Abstract
Arabidopsis expressing the castor bean (Ricinus communis) oleate 12-hydroxylase or the Crepis palaestina linoleate 12-epoxygenase in developing seeds typically accumulate low levels of ricinoleic acid and vernolic acid, respectively. We have examined the presence of a futile cycle of fatty acid degradation in developing seeds using the synthesis of polyhydroxyalkanoate (PHA) from the intermediates of the peroxisomal β-oxidation cycle. Both the quantity and monomer composition of the PHA synthesized in transgenic plants expressing the 12-epoxygenase and 12-hydroxylase in developing seeds revealed the presence of a futile cycle of degradation of the corresponding unusual fatty acids, indicating a limitation in their stable integration into lipids. The expression profile of nearly 200 genes involved in fatty acid biosynthesis and degradation has been analyzed through microarray. No significant changes in gene expression have been detected as a consequence of the activity of the 12-epoxygenase or the 12-hydroxylase in developing siliques. Similar results have also been obtained for transgenic plants expressing the Cuphea lanceolata caproyl-acyl carrier protein thioesterase and accumulating high amounts of caproic acid. Only in developing siliques of the tag1 mutant, deficient in the accumulation of triacylglycerols and shown to have a substantial futile cycling of fatty acids toward β-oxidation, have some changes in gene expression been detected, notably the induction of the isocitrate lyase gene. These results indicate that analysis of peroxisomal PHA is a better indicator of the flux of fatty acid through β-oxidation than the expression profile of genes involved in lipid metabolism.
Membranes of plant cells are composed primarily of five “common” fatty acids, namely stearic, palmitic, oleic, linoleic, and linolenic acids. In contrast, a very large diversity of fatty acids exists in the reserve triacylglycerols (TAG) of seeds. More than 300 naturally occurring fatty acids have been described in seeds to date (Badami and Patil, 1980; van de Loo et al., 1993). The structures of these fatty acids vary in a number of features, including the length of the acyl chains; the number, position, and nature of unsaturated bonds; and the presence of functional groups, such as hydroxy, epoxy, and acetylenic groups. These fatty acids are often referred as “unusual” fatty acids because their structure is different from the common fatty acids found in membranes.
Synthesis of unusual fatty acids has attracted considerable interest, both in fundamental and applied areas of plant biology. Several genes have recently been identified that code for enzymes involved in the synthesis of fatty acids containing unusual groups, such as hydroxy, epoxy, acetylenic, or carbocyclic groups, as well as conjugated unsaturated bonds (for a recent review, see Jaworski and Cahoon, 2003). In the majority of cases, these enzymes were found to be variants of enzymes involved in the synthesis of common fatty acids, such as variants of the soluble stearoyl-acyl carrier protein (ACP) desaturase or of the Δ12-oleic acid desaturase (FAD2; Cahoon et al., 1992; van de Loo et al., 1995). Novel enzymes have also been identified that are thought to play a major role in directing unusual fatty acids toward the storage TAG and away from membrane lipids (Dahlqvist et al., 2000).
Many of the unusual fatty acids have properties that are valuable as renewable feedstocks for the chemical industry. For example, industrial applications of ricinoleic acid (12-hydroxy-octadec-cis-9-enoic acid) or vernolic acid (12,13-epoxy-octadec-cis-9-enoic acid) can be found in paints, lubricants, nylons, plastics, and cosmetics. Because most of the native plants accumulating valuable unusual fatty acids in seeds have inappropriate agronomical features, such as unsynchronized flowering or low seed yield, efforts have been made to transfer the biosynthetic pathway for unusual fatty acids into the present oil crop species, such as rapeseed, as well as in the model oilseed plant Arabidopsis. However, in contrast to the native plants that accumulate normally between 65% to 90% of the unusual fatty acids in their seed oil, the majority of transgenic plants expressing genes involved in the synthesis of unusual fatty acids accumulate only low amounts (<20%) of these fatty acids (Voelker and Kinney, 2001; Jaworski and Cahoon, 2003). Further progress will be dependent on the identification of the factors limiting accumulation of unusual fatty acids in oilseed crops and on the coordinate expression of several transgenes capable of correcting these limitations.
Accumulation of unusual fatty acid in TAG can be divided into two major steps, first the insertion of the functional group into the acyl chain followed by the incorporation of the unusual fatty acids into TAG. In native plants, unusual fatty acids are found almost exclusively in TAG and are excluded from membrane lipids, most likely because they are deleterious to membrane function (Millar et al., 2000). A similar bias has also been found for transgenic plants expressing enzymes for the synthesis of several unusual fatty acids. For example, expression of the lauroyl-ACP thioesterase under the constitutive cauliflower mosaic virus (CaMV) 35S promoter led to accumulation of lauric acid in seeds but not in leaves, despite the demonstration that lauric acid could be produced from isolated leaf chloroplasts (Eccleston et al., 1996). In these plants, as well as in plants expressing constitutively a caproyl-ACP thioesterase, a futile cycle has been demonstrated whereby medium-chain fatty acids synthesized in leaves are rapidly degraded by the peroxisomal β-oxidation cycle (Eccleston and Ohlrogge, 1998; Poirier et al., 1999). Thus, failure to accumulate unusual fatty acids may not only be due to poor synthesis but also by their lack of stable incorporation into lipids such as TAG leading to their rapid degradation through the β-oxidation cycle.
To date, only plants expressing medium-chain acyl-ACP thioesterase have been demonstrated to have a futile cycling of unusual fatty acids toward degradation. It is thus unknown whether the low-level accumulation of other unusual fatty acids in transgenic plants is primarily caused by poor synthesis of the target fatty acids or the failure to efficiently incorporate them into lipids.
This study was aimed at analyzing the potential presence of a futile cycle of vernolic and ricinoleic acid degradation in transgenic Arabidopsis expressing the castor bean (Ricinus communis) oleate Δ12-hydroxylase or the Crepis palaestina linoleate Δ12-epoxygenase in developing seeds. Degradation of unusual fatty acids was determined through the analysis of polyhydroxyalkanoate (PHA) synthesized in the peroxisome. PHA is a polymer of 3-hydroxyacids synthesized by a bacterial PHA synthase from the polymerization of the 3-hydroxyacyl-CoA intermediates generated by the degradation of fatty acids via the β-oxidation cycle (Mittendorf et al., 1998; Poirier, 2002). It has been previously shown that PHA quantity and monomer composition can be used to assess both the quantity and the nature of fatty acids being degraded by the β-oxidation cycle (Mittendorf et al., 1999; Poirier et al., 1999; Poirier, 2002). Microarray analysis was also used to examine the influence of transgene expression and synthesis of unusual fatty acids on the expression of genes involved in lipid biosynthesis and degradation.
RESULTS
Production of Transgenic Plants
The oleate 12-hydroxylase from castor bean and the linoleate 12-epoxygenase from C. palaestina were placed under the control of the napin promoter and were transformed in wild-type Arabidopsis as well as in the transgenic lines (N4.1 or N10.3) that were homozygous for a peroxisomal PHA synthase derived from Pseudomonas aeruginosa that was also expressed under the control of the napin promoter (Poirier et al., 1999). Lines were first selected by identifying hygromycin-segregation ratios that were consistent with the integration of T-DNA at a single locus. From this selected group, the fatty acid composition of mature seeds was determined, and two lines from each were identified for further analysis.
