Abstract
Ribosome-inactivating proteins (RIPs) are N-glycosidases that remove a specific adenine from the sarcin/ricin loop of the large rRNA, thus arresting protein synthesis at the translocation step. In the present study, a protein termed tobacco RIP (TRIP) was isolated from tobacco (Nicotiana tabacum) leaves and purified using ion exchange and gel filtration chromatography in combination with yeast ribosome depurination assays. TRIP has a molecular mass of 26 kD as evidenced by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and showed strong N-glycosidase activity as manifested by the depurination of yeast rRNA. Purified TRIP showed immunoreactivity with antibodies of RIPs from Mirabilis expansa. TRIP released fewer amounts of adenine residues from ribosomal (Artemia sp. and rat ribosomes) and non-ribosomal substrates (herring sperm DNA, rRNA, and tRNA) compared with other RIPs. TRIP inhibited translation in wheat (Triticum aestivum) germ more efficiently than in rabbit reticulocytes, showing an IC50 at 30 ng in the former system. Antimicrobial assays using highly purified TRIP (50 μg mL-1) conducted against various fungi and bacterial pathogens showed the strongest inhibitory activity against Trichoderma reesei and Pseudomonas solancearum. A 15-amino acid internal polypeptide sequence of TRIP was identical with the internal sequences of the iron-superoxide dismutase (Fe-SOD) from wild tobacco (Nicotiana plumbaginifolia), Arabidopsis, and potato (Solanum tuberosum). Purified TRIP showed SOD activity, and Escherichia coli Fe-SOD was observed to have RIP activity too. Thus, TRIP may be considered a dual activity enzyme showing RIP-like activity and Fe-SOD characteristics.
Various plants contain enzymes called ribosomeinactivating proteins (RIPs), officially called rRNA N-glycosidases (EC 3.2.2.22), which catalytically inactivate eukaryotic as well as prokaryotic ribosomes (Barbieri et al., 1993) by removing single adenine residues from the large rRNA (A4324 from rat liver rRNA). RIPs selectively cleave an adenine from the universally conserved sarcin/ricin loop of the large rRNA (Endo and Tsurugi, 1987, Endo et al., 1987; Wool et al., 1992; Barbieri et al.,1993), thus interrupting the interaction of elongation factors I and II with the ribosomes and ultimately arresting protein synthesis at the translocation step (for reviews, see Stirpe et al., 1992; Mehta and Boston, 1998; Tumer et al., 1999; Nielsen and Boston, 2001; Van Damme et al., 2001). In addition to its N-glycosidase activity, some RIPs have DNAse, DNA glycosylase, and apurinic pyrimidinic lyase activities (Li et al., 1991; Roncuzzi and Gasperi-Gampani, 1996; Nicolas et al., 1997, 1998, 2000; Hudak et al., 2000). It has been suggested, however, that the sporadically observed nuclease activities are due to contamination by other enzymes (Day et al., 1998; Barbieri et al., 2000; Van Damme et al., 2001), whereas all RIPs tested remove adenine from DNA and some of them also from poly(A) (Barbieri et al., 1997).
Different RIPs have been reported from about 50 plant species covering 17 families. Some families include many RIP-producing species, particularly Cucurbitaceae, Euphorbiaceae, Poaceae, and families belonging to the superorder Caryophyllales (Grasso and Shepherd, 1978; Stirpe and Barbieri, 1986; Kwon et al., 2000). RIPs are divided into two categories depending upon the conformation of their subunits. Type I RIPs are single-chained proteins with a molecular mass of approximately 30 kD, whereas type II RIPs such as ricin and abrin possess two subunits, a catalytic subunit (A chain) and a lectin subunit (B chain; for review, see Mehta and Boston, 1998; Tumer et al., 1999; Nielsen and Boston, 2001). Generally, type II RIPs are regarded to be highly toxic by virtue of their carbohydrate-binding lectin chain, which interacts with cell membranes and facilitates the uptake of RIP into the cytosol space (Barbieri et al., 1993). However, other type II RIPs have low toxicity (for review, see Ferreras et al., 2000; Van Damme et al., 2001). Apart from these two RIP types, a 60-kD protein (JIP60) has been identified in barley (Hordeum vulgaris) and classified as a type III RIP. Type III RIPs have an N-terminal domain closely related to the A chain of RIPs and linked to an unrelated C-terminal domain with unknown function (Reinbothe et al., 1994).
RIPs have shown broad spectrum antiviral activity against RNA, DNA, and plant and animal viruses (Battelli and Stirpe, 1995; Wang and Tumer, 2000). For instance, the RIP from Mirabilis jalapa has antiviral activity against the tobacco (Nicotiana tabacum) mosaic virus, potato (Solanum tuberosum) virus X, potato virus Y, and viroids such as the potato spindle tuber viroid (Kubo et al., 1990; Kataoka et al., 1991, 1992; Vivanco, 1997). Also, some RIPs have specific DNA nuclease activity against supercoiled, covalently closed, circular plasmid DNA and single-stranded phage DNA (Roncuzzi and Gasperi-Gampani, 1996). A few type I RIPs have been shown to inhibit fungal growth (Roberts and Selitrennikof, 1986; Vivanco et al., 1999; Park et al., 2002a, 2002b). Antifungal activity of the asparagus (Asparagus officinalis) RIP has been reported against Botrytis cinerea (Wang and Ng, 2001). Furthermore, two proteins, reportedly RIPs, from mushrooms Hypsizygus marmoreus and Lyophyllum shimeji, have been shown to be active against an array of fungi (Lam and Ng, 2001a, 2001b). Antibacterial and antifungal activity of type I RIPs have also been reported from Mirabilis expansa RIPs (ME; Vivanco et al., 1999).
The enzymology and bioactivities of RIPs have been well characterized, more so than their biological and physiological functions in plants. As a result, the role of RIPs in planta remains open to speculation (Nielsen and Boston, 2001). Recently, Hudak et al. (2002) have reported that PAP, a type I RIP from Phytolacca americana, recognizes and binds to the cap structure of mRNA and subsequently depurinates the mRNA. In addition, Parikh et al. (2002) showed that PAP can target and destabilize its own mRNA under up-regulated conditions. A critical reason why the biological role of RIPs in planta has been poorly elucidated is the lack of information about this group of enzymes from any model plant system. Identification and characterization of RIPs from a model plant would provide insight into the biological function of this group of enzymes. In this manuscript, we report the identification and characterization of a novel RIP-like enzyme, termed tobacco RIP (TRIP), isolated from tobacco leaves.
RESULTS
Purification of a RIP-Like Protein from Tobacco Leaves
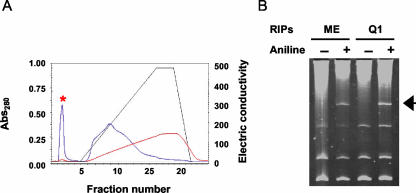
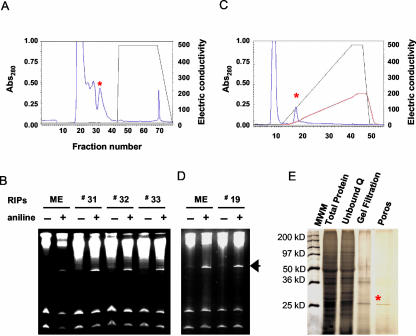
The TRIP was purified from the leaves of 2-month-old tobacco plants using anion exchange, gel filtration, and cation exchange chromatographic techniques as described in “Materials and Methods.” The depurination assay was performed sequentially using yeast (Saccharomyces cerevisiae) ribosomes as a substrate to confirm the presence of RIP activity in the chromatography fractions. The release of a 367-bp “Endo” (Endo et al., 1987) fragment from the large rRNA upon treatment with acidic aniline indicated the presence of RIP activity. The initial separation of the leaf tissue extract with the Q1 anion exchange column resulted in recovery of RIP activity in the flow-through protein fraction (Fig. 1, A and B). This indicated the relatively basic nature of the RIP activity. After gel filtration chromatography of the basic protein fraction, RIP activity was subsequently located in fractions 31 and 32 (Fig. 2, A and B), which further indicated an approximate size of 26 kD under native conditions. Final separation of the active fractions was performed with a POROS-HS strong cation exchange column. Elution of bound proteins with a linear gradient of NaCl (50–1,000 mm) resulted in the recovery of depurination activity in fraction 19 (Fig. 2, C and D). A single protein band of 26 kD was observed in this fraction by SDS-PAGE (Fig. 2E). This protein was named TRIP, and the estimated yield was 0.01% of total starting material. The relative PI for TRIP was found to be greater than pH 10 by pH gradient elution from a cation exchange column (data not shown).
