Abstract
Despite their importance as members of the Roundabout (Robo) family in the control of axonal and vascular patterning, the transcriptional regulation of these genes is poorly understood. In this study, we show that members of the Sry-related high mobility box (Sox) transcription factor family as being transcriptional regulators of roundabout4 (robo4), a Robo gene family member that participates in sprouting angiogenesis in vivo, in zebrafish. Double whole mount in situ hybridization analysis in zebrafish embryos revealed co-localization of the vascular relevant Sox factors sox7 or sox18 mRNA with robo4 transcripts in developing intersomitic vessels. A 3-kb human ROBO4 promoter element was able to drive reporter expression in zebrafish to recapitulate the endogenous temporal intersomitic vessel expression pattern of robo4. EMSA analysis confirmed binding of Sox18 to a canonical Sox binding site (from −1170 bp to −1176 bp) in the ROBO4 promoter (3 kb), and mutation analysis indicated that this site was partially responsible for ROBO4 promoter activity in ECs. A combination of gain- and loss-of-function analysis identified Sox7 and Sox18 co-regulation of robo4 but not fli1a transcripts in zebrafish. Finally, Sox-mediated robo4 transcriptional regulation is conserved across evolution. These studies imply Sox-mediated transcriptional regulation of Robo4 in the developing embryonic vasculature.
Keywords: Cell Migration, Development, Endothelium, Gene Expression, Transcription Promoter, Robo4, Sox Factors, Angiogenesis, Zebrafish
Introduction
In developing vertebrates, neural and vascular patterning generate intricate branching networks that share several similar features (1). However, this connectivity is governed by a limited toolkit of signaling receptor systems. These systems must therefore be subjected to exquisite control to achieve proper patterning and avoid miscues. Recently, members of the axon guidance family have shown both expression and functionality in the developing vasculature. Of the four distinct families of axon guidance signaling partners, namely Slit-Robo, Ephrin-Eph, Netrin-Unc, and Semaphorin-Plexin, our laboratory has focused on the Slit-Robo family members and their role in the vasculature.
Roundabouts (Robos),4 a class of cell surface receptors that were originally identified to function in axon guidance (2), have recently been implicated in providing critical directional information for migration of endothelial cells (ECs) (3, 4). Four mammalian Robos are known, of which the fourth member robo4 is expressed in the intersomitic vessels (ISVs) and is strikingly regulated with peak expression passing in a “wave” along the trunk axis from 19–29 somites (3), suggesting a high degree of transcriptional control of this gene product. Recently, a 3-kb human ROBO4 promoter sequence has been identified that directs endothelial cell-specific expression pattern in vivo and in vitro (5). In addition, a guanine and adenine-binding protein-binding element in the ROBO4 promoter is necessary for endothelial expression in vivo (6). However, to date little is known regarding transcription factors that are involved in regulating robo4 expression during embryonic vascular development. In this study, we provide evidence for Sox7 and Sox18 transcription factors as regulators of robo4 vascular expression during embryonic development in zebrafish. SoxF genes, namely Sox7, Sox17, and Sox18, play pivotal roles in cardiovascular development including the orchestration of endothelial cell fate and cell differentiation (7). During embryonic mouse development, Sox7, Sox17, and Sox18 expression is evident in smaller branching vessels and ISVs (8–10) and zebrafish sox7 and sox18 are expressed in early angioblasts at lateral plate mesoderm and ISVs (11–13). To date, little is known in regard to transcriptional target for SoxF in the developing angiogenesis in vivo, and this study provides evidence that suggests robo4 may indeed serve as one candidate.
EXPERIMENTAL PROCEDURES
Cell Culture and Zebrafish Stocks
HUVECs were purchased from Lonza and maintained in endothelial cell basal medium-2 (EBM-2; Lonza) supplemented with fetal bovine serum (FBS) (2%) and 2 units/ml gentamycin. All HUVEC experiments were performed with cells in passages 3–6. Mouse Robo4 cDNA (probe generation) and human ROBO4 promoter-luciferase construct were kindly provided by Drs. Dean Y. Li (University of Utah) and William Aird (Beth Israel Deaconess Medical Center, Harvard). Dr. Monica Beltrame (Universita' degli Studi di Milano, Italy) provided the zebrafish sox7 and sox18 cDNAs. Zebrafish were grown and maintained at 28.5 °C in a 14-h day and 10-h night cycle. Mating was routinely carried out at 28.5 °C, and all embryos were staged according to established protocols. All zebrafish studies were performed under the Medical College of Wisconsin institutional guidelines (Animal Protocol 312-06-2).
RNA/MO Microinjections and in Situ Hybridization (ISH)
Zebrafish sox7 and sox18 RNA were transcribed by T7 polymerase from linearized vectors containing the respective inserts in pcDNA3.1. For gain-of-function (GOF) experiments, 50–75 pg of capped RNAs were injected into the embryonic cell (1-cell stage). Digoxigenin (DIG)-labeled antisense RNA probes for fli, and robo4 were generated using a DIG RNA labeling kit (Roche Applied Science). The MOs used for sox7 and sox18 were from a previous publication (11) and were injected at a dose of 0.25 pmol/embryo. The specificity and efficacy of the MOs used in this study have been reported previously (11).
