Abstract
Previous work from our laboratory has shown that primary fibroblasts from long-lived Snell dwarf mice display a higher sensitivity to the lethal effects of endoplasmic reticulum (ER) stressors, such as thapsigargin, than cells from normal mice. Here we show that thapsigargin induces higher expression of CHOP, enhanced cleavage of caspase-12, higher caspase-3 activity, and increased phosphorylation of c-JUN, all indicators of enhanced apoptosis, in dwarf fibroblasts. Dwarf and normal fibroblasts show no genotypic difference in up-regulating BiP, GRP94, and ERp72 proteins after exposure to thapsigargin. However, dwarf fibroblasts express lower basal levels of a number of putative XBP1 target genes including Armet, Edem1, Erdj3, p58IPK and Sec61a1, as well as Ire1α itself. Furthermore, when exposed to thapsigargin, dwarf fibroblasts display attenuated splicing of Xbp1, but similar phosphorylation of eIF2α, in comparison to normal fibroblasts. These data support the notion that IRE1/XBP1 signaling is set at a lower level in dwarf fibroblasts. Diminished Xbp1 splicing in dwarf-derived fibroblasts may tilt the balance between prosurvival and proapoptotic signals in favor of apoptosis, thereby leading to higher induction of proapoptotic signals in these cells and ultimately their increased sensitivity to ER stressors. These results, together with recent findings in Caenorhabditis elegans daf-2 mutants, point to a potential interplay between insulin/IGF-1 signals and unfolded protein response signaling.
Keywords: Aging, Apoptosis, C. elegans Genetics, Caspase, ER Stress, Pituitary Gland, Dwarf, IGF-I, XBP1, daf-2
Introduction
Snell dwarf mice (dw/dw) bear a homozygous mutation in the gene encoding the transcription factor Pit-1 (1). This results in abnormal development of the pituitary gland and an altered hormonal profile with primary deficiencies in growth hormone, thyroid-stimulating hormone, and prolactin, and consequent secondary deficiencies in the circulating levels of insulin-like growth factor 1 (IGF-1)4 and thyroxine. These mice live about 40% longer than their littermate controls and display a delay in a number of age-dependent pathologies (2). Previous work from our laboratory has shown that tail skin-derived primary fibroblasts from Snell dwarf mice have an increased resistance to cell death induced by a number of stressors including heat, H2O2, paraquat, cadmium, UV light, and the DNA alkylating agent methylmethanesulfonate (3, 4). In addition, they are relatively more resistant to certain non-lethal forms of stress such as depleting glucose from the growth medium or incubation with rotenone (5). Surprisingly, dwarf-derived fibroblasts are more sensitive to the lethal effects of the endoplasmic reticulum (ER) stressors thapsigargin and tunicamycin; in this respect they resemble fibroblasts derived from naked mole rats, the longest-living rodent species on record (6).
The accumulation of unfolded or misfolded proteins in the endoplasmic reticulum causes ER stress, and initiates a concerted adaptive program known as the unfolded protein response (UPR). The UPR alleviates ER stress by down-regulating the synthesis of secreted proteins, up-regulating ER chaperone and foldase expression levels, and activating ER-associated degradation, thereby easing the burden on the stressed ER by reducing its protein load, increasing its folding capacity, and eliminating irreparably misfolded proteins (7–9). In higher eukaryotes, the proximal transducers of ER stress are PERK, a double-stranded RNA-activated protein kinase-like ER kinase; IRE1, a protein kinase/endoribonuclease; and ATF6, a basic leucine zipper transcription factor. Each of the proximal transducers serves a distinct role in the unfolded protein response. The most rapid outcome is that of translational repression. This is mediated by activated PERK through the phosphorylation of eIF2α and takes effect as early as 30 min after exposure to ER stress (10, 11). Paradoxically, activation of PERK enhances the efficiency of ATF4 translation. ATF4 is the point of convergence of multiple stress pathways. Most importantly, it up-regulates UPR genes involved in amino acid transport and resistance to oxidative stress (12). IRE1 exerts its cytoprotective effect mainly by removing a 26-base intron from the mRNA encoding X-box-binding protein 1 (XBP1) (13, 14). The spliced Xbp1 encodes a potent transcription factor whose targets encode several proteins involved in ER protein folding and the degradation of misfolded ER proteins (15, 16). In response to ER stress, the transmembrane portion of ATF6 is cleaved by S1P and S2P proteases that reside in the Golgi apparatus (17). The cleaved fragment moves to the nucleus and acts as a transcription factor mainly in parallel with the spliced Xbp1 product to up-regulate genes that increase ER chaperone activity and the degradation of misfolded proteins (18, 19).
All ER stressors identified so far initiate both cytoprotective and proapoptotic outputs from the UPR (20). One proapoptotic output of particular importance is the activation of the C/EBP homologous protein (CHOP). Expression of CHOP is mainly regulated at the transcriptional level (21); its promoter can be engaged by all the major inducers of the UPR including ATF4, spliced ATF6, and XBP1 derived from spliced Xbp1 mRNA (22). Although PERK/eIF2α signaling plays the dominant role in this process, maximal induction of CHOP requires the contribution of all three signaling pathways. Upon binding the adaptor molecule TRAF2, IRE1 activates the apoptosis signal-regulating kinase 1. The IRE1-TRAF2-ASK1 trimeric complex then contributes to the apoptotic response by activating c-JUN N-terminal kinase (23, 24). In addition, the binding of IRE1 to TRAF-2 reverses the clustering of caspase-12 and leads to its cleavage (25), which in turn causes the sequential cleavage of caspase-9 and the effector caspase, caspase-3 (26). Finally, alterations in the handling of Ca2+ by the ER can play a role in initiating an apoptotic response (27).
