Abstract
To react to distinct stress situations and to prevent the accumulation of misfolded proteins, all cells employ a number of proteases and chaperones, which together set up an efficient protein quality control system. The functionality of proteins in the cell envelope of Escherichia coli is monitored by the HtrA proteases DegS, DegP, and DegQ. In contrast with DegP and DegS, the structure and function of DegQ has not been addressed in detail. Here, we show that substrate binding triggers the conversion of the resting DegQ hexamer into catalytically active 12- and 24-mers. Interestingly, substrate-induced oligomer reassembly and protease activation depends on the first PDZ domain but not on the second. Therefore, the regulatory mechanism originally identified in DegP should be a common feature of HtrA proteases, most of which encompass only a single PDZ domain. Using a DegQ mutant lacking the second PDZ domain, we determined the high resolution crystal structure of a dodecameric HtrA complex. The nearly identical domain orientation of protease and PDZ domains within 12- and 24-meric HtrA complexes reveals a conserved PDZ1 → L3 → LD/L1/L2 signaling cascade, in which loop L3 senses the repositioned PDZ1 domain of higher order, substrate-engaged particles and activates protease function. Furthermore, our in vitro and in vivo data imply a pH-related function of DegQ in the bacterial cell envelope.
Keywords: Allosteric Regulation, Crystal Structure, Enzyme Mechanisms, Protein Degradation, Serine Protease, Bacterial Cell Envelope, HtrA, Protein Quality Control
Introduction
The accumulation of misfolded and aggregated proteins hampers important biological processes and can lead to cellular malfunctions and even cell death (1, 2). To cope with conditions that interfere with protein structure and function, all cells employ molecular chaperones that support the refolding of non-native polypeptides or cooperate with proteases to eliminate irreversibly damaged proteins (3, 4). The protein quality control system constituted by these factors is not only important for survival under stress but also to perform important housekeeping functions in various cellular compartments. The key factors promoting protein quality control in extracytoplasmic compartments belong to the family of high temperature requirement (HtrA) proteases (5, 6). Prokaryotic HtrAs have been implicated in tolerance to various folding stresses as well as to pathogenicity (7–14), whereas defects in human HtrAs are correlated with protein folding diseases including Alzheimer and Parkinson diseases, arthritis, and neuromuscular disorders (15–22). HtrA proteases have a chymotrypsin-like protease as their catalytic domain and one or two accessory PDZ domains (23) implicated in substrate binding and controlling protease function. HtrA proteases form large molecular assemblies that range from trimers of 100 kDa to 24mers of 1.2 MDa (24–30). They either function as regulatory proteases cleaving specific substrates with pronounced specificity or act as general proteases reducing the levels of misfolded proteins (5, 31).
Even though all HtrA proteases exhibit a similar domain architecture, share a common trimeric building block, and are controlled by a conserved activation mechanism (32), they are involved in diverse biological pathways including protein quality control, outer membrane protein biogenesis, unfolded protein response, apoptosis, cell growth, tumor progression, and the metabolism of amyloid precursor protein (5, 16, 31, 33–34).
The functionality of proteins in the cell envelope, the periplasm, of Escherichia coli is monitored by three HtrA proteases, namely DegS, DegP, and DegQ. DegS is a regulatory protease that is tethered to the cytoplasmic membrane via one transmembrane segment. It senses and binds mislocalized outer membrane proteins. The bound outer membrane proteins function as allosteric activators triggering the DegS-mediated cleavage of RseA, thereby initiating the bacterial unfolded protein response (30, 35–38). The DegP protease chaperone is a heat shock protein that represents the key protein quality control factor in the bacterial cell envelope, eliminating severely damaged proteins (39–41) while in parallel promoting outer membrane protein biogenesis (26). In contrast with the membrane anchored DegS that occurs as a stable trimer (30), DegP can reversibly switch between different oligomeric forms that represent inactive (6-mer) and active (12- and 24-mer) protease states (24, 26). The third HtrA protease of the E. coli, DegQ, is homologous to DegP comprising one protease and two PDZ domains. Both proteases have similar substrate specificities and cleave misfolded protein substrates (42). Consistently, it has been shown that overproduction of DegQ rescues the temperature-sensitive growth defect of a degP null strain (43). Because sequence comparison of active site loops suggests that general HtrA proteases are most closely related to DegQ (31), DegQ appears to be the ideal model system to study the general principles of HtrA protease regulation. To address this point and to delineate the precise function of DegQ in the bacterial cell envelope, we performed a detailed structural and biochemical analysis of DegQ. Our data suggest that HtrA proteases involved in protein quality control are under control of substrate-induced oligomer reassembly, irrespective whether they have one or two PDZ domains. Moreover, we present structural data illustrating the molecular architecture of a catalytically active dodecamer. This DegQ12 structure suggests that the signaling cascade leading to the protease activation of 12- and 24-mer HtrA complexes is conserved and depends on the precise positioning of the PDZ1 domain upon formation of substrate-engaged HtrA particles.
EXPERIMENTAL PROCEDURES
Construct Design
The degQ constructs (DegQ full-length residues 1–438, QProtPDZ1 residues 1–337, and QProt residues 1–237) lacking the native signal sequence were PCR-amplified from the genomic DNA of strain DH5α and cloned into pET26b(+) (Novagen) vector encoding a N-terminal pelB signal sequence for periplasmic localization of the recombinant protein and an additional C-terminal His6 tag for affinity purification. All of the point mutations were introduced using a QuikChange site-directed mutagenesis kit (Stratagene). All of the constructs were verified by DNA sequence analysis.
Protein Expression and Purification
All of the recombinant DegQ variants were overexpressed in the E. coli strain BL21(DE3). Cells were grown at 37 °C in LB medium and induced with 1 mm isopropyl β-d-thiogalactopyranoside for 4 h at an A600 of 0.6. Harvested cells were resuspended in 300 mm NaCl, 50 mm sodium phosphate buffer, pH 8.0, and disrupted by sonication. The cleared lysate was loaded on a nickel-nitrilotriacetic acid column (Qiagen), and DegQ was eluted by applying a stepwise imidazol gradient. The eluate fraction containing 150 mm imidazole was concentrated using VIVASPIN (Vivascience) concentrators (molecular mass cut-off, 50 kDa) and applied to a Superdex 200 column (prep grade; GE Healthcare) equilibrated with 50 mm HEPES/NaOH, pH 7.5, 150 mm NaCl. Protein purity and monodispersity was judged by SDS-PAGE and dynamic light scattering (DynaPro-801; Protein Solutions Inc.), respectively. Recombinant DegP used in the pH-dependent casein degradation assay was purified as described previously (40).
Crystallization and Structure Solution
A deletion construct of DegQ lacking the second PDZ domain (QProtPDZ1) was crystallized at 19 °C using the sitting drop vapor diffusion method by mixing 2.5 μl of a 30 mg/ml protein solution with 2.5 μl of a reservoir solution containing 24% PEG600, 5% PEG1000, 10% glycerol, 0.1 m MES/NaOH,2 pH 5.4. Crystals appeared within a few days and could be directly flash frozen in liquid nitrogen because of the high content of cryoprotectants in the crystallization solution. The diffraction data were collected at the European Synchrotron Radiation Facility (Beamline 14-4, Grenoble, France). The data were integrated using DENZO and scaled with SCALEPACK (44). The crystals belonged to space group P31 with cell parameters a = b = 115.3 Å, c = 287.4 Å and contained 12 protomers in the asymmetric unit. The structure was determined by molecular replacement using the program Phaser (45) and the DegP protease and PDZ1 domain as separate search models (Protein Data Bank code 1ky9). Parts of the loop LA (residues 35–57), the first 10 N-terminal residues, and the last three C-terminal residues were not resolved and omitted in the final structure. In addition, two oligopeptides were observed in the peptide-binding sites of the protease and PDZ1 domain and built-in as six-residue polyalanine models.