A description of the various transgenic plant lines used in this study along with the unusual fatty acid composition of mature seeds is shown in Table I. Among the lines expressing the oleate 12-hydroxylase, the amounts of ricinoleic, densipolic, and lesquerolic acids were found to vary between 1.2 to 5.1 mol %, 1.6 to 3.5 mol %, and 0.7 to 1.1 mol %, respectively. For the transgenic lines expressing the linoleate 12-epoxygenase, the amount of vernolic acid and 12-epoxy-9,15-octadecadienoic acid varied between 0.9 to 2.9 mol %, and from undetectable to 0.2 mol %, respectively. In addition to the accumulation of the oxygenated fatty acids, transgenic lines expressing the 12-hydroxylase or 12-epoxygenase showed increase in the proportion of oleic acid, from approximately 15 mol % in controls up to 22 to 33 mol %. Polyunsaturated fatty acids (linoleic acid + linolenic acid) decreased from approximately 60 mol % in controls down to 30 to 49 mol % in lines expressing the 12-hydroxylase or 12-epoxygenase. Similar changes in unsaturated fatty acids profile have been previously observed for plants expressing the 12-hydroxylase or 12-epoxygenase (Broun and Somerville, 1997; Singh et al., 2001).
Table I.
Transgenic plants and unusual fatty acid composition
| Plant Lines | Transgenes | Unusual Fatty Acidsa | ||||
|---|---|---|---|---|---|---|
| RAd | DAe | LAf | VAg | Epox. FAh | ||
| mol% | ||||||
| N10.3pC3.1 | Napin-PHA synthase + vector pCAMBIA 1300 | ndi | nd | nd | nd | nd |
| N10.3NH1.1 | Napin-PHA synthase + napin-hydroxylase | 1.2 ± 0.2 | 1.6 ± 0.1 | 0.7 ± 0.1 | nd | nd |
| N10.3NH2.2 | Napin-PHA synthase + napin-hydroxylase | 3.0 ± 0.1 | 2.4 ± 0.1 | 0.9 ± 0.1 | nd | nd |
| N10.3NE6.2 | Napin-PHA synthase + napin-epoxygenase | nd | nd | nd | 0.9 ± 0.1 | nd |
| N4.1pC6.1 | Napin-PHA synthase + vector pCAMBIA 1300 | nd | nd | nd | nd | nd |
| N4.1NE15.1 | Napin-PHA synthase + napin-epoxygenase | nd | nd | nd | 2.9 ± 0.1 | 0.2 ± 0.1 |
| WTPC7.3 | Vector pCAMBIA 1300 | nd | nd | nd | nd | nd |
| WTNH8.1 | Napin-hydroxylase | 4.8 ± 0.1 | 3.5 ± 0.1 | 1.1 ± 0.1 | nd | nd |
| WTNH13.1 | Napin-hydroxylase | 5.1 ± 0.2 | 3.4 ± 0.4 | 1.1 ± 0.1 | nd | nd |
| WTNE5.1 | Napin-epoxygenase | nd | nd | nd | 2.8 ± 0.1 | 0.2 ± 0.1 |
| WTNE7.4 | Napin-epoxygenase | nd | nd | nd | 2.7 ± 0.4 | 0.1 ± 0.1 |
| PHAC3.3b | CaMV 35S-PHA synthase | |||||
| N-FatB3c | Napin-caproyl-ACP thioesterase | |||||
a Values presented are means of three values ± sd. b Line previously described by Mittendorf et al. (1998). c Line previously described by Poirier et al. (1999). d 12-Hydroxy-9-octadecenoic acid (ricinoleic acid). e 12-Hydroxy-9, 15-octadecadienoic acid (densipolic acid). f 14-Hydroxy-11-dodecenoic acid (lesquerolic acid). g 12-Epoxy-9-octadecenoic acid (vernolic acid). h 12-Epoxy-9, 15-octadecadienoic acid. i Not detected.
Impact of the Degradation of Ricinoleic and Vernolic Acids on PHA Monomer Composition
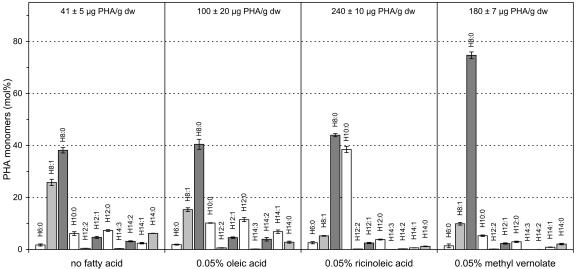
It has been previously demonstrated that the quantity and monomer composition of PHA produced in the peroxisome of transgenic plants can be directly influenced by the quantity and nature of the fatty acids being degraded by the peroxisomal β-oxidation pathway (Mittendorf et al., 1999; Poirier et al., 1999; see Supplementary Figure S1). The impact of the degradation of ricinoleic acid and vernolic acid on PHA quantity and monomer composition was first tested in suspension cultures of transgenic plants expressing the peroxisomal PHA synthase under the CaMV 35S promoter. Addition of oleic acid to the growth medium resulted in an approximate 2.4-fold increase in the quantity of PHA produced as well as an increase in the proportion of the 3-hydroxyacid monomers expected to be generated from the degradation of oleic acid, mainly the unsaturated monomers H14:1 (3-hydroxyacids are denoted by the prefix H) and to a lesser extent the saturated monomers H12:0 and H10:0 (Fig. 1). In contrast, addition of ricinoleic acid to the growth medium resulted in an approximate 6-fold increase in the quantity of PHA produced, whereas the major change in PHA monomer composition was a 6-fold increase in the H10:0 monomer accompanied with smaller increases in the lower saturated monomers H8:0 and H6:0 as well as a decrease in most other monomers and in particular the unsaturated monomers. Addition of methyl vernolate to the plant medium resulted in a 4.3-fold increase in the quantity PHA and a 2-fold increase in the H8:0 monomer, with all other monomers decreasing or remaining unchanged (Fig. 1). Despite the use of different trans-esterifiying methods, no novel monomers could be identified in the PHA produced from the degradation of ricinoleic acid or vernolic acid in plants.
Figure 1.
Analysis of PHA produced in plants from ricinoleic acid and methyl vernolate. Transgenic plant line PHAC3.3, expressing a peroxisomal PHA synthase under the control of the CaMV 35S promoter, was grown in liquid medium supplemented with either oleic acid, ricinoleic acid, or methyl vernolate, all at 0.05% (v/v) final concentration. 3-Hydroxyacyl-CoA monomers are denoted by the prefix H. Values are ± sd. The amounts of PHA are expressed as micrograms of PHA per gram dry weight (dw) of plant material and are indicated at the top.