Figure 1.
Anion exchange chromatography of protein from tobacco leaves. A, The total soluble proteins from the tobacco leaves were extracted, dialyzed, and fractionated on an anion exchange chromatographer. B, Yeast ribosomes were isolated from yeast and incubated with TRIP and ME as described in “Materials and Methods.” rRNAs were extracted, treated with aniline, separated on a 4.5% (w/v) ureapolyacrylamide gel, and stained with ethidium bromide. The presence (+) or absence (-) of aniline is denoted. The arrow shows the presence of the diagnostic 367-nucleotide cleavage product of rRNA.
Figure 2.
Gel filtration and cation exchange chromatography. A, The basic tobacco proteins were separated via gel filtration. B, Gel filtration fractions were checked for depurination activity using yeast ribosomes as a substrate as described in “Materials and Methods.” C, Fractions 31 and 32 were pooled together and fractionated on porous HS (cation exchange column). Bound protein was eluted with the linear gradient of KCl (0–1000 mm). D, Yeast ribosomes were incubated with the bound fraction of the Poros column to confirm the RIP activity of the eluted protein as described in “Materials and Methods.” rRNA was extracted, treated with aniline, separated on 4.5% (w/v) urea gel, and visualized by staining with ethidium bromide. An arrow mark shows the diagnostic fragment released due to the treatment of aniline. The presence and absence of aniline is denoted. E, Analysis by SDS-PAGE of protein fractions exhibiting depurination activity at various stages of purification. Molecular weight marker, Mr marker proteins, anion exchange chromatography, gel filtration fraction, and cation exchange fractions were fractionated on 12% (w/v) SDS-PAGE. Approximately 10 μg of the partially purified fractions and 0.25 μg of the purified protein was loaded on the gel. The protein band was visualized by silver staining after electrophoresis.
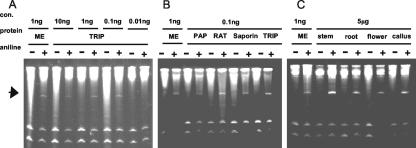
We screened various tobacco tissues for RIP activity. Partially purified protein fractions (basic protein enriched by cation exchange chromatography) were used to concentrate any depurination activity, because TRIP was found in such low amounts in leaf tissue. As shown in Figure 3C, TRIP was detected in the roots, stems, flowers, and callus tissue of tobacco, but the yield appeared similar to that of leaf tissue.
Figure 3.
Detection of TRIP in different organs of tobacco, titers of TRIP, and a comparison of TRIP activity and that of different RIPs. A, RIP activity was tested in the different parts of the tobacco plant, i.e. roots, stem, flower, and callus culture. Partially purified proteins from the different parts of the tobacco plant were incubated with yeast ribosomes, and the rRNA was analyzed after aniline treatment on urea gel as described in “Materials and Methods.” Depurination activity of TRIP was tested ranging from 0.01 to 10 ng mL-1 (B) and was compared at 0.1 ng mL-1 concentration to the other known RIPs like ME, PAP-H (from P. americana), ricin (castor bean [Ricinus communis]), and saporin (Saponaria officinalis; C).
The approximate titer for TRIP RNA N-glycosidase activity was further tested on yeast ribosomes, testing at concentrations ranging from 0.01 to 10 ng mL-1. The characteristic 367-nucleotide “Endo” fragment was observed with as little as 0.1 ng mL-1 (Fig. 3A). This level of activity was comparable with other RIPs, such as MEs, saporin (from S. officinalis), PAP-H (from P. americana), and ricin (from castor bean; Fig. 3B).
Adenine Release by TRIP from Ribosomal and Non-Ribosomal Animal Substrates
We examined the ability of TRIP to release adenine from different ribosomal and non-ribosomal animal sources. The adenine release was measured according to the HPLC method of Barbieri et al. (2003). As evidenced from Table I, adenine residues were released from different sources such as herring sperm DNA (hsDNA), rRNA, and tRNA. This experiment was performed under two different pH conditions (pH 4.0 and 7.0). At pH 7.0, the adenine released from tRNA was almost double that released at pH 4.0 (0.32–0.67 pmol). Similarly, the ability of TRIP to use different ribosomal substrates was also studied. The ribosomes from Artemia sp. and rat were incubated with TRIP and momordin II (10 pmol each), and the adenine-releasing capacity of these RIPs was compared. As evidenced in Table II, TRIP released less adenine than momordin. The release of adenine by TRIP was also low compared with the activity reported for other known RIPs (Barbieri et al., 1997). Although able to damage ribosomes, TRIP appears less potent than other type I RIPs.
Table I.
Adenine release from herring sperm DNA, Escherichia coli, and tRNA by TRIPs
Adenine release from hs DNA, from E. coli rRNA, and from phe-specific tRNA was measured after separation by HPLC. The reaction mixture (50 μL) contained 50 mm acetate buffer, pH 4.0 and pH 7.0, 10 μg of nucleic acid, and 10 pmol of an appropriate enzyme and was incubated at 37°C for 40 min. The reaction was stopped in ice by the addition of 200 μL of cold BA buffer (50 mm sodium acetate, pH 4.0 and pH 7.0, and 20 mm KCl), and adenine was measured by LC/MS on a waters Alliance/zq.
| TRIP (10 pmol) | Hs DNA (10 μg) Adenine Released | rRNA (10 μg) Adenine Released | tRNA (10 μg) Adenine Released |
|---|---|---|---|
| pmol | |||
| pH 7.0 | 0.01 | 0.03 | 0.67 |
| pH 4.0 | 0.01 | 0.1 | 0.32 |
Table II.