For sox7 and sox18, chromogenic detection of single transcripts was carried out as described (14). For fluorogenic detection of two transcripts in co-expression studies, a DNP-labeled robo4 probe was used together with DIG-labeled sox7 and sox18 probes, respectively. Hybridized probes were visualized using peroxidase conjugated anti-DIG/DNP antibodies (1:1000; Roche Applied Science), FITC- and Cy3 tyramides (1:100) from the TSA system (PerkinElmer Life Sciences) according to the manufacturer's instructions. Pictures were acquired using a Zeiss Observer Z1 inverted microscope (single staining) or a Zeiss LSM 501 confocal microscope. Confocal data were processed with AxioVision 6.8, and linear level adjustment was carried out with Adobe Photoshop CS. Details on double stainings are available upon request. The mouse probe for Robo4 was made as described previously (15). Mouse section in situ hybridization was performed on 7-μm sections of paraformaldehyde-fixed, paraffin-embedded embryos. Sections were de-waxed, rehydrated, and incubated in 5 mg/ml proteinase K for 20 min at room temperature. After washing in phosphate buffered saline, sections were refixed with 4% paraformaldehyde for 10 min at room temperature, acetylated, and prehybridized with hybridization solution (50% formamide, 5× SSC, 5× Denhardt's, 250 mg/ml yeast RNA, 500 mg/ml herring sperm DNA) for 2 h at room temperature. Hybridization (hybridization solution + 0.5 μg/ml probe) was performed overnight at 60 °C. Slides were washed in 5× SSC for 5 min, 0.2× SSC for 1 h at 60 °C, 0.2× SSC for 5 min at room temperature and NT buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.5) for 5 min at room temperature, before incubating for 2 h with blocking solution (0.5% blocking powder (Roche Applied Science) in NT buffer) in a humidified chamber. Anti-DIG antibody (Roche Applied Science) at a 1:500 dilution in blocking solution was added to the slides and incubated overnight at 4 °C. Unbound antibodies were removed by washing three times in NT buffer supplemented with 0.05% Tween 40 (Sigma). Sections were equilibrated in detection buffer (0.1 m Tris, pH 8.0, 0.1 m NaCl, 10 mm MgCl2) for 15 min at room temperature and incubated with color solution (BM purple; Roche Applied Science) according to the manufacturer's recommendations. Finally, sections were washed 2 × 5 min in phosphate buffered Tween and mounted in 80% glycerol in PBS. Stained sections were examined with an Olympus BX-51 microscope (DP-70 12Mp color camera).
Electrophoretic Mobility Shift Assay (EMSA)
Oligonucleotides synthesized by Integrated DNA Technologies (Coralville, IA) were used for DNA binding assays. Sequence information is provided in the supplemental Methods. Double-stranded probes were generated by heating equal molar amounts of each of the 5′→3′ oligonucleotide with its respective complementary oligonucleotide at 95 °C for 10 min followed by cooling to room temperature for 1 h. Next, double-stranded oligonucleotides were labeled with DIG-11-ddUTP using recombinant terminal transferase (20 units/ml) in a final volume of 25 μl according to the DIG Gel Shift Kit, Second Generation instructions (Roche Applied Science). EMSA was performed as we have previously described in detail (16). Briefly, DNA binding reactions were set up with 1 μg of nuclear or cytoplasmic proteins (HUVEC passage 3) and 0.08 pmol of the DIG-labeled wild-type or mutant SOX18 probe in our modified DNA binding buffer (20 mm Hepes, pH 7.6, 10 mm (NH4)2SO4, 0.2% Tween 20, 30 mm KCl) (17) containing 1 μg of poly(dI-dC) and 0.1 μg of poly-l-lysine in a final reaction volume of 20 μl. For supershift assays, 2–3 μl of Sox18 antibody (4 μg, EMSA certified; Santa Cruz Biotechnology) was added to the nuclear or cytoplasmic proteins prior to addition of the probe. For competition experiments, unlabeled double-stranded SOX18 WT oligonucleotide at final concentrations of 0.08, 0.8, 4.0, 8.0, 16.0, and 32.0 pmol were added simultaneously with 0.08 pmol of DIG-labeled SOX18 WT probe to the binding reaction with 1 μg of nuclear protein. Control reactions were performed with the probe alone. Reactions were incubated at room temperature for 15 min after which samples were subjected to electrophoresis on a 6% DNA Retardation Gel (Invitrogen). Blotting on to a positively charged nylon membrane was followed by UV cross-linking. Blots were then incubated with AP-conjugated to anti-DIG antibody followed by addition of substrate disodium 3-chloro-3-(methoxyspiro{1,2-dioxetane-3-2′-(5′-chloro)tricyclo[3.3.3.3]decan}-4-yl) phenyl phosphate, chemiluminescent alkaline phosphatase substrate (CSPD). Fluor Chem HD from Alpha Innotech was used for chemiluminescence detection and quantification of bands was done using ImageJ software (National Institutes of Health).
Statistical Analysis
Statistical analysis was performed using the Student's t test with Graph Pad Prism (GraphPad Software, La Jolla, CA) and Microsoft Office Excel 2010 software package. All data are presented as mean ± S.E. (n and p value are provided in each figure or legend).