The present study takes our previous findings concerning the higher sensitivity of primary fibroblasts from Snell dwarf mice to ER stress-induced cell death (6) as its point of departure, and uses immunoblotting, luminescence assays, measurement of cytosolic Ca2+, as well as conventional and quantitative PCR to explore the basis for this phenomenon at the RNA and protein levels.
EXPERIMENTAL PROCEDURES
Animals
Snell dwarf (Pitdw/dw) mice and their heterozygotic (Pitdw/+) littermate controls were bred as the progeny of (DW/J × C3H/HeJ)-dw/+ females and (DW/J × C3H/HeJ)F1-dw/dw males. Their sires had been previously treated with growth hormone and thyroxine to increase body size and fertility. Littermates with the (+/dw) genotype were used as controls. Tail skin biopsies were taken from 3–4-month-old male mice (4).
Antibodies
Antibodies against β-actin (clone 13E5), caspase-12, CHOP (clone L63F7), phospho-eIF2α (clone 119A11), eIF2α (clone L57A5), ERp72, GAPDH (clone 14C10), GRP94, and PDI were obtained from Cell Signaling Technology (Danvers, MA). Anti-BiP (clone 40) was from BD Biosciences. Anti-phospho-c-JUN (clone KM1), and alkaline phosphatase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Exposure to Thapsigargin
Fibroblasts were obtained from tail skin biopsies of 3–4-month-old male dwarf mice and their normal counterparts, grown in vitro to the third passage (4), and plated in 6-well tissue culture plates (5 × 105 cells per well) in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum, 100 units/ml of penicillin, 100 μg/ml of streptomycin, 0.25 μg/ml of fungizone, 1 mm sodium pyruvate, and 10 mm HEPES at 37 °C in a humidified incubator with 10% CO2 in the air. After an 18-h overnight incubation, the cells were either left untreated or exposed to 100 nm thapsigargin (EMD Chemicals, Gibbstown, NJ) for varying time periods. Exposures lasted 10 min, 30 min, 1 h, 2 h, 4 h, 8 h, or 24 h for samples destined for Western blot analysis; and 30 min, 1 h, 2 h, 4 h, or 8 h for the Xbp1 splicing assay. Although the period of exposure to the stressor varied between different samples, the experiments were planned so that all samples were in culture for the same length of time.
Western Blot Analysis
At the end of the indicated treatments, fibroblasts were washed in ice-cold PBS (2 times), and subsequently lysed in ice-cold modified RIPA buffer (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS) supplemented with a protease inhibitor mixture comprising 500 μm 4-(2-aminoethyl)benzenesulfonyl fluoride, 500 nm aprotinin, 20 μm elastatinal, 1 μm E-64, 1 μm leupeptin, and 1 μm Glu-Gly-Arg-Chlormethyl ketone (EMD Chemicals). The cell lysates were subjected to 2 rounds of centrifugation at 12,000 × rpm for 10 min. The BCA protein assay (Thermo Fisher) was used to determine the protein concentration of each of the cleared lysates. Fibroblast cell lysates were boiled for 5 min in reducing sample buffer containing DTT and resolved by SDS-PAGE electrophoresis, transferred to PVDF membranes, and probed with the indicated antibodies. The membranes were exposed to enhanced chemifluorescence substrate (GE Healthcare), followed by scanning on a Storm 840 imaging system (GE Healthcare) to obtain a digital image of the probed protein. The bands were then quantified with ImageQuant software (GE Healthcare).
Detection of Xbp1 mRNA Splicing
Xbp1 mRNA splicing was assessed as previously described (28), with a slight modification. Briefly, the SuperScript III RT-PCR kit (Invitrogen) was used according to the manufacturer's instructions to amplify both unspliced and spliced Xbp1 in mRNA samples prepared at the end of the indicated treatments. The primers used in the assay flanked the Xbp1 intron and had the following sequences: upstream, ttgtggttgagaaccagg; downstream, tccatgggaagatgttctgg. As in the case of Western blots, the bands were quantified with ImageQuant software.
Assessment of Caspase-3/-7 Activity
Fibroblasts from dwarf and normal mice grown to the third passage were plated in 96-well tissue culture plates (3 × 104 cells per well) in complete DMEM. After an 18-h overnight incubation, the cells were either left untreated or exposed to 0.3, 1, or 3 μm thapsigargin for 6 h. The Caspase-Glo 3/7 homogenous luminescent assay system (Promega, Madison, WI) was used in accordance with the manufacturer's instructions to measure caspase-3/-7 activity.
Measurement of Cytosolic Ca2+
Cytosolic Ca2+ measurement was performed as previously described (28), with some modifications. Mouse fibroblasts were loaded with 2.5 μm Indo-1 AM (EMD Chemicals) at 37 °C for 30 min, washed, and then re-suspended in Hanks' balanced salt solution (HBSS), either containing Ca2+ (for the full response) or devoid of it (to evaluate release of Ca2+ from internal stores). 1 mm EGTA (Anaspec, San Jose, CA) was added to each of the samples evaluated for internal Ca2+ release 1 min prior to starting the Ca2+ mobilization assay. The Ca2+ mobilization assay was conducted at 37 °C on a FACSVantage flow cytometer using FACSDiva software (BD Biosciences). Baseline levels were collected for 30 s, after which the cells were stimulated with 100 nm thapsigargin. Each assay was performed for a 5-min period. The acquired data were analyzed using a FlowJo kinetic platform (TreeStar, Ashland, OR). Each trace displays the mean of the violet:blue ratio as a function of time with smoothing for moving average.