The QProt crystals were grown using the same method by mixing 3 μl of a 35-mg/ml protein solution with 1.5 μl of a reservoir solution containing 1.6 m ammonium sulfate, 3% PEG400, 0.1 m HEPES/NaOH, pH 7.0. The data were collected in-house on a MarResearch image plate at room temperature with crystals mounted in a glass capillary. Data were integrated using DENZO and scaled with SCALEPACK (44). Crystals belonged to space group P31 with cell parameters of a = b = 70.9 Å and c = 152.0 Å and three QProt protomers per asymmetric unit. The crystal structure was solved by molecular replacement using the program Phaser (45) and the DegP protease domain (Protein Data Bank code 1ky9) as a search model. Because of the lack of interpretable electron density, parts of the loop LA (residues 30–63), loop L2 (residues 207–212), and the first ten N-terminal residues are absent in the final model.
In both cases, the models were built with O (46) and refined with CNS (47). Data collection, phasing, and refinement statistics are summarized in Table 1. Accordingly, the two protein structures exhibit good stereochemistry and have no outliers in the Ramachandran plot (48). Coordinates of the QProt and QProtPDZ1 crystal structures have been deposited at the PDB Data Bank with accession codes 3sti and 3stj, respectively. All of the graphical presentations were prepared using the program Pymol (49).
TABLE 1.
Data collection and refinement statistics
| QProtPDZ1 | QProt | |
|---|---|---|
| Data collection | ||
| Space group | P31 | P31 |
| Unit cell parameters (Å) | a = 115.3 | a = 70.9 |
| b = 115.3 | b = 70.9 | |
| c = 287.4 | c = 152.0 | |
| Resolution (Å)a | 20-2.6 | 20-2.5 |
| Completeness (%) | 95.4 (87.8) | 96.4 (93.8) |
| Rsym (%)b | 8.0 (33.9) | 9.3 (67.8) |
| I/σ(I) | 15.1 (1.5) | 11.2 (1.2) |
| Redundancy | 2.3 (1.9) | 3.7 (3.2) |
| Refinement | ||
| Resolution | 20-2.6 | 20-2.6 |
| Number of reflections Rwork/Rfree | 118,403/6,289 | 22,399/1,204 |
| Number of protein atoms | 26,507 | 4023 |
| Number of ligand atoms | ||
| Rcryst/Rfreec | 18.5/21.2 | 22.2/25.7 |
| Average B-factor (Å2) | 60.3 | 51.9 |
| Root mean square deviations of bond length (Å)/angles (°) | 0.010/1.3 | 0.010/1.3 |
| Ramachandran statistics (%) most favored, allowed, and disallowed regiond | 91.7, 8.3,0.0,0.0 | 91.3,7.8,0.4,0.4 |
a Numbers in parentheses refer to the highest resolution shell.
b Rsym is the unweighted R value on I between symmetry mates.
c Rcryst = Σhkl||Fobs (hkl)| − k|Fcalc (hkl)||/Σhkl|Fobs (hkl)| for the working set of reflections; Rfree is the R value for 5% of the reflections excluded from refinement.
d The stereochemistry of the model was validated with PROCHECK (48).
Characterization of DegQ and DegQ-Substrate Complexes by Gel Filtration
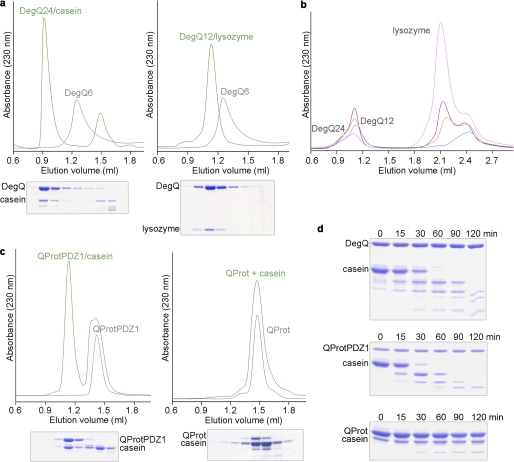
Complex formation was analyzed using the proteolytically inactive mutants (S187A) of all DegQ variants tested. We incubated 50 μm of full-length DegQ variants, 120 μm QProtPDZ1 or 120 μm QProt with either 160 μm casein or 200 μm lysozyme in 50 mm HEPES/NaOH, pH 7.5, 150 mm NaCl supplied with 10 mm DTT in the case of the lysozyme assay. Prior to the experiment lysozyme was denatured by preparing a 50 mg/ml protein solution in 4 m urea and 10 mm DTT. Assays were incubated for 10 min at 37 °C before samples were injected on a Superdex 200 gel filtration column (PC3.2/30; GE Healthcare). Comparison with marker proteins and SDS-PAGE analysis revealed the size and composites of the individual complexes.
To follow the dose-dependent DegQ12 and DegQ24 formation, DegQ (50 μm) was incubated with various amounts of lysozyme (120, 300, 450, or 950 μm) at 37 °C for 10 min. The resulting complexes were analyzed by size exclusion chromatography (SEC).
To survey the complex formation in the presence of the activating peptide, 30 μm DegQ was incubated with 200 μm SPMFKGVLDMMYGGMRGYQV peptide in 50 mm HEPES/NaOH, pH 7.5, 150 mm NaCl buffer for 15 min at 37 °C. Subsequently, the sample was injected on the gel filtration column pre-equilibrated with corresponding buffer supplied with 200 μm of the peptide activator.
To analyze the pH-dependent change in the oligomeric state of DegQ, we dialyzed 50 μm DegQ aliquots against 50 mm MES/NaOH, pH 5.5, 150 mm NaCl, or 50 mm HEPES/NaOH, pH 7.5, 150 mm NaCl at 4 °C with slow stirring. After 3 h, the samples were directly applied on the gel filtration column pre-equilibrated with the respective buffer.
Isothermal Titration Calorimetry
The thermodynamic values of the interaction between DegQ and the activating peptide were determined using an isothermal titration microcalorimeter (MCS-ITC; Microcal). All of the experiments were conducted in overflow mode at 30 °C. 1.8 ml of solution of 20 μm DegQ was placed in the temperature-controlled sample cell and titrated with 200 μm peptide loaded in the 300-μl mixing syringe. For the experiment 50 mm HEPES/NaOH, pH 7.5, 150 mm NaCl was used as isothermal titration calorimetry (ITC) buffer. Injections of 10 μl peptide were dispensed into the sample cell using a 120-s equilibration time between injections and stirring at 300 rpm. Control experiments were carried out to measure and correct the heat of dilution upon buffer addition. Finally, the data were analyzed using the program ORIGIN following the instructions of the manufacturer.
Characterization of the Proteolytic Activity
The effect of the peptide activator on the activity of DegQ was measured using the pNA-chromogenic peptide substrate (SPMFKGV-pNA). The 0.8-ml reaction mixtures containing 0.5 mm pNA substrate in 50 mm HEPES/NaOH, pH 7.5, 150 mm NaCl, and the 0.3 mm of activating peptide were preincubated for 15 min at 37 °C. The cleavage reaction was initiated by the addition of DegQ to a final concentration of 5 μm. The continuous increase in the absorbance at 405 nm at 37 °C was monitored.
