., Fowler T. J., Gathman A. C., Lombard V., Henrissat B., Knabe N., Kües U., Lilly W. W., Lindquist E., Lucas S., Magnuson J. K., Piumi F., Raudaskoski M., Salamov A., Schmutz J., Schwarze F. W., vanKuyk P. A., Horton J. S., Grigoriev I. V., Wösten H. A. (2010) Nat. Biotechnol. 28, 957–963 [DOI] [PubMed] [Google Scholar]
- 26. Zhang B., Palcic M. M., Mo H., Goldstein I. J., Hindsgaul O. (2001) Glycobiology 11, 141–147 [DOI] [PubMed] [Google Scholar]
- 27. Tateno H., Winter H. C., Goldstein I. J. (2004) Biochem. J. 382, 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pechan T., Ye L., Chang Y., Mitra A., Lin L., Davis F. M., Williams W. P., Luthe D. S. (2000) Plant Cell 12, 1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pechan T., Cohen A., Williams W. P., Luthe D. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13319–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mohan S., Ma P. W., Pechan T., Bassford E. R., Williams W. P., Luthe D. S. (2006) J. Insect Physiol. 52, 21–28 [DOI] [PubMed] [Google Scholar]
- 31. van der Hoorn R. A., Jones J. D. (2004) Curr. Opin. Plant Biol. 7, 400–407 [DOI] [PubMed] [Google Scholar]
- 32. Mohan S., Ma P. W., Williams W. P., Luthe D. S. (2008) PLoS One 3, e1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McGhee J. D. (2007) WormBook, 1–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lencer W. I., Saslowsky D. (2005) Biochim. Biophys. Acta 1746, 314–321 [DOI] [PubMed] [Google Scholar]
- 35. Beddoe T., Paton A. W., Le Nours J., Rossjohn J., Paton J. C. (2010) Trends Biochem. Sci. 35, 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Warner R. L., Winter H. C., Speyer C. L., Varani J., Oldstein I. J., Murphy H. S., Johnson K. J. (2004) Exp. Mol. Pathol. 77, 77–84 [DOI] [PubMed] [Google Scholar]
- 37. Kim J. H., Johannes L., Goud B., Antony C., Lingwood C. A., Daneman R., Grinstein S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2997–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ong D. S., Mu T. W., Palmer A. E., Kelly J. W. (2010) Nat. Chem. Biol. 6, 424–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Halachmi D., Eilam Y. (1989) FEBS Lett. 256, 55–61 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.