RESULTS
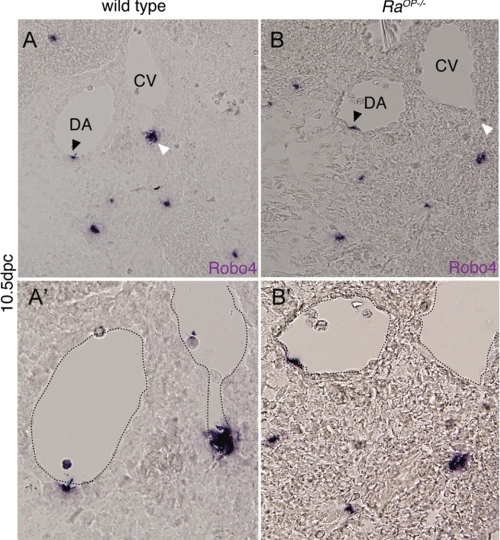
In a microarray transcriptional profile study comparing lymphatic ECs isolated from ragged-opossum (RaOp) mutant mice (RaOp−/− mice) (18, 19), which carries a mutation in Sox18 to lymphatic ECs obtained from WT mice, we observed an 8.5-fold decrease in Robo4 transcript levels with no detectable change in Robo1 and Robo2 transcript levels detectable. To validate the microarray result, we performed robo4 ISH on embryonic day 10.5 sections of RaOp−/− mice and WT mice. RaOp−/− mice showed diminished robo4 expression in caudal vein ECs (Fig. 1B, white arrowhead) compared with WT mice (Fig. 1A). No change in Robo4 expression was observed in dorsal aorta (DA) EC (Fig. 1A, black arrowhead), suggesting preferential venous loss of Robo4 expression in Sox18 KO mice. These results led us to hypothesize that Sox18 transcription factor affects transcription of Robo4 gene during embryonic vascular development in vivo. To test this hypothesis, we investigated the Sox-mediated transcriptional regulation of robo4 gene in embryonic zebrafish development.
FIGURE 1.
Sox18 knock-out mice show polar loss of Robo4 expression in caudal vein (CV). A and B are transverse section of Robo4 in situ 10.5 days postcoitus (dpc) embryos (WT and RaOp−/−). The expression of Robo4 is detected in the endothelium of both DA and caudal vein in dorsolateral polarized fashion. The black arrowhead shows Robo4 expression in the DA, and the white arrowhead shows the expression in the caudal vein. A′ and B′ are, respectively, high power images of the A and B, respectively, with the dotted line outlining the DA and CV. ISH was performed on 3 WT and 3 RaOp−/− homozygous embryos.
Sox 7 and Sox 18 Transcripts Are Expressed Prior to Robo4 Transcript Expression and Co-localize during Embryonic Zebrafish ISV Development
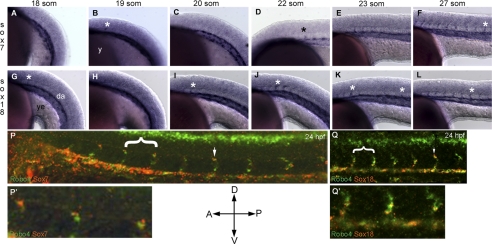
In zebrafish, two Sox factors, namely Sox7 and Sox18, show redundant function during embryonic vascular development (11–13). We next performed whole mount ISH for sox7 (Fig. 2, A–F) and sox18 (Fig. 2, G–L) in zebrafish embryos ranging from 18 to 27 som compared with robo4 between 19 and 24 som (supplemental Fig. S1, A–C). At 18–19 som, both sox7 (Fig. 2A) and sox18 (Fig. 2G, asterisk) expression was observed in rostral ISV sprouts as they began to emerge and in axial vessel, DA. The robo4 expression at this time point was strong in notochord and was observed in angioblasts (supplemental Fig. S1A). As ISV development progresses, the expression of sox7 (Fig. 2, D–F) and sox18 (Fig. 2, I–L) followed a rostral-caudal temporal pattern resembling the pattern observed previously for robo4 (3) although the sox expression appears a somite or two early than robo4. We next investigated whether robo4, sox7, and sox18 transcripts were co-expressed in the vasculature during embryonic zebrafish development stages (19–24 som) where robo4 is expressed in ISVs (3). Confocal analysis of two-color fluorescence ISH of the zebrafish trunk regions shows that both sox7 (Fig. 2P) and sox18 (Fig. 2Q) transcripts were co-localized with robo4 transcript in the developing zebrafish vasculature. At this time point (24 hpf), sox7 expression was noticed predominantly in the leading ISV cell (Fig. 2P′) whereas sox18 expression was predominant in the axial vessels and the cell immediately ventral to the ISV leading cell (Fig. 2Q′). Our double ISH analysis clearly shows that both robo4 and individual sox (sox7/sox18) transcripts are co-localized on ISVs. Additional inverted microscope images are provided in supplemental Fig. S2, A–F along with three-dimensional surface rendering of the confocal picture that capture the regions of co-localization (supplemental Fig. S2G). The ISV expression data together suggest that sox7 and sox18 are expressed in the rostral ISV sprouts prior to robo4 and could potentially influence the highly dynamic robo4 ISV expression observed in these stages.
FIGURE 2.
Montage of robo4, sox7, and sox18 endogenous expression across embryonic zebrafish development. Whole mount for sox7 (A–F), and sox18 (G–L) ISH embryos were performed as indicated under “Experimental Procedures.” Embryos were positioned with anterior (A) to the left, posterior (P) to the right and dorsal (D) to the top and ventral (V) to the bottom as indicated by the orientation bars. Embryos were staged according to the somite numbers as indicated in the respective panels. Asterisks (black and white) indicate ISVs in the zebrafish trunk region. da, dorsal aorta; y, yolk; ye, yolk extension. P and Q are whole mount two-color confocal fluorescent sox7 or sox18 (red) with robo4 (green) ISH images of 24 hpf zebrafish trunk. White arrows indicate ISVs co-localized for robo4 and sox7/18 transcript. P′ and Q′ are higher magnification of regions highlighted by white brackets in P and Q, respectively.
