Abstract
We examined over 50 mutations in the Drosophila βPS integrin subunit that alter integrin function in situ for their ability to bind a soluble monovalent ligand, TWOW-1. Surprisingly, very few of the mutations, which were selected for conditional lethality in the fly, reduce the ligand binding ability of the integrin. The most prevalent class of mutations activates the integrin heterodimer. These findings emphasize the importance of integrin affinity regulation and point out how molecular interactions throughout the integrin molecule are important in keeping the integrin in a low affinity state. Mutations strongly support the controversial deadbolt hypothesis, where the CD loop in the β tail domain acts to restrain the I domain in the inactive, bent conformation. Site-directed mutations in the cytoplasmic domains of βPS and αPS2C reveal different effects on ligand binding from those observed for αIIbβ3 integrins and identify for the first time a cytoplasmic cysteine residue, conserved in three human integrins, as being important in affinity regulation. In the fly, we find that genetic interactions of the βPS mutations with reduction in talin function are consistent with the integrin affinity differences measured in cells. Additionally, these genetic interactions report on increased and decreased integrin functions that do not result in affinity changes in the PS2C integrin measured in cultured cells.
Keywords: Cell Adhesion, Drosophila, Extracellular Matrix, Integrin, Ligand-binding Protein, Protein Structure
Introduction
Integrin cell surface receptors are important for morphogenetic events in normal development and are also critical for adult and many disease-related processes as well (1). The integrin αβ heterodimer is a dynamic receptor, whose activity is regulated by interactions with extracellular, intracellular, and other transmembrane proteins. This regulation can change the affinity of individual integrin molecules or alter the clustering of integrins such that their avidity is altered. Experiments utilizing conformation-specific antibodies, high resolution x-ray, NMR, small angle neutron scattering, molecular dynamics, steered molecular dynamics, and electron microscopy are elucidating how the α and β subunits are organized and change in response to activating conditions and ligand binding. One goal of the current work on integrin biology is to understand how the structural information gained on isolated integrins relates to integrin function in a cellular context. Therefore, models for integrin activation are being tested by site-directed mutagenesis (reviewed in Refs. 2–4).
We have used a complementary approach of first identifying amino acids in the Drosophila integrin βPS subunit that alter its function in the whole organism and then examining the effects of these mutations at the cellular and molecular level. Previous work demonstrated that myospheroid (mys, encoding βPS integrin) hypomorphic mutations displayed stronger phenotypes at higher temperatures (5). Therefore, we screened for mutations in mys that produce viable flies at low temperatures but lethality at high temperatures. Because null alleles of mys are completely lethal at all temperatures, we know that these mutations do not completely inactivate the function of the integrin but rather compromise its function. Due to unique properties of one of the mutants, mysb58, we examined it in detail and found that it is an activating mutation that results in an integrin with higher affinity for a soluble extracellular matrix ligand (6, 7). At that time, we suspected that mysb58 was unique among the mutants in our screen.
We have now tested the ligand binding ability of all 50 of the mutants generated in our previous screen for function-altering alleles as well as a number of lethal mutants that produce mutant integrin subunits that are expressed on the cell surface and site-directed mutations suggested by our initial studies or results obtained in work on vertebrate integrins. The ligand binding assay that we used measures the ability of the cell surface integrin to bind to a monovalent ligand and therefore probes only the affinity of integrins for one ligand. The assay reports on the effect a mutation has on the regulation of integrin affinity, presumably by altering an amino acid contacting the ligand or one regulating integrin conformation. This is in contrast to the testing of numerous mutations in the human integrin β3 subunit generated by site-directed mutagenesis or identified in Glanzmann thrombasthenia patients. Typically, these mutations were tested for their ability to bind soluble multivalent ligands like PAC-1 IgM, PAC-1 Fab fragments in combination with polyclonal secondary antibodies, or fibrinogen. The activity of these mutant integrins as measured by these multivalent ligands did not distinguish between effects on affinity or clustering. Our results, specifically addressing affinity changes, do generally support the conclusions drawn from the use of multivalent ligands.
To confirm that affinity changes measured in our cell culture assay have similar effects in the whole organism, we tested their effects in flies that expressed reduced levels of talin. Because talin increases integrin activity in flies (8, 9), reducing talin levels would be predicted to increase phenotypes (lethality in our tests) of mutations that we find reduce integrin affinity and rescue phenotypes of mutations that increase integrin affinity. In general this is what we find. Using this genetic interaction test, we also tested those mutants that did not show an alteration in affinity of PS2C integrins for soluble ligand. Most of these mutants showed increased integrin function in the organism.
The surprising finding from our work is that most of our integrin mutations do not reduce the ability to bind ligand but actually enhance it. They are activating mutations. Expression, in flies, of activating deletion mutants of the αPS2 subunit was previously found to be unable to rescue the lethality of a null mutation in that subunit, pointing out in a different context the importance of proper regulation of integrin activity (10). We discuss our mutant results in the context of current structures available for the human αIIbβ3 integrin. We have additionally used site-directed mutagenesis to test the deadbolt hypothesis of integrin affinity regulation (11). This hypothesis predicts that the CD loop in the βTD (tail domain) contacts the β I domain in the bent conformation and restrains it in a low affinity state. Work on αIIbβ3 and αVβ3 integrins has yielded contradictory results (12, 13). Our results on the Drosophila PS2C integrin strongly support the deadbolt hypothesis. Finally, site-directed mutagenesis suggests that a conserved cysteine in the cytoplasmic domain of αPS2C that is palmitoylated in human α3 and α6 is involved in affinity regulation.
EXPERIMENTAL PROCEDURES
Cell Culture
Drosophila S2/M3 cells were cultured in Shields and Sang M3 medium supplemented with 12% heat-inactivated fetal calf serum as described previously (14). Cells were co-transfected with plasmids expressing an αPS2C subunit and a βPS subunit, both under the regulation of the heat shock protein 70 promoter, and the bacterial dihydrofolate reductase selectable marker as described previously (15). Transformed cells were selected in 2 × 10−7 m methotrexate.
Prior to performing TWOW-1 assays, cells were treated with dsRNA targeting the 3′-untranslated sequence of mys for 3 days to remove expression of endogenous βPS integrin as described earlier (7). This 3′-untranslated sequence is not present in the βPS transgenes.
TWOW-1 Binding Assay
This assay has been described in detail (7). TWOW-1 is an Fab molecule whose heavy chain complementarity-determining region 3 was replaced with a sequence encoding 53 amino acids found in Drosophila Tiggrin. These 53 amino acids contain the Tiggrin RGD sequence and 24 amino acids prior to and 26 amino acids following the RGD (7).
To remove preexisting integrins and matrix and to induce high expression from the heat shock-regulated integrin transgenes, cells were incubated at 36 °C with collagenase/dispase. Cells were washed and allowed to recover for 4 h at 23 °C. Cells were incubated with TWOW-1 for 10 min at room temperature. This was done in the presence of Ca2+ and Mg2+ (standard binding); Ca2+, Mg2+, and Mn2+ (Mn2+-activated binding); or EDTA (non-binding). Buffer containing formaldehyde was added to fix the bound TWOW-1 to the cells. Bound TWOW-1 was detected with AlexaFluor488-labeled secondary antibody. Levels of PS2C integrin on the surface of cells were determined by staining with a biotin-labeled anti-αPS2 antibody (CF2C7) followed by detection with phycoerythrin-streptavidin. Fluorescence levels for both TWOW-1 and PS2 levels were analyzed by flow cytometry.
Specific TWOW-1 binding is the mean fluorescence intensity (MFI)2 of TWOW-1 binding minus the MFI of the same cells in the presence of EDTA. Typically, specific binding was greater than 90% of total binding. For our analyses, this specific binding is expressed relative to surface integrin expression (TWOW-1 MFI/anti-αPS2 MFI). Finally, because assays were conducted over a period of more than 2 years, expression and TWOW-1 MFI/anti-αPS2 MFI were normalized to a control cell line (untagged wild type line 1 or WT1) expressing wild type PS2C integrins that was included in every assay. Z scores are the number of S.D. values from the average binding of wild type PS2C integrins taken from 10 independent lines expressing Myc-tagged βPS and αPS2C.
Nomenclature
We discuss the Drosophila βPS functional mutants as they relate to crystal structures of β3 integrins. Therefore, the mutants generated in fly genetic screens are described as follows: mysallele(single letter designation of the amino acid change in βPS; corresponding amino acid in β3; z score for TWOW-1 binding). Site-directed mutations not obtained from genetic screens are designated as follows: βPS amino acid change (corresponding amino acid in β3). The βPS numbering begins with the start methionine as in our previous descriptions of these mutants (6). The β3 numbering system begins with the first amino acid (G) in the mature, processed subunit and corresponds to the common β3 numbering used in the structure, Protein Data Bank entry 3FCS (16), that we have used as a reference to assess likely consequences of our mutations. Unless otherwise noted, amino acid interactions are discussed using this structure as a reference, and the αIIb and β3 amino acids are presented in boldface type.
βPS-Talin Interaction Tests
To test mutant mys alleles for genetic interactions with talin, w mys/w mys (or w mys/FM7c);rhea+/rhea+ females were crossed to w mys+; rhea2 (w+)/TM3 Ser males. The mys alleles, located on the X chromosome, were generated in previous screens (6, 17). rhea2, located on chromosome 3, is a strong allele of the gene encoding talin (8, 18). Crosses were done at a temperature (18, 22, 25, 28, or 31 °C) that resulted in reduced viability in mutant mys male progeny. This temperature differed for different mys mutant alleles and is indicated in supplemental Table S5. Due to sterility at 31 °C, tests at this temperature involved egg laying at 28 °C for 24–48 h, and then eggs and larvae were transferred to 31 °C. Progeny bearing one mutant copy of rhea were identified by the presence of a w+ transgene on the rhea2 chromosome (and the absence of Ser in the case of mysb3, mysb4, and mysts2, which contained a w+ gene on the chromosome containing the mys mutation). All male progeny were mutant for the mys allele, and females were heterozygous for the mys mutation (Fm7c males and females were discarded).