To follow the degradation of the model substrate casein, we mixed 2.5 μm of DegQ or derivatives thereof with 20 μm casein in 50 mm Hepes/NaOH, pH 7.5, 150 mm NaCl buffer or 4 μm of DegQ with 20 μm lysozyme in the same buffer supplied with 10 mm DTT. Prior to the assay, lysozyme was denatured in the 4 m urea and 10 mm DTT. The samples were incubated at 37 °C. The reaction was stopped at the indicated time points by adding SDS loading buffer and boiling the samples for 10 min at 95 °C. Subsequently, the aliquots were analyzed by SDS-PAGE followed by Coomassie Blue staining.
The pH-dependent proteolytic activity of DegQ and DegP was determined with resorufin-labeled casein (Roche Applied Science). 15 μl of 0.4% (w/v) resorufin-labeled casein was added to 100 μl of incubation buffer of a respective pH containing 3 μm of DegQ or DegP and incubated at 37 °C for 3 h (DegP) and 12 h (DegQ). The reaction was stopped by precipitating casein with 480 μl of 10% (w/v) TCA. The samples were again incubated for 10 min at 37 °C and subsequently centrifuged (10 min, 10000 × g, room temperature). 400 μl of the supernatant was mixed with 600 μl of 1 m Tris/HCl, pH 8.8, and the absorbance at 574 nm was determined. In the pH screen, we used the following 50 mm buffers supplemented with 150 mm NaCl: acetic acid (pH levels 4.5 and 5.0), MES/NaOH (pH levels 5.5, 6.0, and 6.5), HEPES/NaOH (pH levels 7.0 and 7.5), Tris/HCl (pH levels 8.0, 8.5, and 9.0). The pH was adjusted at 37 °C.
Recording Bacterial Growth Curves
The unbuffered overnight cultures of degP null (CLC198, degP::Tn10) (40), degQ null (MG1655 degQ::Tn5 KanR, strain ordered from E. coli Genome Project) and their parental strains were standardized to equal A600 and used to inoculate 100 ml of fresh LB medium. In the case of degQ null strain, kanamycin was added to a final concentration of 25 μg/ml. After 30 min of growth (37 °C, 220 rpm), LB media were buffered by a direct addition of sterile filtered 10 ml 1 m HEPES/NaOH, pH 7.5, or 10 ml of 0.5 m MES/NaOH, pH 5.5. Cells growth and pH were monitored at 30-min time points until the stationary phase was reached.
RESULTS
Oligomer Conversion and Activation of DegQ Does Not Depend on the Second PDZ Domain
Recent studies with DegP from E. coli suggested that substrate-induced oligomer conversion and activation is of central importance for HtrA protease regulation (24, 26). To explore whether DegQ employs a similar mechanism, we first analyzed complex formation with the unfolded model substrates lysozyme and β-casein. For this purpose, we used a catalytically inactive DegQ mutant, in which the active site serine Ser-187 was replaced by alanine. SEC and SDS-PAGE analysis revealed that incubation of DegQ with lysozyme and casein leads to the formation of higher order, substrate-engaged DegQ12 and DegQ24 multimers, respectively (Fig. 1a), with the size of the generated particle depending on the amount of substrate. At lower substrate concentration, the 12-mer is predominantly formed, whereas substrate at increasing concentration is preferentially captured in DegQ24 (Fig. 1b).
FIGURE 1.
Substrate-induced oligomer reassembly of DegQ. a, full-length DegQ (gray line) was incubated with casein (left panel) or unfolded lysozyme (right panel), and complex formation was monitored by SEC and SDS-PAGE. Substrate binding transforms the DegQ hexamer (DegQ6) into DegQ24-casein and DegQ12-lysozyme complexes, respectively. b, incubation of DegQ6 (50 μm) with increasing lysozyme concentrations (120 μm, blue; 300 μm, red; 450 μm, purple; 950 μm, magenta) reveals that the distinct higher order oligomers are formed depending on the amount of substrate. For lysozyme concentrations higher than 1 mm, the 24-mer starts to be preferentially formed. The absorbance peak at 2.4 ml can be attributed to the reducing agent used to facilitate lysozyme unfolding. c, the QProtPDZ1 mutant occurs in its latent state as a trimer (left, gray line) as well as the QProt mutant (right, gray line). Incubation with an unfolded substrate (casein) triggers formation of a dodecameric QProtPDZ1-substrate complex (blue line), whereas no higher order oligomer is formed with QProt. d, SDS-PAGE analysis of the proteolyic activity of the different DegQ variants (DegQ wild type, top panel; QProtPDZ1, middle panel; QProt, bottom panel) against casein at distinct time points.
To extend these studies to HtrAs containing a single PDZ domain, we performed complex formation analyses with a DegQ variant lacking the second PDZ domain (QProtPDZ1). In the presence of substrate, we could observe the formation of higher order assemblies, however exclusively of 12-meric particles (Fig. 1c). The inability to form 24-mers is not surprising because the PDZ2 domain is essential to mediate intertrimer contacts in this higher order oligomer as suggested by the architecture of DegP24 (24, 26). Moreover, in the absence of substrate, QProtPDZ1 occurred as a trimer, which should represent the resting state of the mutant. With regard to protease activity, QProtPDZ1 retained the ability to degrade casein, demonstrating that PDZ2 is dispensable for protease activity (Fig. 1d). Conversely, removal of both PDZ domains (the QProt variant) resulted in proteolytically inactive trimers that are incapable of forming higher order oligomers (Fig. 1, c and d). Together, these data indicate that only PDZ1 is essential to couple substrate binding with the formation of proteolytically active higher order DegQ oligomers.
Crystal Structure of the QProtPDZ1 Dodecamer and the QProt Trimer
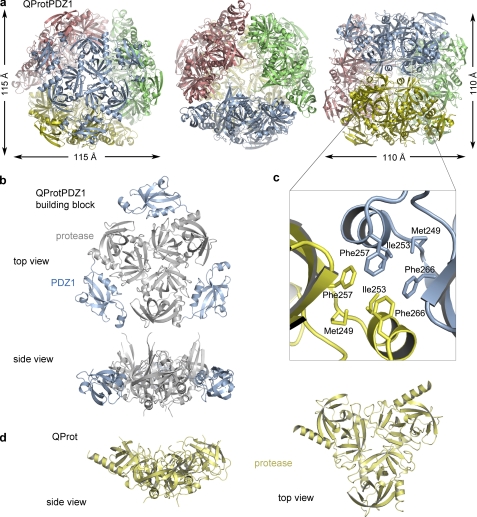
Because of the failure to crystallize full-length DegQ (i.e. DegQ6 and DegQ12/24 complexes with substrate), we crystallized suitable deletion variants including the QProtPDZ1 mutant that retained the capability to form 12-mer complexes in the presence of substrate. This dodecamer is also seen in the crystal structure, which was solved by molecular replacement at 2.6 Å resolution (Table 1). The crystal structure of the QProtPDZ1 dodecamer shows a 400-kDa hollow particle with dimensions of 115 Å × 115 Å × 110 Å (Fig. 2a). The four trimers are located at the vertices of a tetrahedron and assemble a protein shell that encloses an internal cavity of ∼50 Å diameter. The contacts between the trimers are mainly mediated by the PDZ1 domains yielding a rigid molecular cage. The proteolytic sites are sequestered within this cage and open up into the interior. Therefore cleavage products have to leave the particle through one of the four ∼20 Å-wide pores, which are bordered by the protease domains of adjacent trimers. The spatial organization of the trimers resembles a planar triangle with centered protease and PDZ domains at the vertices (Fig. 2b). The peripheral PDZ1 domains contact each other via the interaction clamp, an HtrA signature motif that is important to form higher order oligomers by mediating contacts between juxtaposed trimers (23, 25–26). The interaction clamp of QProtPDZ1 comprises a hydrophobic region (residues 249–266) that interacts with the corresponding region of PDZ1* (the asterisk denotes a neighboring molecule) of the adjacent trimer, thereby constituting the hydrophobic core of the 12-mer interface (Fig. 2c).