Analysis of the Degradation of Fatty Acids in Developing Seeds of Transgenic Plants Expressing the Hydroxylase and Epoxygenase
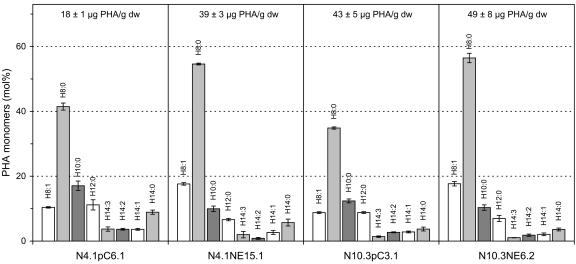
The quantity and monomer composition of PHA accumulated in seeds of transgenic lines expressing both the peroxisomal PHA synthase and either the 12-hydroxylase or 12-epoxygenase were analyzed. For the transgenic lines N10.3NH1.1 and N10.3NH2.2 expressing the 12-hydroxylase, a statistically significant (Student t test, P < 0.05) 1.6-fold increase in PHA quantity was observed in both hydroxylase-expressing plants compared with control plants, indicating a significant increase in the degradation of fatty acids in the developing seeds of plants producing hydroxy fatty acids relative to control plants (Fig. 2). The PHA monomer composition was also changed in the 12-hydroxylase-expressing plants, with a significant increase in the H8:1 (1.3- to 1.6-fold) and H10:0 (1.4- to 1.8-fold) monomer. Although the increase of the H10:0 monomer agreed well with the degradation of ricinoleic acid, the increase in H8:1 was unexpected. It is speculated that the H8:1 monomer corresponds to the degradation of densipolic acid (see “Discussion”).
Figure 2.
Analysis of PHA produced in mature seeds of transgenic plants expressing the castor bean 12-hydroxylase. Line N10.3pC3.1 was transformed with the control vector, whereas lines N10.3NH1.1 and N10.3NH2.2 were transformed with a napin-hydroxylase construct. Values are ± sd.
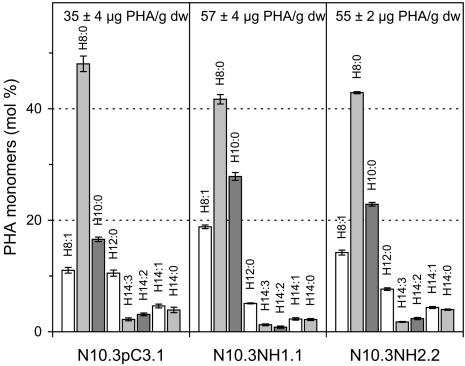
Transgenic line N4.1NE15.1 was found to have a 2-fold increase in the amount of PHA in mature seeds compared with its control (N4.1pC6.1; Fig. 3). In contrast, line N10.3NE6.2, which accumulated a lower amount of epoxy fatty acids compared with N4.1NE15, did not show a significant change in PHA quantity compared with its control (N10.3pC3.1; Fig. 3). For both N4.1NE15.1 and N10.3NE6.2, a significant increase in the monomers H8:1 (1.7- to 2.0-fold), and H8:0 (1.3- to 1.6-fold) were observed. Although the increase of the H8:0 monomer agreed well with the degradation of vernolic acid, the increase in H8:1 was unexpected. It is speculated that the H8:1 monomer corresponds to the degradation of 12-epoxy-9,15-octadecadienoic acid (see “Discussion”).
Figure 3.
Analysis of PHA produced in mature seeds of transgenic plants expressing the C. palaestina 12-epoxygenase. Lines N4.1pC6.1 and N10.3pC3.1 were transformed with the control vector, whereas lines N4.1NE15.1 and N10.3NE6.2 were transformed with a napin-epoxygenase construct. Line N4.1pC6.1 is used as the control for line N4.1NE15.1, whereas line N10.3pC3.1 is used as the control for line N10.3NE6.2. Values are ± sd.
Expression Profile of Genes Involved in Lipid Biosynthesis and Degradation during Germination and Silique Development
To assess the impact of the production of unusual fatty acids in transgenic plants on lipid metabolism, we have analyzed the expression profile of genes involved in the synthesis and degradation of lipids. For this purpose, a microarray was constructed containing nearly 200 genes encoding proteins involved in lipid biosynthesis as well as degradation, including genes involved in peroxisome metabolism and function, the peroxisome being the main site of fatty acid degradation in plant cells. The microarray also contained more than 80 genes representing various proteins involved in primary carbon metabolism, including gluconeogenesis, starch synthesis and degradation, and amino acid synthesis, as well as genes involved in secondary metabolism and oxidative stress. Also included was a set of 19 genes used as controls for constitutive expression. Details on the design of the microarray and on the quality controls are included in “Materials and Methods” and the supplementary data.
To test the use of the microarray for the detection of changes in the expression of genes involved in lipid biosynthesis and degradation, we have first examined differences in gene expression between germinating seeds 48 h after imbibition of seeds on agar plates containing mineral salts and placed under continuous light and plants grown for 12 d under the same conditions. Results are summarized in Table II. Ten of the 18 most induced genes, or 12 of a total of 30 genes induced 3.2-fold or more in germinating seeds, are encoding peroxisomal proteins involved directly or indirectly in fatty acid degradation, such as the enzymes of the β-oxidation cycle, the accompanying glyoxylate cycle, as well as the peroxisomal membrane proteins (peroxins), such as PEX5. Other genes found induced in germinating seeds include genes involved in gluconeogenesis, starch and amino acid biosynthesis, fatty acid desaturation, and lipid biosynthesis.
Table II.
Summary of genes induced in germinating seeds compared with 12-d-old plants
| Gene | Gene ID | Ratioa |
|---|---|---|
| Malate synthase | AT5G03860 | 77 |
| Isocitrate lyase | AT3G21720 | 47 |
| 3-Ketoacyl thiolase (PED1) | AT2G33150 | 15 |
| Oleosin | AT5G40420 | 11 |
| Multifunctional protein (MFP2) | AT3G06860 | 10 |
| Phosphoenolpyruvate carboxykinase (PCK3) | AT2G14840 | 10 |
| Acyl-CoA oxidase (ACX3) | AT1G06290 | 8.7 |
| Phosphoenolpyruvate carboxykinase | AT4G37870 | 8.6 |
| Asp aminotransferase (Asp3) | AT5G11520 | 8.2 |
| 3,2-Enoyl-CoA isomerase | AT4G16210 | 7.8 |
| Lipid transfer protein (LTP3) | AT5G59320 | 7.2 |
| ADP-Glc pyrophosphorylase large subunit (ApL3) | AT4G39210 | 6.5 |
| 3-Ketoacyl thiolase | AT1G04710 | 6.3 |
| Peroxin (PEX5) | AT5G56290 | 6.1 |
| Elongation factor 1-α | AT1G07920 | 5.6 |
| Acyl-CoA oxidase (ACX2) | AT5G65110 | 5.4 |
| ADP-Glc pyrophosphorylase large subunit (ApL2) | AT1G27680 | 5.1 |
| Catalase | AT4G35090 | 5.0 |
| Ascorbate peroxidase (APX3) | AT4G35000 | 4.8 |
| Phosphoenolpyruvate carboxykinase (PCK2) | AT5G65690 | 4.6 |
| Plastidial linoleate desaturase (FAD8) | AT5G05580 | 4.4 |
| ER linoleate desaturase (FAD3) | AT2G29980 | 4.4 |
| Gln synthetase | AT1G48470 | 3.7 |
| Ferredoxin | AT1G10960 | 3.7 |
| Starch synthase | AT5G24300 | 3.7 |
| Peroxin (PEX6) | AT1G03000 | 3.7 |
| Oleosin | AT4G25140 | 3.3 |
| Peroxin (PEX10) | AT2G26350 | 3.3 |
| Oleosin | AT3G27660 | 3.3 |
| Epoxide hydrolase | AT3G05600 | 3.2 |
Ratio of the gene expression level from germinating seeds 48 h after imbibition over the gene expression level from 12-d-old seedlings. Each ratio is an average of four values. Further details on the calculations of ratios can be found in the supplementary materials.