Quantification of mRNA Levels Using Multiplex Quantitative RT-PCR
The RNeasy Mini kit (Qiagen, Valencia, CA) was used to isolate RNA from the fibroblast samples. An Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA) was used to determine the concentration and quality of each RNA isolate. Complementary DNA (cDNA) was generated from each RNA sample using the High Capacity cDNA reverse transcription kit (Applied Biosystems). 100 μl of master mixture containing 0.5 μg of each cDNA reaction product was loaded onto one port of Applied Biosystems multiplex (microfluidics-based) reverse transcription-polymerase chain reaction (RT-PCR) cards custom-made to contain replicate sets of 48 primer pairs. The cards were run on the ABI Prism 7900HT Sequence Detection System and analyzed with version 2.1.1 of the software. The relative RNA expression levels were inferred from the Ct values.
Statistical Analyses
Differences in protein expression or phosphorylation between dwarf and normal fibroblasts, as well as the extent of Xbp1 splicing and caspase-3 activity, were evaluated by using repeated measures analysis of variance at p = 0.05 with genotype and time (or genotype and dose for caspase-3 activity) as the two factors. The performed analysis of variances excluded the baseline. The analysis determines the significance of difference in response between dwarf and normal fibroblasts (G = genotype) after exposure to thapsigargin. It also tests the significance of time (T) after exposure. Lastly, the significance of the interaction term (I) evaluates the extent to which the difference in response to thapsigargin between dwarf and normal cells is time-dependent. In the case of Ca2+ assays, the mean response was calculated for each sample, with the data then subjected to a two-tailed Mann-Whitney test at p = 0.05. The multiplex quantitative RT-PCR data were normalized as previously described (29). Briefly, the variation among the cards was adjusted by defining a normalization constant for each card based upon the mean Ct value of the 16 mRNAs that had the highest mRNA abundance (lowest Ct values) across the entire series of cards. Each individual Ct value was then adjusted by adding in this card-specific normalization factor, so that each card had the same average estimate of mRNA for the 16 most abundant mRNAs. The statistical analysis for microarray software developed by Tusher and colleagues (30) was then used to compare the basal expression levels of each gene between the two genotypes. In each case, genes for which the false discovery rate ≤0.01 were considered significant.
RESULTS
Exposure to Thapsigargin Leads to Higher Induction of Proapoptotic Signals in Dwarf Fibroblasts
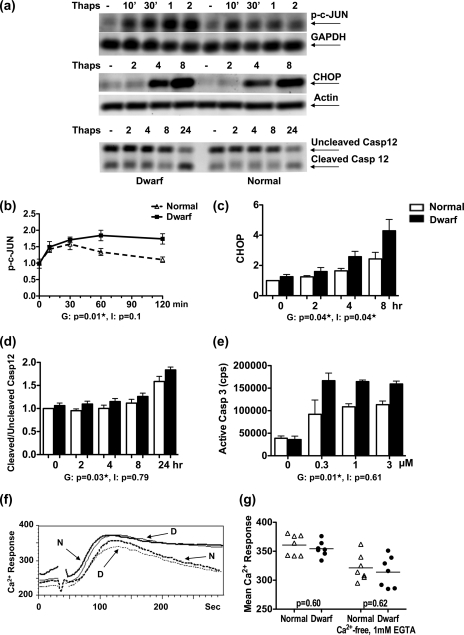
As a first step towards elucidating the underlying mechanism for the higher sensitivity of dwarf fibroblasts to ER stressors, fibroblasts from dwarf and normal mice were examined for their proapoptotic output in response to thapsigargin. These included assessment of the time course for c-JUN phosphorylation, CHOP expression, and caspase-12 cleavage with Western blots; caspase-3 activity with a luminescence assay; and Ca2+ mobilization and influx using flow cytometry. c-JUN phosphorylation could be detected as early as 10 min after exposure to thapsigargin in both sets of fibroblasts (Fig. 1, a and b). In normal cells, c-JUN phosphorylation peaked at 30 min and returned to baseline levels by 2 h after exposure to thapsigargin. In contrast, cells from dwarf mice reached a higher peak level of phosphorylation, and maintained this level through the 2-h period of assessment. This genotype effect was significant (p = 0.01). In the case of CHOP, induction of the protein could be seen in both genotypes at the 4-h time point, and increased still further by 8 h after exposure to thapsigargin (Fig. 1, a and c). Here, too, the dwarf cells showed a significantly higher response than cells from littermate controls (genotype, p = 0.04; time, p < 0.0001; interaction, p = 0.04).
FIGURE 1.
Heightened induction of proapoptotic signals in dwarf fibroblasts after exposure to thapsigargin. Dwarf and normal fibroblasts were either left untreated or exposed to 100 nm thapsigargin (Thaps) for the indicated times and evaluated for phosphorylation of c-JUN (n = 4 pairs), expression of CHOP (n = 6 pairs), and cleavage of caspase-12 (n = 6 pairs) by immunoblotting. a shows a representative image for each of these assays. The graphs in b-d represent the mean ± S.E. of the response for each of these molecules. e shows the amount of active caspase-3 in dwarf and normal fibroblasts after 6 h of exposure to 0.3, 1, and 3 μm thapsigargin (n = 3). f shows the Ca2+ response of one pair of dwarf and normal fibroblasts to 100 nm thapsigargin when incubated in HBSS containing calcium (continuous traces) or in calcium-free HBSS supplemented with EGTA (dotted traces). g shows the Ca2+ response for all the tested pairs of dwarf and normal fibroblasts. The result for the analysis of variance for each molecule can be seen under its graph. G stands for genotype and pertains to the difference in response between normal and dwarf cells. I is short for interaction and shows that the extent of the difference in response to thapsigargin between dwarf and normal cells is time-dependent (or dose-dependent in the case of caspase-3). A p value of ≤0.05 indicates a significant difference between dwarf and normal cells for each term and is marked with a ★.
