Abstract
Recently, a new member of the ABC transporter superfamily of Arabidopsis, AtMRP5, was identified and characterized. In the present work, we found that AtMRP5 can bind specifically to sulfonurea when it is expressed in HEK293 cells. We also present evidence for a new role of AtMRP5 in the salt stress response of Arabidopsis. We used reverse genetics to identify an Arabidopsis mutant (atmrp5-2) in which the AtMRP5 gene was disrupted by transferred DNA insertion. In root-bending assays using Murashige and Skoog medium supplemented with 100 mm NaCl, root growth of atmrp5-2 was substantially inhibited in contrast to the almost normal growth of wild-type seedlings. This hypersensitive response of the atmrp5-2 mutant was not observed during mannitol treatment. The root growth of the wild-type plant grown in Murashige and Skoog medium supplemented with the MRP inhibitor glibenclamide and NaCl was inhibited to a very similar extent as the root growth of atmrp5-2 grown in NaCl alone. The Na+-dependent reduction of root growth of the wild-type plant in the presence of glibenclamide was partially restored by diazoxide, a known K+ channel opener that reverses the inhibitory effects of sulfonylureas in animal cells. Moreover, the atmrp5-2 mutant was defective in 86Rb+ uptake. When seedlings were treated with 100 mm NaCl, atmrp5-2 seedlings accumulated less K+ and more Na+ than those of the wild type. These observations suggest that AtMRP5 is a putative sulfonylurea receptor that is involved in K+ homeostasis and, thus, also participates in the NaCl stress response.
The ATP-binding cassette (ABC) transporter superfamily is the largest known membrane transporter protein family, and its members are capable of a multitude of transport functions (Higgins, 1992, 1995). These proteins are highly interesting because of their involvement in numerous pathologies, such as cystic fibrosis, diabetes, and multidrug resistance (Demolombe and Escande, 1996). In animals, two ABC proteins are directly involved in regulating ion channels in the plasma membrane, specifically ATP-sensitive potassium channels (KATP channels) and the cystic fibrosis transmembrane conductance regulator (CFTR). KATP channels were initially identified in the heart (Noma, 1983) and are complexes composed of an inward rectifier K+ channel and an ABC protein, the sulfonylurea receptor (Babenko et al., 1998; Miki et al., 1999). KATP channels are highly specific for K+ and are inhibited by micromolar concentrations of intracellular ATP. CFTR is a chloride channel expressed by a variety of secreting epithelial cells that is regulated by cAMP-dependent phosphorylation and ATP (Anderson et al., 1991; Bear et al., 1992). These channels serve to link the electrical activity of cell membranes with cellular metabolism.
The presence of ABC proteins in plants was established by the cloning of several genes encoding members of this group in Arabidopsis and other species (Dudler and Hertig, 1992; Smart and Fleming, 1996; Davies et al., 1997; Lu et al., 1997, 1998; Tommasini et al., 1997, 1998; Marin et al., 1998; Rea et al., 1998; Sánchez-Fernández et al., 1998; Rea, 1999; Theodoulou, 2000; Gaedeke et al., 2001). After the completion of the genomic sequencing of Arabidopsis (Arabidopsis Genome Initiative, 2000), the complete inventory of ABC protein superfamily of Arabidopsis was described (Sánchez-Fernández et al., 2001; Martinoia et al., 2002). Of these, the Arabidopsis multidrug resistance-related proteins (AtMRP) are the most extensively characterized plant ABC transporters to date. These proteins function as vacuolar sequesters of glutathionated compounds, malonylated chlorophyll catabolites, and glucuronides (for review, see Rea et al., 1998; Rea, 1999; Theodoulou, 2000; Martinoia et al., 2002).
Recent results show that the plant ABC transporters are not only implicated in detoxification and ion regulation processes but also in plant growth processes. Sidler et al. (1998) demonstrated that AtPGP1 (Arabidopsis P-glycoprotein1) is involved in a developmental pathway that regulates hypocotyl cell elongation under low light. In addition, more recent studies by Noh et al. (2001) revealed that two MDR-like genes of Arabidopsis encode 1-naphthylphthalamic acid-binding proteins that are required for normal auxin distribution and auxin-mediated development.