Flies were raised on cornmeal, sugar, and yeast medium. Marker mutations are described in FlyBase (available on the World Wide Web) and described by Lindsley and Zimm (19).
RESULTS AND DISCUSSION
TWOW-1 Binding Is Consistent between Independent Cell Lines
We have generated stable cell lines expressing more than 50 different function-altering mutations in the βPS integrin subunit. All of the mutant βPS integrin subunits were labeled with a Myc tag inserted into the large serine-rich loop (present only in the Drosophila βPS subunit) that is located between the X and A strands of the Hybrid domain.
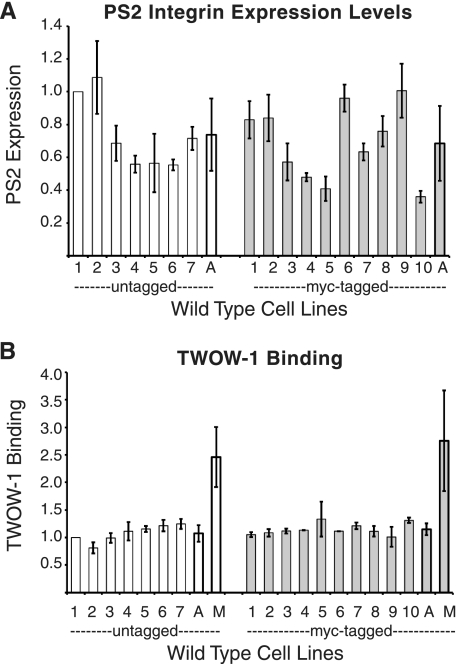
To control for experiment-to-experiment variability over the 2 years that these experiments were performed, each group of cell lines being assayed for integrin expression and TWOW-1 binding always included line WT1 (wild type 1; expressing untagged βPS and αPS2C), and values were normalized to its levels. The level of TWOW-1 used in all binding assays was 40 μg/ml. For wild type PS2C integrins, this is a subsaturating concentration resulting in ∼50% of the binding levels seen for integrins activated with Mn2+ (7) (Fig. 1B). At this concentration, we are able to detect both increases and decreases in affinity.
FIGURE 1.
Integrin expression levels and TWOW-1 binding of multiple lines expressing wild type PS2C integrins. A, expression of cell surface PS2C integrins on the surface of 17 independent S2 lines transformed with genes expressing αPS2C and βPS was determined by staining with an αPS2 antibody and analyzed by flow cytometry. Seven lines expressed unmodified βPS (white bars), and 10 lines expressed βPS containing a Myc tag inserted into the serine-rich loop (gray bars). B, TWOW-1 binding of the same lines normalized to integrin expression levels. TWOW-1 binding is fairly consistent between independent lines. The average TWOW-1 binding for both groups of cell lines in the presence of 1 mm Mn2+ (M) is also shown. Values shown for individual lines are the median ± S.E. (error bars) from three independent experiments with each line. Averages (A) for cell lines expressing tagged and untagged are the mean ± S.D. (error bars) (for this value, we show S.D. instead of S.E. because the S.D. of the wild type lines was used in calculation of z scores for the ligand binding properties of mutant integrins). Untagged line 1 (also referred to as WT1) values do not have error bars because they were used to normalize each experiment and therefore always have a value of 1.
Variability in integrin expression levels and their ability to bind TWOW-1 is due to either the specific βPS mutant subunit being expressed or cell line variability. To determine cell line variability, we examined the integrin expression levels and TWOW-1 binding of 17 transformed cell lines expressing wild type PS2C integrins. Seven lines expressed untagged βPS, and 10 expressed Myc-tagged βPS. All lines also expressed the αPS2C integrin subunit.
Fig. 1A demonstrates that independent cell lines transfected with wild type PS2C integrin transgenes express variable levels of cell surface integrin. The average levels are not significantly different between those lines expressing untagged or Myc-tagged βPS (p = 0.94). In contrast to the variable integrin expression levels, the amount of TWOW-1 bound per integrin shows much less variability (Fig. 1B). The range of TWOW-1 binding varies between 0.81 and 1.25 (mean = 1.08 ± 0.15) for the lines expressing untagged integrins and between 1.01 and 1.33 (mean = 1.15 ± 0.11) for tagged integrins, relative to that observed for WT1. Mn2+ activates the affinity of PS2C integrins for TWOW-1 (7) (Fig. 1B). For untagged and tagged integrins, levels of TWOW-1 binding in the absence of Mn2+ were 44 ± 10 and 42 ± 14%, respectively, of that measured in the presence of Mn2+. These data demonstrate that the presence of the 15-amino acid Myc-tag in the serine-rich loop does not alter the ligand binding ability of the PS2C integrin or its affinity modulation by Mn2+. Second, the data demonstrate that line-to-line variability is to be expected for expression levels but not for TWOW-1 binding normalized to PS2C integrin expression levels.
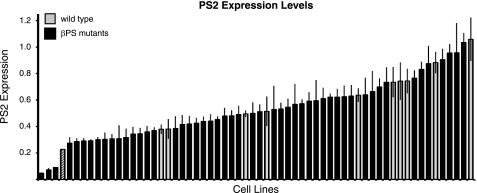
Most Mutant βPS Subunits Are Well Expressed
As with different cell lines expressing wild type PS2C integrins, expression levels of cell lines expressing mutant integrins vary widely (Fig. 2 and supplemental Table S1). Only three mutants, mysXN101(C629S;C549), mysb47(A293T;A225), and mysG12(D356N;D288), expressed levels of integrins 2 S.D. values below the average of the wild type lines. mysXN101 was already known to be poorly expressed in flies, whereas mysb47 and mysG12 did not show reduction in expression upon limited examination in either wing discs or muscle attachment sites (5, 6, 15). To ensure that low expression of mysb47(A293T;A225) and mysG12(D356N;D288) was not due to line-to-line variability, a second stably transformed line was obtained and tested. Again, both showed dramatically reduced PS2 integrin staining (supplemental Table S1).
FIGURE 2.
Distribution of αPS2CβPS expression levels in cells expressing wild type or mutant βPS. Expression of cell surface PS2C integrins was determined by staining with an αPS2 antibody and analyzed by flow cytometry. Ten independent lines transformed with αPS2C and Myc-tagged wild type βPS are shown (gray). Two S.D. values below the mean of the wild type lines is denoted by a crosshatched bar. None of the mutants (black) expressed levels greater than 1 S.D. value above wild type levels. Mutations and specific values and tests of duplicates for certain mutants can be found in supplemental Table S1. Values are the median ± S.E. (error bars).
The extremely low levels of expression of the lethal mutation mysG12(D356N;D288) and the viable mysb47(A293T;A225) are consistent with observations of mutations of β1 amino acids Asp-295 (Asp-288) and Asp-233 (adjacent to β1 Ala-234; β3 Ala-225). Both of these were poorly expressed on the surface of the cell and appeared to be defective in α/β association (20). In the crystal structure (supplemental Fig. S1), Asp-288 hydrogen-bonds with Leu-258, which is itself hydrogen-bonded to αIIb Tyr-237. Leu-258 is in the 310 helix that contains other residues interacting with the α subunit, including Asp-259, Arg-261, and Leu-262. Mutation of Asp-288 may therefore affect not only Leu-258 but other nearby α/β contacts as well. Finally, Asp-288 is immediately followed by helix α5, which contains Ser-291 and Glu-297, which are within 3 Å of αIIb. The effect of mysb47(A293T;A225) is less direct. Ala-225 is closest to (within 3.5 Å of) Leu-117, which contacts (within 3 Å) Val-247 and Thr-249 in β4, which precedes the β4–310 helix loop containing the close α/β contact Lys-253. Mutation of alanine to threonine probably alters the positioning of β4-310 helix loop and α/β contacts. Mutation of Cys-549 in β3 integrin (corresponding to mysXN101) showed reduced expression (21). Cys-549 stabilizes a loop in I-EGF3 that contacts the Hybrid domain in the closed conformation of β3 integrin (supplemental Fig. S1C), and its mutation activates the integrin. This activation probably alters the position of the Hybrid domain and breaks the hydrogen bond between α subunit (Asp-319) and the β Hybrid domain (Leu-362). Additional separation of the α and β leg regions due to Hybrid swing-out may also be an additional consequence (22). Failure of these associations are the likely causes of poor α/β association.
Ligand binding assays with the lethal mysG12 mutant were inconsistent between separate lines, due to the extremely low levels of expression, and we therefore do not report this. mysb47(A293T;A225) and mysXN101(C629S;C549), although also expressed at very low levels, gave very consistent ligand binding data, and these are presented but should also be viewed with more caution than the data from the other mutants. Finally, we have done duplicate transformations of some mutant integrins that expressed at or more than 2 S.D. values lower than wild type levels, and most again expressed at levels between 1 and 2 S.D. values below wild type, which suggests that they too may affect expression levels to some extent (supplemental Table S1).