Cleavage of caspase-12 was evident after 24 h of exposure to thapsigargin (Fig. 1, a and d) and was significantly higher in dwarf-derived fibroblasts (genotype, p = 0.03; time, p = <0.0001; interaction, p = 0.79). The activity of caspase-3/-7 was studied in dwarf and normal fibroblasts that were exposed to 0, 0.3, 1, or 3 μm thapsigargin for 6 h (Fig. 1e). Fibroblasts from both dwarf and normal mice showed an increase in caspase-3/-7 activity after exposure to each dose of the stressor, with dwarf-derived fibroblasts displaying significantly higher caspase-3/-7 activity than their normal counterparts (genotype, p = 0.01; dose, p = 0.84; interaction, p = 0.61).
We also assessed the internal release of Ca2+, as well as its influx, in response to thapsigargin in dwarf and normal fibroblasts (Fig. 1, f and g). Fibroblasts loaded with Indo-1 were stimulated with thapsigargin while incubated either in HBSS containing calcium or in calcium-free HBSS supplemented with EGTA. The cells from both genotypes reached similar peak Ca2+ levels in each of the two media. The cells in HBSS containing calcium were able to maintain the peak during the 5-min period of evaluation, whereas cells placed in calcium-free HBSS supplemented with EGTA showed a gradual decline in their Ca2+ levels as intracellular stores were used up. There was no genotypic difference between the two sets of fibroblasts in either intracellular release or influx of Ca2+ (internal release, p = 0.62; influx, p = 0.60).
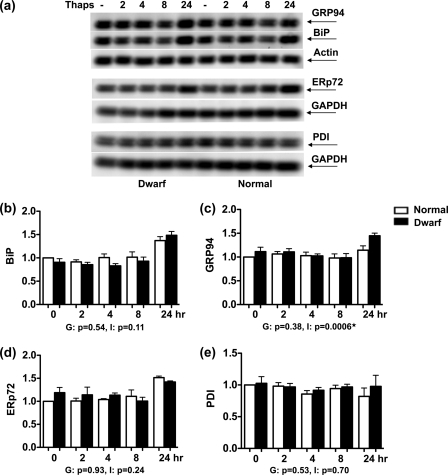
Enhanced Sensitivity of Dwarf Fibroblasts Occurs Despite Unimpaired Chaperone Up-regulation
Up-regulation of ER chaperones is the hallmark of UPR activation, and impaired chaperone up-regulation sensitizes cells to ER stress. Thus, the induction of BiP, GRP94, ERp72, and PDI in response to thapsigargin was studied over a 24-h time span. Of these four proteins, BiP and GRP94, the endoplasmic reticulum paralogue of HSP90, act as chaperones, whereas PDI and ERp72 are members of the ER oxidoreductase family (31). Dwarf- and normal-derived fibroblasts expressed similar baseline levels of all 4 proteins, i.e. at time 0. In both types of fibroblasts, the induction of BiP, GRP94, and ERp72 was only observed at the 24-h time point (Fig. 2, a and b–d). There was no genotypic difference between the two sets of fibroblasts in their production of BiP (genotype, p = 0.54; time, p < 0.0001; interaction, p = 0.11) or ERp72 (genotype, p = 0.93; time, p < 0.0001; interaction, p = 0.24). In the case of GRP94, there was no significant effect of genotype (p = 0.38) when all time points after thapsigargin exposure were considered together, but the significant interaction term (p = 0.0006) indicated a stronger induction of GRP94 in dwarf fibroblasts at the 24-h time point. There was no induction of PDI in response to thapsigargin at any time point (genotype, p = 0.53; time, p = 0.58; interaction, p = 0.70) (Fig. 2, a and e). Overall, these data rule out any gross impairment of chaperone induction in either genotype.
FIGURE 2.
Induction of the unfolded protein response in dwarf and normal fibroblasts upon exposure to thapsigargin. Dwarf and normal fibroblasts were either left untreated or exposed to 100 nm thapsigargin (Thaps) for the indicated times and evaluated for the expression of BiP (n = 4 pairs), GRP94 (n = 4 pairs), ERp72 (n = 4 pairs), and PDI (n = 4 pairs) by immunoblotting. a shows a representative image for each of these assessments. The graphs in b–e depict the mean ± S.E. of the response for each of the evaluated molecules. The result for the analysis of variance for each molecule can be seen under its graph. G and I stand for genotype and interaction, respectively. A p value of ≤0.05 indicates a significant difference between dwarf and normal cells for each term and is marked with a ★.
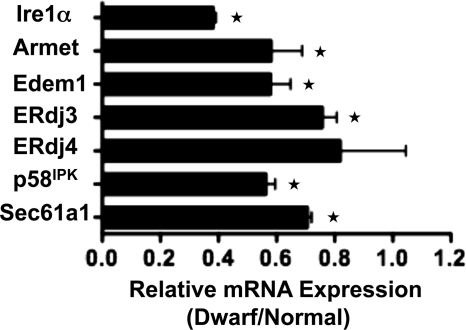
Dwarf-derived Fibroblasts Show Lower Basal Expression of Putative IRE1/XBP1 Targets
The UPR output goes beyond up-regulation of chaperones. In fact, there is accumulating evidence that other cellular signaling pathways can modulate UPR output even in the absence of overt ER stress. We thus used a multiplex quantitative RT-PCR approach to examine the baseline expression of ER stress-related genes in dwarf and normal fibroblasts. The list of selected genes included proximal ER stress sensors, ER-resident molecular chaperones, and co-chaperones, lectins, foldases of the disulfide isomerase subclass, bZIP transcription factors, and some of their interaction partners, genes involved in ER-associated degradation, Ca2+ homeostasis and membrane integrity, cell cycle and apoptosis-related molecules, as well as luminal transporters (see Table 1). A false discovery rate method that adjusts for the number of different mRNAs examined simultaneously was used to evaluate any significant difference in the basal expression of the genes under study between dwarf and normal fibroblasts. A number of the examined genes were expressed at a significantly lower level in dwarf fibroblasts. These included Atf3, Armet, Edem1, Erdj3, Erol1, Gclc, Gclm, Gadd45b, Ire1α, p58IPK, Pdia5, Sec61a1, Sec63a, and Wfs1. Although these genes fall into diverse groups, several of them, i.e. Armet, Edem1, Erdj3, p58IPK, and Sec61a1, have been identified as putative targets of XBP1 (15, 32, 33) (Fig. 3). XBP1 is itself the most proximal target of IRE1, whose basal mRNA expression level is significantly reduced in the dwarf fibroblasts (Fig. 3).