In contrast to the studies with animal cells, currently, there is little known about the involvement of ABC proteins in the control of plant ion channels. Electrophysiological studies using Vicia faba guard cell protoplasts suggest that plants may have a sulfonylurea receptor-like protein that modulates stomatal movements and transmits the signals from sulfonylureas and potassium channel openers to potassium and/or anion channels on guard cells (Leonhardt et al., 1997, 1999). However, it is not known yet what kind of molecule(s) is (are) responsible for these sulfonylurea-sensitive currents involved in stomatal movements.
Of the 15 MRPs identified in the Arabidopsis genome, AtMRP5 has been studied most extensively because of the existence of an AtMRP5 transferred DNA (T-DNA) insertional knockout mutant (mrp5-1). Gaedeke et al. (2001) showed that AtMRP5 controls root development because the phenotypic characterization of mrp5-1 revealed that this mutant exhibits decreased root growth and increased lateral root formation in reduced-strength (0.25×) Murashige and Skoog medium. They also showed that AtMRP5 participates in stomatal movement by comparing the stomatal movements of the wild-type and mrp5-1 plants (Klein et al., 2003). AtMRP5 also regulates stomatal movements that are sensitive to the sulfonylurea analog glibenclamide (Gaedeke et al., 2001). Thus, AtMRP5 is also involved in guard cell signaling and water use. Promoter expression analyses by this group with the aid of a promoter-β-glucuronidase (GUS) construct revealed that At-MRP5 is expressed in the vascular bundle and the epidermis, especially in guard cells. Based on these results, it was concluded that AtMRP5 may work: (a) as an auxin conjugate transporter or as a component in the ion homeostasis affecting auxin concentration, and/or (b) as an ion channel regulator (Gaedeke et al., 2001), and/or (c) as an important component of guard cell functioning (Klein et al., 2003). However, it was not determined whether AtMRP5 is an ion channel, or an ion channel regulator, or another signaling component involved in guard cell function.
Of the many plant research areas, considerable effort has been devoted to elucidating the mechanisms of plant salt tolerance. A widely used approach to unravel this tolerance mechanism involves the identification of the cellular processes and genes whose activity or expression is regulated by salt stress (for review, see Hasegawa et al., 1987; Cushman et al., 1990; Skriver and Mundy, 1990; Bray, 1993; Bohnert et al., 1995; Zhu et al., 1997; Zhu, 2000). Studies by Zhu et al. provide clear evidence of a signal transduction pathway that mediates salt tolerance in plants by controlling ion homeostasis. Their results suggest that the capacity of plants to counteract salinity stress strongly depends on the status of their K+ nutrition and external Ca2+, as has already been postulated long ago (Epstein, 1969; LaHaye and Epstein, 1969). Although one outcome of the regulatory pathway in salt stress involves the up-regulation of SOS1 Na+/K+ antiporter gene expression (Shi et al., 2000), it is suggested that the SOS3/SOS2 regulatory pathway may also modulate the abundance and/or activity of certain K+ and Na+ transporters (Zhu, 2000).
In this report, we reveal new biochemical and physiological functions of AtMRP5. We found that AtMRP5 heterologously expressed in HEK293 cells bound sulfonylurea with high affinity. An AtMRP5 knockout mutant (atmrp5-2) was isolated and found to be hypersensitive to salt stress. This mutant phenotype was mimicked by wild-type plants when they were treated with the MRP inhibitor glibenclamide in the presence of high salt concentrations. The atmrp5-2 mutant also displayed defects in root growth, possibly because of reduced K+ uptake and K+ accumulation. These observations suggest that AtMRP5 plays novel functions in regulating K+ uptake and ion homeostasis under salt stress and is a sulfonylurea receptor protein that functions in the inhibition of root growth because of salt stress. Our results strongly support the hypothesis that AtMRP5 acts as an ion channel regulator in root growth. This function of AtMRP5 also has been proposed previously to regulate guard cell activity (Gaedeke et al., 2001; Klein et al., 2003).