Gene expression was also analyzed in developing siliques 9 to 13 d after fertilization and compared with plants grown for 12 d in medium as described above (Table III). Of the 28 genes induced 3.2-fold or more in developing siliques, 22 are involved in some aspect of lipid synthesis and oil body formation. The remaining genes are involved in carbohydrate or nitrogen metabolism, flavonoid biosynthesis, or an unknown function relating to peroxisome membranes. Comparison of the present data with previously published results analyzing 8- to 11-d-old developing seeds of Arabidopsis versus leaves of flowering plants (Girke et al., 2000) revealed that of 10 genes found on both microarrays and shown to have an increased expression in developing seeds, eight also showed an increase in expression in the developing silique. Furthermore, several genes involved in fatty acid biosynthesis which have been previously described as being up-regulated during seed development by either northern blot or microarray analysis are also found induced in the present microarray, including the components of the fatty acid synthase, acyl-ACP thioesterase, and stearoyl-ACP desaturase (de Boer et al., 1998; O'Hara et al., 2002; Ruuska et al., 2002). These data indicate that despite a potential dilution effect from RNA derived from the silique wall and other non-seed tissues, changes in gene expression profile observed in developing siliques is representative of changes occurring during seed development.
Table III.
Summary of genes induced in developing siliques compared with 12-d-old-plants
| Gene | Gene ID | Ratioa |
|---|---|---|
| Oleosin | AT5G40420 | 117 |
| Oleosin | AT3G27660 | 99 |
| Lipid transfer protein (LTP4) | AT5G59310 | 79 |
| Lipid transfer protein (LTP3) | AT5G59320 | 77 |
| Oleosin | AT4G25140 | 47 |
| Oleosin | AT5G51210 | 15 |
| Suc synthase | AT5G49190 | 13 |
| Gln synthetase | AT1G48470 | 10 |
| Flavonoid 3′-hydroxylase | AT5G07990 | 7.4 |
| β-Amylase | AT4G15210 | 6.9 |
| Acyl-CoA synthase (AtLACS1) | AT2G47240 | 5.5 |
| Putative stearoyl-ACP desaturase | AT3G02630 | 5.2 |
| ER linoleate desaturase (FAD3) | AT2G29980 | 4.7 |
| ACPs (plastidial) | AT1G54580 | 4.7 |
| Peroxin (Pmp22.1) | AT4G04470 | 4.6 |
| Ketoacyl-ACP synthase (KAS2) | AT1G74960 | 4.3 |
| Stearoyl-ACP desaturase | AT2G43710 | 4.2 |
| 3-Hydroxyacyl-ACP dehydrase 2 | AT5G10160 | 4.2 |
| Epoxide hydrolase | AT3G05600 | 4.0 |
| Lipoxygenase (LOX5) | AT3G22400 | 3.8 |
| Epoxide hydrolase | AT3G51000 | 3.8 |
| Peroxin (Pmp22.1) | AT4G04470 | 3.8 |
| Ketoacyl-ACP synthase (KAS1) | AT5G46290 | 3.8 |
| Ferredoxin | AT1G10960 | 3.6 |
| δ-8 Sphingolipid desaturase | AT2G46210 | 3.6 |
| Ketoacyl-ACP reductase | AT1G24360 | 3.5 |
| Acyl-ACP thioesterase FatA | AT3G25110 | 3.5 |
| Enoyl-ACP reductase | AT2G05990 | 3.5 |
Ratio of the gene expression level from developing siliques 9 to 13 d post-fertilization over the gene expression level from 12-d-old seedlings. Each ratio is an average of four values. Further details on the calculations of ratios can be found in the supplementary materials.
Expression Profile of Genes in Developing Siliques of Transgenic Plants Accumulating Ricinoleic Acid or Vernolic Acid
The expression profile of genes in developing siliques of transgenic plants producing vernolic and ricinoleic acids were performed on two independent lines that accumulated the highest and similar amounts of unusual fatty acids. These lines were WTNH8.1 and WTNH13.1 for the 12-hydroxylase-expressing plants and WTNE5.1 and WTNE7.4 for 12-epoxygenase-expressing plants (Table I). The control plant line WTPC7.3 was transformed with only the pCAMBIA 1300 vector.
The expression profile of genes was studied for developing siliques 9 to 13 d after fertilization. This time window included stages in seed development where the rate of oil accumulation was at its maximum and where the expression of the napin promoter was high. Comparison of lines accumulating ricinoleic acid or vernolic acid with the control confirmed strong expression of the 12-hydroxylase and 12-epoxygenase genes in developing siliques (>60-fold increase compared with control). However, in all of the lines tested, no other genes were found to change expression level above the cutoff level of 2-fold. The only exception was the 2.6-fold reduction in the expression of the GST11 gene in transgenic line N10.3NH1.1 detected with the use of a full-length cDNA fragment. Further analysis of the expression profile of all the members of the glutathione s-transferase (GST) gene family with smaller gene-specific fragments did not reveal any change in the expression of GST-encoding genes, including GST11 (E. Rezzonico, Y. Poirier, and F. Mauch, unpublished data). No change in GST11 expression was detected in other transgenic lines.
Expression Profile of Genes in Developing Siliques of Transgenic Plants Producing Medium-Chain Fatty Acids and in the tag1 Mutant
The profile of gene expression was also examined in plants that had been previously shown, through PHA analysis, to have a high futile cycling of fatty acids toward β-oxidation. Transgenic plants expressing the Cuphea lanceolata FatB3 gene encoding the caproyl-ACP thioesterase under the control of either the CaMV 35S promoter or the napin promoter were previously shown to accumulate capric acid in seeds up to 2.5 and 19 mol %, respectively (Mittendorf et al., 1999; Poirier et al., 1999). Flux of medium-chain fatty acids toward the peroxisomal β-oxidation cycle has been previously demonstrated in leaves, for plants expressing FatB3 under the CaMV 35S promoter, and in developing seeds for plants expressing FatB3 under the napin promoter. Analysis of the profile of gene expression in 9- to 13-d-old siliques of plants expressing the napin-FatB3 construct did not reveal any significant changes except for the FatB3 gene itself (>100-fold induction). Similarly, no significant changes in expression were detected in leaves of plants expressing the CaMV-35S-FatB3 construct (data not shown).