Depurination of Artemia sp. ribosomes and rat ribosomes by TRIPs
Two picomoles each of Artemia sp. ribosomes and rat ribosomes was used for the depurination by TRIP and momordin II. The appropriate ribosomes were incubated with the enzyme for 40 min at 37°C. The adenine was measured by the LC/MS on a waters/zq apparatus. Other experimental conditions are described in detail in the text.
| Treatment (10 pmol) | Artemia sp. Ribosomes (2 pmol) | Rat Ribosomes (2 pmol) |
|---|---|---|
| Momordin II | 1.78 | 0.63 |
| TRIP | 0.45 | 0.25 |
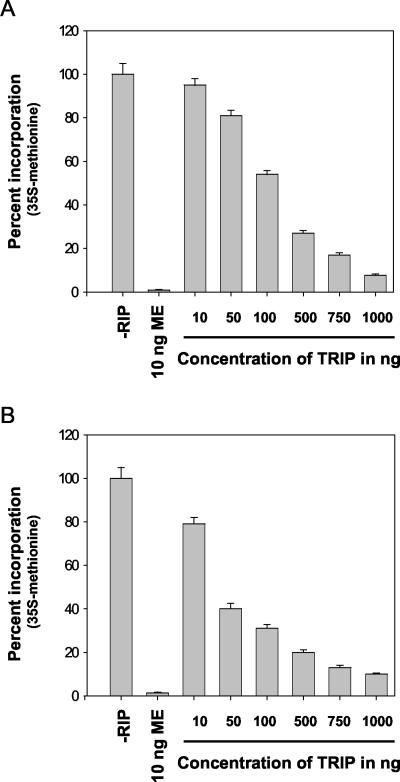
Translation Inhibition of Rabbit Reticulocytes and Wheat Germ Lysate by TRIP
The in vitro translation inhibitory effect of TRIP was compared for rabbit reticulocytes and wheat germ lysates. Rabbit reticulocytes and wheat germ lysates were incubated with increasing concentrations of TRIP (10–1000 ng) for 30 min at 30°C before translation was initiated. Untreated rabbit reticulocytes and wheat germ lysates were used as the negative control and ME-treated reticulocytes and lysates were used as a positive control. The reaction was started by adding Brome mosaic virus mRNA to the TRIP and ME-treated reticulocytes and wheat germ lysates. As observed in Figure 4, A (rabbit reticulocytes) and B (wheat germ lysates), TRIP inhibited the wheat germ translation system more efficiently and at a lower concentration than it inhibited the rabbit reticulocyte translation system. The IC50 for the wheat germ was found to be 30 ng compared with 100 ng in rabbit reticulocytes. Further comparison of in vitro translation inhibition of TRIP with ME showed that TRIP is at least 1,000-fold less efficient at inhibiting both systems than ME. The ability of RIPs to inhibit in vitro translation generally varies in RIPs isolated from different plant systems, and some RIPs require a cofactor like ATP for their maximal translational inhibitory activity (Nielsen and Boston, 2001). As we noted above, TRIP is less efficient at inhibiting translation compared with ME, and thus it may be possible that TRIP needs a cofactor or other biochemical optimization agent for its maximal inhibitory activity. This result was another indication of the RIP-like nature of TRIP.
Figure 4.
Inhibition of in vitro protein synthesis by TRIP. The rabbit reticulocytes (A) and wheat germ lysates (B) were treated with different concentrations of TRIP (10–1,000 ng) at 30°C for 30 min. The treated wheat germ lysate and rabbit reticulocytes were incubated with Brome mosaic virus mRNA. The 35S-labeled Met was used to label the product. The newly synthesized protein was treated with 1 n NaOH and 0.2% (v/v) H2O2 and incubated at 37°C for 30 min. The protein was then precipitated by 25% (w/v) chilled trichloroacetic acid (TCA) and incubated on ice for 20 min. The protein was harvested on 25-mm glass fiber filters. The filters were then washed with three washes of 5% (w/v) chilled TCA with a final wash of cold acetone. The radioactivity was measured by using a liquid scintillation counter. Inhibition of the protein synthesis was expressed as the percent incorporation of radioactivity in the control where no TRIP was added. Wheat germ lysate and rabbit reticulocytes without any treatment served as the negative control, and ME was taken as the positive control.
Antimicrobial Activity of TRIP
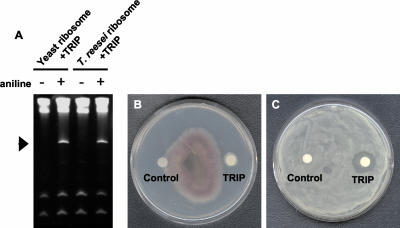
To explore the biological significance of the enzymatic activity of TRIP on the growth of different fungi and bacteria, we examined the N-glycosidase activity of TRIP on ribosomes isolated from Trichoderma reesei (Fig. 5A) and P. solancearum (data not shown). The depurination assay was conducted in the same manner as the depurination assay of yeast ribosomes. TRIP showed depurination activity on the purified fungal and bacterial ribosomes tested at 10 ng mL-1. As a consequence of this finding, TRIP (50 μg mL-1) was tested on a number of pathogenic fungi and bacteria. The purified TRIP was found to inhibit a number of these pathogens (Table III). For fungi, the growth inhibition due to TRIP activity was observed as a crescent-shaped zone of inhibition at the mycelial front. Out of many fungi tested, TRIP was found to be most active on T. reesei. The time-course assay with T. reesei showed a strong mycelial growth inhibition with 50 μg mL-1 purified TRIP (Fig. 5B). Among all of the bacteria tested, P. solancearum showed the greatest inhibition by TRIP at a 50 μg mL-1 concentration (Fig. 5C).
Figure 5.
Antimicrobial activity of the purified TRIP. A, Depurination of T. reesei in vitro. Ribosomes were isolated from both of the organisms and incubated with ME and TRIP. rRNA was isolated after the treatment and treated with aniline. The presence and absence of the aniline is denoted. B, Radial growth inhibition assay of TRIP against T. reesei. C, The plate inhibition assay of TRIP against P. solancearum. Sodium-phosphate buffer (20 mm), pH 7.0 (control) and 50 μg of filter-sterilized TRIP were applied to the sterilized disc and tested for antimicrobial activity.
Table III.
Antimicrobial activity of TRIP
Antifungal activity of TRIP against various fungi is shown. Fifty micrograms of filter-sterilized purified TRIP in 100 μL of sterile 20 mm phosphate buffer, pH 7.0, was used for determination of antifungal activity. The effect on fungal growth is expressed in qualitative terms according to the method of Schlumbaum et al. (1986), in which +++ stands for strong inhibition, + for just detectable inhibition, and - for no inhibition under similar conditions. Antibacterial activity of TRIP against several bacteria is also shown. Fifty micrograms of filter-sterilized TRIP in 100 μL of 20 mm phosphate buffer, pH 7.0, was used for determination of antibacterial activity. The results shown here represent the mean of three replicates.
| Tested Pathogen | Inhibition of Growth |
|---|---|
| Bacteria | |
| P. solancearum | ++++ |
| Ervinia amylovora | +++ |
| Shigella asonei | +++ |
| Salmonella typhimurium | ++ |
| Rhizobium leguminosarum | ++ |
| Enterobacter aerogenes | + |
| Agrobacterium radiobacter | + |
| Bacillus subtilis | + |
| Erwinia herbicola | + |
| Fungi | |
| B. cinerea | — |
| Trichoderma viride | — |
| T. reesei | +++ |
| Cytospora cankar | ++ |
| Pestalotia sp. | + |
| Fusarium oxysporum | + |
| Phoma sp. | + |
| Cochlioboluo heterostrophus | + |
Internal Sequence and Comparison of Sequence with Iron-Superoxide Dismutase (Fe-SOD)
Because the N-terminal amino acid sequence of TRIP was blocked, a partial internal amino acid sequence was obtained for TRIP (Fig. 6). A BLAST database sequence search indicated that the internal 15 amino acid sequence of TRIP was identical with the Fe-SOD from wild tobacco (Nicotiana plumbaginifolia; gi-100284), potato (gi-27543371), and Arabidopsis (gi-2072628). The Fe-SODs from Arabidopsis (gi-15241373) and the SOD gene of chloroplasts (gi-134642) had three amino acid variations compared with the 15-amino acid region of TRIP. TRIP (26 kD) isolated from tobacco was larger in size than the Fe-SOD (21 kD) from wild tobacco.
Figure 6.
Comparison of the internal amino acid sequence of TRIP with different Fe-SODs. The internal sequence of the TRIP was compared with Fe-SOD from wild tobacco (GenBank accession no. gi-2072628), potato (gi-2754337), Arabidopsis (gi-134642), Arabidopsis (gi-100284), and chloroplast (gi-15241373).