FIGURE 2.
Crystal structures of the QProtPDZ1 dodecamer and the QProt trimer. a, ribbon plot of the QProtPDZ1 particle illustrating its overall dodecameric architecture. The constituting trimers (colored differently) occupy the vertices of a tetrahedron yielding a proteolytic cavity of ∼50 Å in diameter and defined exit pores. The particle is shown along the molecular 3-fold (left and middle panels) and the 2-fold axis (right panel). b, side and top view of one QProtPDZ1 trimer of the dodecamer with the protease domain colored gray and the PDZ1 domain colored blue. c, the intertrimeric contacts in the dodecamer are mediated mainly by the hydrophobic “interaction clamp” of the PDZ1 domain. The clamp residues involved in the interactions between two adjacent PDZ1 domains (yellow and blue) are depicted in stick mode. d, side and top view of the QProt trimer.
In addition, we determined the crystal structure of the protease domain alone (QProt) representing the inactive state. This structure enabled us to delineate the function of the PDZ domains in mediating the switch in activity. In contrast with QProtPDZ1, the QProt mutant could not form higher order complexes and remained as trimer in solution under all of the conditions tested. Consistently, QProt was observed as trimer in the crystal lattice (Fig. 2d). The assembly of the protease trimer is similar to QProtPDZ1, in which intersubunit contacts are exclusively established between residues of the protease domain, most of which originate from the N-terminal α-helix.
QProtPDZ1 and QProt Reflect the Proteolytically Active and Inactive States of DegQ
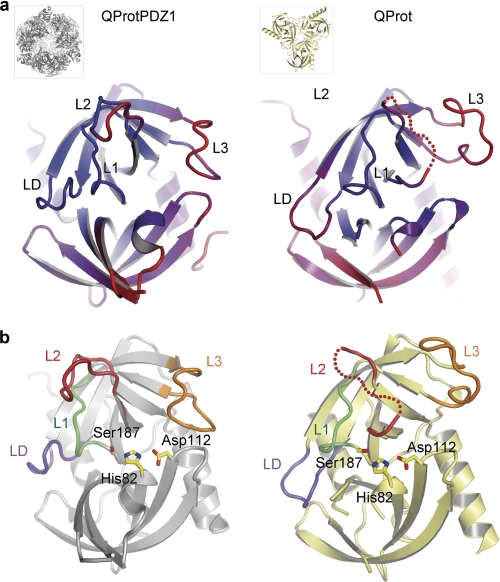
Comparison of the two DegQ crystal structures (root mean square deviation value of 1.9 Å for 175 aligned Cα atoms) revealed that QProtPDZ1 and QProt show characteristic differences in their active site architecture. In QProt, the activation domain (L1, L2, and LD) as well as loop L3 (loop that typically mediates interaction with the PDZ domain) are highly flexible as indicated by the elevated crystallographic temperature factors (Fig. 3a) and the absence of interpretable electron density for residues 207–212 of loop L2. The conformational flexibility within the active site impedes proper adjustment of the catalytic triad, oxyanion hole, and S1 specificity pocket and thus explains the drastically reduced catalytic activity of the PDZ-less mutant. Conversely, in QProtPDZ1, the activation domain is well defined by electron density and adopts a strikingly different conformation (Fig. 3b, with detailed views shown in supplemental Fig. S1): First, a functional catalytic triad is set up between His-82, Asp-112, and Ser-187 with the hydroxyl, imidazol, and carboxylate group being properly aligned to hydrogen bond each other. Second, a peptide flip of the amide linkage between residues Gly-185 and Arg-184 enables the Arg-184 carbonyl oxygen to interact with the amide nitrogen of Phe-148 of loop LD, thereby allowing formation of the oxyanion hole constituted by residues 184–187 of loop L1. Third, the residue Ile-182 together with Ile-205 and Ala-204 of loop L2 are properly oriented to establish the S1 specificity pocket, whereas residues Thr-203, Ala-204, Ile-205, Leu-206, and Ala-207 adopt a β-strand conformation required to bind the main chain of the incoming protein substrate by β-augmentation. The shallow S1 hydrophobic pocket selects for small hydrophobic residues, which is consistent with the previously described specificity of DegQ cleaving model substrates at discrete Val/Xaa or Ile/Xaa sites (42). Based on our structural data, we conclude that QProt and QProtPDZ1 represent the inactive and active states of DegQ. Therefore, protease activity depends on the regulated folding of the activation domain, a process that is under control of loop L3 and its interaction with the PDZ1 domain. Similarly to other serine proteases of the chymotrypsin family, this activation process is connected with a disorder-to-order transition of the activation domain (Fig. 3b).
FIGURE 3.
Comparison of the active QProtPDZ1 and the inactive QProt protease folds. Ribbon plots of the protease domains of QProtPDZ1 (left panels) and QProt (right panels) are shown. a, structures are color-coded by the crystallographic temperature factors (rigid portions, blue; disordered regions, red) with the active site loops being labeled. b, highlighted active site loops that adopt strikingly different conformations in the two DegQ variants representing the active (QProtPDZ1) and the inactive state (QProt). The active site residues (His-82, Asp-112, and Ser-187) are shown in stick mode.
Peptide Binding to PDZ1 Triggers Formation of Proteolytically Active Higher Order Oligomers
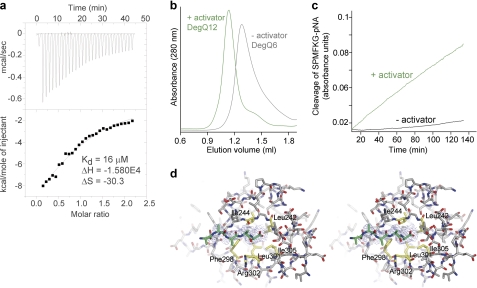
The activation of HtrA proteases is known to be a reversible process that can be triggered by distinct molecular cues. For example in DegS, peptides that signal folding stress are recognized and bound by the PDZ domain. These bound peptides are capable of inducing rearrangement of the sensor loop L3, which in turn triggers the remodeling of the activation domain into its functional state capable of cleaving the substrate protein RseA (30, 36, 38). In contrast with this transactivation mechanism, allosteric regulation of DegP depends on the substrate itself. Substrate binding to the first PDZ domain of DegP, PDZ1, induces oligomer conversion from DegP6 to DegP24 that leads to a repositioning and immobilization of the PDZ domains such that they can induce rearrangement of loop L3, thereby activating protease function (26, 32, 50). To test which activation mechanism is employed by DegQ, we assessed its interactions with various synthetic peptides. Similarly to DegP, DegQ showed the highest affinity to peptides having a C-terminal valine residue, whereas an interaction with the preferred peptide ligand of DegS having a C-terminal phenylalanine could not be observed by means of ITC. For the SPMFKGVLDMMYGGMRGYQV peptide, a described allosteric activator of DegP (32, 51), ITC measurements revealed a KD value of 16 μm (Fig. 4a). Further SEC analysis demonstrated that this peptide is capable of inducing the transformation of DegQ6 into DegQ12 (Fig. 4b), which is correlated with an enhanced proteolytic activity against a chromogenic model substrate (Fig. 4c). In contrast, effector peptides with a C-terminal glutamate residue did not bind to DegQ and did not stimulate protease activity (data not shown).