The tag1 mutant of Arabidopsis is deficient in the diacylglycerol acyltransferase and was shown to have a strongly reduced level of TAG in seeds as well as an increase in the proportion of linolenic acid in seed lipids (Katavic et al., 1995). Analysis of PHA synthesized in developing seeds of the tag1 mutant revealed the presence of a strong futile cycling of long-chain fatty acids, including of linolenic acid (Poirier et al., 1999). Analysis of gene expression in the developing silique of the tag1 mutant SK353 revealed only three genes that were induced above 2-fold, namely the isocitrate lyase (5.8-fold) and two oleosins (2.0- and 2.4-fold; see supplementary material).
DISCUSSION
Synthesis of PHA in the peroxisome of plants has previously been demonstrated to be a useful tool to study the flow of fatty acids through the peroxisomal β-oxidation cycle (Mittendorf et al., 1999; Poirier et al., 1999; Poirier, 2002). Because degradation of a fatty acid through β-oxidation generates a series of 3-hydroxyacyl-CoA intermediates that can be trapped into PHA (the PHA synthase from P. aeruginosa typically accepts 3-hydroxyacyl-CoA from six to 14 carbons), the PHA monomer composition can be used to reveal the type of fatty acid being degraded. For example, degradation of linolenic acid yields the 3-hydroxyacyl-CoA intermediates H14:3, H12:2, H10:2, H8:1, and H6:0, whereas degradation of oleic acid yields the 3-hydroxyacyl-CoA intermediates H14:1, H12:0, H10:0, H8:0, and H6:0 (Mittendorf et al., 1999). Thus, although certain monomers are generated by the degradation of a specific fatty acid (e.g. H14:3 for linolenic acid, H14:2 for linoleic acid, and H14:1 for oleic acid), other monomers are generated by a broad range of fatty acids (e.g. H8:0 by linoleic, oleic, palmitic, and stearic acids; see fig. 1 of Mittendorf et al., 1999). It is thus important to consider both the quantity of PHA as well as the monomer composition of the polymer when interpreting results obtained from PHA.
To use PHA to assess the presence of a futile cycle of ricinoleic acid or vernolic acid in transgenic plants, the shift in PHA monomer composition triggered by the degradation of these fatty acids was first analyzed in plants expressing a constitutive peroxisomal PHA synthase grown in liquid cultures supplied with or without an external source of fatty acids. The addition of oleic acid yielded an increase in the H14:1, H12:0, and H10 monomers compared with plants grown without fatty acids. These results are in agreement with previous studies looking at the impact of the degradation of oleic acid on PHA in plants, yeast, and bacteria (de Waard et al., 1993; Mittendorf et al., 1999; Poirier et al., 2001).
Addition of ricinoleic acid to the growth medium of plants producing PHA resulted primarily in a large increase in the H10 monomer (Fig. 1). No novel monomers containing hydroxy side chain could be detected. Similar results have been obtained from the degradation of ricinoleic acid in the PHA-producing bacterium Pseudomonas putida (D. Caldelari, L. Moire, and Y. Poirier, unpublished data). According to the pathway proposed by Gerhart (1993), the degradation of ricinoleic acid through the plant β-oxidation pathway should, in theory, yield the CoA esters of 3,8-dihydroxytetradec-5-cis-ecenoic acid, 3,6-dihydroxydodecanoic acid, and 3,4-dihydroxydecanoic acid, followed by the generation of 3-hydroxyheptanoyl-CoA via one cycle of α-oxidation. Although the bacterial PHA synthases have a broad substrate specificity and can incorporate 3-hydroxyacids having a variety of functional groups, it is known to be very inefficient at incorporating 3-hydroxyacids containing hydrophilic moieties in the middle of the chain, such as hydroxy or epoxy groups (Lenz et al., 1992). Thus, no PHA containing significant amount of monomers with epoxy or hydroxy side chains have been synthesized in bacteria through feeding experiments. The failure to detect the dihydroxy intermediates in the plant was thus in line with results obtained in bacteria. It is nevertheless striking that 3-hydroxydecanoic acid is the main monomer generated by the degradation of ricinoleic acid, because the only 10 carbon intermediate expected from the proposed pathway is the dihydroxy intermediate 3,4-dihydroxydecanoyl-CoA. Furthermore, the predicted generation of 3-hydroxyheptanoyl-CoA is not substantiated by an increase in the 3-hydroxyheptanoic acid monomer in PHA generated in bacteria or plants from ricinoleic acid. Although it is not possible to deduce the pathway of ricinoleic acid degradation from these results alone, they nevertheless indicate that at least part of the proposed pathway needs to be revised. Despite this, the strong increase in the H10 monomer provides a reliable marker for the degradation of ricinoleic acid in transgenic plants.
Addition of methyl vernolate to the growth medium of plants producing PHA resulted primarily in a large increase in the H8 monomer. No novel monomers containing epoxy side chain could be detected. Similar results have been obtained with vernolic acid (L. Moire and Y. Poirier, unpublished data). In a manner similar to ricinoleic acid, the failure to detect 3-hydroxyacid monomers containing epoxy groups is in line with the poor affinity of the PHA synthase for these substrates. Thus, although the pathway of degradation of vernolic acid cannot be easily deduced from the PHA monomer composition, the increase of the H8 monomer provides a reliable marker for the degradation of vernolic acid in transgenic plants.
Transgenic plants expressing the PHA synthase and either the castor bean 12-hydroxylase or the C. palaestina linoleate 12-epoxygenase in developing seeds were used to assess the presence of futile cycling of ricinoleic or vernolic acids. The amount of hydroxy and epoxy fatty acids produced in the transgenic plants that were chosen for analysis are in the low- to mid-range when compared with levels reported in other studies of transgenic Arabidopsis expressing the same transgenes (Broun and Somerville, 1997; Broun et al., 1998; Lee et al., 1998; Singh et al., 2001; Thomaeus et al., 2001). It is likely that the selection of lines showing a hygromycin segregation ratio indicative of T-DNA integration at a single locus favored the isolation of plants having few copies of the transgenes and thus lower amount of transgene expression. This screening was done to limit the likelihood of gene silencing and unstable expression of either the PHA synthase, 12-hydroxylase, or 12-epoxygenase in subsequent generations. A previous study of transgenic canola (Brassica napus) expressing a lauroyl-ACP thioesterase has shown a positive correlation between the number of transgenes and the level of accumulated laurate (Voelker et al., 1996).
Analysis of PHA produced in developing seeds of two independent lines accumulating between 3.5 and 6.3 mol % hydroxy fatty acids revealed a significant 1.4- to 1.8-fold increase in the H10 monomer as well as a 1.3- to 1.6-fold increase in H8:1 monomer, accompanied by an overall 1.6-fold increase in the total amount of PHA, when compared with control parental plants expressing only the PHA synthase (Fig. 2). Although the H10 monomer could be generated by the degradation of stearic, palmitic, or oleic acids, the fact that other monomers generated by the degradation of the same fatty acids, such as H8, H12, H14, or H14:1 are either stable or decrease, indicates that the increase in H10 cannot be caused by the turnover of these common fatty acids. Rather, the increase in H10 is strongly indicative of the degradation of ricinoleic acid and also very likely of lesquerolic acid, because both of these fatty acids would generate the same β-oxidation intermediates that are 18 carbons or less. Although the H8:1 monomer could be generated by the degradation of linolenic acid, the H14:3 monomer, which is also specifically generated by the degradation of this fatty acid, is slightly decreased, thus excluding this possibility. The increase of H8:1 could, however, be explained by the degradation of densipolic acid, which contains an additional unsaturated bond three carbons beyond the hydroxyl group compared with ricinoleic acid. Together, these results indicate the presence of a futile cycle of hydroxy fatty acids in plants expressing the oleate 12-hydroxylase.