Effects of βPS Mutations on TWOW-1 Binding
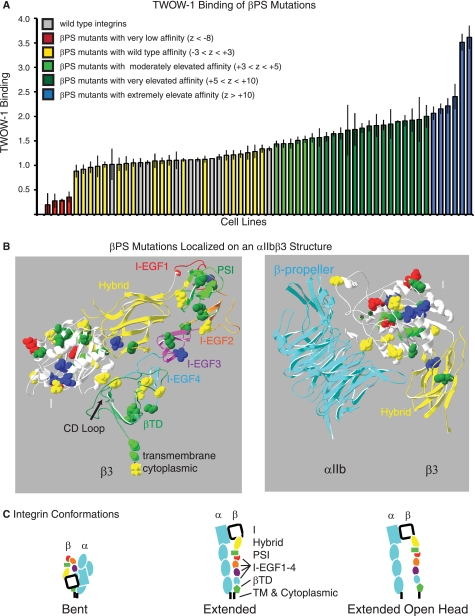
The affinity for the soluble monovalent ligand, TWOW-1, of PS2C integrins with different mutant βPS subunits was examined and compared with that of wild type integrins expressed on 10 independent cell lines. Results of this assay are arranged in order of binding ability in Fig. 3A (and supplemental Table S2). Of the 51 mutants generated in genetic screens, five displayed severely reduced binding levels that were more than 8 S.D. values below the mean of the wild type integrins (z score <−8). Seventeen lines showed no significant changes in TWOW-1 binding. Twenty-two of the mutant integrins displayed increased binding with z scores greater than 5, and an additional seven bound with z scores of 2.4–5.0.
FIGURE 3.
TWOW-1 binding levels of mutant integrins and localization of mutations within the integrin β subunit. A, distribution of TWOW-1 binding levels in cells expressing PS2C integrins with mutant βPS subunits. Ten independent lines transformed with αPS2C and wild type βPS are also shown (gray). Binding levels have been color-coded based on z scores (negative values are S.D. below and positive values are S.D. above the median of the wild type lines) as indicated. Values shown are the median ± S.E. (error bars) from 3–9 independent experiments. B, βPS mutants are mapped onto the bent αIIbβ3 ribbon structure (from Ref. 16). In the left panel, only β3 is depicted. Individual domains are labeled and color-coded. The mutated amino acid side chains are shown on the ribbon diagram. They are color-coded (the same as in A) according to the effects they have on TWOW-1 binding. Thus, all mutations that reduce binding (red) are in the I domain, and mutations that increase binding are found in all domains. The transmembrane and cytoplasmic tails were not present in the crystal structure and therefore have been depicted as a straight black line added to the βTD. In the right panel, the β integrin I and Hybrid domains have been rotated and shown together with the αIIb β-propeller domain. In this view, the SyMBS, MIDAS, and ADMIDAS divalent cations (*) can be seen. Details of individual mutations, corresponding β3 amino acids, z scores, and p values, can be found in supplemental Table S2 (sorted by binding levels) and in supplemental Table S3 (sorted by position in the βPS subunit). C, the bent, extended, and extended with open head integrin conformations are depicted in schematic form. α integrin subunit domains are light blue, whereas the β integrin domains are as follows: white, I domain; yellow, Hybrid domain; green, PSI domain; red, I-EGF1 domain; orange, I-EGF2 domain; purple, I-EGF3 domain; light blue, I-EGF4 domain; green, βTD; black, transmembrane and cytoplasmic domains.
In addition to comparing ligand binding levels of mutant integrins normalized to the amount of integrin on the surface, we also compared their activation indexes (supplemental Table S2, column I). The activation index is the binding of TWOW-1 in the absence of Mn2+ divided by the binding in the presence of activating Mn2+. Although this approach removes the dependence on an antibody to measure surface integrin levels, it has its own limitations, including the observation that effects of mutants and Mn2+ can be additive (7), and mutants may influence the ability of Mn2+ to activate the integrins. For the mutants we analyzed, the activation index data give almost identical results as binding normalized to integrin levels. Of the 22 strongly activating mutants (z scores above 5), only two did not give similarly significant z scores when normalized by Mn2+-activated values (supplemental Table S2). The remaining two strongly activating mutants and six of the seven moderately activating (z scores 2.4–4.9) did not show significant activation index increases due to the mutations increasing binding both in the absence and in the presence of Mn2+ to similar extents.
Examination of the overall distribution of the mutations within the βPS subunit and their effects on ligand binding is interesting (Fig. 3B and summarized in Table 1). All mutations that reduce ligand binding are found in the I domain and specifically in the metal ion-dependent adhesion site (MIDAS) and synergistic metal binding site (SyMBS). This confirms the previously recognized importance of these domains in ligand binding. Mutations that increase ligand binding are distributed in all domains, illustrating the importance of all domains in restraining the integrin in the low affinity (bent, head closed) conformation (Fig. 3C). Activating mutations are postulated to disrupt this conformation and push the equilibrium toward the unbent and/or head open conformations that result in increased affinity for ligand (Fig. 3C; reviewed in Refs. 2–4).
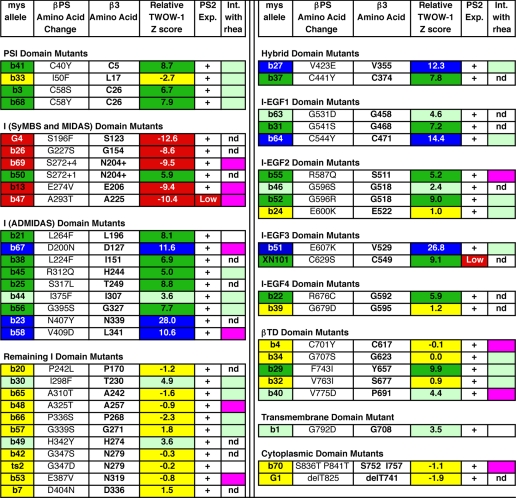
TABLE 1.
Summary of mys mutant TWOW-1 binding, expression, and genetic interactions with rhea
This is a summary of the detailed data that can be found in supplemental Tables S1–S3 and S5. Mutants are grouped by domains. Color coding of the mys allele and TWOW-1 z score boxes is the same as in Fig. 1: red, very low affinities for TWOW- 1 (z scores <−8); yellow, wild type affinities (z scores between −3 and 3); light green, moderately elevated affinities (z scores between 3 and 5); dark green, very elevated affinities (z scores between 5 and 10); blue, extremely elevated affinities (z scores >10). The one exception to this color coding is mysb46. Its p value was significant (<0.001), so it was shaded light green, although its z score was only 2.4. For mysb69 and mysb50, S272+4 and S272+1 indicate insertions of VRQ or Q following amino acid 272. For mysG1, de1T825 refers to the carboxyl-terminal 21 amino acids deleted and replaced by 25 essentially random amino acids (due to a splice site mutation). mysb24 is a mutation of the single amino acid between I-EGF2 and I-EGF3. (For convenience, it has been placed in the I-EGF2 group). For the genetic interaction tests with rhea (Int. with rhea), magenta indicates an enhancing interaction; flies with the βPS mutation are more lethal with only one wild type copy of rhea than when they contain two wild type copies of rhea. Light green indicates a rescuing interaction; flies with the βPS mutation are less lethal with only one wild type copy of rhea than when they contain two wild type copies of rhea. No shading indicates a lack of significant difference in lethality. In some cases, the interaction test was not done (nd) due to too much or too little lethality of the mys allele.
Detailed data describing TWOW-1 binding arranged by mutant position in the βPS integrin, including references to similar mutations found in other integrins, is shown in supplemental Table S3. In the following sections, we group the mutations by the domains they are likely to affect and discuss the probable consequences for integrin structures. Supplemental figures are provided for each set of mutants that focus on their positions and interactions with important structural components (supplemental Figs. S1–S9).
Mutations in the Plexins-Semaporin-Integrin (PSI) Domain Activate Integrins
The three mutations in the PSI domain that disrupt disulfide bridges, mysb3(C58S;C26;z = 6.7), mysb68(C58Y;C26;z = 7.9), mysb41(C40Y;C5;z = 8.7) activate the integrins. mysb3 and mysb68 both mutate the same cysteine residue and have similar effects on activating the integrin. Cys-26 is located between Trp-25 and Ser-27 that along with Glu-29 and Arg-37 are at the interface with the I-EGF1 domain residues Gly-458 and Cys-457 (supplemental Fig. S2). One of these I-EGF1 residues is mutated in mysb63(G531D;G458;z = 4.6) and is also activating (see “I-EGF Domain Mutants”). It is likely that defects in all three of these mutants perturb the PSI-I-EGF1 interaction. Activating mysb41(C40Y;C5;z = 8.7) probably disrupts PSI-Hybrid interactions. In β2 integrin, the Cys-3 residue (Cys-5 in β3) is adjacent to Thr-4 (Thr-6 in β3) that has been implicated in restraining the activation state of the integrin (23). Also in β2, Cys-3 (Cys-5) forms a disulfide bridge with Cys-21 (Cys-23), which is part of the PSI-Hybrid interaction domain (24). In β3, Cys-23 forms a hydrogen bond with Hybrid residue Arg-93 (supplemental Fig. S2). Breaking of this disulfide bridge in mysb41 probably alters the position of Thr-6 and Cys-23 and thereby weakens the PSI-Hybrid interaction.
Integrin crystal structures suggest that the PSI domain is important in stabilizing the Hybrid-I-EGF1 connections. This could be important for restraining the integrin in an inactive state (23) and/or executing the activation of the integrin via a Hybrid swing-out mechanism (24–26). Our mutations, including complete deletion of the PSI domain (7), point to a role for it in restraint of integrin affinity. Activation of the integrin by mutation of the long range disulfide bridge linking the PSI domain to the junction of the Hybrid and I-EGF (supplemental Fig. S2) also suggests that this is the case (27). By mutating only one cysteine at a time, there is the potential that the remaining cysteine of the pair inappropriately paired with nearby cysteine and thereby disrupted more than just the long range disulfide bridge. We therefore tested the function of this disulfide bridge in βPS by the site-directed double mutant βPS(C46A,C505A;C13,C435). This mutation strongly increased the affinity of the integrin (z = 11.9; supplemental Table S4).