FIGURE 4.
Oligomer conversion and proteolytic activity of DegQ is regulated by peptide binding to the PDZ1 domain. a, ITC measurement of the binding of the activating peptide to DegQ6. The area under each peak was integrated and plotted against the molar ratio of peptide to DegQ inside the sample cell (lower panel). The black line represents the fit to a binding isotherm, assuming one binding site per protomer. The indicated thermodynamic values were calculated using the protomer concentration of DegQ. b, SEC analysis illustrating conversion of DegQ6 (Q6, gray) into DegQ12 (Q12, green) in the presence of the activating peptide (200 μm). c, cleavage of the chromogenic SPMFKGV-pNA substrate by DegQ in the absence (black) and presence of the activating peptide (green). d, cartoon representation of the peptide binding to PDZ1 in stereo view. The polyalanine chain (green) binds to the PDZ1 domain (gray) via β-augmentation. The 2Fo − Fc electron density map, which was calculated at 2.6 Å resolution without contribution of the peptide ligand, is contoured at 1.2 σ. Residues of the PDZ1 domain accommodating the C-terminal side chain of the peptide are highlighted in yellow and labeled.
Consistent with these biochemical data, two peptide ligands were observed in the electron density map of the QProtPDZ1 dodecamer. One peptide is accommodated in the binding groove of the PDZ1 domain. The built-in polyalanine model illustrates that the peptide is attached via β-augmentation to the core of the PDZ1 domain allowing the C-terminal residue to penetrate a shallow hydrophobic pocket lined by residues Phe-298, Leu-242, Ile-244, Leu-301, Arg-302, and Ile-305 (Fig. 4d). The second peptide is accommodated in the proteolytic site, where it is tethered to the β-strand formed by residues Thr-203, Ala-204, Ile-205, Leu-206, and Ala-207 (in orange in supplemental Fig. S2), thus allowing the side chain of the P1 residue to protrude in the S1 specificity pocket (supplemental Fig. S2). Because QProtPDZ1 is not subject to autodegradation, the bound peptides cannot result from autocleavage, as seen for example in Mycobacterium tuberculosis HtrA (28), and should thus represent co-purified and co-crystallized oligopeptides that mimic potential cleavage intermediates. A similar scenario has been reported for DegP, in which a variety of oligopeptides were captured in the PDZ1 binding groove of the functionally active DegP24, but not in the resting DegP6. These bound peptides appear to rearrange the carboxylate binding loop of the PDZ1 domain and the adjacent interdomain linker segment, thereby inducing a domain rearrangement such that DegP6 is transformed into DegP12/24 (32). Given the similarity of DegP and DegQ in their peptide binding modes and activation mechanism, we thus presume that the peptides bound to QProtPDZ1 orient and immobilize the PDZ1 domain, thereby allowing crystallization of the functionally active dodecamer. Taken together, our biochemical and structural data demonstrate that peptide binding to PDZ1 stimulates the protease activity of DegQ by triggering formation of catalytically active higher order oligomers.
The PDZ1 Domain and Loop L3 Constitute a Molecular Switch Regulating Protease Function
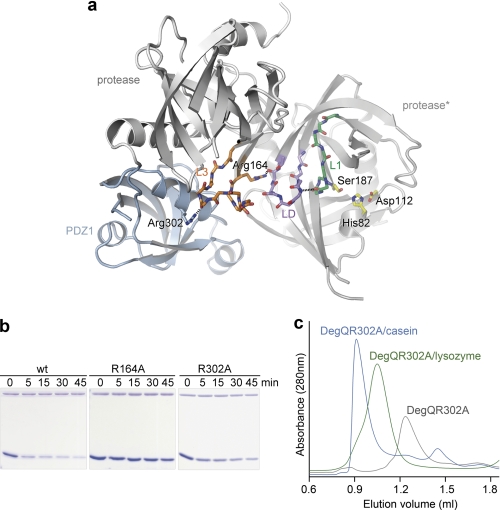
As shown for the DegP protease, the PDZ1 domain plays an essential role in the activation process. Upon substrate binding and DegP12/24 oligomer formation, the repositioned PDZ1 domain induces rearrangement of the protease loop L3, which in turn stabilizes the functional state of the proteolytic site. The high resolution structure of QProtPDZ1 enabled us to explore whether a similar PDZ1 → L3 → LD/L1/L2 protease activation cascade occurs in DegQ (Fig. 5a). To test whether the interplay between PDZ1 domain and the L3 loop is critical to activate protease function in the dodecameric scaffold, we disrupted this interaction by introducing the R302A mutation (Arg-302 of PDZ1 forms a hydrogen bond with the carbonyl oxygen of Gly-171 in loop L3; Fig. 5a). Indeed, when we assayed the catalytic activity of the mutant, we could observe that the degradation of lysozyme was strongly reduced (Fig. 5b). To show that the abolished activity results from the disrupted PDZ1-L3 communication and not from impaired substrate binding or hindered 12/24-mer formation, the R302A mutant was subjected to SEC analysis. The R302A mutant retained the ability form higher order complexes in the presence of substrate (Fig. 5c), indicating that the reduced protease activity is the direct consequence of the abrogated PDZ1-L3 interaction. Once initiated by loop L3, the signaling cascade results in the remodeling of the activation domain of the protease. This step is mediated by a conserved arginine residue (Arg-164 in DegQ) that is located on the N-terminal stem segment of loop L3 (Fig. 5a). To test the importance of Arg-164 for transferring the activation signal from the PDZ1 domain to the proteolytic site, we exchanged it to an alanine, thereby preventing its interaction with the main-chain carbonyl of Gln-152 and the hydroxyl group of Thr-153 of loop LD. In the SDS-PAGE assay, the proteolytic activity of the R164A mutant against unfolded lysozyme was significantly reduced, highlighting the importance of the L3-LD interaction for protease activation (Fig. 5b). Moreover, comparison with the inactive QProt structure revealed that a flexible loop L3 that is not tethered by PDZ1 is not capable of interacting with loop LD. As a consequence, the remodeling of the activation domain into the active state is prevented, explaining the abolished protease activity of the QProt mutant. In sum, these data indicate the preservation of the intramolecular PDZ1 → L3 → LD/L1/L2 signaling module, suggesting that loop L3 functions as a conserved molecular switch in regulating HtrA proteases in both 12- and 24-meric HtrA oligomers.
FIGURE 5.
Activation cascade in DegQ. a, illustration of the key interactions involved in signaling from loop L3 to the proteolytic site as seen in the QProtPDZ1 crystal structure. The fixed position of PDZ1 (blue) orients loop L3 (orange) by interactions mediated via Arg-302 (depicted in stick mode). As a result, Arg-164 of the loop L3 interacts with the main chain carbonyl of residue Gln-152 and the hydroxyl group of Thr-153 of loop LD (lilac) in the adjacent protomer (protease*), which in turn induces remodeling of the proteolytic sites (loop L1 shown in green and functional catalytic triad in yellow). b, SDS-PAGE analysis of the cleavage of unfolded lysozyme by DegQ wild type and R164A or R302A mutants indicating that both mutations render the DegQ protease inactive. c, SEC analysis reveals that, although proteolytically inactive, both mutants (R302A mutant shown here as representative) can form higher order oligomers with casein (24-mer, blue) and lysozyme (12-mer, green).