RESULTS
Expression of AtMRP5 in HEK293 Cells and Specific Binding of [3H]Glibenclamide
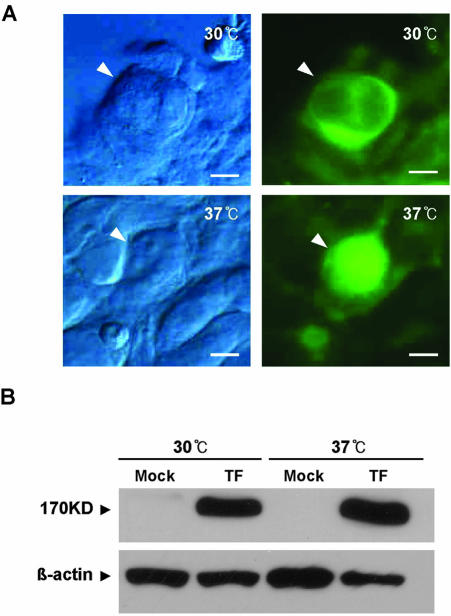
To determine the biochemical activity of AtMRP5 in a heterologous expression system, full-length AtMRP5 cDNA (GenBank/EMBL accession no. Y11250) fused to green fluorescence protein (GFP; AtMRP5:GFP) was transiently expressed in HEK293 cells using the pcDNA3.1 mammalian expression vector. The transfection efficiency was assessed with a β-galactosidase assay, which demonstrated that approximately 90% cells were transfected. Strong GFP signals were observed in the plasma membrane when transfected cells were grown at 30°C (Fig. 1A). However, when the cells were grown at 37°C, GFP was found in the cytoplasm (Fig. 1A), even though the transfected cells grown at each temperature expressed approximately the same amount of the AtMRP:GFP fusion protein (Fig. 1B). This result indicated that the heterologous expression system formed a basis for the binding studies.
Figure 1.
Cellular localization of the AtMRP5:GFP fusion protein in HEK293 cells. A, HEK293 cells were transfected with the AtMRP5: GFP fusion protein and incubated at 30°C or 37°C. Cells were fixed and examined by light microscopy (left) and fluorescence microscopy (right). At 30°C, green fluorescence was observed throughout the cell and close to or at the plasma membrane, whereas at 37°C, the protein was clearly blocked at the ER. B, Immunoblot analysis of AtMRP5:GFP in HEK293 cells. The AtMRP5:GFP fusion proteins were extracted from cells grown at 30°C and 37°C. Protein (50 μg) was electrophoresed on a 6% (w/v) SDS-polyacrylamide gel, electroeluted onto filters, and probed first with living color peptide A.v. antibody and then with donkey anti-rabbit IgG-horseradish peroxidase. Filters were then washed, and antibody binding was visualized with ECL. Mock, Mock transfected with vector alone; TF, transfected with AtMRP5:GFP fusion construct.