Sox18 Induces ROBO4 Promoter Activity via Specific Sox Binding Site in Vitro in ECs
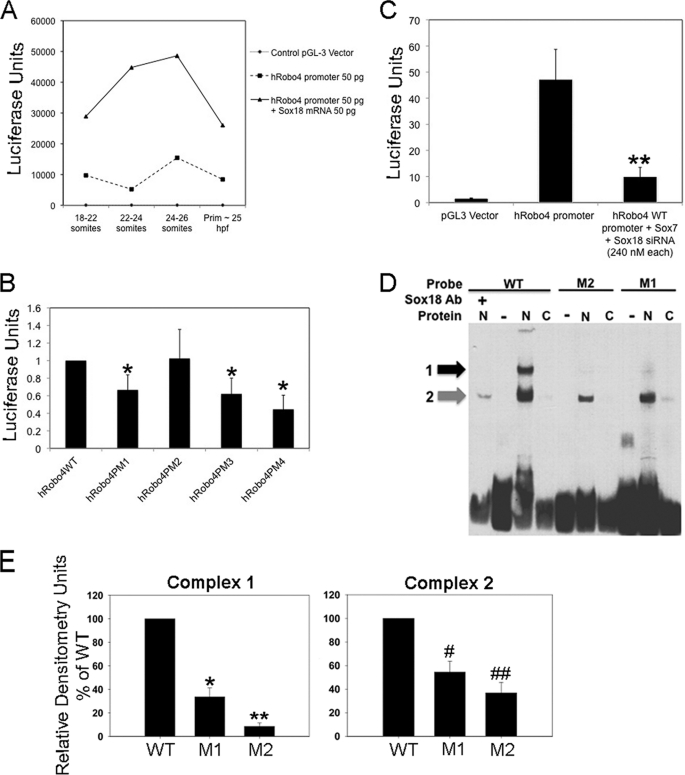
Because sox7 and sox18 were expressed prior to robo4 during ISV development, we investigated whether Sox TFs regulated robo4 ISVs expression level in zebrafish. To investigate this question, we utilized the 5′-flanking sequence element of the human ROBO4 promoter, which was already shown to direct EC-specific expression in vivo (5, 6). Further, bioinformatic analysis of this promoter indicated the presence of putative Sox18 TF binding consensus sequences (A/TA/TCAAA/TG) between −1170 and −1176 bp, which is conserved in mouse Robo4 promoter between −1339 and −1405 bp as well (supplemental Fig. S1E). We injected the 3-kb human ROBO4 promoter-luciferase construct alone (50 pg) or in combination with sox18 mRNA (50 pg) at 1-cell stage into the embryonic cell and collected embryos at 18–21 som, 21–26 som, 26 som, and 24-prim6 stages for luciferase assays (Fig. 3A), developmental stages in which endogenous robo4 expression is highly dynamic. Embryos from each stage were lysed and assayed for luciferase activity. Remarkably, the exogenous human ROBO4 promoter activity drives luciferase in a pattern reminiscent of endogenous robo4 expression in ISVs. We observed a bell-shaped curve over the time course with maximal reporter activity at around 24–26 som stage and returning to base line at 25 hpf-prim6 stage (Fig. 3A, dotted line). Interestingly, in sox18 mRNA co-injected embryos (Fig. 3A, black line), we found that the ROBO4 promoter activity was increased at all time intervals compared with basal levels (Fig. 3A, diamond bars), in essence replicating the bell-shaped curve of promoter alone injected embryos but with higher luciferase values. Next, we generated four point mutants (SoxPM1, SoxPM2, SoxPM3, SoxPM4) (supplemental Fig. S1E) that had single nucleotide (SoxPM1–3) or multiple nucleotide (SoxPM4) substitutions at the putative Sox18 binding site in the ROBO4 promoter. These constructs were individually transfected in HUVECs (Fig. 3B), and their activity was compared with WT human ROBO4 promoter (supplemental Fig. S1D). All constructs were transfected via electroporation or Lipofectamine 2000 into HUVEC cells, and luciferase assays were performed from HUVEC lysates. Interestingly, two (SoxPM1 and SoxPM3) of the three point mutants in addition to SoxPM4 showed reduction in luciferase output (Fig. 3B), indicating that the putative Sox binding site on −1169 bp site is responsible in part for ROBO4 promoter activity in ECs. All groups except SoxPM2 were statistically significant (p < 0.05) compared with the WT human ROBO4 promoter group.
FIGURE 3.