DegQ Is a pH-sensitive HtrA Protease
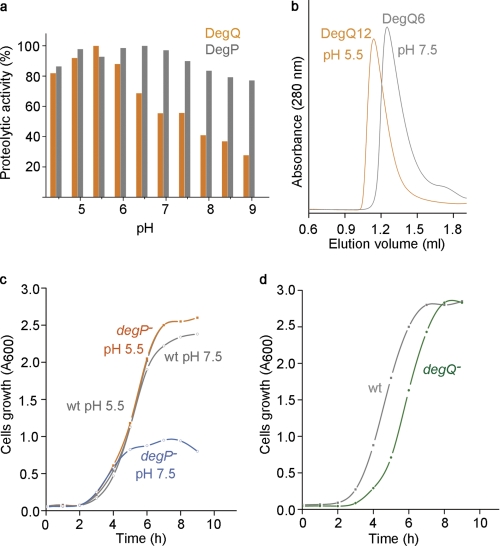
Because it is known that HtrA proteases are regulated by different molecular cues (38, 40, 52, 53), we systematically analyzed the effect of different physical and chemical stimuli on the activity of DegQ and found that DegQ digests substrates in a pH-dependent manner. We monitored the proteolytic activity of DegQ at different pHs in a colorimetric assay using resorufin-labeled casein and observed that the degradation is most efficient at pH values between 4.5 and 6 with an optimum at pH 5.5 (Fig. 6a). Remarkably, SEC analysis revealed that pH had an additional effect on the oligomeric state of DegQ. Whereas the hexamer is the dominant form at pH 7.5, the equilibrium shifts to the dodecamer at pH 5.5 (Fig. 6b). Despite these different oligomeric states, casein was encapsulated at both pH 5.5 and pH 7.5 in the 24-mer particle, suggesting that the pH does not interfere with substrate engagement (data not shown). Although the exact molecular mechanism of the pH effect is still elusive, it is apparent that the distinct oligomeric states of DegQ occur in a dynamic, pH-dependent equilibrium. We presume that at slightly acidic pH, the activation barrier to transform the 6-mer into the 12/24-mer is decreased. Accordingly, substrate encapsulation and degradation in DegQ12/24 should be facilitated.
FIGURE 6.
DegQ function is affected by pH. a, effect of pH on the proteolytic activity of DegQ (orange) and DegP (gray) against casein in the pH range from 4.5 to 9. The relative proteolytic activities were calculated by standardization to the highest obtained value, which was regarded as 100%. b, SEC analysis of the oligomeric state of DegQ in pH 5.5 (orange line) and 7.5 (gray line) revealed pH-dependent DegQ6 to DegQ12 conversion. c, growth of degP-null and wild type strains in LB medium buffered to either pH 5.5 or 7.5. The growth rate of the wild type strain was not affected by pH (gray lines), whereas degP-null cells cultured in pH 7.5 (blue line) entered the stationary phase 3.5 h earlier compared with pH 5.5 (orange line) and grow to lower cell density. d, growth of degQ-null and wild type strains in LB media buffered to pH 7.5. The mutant strain (green line) shows extended lag time after inoculation compared with the wild type (gray line).
To examine the physiological importance of the pH-dependent protease activity, we compared the growth rate of a degP null mutant exposed to slightly acidic (pH 5.5) and neutral (pH 7.5) medium and observed a striking correlation with the pH-dependent activity of DegQ. At pH 5.5, the growth of the degP null mutant was identical to wild type, whereas at pH 7.5, cells stopped dividing and entered stationary phase 3.5 h earlier (Fig. 6c). These data indicate that DegQ is capable of taking over the function of DegP in the degP null mutant strain. However, it can only reconstitute the wild type situation at slightly acidic pH values, at which the protease activity of DegQ is the highest. Analyzing the growth rates of a degQ null strain did not reveal a similar pH dependence (data not shown), thus confirming that the activity of DegP is largely pH-independent in the range analyzed. However, when we compared the initial growth phase of the degQ null mutant with the wild type strain, we noticed that the adaptation of the mutant cells was prolonged (Fig. 6d). In the absence of DegQ, the degP expression has to be up-regulated in response to the accumulation of unfolded proteins in the cell envelope. Because of the time required to sense the stress situation and to trigger the corresponding unfolded protein response (43), the initial growth phase appears to be delayed until DegP is produced in sufficient amounts. Together, these findings reveal that DegQ functions as pH-sensitive protease in the cellular envelope, establishing the initial proteolytic response against misfolded proteins.
DISCUSSION
HtrA Proteases with a Single PDZ Domain Are Capable of Forming Proteolytically Active Higher Order Particles
Our biochemical and structural analysis revealed that a DegQ mutant lacking the second PDZ domain can form higher order oligomers to encapsulate and degrade substrate proteins. In analogy to DegP (24, 26), binding of an unstructured polypeptide to the PDZ1 domain induces the conversion of the resting (DegQ-hexamer and QProtPDZ1-trimer) into the catalytically active state (DegQ-12/24mer and QProtPDZ1–12mer). Consistently, it was recently shown that human HtrA1, an HtrA protease containing a single PDZ domain and carrying out protein quality control in the extracellular matrix (34), encapsulates misfolded proteins in higher order complexes (54). Given that the majority of HtrA proteases encompass only a single PDZ domain, the reassembly of trimers into higher molecular weight complexes can be considered as a conserved mechanism regulating the activity of most HtrA proteases. However, it should be noted that activation by oligomer conversion is only relevant for soluble HtrAs, because membrane anchored HtrA proteases like DegS from E. coli (30) do not form oligomers larger than a trimer. Moreover, the membrane-anchored HtrA family members are often regulatory proteases, in which activation and proteolytic cleavage occur separately (30, 36). Therefore, the regulatory mechanism linking substrate binding with protease activation and oligomer conversion should be relevant for all HtrA proteases having one or two PDZ domains and acting on a broad range of misfolded proteins during protein quality control. More specialized members like the DegS stress sensor appear to be under control of more specific regulatory mechanisms that act in trans and that are not directly coupled with substrate binding.
The Activation Cascade PDZ → L3 → LD/L1/L2 Is Conserved in 12- and 24-meric HtrA Oligomers
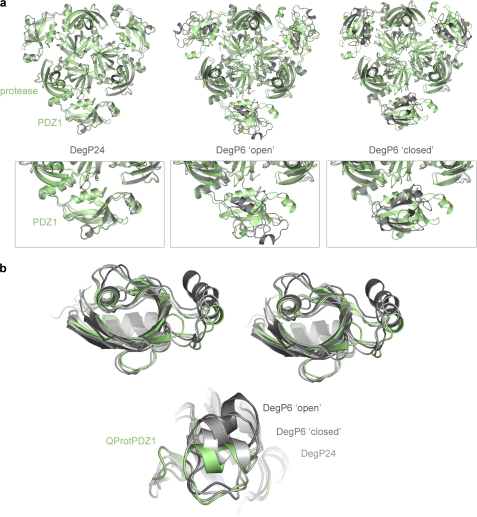
Recent structural work on DegP24 uncovered key aspects of how HtrA proteases involved in protein quality control recognize, bind, and processively cleave substrate and how they are regulated by the mechanism of substrate-induced oligomer conversion (24, 26). However, to fully understand the regulation of HtrA proteases, which form distinct substrate engaging oligomers (24, 26, 29), high resolution data of a dodecameric form of a HtrA protease is required. Our structural studies of the truncated version of DegQ (QProtPDZ1) provide this information and present a detailed view of the molecular architecture of a functional HtrA dodecamer. Based on the structural information, we could verify that the molecular mechanism underlying regulation of HtrA 24-mers (with DegP24 as best characterized representative) (32, 50) is conserved in substrate engaged 12-mer particles (QProtPDZ1; this work). A structural alignment demonstrates that the relative position of PDZ1 and protease domains observed in QProtPDZ1 fits remarkably well to DegP24 (root mean square deviation 1.1 Å for 298 Ca atoms of protease and PDZ1 domain), whereas it is strikingly different from the domain arrangement in the resting DegP6 (Fig. 7a). The similar domain orientation in the 12- and 24-mer particles is even more surprising, because the two states were derived from two different proteins, DegQ and DegP, respectively, and from protein variants that differ in their domain composition. Therefore, the present data implicate that a precisely aligned PDZ1 domain is key to trigger protease activation in both HtrA 12- and 24-multimers. These multimers are hold together by the PDZ interaction clamp that mediates contacts between adjacent trimers. Because of the en-bloc mobility of this motif within the PDZ fold (Fig. 7c), HtrA proteases can form different oligomeric assemblies while maintaining the critical loop L3-PDZ1 interaction required for protease activation.