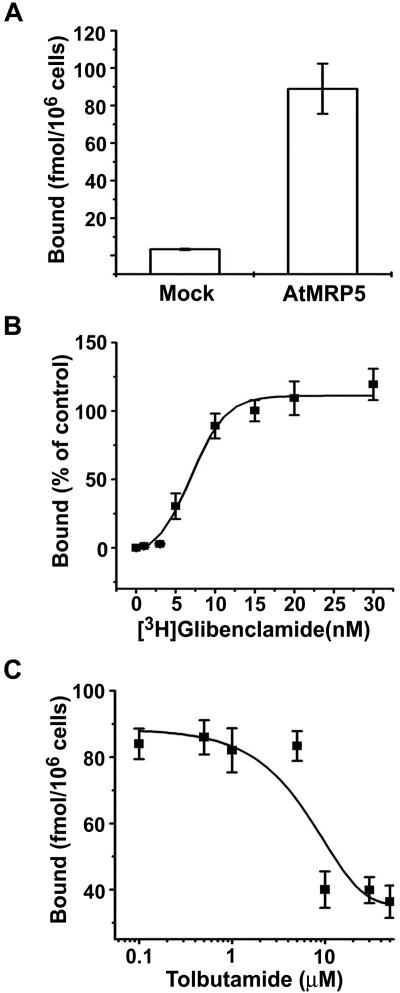
The expression of AtMRP5 in HEK293 cells was accompanied by a dramatic increase in specific binding of [3H]glibenclamide (Fig. 2A). The specific binding was determined by subtracting the nonspecific binding measured in the presence of 1 μm unlabeled glibenclamide. HEK293 cells expressing GFP alone as a control did not show any significant specific binding activity (n = 2). The mean Kd value for binding to whole cells was 7.2 ± 1.3 nm (n = 3; Fig. 2B), which is comparable with that of native β-cells (range of 0.3-7 nm; Ashcroft and Ashcroft, 1992; Ämmälä et al., 1996). The mean maximum binding capacity was 190 ± 60 fmol per 106 cells (n = 3). The affinity for another sulfonylurea drug, tolbutamide, was lower because the inhibition coefficient (Ki) for the displacement of [3H]glibenclamide binding by tolbutamide was 28 ± 11 μm (n = 3; Fig. 2C). Similar values are reported for native β-cells (Ashcroft and Ashcroft, 1992). Thus, At-MRP5 expressed in HEK293 cells specifically binds glibenclamide.
Figure 2.
Sulfonylurea-binding activity of AtMRP5 expressed in HEK293 cells. A, [3H]glibenclamide binding (fmol per 106 cells) to whole HEK293 cells that have been mock transfected (n = 3) or transfected with AtMRP5 (n = 3) was measured 48 h after transfection. Mock, As in Figure 1A. B, Dissociation constant (Kd) value of [3H]glibenclamide binding to whole HEK293 cells transfected with AtMRP5. The amount of specific [3H]glibenclamide bound (fmol per 106 cells) was plotted against the free [3H]glibenclamide concentration. Specific binding was determined by subtracting nonspecific binding measured in the presence of 1 μm unlabeled glibenclamide. Each point indicates the mean of three replicates. The line represents the best fit of a single-binding site model to the data. C, Inhibition of [3H]glibenclamide binding by tolbutamide. This was measured in the presence of 10 nm [3H]glibenclamide.
Isolation of a T-DNA-Tagged Plant with a Disrupted AtMRP5 Gene
The function of AtMRP5 in vivo was analyzed by isolating a plant with a disruption in this gene caused by T-DNA insertion. The T-DNA insertional knockout mutant was isolated by using a reverse genetics approach described by Krysan et al. (1996). From a population (provided by Arabidopsis Biological Resource Center) of 12,940 different T-DNA lines that contains about 18,000 independent insertional events, we identified and isolated a single mutant plant with a T-DNA insertion in AtMRP5 by PCR using primers corresponding to the left and right borders of T-DNA and the AtMRP5 gene, respectively (Krysan et al., 1996). This mutant was designated atmrp5-2 to indicate that it is different from the AtMRP5 mutant resulting from T-DNA insertion that was isolated by Gaedeke et al. (2001). The position of the disrupting T-DNA in the AtMRP5 gene was determined by sequencing PCR-amplified fragments. Several lines of evidence described below suggest that the mutant is homozygous for the T-DNA insertion and carries a T-DNA insertion at a single insertion locus. First, all PCRs with a combination of primers specific for AtMRP5 and T-DNA that were performed using genomic DNA isolated from >10 individuals of the offspring of the mutant plant resulted in PCR products of the expected size (data not shown). In contrast, all PCRs performed with the AtMRP5 forward/reverse primer combination did not yield fragment amplification. This is expected because a single T-DNA element is approximately 14 kb in length, most insertions contain multimers of the repeat unit, and the PCR conditions used in this study did not allow for the formation of such large products (Feldman, 1991). Second, on selective medium, the F2 generation of the AtMRP5-2/Ws-0 backcross segregated in a ratio of approximately 3:1 for the kanamycin marker (resistant:sensitive = 78:28). This result suggests that the T-DNA insertion is inherited in a Mendelian manner and indicates a single T-DNA insertion in the AtMRP5 gene. Third, Southern-blot analysis using the DNA isolated from atmrp5-2, the progeny of atmrp5-2/Ws-0, and wild-type plants indicated that a probe specific for T-DNA left border sequences hybridized only in atmrp5-2 and the atmrp5-2/Ws-0 heterozygous plants (data not shown). Thus, atmrp5-2 plants carry a single T-DNA insertion in the AtMRP5 gene.