Robo4:Sox7/18 transcriptional regulation. A, graphical representation of Sox18-induced ROBO4 promoter activity in vivo. Details of the experimental design are provided under “Experimental Procedures.” Dotted line denotes the base-line exogenous ROBO4 promoter activity. Black line denotes exogenous ROBO4 promoter activity in sox18 overexpression (sox18 mRNA) embryos. Line along the x axis indicates empty control pGL3 vector-injected embryos. The graph is a representative experimental data set, and each time interval contained 20–25 embryos. This experiment was performed twice with identical trends, and error bars have not been provided due to the high variation in the luciferase values from one experiment to the next. B, in vitro luciferase assays in HUVECs for ROBO4 promoter point mutants (PM1–4) compared with h ROBO4 WT promoter. Error bars represent S.E. from three independent experiments, and all luciferase values are shown as -fold compared with h ROBO4 WT promoter sample. All sample groups were compared with h ROBO4 WT promoter groups, and all samples except h ROBO4PM2 were statistically significant at *p < 0.05. C, ROBO4 promoter activity in control (lacZ siRNA) and Sox7 and Sox18 knockdown (Sox7 + Sox18 siRNA) ECs. B and C, HUVECs transiently transfected with WT-Robo4 and Robo-4 PM1–4 (four mutants) constructs or WT h ROBO4 promoter (1.5 μg) and lacZ and Sox7 + Sox18 siRNA (240 nm each) for 36 h and promoter activity determined as a function of luciferase activity. Luciferase (firefly) readings were normalized to the control, and data from three independent experiments are compiled together. **, p < 0.05 was determined by two-tailed statistical analysis. D, EMSA of DNA binding reactions with Sox18 mutant (M1 and M2) and WT probes incubated with nuclear (N) or cytoplasmic (C) proteins from ECs, in the presence or absence of SOX18 antibody (Ab). The black and gray arrows indicate the shifted complex 1 and 2, respectively. Complex 1, top band (black arrow), is diminished in intensity in mutant M1 probe N lanes and is absent in mutant M2 probe and wild-type probe plus Ab lanes. E, chemiluminescence values of the complex 1 and 2 bands (arbitrary unit) measured by Fluor Chem HD and band intensities of the two complexes in M1 and M2 probe EMSA relative to WT probe EMSA. Data are from three independent EMSA reactions, and error bars represent S.E. (Complex 1 M1: *, p = 0.0009; M2: **, p = 0.0001; Complex 2 M1: #, p = 0.0076; M2: ##, p < 0.0018).
To investigate whether the endogenous Sox7 or Sox18 proteins are responsible for the ROBO4 promoter activity observed in human ECs, we utilized sox7 or sox18 gene-specific efficacy-confirmed siRNAs (Santa Cruz Biotechnology) (supplemental Fig. S2I) to knock down endogenous Sox proteins and measured luciferase activity output of the hROBO4 promoter. The luciferase activity is greatly reduced from the ROBO4 promoter co-transfected with Sox7/18 double siRNA sample compared with ROBO4 promoter-luciferase construct alone (Fig. 3C, p < 0.05). To determine conclusively whether Sox7/18 proteins bind to putative Sox binding site in ROBO4 promoter, we performed an EMSA (Fig. 3D) with nuclear extracts from ECs using a WT ROBO4 promoter oligonucleotide probe and Sox18 mutant ROBO4 promoter oligonucleotide probes (M1 and M2). As observed in the EMSA blot, the nuclear proteins from HUVECs in the WT probe lane clearly formed two complexes: complex 1 (slower migrating) and complex 2 (faster migrating) band (Fig. 3D). Interestingly, the M1 mutant ROBO4 promoter oligonucleotide probe also showed the presence of the two complexes albeit of lower intensity (Fig. 3D, M1 lane). Because the M1 mutant probe has additional Sox18 binding sites, which likely serves as alternate site for interaction, we generated a second mutant probe M2, where we mutated this site (for mutant probe sequences, see supplemental Methods). Clearly, the M2 probe shows lower intensities in complex 2 (Fig. 3D, M2 lane). When the complex intensities in M1 and M2 probe lanes for nuclear protein extracts (N) were compared across three independent experiments with WT probe bands, we noticed a 70 and 90% decrease in complex 1 (M1: *, p < 0.001; M2: **, p < 0.0005), and a 50 and 70% decrease in complex 2 (M1: #, p < 0.001; M2: ##, p < 0.005) for M1 and M2 probes (Fig. 3E). The M2 probe showed lower intensities for both complexes compared with the M1 probe. Both complexes also showed a dose-dependent reduction in intensity when unlabeled competitor probes was included in the EMSA reaction (supplemental Fig. S3B).
To determine whether SOX18 protein was present in the shifted complexes, we performed supershift assays with SOX18 antibody (Fig. 3D, Ab lane). Supershift results indicated the presence of SOX18 in both complex 1 (black arrow), and 2 (gray arrow) (Fig. 3D, lanes N and Ab). In addition, when binding reactions with the mutant (M1, data not shown) and WT probes (Fig. 3D, lane +SOX18 Ab) were performed in the presence of SOX18 antibody, the complex 1 band completely disappeared in WT probe lane (data not shown for M1), and the intensity of the complex 2 band decreased by 49 and 65% in reactions with M1 and WT probes, respectively. Collectively, these data suggest that Sox18 is part of the complex that binds to ROBO4 promoter sequences and, in addition other co-factors may also be responsible in part for ROBO4 promoter activity in ECs.
Taking the in vivo human ROBO4 promoter activity across ISV development, the ability of Sox18 to induce the ROBO4 promoter robustly in vivo, point mutation analysis and Sox siRNA experiments in vitro, and the EMSA analysis data together we conclude that Sox7/18 are responsible in part for inducing ROBO4 promoter activity in ECs in vivo and in vitro.