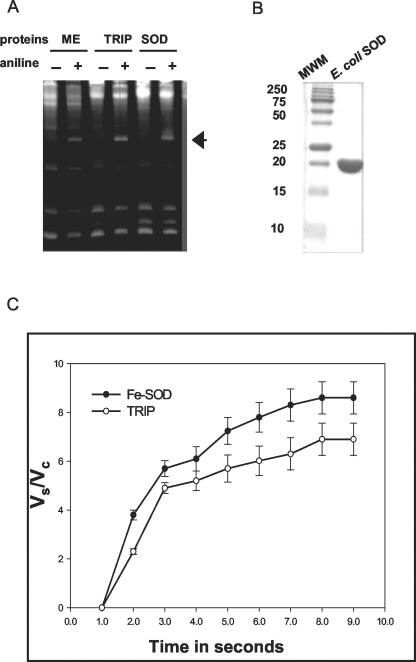
Because TRIP showed an exact internal amino acid homology when compared with Fe-SOD, we tested a commercially available Fe-SOD from E. coli for depurination activity. Surprisingly, the Fe-SOD from E. coli (0.1 ng mL-1) showed depurination activity against yeast rRNA identical to the activity of TRIP (Fig. 7A). To rule out any contamination as the cause of this surprising result, the E. coli Fe-SOD protein was examined on SDS-PAGE (Fig. 7B), where it showed a single band of 21 kD without any contaminating proteins. TRIP did not show sequence similarity with known type I RIPs, further indicating a unique RIP type.
Figure 7.
Depurination activity of Fe-SOD, SDS-PAGE of Fe-SOD, and comparison of SOD activity of TRIP with Fe-SOD. A, The Fe-SOD from E. coli was checked for RIP activity. E. coli Fe-SOD (Sigma-Aldrich) was solubilized in 20 mm sodium-phosphate, pH 7.0, buffer. Yeast ribosomes were incubated with 0.1 ng mL-1 ME, TRIP, and E. coli Fe-SOD as described in “Materials and Methods.” rRNAs were extracted, treated with aniline, separated on a 4.5% (w/v) urea-polyacrylamide gel, and stained with ethidium bromide. The presence (+) or absence (-) of aniline is denoted. The arrow shows the presence of the diagnostic 367-nucleotide cleavage product of rRNA. B, E. coli Fe-SOD was fractionated on 15% (w/v) SDS-PAGE. High Mr standard protein markers (Bio-Rad Laboratories) were fractionated in a parallel lane. C, The SOD assay was performed according to the manufacturer's instructions (Calbiochem). Different enzymes, Fe-SOD from E. coli, and highly purified TRIP were incubated in buffer containing 50 mm 2-amino-2methyl-1,3-propanediol, 3.3 mm boric acid, and 0.11 mm diethylenetriaminepentaacetic acid (DTPA), pH 8.8, at 37°C. Mercapto scavenger 1,4,6-trimethyl-2-vinylpyridinium trifluoromethanosulfonate, 33.3 mm in 1 mm HCl was added and incubated for 1 h at 37°C. Chromogenic reagent 5,6,6a,11b-tetrahydro-3,9,10-trihydroxybenzol[c]fluorine, 0.66 mm in 32 mm HCl containing 0.5 mm DTPA and 2.5% (v/v) ethanol was added and A525 was monitored for 90 s. The A525 was recorded at 10-s intervals and was monitored up to 90 s. The data shown here represent the mean of three individual experiments. Versus is the A525 with sample, and Vc is the A525 without sample.
Comparison of SOD Activity of E. coli Fe-SOD and TRIP
Because TRIP showed high homology with the Fe-SOD from different plant sources and E. coli Fe-SOD exhibited depurination activity on yeast ribosomes, we tested highly purified TRIP and E. coli Fe-SOD for SOD activity (Fig. 7C). We first checked the E. coli Fe-SOD enzyme activity at concentrations ranging from 5 to 20 μg, and we found a linear increase in the oxidation of 5,6,6a 11b-tetrahydro-3,910-trihydroxybenzol[c]fluorine at a concentration of 15 μg mL-1. Similarly, TRIP (15 μg mL-1) showed SOD activity in the same range as E. coli Fe-SOD. The SOD activity of TRIP was further confirmed by a second SOD activity assay consisting of reduction of nitroblue tetrazolium (NBT) by xanthine oxidase (Beauchamp and Fridovich, 1971) and subsequent inhibition of reduction by SODs. We used E. coli Fe-SOD as standard, and the SOD activity of TRIP was compared. Different concentrations of TRIP were tested for the ability to inhibit NBT (250 and 500 ng mL-1). TRIP caused a 60% reduction of NBT at 500 ng mL-1 concentration (Supplementary Table S1).
DISCUSSION
In this manuscript, we describe the isolation and characterization of a new RIP-like protein from the leaves of tobacco. The purification and general properties of this enzyme are typical for type I RIPs. TRIP damages ribosomes, causing the formation of an Endo fragment (Endo et al., 1987). However, we found that TRIP is expressed at very low levels in tobacco (0.01% of total starting material). These levels are in contrast to the expression levels typical for other RIPs. For instance, ME showed an organ-specific high level of expression, accounting for almost 20% of the total soluble proteins in the storage roots (Vivanco et al., 1999; Vepachedu et al., 2003). Similarly, the RIP from iris (Iris hollandica) is the most abundant protein in the bulbs (Van Damme et al., 1997). TRIP was expressed at very low levels in tobacco, and it was detected in several organs of this plant species (Fig. 3A).
We have found several unique characteristics in TRIP that distinguish it from known type I RIPs. For example, we found that TRIP inhibits the wheat germ translation system more efficiently than that of rabbit reticulocytes. However, the efficiency of TRIP was 1,000-fold less than the efficiency of ME (Vepachedu et al., 2003). Because TRIP showed many of the characteristics of RIPs, we assume that the lower efficiency of TRIP at inhibiting protein synthesis might be due to the requirement of a cofactor like ATP (Nielsen and Boston, 2001). Similarly, when the capacity to release adenine from non-ribosomal and animal ribosomal substrates was compared with that of momordin II, TRIP released less adenine. As reported by Barbieri et al. (1997), most of the RIPs release adenine from DNA, RNA, and poly(A) in very high amounts. However, release amounts do vary widely: Whereas saporin releases 6,257 pmol of adenine, native ricin releases 689 pmol of adenines under the same assay conditions; the A-chain (the toxic chain of ricin) releases 185 pmol of adenine, and TRIP falls at the low end, releasing just 0.45 and 0.25 pmol of adenines from Artemia sp. ribosomes and rat ribosomes (animal ribosomes) and 0.01 pmol from hsDNA, 0.01 pmol from rRNA, and 0.32 pmol from tRNA, respectively. Further studies with saporin L-1 on various forms of mammalian DNA showed that even the amounts of adenine released by a single RIP, saporin, vary significantly depending on the substrate (Barbieri et al., 2000). Thus, these results allowed us to classify TRIP as a RIP-like protein.
RIPs have been implicated in plant defense roles (Stirpe et al., 1992) by virtue of their antiviral, antifungal, and antibacterial activities (Barbieri et al., 1993; Girbés et al., 1996; Vivanco et al., 1999). Accordingly, we found that TRIP showed antimicrobial activity against a number of bacteria and fungi tested (Table III) along with its depurination activity on yeast ribosomes (Fig. 1B). It has been hypothesized that once pathogens infect plant cells, RIPs may depurinate the host plant ribosomes, arresting protein synthesis and stopping the spread of the pathogen in a so-called “defensive suicide mechanism” (Ready et al., 1986). Previous studies have shown that expression of PAP mutants in transgenic tobacco plants leads to resistance against viral and fungal pathogens and that this resistance is associated with the activation of the salicylic acid-independent defense pathway (Lodge et al., 1993; Zoubenko et al., 1997, 2000). The participation of TRIP in a similar defense mechanism in tobacco has not been investigated in the present study; however, these studies suggest that TRIP, like other RIPs, might be involved in the activation of defense pathways and that the direct antimicrobial activity is a secondary function (Wang et al., 2003).