Mutations in the MIDAS and SyMBS Domains Impair Ligand Binding
In the β integrin subunit, two domains, MIDAS and SyMBS (or ligand-associated metal binding site (LIMBS)), have been identified as contributing to ligand binding. The MIDAS domain coordinates a metal ion, typically Mg2+, which directly interacts with ligand. The SyMBS domain and its associated divalent cation, Ca2+, are close to the MIDAS cation and positively regulate it (reviewed in Refs. 3, 4, 11, and 28). Mutations that compromise these two domains either by mutating metal ion-coordinating amino acids or altering their specific location would be predicted to reduce ligand binding, and indeed the five mutations that reduce ligand binding by 29% or more are all found in or near these two domains (supplemental Fig. S3). mysG4(S196F;S123;z = −12.6) is in the MIDAS coordination site DXSXS (192–196;119–123). This lethal mutation has the most dramatic negative effect on ligand binding and is consistent with similar mutations examined in the β3 subunit (29, 30). The viable mutation that most severely reduces ligand binding, mysb47(A293T;A225;z = −10.4), is located in a position that suggests that it alters the positions of loops containing MIDAS and SyMBS-coordinating residues. Ala-225 in helix α3 is 5 amino acids from the DAPEG motif (217–221) that contributes to the MIDAS and SyMBS. This helix α3 and Ala-225 in particular are next to (within 4 Å of) β1 and β2 strands prior to loops containing the DXSXS (192–196;119–123) MIDAS residues and the SyMBS Asp-158. Helix α2 is just prior to the DAPEG sequence and is the site of the two negative ligand binding mutations mysb13(E274V;E206;z = −9.4) and the insertion of SVRQ following amino acid Ser-272 in mysb69(S272+SVRQ;N204+;z = −9.5). Interestingly, a smaller insertion is found in mysb50(S272+Q;N204+;z = 5.9), and this mutant is activating for ligand binding. Different effects of mutations at this same residue have been observed before in the human β3 integrin, where E206T was activating but E206I was not (31). The helix α2 undergoes a small twist in the transition from closed (non-activated) to the open conformation (26, 31). This would suggest that these mutations are not removing critical amino acids but perhaps instead repositioning the SyMBS domain. The remaining mutation with a negative effect on binding is mysb26(G227S;G154;z = −8.6) in the β2 strand that contributes to positioning the SyMBS domain Asp-158 residue. We confirmed the positive contribution to ligand affinity of the SyMBS residues in the βPS subunit by site-directed mutagenesis of Asp-158 (D231A) and Asn-215 (N283A). Both of these mutations showed essentially no ligand binding (z <−11; supplemental Table S4). In conclusion, the mutations isolated from our genetic screen that impair ligand binding are associated with MIDAS and SyMBS domains already recognized as being important for ligand binding.
In summary, all of the mutants that negatively affect soluble binding are found in the domain binding the ligand-coordinating metal ion, MIDAS, or the SyMBS domain that exerts a positive influence on it. Our mutagenesis survey has not detected any new regions in the βPS integrin subunit necessary for efficient soluble monovalent ligand binding.
Mutations in ADMIDAS Activate βPS
The ADMIDAS site also binds metal ions, typically Ca2+, and mutations of ADMIDAS residues have been observed to exert either negative (32, 33), positive (34, 35), or no (29, 36) effect on integrin activity. In these experiments, integrin activity was measured in a variety of assays, with none measuring cell surface integrin binding to a soluble monovalent ligand. Interestingly, αLβ2 binding to soluble ICAM-1 (intercellular adhesion molecule 1) was increased when ADMIDAS residues were mutated but cell spreading was abolished. This suggested that ADMIDAS negatively regulates affinity but is required for outside-in signaling required for cell spreading (35). Finally, a direct interaction between the αIIbβ3 ADMIDAS Ca2+ and valine in the KQAGDV fibrinogen peptide has been observed (37). In Drosophila βPS integrin, a number of mutants suggest that the ADMIDAS domain has a negative regulatory role in affinity regulation. The mutations mysb67, mysb58, mysb38, mysb21, mysb45, mysb44, mysb23, mysb25, and mysb56 change amino acids that either contribute a coordinating side chain or appear to be important in positioning coordinating residues so that ADMIDAS restrains the integrin in a low affinity state. All of these mutants activate the PS2C integrins (Table 1 and supplemental Tables S2 and S3).
ADMIDAS-coordinating residues derive from two separate regions of the β I domain (supplemental Figs. S4 and S5). In the unliganded (closed) structure, the α1,1′ helix provides coordinating residues Ser-123, Asp-126, and Asp-127, and the β6-α7 loop positions Met-335. The β6-α7 loop contacts the α1,1′ helix at the top of α7 just following the β6-α7 loop. Ligand binding results in the open structure where α7 moves downward, the coordination of the ADMIDAS metal ion by the carbonyl of Met-335 is lost, and the shape of α1,1′ changes bring it and the ADMIDAS metal ion toward the MIDAS (supplemental Fig. S4). This shift allows the movement of the β1-α1 loop, containing the DXSXS motif, toward the MIDAS Mg2+ and ligand, where it forms two stabilizing hydrogen bonds with the ligand aspartate. As the bottom α7 connects directly to the Hybrid domain, its downward movement is accompanied by rearrangement of the I, Hybrid, and PSI domains, resulting in the swinging out of the Hybrid domain by 60° (16, 26, 37).
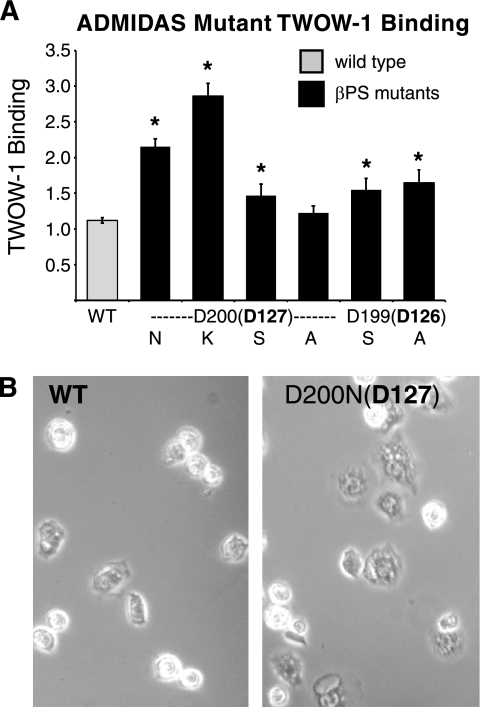
Asp-127 in helix α1,1′ directly coordinates the ADMIDAS metal Ca2+ (16) and is mutated in the strongly activating mysb67(D200N;D127;z = 11.6). Mutations of this amino acid in β1, β2, β3, and β7 integrins give widely varying results from activation to inactivation (29, 32–36, 38). This might result from the removal of the wild type residue and/or the presence of the new residue, asparagine. We further tested the requirement for Asp-200 (Asp-127) by mutating it to lysine, serine, or alanine. In none of these mutations was the binding decreased. Replacement with lysine (D200K;D127;z = 19.6) resulted in even stronger activation. Alanine (D200A;D127;z = 3.9) resulted in moderate activation, whereas serine resulted in no change in binding ability (D200S;D127;z = 1.2) (Fig. 4A and supplemental Table S4). Site-directed mutation of the neighboring aspartate, Asp-199 (Asp-126), that also contributes an ADMIDAS-coordinating residue increased ligand binding as well (Fig. 4A and supplemental Table S4). mysb67(D200N;D127;z = 11.6) and most of the site-directed mutants are similar to mutations in αLβ2 that increase ligand binding (35) but differ in that they do not abolish cell spreading (Fig. 4B).
FIGURE 4.
Mutations in ADMIDAS residues increase TWOW-1 binding levels and enhance cell spreading. A, binding levels of cells expressing mutations of ADMIDAS Asp-200 (Asp-127) to Asn (as in mysb67(D200N;D127;z = 11.6)); Lys, Ser, or Ala and mutations of ADMIDAS Asp-199 (Asp-126) to Ser and Ala are shown. Values shown are the median ± S.E. (error bars) (n = 30 for WT (10 different lines done three times each); n = 6 for D200N, D199S, and D199A; n = 3 for D200K, D200S, or D200A). *, p < 0.004. B, phase-contrast images of cells expressing WT or mysb67(D200N;D127) integrins are shown spreading on tiggrin ligand. Counting over 300 cells from three different fields demonstrated that twice as many cells bearing the mysb67 mutation spread compared with those expressing wild type integrins (57% versus 23%).
Asp-127 and the neighboring ADMIDAS Asp-126 are very well conserved with the exception of β8 integrin, where both are asparagine. The presence of potentially activating substitutions in β8 ADMIDAS has been noted before with regard to β8 containing an asparagine at the position Leu-341 (the connection between α7 and α1,1′). All other human and Drosophila β subunits contain a hydrophobic leucine, isoleucine, or valine, and mutation to the charged aspartic acid activates the integrin (6, 7). We additionally find that deletion of the CD loop in the β tail domain, which is not present in β8, strongly activates the PS2 integrin (see below). These four activating mutations strongly suggest that β8 evolved to be constitutively present on the cell surface in a high affinity state.