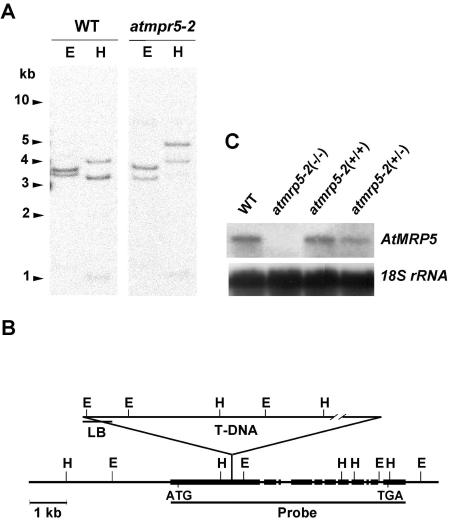
Southern-blot analyses of Arabidopsis DNA revealed that AtMRP5 is encoded by a single gene (Fig. 3A). The AtMRP5 locus is localized on chromosome 1 (between cer1 and axr1 [0846A marker]; bacterial artificial chromosome clone no. F20D22; GenBank/EMBL accession no. Y11250). Southern blotting of genomic DNA from both wild-type and atmrp5-2 mutant plants using radiolabeled DNA corresponding to the AtMRP5 coding region revealed a T-DNA insertion within the first exon of the coding region (Fig. 3B). Sequence analysis revealed that the T-DNA insertion site was 1,629 bp downstream of the start codon. Northern blotting using the 3′-half of AtMRP5 cDNA (3.2 Kb) as a probe revealed no detectable AtMRP5 mRNA in the atmrp5-2 mutant (Fig. 3C). This suggests that the T-DNA insertional knockout mutants cannot generate a functional stable transcript. With regard to the allele denoted as atmrp5-1 that was isolated by Gaedeke et al. (2001), who used an early collection of 4,120 Feldmann's T-DNA-transformed lines (Forsthoefel et al., 1992), the T-DNA insertion in the AtMRP5 locus of this mutant occurred at position +1,471.
Figure 3.
Analysis of the AtMRP5 gene in wild-type and atmrp5-2 plants and measurement of steady-state AtMRP5 transcript levels in wild-type and atmrp5-2 plants. A, Southern-blot analysis of genomic DNA from wild-type and atmrp5-2 mutant Arabidopsis digested with EcoRI (lane E) or HindIII (lane H) that was hybridized with radiolabeled DNA corresponding to the AtMRP5 coding region. B, Restriction map of the AtMRP5 genomic DNA of the atmrp5-2 mutant based on Southern analysis (A) and the location of the T-DNA insertion in the AtMRP5 gene. Boxes, Exons. T-DNA was inserted into the first exon between the HindIII and EcoRI sites. C, Steady-state levels of AtMRP5 transcripts in wild-type plants and homozygous and heterozygous atmrp5-2 mutant plants.