Interestingly, the internal sequence of TRIP showed exact sequence homology with the Fe-SOD of wild tobacco, Arabidopsis, and potato (Fig. 7). To explore this observation, we checked a commercially available Fe-SOD from E. coli for RIP activity and found that this Fe-SOD depurinated the yeast rRNA at a very low concentration. It was believed that most plant species lack Fe-SOD until Camp et al. (1990) showed this enzyme in two diverse species, Arabidopsis and wild tobacco. Similarly, it is believed that RIP-like proteins are not found in all plants. In our studies, young tobacco seedlings showed no RIP activity (data not shown), whereas we detected RIP activity in the basic protein of 2-month-old tobacco leaves (Fig. 1B). Interestingly, Kurepa et al. (1997) detected the Fe-SOD transcript in leaves of young tobacco, but they could only detect the enzymatic activity in the leaves and stems of mature plants. This report is in accordance with our observation that RIP activity is detected in different organs of 2-month-old tobacco plants.
Fe-SOD in tomato (Lycopersicon esculentum) chloroplasts can be strongly induced by some stress conditions (Kwiatowski et al., 1985). Similarly, there are also reports that Mn-SOD, which is very similar to Fe-SOD in its primary, secondary, and tertiary structure (Parker and Blake, 1988), is induced dramatically during stress conditions, most notably metabolic stress under tissue culture conditions and during the pathogenesis response (Bowler et al., 1989; Kurepa et al., 1997). Thus, it is suggested that SOD and TRIP share a protective role in plants, and in many cases both are induced by similar stresses. Even though the present observation is new and surprising, the role of SODs as free radical scavengers and the role of RIPs as defense proteins protecting the plant from stress and pathogens are well documented (Stirpe et al., 1992; William and Silverman, 2002).
In plants, resistance to air pollutants such as ozone and SO2 has been shown to correlate with increased levels of enzymes involved in superoxide detoxification (Bowler et al., 1991). More recently, Mn-SOD from tobacco and Cu/Zn-SOD from porcine erythrocyte and bovine erythrocyte have been shown to have in vitro-cleaving activity of DNA, specifically to circular supercoiled double-stranded DNA (Ling et al., 1998). Many known RIPs, like trichosanthin, α-sarcin, dianthin 30, saporin-6, and gelonin have DNA-cleaving activity. Apparently, this activity might be shared by some SODs. We suggest that TRIP may have dual enzymatic activity as an RIP-like protein and as a SOD. However, at this point, we are not inferring which activity may be more prominent in tobacco.
Although not many proteins are known to have dual enzymatic activity, more and more proteins with dual enzymatic activity have been found. For example, MazG protein from Thermotoga maritime, which has both nucleoside triphosphate pyrophosphohydrolase and pyrophosphatase activities (Zhang et al., 2003), and MJ0109, a protein from Methanococcus jannaschii, which has both inositol monophosphatase and Fru-1,6-bisphosphatase activity (Stec et al., 2000). Similarly, a recent report identifies firefly luciferase as a bifunctional enzyme; it has ATP-dependent monooxygenase and a long-chain fatty acyl-CoA synthetase activity (Oba et al., 2003). In this case, both the activities are very different from each other. In the present paper, we describe a RIP-like protein, which shows ribosome-damaging activity typical of RIPs, and also shows SOD activity under in vitro conditions. The findings that TRIP shows all of the characteristics of RIPs except sequence homology with known type I RIPs and that E. coli Fe-SOD showed one of the characteristics of RIPs (that of releasing the classical “Endo” fragment from 28S rRNA) combined with our other observations allow us to suggest that some RIP-like proteins and Fe-SODs may share enzymatic activity. Interestingly, in plants, both enzymes play a central role in defense responses. We propose that TRIP may act as a SOD under certain environmental/biological conditions and as an RIP-like protein under other conditions depending upon the requirement of the system.
MATERIALS AND METHODS
Plant Material
Seeds of tobacco (Nicotiana tabacum cv Samsun) were obtained from Dr. Nilgun Tumer (Rutgers University, New Brunswick, New Jersey). Seeds were washed five times with sterile water and germinated on filter papers on a petri dish. The 2-week-old seedlings were then transferred to pots and grown under greenhouse conditions.
Protein Extraction from Tobacco Leaves
Leaves from 2-month-old tobacco plants were ground in liquid N2, homogenized in 3 volumes of extraction buffer (25 mm NaPO4, pH 7.0, with 250 mm NaCl, 10 mm EDTA, 5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1.5% [w/v] polyvinylpolypyrrolidone), and then centrifuged for 30 min at 10,000g. The supernatant was brought to 20% (w/v) ammonium sulfate by continuous stirring. The mixture was chilled for 1 h and then centrifuged again for 30 min at 10,000g. The supernatant was precipitated with 20% to 80% (w/v) ammonium sulfate and centrifuged at 14,000g for 30 min. The pellet was suspended in 25 mm HEPES/NaOH, pH 8.0, containing 50 mm NaCl and then dialyzed against 25 mm HEPES/NaOH (pH 8.0) until it was free from the sulfate ion. All extraction procedures were conducted at 4°C, and the ammonium sulfate fraction was stored at -80°C until use.
Chromatographic Separations
Total protein from tobacco was initially fractionated using a Uno-Q1 column attached to an FPLC (Bio-Rad Laboratories, Hercules, CA). Equilibration and loading of the protein solution was carried out using 25 mm HEPES/NaOH (pH 8.0) containing 50 mm NaCl at a flow rate of 1 mL min-1. The flow-through protein (basic protein fraction) was collected, and the protein mixture was then further fractionated using gel filtration chromatography. The gel filtration column (7.8 × 300 mm; Bio Sil SEC 250-5, Bio-Rad Laboratories) was equilibrated with 25 mm HEPES/NaOH (pH 8.0) containing 100 mm NaCl. The basic protein (1 mL) was loaded on the gel filtration column, and the proteins were eluted with an isocratic gradient of 25 mm HEPES/NaOH (pH 8.0) and 100 mm NaCl at a flow rate of 0.5 mL min-1. Each fraction was assayed for RIP activity, and the active fractions were pooled, concentrated, and desalted using 5-kD cutoff ultrafiltration membranes (Millipore, Bedford, MA). The desalted proteins were further resolved by cation exchange chromatography with a Poros-HS column (4.6 × 100 mm; PE Biosystems, Foster City, CA) attached to an FPLC (Bio-Rad Laboratories). Equilibration, loading, and washing were carried out in 20 mm HEPES/NaOH (pH 8.0) containing 50 mm NaCl at a flow rate of 3 mL min-1. The target protein was eluted with a linear gradient of NaCl (50–1,000 mm). Individual peaks were collected, concentrated, and desalted using 10-kD ultrafiltration devices (Millipore).
SDS-PAGE
SDS-PAGE gel electrophoresis was performed with 12% (v/v) acrylamide discontinuous gels (Laemmli, 1970) using an electrophoresis cell (Bio-Rad Laboratories) according to the manufacturer's instructions. High Mr protein markers (from Bio-Rad Laboratories) were run simultaneously in the electrophoresis gel. The protein bands were visualized by silver staining (Bio-Rad Laboratories). The protein at all of the steps of purification was estimated according to the method of Bradford (1976) using a Bio-Rad protein assay kit.