The β6-α7 loop is critical for ADMIDAS function, and a number of our activating mutations are located in this loop or the flanking β6 or α7 (supplemental Fig. S4). Positioning of the loop by α7-α1,1′ interaction is disrupted in mysb58(V409D;L341;z = 10.6). Leu-341 stabilizes the association of α7 and α1,1′ through interactions with Leu-134 and Leu-138, and we have previously demonstrated that it activates the PS2C integrins for cell spreading, adhesion, and monovalent ligand binding (6, 7). Also directly in contact with α7 in the closed conformation is the weakly activating mysb44(I375F;I307;z = 3.6). The nearest neighbors of Ile-307, within 3.9 Å, are Ala-347 and Ile-351 on one face of α7. Near the α7 Ile-351 is Gly-327 located at the turn following β6, and this residue is mutated in the strongly activating mysb56(G395S;G327;z = 7.7). The carbonyl group of Gly-327 and the moderately activating mysb45(R312Q;H244;z = 5.0) hydrogen bond with Asn-305 that does, like Ile-307 (mysb44), contact α7 Ile-351 and additionally Arg-352. In the open conformation, Ile-351 and Arg-352 contacts with Asn-305 and Ile-307 are lost as α7 shifts downward. These interactions with Ile-351, and Arg-352, like Leu-341 interactions with α1,1′, probably stabilize α7 in the inactive conformation. Finally, near mysb58, the strongly activating mysb23(N407Y;N339;z = 28) is key to the positioning of the β6-α7 loop. Asn-339 forms hydrogen bonds with Ser-334 and Val-332, stabilizing the loop and the position of ADMIDAS Met-335. This is supported by modeling and N339S mutations in β3 and the corresponding N329S in β2 (39, 40).
Next to Met-335 in the loop following β4 is MIDAS/ADMIDAS-coordinating Asp-251, whose interactions shift from polarizing with MIDAS in the closed state to polarizing with ADMIDAS in the open state (16). Activating mysb25(S317L;T249;z = 8.8), separated by a single residue from Asp-251, may result in subtle shifts in its position favoring the open state (supplemental Fig. S4).
The shape and contacts of α1,1′ also change dramatically in the transition from the closed to the open state. A number of our mutants change amino acids involved in interactions that stabilize α1,1′ in the closed state, thereby increasing the affinity of the integrin. In the closed state, α1′ Leu-134 and Leu-138 side chains are within 4 Å of Leu-341 (mysb58; discussed above). α1′ Leu-138 also contacts Ile-151 in the closed but not the active conformation (3.7 Å versus 4.6 Å; supplemental Fig. S5). Ile-151 is mutated in the activating mysb38(L224F;I151;z = 6.9). Ile-151 also is within 3.8 Å of Phe-203, the residue neighboring Asn-204 that hydrogen-bonds with α1,1′ in the closed conformation. Thus, Ile-151 appears to be involved in stabilizing the closed conformation. Activating mysb21(L264F;L196;z = 8.1) mutates a residue that is just prior to helix α2 containing the aforementioned Phe-203 and Asn-204 as well as Val-200, Thr-201, and Lys-208, which make contacts with α1,1′ (supplemental Fig. S5). Lys-196 is also within 3.5 Å of Ile-151 (mysb38 just discussed). Both mysb38 and mysb21 may destabilize the closed conformation of α1,1′ and shift the β integrin to the high affinity state. In β3 integrins, mutation of Leu-196 to proline differentially affected αIIbβ3 and αVβ3 integrin expression and ability to interact with immobilized ligands. αVβ3 integrin-mediated spreading on fibrinogen was blocked by this mutation, whereas αIIbβ3 activation by dithiothreitol, antibody, or platelet activation was blocked (41, 42). Given that mutation of this residue in βPS to phenylalanine has the opposite effect of mutation in αIIbβ3 to proline, one of these substitutions probably does more than simply remove the well conserved leucine at this position, so its contribution to integrin function is still in question and will require further analysis.
Remaining I Domain Activating Mutants
There are two remaining activating I domain mutations, mysb49(H342Y;H274;z = 3.6) and mysb30(I298F;T230;z = 4.9). The location of His-274 and previous observations on nearby Val-275–His-280 (43) suggest that this residue may be important in positioning β integrin residues for interaction with the α integrin subunit (supplemental Fig. S6). His-274 forms hydrogen bonds with the side chain of Asp-270. Asp-270 is within 3.7 Å of Met-287, the residue adjacent to Asp-288 (mutated in mysG12) that is known to be important for α/β association and integrin expression (discussed above). Asp-288 (mysG12) hydrogen-bonds with Gln-267 adjacent to Val-266, which is within 3.5 Å of αIIb Tyr-440. The proximity of this region of β3 integrin to αIIb and the α/β interaction mutant Asp-288 suggests that His-274 plays a role in restraining integrin activation by stabilizing the α/β interaction in the closed conformation. Whether Asp-288 (mysG12) is activating or not could not be determined due to its extremely low expression level. Further supporting this proposition is the observation that hydrogen bonds with αIIb Arg-320 and Glu-324 by the β3 integrin Glu-297 and Ser-291 appear to be lost upon transition to the open conformation (supplemental Fig. S6) (16, 22). mysb30(I298F;T230;z = 4.9) may increase integrin activity by indirectly disrupting Hybrid-I domain interactions that are needed to maintain the closed conformation (supplemental Fig. S7, C and F). Located at the base of helix α3, Thr-230 contacts (is within 3.5 Å of) Leu-299 of helix α5. At the base of helix α5 is Asn-303, which is within 3.0 and 3.8 Å of Hybrid residues Lys-417 and Lys-422 in the closed conformation. In the open conformation, these distances increase to greater than 20 Å. If the mysb30 mutation disrupts positioning of helix α5, this would destabilize the Hybrid-I domain contacts and favor the open, high affinity conformation.
Hybrid Domain Activation Mutants
Downward movement of α7 is key to the transition from the closed to the open conformation. In the closed conformation at the bottom of α7 just into the Hybrid domain (Hybrid strand C) is Val-355, which is mutated in the strongly activating mysb27(V423E; V355;z = 12.3). In the closed conformation, Val-355 interacts with (is within 3.4 Å of) Val-104 and Gln-106, which are located in the other junction of the Hybrid domain with the I domain just prior to β1 (supplemental Fig. S7). It is very likely that these interactions anchor α7 in the closed conformation and that mutation of Val-355 to glutamic acid interferes with the proper association of Hybrid strand C with the rest of the Hybrid domain and thereby allows α7 to move downward and activate the integrin, similar to mutation of Leu-341 (to aspartic acid in mysb58) at the opposite end of α7. Although not as dramatically as Leu-341, Val-355 does change its interactions between closed and open conformations (supplemental Fig. S7). In the closed conformation, it is within 3.4 Å of Val-104 and Gln-106 and is separated from Leu-389 by 3.9 Å. Val-395 and Phe-397 are further away at 4.6 and 4.5 Å. In the open state, Val-355 shifts away from Gln-106 (now separated by 5.9 Å) and moves closer to Val-395 and Phe-397 (now within 3.9 Å). Leu-389 hydrogen bonding with Ser-353 is also altered, and this brings it closer to Val-355 in the open conformation (3.3 Å versus 3.9 Å). Because this shift in the position is subtle, we also examined the closed and ligand-bound states reported for αVβ3 (44, 45). Val-355 shifts here as well, with the distance from Phe-397 narrowing from 4.0 to 3.4 Å (not shown). The importance of the side chain of Val-355 is further suggested by the high degree of conservation. Valine is found in this position in seven of the eight human β subunits (with β4 having a leucine) as well as in the fly βPS. Phe-397 is similarly conserved, being phenylalanine in all but β4, where it is a leucine. mysb27 illustrates the importance of firmly anchoring both ends of α7 in the closed conformation.
The activating mysb37(C441Y; C374;z = 7.8) disrupts a cysteine that is probably necessary to stabilize the Hybrid domain structure, and this is important in stabilizing the closed conformation. The same mutation, in β3 integrin, has been observed in a case of Glanzmann thrombasthenia. Testing of the mutation showed much reduced expression but increased ligand interactions when normalized for expression differences (46).
I-EGF Domain Mutants
The four I-EGF repeats appear to stabilize the inactive state of αIIbβ3 integrins. Specifically, the I-EGF domains 2, 3, and 4 extend in an almost straight orientation to comprise the lower β leg that is located, in the bent conformation, in a narrow crevice formed by the upper β leg (I-EGF1), the β I, α leg, and β propeller domains. Movement of the lower leg out of this crevice appears to be key to integrin extension and increased activity (16). Ten of 11 mutations in the I-EGF domains increase the affinity of the integrin for ligand (supplemental Table S3). These mutants disrupt the conserved final disulfide loops in the I-EGF domains (supplemental Fig. S8). The remaining mutant may interfere with proper localization of the PSI domain next to I-EGF1.