atmrp5-2 Plants Are Hypersensitive to Na+ But Not Mannitol Stress
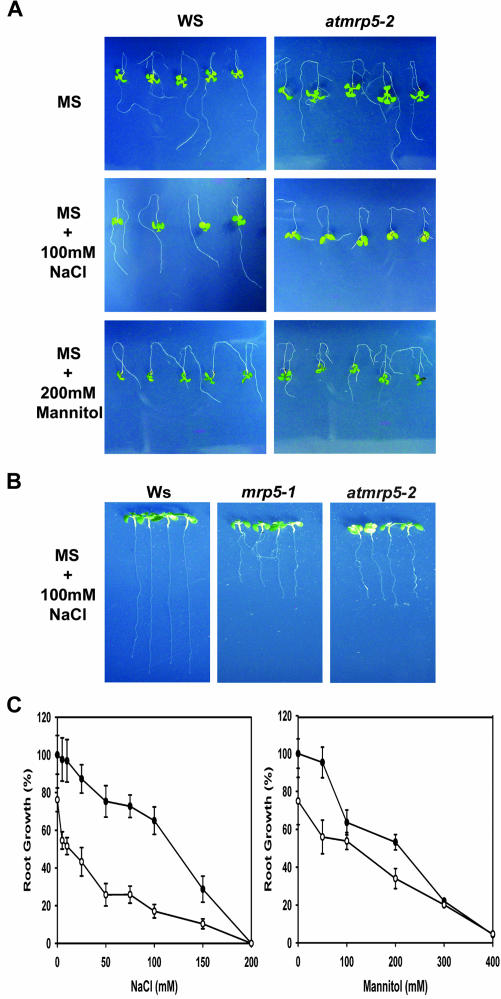
When we used a AtMRP5 2.4-kb promoter-GUS fusion construct, we found that AtMRP5 is mainly expressed in the roots (data not shown). This was also shown previously by Gaedeke et al. (2001). Then, we measured the root growth of wild-type and atmrp5-2 seedlings by using a root-bending assay that had been described previously by Zhu and colleagues (Wu et al., 1996). Both atmrp5-2 mutant and wild-type plants grew relatively well and appeared healthy on control Murashige and Skoog (1×) medium that lacked added NaCl (Fig. 4A, upper row). With full-strength Murashige and Skoog medium supplemented with 100 mm NaCl, atmrp5-2 growth was substantially inhibited, in contrast to the essentially normal growth of wild-type plants (Fig. 4A, middle row). The inhibition of root growth by NaCl stress occurred in a concentration-dependent manner, with IC50 values of approximately 100 mm for the wild-type and 40 mm for the atmrp5-2 mutant (Fig. 4C). The atmrp5-2 seedlings were not significantly hypersensitive to the osmotic stress caused by mannitol, with an observed I50 value of approximately 200 mm for both wild-type and atmrp5-2 mutant seedlings (Fig. 4, A, bottom row, and C). When wild-type and atmrp5-2 seedlings were treated with Murashige and Skoog supplemented with various concentrations of Li+ (a toxic cation closely related to Na+), the atmrp5-2 mutant displayed hypersensitivity to Li+ (data not shown). Interestingly, atmrp5-2 was not hypersensitive to stress because of Cs+ (also a toxic cation related to Na+), unlike the wild-type plant (data not shown). These observations collectively indicate that the atmrp5-2 mutation does not cause a defective osmotic stress response. Rather, the defect is restricted to Na+ tolerance.
Figure 4.
Salt stress sensitivity of wild-type and atmrp5-2 plants grown on vertical plates. Root-bending assays were performed as described in “Materials and Methods.” Four-day-old seedlings were transferred from Murashige and Skoog medium (1×) to Murashige and Skoog media containing 0.1 m NaCl or 0.2 m mannitol, and seedlings were allowed to grow for 7 d. A, Root growth of wild-type (left) and atmrp5-2 (right) plants in Murashige and Skoog medium supplemented with 100 mm NaCl or 200 mm mannitol. B, Root growth of wild-type plants and the atmrp5-1 and atmrp5-2 mutant plants in Murashige and Skoog medium were supplemented with 100 mm NaCl. C, IC50 values showing the tolerance of wild-type and atmrp5-2 plants to NaCl and mannitol. Black circle, Wild type; white circle, atmrp5-2.