Analysis of PHA produced in developing seeds of two independent lines accumulating between 0.9 and 3.1 mol % epoxy fatty acids revealed a significant 1.3- to 1.6-fold increase in the H8 monomer as well as a 1.7- to 2.0-fold increase in H8:1 monomer. Although the line accumulating only 0.9 mol % epoxy fatty acids did not show an increase in PHA amounts compared with control, the line accumulating 3.1 mol % epoxy fatty acid showed a 2-fold increase in the total amount of PHA. Using the same reasoning as for the analysis of PHA from the plants expressing the 12-hydroxylase, the conclusion drawn from these results is that the increase in H8:0 is derived from the degradation of vernolic acid, whereas the increase in H8:1 most likely reflects the degradation of 12-epoxy-9,15-octadecadienoic acid. Thus, a futile cycling of epoxy fatty acid also occurs in plants expressing the linoleate 12-epoxygenase in developing seeds.
Futile cycling of medium-chain fatty acids has previously been described in developing seeds of transgenic Arabidopsis expressing either a lauroyl-ACP thioesterase from California bay or a caproyl-ACP thioesterase from C. lanceolata (Eccleston and Ohlrogge, 1998; Poirier et al., 1999). In Arabidopsis co-expressing the peroxisomal PHA synthase and plastidial caproyl-ACP thioesterase, both under the napin promoter, there was an 18-fold increase in PHA in the thioesterase-expressing plants compared with control as well as a 4-fold increase in the H10 monomer. Although the increase in PHA as well as the shift in monomer composition were larger in plants expressing the caproyl-ACP thioesterase compared with the plants of this study expressing the 12-hydroxylase or 12-epoxygenase, it must be noted that the amount of medium-chain fatty acids accumulated in seeds of the thioesterase-expressing plants reached 25 mol %, whereas the amount of unusual fatty acids in the plants used for this study only reached 6.3 mol % for hydroxy fatty acids and 3.1 mol % for epoxy fatty acids. Although PHA analysis cannot give an absolute quantitative value of the proportion of fatty acid being degraded, the fact that there is significant increase in the monomers generated by the degradation of hydroxy and epoxy fatty acids despite their relative low abundance at steady-state levels in lipids indicates that the proportion of synthesized hydroxy and epoxy fatty acids that is directed toward peroxisomal β-oxidation is significantly higher than it is for the common fatty acids.
The oleate 12-hydroxylase and linoleate 12-epoxygenase act on fatty acids present on phospholipids on the endoplasmic reticulum. The newly synthesized unusual fatty acids are then transferred from phospholipids to be mobilized into TAG via a number of potential routes implicating either the action of specific phospholipases or acyltransferases, including the phosphatidylcholine:diacylglycerol acyltransferase (Bafor et al., 1991; Ståhl et al., 1995; Banas et al., 2000; Dahlqvist et al., 2000). The same pathways are also likely to be involved in the exclusion of epoxy and hydroxy fatty acids from membrane lipids in both native and transgenic plants (Thomaeus et al., 2001). Because fatty acids or fatty acyl-CoAs cannot accumulate in plants cells, the exclusion of unusual fatty acids from lipids would likely lead to their degradation via the peroxisomal β-oxidation cycle. In this perspective, the futile cycle of hydroxy or epoxy fatty acids observed in this study likely reflects their inefficient stable incorporation into lipid, including TAG.
Analysis of the expression profile, during silique development, of genes involved in several aspects of lipid biosynthesis and degradation revealed no change in gene expression as a result of the synthesis of hydroxy or epoxy fatty acids. Thus, despite the evidence of futile cycling of unusual fatty acids, no genes involved in fatty acid catabolism, gluconeogenesis, or peroxisome function or assembly were up-regulated. There was also no induction of genes involved in fatty acid or lipid biosynthesis that might have been involved in compensating for the loss of fatty acids through β-oxidation or to correct the imbalance in the amount of monounsaturated and polyunsaturated fatty acids caused by the expression of the 12-hydroxylase and the 12-epoxygenase. The same results were found for plants expressing the caproyl-CoA thioesterase in developing seeds and leaves, where a strong futile cycle has been demonstrated (Mittendorf et al., 1999; Poirier et al., 1999). Although the microarray used in these experiments contains only a small subset of the genes present in the Arabidopsis genome, raising the possibility that other genes not included on the array may be up- or down-regulated in transgenic plants, the majority of the key genes involved in lipid synthesis and degradation were included on the microarray. Together, these results indicate that substantial changes can occur in fatty acid composition as well as carbon flux through lipid metabolic pathways as a result of transgene expression without impacting the level of expression of genes involved in lipid metabolism.
Only for the tag1 mutant, deficient in the accumulation of TAG as a result of a mutation in a gene encoding a diacylglycerol acyltransferase, have a few changes in gene expression been observed. Analysis of PHA synthesis in the tag1 mutant had previously shown a strong futile cycling of common fatty acids (Poirier et al., 1999). In line with these results, the gene found most highly up-regulated was the isocitrate lyase gene with a 5.8-fold-increase in gene expression. Analysis of gene expression during seed germination revealed that the genes encoding the isocitrate lyase and malate synthase, two key enzymes of the glyoxylate cycle, are the most strongly up-regulated genes found on the array (Table II). These genes thus appear to be the most sensitive markers of fatty acid catabolism at the level of gene expression. Together, these results would indicate that among the various Arabidopsis lines analyzed in this study, the tag1 mutant would have the strongest flux of fatty acids toward the β-oxidation cycle.
Analysis of transgenic canola expressing the lauroyl-ACP thioesterase in leaves or seeds revealed an increase in isocitrate lyase activity in leaves and of both isocitrate lyase and acyl-CoA oxidase activity in seeds (Eccleston et al., 1996; Eccleston and Ohlrogge, 1998). In contrast, no changes in gene expression or enzyme activity for several enzymes involved in fatty acid catabolism have been detected in leaves of Arabidopsis plants expressing the lauroyl-ACP thioesterase (Hooks et al., 1999), whereas a weak increase in medium-chain and long-chain acyl-CoA oxidase activity was detected in plants expressing the caproyl-ACP thioesterase in leaves (Mittendorf et al., 1999). Together, these data indicate that the increase degradation of fatty acid through the peroxisomal β-oxidation cycle observed in leaves or seeds of transgenic plants accumulating unusual fatty acids cannot be reliably detected through the analysis of gene expression or enzyme activities. In this context, analysis of PHA synthesized from the intermediates of β-oxidation is a better and more sensitive marker of fatty acid futile cycling. Analysis of PHA would thus be a valuable tool to determine whether stable incorporation of synthesized fatty acid into lipids is also limiting the accumulation of other unusual fatty acids in transgenic plants.