TABLE 1.
List of evaluated ER stress-related genes
The groupings of the genes are based on either the structural relationship of the particular set of genes or their established (or purported) functions.
| ER stress-related genes | |
|---|---|
| Proximal sensors | Atf4, Ire1α, Atf6, Ire1βa |
| Chaperones and co-chaperones | Bip, Grp94, p58IPK, Erdj3, Erdj4, Sil1 |
| eIF2α–ATF4 pathway | Atf3, Chop, Erol1, Wars |
| Luminal transporters | Sec61a1, Sec23b, Sec63, Slc39a6 |
| ER-associated degradation | Edem1, Herpud |
| Disulfide isomerase foldases | Pdi, Pdia5, Pdia6, Erp18, Erp29, Erp44, Erp57, Erp72 |
| Calcium homeostasis and membrane integrity | Bak1, Bax, Calr, Canx, Cherp, Wfs1 |
| Cell cycle related | Armet, Gadd45a, Gadd45b |
| Nrf2 related | Nrf2, Gclc, Gclm, Hmox1, Hsf1 |
| Miscellaneous | Nfil3, Sirt1, Nrf1, 18S, Actb, Gapdh |
a Ire1β was undetectable in fibroblasts of either genotype.
FIGURE 3.
Lower baseline expression of Ire1α and putative XBP1 target genes in dwarf-derived fibroblasts. Fibroblast cultures from three pairs of dwarf and normal mice were evaluated for the expression of ER stress-related genes using custom-made multiplex RT-PCR cards. Each bar represents the mean ± S.E. of the relative expression of the depicted gene. ★ denotes a significant difference in baseline expression levels between dwarf and normal fibroblasts, i.e. false discovery rate ≤0.01.
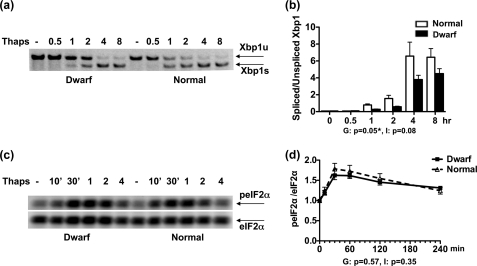
IRE1 Activation Is Specifically Impaired in Dwarf Fibroblasts
The lower baseline expression of the mRNAs for Ire1α and a number of putative XBP1 target genes led us to speculate that the “set point” of IRE1 signaling in dwarf fibroblasts should be lower than in normal fibroblasts. This means that the most proximal target of IRE1, i.e. splicing of Xbp1 mRNA, should be impaired in dwarf fibroblasts, whereas other proximal UPR pathways should remain intact. To test this hypothesis, we examined thapsigargin-induced Xbp1 splicing in dwarf and normal fibroblasts over an 8-h period. Spliced Xbp1 was first detected at the 1-h time point in both genotypes (Fig. 4, a and b). Later time points showed an increasing ratio of the spliced form in each set of fibroblasts. However, dwarf-derived fibroblasts spliced significantly lower amounts of Xbp1 than their normal counterparts (genotype, p = 0.05; time, p < 0.0001; interaction, p = 0.08), a finding consistent with the lower expression level of the mRNA for Ire1α.
FIGURE 4.
Xbp1 splicing and eIF2α phosphorylation in dwarf and normal fibroblasts upon exposure to thapsigargin. Dwarf and normal fibroblasts were either left untreated or exposed to 100 nm thapsigargin (Thaps) for the indicated times and evaluated for IRE1-mediated splicing of Xbp1 (n = 4 pairs) using RT-PCR, and for phosphorylation of eIF2α (n = 6 pairs) by immunoblotting. a and c show representative images for each of these assessments, respectively. The graphs in b and d depict the mean ± S.E. of the response of dwarf and normal fibroblasts for the evaluated molecules. The result for the analysis of variance for each molecule can be seen under its graph. G and I stand for genotype and interaction, respectively. A p value of ≤0.05 indicates a significant difference between dwarf and normal cells for each term and is marked with a ★.
Next we examined the effect of thapsigargin on the PERK-mediated phosphorylation of eIF2α, one of the earliest outcomes of exposure to ER stress. We monitored the phosphorylation of eIF2α in response to thapsigargin in dwarf- and normal-derived fibroblasts over a 4-h period. As shown in Fig. 4, c and d, thapsigargin induced the swift phosphorylation of eIF2α in both groups of fibroblasts, i.e. as early as 10 min. The response was at its peak at the 30–60-min time points after which it started to diminish. However, it was still above baseline after 4 h. The response was not significantly different between the two genotypes (genotype, p = 0.57; time, p < 0.0001; interaction, p = 0.35).