To confirm that atmrp5-2 is another allele of the AtMRP5 locus, along with mrp5-1, we compared the salt-hypersensitive phenotypes of atmrp5-1 (kindly provided by Dr. Markus Klein, University of Zurich) and atmrp5-2. As shown in Figure 4B, the seedlings of both mutant plants displayed equivalent reduced root growth in full-strength Murashige and Skoog medium that had been supplemented with 100 mm NaCl compared with their root growth in unaltered Murashige and Skoog medium. In addition to this salt-hypersensitive phenotype, we also observed that the atmrp5-2 mutant showed reduced root growth of modified 0.25× Murashige and Skoog medium. That atmrp5-1 plants also show this phenotype has been described previously (Gaedeke et al., 2001; data not shown). These results demonstrate that atmrp5-2 is the second allele of AtMRP5 locus. In addition, they show that the salt-hypersensitive phenotype of atmrp5-2 is because of the T-DNA disruption of At-MRP5 gene, indicating that mutation of the AtMRP5 gene confers a salt-hypersensitive phenotype.
Glibenclamide Mimics the Effect of the Mutation in AtMRP5 during Na+ Stress
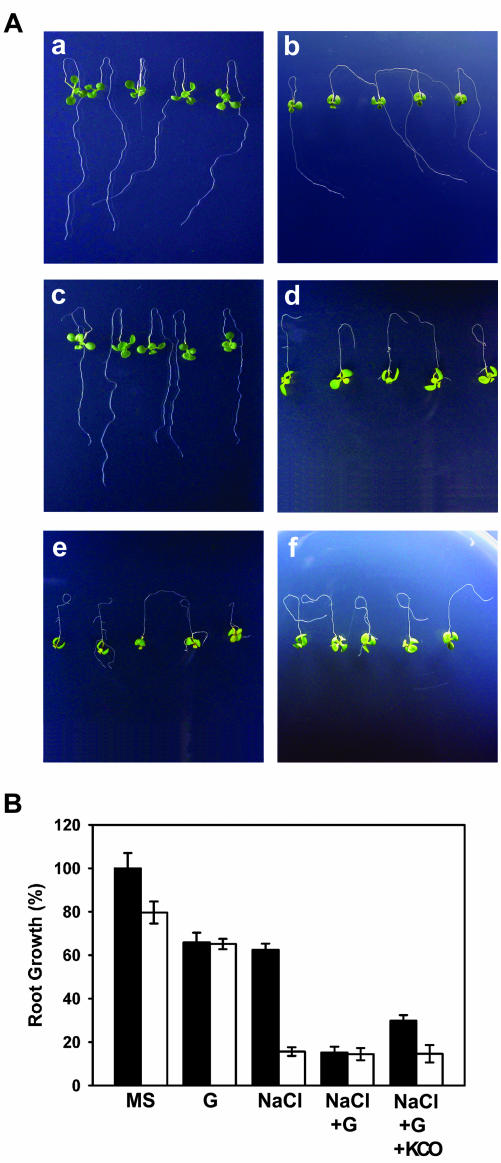
To examine whether modulation of AtMRP5 function with a sulfonylurea compound mimics the effect of the atmrp5-2 mutation during salt stress, wild-type seedlings were treated with 10 μm glibenclamide during Na+ stress, and the root growth phenotypes were compared with atmrp5-2. As shown in Figure 5, wild-type seedlings displayed a decrease in root length of approximately 25% in the presence of 100 mm Na+, as was also shown previously (Fig. 4). When wild-type seedlings were treated with 10 μm glibenclamide in conjunction with the same Na+ stress (100 mm), root growth was further reduced to a considerable degree that was comparable with that observed in atmrp5-2 (Fig. 5, A and B). When the wild-type seedlings were treated with 100 μm diazoxide (a well-known K+ channel opener, KCO) in addition to 10 μm glibenclamide and Na+ stress, there was a small but significant restoration of the Na+- and glibenclamide-induced inhibition of root growth (Fig. 5, A and B). In the presence of either Na+ plus glibenclamide or Na+ plus glibenclamide plus KCO, atmrp5-2 did not display further inhibition in root growth compared with that in Na+ alone. These results suggest that of the 15 AtMRPs in Arabidopsis, AtMRP5 may be responsible for the sensitivity of wild-type root growth to glibenclamide and diazoxide during salt stress.