MATERIALS AND METHODS
DNA Constructs
The castor bean (Ricinus communis) oleate 12-hydroxylase cDNA (U22378) and Crepis palaestina linoleate 12-epoxygenase cDNA (Y16283; both kindly provided by S. Stymne, Swedish University of Agricultural Sciences, Alnarp, Sweden) were cloned in the pART7-napin vector (Gleave, 1992; Poirier et al., 1999) as BamHI or EcoRI fragments, respectively, placing the cDNAs under the control of the napin promoter from canola (Brassica napus) and the octopine synthase terminator. The cDNAs with the upstream and downstream regulatory elements were cloned as SacI-PstI fragments in the pCAMBIA 1300 vector (http://www.cambia.org.au) containing the hygromycin resistance gene under the control of CaMV 35S promoter. All binary vectors were transferred into Agrobacterium tumefaciens pGV3101 by electroporation.
Bacterial and Plant Material
Wild-type Arabidopsis, as well as the transgenic Arabidopsis lines N4.1 and N10.3 (formerly described as N-PHA-4.1 and N-PHA-10.3) expressing the Pseudomonas aeruginosa PHAC1 synthase modified for peroxisome targeting under the control of the 1.1-kb canola napin promoter (Poirier et al., 1999), were transformed with the pCAMBIA 1300 vector or constructs containing the 12-hydroxylase or 12-epoxygenase genes, by the floral dip method (Clough and Bent, 1998). T1 transformants were isolated by plating seeds on medium containing one-half-strength Murashige and Skoog salts, 1% (w/v) Suc, 0.7% (w/v) agar, and 30 μg mL-1 hygromycin. Hygromycin-resistant plants were subsequently transferred to soil and grown under continuous fluorescent light at 19°C.
Transgenic Arabidopsis line C-PHA-3.3 expressing the peroxisome-targeted PHAC1 synthase under the control of a CaMV 35S promoter has been previously described (Mittendorf et al., 1998, 1999). In experiments aimed at analyzing the degradation of ricinoleic acid and vernolic acid, C-PHA-3.3 plants were grown under constant agitation (100 rpm) and continuous fluorescent light at 19°C in liquid medium (one-half-strength Murashige and Skoog salts and 1% [w/v] Suc). After 7 d of culture, the liquid medium was replaced by the same medium supplemented with 2% (w/v) Pluronic F-127 (Sigma-Aldrich, St. Louis) alone or in combination with 0.05% (w/v) fatty acids. Plants were harvested after 5 d of culture, washed in water, and lyophilized.
The fatty acids oleic acid and ricinoleic acid were purchased from Sigma-Aldrich and Nu-Chek-Prep (Elysian, MN), respectively. Methyl ester of vernolic acid was kindly provided by J.W. Veldsink, ATO-DLO (Wageningen, The Netherlands).
PHA and Fatty Acid Analysis
Extraction of PHA from plant material or bacteria and analysis by gas-chromatography and mass spectrometry was done as previously described (Mittendorf et al., 1998). The presence of contaminants in the PHA extract from seeds prevented the determination of all potential PHA monomers. Analysis was thus focused on PHA monomers that could be reliably analyzed in all samples and included the monomers H14:0, H14:1, H14:2, H14:3, H12:0, H10:0, H8:0, H8:1, and H6:0.
Seed fatty acid methyl-esters were prepared by acid-catalyzed (1 n HCL in methanol, 2 h, 80°C) or base-catalyzed (0.1 m sodium methoxide in methanol, 1 h, 60°C) transesterification. After reaction, the fatty acid methylesters were extracted with hexane and water, and the organic phase was transferred to vials. Analysis was performed using a gas chromatograph equipped with a glass capillary column (model SP230, Supelco, Bellefonte, PA) and a flame ionization detector.
Statistical analyses of the amount and composition of PHA and fatty acids present in various transgenic lines were measured by a Student's t test with a level of significance set at 0.05.
cDNA Microarray Experiments
Plasmids of selected expressed sequence tags were obtained from the Arabidopsis Biological Resource Center, The Kazusa DNA Research Institute, the Institut National de la Recherche Agronomique, and other colleagues. All clones have been sequenced to ensure accuracy. Inserts were amplified by the PCR from the cDNA clones using universal primers. For some genes, DNA fragments generated by PCR from genomic DNA were used. The quantity and quality of the amplification were checked by gel electrophoresis.
PCR products were purified using QIAquick-96 columns (Qiagen, Basel), lyophilized, and resuspended in 3× SSC, 1,5 m betaine (N,N,N-trimethyl-Gly, Sigma-Aldrich) in a 384-well microtiter plate. Several negative controls were also included, namely a set of 10 non-related human genes provided by the Arabidopsis Functional Genomics Consortium and 10 spiking control from Stratagene (SpotReport Alien cDNA Array Validation System). Amplicons were spotted, in duplicate, on SuperAldehyde Substrates microarray slides (TeleChem International, San Jose, CA) using a high-precision gridding robot (GeneMachines, San Carlos, CA) equipped with four printing tips (TeleChem International). After printing, slides were processed before hybridization as described by Reymond et al. (2000).
Total RNA was extracted from germinating seeds, 12-d-old seedlings, and mature leaves by the Trizol method (Chomczynski and Sacchi, 1987), whereas a hot borate protocol (Wan and Wilkins, 1994) was used to extract RNA from siliques, 9 to 13 d after fertilization. In both cases, the obtained RNA was further purified using the RNeasy midi kit (Qiagen) before labeling. One hundred micrograms of total RNA was converted into fluorescently labeled single-stranded cDNAs by direct incorporation of either Cy3-dCTP or Cy5-dCTP and hybridized for 12 to 16 h in a humidified hybridization chamber as described by Reymond et al. (2000). Microarray slides were scanned using the ScanArray4000 (Packard BioChip Technologies, Billerica, MA) scanning laser microscope with its accompanying scanning software. Intensities of the fluorescent signals were quantified with the Imagene 5.1 (BioDiscovery, Inc., Marina del Rey, CA) software.
Further details on the microarray experiments, including quality controls and data analysis, are provided in the supplementary material.
Supplementary Material
Acknowledgments
We are very grateful to Edward Farmer and Philippe Reymond for their involvement in setting up the cDNA microarray at the University of Lausanne and to Nadine Erard for her technical assistance. We extend our thanks also to the members of the CONFAB consortium for their support.
This work was funded by the Office Fédéral de l'Education et de la Science (grant no. 99.0692 to Y.P.) under the 5th Framework of the European Union (CONFAB project no. GLRT-1999-00213). Contributions are also acknowledged from the University of Lausanne and the Canton de Vaud.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.032938.