DISCUSSION
The current study sought to determine the biochemical basis for the higher sensitivity of primary skin fibroblasts from long-lived Snell dwarf mice to the lethal effects of ER stress. Results presented here show a higher induction of CHOP, increased cleavage of caspase-12 and caspase-3, and enhanced phosphorylation of c-JUN in dwarf-derived fibroblasts after exposure to thapsigargin, all indicators of a higher proapoptotic output in dwarf fibroblasts, and consistent with the higher sensitivity of these cells to cell death after ER stress. The two genotypes did not differ in either Ca2+ mobilization from internal stores, or in Ca2+ influx after exposure to thapsigargin. This rules out a similar role for Ca2+ signaling in the current model as the one it serves in the Bax inhibitor-1 (BI-1) knock-out model of sensitivity to ER stress (34, 35); yet it allows us to address an important mechanistic possibility. It can be argued that the differential sensitivity to thapsigargin between dwarf and normal fibroblasts is simply due to an altered pharmacokinetic response on the part of dwarf-derived fibroblasts. However, the Ca2+ data suggest that the differential sensitivity to ER stress between the two genotypes is caused by differences in their unfolded protein response, rather than a pharmacokinetic difference in the response to thapsigargin.
All ER stressors induce both prosurvival and proapoptotic outputs, with the fate of the cell depending on which set of signals prevails. Dwarf and normal fibroblasts both displayed an increased expression of the BiP, GRP94, and ERp72 proteins after exposure to thapsigargin. These responses showed no significant genotypic difference, and ruled out any gross impairment of the UPR translational output in either genotype. Furthermore, thapsigargin induced similar levels of eIF2α phosphorylation in dwarf and normal fibroblasts, whereas dwarf fibroblasts spliced lesser amounts of Xbp1 than their normal counterparts at the examined time points.
The lower basal expression of mRNAs for Ire1α, and the putative XBP1 target genes Armet, Edem1, ERdj3, p58IPK, and Sec61a1 (15, 32, 33), merits particular emphasis. These, together with the lesser splicing of Xbp1 in dwarf fibroblasts, suggest that the IRE1/XBP1 pathway is set at a lower level in dwarf fibroblasts than in their normal counterparts, a finding in concordance with recent observations in Caenorhabditis elegans insulin/IGF-1 (daf-2) signaling mutants (36). By contrast, similar levels of eIF2α phosphorylation in fibroblasts from dwarf and normal mice rules out any difference in the induction of PERK in the two genotypes. This, too, mirrors the findings in the daf-2 mutants.
The level of IRE1/XBP1 signaling plays an important role in the outcome of ER stress. Enhanced IRE1/XBP1 signaling confers resistance against ER stress-induced apoptosis (37), whereas a decline in its activity can lead to cell death as the outcome (38). Similarly, knocking down IRE1 or XBP1 enhances cell death after exposure to ER stress (39). On this basis, one could plausibly suggest that the lower baseline expression of Ire1α in dwarf-derived fibroblasts, together with diminished Xbp1 splicing in these cells, may tilt the balance between the prosurvival and proapoptotic signals in dwarf cells in favor of apoptosis, therefore leading to the higher induction of proapoptotic signals in dwarf-derived fibroblasts and ultimately their increased sensitivity to the cytotoxic effects of ER stressors.
It is becoming increasingly evident that the UPR plays a central homeostatic role in the normal physiology of vertebrates. By utilizing its multiple sensors and their respective downstream pathways, as well as cell- and condition-specific interactions with components of pathways other than the UPR, the unfolded protein response can generate nuanced and at times disparate responses tailored toward the physiologic functions of particular cells and organs and the different range of environmental parameters that they encounter (40). The control of diverse tissue- and condition-specific targets by key UPR effector(s) (41), and the formation of unique protein complexes through association of different adaptor and modulator proteins (37) are potential means of generating diverse outputs by the UPR in different tissues. Recent work from a number of laboratories has documented specific examples of how interaction with parallel signaling pathways can modulate the UPR output in a context-specific manner. Exposure to lipopolysaccharide, and the consequent activation of Toll-like receptor 4, leads to prolonged ER stress. Remarkably, through its cross-talk with the UPR, Toll-like receptor 4 signaling simultaneously leads to a significant increase in eIF2α phosphorylation and the selective suppression of CHOP, therefore allowing the cell to take advantage of the beneficial effects of prolonged physiological ER stress while avoiding its undesirable apoptotic consequences (42). The interaction of phosphoinositide 3-kinase (PI3K) with the UPR is another important example (43). The p85α subunit of PI3K can form a physical complex with XBP1, thereby promoting its stability and the accumulation of spliced XBP1 in the nucleus. The absence of p85α leads to lower ER stress-dependent accumulation of spliced XBP1 in the nucleus as well as increased rates of apoptosis.