Sox GOF or LOF Embryos Show Complementary Gain or Loss of robo4 Expression
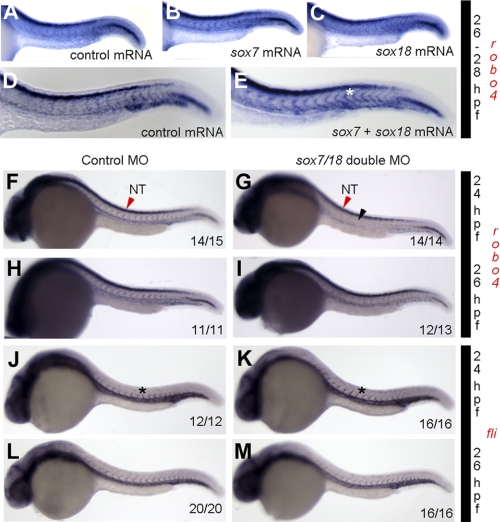
To investigate whether endogenous robo4 expression is modulated by Sox transcription factors, we performed GOF and LOF for sox7 and sox18 in zebrafish embryos. For GOF experiments, we injected sox7 or sox18 mRNA alone or in combination into 1-cell embryo and checked robo4 expression by ISH at 26–28 hpf. The robo4 ISV expression is enhanced in sox7 + sox18 mRNA (Fig. 4E, white asterisk)-injected embryos compared with sox7 (Fig. 4B) or sox18 (Fig. 4C) or control mRNA (Fig. 4D) alone injected embryos at 24 hpf, which suggests co-operative interaction between the two factors in inducing robo4 ISV expression. Conversely, in LOF experiments, double knockdown of sox18/sox7 using MOs resulted in a diminished robo4 expression at 24 hpf (Fig. 4G) and 26 hpf (Fig. 4I) compared with control MO (Fig. 4, F and H) injected embryos. At 24 hpf, 40 of 42 (95%) sox18/sox7 double knockdown embryos show the phenotype depicted in Fig. 4G. At 26 hpf, 26 of 27 double morphants showed diminished robo4 expression (Fig. 4I). Interestingly, the diminished expression was selectively observed in the ISVs (Fig. 4G, black arrowhead) with little to no qualitative change detected in the neural tube (Fig. 4, G and F, NT, red arrowhead). We also checked fli1a expression in control MO (Fig. 4, J and L) and sox7/18 double MO injected (Fig. 4, K and M) 24 or 26 hpf embryos and observed no change (Figs. 4, J and K, black asterisk) in control (21 embryos) and double morphants (32 embryos), suggesting specificity of Sox regulation of robo4 ISV expression. Further, quantitative PCR for robo4 and robo1 transcripts in Sox knockdown embryos shows selective down-regulation of robo4 versus robo1 transcripts (supplemental Fig. S3C). This also suggests some level of Robo specificity for Sox-mediated transcriptional regulation during development.
FIGURE 4.
Sox GOF and LOF show reciprocal change in robo4 transcript expression but no changes in fli expression. A–E, robo4 ISH trunk expression between 26 and 28 hpf zebrafish embryo microinjected with control (A and D), sox7 (B), sox18 (C), and sox7 + sox18 mRNA (E) (50 pg each). A–E show strong robo4 expression in the neural tube. However, sox7 + sox18 double mRNA injected embryos show strong robo4 expression in the ISV (asterisk). Experiments were repeated three independent times, and pictures are representative of 15 embryos/injection group. Additional quantification is provided in supplemental Fig S2F. F–M, robo4 ISH trunk expression in 24 hpf (F and G) and 26 hpf (H and I) and flia at 24 hpf (J and K) and 26 hpf (L and M) zebrafish embryos injected with control MO (F, H, J, and L) and sox7 + sox18 double MO (G, I, K, and M). The inset numbers in F, G, H, I, J, K, L, and M indicate number of embryos that show phenotype similar to that in the image. Asterisks in J and K and black arrowhead in G indicate ISV expression. Red arrowheads in F and G indicate NT robo4 expression.
Because robo4 expression is also observed prior to 24 hpf in nonvascular tissues such as neural tube (NT) and notochord, we investigated the effect of sox7 or sox18 or double (sox7 + sox18) mRNA-injected embryos for robo4 transcript expression at 18 hpf (supplemental Fig. S4). At 18 hpf, little to no change was observed in robo4 expression in notochord. However, in NT (supplemental Fig. S4, B–D, black asterisk) and midbrain-hindbrain boundary (supplemental Fig. S4, B–D, red arrow), we observed an expansion in the robo4 expression domain in sox7 (supplemental Fig. S4B) or sox18 (supplemental Fig. S4C) or sox7 + sox18 (supplemental Fig. S4D) mRNA-injected embryos. Quantitation shows >60% of injected embryos show strong overall robo4 induction qualitatively in midbrain-hindbrain boundary and NT (supplemental Fig. S4E) at 18 hpf. This result is not totally unexpected because Sox proteins are well known to function in neural development (20, 21). At 24 hpf, quantitation was performed for robo4 induction in ISVs (supplemental Fig. S4F). The Sox GOF and LOF results when taken together show complementary changes in robo4 ISV expression in vivo, which is in agreement with the redundant function of Sox7 and Sox18 function in zebrafish vascular development (11). Taking the mouse data together from Fig. 1, these data also suggest that Sox-mediated transcriptional regulation of Robo4 is conserved across evolution.
DISCUSSION
This study identifies members of the Sox protein transcription factor family as putative regulators of Robo4 gene expression in vivo. The primary findings of this study include the following: (i) early temporal expression of sox7 and sox18 transcripts in DA prior to robo4 expression; (ii) remarkably conserved human ROBO4 promoter activity in its behavior in zebrafish, correlating well with vascular robo4 expression pattern in vivo; (iii) polar regulation of robo4 expression via Sox in zebrafish and mice; (iv) Sox7 and Sox18 along with temporal and spatial specific co-factors in regulation of robo4 transcript expression in the vasculature.