References
- Badami RC, Patil KB (1980) Structure and occurrence of unusual fatty acids in minor seed oils. Prog Lipid Res 19: 119-153 [DOI] [PubMed] [Google Scholar]
- Bafor M, Smith MA, Jonsson L, Stobart K, Stymne S (1991) Ricinoleic acid biosynthesis and triacylglycerol assembly in microsomal preparations from developing castor-bean (Ricinus communis) endosperm. Biochem J 280: 507-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas A, Dahlqvist A, Stahl U, Lenman M, Stymne S (2000) The involvement of phospholipid:diacylglycerol acyltransferases in triacylglycerol production. Biochem Soc Trans 28: 703-705 [PubMed] [Google Scholar]
- Broun P, Boddupalli S, Somerville C (1998) A bifunctional oleate 12-hydroxylase: desaturase from Lesquerella fendleri. Plant J 13: 201-210 [DOI] [PubMed] [Google Scholar]
- Broun P, Somerville C (1997) Accumulation of ricinoleic, lesquerolic, and densipolic acids in seeds of transgenic Arabidopsis plants that express a fatty acyl hydroxylase cDNA from castor bean. Plant Physiol 113: 933-942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Shanklin J, Ohlrogge JB (1992) Expression of a coriander desaturase results in petroselinic acid production in transgenic tobacco. Proc Natl Acad Sci USA 89: 11184-11188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156-159 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735-743 [DOI] [PubMed] [Google Scholar]
- Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S (2000) Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci USA 97: 6487-6492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer G-J, Kater MM, Fawcett T, Slabas AR, Nijkamp HJJ, Stuitje AR (1998) The NADH-specific enoyl-acyl carrier protein reductase: characterization of a housekeeping gene involved in storage lipid synthesis in seeds of Arabidopsis and other plant species. Plant Physiol Biochem 36: 473-486 [Google Scholar]
- de Waard P, van der Wal H, Huijberts GN, Eggink G (1993) Heteronuclear NMR analysis of unsaturated fatty acids in poly(3-hydroxyalkanoates): study of beta-oxidation in Pseudomonas putida. J Biol Chem 268: 315-319 [PubMed] [Google Scholar]
- Eccleston VS, Cranmer AM, Voelker TA, Ohlrogge JB (1996) Medium-chain fatty acid biosynthesis and utilization in Brassica napus plants expressing lauroyl-acyl carrier protein thioesterase. Planta 198: 46-53 [Google Scholar]
- Eccleston VS, Ohlrogge JB (1998) Expression of lauroyl-acyl carrier protein thioesterase in Brassica napus seeds induces pathways for both fatty acid oxidation and biosynthesis and implies a set point for triacylglycerol accumulation. Plant Cell 10: 613-622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart B (1993) Catabolism of fatty acids (α- and β-oxidation). In TS Moore, ed, Lipid Metabolism in Plants. CRC Press, Boca Raton, FL, pp 527-565
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J (2000) Microarray analysis of developing Arabidopsis seeds. Plant Physiol 124: 1570-1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203-1207 [DOI] [PubMed] [Google Scholar]
- Hooks MA, Fleming Y, Larson TR, Graham IA (1999) No induction of beta-oxidation in leaves of Arabidopsis that over-produce lauric acid. Planta 207: 385-392 [DOI] [PubMed] [Google Scholar]
- Jaworski J, Cahoon EB (2003) Industrial oils from transgenic plants. Curr Opin Plant Biol 6: 178-184 [DOI] [PubMed] [Google Scholar]
- Katavic V, Reed DW, Taylor DC, Giblin EM, Barton DL, Zou J, Mackenzie SL, Covello PS, Kunst L (1995) Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol 108: 399-409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Lenman M, Banas A, Bafor M, Singh S, Schweizer M, Nilsson R, Liljenberg C, Dahlqvist A, Gummeson PO et al. (1998) Identification of non-heme diiron proteins that catalyze triple bond and epoxy group formation. Science 280: 915-918 [DOI] [PubMed] [Google Scholar]
- Lenz RW, Kim YB, Fuller RC (1992) Production of unusual bacterial polyesters by Pseudomonas oleovorans through cometabolism. FEMS Microbiol Lett 103: 207-214 [Google Scholar]
- Millar AA, Smith MA, Kunst L (2000) All fatty acids are not equal: discrimination in plant membrane lipids. Trends Plant Sci 5: 95-101 [DOI] [PubMed] [Google Scholar]
- Mittendorf V, Bongcam V, Allenbach L, Coullerez G, Martini N, Poirier Y (1999) Polyhydroxyalkanoate synthesis in transgenic plants as a new tool to study carbon flow through beta-oxidation. Plant J 20: 45-55 [DOI] [PubMed] [Google Scholar]
- Mittendorf VV, Robertson EJ, Leech RM, Kruger N, Steinbuchel A, Poirier Y (1998) Synthesis of medium-chain-length polyhydroxyalkanoates in Arabidopsis thaliana using intermediates of peroxisomal fatty acid beta-oxidation. Proc Natl Acad Sci USA 95: 13397-13402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara P, Slabas AR, Fawcett T (2002) Fatty acid and lipid biosynthetic genes are expressed at constant molar ratios but different absolute levels during embryogenesis. Plant Physiol 129: 310-320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y (2002) Polyhydroxyalknoate synthesis in plants as a tool for biotechnology and basic studies of lipid metabolism. Prog Lipid Res 41: 131-155 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Erard N, Petetot JM (2001) Synthesis of polyhydroxyalkanoate in the peroxisome of Saccharomyces cerevisiae by using intermediates of fatty acid beta-oxidation. Appl Environ Microbiol 67: 5254-5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Ventre G, Calelari D (1999) Increased flow of fatty acids toward β-oxidation in developing seeds of Arabidopsis deficient in diacylglycerol acyltransferase activity or synthesizing medium-chain-length fatty acids. Plant Physiol 121: 1359-1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707-719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191-1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Thomaeus S, Lee M, Stymne S, Green A (2001) Transgenic expression of a delta 12-epoxygenase gene in Arabidopsis seeds inhibits accumulation of linoleic acid. Planta 212: 872-879 [DOI] [PubMed] [Google Scholar]
- Ståhl U, Banas A, Stymne S (1995) Plant microsomal phospholipid acyl hydrolases have selectivities for uncommon fatty acids. Plant Physiol 107: 953-962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomaeus S, Carlsson AS, Stymne S (2001) Distribution of fatty acids in polar and neutral lipids during seed development in Arabidopsis thaliana genetically engineered to produce acetylenic, epoxy and hydroxy fatty acids. Plant Sci 161: 997-1003 [Google Scholar]
- van de Loo FJ, Broun P, Turner S, Somerville C (1995) An oleate 12-hydroxylase from Ricinus communis L. is a fatty acyl desaturase homolog. Proc Natl Acad Sci USA 92: 6743-6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Loo FJ, Fox BG, Somerville C (1993) Unusual fatty acids. In TS Moore, ed, Lipid Metabolism in Plants. CRC Press, Boca Raton, FL, pp 91-126
- Voelker T, Kinney AJ (2001) Variations in the biosynthesis of seed-storage lipids. Annu Rev Plant Physiol Plant Mol Biol 52: 335-361 [DOI] [PubMed] [Google Scholar]
- Voelker TA, Hayes TR, Cranmer AM, Turner JC, Davies HM (1996) Genetic engineering of a quantitative trait: metabolic and genetic parameters influencing the accumulation of laurate in rapeseed. Plant J 9: 229-241 [Google Scholar]
- Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223: 7-12 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.