Isolation of Yeast Ribosomes and rRNA Depurination Assay
Yeast (Saccharomyces cerevisiae) strain YPH500 (Sikorski and Hieter, 1989) was grown in yeast peptone dextrose medium. Yeast cells were then pelleted by centrifugation. To isolate ribosomes, 10 g of pelleted yeast cells was ground in a mortar with liquid N2 and dissolved in 100 mL of extraction buffer (200 mm KCl, 25 mm MgCl2, 25 mm EGTA, 200 mm Suc, and 25 mm β-mercaptoethanol in 200 mm Tris-HCl, pH 9.0). The supernatant, after centrifugation at 10,000g for 20 min at 4°C, was pipetted onto a Suc cushion (1 m Suc, 20 mm KCl, and 5 mm MgCl2 in 25 mm Tris-HCl, pH 7.6) in 70-Ti tubes (Beckman Coulter, Fullerton, CA) and centrifuged at 55,000 rpm for 4 h at 4°C (l-70 Ultracentrifuge, Beckman Coulter). The pellets were resuspended in 25 mm Tris-HCl buffer (pH 7.6) with 25 mm KCl and 5 mm MgCl2 and stored at -80°C until use.
The depurination assay was conducted according to Tumer et al. (1997). In brief, ribosomes were resuspended in RIP buffer (167 mm KCl, 100 mm MgCl2, and 100 mm Tris-HCl, pH 7.2) and incubated with RIPs at 30°C for 30 min in a total volume of 100 μL. rRNA incubated in the absence of RIPs served as a negative control. After incubation, the RIPs were removed from the mixture by phenol-chloroform extraction, and the RNA was divided in half. One-half of the extracted RNA was incubated on ice for 30 min with 1 m aniline acetate (pH 4.5) and precipitated with ethanol. Both anilinetreated and untreated rRNAs were subjected to electrophoresis in a 7 m urea/4.5% (w/v) polyacrylamide gel and stained with ethidium bromide.
In Vitro Protein Synthesis Inhibition by TRIP
TRIP was tested for in vitro protein synthesis inhibition activity by using Flexi rabbit reticulocytes and wheat germ lysate systems (Promega, Madison, WI). The translation was performed according to the manufacturer's protocol in the presence of [35S]Met to label the products. The reaction was carried out at 30°C and 25°C for 90 min for rabbit reticulocytes and wheat germ lysate, respectively. The reaction was terminated by adding 0.25 mL of 1 n NaOH containing 0.2% (v/v) H2O2, followed by incubation at 37°C for 10 min. The protein was precipitated with 25% (w/v) tricholoacetic acid on ice for 30 min and harvested on 26-mm glass fiber filters. The filters were washed three times with 5% (w/v) chilled TCA with a final wash of chilled acetone. The filters were dried at room temperature for 10 min and were counted using liquid scintillation fluid. Activity was expressed as a percentage of the control where no TRIP was added. ME was used as a positive control and different concentrations of TRIP (10–1,000 ng) were used to study the protein synthesis inhibition. ID50 represents the concentration of TRIP that inhibited in vitro protein synthesis by 50%.
Determination of Glycosylase Activity
RNA N-glycosylase activity was determined as described by Barbieri et al. (2003). In brief, adenine released from hsDNA (Sigma-Aldrich, St. Louis), from Escherichia coli rRNA (Roche Diagnostics, Mannheim, Germany), and from Phe-specific tRNA (Sigma-Aldrich) was measured after separation by HPLC. The reaction was performed in a final volume of 50 μL containing 50 mm sodium-acetate buffer, pH 4.0, 10 μg of nucleic acid, and the appropriate amount of enzyme. The reaction was stopped in ice by addition of 200 μL of cold buffer (50 mm sodium-acetate, pH 4.0, and 20 mm KCl), and then the reaction products were applied to sample preparation columns (Bond Elut NH2, Varian, Harbor City, CA) previously equilibrated at 2°C with BA buffer. Adenine was eluted in 400 μL of BA buffer by briefly centrifuging at 200g, while phosphate containing nucleic acids was retained; samples were in duplicate. Adenine was measured by liquid chromatography and mass spectroscopy on a Alliance/zq apparatus (Waters, Milford, MA) as described by Barbieri et al. (2003).
Antimicrobial Assay
The antifungal activity of the purified TRIP was determined using a radial growth inhibition assay according to the method of Schlumbaum et al. (1986). A mycelial plug was placed in the center of a potato-dextrose-agar plate. Sterile paper discs were placed at the two sides of the plate. Protein and buffer solutions were filter sterilized using 0.22-μm filters (Millipore). Fifty micrograms of purified TRIP was plated onto one of the paper discs, and buffer alone was placed on the other paper disc as a control. The plates were incubated in the dark at 23°C. Antifungal activity was observed as a crescent-shaped zone of inhibition at the mycelial front. The effect on fungal growth was expressed qualitatively according to the method of Schlumbaum et al. (1986).
Antibacterial activity was screened using an inhibition-halo-plate assay. Bacterial cultures were plated on liquid Luria-Bertani medium and incubated at 37°C for 12 h. The A600 of bacterial cultures was measured and adjusted to an optical density of 0.2 for the antibacterial experiments. Bacterial plates were prepared by spreading 100 μL of the bacteria onto the plate. Two sterile paper discs were placed in opposite positions. The protein and buffer solutions were filter sterilized by using 0.22-μm filters (Millipore) and applied to the paper discs. The petri plates were incubated in the dark at 23°C for 24 h. Antibacterial activity was measured as the radius of inhibition from the border of the paper disc.
Internal Amino Acid Sequencing of Purified TRIP
The protein was digested by trypsin, and the fragment generated due to trypsin digestion was sequenced. The purified TRIP was sequenced on a Precise Protein Sequencer System (Applied Biosystems, Foster City, CA) at the Protein Sequencing Core Facility, University of Nebraska Medical Center (Omaha, Nebraska).
Comparison of SOD Activity of E. coli Fe-SOD and TRIP
The SOD assay was performed according to the manufacturer's instruction manual (Calbiochem, San Diego). Different enzymes, Fe-SOD from E. coli, and highly purified TRIP (15 μg mL-1) were incubated in buffer containing 50 mm 2-amino-2 methyl-1,3propanediol, 3.3 mm boric acid, and 0.11 mm DTPA, pH 8.8, at 37°C. Mercapto scavenger 1,4,6-trimethyl-2-vinylpyridinium trifluoromethanosulfonate, 33.3 mm in 1 mm HCl was added and incubated for 1 h at 37°C. Chromogenic reagent 5,6,6a,11btetrahydro-3,9,10-trihydroxybenzol[c]fluorine, 0.66 mm in 32 mm HCl containing 0.5 mm DTPA and 2.5% (v/v) ethanol was added and A525 was monitored for 90 s. Data were collected at 10-s intervals up to 90 s. Versus represent the A525 with sample and Vc is the A525 without sample. SOD activity of TRIP was further tested by the more sensitive method described by Beauchamp and Fridovich (1971). The reaction mixture contained 0.1 mm xanthine, 0.1 mg NBT, and 0.002 unit of xanthine oxidase in 1 mL (Roche Diagnostics). Different concentrations of TRIP (250 and 500 ng mL-1) were used to determine the percent inhibition of reduction of NBT. Fe-SOD (250 ng mL-1) from E. coli was used as a control.
Supplementary Material
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.029389.
This work was supported by a CAREER award from the National Science Foundation (grant no. MCB-0093014 to J.M.V.) and by the Colorado State University Agricultural Experiment Station (to J.M.V.). The work done in Bologna was supported by grants from the University of Bologna (the Ministero dell'Istruzione), Università e Ricerca (the Ministero della Salute). and the Pallotti's Legacy for Cancer Research.