FIGURE 7.
Immobilization of the PDZ1 domain in higher order HtrA oligomers is crucial for protease activation. a, structural alignment of one QProtPDZ1 (green) trimer to trimers of DegP24 (left panel, gray), DegP6 open conformation (middle panel, gray), and DegP6 closed conformation (right panel, gray). The enlarged picture of the protease/PDZ1 interface illustrates that QProtPDZ1 and DegP24 have strikingly similar domain orientations, whereas the PDZ1 domain of QProtPDZ1 and the two DegP6 forms is oriented differently relative to the protease domain. b, structural alignment of the PDZ1 domain of QProtPDZ1 (green), DegP24 (light gray), DegP6 closed (medium gray), and DegP6 open (dark gray) illustrating that the interaction clamp (helix on right side) is the structural element showing the highest en-bloc flexibility within the PDZ scaffold of HtrA proteases.
DegQ Is a pH-sensitive Protease in the Bacterial Cell Envelope
Our functional in vitro and in vivo studies reveal that DegQ is a pH-related protease that maintains protein homeostasis in the bacterial cell envelope. Given the porous and thus highly permeable character of the outer membrane, all periplasmic proteins are exposed to rapid environmental changes such as changes in the pH. It is evident that resultant protein damage has to be counteracted immediately. Because of its pH-dependent activity, DegQ appears to be the “first-in-place” protease to react on pH-mediated protein misfolding. Only when the protease-chaperone system of the periplasm is overloaded and damaged proteins accumulate, is the production of DegP up-regulated. Under such stress conditions, DegP would function as the primary protease reducing the levels of misfolded proteins (55, 56). Therefore, DegQ and DegP appear to closely collaborate with each other in the bacterial cell envelope, ensuring high fidelity protein quality control under mild and severe stress conditions, respectively.
DegQ is not the only HtrA protease whose proteolytic activity is affected by pH. Deg1 and Deg2 from Arabidopsis thaliana chloroplasts also cleave substrates in a pH-dependent manner. The distinct pH optima of Deg1 and Deg2 appear to reflect the adaptation of the two proteases to their individual compartments (52, 53). Deg1 resides in the thylakoid lumen (57), whereas Deg2 is located at the stromal side of the thylakoid membrane (53). The light-induced pH gradient between the two adjacent compartments sustains a low pH (pH 4.5–6.0) in the lumen and an alkaline pH (above 8.0) in the stroma (58, 59). Consistently, the luminal Deg1 protease has an optimum at pH 6, whereas the stromal Deg2 protease most efficiently degrades proteins at pH 8 (52, 53). Accordingly, the regulatory mechanism employed by DegQ could be associated with the changes of external pH in the enteric habitat of E. coli, which can vary between pH 5 and 8 (60). Evolving a pH-regulated protease such as DegQ would help bacteria to deal with mild pH alterations and to avoid energy- and time-consuming processes required to up-regulate the stress response machinery.
Supplementary Material
This work was supported by an ERA-Net (NEURON, FWF I 235-B09) studentship (to F. C.). The Institute of Molecular Pathology is funded by Boehringer Ingelheim.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (codes 3sti and 3stj) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MES
- 4-morpholineethanesulfonic acid
- SEC
- size exclusion chromatography
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Macario A. J., Conway de Macario E. (2005) N. Engl. J. Med. 353, 1489–1501 [DOI] [PubMed] [Google Scholar]
- 2. Selkoe D. J. (2003) Nature 426, 900–904 [DOI] [PubMed] [Google Scholar]
- 3. Gottesman S., Wickner S., Maurizi M. R. (1997) Genes Dev. 11, 815–823 [DOI] [PubMed] [Google Scholar]
- 4. Wickner S., Maurizi M. R., Gottesman S. (1999) Science 286, 1888–1893 [DOI] [PubMed] [Google Scholar]
- 5. Clausen T., Southan C., Ehrmann M. (2002) Mol. Cell 10, 443–455 [DOI] [PubMed] [Google Scholar]
- 6. Pallen M. J., Wren B. W. (1997) Mol. Microbiol. 26, 209–221 [DOI] [PubMed] [Google Scholar]
- 7. Antelmann H., Darmon E., Noone D., Veening J. W., Westers H., Bron S., Kuipers O. P., Devine K. M., Hecker M., van Dijl J. M. (2003) Mol. Microbiol. 49, 143–156 [DOI] [PubMed] [Google Scholar]
- 8. Cortés G., de Astorza B., Benedí V. J., Albertí S. (2002) Infect. Immun. 70, 4772–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ibrahim Y. M., Kerr A. R., McCluskey J., Mitchell T. J. (2004) Infect. Immun. 72, 3584–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jones C. H., Bolken T. C., Jones K. F., Zeller G. O., Hruby D. E. (2001) Infect. Immun. 69, 5538–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lewis C., Skovierova H., Rowley G., Rezuchova B., Homerova D., Stevenson A., Spencer J., Farn J., Kormanec J., Roberts M. (2009) Microbiology 155, 873–881 [DOI] [PubMed] [Google Scholar]
- 12. Mo E., Peters S. E., Willers C., Maskell D. J., Charles I. G. (2006) Microb. Pathog. 41, 174–182 [DOI] [PubMed] [Google Scholar]
- 13. Raivio T. L. (2005) Mol. Microbiol. 56, 1119–1128 [DOI] [PubMed] [Google Scholar]
- 14. Wilson R. L., Brown L. L., Kirkwood-Watts D., Warren T. K., Lund S. A., King D. S., Jones K. F., Hruby D. E. (2006) Infect. Immun. 74, 765–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baldi A., Mottolese M., Vincenzi B., Campioni M., Mellone P., Di Marino M., di Crescenzo V. G., Visca P., Menegozzo S., Spugnini E. P., Citro G., Ceribelli A., Mirri A., Chien J., Shridhar V., Ehrmann M., Santini M., Facciolo F. (2008) Pharmacogenomics 9, 1069–1077 [DOI] [PubMed] [Google Scholar]
- 16. Bhuiyan M. S., Fukunaga K. (2009) Curr. Drug Targets 10, 372–383 [DOI] [PubMed] [Google Scholar]
- 17. Campioni M., Severino A., Manente L., Tuduce I. L., Toldo S., Caraglia M., Crispi S., Ehrmann M., De Falco M., de Luca A., He X., Maguire J., Shridhar V., Baldi A. (2010) Mol. Cancer Res. 8, 1248–1260 [DOI] [PubMed] [Google Scholar]