Whole mount ISH for sox7 and sox18 during zebrafish ISV development shows a preponderance of sox7 transcript in the tip of the leading sprout and sox18 transcript in the base of the sprout with prominent expression in axial vessels. Further, both sox7 and sox18 transcripts appear in the rostral sprouts earlier than robo4 expression, suggesting regulatory mechanisms of Sox and Robo in ISV sprouting process. In terms of robo4, ISV expression is tightly controlled across a short temporal window, and the expression is observed along the length of the entire sprout as well as the DA. These data argue that different Robo-Sox combinations are involved in ISV cell development in vivo. Perhaps Robo4 and Sox7 in the tip and Robo4 and Sox18 in the base of the sprout function together to direct and maintain ISV sprout formation. Whether the expression levels of sox transcripts correlate with protein expression and function is not known. Interestingly, the polar distribution of sox transcripts in zebrafish ISVs suggests polar regulation of robo4 transcript, which is curiously observed in mice. The Robo4 expression in RaOp mutant mice is selectively down-regulated in vein but not in artery, suggesting selective regulatory function of Sox-mediated Robo4 induction in vein. It is worthwhile to note that the site of Robo4 expression in vein is the area where emergence of Sox18-mediated lymphatic Prox1+ cells is noted (22), postulating a role of Robo4 in lymphatic ECs cell directional sprouting. Because lymphatic ECs derive from venous ECs (23) it is tempting to speculate that the selective regulation of Sox-mediated Robo4 induction in venous ECs is potentially the event that triggers the directional migration of Prox1 LECs from vein.
In terms of evolution, both mouse and human ROBO4 promoter share a Sox18 binding site proximal to the start site (−1170 in human and −1339 in mouse). Because robo4 ISV expression is dynamically controlled across a short temporal window (18–29 hpf), this would suggest tight control of ROBO4 promoter activity during this time frame. In fact, a previously published 3-kb human ROBO4 promoter (5, 6) that shows endothelial-restricted expression in mice behaves similarly in zebrafish across the short temporal window of robo4 ISV expression. This remarkable correlative behavior is enhanced when exogenous sox18 mRNA is provided and is diminished when the Sox18 binding site is mutated. Although the ROBO4 promoter activity is not lost completely in the absence of Sox18 binding site, which is expected because other sites on the promoter such as guanine and adenine-binding protein-binding element has also been shown previously to be necessary for endothelial expression in vivo (6). In fact, at least three pieces of distinct experimental evidence point to the role of co-factors that participate with Sox in regulating robo4 ISV expression. (i) Mutation of Sox binding site on ROBO4 promoter (Fig. 3B) does not result in complete loss of ROBO4 promoter activity in ECs. (ii) The EMSA study shows that Sox18 mutant probe (M1) clearly binds to non-Sox nuclear proteins, which is not blocked by Sox18 antibody (data not shown). (iii) Sox GOF single mRNA and double mRNA-injected embryos (Fig. 4, A–D) show differential robo4 transcript regulation at 18 hpf (individual Soxs and double Soxs) (supplemental Fig. S4) and 24 hpf (only double Soxs) (Fig. 4) presumably mediated by different sets of co-factors expressed at these time points during embryonic development. The 24 hpf double mRNA data are in agreement with redundant function of Soxs in vascular development in zebrafish (11).
In this study, we provide convincing data in three species, each of which independently provides evidence that Sox transcription factors putatively regulate robo4 gene expression. In each of these systems there is ample published evidence of conservation of the molecular mechanisms of vascular development for Sox and Robo (3, 7, 9, 11, 13, 15, 18, 24). In addition, the genes investigated in our study are well conserved at the sequence level between the species. In sum, the presented zebrafish, mouse, and human EC data make a substantive body of evidence for conservation of this observation in development. This study for the first time provides a molecular link between Sox and Robo family in vascular development, a paradigm recently shared by another axon guidance gene of the ephrin-Eph family (25).
Supplementary Material
Acknowledgments
We thank members of the Vascular Biology Affinity Group and the Developmental Biology Group for invaluable input in these studies. We are grateful to Marc Achen, Steven Stacker, and Tara Karnezis for generously providing unpublished microarray data on the transcriptional profile study comparing lymphatic ECs from Sox18 KO and WT mice. We thank Suresh Kumar of the Medical College of Wisconsin/Children's Research Institute confocal facility with assistance in obtaining zebrafish trunk ISV images.
This work was supported, in whole or in part, by National Institutes of Health Grant HL090712 and an administrative supplement to HL090712 (to R. R.). This work was also supported by an Advancing Healthier Wisconsin grant and Children's Research Institute seed funds (to R. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4, Methods, and additional references.
- Robo
- human roundabout
- robo4
- zebrafish roundabout4
- DA
- dorsal aorta
- DIG
- digoxigenin
- fli
- friend leukemia virus integration
- GOF
- gain-of-function
- hpf
- hours postfertilization
- ISH
- in situ hybridization
- ISV
- intersomitic vessel
- HUVEC
- human umbilical vein endothelial cell
- LOF
- loss-of-function
- MO
- morpholino
- NT
- neural tube
- Op
- opossum
- som
- somite.