Research conducted on various model organisms in the past two decades has identified a number of genetic mechanisms for lifespan control. The insulin/IGF-1 signaling pathway was the first of these to be discovered (44). Inhibiting insulin/IGF-1 signaling can extend lifespan and delay age-related disease in species throughout the animal kingdom. When comparing the data published on the ER stress response in daf-2 mutants (36) with our current findings in Snell dwarf mice, two points need particular emphasis. First, and more importantly, in C. elegans daf-2 mutants, which bear a mutation in insulin/IGF-1 signaling, IRE1/XBP1 activity is set at a lower level than in non-mutant nematodes. This finding is analogous to our finding in Snell dwarf mice in the current study. Secondly, despite the lower activity of IRE1/XBP1 in daf-2 mutants, these worms have an increased resistance to ER stress. The latter is contrary to the authors' own expectation, as well as the current literature on IRE1/XBP1 signaling, which associates lower IRE1/XBP1 activity with lower resistance to ER stress. They reconcile this inconsistency by showing that in daf-2 mutants, the increased resistance to ER stress is due to an interaction, directly or indirectly, between XBP1 and DAF-16. By contrast, in fibroblasts from Snell dwarf mice, lower IRE1/XBP1 signaling coincides with a lower resistance to ER stress, which is in line with the prevailing view on this issue. Reduced IRE1/XBP1 signaling in daf-2 mutants and Snell dwarf mice, two evolutionarily distant species, is indicative of a potential interplay between insulin/IGF-1 signaling and the unfolded protein response. The diametrically opposed effect of this lower activity on ER stress resistance in daf-2 mutants, where resistance to ER stress is unexpectedly increased, and Snell dwarf mice, where resistance to ER stress is decreased as expected, could be due to the previously unsuspected interaction of XBP1 and DAF-16 in the daf-2 mutants.
The use of primary fibroblasts as the in vitro component of an overall experimental approach in addressing the biology of aging has gained increasing recognition and momentum in the past few years (see Ref. 45 and references therein). Although it may seem implausible at first to think that the study of skin-derived fibroblasts in vitro could provide any useful insight about the mechanisms of aging in whole animals, there is increasing evidence that the properties of fibroblasts in culture do partly reflect the key characteristics of their respective species of origin. This concept served as our rationale for using skin-derived fibroblasts to study the response to ER stress in Snell dwarf mice and their wild-type counterparts. One prevailing theory proposes that the precise tuning of the threshold for the ER stress response could correlate with life span in organisms of different species, or among members of a cohort (46). In other words, a lower set point for entering apoptosis in response to ER stress in cells from long-lived mammals may allow them to exclude damaged cells more swiftly through programmed cell death, thereby providing them a survival advantage over their short-lived counterparts (47).
Nonetheless, all in vitro models have their inherent limitations. This is because, in strict terms, they reflect the characteristics of a single cell type, whereas different cell types can potentially respond in variegated and unique ways to the same stimulus in vivo. Therefore, as in any model, the relevance of these in vitro findings to the whole organism has to be assessed at the in vivo level. It is important to establish the extent to which insulin/IGF-1 signaling contributes to setting the threshold of sensitivity to ER stress and whether this is done differentially, with or without interaction with disparate targets, in different tissues and/or organisms. To this end, our future efforts will first focus on determining the degree to which the in vitro fibroblast findings are recapitulated in vivo in different organs of the Snell dwarf mice. This could lead to a better understanding of the interplay of insulin/IGF-1 signaling with the ER stress response and provide valuable information about the role of UPR in Snell dwarf longevity.
Acknowledgments
We thank David Adams of the University of Michigan Flow-Cytometry Core for invaluable help in performing the Ca2+ assays; Kathleen Welch from the University of Michigan Center for Statistical Consultation and Research (CSCAR) for advice on statistical analyses; and Margaret Lauderdale and Lisa Burmeister for breeding the Snell dwarf mice and their normal counterparts.
This work was supported, in whole or in part, by National Institutes of Health Grants AG031736, AG024824, AG019899 (to R. A. M.), and DK084058 and a Carver Trust Medical Research Initiative award (to D. T. R.), a University of Michigan Nathan Shock Center Pilot grant (to J. M. H.), and a University of Michigan Claude Pepper Older Americans Independence Center RCDC Junior faculty award (to A. A. S. A.).
- IGF-1
- insulin-like growth factor 1
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response
- CHOP
- C/EBP homologous protein
- PDI
- protein-disulfide isomerase
- HBSS
- Hanks' balanced salt solution
- XBP1
- X-box-binding protein 1.
REFERENCES
- 1. Li S., Crenshaw E. B., 3rd, Rawson E. J., Simmons D. M., Swanson L. W., Rosenfeld M. G. (1990) Nature 347, 528–533 [DOI] [PubMed] [Google Scholar]
- 2. Flurkey K., Papaconstantinou J., Miller R. A., Harrison D. E. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6736–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murakami S., Salmon A., Miller R. A. (2003) FASEB J. 17, 1565–1566 [DOI] [PubMed] [Google Scholar]
- 4. Salmon A. B., Murakami S., Bartke A., Kopchick J., Yasumura K., Miller R. A. (2005) Am. J. Physiol. Endocrinol. Metab. 289, E23–29 [DOI] [PubMed] [Google Scholar]