The online version of this article contains Web-only data.
References
- Barbieri L, Balteli MG, Stirpe F (1993) Ribosome inactivating proteins from plants Biochim Biochem Biophys Acta 1154: 237-284 [DOI] [PubMed] [Google Scholar]
- Barbieri L, Brigotti M, Perocco P, Carcnicelli D, Ciani M, Mercatali L, Stirpe F (2003) Ribosome-inactivating proteins depurinate poly (ADPribosyl)ated poly (ADP-ribose) polymerase and have transforming activity for 3T3 fibroblasts. FEBS Lett 538: 178-182 [DOI] [PubMed] [Google Scholar]
- Barbieri L, Valbonesi P, Bonora E, Gorini P, Bolognesi A, Stirpe F (1997) Polynucleotide:adenosine glycosidase activity of ribosome-inactivating proteins: effect on DNA, RNA and poly(A). Nucleic Acids Res 25: 518-522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri L, Valbonesi P, Righi F, Zuccheri G, Monti F, Gorini P, Samorì B, Stirpe F (2000) Polynucleotide:adenosine glycosidase is the sole activity of ribosome-inactivating proteins on DNA. J Biochem 128: 883-889 [DOI] [PubMed] [Google Scholar]
- Battelli MG, Stirpe F (1995) Ribosome-inactivating proteins from plants. In M Chessin, D Deborde, A Zipf, eds, Antiviral Proteins in Higher Plants. CRC Press, Boca Raton, FL, pp 39-64
- Beauchamp C, Fridovich I (1971) Superoxide dismutase: an improved assay and an assay applicable to acrylamide gels. Anal Biochem 44: 276-287 [DOI] [PubMed] [Google Scholar]
- Bowler C, Thierry A, Loose MD, Montagu MV, Inzé D (1989) The induction of manganese superoxide dismutase in response to stress in Nicotiana plumbaginifolia. EMBO 8: 31-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Stoolen L, Vandebranden S, Ryeke ED, Bolterman J, Sybesma C, Montagu MV, Inzé D(1991) Manganese superoxide dismutase can reduce cellular damage mediated by oxygen radicals in transgenic plants. EMBRO J 10: 1723-1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254 [DOI] [PubMed] [Google Scholar]
- Camp WV, Bowler C, Villarroel E, Tsang EWT, Montagu MV, Inzé D (1990) Characterization of iron superoxide dismutase cDNA from plants obtained by genetic complementation in Escherichia coli. Proc Natl Acad Sci USA 87: 9903-9907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day PJ, Lord JM, Roberts LM (1998) The deoxyribonuclease activity attributed to ribosome-inactivating proteins is due to contamination. Eur J Biochem 258: 540-545 [DOI] [PubMed] [Google Scholar]
- Endo Y, Mitsui K, Tsurugi K (1987) The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes: the site and the characteristics of the modification in 28S ribosomal RNA caused by the toxins. J Biol Chem 262: 5908-5912 [PubMed] [Google Scholar]
- Endo Y, Tsurugi K (1987) RNA N-glycosidase activity of ricin A-chain: mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J Biol Chem 263: 8735-8739 [PubMed] [Google Scholar]
- Ferreras JM, Citores L, de Benito FM, Arias FJ, Rojo MA, Muñoz R, Iglesias R, Girbés T (2000) Ribosome-inactivating proteins and lectins from Sambucus. Curr Top Phytochem 3: 113-128 [Google Scholar]
- Girbés T, de Torre C, Iglesias R, and Ferreras JM, Mendez E (1996) RIP for viruses. Nature 379: 777-7788587600 [Google Scholar]
- Grasso S, Shepherd RJ (1978) Isolation and partial characterization of virus inhibitors from plant species taxonomically related to Phytolacca. Physiol Biochem 68: 199-205 [Google Scholar]
- Hudak KA, Bauman JD, Tumer NE (2002) Pokeweed antiviral protein binds to the cap structure of eukaryotic mRNA and depurinates the mRNA downstream of the cap. RNA 8: 1148-1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudak KA, Wang P, Tumer NE (2000) A novel mechanism for inhibition of translation by pokeweed antiviral protein: depurination of the capped RNA template. RNA 6: 369-380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka J, Habuka N, Miyano M, Masuta C, Koiwai A (1992) Adenine depurination and inactivation of plant ribosomes by an antiviral protein of Mirabilis jalapa (MAP). Plant Mol Biol 20: 1111-1119 [DOI] [PubMed] [Google Scholar]
- Kataoka J, Habuka N, Miyano M, Takanami Y, Koiwai A (1991) DNA sequence and of Mirabilis antiviral protein (MAP), a ribosomeinactivating protein with antiviral property, from Mirabilis jalapa L. and its expression in E. coli. J Biol Chem 266: 8426-8430 [PubMed] [Google Scholar]
- Kubo S, Ikeda T, Imaizumi S, Takanami Y, Mikami Y (1990) A potent plant virus inhibitor found in Mirabilis jalapa L. Ann Phytopathol Soc Jpn 56: 481-487 [Google Scholar]
- Kurepa J, Herouart D, Montagu MV, Inzé D (1997) Differential expression of CuZn-and Fe-superoxide dismutase genes of tobacco during development, oxidative stress, and hormonal treatment. Plant Cell Physiol 38: 463-470 [DOI] [PubMed] [Google Scholar]
- Kwon SY, An CS, Liu JR, Kwak S-S, Lee HS, Lim JK, Paek KH (2000) Molecular cloning of a cDNA encoding ribosome-inactivating protein from Amaranthus viridis and its expression in E. coli. Mol Cells 10: 8-12 [DOI] [PubMed] [Google Scholar]
- Kwiatowski J, Safianowska A, Kaniuga Z (1985) Isolation and characterization of an iron-containing superoxide dismutase from tomato leaves, Lycopersicon esculentum. Eur J Biochem 146: 459-466 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685 [DOI] [PubMed] [Google Scholar]
- Lam SK, Ng TB (2001a) Hypsin, a novel thermostable ribosome-inactivating protein with antifungal and antiproliferative activities from fruiting bodies of the edible mushroom Hypsizigus marmoreus. Biochem Biophys Res Commun 285: 1071-1075 [DOI] [PubMed] [Google Scholar]
- Lam SK, Ng TB (2001b) First simultaneous isolation of a ribosomeinactivating protein and an antifungal protein from a mushroom (Lyophyllum shimeji) together with evidence for synergism of their antifungal effects. Arch Biochem Biophys 393: 271-280 [DOI] [PubMed] [Google Scholar]
- Li MX, Yeung HW, Pan LP, Chan SI (1991) Trichosanthin, a potent HIV-1 inhibitor, can cleave supercoiled DNA in vitro. Nucleic Acid Res 19: 6309-6312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Gao X, Liu WY, Ruan H (1998) DNA-cleaving activity of superoxide dismutase specific for circular supercoiled double-stranded DNA in vitro. Int J Biochem Cell Biol 30: 1123-1127 [DOI] [PubMed] [Google Scholar]
- Lodge JK, Kaniewski WK, Tumer NE (1993) Broad spectrum virus resistance in transgenic plants expressing pokeweed antiviral protein. Proc Natl Acad Sci USA 90: 7089-7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta AD, Boston RS (1998) Ribosome-inactivating protein. In J Bailey-Serres, DR Gallie, eds, A Look beyond Transcription: Mechanisms Determining mRNA Stability and Translation in Plants. American Society of Plant Physiologists, Rockville MD, pp 145-152