EGF repeats 2–4 contain 8 cysteines that form disulfide bonds Cys-1–Cys-5, Cys-2–Cys-4, Cys-3–Cys-6, and Cys-7–Cys-8. EGF repeat 1 has eliminated the Cys-2–Cys-4 pair and contains only 6 cysteines. The final pair of cysteines forms a loop separated by a single amino acid from the next I-EGF repeat, resulting in a gimbal-like connection (16). In order for the final pair of cysteines to pair and take on the correct looped structure, two turns must occur with one just prior to the final cysteine. This final turn necessitates a glycine positioned 3 residues prior to the final cysteine. This glycine is almost completely conserved between all EGF repeats, including those found in integrins as well as in EGF itself, the 10 repeats in the TIED (ten β-integrin EGF-like repeat domains) protein, tenascin, and many others (47). Six of the 11 I-EGF mutants change either one of the final pair of cysteines or the conserved glycine residue (supplemental Fig. S8): mysb31(G541S;G468;z = 7.2), mysb64(C544Y;C471;z = 14.4), mysb46(G596S;G518;z = 2.4), mysb52(G596R;G518;z = 9.0), mysXN101(C629S;C549;z = 9.1), and the non-activating mysb39(G679D; G595;z = 1.2). One additional mutation found in the Cys-7–Cys-8 loop is mysb22(R676C;G592;z = 5.9). Inclusion of a cysteine in this loop may interfere with turns necessary to bring Cys-7 and Cys-8 together, or it may form inappropriate disulfide bridges. These mutants probably prohibit the correct formation of the Cys-7–Cys-8 disulfide bridge and loop.
Two additional activating mutations probably do not prevent formation of Cys-7–Cys-8 loops but disrupt the positioning of them. In EGF and the integrin I-EGF repeats, the residue in the turn just prior to the final cysteine is close to the residue following Cys-2 (48). In repeat 3, this brings Asn-637 (Tyr-557) within 3.1 Å of Glu-607 (Val-529). mysb51(E607K;V529;z = 26.8) would clearly be expected to interfere with this contact and is strongly activating (supplemental Fig. S8). mysb55(R587Q;S511;z = 5.2) is located in the first turn between Cys-7–Cys-8 in I-EGF2 and, together with Phe-513, is closely associated with the beginning of I-EGF3 because both are within 3.5 Å of Phe-526 (supplemental Fig. S8). Thus, correct positioning of the I-EGF2/I-EGF3 junction may also be critical to the stabilization of the inactive conformation burying the lower β-leg, I-EGF2–4, in the cleft between the lower α-leg and upper β-leg.
A number of naturally occurring and in vitro generated mutations in the I-EGF repeats have also been observed to increase αIIbβ3 integrin activity (21, 49–52). β3 C549S, analogous to mysXN101(C629S;C549;z = 9.1) (supplemental Fig. S8), is strongly activating (21, 51). Residues within this loop make direct contact via hydrogen bonds with the Hybrid domain (via Ser-367 and Leu-368) and probably contribute to restraining it in the closed inactive state. In the closed state, the nearby Hybrid domain residue Leu-362 forms a hydrogen bond with αIIb Asp-319, and disruption of this by activation may explain low expression levels of Cys-549 mutants (20) such as mysXN101.
The only mutation not in or contacting the final disulfide loop in the I-EGF repeats is mysb63(G531D;G458;z = 4.6). The carbonyl oxygen of the corresponding glycine in β2 (Gly-446) was previously identified as being part of the PSI-Hybrid interface, forming a hydrogen bond with Arg-39 (in β2). The carbonyl group of the neighboring Cys-445 (in β2) was part of this interface and similarly hydrogen-bonded to Arg-39 (24). In β3, these two residues (Gly-458 and Cys-457) are within 3.7 Å of the PSI domain residue Arg-37 (corresponding to Arg-39 in β2) as well as Trp-25 and Ser-27 (supplemental Fig. S2). Cys-457 is also disulfide-bonded to Cys-437 just 2 residues from the Cys-435 that bridges the PSI domain (via Cys-13) to the junction between the Hybrid and I-EGF1. Substituting aspartic acid for glycine probably interferes with many of these interactions, including perhaps the Cys-13–Cys-435 bridge, and failure to make this connection has been demonstrated to increase integrin activity (27) (this report).
βTD Mutants
The βTD appears to be highly flexible (16), and there is controversy regarding its potential function in regulating integrin affinity. It has been proposed that the βTD provides contacts with the α7 helix via a CD loop (the “deadbolt”) (11), whereas others find no evidence for such regulation (12, 16). We obtained five mutations in this domain. Three had no effect on ligand binding, and two increased the integrin affinity (supplemental Table S3). mysb40(V775D;P691;z = 4.4) is activating and is found just 2 residues outside of the predicted transmembrane domain. Current crystal structures do not allow us to make strong predictions regarding the consequences of this mutation, but the nearby residues Pro-688–Gly-690 are close and parallel to αV residues Ile-955–Pro-959 (13), so it is reasonable to predict that this mutation at Pro-691 may alter α-β interaction just outside the transmembrane, thereby activating the integrin. mysb29(F743I;Y657;z = 9.9) is more strongly activating. Tyr-657 is at the end of a β sheet strand and is within 4 Å of Lys-612 and Glu-616, which precede it, and Val-665 and Phe-667 in the next β strand. This second β strand is just prior to the CD loop. Therefore, it is possible that the mutation disrupts the positioning of the CD loop and thereby activates the βPS integrin.
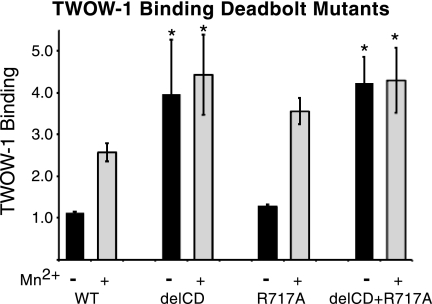
No effect on ligand binding was observed by removal of the CD loop (Asp-672–Lys-676) in either αVβ3 or αIIbβ3 (12). Removal of the CD loop in αVβ3 together with an R633A mutation (disrupting βTD-Hybrid interactions) did result in a small but significant activation (13). Unfortunately, the effect of the R633A mutation alone was not given, so the contribution of the CD loop removal is unclear. The R633A mutation alone has been seen to partially activate αIIbβ3 (53). To explore the potential function of the CD loop further, we made mutations in this loop of βPS analogous to those constructed previously in β3 (12, 13). Cell lines were generated that expressed βPS lacking CD loop residues Gln-758–His-762 (Asp-672–Lys-676), R717A(R633), and the combined deletion Gln-758–His-762 (Asp-672–Lys-676) together with R717A(R633) (supplemental Fig. S9). The results of these mutations strongly support the deadbolt hypothesis for the βPS integrin subunit (Fig. 5 and supplemental Table S4). Deletion of the 5 CD loop residues Gln-758–His-762 (Asp-672–Lys-676) completely activates the integrin with no further binding seen in the presence of Mn2+. The R717A(R633) mutation did not show significant activation, and together they show the complete activation that was observed for the CD loop deletion alone.
FIGURE 5.
TWOW-1 binding levels in cells expressing site-directed deadbolt mutations. Deletion of the CD loop residues Gln-758–His-762 (Asp-672–Lys-676) in the βTD (delCD) results in increased TWOW-1 binding that is not further increased by Mn2+. The R717A(R633) mutation does not activate TWOW-1 binding. The double mutant (delCD+R717A) displays binding levels similar to those of the delCD alone. All values are relative to binding levels for the reference cell line, untagged wild type line 1, in the absence of Mn2+ and are the median ± S.E. (error bars) (n = 30 for WT (10 different lines done three times each), n = 3 for delCD and R717A, and n = 6 (two different lines done three times each) for delCD+R717A). *, p < 0.005.
Transmembrane and Cytoplasmic Domain Mutants
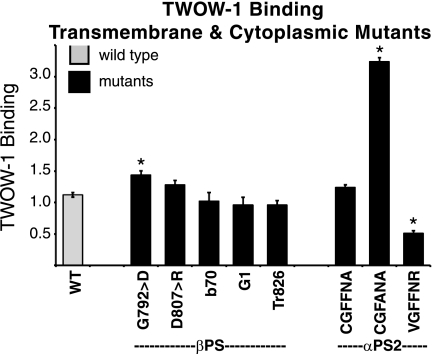
We isolated one mutation in the transmembrane domain, mysb1(G792D;G708;z = 3.5). This mutation increased the affinity (Fig. 6 and supplemental Table S4) for our monovalent ligand, consistent with observations that mutation of Gly-708 activates β3 integrins (54–56). In one study, this increase in activation was demonstrated at the affinity level through the use of a monovalent soluble ligand, and activation due to some mutants was cell line-dependent (55).
FIGURE 6.
TWOW-1 binding levels in cells expressing transmembrane or cytoplasmic domain mutations in βPS or αPS2C. mysb1(G792D;G708) binds significantly more TWOW-1 (n = 9; p = 2.2 × 10−6) as does the CGFFNR → CGFANA αPS2C mutation (n = 3; p = 3.28 × 10−17), and the CGFFNR → VGFFNR binds less (n = 6; p = 4.01 × 10−8) than wild type integrins (WT; n = 30). Neither of the α-β hinge mutations D807R(D723) in βPS nor Arg → Ala in αPS2C (CGFFNA) alter binding significantly (n = 6; p > 0.17). Mutations removing the final 21 amino acids of βPS, mysG1 and Tr 826–846, and mysb70(S836T/P841T;S752/I757) also do not affect TWOW-1 binding (n = 3–6; p > 0.2) Values shown are the median ± S.E. (error bars).