- 5. Leiser S. F., Salmon A. B., Miller R. A. (2006) Mech. Ageing Dev. 127, 821–829 [DOI] [PubMed] [Google Scholar]
- 6. Salmon A. B., Sadighi Akha A. A., Buffenstein R., Miller R. A. (2008) J. Gerontol. A Biol. Sci. Med. Sci. 63, 232–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schröder M., Kaufman R. J. (2005) Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 8. Marciniak S. J., Ron D. (2006) Physiol. Rev. 86, 1133–1149 [DOI] [PubMed] [Google Scholar]
- 9. Lin J. H., Walter P., Yen T. S. (2008) Annu. Rev. Pathol. 3, 399–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harding H. P., Zhang Y., Bertolotti A., Zeng H., Ron D. (2000) Mol. Cell 5, 897–904 [DOI] [PubMed] [Google Scholar]
- 11. Novoa I., Zhang Y., Zeng H., Jungreis R., Harding H. P., Ron D. (2003) EMBO J. 22, 1180–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 13. Sidrauski C., Walter P. (1997) Cell 90, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 14. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 15. Lee A. H., Iwakoshi N. N., Glimcher L. H. (2003) Mol. Cell. Biol. 23, 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oda Y., Okada T., Yoshida H., Kaufman R. J., Nagata K., Mori K. (2006) J. Cell Biol. 172, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ye J., Rawson R. B., Komuro R., Chen X., Davé U. P., Prywes R., Brown M. S., Goldstein J. L. (2000) Mol. Cell 6, 1355–1364 [DOI] [PubMed] [Google Scholar]
- 18. Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. (2007) Dev. Cell 13, 365–376 [DOI] [PubMed] [Google Scholar]
- 19. Wu J., Rutkowski D. T., Dubois M., Swathirajan J., Saunders T., Wang J., Song B., Yau G. D., Kaufman R. J. (2007) Dev. Cell 13, 351–364 [DOI] [PubMed] [Google Scholar]
- 20. Kim I., Xu W., Reed J. C. (2008) Nat. Rev. Drug Discov. 7, 1013–1030 [DOI] [PubMed] [Google Scholar]
- 21. Oyadomari S., Mori M. (2004) Cell Death Differ. 11, 381–389 [DOI] [PubMed] [Google Scholar]
- 22. Ma Y., Brewer J. W., Diehl J. A., Hendershot L. M. (2002) J. Mol. Biol. 318, 1351–1365 [DOI] [PubMed] [Google Scholar]
- 23. Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H. P., Ron D. (2000) Science 287, 664–666 [DOI] [PubMed] [Google Scholar]
- 24. Nishitoh H., Matsuzawa A., Tobiume K., Saegusa K., Takeda K., Inoue K., Hori S., Kakizuka A., Ichijo H. (2002) Genes Dev. 16, 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoneda T., Imaizumi K., Oono K., Yui D., Gomi F., Katayama T., Tohyama M. (2001) J. Biol. Chem. 276, 13935–13940 [DOI] [PubMed] [Google Scholar]
- 26. Rao R. V., Castro-Obregon S., Frankowski H., Schuler M., Stoka V., del Rio G., Bredesen D. E., Ellerby H. M. (2002) J. Biol. Chem. 277, 21836–21842 [DOI] [PubMed] [Google Scholar]
- 27. Demaurex N., Distelhorst C. (2003) Science 300, 65–67 [DOI] [PubMed] [Google Scholar]
- 28. Rutkowski D. T., Arnold S. M., Miller C. N., Wu J., Li J., Gunnison K. M., Mori K., Sadighi Akha A. A., Raden D., Kaufman R. J. (2006) PLoS Biol. 4, e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun L., Sadighi Akha A. A., Miller R. A., Harper J. M. (2009) J. Gerontol. A Biol. Sci. Med. Sci. 64, 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tusher V. G., Tibshirani R., Chu G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mazzarella R. A., Srinivasan M., Haugejorden S. M., Green M. (1990) J. Biol. Chem. 265, 1094–1101 [PubMed] [Google Scholar]
- 32. Shaffer A. L., Shapiro-Shelef M., Iwakoshi N. N., Lee A. H., Qian S. B., Zhao H., Yu X., Yang L., Tan B. K., Rosenwald A., Hurt E. M., Petroulakis E., Sonenberg N., Yewdell J. W., Calame K., Glimcher L. H., Staudt L. M. (2004) Immunity 21, 81–93 [DOI] [PubMed] [Google Scholar]
- 33. Lee A. H., Scapa E. F., Cohen D. E., Glimcher L. H. (2008) Science 320, 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chae H. J., Kim H. R., Xu C., Bailly-Maitre B., Krajewska M., Krajewski S., Banares S., Cui J., Digicaylioglu M., Ke N., Kitada S., Monosov E., Thomas M., Kress C. L., Babendure J. R., Tsien R. Y., Lipton S. A., Reed J. C. (2004) Mol. Cell 15, 355–366 [DOI] [PubMed] [Google Scholar]
- 35. Xu C., Xu W., Palmer A. E., Reed J. C. (2008) J. Biol. Chem. 283, 11477–11484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Henis-Korenblit S., Zhang P., Hansen M., McCormick M., Lee S. J., Cary M., Kenyon C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 9730–9735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gupta S., Deepti A., Deegan S., Lisbona F., Hetz C., Samali A. (2010) PLoS Biol. 8, e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin J. H., Li H., Yasumura D., Cohen H. R., Zhang C., Panning B., Shokat K. M., Lavail M. M., Walter P. (2007) Science 318, 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lisbona F., Rojas-Rivera D., Thielen P., Zamorano S., Todd D., Martinon F., Glavic A., Kress C., Lin J. H., Walter P., Reed J. C., Glimcher L. H., Hetz C. (2009) Mol. Cell 33, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rutkowski D. T., Hegde R. S. (2010) J. Cell Biol. 189, 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Acosta-Alvear D., Zhou Y., Blais A., Tsikitis M., Lents N. H., Arias C., Lennon C. J., Kluger Y., Dynlacht B. D. (2007) Mol. Cell 27, 53–66 [DOI] [PubMed] [Google Scholar]
- 42. Woo C. W., Cui D., Arellano J., Dorweiler B., Harding H., Fitzgerald K. A., Ron D., Tabas I. (2009) Nat. Cell Biol. 11, 1473–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Winnay J. N., Boucher J., Mori M. A., Ueki K., Kahn C. R. (2010) Nat. Med. 16, 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kenyon C. (2011) Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miller R. A., Williams J. B., Kiklevich J. V., Austad S., Harper J. M. (2011) Ageing Res. Rev. 10, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sierra F. (2006) J. Gerontol. A Biol. Sci. Med. Sci. 61, 557–561 [DOI] [PubMed] [Google Scholar]
- 47. Miller R. A. (2009) J. Gerontol. A Biol. Sci. Med. Sci. 64, 179–182 [DOI] [PMC free article] [PubMed] [Google Scholar]