- Nicolas E, Beggs JM, Haltiwanger BM, Taraschi TF (1997) Direct evidence for the deoxyribonuclease activity of the plant ribosome inactivating protein gelonin. FEBS Lett 406: 162-164 [DOI] [PubMed] [Google Scholar]
- Nicolas E, Beggs JM, Haltiwanger BM, Taraschi TF (1998) A new class of glycosylase/apurinic/apyrimidinic lyase that act on specific adenines in single stranded DNA. J Biol Chem 273: 17216-17220 [DOI] [PubMed] [Google Scholar]
- Nicolas E, Beggs JM, Taraschi TF (2000) Gelonin is an unusual DNA glycosylase that removes adenine from single-stranded DNA, normal base pairs and mismatches. J Biol Chem 275: 31399-31406 [DOI] [PubMed] [Google Scholar]
- Nielsen K, Boston RS (2001) Ribosome-inactivating proteins: a plant perspective. Annu Rev Plant Physiol Plant Mol Biol 52: 785-816 [DOI] [PubMed] [Google Scholar]
- Oba Y, Ojika M, Inouye S (2003) Firefly luciferase is a bifunctional enzyme: ATP-dependent monooxygenase and a long Caín fatty acyl-CoA synthetase. FEBS Lett 540: 251-254 [DOI] [PubMed] [Google Scholar]
- Parikh BA, Coetzer C, Tumer NE (2002) Pokeweed antiviral protein regulates the stability of its own mRNA by a mechanism that requires depurination, but can be separated from depurination of the alpha-sarcin/ricin loop of rRNA. J Biol Chem 277: 41428-41437 [DOI] [PubMed] [Google Scholar]
- Park SW, Lawrence CB, Linden JC, Vivanco JM (2002a) Isolation and characterization of a novel ribosome-inactivating protein from root cultures of pokeweed and its mechanism of secretion from roots. Plant Physiol 130: 164-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Stevens NA, Vivanco JM (2002b) Enzymatic specificity of three ribosome-inactivating proteins against fungal ribosomes, and correlation with antifungal activity. Planta 216: 227-234 [DOI] [PubMed] [Google Scholar]
- Parker MW, Blake CC (1988) Iron and manganese containing superoxide dismutase can be distinguished by analysis of their primary structures. FEBS Lett 229: 377-382 [DOI] [PubMed] [Google Scholar]
- Ready MP, Brown DT, Robertus JD (1986) Extracellular localization of pokeweed protein. Proc Natl Acad Sci USA 84: 5053-5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C, Lehmann J, Becker W, Apel K, Partheir B (1994) JIP60, a methyl jasmonates-induced ribosome-inactivating protein involved in plant stress reactions. Proc Natl Acad Sci USA 91: 7012-7016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts WK, Selitrennikof CP (1986) Isolation and characterization of two antifungal proteins from barley. Biochim Biophys Acta 880: 161-170 [DOI] [PubMed] [Google Scholar]
- Roncuzzi L, Gasperi-Gampani A (1996) DNA-nuclease activity of the single-chain ribosome-inactivating proteins dianthin 30, saporin 6 and gelonin. FEBS Lett 392: 16-20 [DOI] [PubMed] [Google Scholar]
- Schlumbaum A, Mauch F, Vogeli U, Boller T (1986) Plant chitinases are potent inhibitors of fungal growth. Nature 324: 365-367 [Google Scholar]
- Sikorski RS, Heiter PC (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in saccharomyces cerevisiae. Genetics 122: 12-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec B, Yang H, Johnson KA, Chen L, Roberts MF (2000) MJ0109 is an enzyme that is both an inositol monophosphatase and the “missing” archael fructose-1,6-bisphosphatase. Nature 7: 1046-1050 [DOI] [PubMed] [Google Scholar]
- Stirpe F, Barbieri L (1986) Ribosome-inactivating proteins up to date. FEBS Lett 195: 1-8 [DOI] [PubMed] [Google Scholar]
- Stirpe F, Barbieri L, Battelli MG, Soria M, Lappi DA (1992) Ribosomeinactivating proteins from plants: present status and future prospects. Bio/Technology 10: 405-412 [DOI] [PubMed] [Google Scholar]
- Tumer NE, Hudak K, Di R, Coetser C, Wang R, Zoubenko O (1999) Pokeweed antiviral protein and its applications. Curr Top Microbiol Immunol 240: 139-158 [DOI] [PubMed] [Google Scholar]
- Tumer NE, Hwang DJ, Bonness M (1997) C-terminal deletion mutant of pokeweed antiviral protein inhibits viral infection but does not depurinate host ribosomes. Proc Natl Acad Sci USA 94: 3866-3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJM, Barre A, Barbieri L, Valbonesi P, Rougé P, Van Leuven F, Stirpe F, Peumans WJ (1997) Type I ribosome-inactivating proteins are the most abundant proteins in iris (Iris hollandica var. Professor Blaauw) bulbs: characterization and molecular cloning. Biochem J 324: 963-970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJM, Hao Q, Barre A, Vandenbussche F, Desmyter S, Rougé P, Peumans WJ (2001) Ribosome-inactivating proteins: a family of plant proteins that do more than inactivate ribosomes. Crit Rev Plant Sci 20: 395-465 [Google Scholar]
- Vepachedu R, Bais HP, Vivanco JM (2003) Molecular characterization and post-translational regulation of ME1, a type I ribosome-inactivating protein from Mirabilis expansa. Planta 217: 498-506 [DOI] [PubMed] [Google Scholar]
- Vivanco JM (1997) Efecto inhibitorio de los extracto de Mirabilis ialapa en contra de PVX, PVY y PSTVd. PhD Thesis. Universidad Nacional Agraria La Molina, Lima, Peru
- Vivanco JM, Savary BJ, Flores HE (1999) Characterization of two novel type I ribosome-inactivating proteins from the storage roots of the Andean crop Mirabilis expansa. Plant Physiol 119: 1447-1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HX, Ng TB (2001) Isolation of a novel deoxyribonuclease with antifungal activity from Asparagus officinalis seeds. Biochem Biophys Res Commun 289: 120-124 [DOI] [PubMed] [Google Scholar]
- Wang J-H, Nie H-L, Huang H, Tam S-C, Zheng Y-T (2003) Independency of anti-HIV-1 activity from ribosome-inactivating activity of trichosanthin. Biochem Biophys Res Commun 302: 89-94 [DOI] [PubMed] [Google Scholar]
- Wang P, Tumer NE (2000) Virus resistance mediated by ribosome inactivating proteins. Adv Virus Res 55: 325-355 [DOI] [PubMed] [Google Scholar]
- William GB, Silverman DN (2002) Activation of the proteon transfer pathway in catalysis by iron superoxide dismutase. J Biol Chem 277: 49282-49286 [DOI] [PubMed] [Google Scholar]
- Wool IG, Gluck A, Endo Y (1992) Ribotoxin recognition of ribosomal RNA and a proposal for the mechanism of translocation. Trends Biochem Sci 17: 266-269 [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang Y, Inouye M (2003) Thermotoga maritime MazG protein has both nucleoside triphosphate pyrophosphohydrolase and pyrophosphatase activity. J Biol Chem 278: 32300-32306 [DOI] [PubMed] [Google Scholar]
- Zoubenko O, Hudak K, Tumer NE (2000) A non-toxic pokeweed antiviral protein mutant inhibits pathogen infection via a novel salicylic acid-independent pathway. Plant Mol Biol 44: 219-229 [DOI] [PubMed] [Google Scholar]
- Zoubenko O, Uckun F, Hur Y, Chet I, Tumer NE (1997) Plant resistance to fungal infection induced by nontoxic pokeweed antiviral protein mutants. Nat Biotechnol 15: 1992-1996 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.