- 18. Grau S., Baldi A., Bussani R., Tian X., Stefanescu R., Przybylski M., Richards P., Jones S. A., Shridhar V., Clausen T., Ehrmann M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6021–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones J. M., Datta P., Srinivasula S. M., Ji W., Gupta S., Zhang Z., Davies E., Hajnóczky G., Saunders T. L., Van Keuren M. L., Fernandes-Alnemri T., Meisler M. H., Alnemri E. S. (2003) Nature 425, 721–727 [DOI] [PubMed] [Google Scholar]
- 20. Plun-Favreau H., Klupsch K., Moisoi N., Gandhi S., Kjaer S., Frith D., Harvey K., Deas E., Harvey R. J., McDonald N., Wood N. W., Martins L. M., Downward J. (2007) Nat. Cell Biol. 9, 1243–1252 [DOI] [PubMed] [Google Scholar]
- 21. Shridhar V., Sen A., Chien J., Staub J., Avula R., Kovats S., Lee J., Lillie J., Smith D. I. (2002) Cancer Res. 62, 262–270 [PubMed] [Google Scholar]
- 22. Yang Z., Camp N. J., Sun H., Tong Z., Gibbs D., Cameron D. J., Chen H., Zhao Y., Pearson E., Li X., Chien J., Dewan A., Harmon J., Bernstein P. S., Shridhar V., Zabriskie N. A., Hoh J., Howes K., Zhang K. (2006) Science 314, 992–993 [DOI] [PubMed] [Google Scholar]
- 23. Kirk R., Clausen T. (2010) in Sensory Mechanisms in Bacteria: Molecular Aspects of Signal Recognition (Spiro S., Dixon R. eds) Caister Academic Press, Norwich, U.K [Google Scholar]
- 24. Jiang J., Zhang X., Chen Y., Wu Y., Zhou Z. H., Chang Z., Sui S. F. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11939–11944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krojer T., Garrido-Franco M., Huber R., Ehrmann M., Clausen T. (2002) Nature 416, 455–459 [DOI] [PubMed] [Google Scholar]
- 26. Krojer T., Sawa J., Schäfer E., Saibil H. R., Ehrmann M., Clausen T. (2008) Nature 453, 885–890 [DOI] [PubMed] [Google Scholar]
- 27. Li W., Srinivasula S. M., Chai J., Li P., Wu J. W., Zhang Z., Alnemri E. S., Shi Y. (2002) Nat. Struct. Biol. 9, 436–441 [DOI] [PubMed] [Google Scholar]
- 28. Mohamedmohaideen N. N., Palaninathan S. K., Morin P. M., Williams B. J., Braunstein M., Tichy S. E., Locker J., Russell D. H., Jacobs W. R., Jr., Sacchettini J. C. (2008) Biochemistry 47, 6092–6102 [DOI] [PubMed] [Google Scholar]
- 29. Shen Q. T., Bai X. C., Chang L. F., Wu Y., Wang H. W., Sui S. F. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilken C., Kitzing K., Kurzbauer R., Ehrmann M., Clausen T. (2004) Cell 117, 483–494 [DOI] [PubMed] [Google Scholar]
- 31. Kim D. Y., Kim K. K. (2005) J. Biochem. Mol. Biol. 38, 266–274 [DOI] [PubMed] [Google Scholar]
- 32. Krojer T., Sawa J., Huber R., Clausen T. (2010) Nat. Struct. Mol. Biol. 17, 844–852 [DOI] [PubMed] [Google Scholar]
- 33. Chien J., Campioni M., Shridhar V., Baldi A. (2009) Curr. Cancer Drug Targets 9, 451–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clausen T., Kaiser M., Huber R., Ehrmann M. (2011) Nat. Rev. Mol. Cell Biol. 12, 152–162 [DOI] [PubMed] [Google Scholar]
- 35. Hasenbein S., Merdanovic M., Ehrmann M. (2007) Genes Dev. 21, 6–10 [DOI] [PubMed] [Google Scholar]
- 36. Hasselblatt H., Kurzbauer R., Wilken C., Krojer T., Sawa J., Kurt J., Kirk R., Hasenbein S., Ehrmann M., Clausen T. (2007) Genes Dev. 21, 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meltzer M., Hasenbein S., Mamant N., Merdanovic M., Poepsel S., Hauske P., Kaiser M., Huber R., Krojer T., Clausen T., Ehrmann M. (2009) Res. Microbiol. 160, 660–666 [DOI] [PubMed] [Google Scholar]
- 38. Walsh N. P., Alba B. M., Bose B., Gross C. A., Sauer R. T. (2003) Cell 113, 61–71 [DOI] [PubMed] [Google Scholar]
- 39. Lipinska B., Fayet O., Baird L., Georgopoulos C. (1989) J. Bacteriol. 171, 1574–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spiess C., Beil A., Ehrmann M. (1999) Cell 97, 339–347 [DOI] [PubMed] [Google Scholar]
- 41. Strauch K. L., Beckwith J. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 1576–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kolmar H., Waller P. R., Sauer R. T. (1996) J. Bacteriol. 178, 5925–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Waller P. R., Sauer R. T. (1996) J. Bacteriol. 178, 1146–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Otwinowski Z., Minor W. (1997) Methods in Enzymology 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 45. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 47. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 48. Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 49. DeLano W. L. (2002) The PyMol Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 50. Merdanovic M., Mamant N., Meltzer M., Poepsel S., Auckenthaler A., Melgaard R., Hauske P., Nagel-Steger L., Clarke A. R., Kaiser M., Huber R., Ehrmann M. (2010) Nat. Struct. Mol. Biol. 17, 837–843 [DOI] [PubMed] [Google Scholar]
- 51. Krojer T., Pangerl K., Kurt J., Sawa J., Stingl C., Mechtler K., Huber R., Ehrmann M., Clausen T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7702–7707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chassin Y., Kapri-Pardes E., Sinvany G., Arad T., Adam Z. (2002) Plant Physiol. 130, 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Haussühl K., Andersson B., Adamska I. (2001) EMBO J. 20, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Truebestein L., Tennstaedt A., Mönig T., Krojer T., Canellas F., Kaiser M., Clausen T., Ehrmann M. (2011) Nat. Struct. Mol. Biol. 18, 386–388 [DOI] [PubMed] [Google Scholar]
- 55. Danese P. N., Snyder W. B., Cosma C. L., Davis L. J., Silhavy T. J. (1995) Genes Dev. 9, 387–398 [DOI] [PubMed] [Google Scholar]
- 56. Erickson J. W., Gross C. A. (1989) Genes Dev. 3, 1462–1471 [DOI] [PubMed] [Google Scholar]
- 57. Itzhaki H., Naveh L., Lindahl M., Cook M., Adam Z. (1998) J. Biol. Chem. 273, 7094–7098 [DOI] [PubMed] [Google Scholar]
- 58. Hauser M., Eichelmann H., Oja V., Heber U., Laisk A. (1995) Plant Physiol. 108, 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pfundel E. E., Renganathan M., Gilmore A. M., Yamamoto H. Y., Dilley R. A. (1994) Plant Physiol. 106, 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Evans D. F., Pye G., Bramley R., Clark A. G., Dyson T. J., Hardcastle J. D. (1988) Gut 29, 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.