REFERENCES
- 1. Carmeliet P., Tessier-Lavigne M. (2005) Nature 436, 193–200 [DOI] [PubMed] [Google Scholar]
- 2. Kidd T., Brose K., Mitchell K. J., Fetter R. D., Tessier-Lavigne M., Goodman C. S., Tear G. (1998) Cell 92, 205–215 [DOI] [PubMed] [Google Scholar]
- 3. Bedell V. M., Yeo S. Y., Park K. W., Chung J., Seth P., Shivalingappa V., Zhao J., Obara T., Sukhatme V. P., Drummond I. A., Li D. Y., Ramchandran R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6373–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaur S., Castellone M. D., Bedell V. M., Konar M., Gutkind J. S., Ramchandran R. (2006) J. Biol. Chem. 281, 11347–11356 [DOI] [PubMed] [Google Scholar]
- 5. Okada Y., Yano K., Jin E., Funahashi N., Kitayama M., Doi T., Spokes K., Beeler D. L., Shih S. C., Okada H., Danilov T. A., Maynard E., Minami T., Oettgen P., Aird W. C. (2007) Circ. Res. 100, 1712–1722 [DOI] [PubMed] [Google Scholar]
- 6. Okada Y., Jin E., Nikolova-Krstevski V., Yano K., Liu J., Beeler D., Spokes K., Kitayama M., Funahashi N., Doi T., Janes L., Minami T., Oettgen P., Aird W. C. (2008) Blood 112, 2336–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Francois M., Koopman P., Beltrame M. (2010) Int. J. Biochem. Cell Biol. 42, 445–448 [DOI] [PubMed] [Google Scholar]
- 8. Pennisi D., Bowles J., Nagy A., Muscat G., Koopman P. (2000) Mol. Cell. Biol. 20, 9331–9336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sakamoto Y., Hara K., Kanai-Azuma M., Matsui T., Miura Y., Tsunekawa N., Kurohmaru M., Saijoh Y., Koopman P., Kanai Y. (2007) Biochem. Biophys. Res. Commun. 360, 539–544 [DOI] [PubMed] [Google Scholar]
- 10. Young N., Hahn C. N., Poh A., Dong C., Wilhelm D., Olsson J., Muscat G. E., Parsons P., Gamble J. R., Koopman P. (2006) J. Natl. Cancer Inst. 98, 1060–1067 [DOI] [PubMed] [Google Scholar]
- 11. Cermenati S., Moleri S., Cimbro S., Corti P., Del Giacco L., Amodeo R., Dejana E., Koopman P., Cotelli F., Beltrame M. (2008) Blood 111, 2657–2666 [DOI] [PubMed] [Google Scholar]
- 12. Herpers R., van de Kamp E., Duckers H. J., Schulte-Merker S. (2008) Circ. Res. 102, 12–15 [DOI] [PubMed] [Google Scholar]
- 13. Pendeville H., Winandy M., Manfroid I., Nivelles O., Motte P., Pasque V., Peers B., Struman I., Martial J. A., Voz M. L. (2008) Dev. Biol. 317, 405–416 [DOI] [PubMed] [Google Scholar]
- 14. Oxtoby E., Jowett T. (1993) Nucleic Acids Res. 21, 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park K. W., Morrison C. M., Sorensen L. K., Jones C. A., Rao Y., Chien C. B., Wu J. Y., Urness L. D., Li D. Y. (2003) Dev. Biol. 261, 251–267 [DOI] [PubMed] [Google Scholar]
- 16. Tabatabai N. M., Blumenthal S. S., Petering D. H. (2005) Toxicology 207, 369–382 [DOI] [PubMed] [Google Scholar]
- 17. Kothinti R. K., Blodgett A. B., Petering D. H., Tabatabai N. M. (2010) Toxicol. Appl. Pharmacol. 244, 254–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. François M., Caprini A., Hosking B., Orsenigo F., Wilhelm D., Browne C., Paavonen K., Karnezis T., Shayan R., Downes M., Davidson T., Tutt D., Cheah K. S., Stacker S. A., Muscat G. E., Achen M. G., Dejana E., Koopman P. (2008) Nature 456, 643–647 [DOI] [PubMed] [Google Scholar]
- 19. James K., Hosking B., Gardner J., Muscat G. E., Koopman P. (2003) Genesis 36, 1–6 [DOI] [PubMed] [Google Scholar]
- 20. Cheung M., Briscoe J. (2003) Development 130, 5681–5693 [DOI] [PubMed] [Google Scholar]
- 21. Honoré S. M., Aybar M. J., Mayor R. (2003) Dev. Biol. 260, 79–96 [DOI] [PubMed] [Google Scholar]
- 22. Srinivasan R. S., Geng X., Yang Y., Wang Y., Mukatira S., Studer M., Porto M. P., Lagutin O., Oliver G. (2010) Genes Dev. 24, 696–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Srinivasan R. S., Dillard M. E., Lagutin O. V., Lin F. J., Tsai S., Tsai M. J., Samokhvalov I. M., Oliver G. (2007) Genes Dev. 21, 2422–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pennisi D., Gardner J., Chambers D., Hosking B., Peters J., Muscat G., Abbott C., Koopman P. (2000) Nat. Genet. 24, 434–437 [DOI] [PubMed] [Google Scholar]
- 25. Parrinello S., Napoli I., Ribeiro S., Digby P. W., Fedorova M., Parkinson D. B., Doddrell R. D., Nakayama M., Adams R. H., Lloyd A. C. (2010) Cell 143, 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.