In the cytoplasmic domain, our screens identified mysb70, which altered two residues, mysb70(S836T and P841T;S752 and I757;z = −1.1). Soluble ligand binding of this mutant was not different from wild type integrins (Fig. 6 and supplemental Table S4). In case the two mutations somehow compensated for the function of the other, we also made cell lines expressing each of the mutations individually. These also displayed wild type TWOW-1 binding (not shown). Mutation of Ser-752 to proline in β3 has dramatic effects on integrin function and less when mutated to alanine (57–60). In β1 and β2, the residue is threonine (Thr-788 and Thr-759, respectively), as it is in mysb70. The second amino acid change in mysb70, P841T(I757), is the proline of the second NPXY motif residue, and mutation of this proline in β1 resulted in poorly expressed integrins (61). This second NPXY motif is involved in binding cytoplasmic proteins that regulate integrin activity (reviewed in Ref. 62). In flies, we find that the mysb70 mutation shows a strong genetic interaction with reduction of talin function (supplemental Table S5; mentioned in Ref. 6). mysG1(del T825; del T741;z = −1.9) is a lethal antimorphic allele that arises due to a splicing/frameshift lesion. The mature protein has lost the final, carboxyl-terminal, 21 amino acids that include both NPXY motifs and contains instead 25 essentially random amino acids. These 21 amino acids that have been deleted contain multiple binding sites for proteins known to interact with β integrins, including talin (reviewed in Ref. 62). We have tested this mutant along with a simple deletion of the final 21 amino acids and find that TWOW-1 binding is essentially normal (Fig. 6 and supplemental Table S4) although these mutants reduce cell spreading (15). These results are consistent with our previous findings that although talin binding is important for integrin activity, it may not be required for monovalent ligand affinity modulation (63, 64). Finally, in some but not all integrins, a salt bridge between the arginine of the α-cytoplasmic domain GFFKR motif and the β3 integrin Asp-723, is required to maintain the integrin in an inactive state (65, 66). We have mutated this aspartic acid to arginine in βPS (D807R) and find no increase in affinity for TWOW-1. Mutation of the PS2C arginine (GFFNR → GFFNA) also does not affect soluble ligand binding (Fig. 6 and supplemental Table S4). Thus, in this system, breaking the arginine-aspartic acid interaction in the cytoplasmic membrane-proximal region is not sufficient to change integrin affinity for ligand.
Given our failure to detect affinity changes in mutations of the βPS cytoplasmic domain and the αPS2C (GFFNR → GFFNA) mutant, we have considered the possibility that our assay is not sensitive enough to detect changes in integrin affinity transmitted from inside the cell to outside. However, our results with additional cytoplasmic αPS2C mutants suggest that this is not the case.
αPS2C Cytoplasmic Mutations Modulate Affinity
In contrast to the results with βPS and αPS2C GFFNR → GFFNA, mutation of the αPS2C cytoplasmic domain GFFNR sequence to GFANA does significantly increase integrin affinity (7) (Fig. 6 and supplemental Table S4). Additionally, just prior to the GFFNR in αPS2C is a cysteine residue. The human integrin α subunits α3, α6, and αE also contain a cysteine at this location. In α3 and α6, this cysteine is palmitoylated (67). Affinity regulation by the cysteine in any α subunit has not been tested, but palmitoylation of β4 does influence integrin-mediated cell morphology and signaling (67). We have mutated this cysteine in αPS2C to a valine. Valine was chosen because it is the amino acid found in some human α subunits (αIIB, αX, and αL), but more importantly, it is the amino acid found in the αPS1 integrin, another binding partner for βPS. We find that this mutation significantly reduces the ability of the integrin to bind soluble ligand (Fig. 6 and supplemental Table S4). In conclusion, changes in ligand binding ability due to cytoplasmic domain mutations can be detected by our assay, and the mutations in βPS subunit that were expected to affect the integrin affinity may not.
Mutations Not Affecting Integrin Binding to Ligand
A number of our mutants do not result in significant alterations in the ability of the PS2C integrins to bind TWOW-1. These mutants may influence the activity of the integrin by a wide variety of other mechanisms that need to be tested with different assays. They may alter the integrin clustering, signaling capabilities, stability, or critical changes in conformation when traction forces are applied due to the integrins engaging with insoluble ligand bound to a matrix and the cytoskeleton in a three-dimensional context. Finally, βPS integrin is a subunit in other integrins besides the PS2C integrin, and the PS2C integrin itself can bind a variety of RGD-containing ligands. Therefore, it may be that those mutants not showing altered TWOW-1 binding may still affect the affinity for different ligands in different integrins.
In Vivo Test of Mutant Integrin Activity
Genetic experiments in the whole organism are unable to directly probe the affinity of individual integrin receptors, but they can assess the general functioning of the integrins. Therefore, we tested the viable mys mutants for genetic interactions with rhea, the gene encoding talin. Talin is important for promoting integrin activity in Drosophila (8, 9), and genetic interactions between rhea and mys mutants have been reported (6, 8, 18). We raised mys mutant flies at temperatures that reduced their viability and tested the effect of reducing talin levels by making the flies heterozygous for rhea2, a strong allele (supplemental Table S5). Results showed that temperature had no effect on flies heterozygous for rhea without the presence of a mutation in mys. Supporting the validity of the cell culture assay, the three viable mutations that reduced integrin function (by reduced affinity) showed enhanced lethality when heterozygous for rhea. Of the mutants with increased affinity, 18 could be tested for interactions with rhea. For 12 of the mutants, heterozygous rhea reduced lethality; for two, there was no interaction; and for four, heterozygous rhea enhanced lethality. Reduced lethality (12 cases) when talin function is reduced is entirely consistent with lethality of the mys mutant attributed to increased integrin activity, validating the cell culture binding assay results reporting on affinity changes. In the whole fly, integrin functions are likely to be complex, and a mutation increasing integrin affinity in our cell culture assay may also have the effect in the fly of reducing integrin function. This could be due to effects on an integrin other than PS2C (βPS paired with a different α subunit), interactions with a different ligand, or effects on clustering, turnover, or signaling. This would appear to be the case for mysb58 (and three others), which clearly increases integrin affinity and ability to promote cell adhesion and spreading (6,7) (this report) but shows increased lethality in combination with rhea. Seventeen of our βPS mutants did not alter binding to TWOW-1, and we therefore had no information on whether they increased or decreased integrin activity. Twelve of these could be tested for interactions with rhea. Seven showed reduced lethality, suggesting that increased integrin function by means other than affinity of PS2C integrins for a tiggrin ligand was the cause of their lethality. Four showed increased lethality, indicating that reduction of integrin function at some level was the cause of their lethality. Details of the genetic interactions can be found in supplemental Table S5, and the qualitative results are included in Table 1 and supplemental Tables S2 and S3.
Integrin Structure Is Devoted to Restraint
Our genetic screens in flies were designed to yield mutants that completely inactivated the βPS integrin subunit (15) or decreased its activity (6). Here we have determined that very few of the mutations render the integrin less capable of binding a soluble monovalent ligand, whereas many more activate the integrin. This result can be explained by considering that the region of the molecule devoted to positive interactions between integrin and ligand is restricted to the MIDAS domain (and indirectly the SyMBS domain). All of our mutants that decrease ligand binding, with the exception of the in vitro mutant in the cysteine residue preceding the GFFNR sequence in the αPS2C cytoplasmic domain, are indeed in the MIDAS or SyMBS domains (Fig. 3C and Table 1). Our results would suggest that the remainder of the molecule has evolved multiple interactions designed to keep the binding domain in a low affinity state but poised to shift to high affinity. Disruption of these interactions by mutations in the PSI, I, Hybrid, four I-EGF repeats, βTD, and the cytoplasmic domains (at least in the case of αPS2C), results in increased affinity of the integrin for ligand.
Supplementary Material
Acknowledgments
We thank Paula Campbell and Junesse Farley of the Arizona Cancer Center/Arizona Research Laboratories-Division of Biotechnology Cytometry Core Facility. We also thank Candida Morris, who assisted with manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM42474 and CA 023074 (Cancer Center support grant from NCI).
This work is dedicated to the memory of our close colleague and friend, Danny Brower, whose untimely death happened just prior to the beginning of the work reported here. He provided helpful discussions regarding this project and secured the funding that made this work possible.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S5 and Figs. S1–S9.
- MFI
- mean fluorescence intensity
- βTD
- β tail domain
- I-EGF
- integrin-EGF
- PSI
- plexin-semaphorin-integrin
- MIDAS
- metal ion-dependent adhesion site
- ADMIDAS
- adjacent to MIDAS
- SyMBS
- synergistic metal binding site.
REFERENCES
- 1. Hynes R. O. (2002) Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 2. Shattil S. J., Kim C., Ginsberg M. H. (2010) Nat. Rev. Mol. Cell Biol. 11, 288–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo B. H., Carman C. V., Springer T. A. (2007) Annu. Rev. Immunol. 25, 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arnaout M. A., Goodman S. L., Xiong J. P. (2007) Curr. Opin. Cell Biol. 19, 495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bunch T. A., Salatino R., Engelsgjerd M. C., Mukai L., West R. F., Brower D. L. (1992) Genetics 132, 519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jannuzi A. L., Bunch T. A., West R. F., Brower D. L. (2004) Mol. Biol. Cell 15, 3829–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bunch T. A., Helsten T. L., Kendall T. L., Shirahatti N., Mahadevan D., Shattil S. J., Brower D. L. (2006) J. Biol. Chem. 281, 5050–5057 [DOI] [PubMed] [Google Scholar]
- 8. Brown N. H., Gregory S. L., Rickoll W. L., Fessler L. I., Prout M., White R. A., Fristrom J. W. (2002) Dev. Cell 3, 569–579 [DOI] [PubMed] [Google Scholar]
- 9. Tanentzapf G., Brown N. H. (2006) Nat. Cell Biol. 8, 601–606 [DOI] [PubMed] [Google Scholar]
- 10. Martin-Bermudo M. D., Dunin-Borkowski O. M., Brown N. H. (1998) J. Cell Biol. 141, 1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arnaout M. A., Mahalingam B., Xiong J. P. (2005) Annu. Rev. Cell Dev. Biol. 21, 381–410 [DOI] [PubMed] [Google Scholar]
- 12. Zhu J., Boylan B., Luo B. H., Newman P. J., Springer T. A. (2007) J. Biol. Chem. 282, 11914–11920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xiong J. P., Mahalingham B., Alonso J. L., Borrelli L. A., Rui X., Anand S., Hyman B. T., Rysiok T., Müller-Pompalla D., Goodman S. L., Arnaout M. A. (2009) J. Cell Biol. 186, 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bunch T. A., Brower D. L. (1992) Development 116, 239–247 [DOI] [PubMed] [Google Scholar]
- 15. Jannuzi A. L., Bunch T. A., Brabant M. C., Miller S. W., Mukai L., Zavortink M., Brower D. L. (2002) Mol. Biol. Cell 13, 1352–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu J., Luo B. H., Xiao T., Zhang C., Nishida N., Springer T. A. (2008) Mol. Cell 32, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wright T. R. (1968) in Proceedings of the 12th Annual Congress on Genetics, Tokyo, Japan, Vol. 1, p. 41 [Google Scholar]
- 18. Prout M., Damania Z., Soong J., Fristrom D., Fristrom J. W. (1997) Genetics 146, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lindsley D. L., Zimm G. G. (1992) Annu. Rev. Genomics Hum. Genet. 4, 1133 [Google Scholar]
- 20. Puzon-McLaughlin W., Takada Y. (1996) J. Biol. Chem. 271, 20438–20443 [DOI] [PubMed] [Google Scholar]
- 21. Mor-Cohen R., Rosenberg N., Peretz H., Landau M., Coller B. S., Awidi A., Seligsohn U. (2007) Thromb. Haemost. 98, 1257–1265 [PubMed] [Google Scholar]
- 22. Zhu J., Luo B. H., Barth P., Schonbrun J., Baker D., Springer T. A. (2009) Mol. Cell 34, 234–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zang Q., Springer T. A. (2001) J. Biol. Chem. 276, 6922–6929 [DOI] [PubMed] [Google Scholar]
- 24. Shi M., Sundramurthy K., Liu B., Tan S. M., Law S. K., Lescar J. (2005) J. Biol. Chem. 280, 30586–30593 [DOI] [PubMed] [Google Scholar]
- 25. Luo B. H., Springer T. A. (2006) Curr. Opin. Cell Biol. 18, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiao T., Takagi J., Coller B. S., Wang J. H., Springer T. A. (2004) Nature 432, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun Q. H., Liu C. Y., Wang R., Paddock C., Newman P. J. (2002) Blood 100, 2094–2101 [DOI] [PubMed] [Google Scholar]
- 28. Murcia M., Jirouskova M., Li J., Coller B. S., Filizola M. (2008) Proteins 71, 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bajt M. L., Loftus J. C. (1994) J. Biol. Chem. 269, 20913–20919 [PubMed] [Google Scholar]
- 30. Nair S., Ghosh K., Shetty S., Mohanty D. (2004) Haematologica 89, 1529–1530 [PubMed] [Google Scholar]
- 31. Luo B. H., Karanicolas J., Harmacek L. D., Baker D., Springer T. A. (2009) J. Biol. Chem. 284, 3917–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mould A. P., Barton S. J., Askari J. A., Craig S. E., Humphries M. J. (2003) J. Biol. Chem. 278, 51622–51629 [DOI] [PubMed] [Google Scholar]
- 33. Valdramidou D., Humphries M. J., Mould A. P. (2008) J. Biol. Chem. 283, 32704–32714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen J., Salas A., Springer T. A. (2003) Nat. Struct. Biol. 10, 995–1001 [DOI] [PubMed] [Google Scholar]
- 35. Chen J., Yang W., Kim M., Carman C. V., Springer T. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13062–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krukonis E. S., Isberg R. R. (2000) Cell Microbiol. 2, 219–230 [DOI] [PubMed] [Google Scholar]
- 37. Springer T. A., Zhu J., Xiao T. (2008) J. Cell Biol. 182, 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raborn J., Wang W., Luo B. H. (2011) Biochemistry 50, 2084–2091 [DOI] [PubMed] [Google Scholar]
- 39. Hato T., Yamanouchi J., Yakushijin Y., Sakai I., Yasukawa M. (2006) J Thromb. Haemost. 4, 2278–2280 [DOI] [PubMed] [Google Scholar]
- 40. Cheng M., Foo S. Y., Shi M. L., Tang R. H., Kong L. S., Law S. K., Tan S. M. (2007) J. Biol. Chem. 282, 18225–18232 [DOI] [PubMed] [Google Scholar]
- 41. Morel-Kopp M. C., Melchior C., Chen P., Ammerlaan W., Lecompte T., Kaplan C., Kieffer N. (2001) Thromb. Haemost. 86, 1425–1434 [PubMed] [Google Scholar]
- 42. Nurden A. T., Ruan J., Pasquet J. M., Gauthier B., Combrié R., Kunicki T., Nurden P. (2002) Platelets 13, 101–111 [DOI] [PubMed] [Google Scholar]
- 43. McKay B. S., Annis D. S., Honda S., Christie D., Kunicki T. J. (1996) J. Biol. Chem. 271, 30544–30547 [DOI] [PubMed] [Google Scholar]
- 44. Xiong J. P., Stehle T., Diefenbach B., Zhang R., Dunker R., Scott D. L., Joachimiak A., Goodman S. L., Arnaout M. A. (2001) Science 294, 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xiong J. P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S. L., Arnaout M. A. (2002) Science 296, 151–155 [DOI] [PubMed] [Google Scholar]
- 46. Grimaldi C. M., Chen F., Scudder L. E., Coller B. S., French D. L. (1996) Blood 88, 1666–1675 [PubMed] [Google Scholar]
- 47. Takagi J., Beglova N., Yalamanchili P., Blacklow S. C., Springer T. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11175–11180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Campbell I. D., Bork P. (1993) Curr. Opin. Struct. Biol. 3, 385–392 [Google Scholar]
- 49. Kashiwagi H., Tomiyama Y., Tadokoro S., Honda S., Shiraga M., Mizutani H., Handa M., Kurata Y., Matsuzawa Y., Shattil S. J. (1999) Blood 93, 2559–2568 [PubMed] [Google Scholar]
- 50. Ruiz C., Liu C. Y., Sun Q. H., Sigaud-Fiks M., Fressinaud E., Muller J. Y., Nurden P., Nurden A. T., Newman P. J., Valentin N. (2001) Blood 98, 2432–2441 [DOI] [PubMed] [Google Scholar]
- 51. Kamata T., Ambo H., Puzon-McLaughlin W., Tieu K. K., Handa M., Ikeda Y., Takada Y. (2004) Biochem. J. 378, 1079–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vanhoorelbeke K., De Meyer S. F., Pareyn I., Melchior C., Plançon S., Margue C., Pradier O., Fondu P., Kieffer N., Springer T. A., Deckmyn H. (2009) J. Biol. Chem. 284, 14914–14920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Matsumoto A., Kamata T., Takagi J., Iwasaki K., Yura K. (2008) Biophys. J. 95, 2895–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li R., Mitra N., Gratkowski H., Vilaire G., Litvinov R., Nagasami C., Weisel J. W., Lear J. D., DeGrado W. F., Bennett J. S. (2003) Science 300, 795–798 [DOI] [PubMed] [Google Scholar]
- 55. Luo B. H., Carman C. V., Takagi J., Springer T. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3679–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Partridge A. W., Liu S., Kim S., Bowie J. U., Ginsberg M. H. (2005) J. Biol. Chem. 280, 7294–7300 [DOI] [PubMed] [Google Scholar]
- 57. Chen Y. P., Djaffar I., Pidard D., Steiner B., Cieutat A. M., Caen J. P., Rosa J. P. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10169–10173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen Y. P., O'Toole T. E., Ylänne J., Rosa J. P., Ginsberg M. H. (1994) Blood 84, 1857–1865 [PubMed] [Google Scholar]
- 59. Perrault C., Mekrache M., Schoevaert D., Kieffer N., Melchior C., Warszawski J., Baruch D. (1998) Cell Adhes. Commun. 6, 335–348 [DOI] [PubMed] [Google Scholar]
- 60. Kieffer N., Melchior C., Guinet J. M., Michels S., Gouon V., Bron N. (1996) Cell Adhes. Commun. 4, 25–39 [DOI] [PubMed] [Google Scholar]
- 61. Sakai T., Zhang Q., Fässler R., Mosher D. F. (1998) J. Cell Biol. 141, 527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Legate K. R., Fässler R. (2009) J. Cell Sci. 122, 187–198 [DOI] [PubMed] [Google Scholar]
- 63. Helsten T. L., Bunch T. A., Kato H., Yamanouchi J., Choi S. H., Jannuzi A. L., Féral C. C., Ginsberg M. H., Brower D. L., Shattil S. J. (2008) Mol. Biol. Cell 19, 3589–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bunch T. A. (2010) J. Biol. Chem. 285, 1841–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hughes P. E., Diaz-Gonzalez F., Leong L., Wu C., McDonald J. A., Shattil S. J., Ginsberg M. H. (1996) J. Biol. Chem. 271, 6571–6574 [DOI] [PubMed] [Google Scholar]
- 66. Ahrens I. G., Moran N., Aylward K., Meade G., Moser M., Assefa D., Fitzgerald D. J., Bode C., Peter K. (2006) Exp. Cell Res. 312, 925–937 [DOI] [PubMed] [Google Scholar]
- 67. Yang X., Kovalenko O. V., Tang W., Claas C., Stipp C. S., Hemler M. E. (2004) J. Cell Biol. 167, 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.