Figure 5.
Chemical phenocopying with glibenclamide of the effect of the atmrp5-2 mutation during salt stress. Root-bending assays were performed as described in “Materials and Methods.” A, Root growth of wild-type and atmrp5-2 plants. a, Wild-type plants in Murashige and Skoog medium. b. Wild-type plants in Murashige and Skoog medium supplemented with 100 mm NaCl. c, atmrp5-2 plants in Murashige and Skoog medium. d, atmrp5-2 plants in Murashige and Skoog medium supplemented with 100 mm NaCl. e, Wild-type plants in Murashige and Skoog medium supplemented with 100 mm NaCl plus 10 μm glibenclamide. F, Wild-type plants in Murashige and Skoog medium supplemented with 100 mm NaCl plus 10 μm glibenclamide and 100 μm diazoxide. B, Quantitation of the root growth of the wild-type and atmrp5-2 plants shown in A.
AtMRP5 Is Involved in K+ Uptake
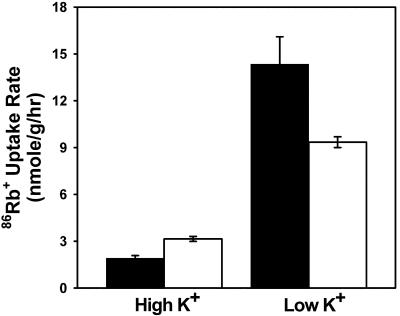
We investigated whether atmrp5-2 is defective in ion homeostasis. To determine the effect of the mutation in the AtMRP5 gene on K+ absorption by the roots, we performed 86Rb+ tracer flux analyses on roots obtained from plants grown in high and low concentrations of external K+ (Murashige and Skoog salts and modified Murashige and Skoog salts containing 100 μm K+, respectively). When plants were grown in media containing high external K+ (Fig. 6, black bars), 86Rb+ uptake rates of wild-type and atmrp5-2 plants were almost identical and low compared with those of seedlings grown in low K+. However, when plants were grown in media containing low external K+ (Fig. 6, white bars), the atmrp5-2 mutant displayed considerably lower 86Rb+ uptake than its wild-type counterpart.
Figure 6.
Radioactive tracer flux analysis of Rb+ uptake in atmrp5-2 and wild-type roots. Seedlings were grown in normal Murashige and Skoog medium on vertical plates for 4 d, transferred to either normal or modified Murashige and Skoog medium containing 100 μm KCl and allowed to grow for 3 d. 86Rb+ uptake assays were performed in 1/20-strength major Murashige and Skoog salts with normal amounts of micronutrients supplemented with 100 μm KCl and 0.5 μCi mL-1 86Rb+. Black bars, seedlings grown in normal Murashige and Skoog medium for 4 d, transferred to the same medium, and grown for 3 d. White bars, Seedlings grown in normal Murashige and Skoog medium for 4 d and transferred to modified Murashige and Skoog medium with 100 μm KCl and grown for 3 d.
The K+ and Na+ contents of wild-type and atmrp5-2 seedlings were measured after being grown in the absence or presence of 100 mm NaCl for 7 d. Although there was approximately 37% decrease in K+ content in atmrp5-2 seedlings in the absence of 100 mm NaCl, atmrp5-2 seedlings treated with NaCl were found to have even lower K+ than that of the wild type, which is approximately 27% of that of the wild type in the absence of NaCl (Table I). Na+ content in atmrp5-2 seedlings treated with 100 mm NaCl was dramatically higher than those in the wild type either with or without NaCl treatment and atmrp5-2 without NaCl treatment. The results indicate that salt sensitivity of the atmrp5-2 mutant is correlated with their cellular Na+ content.
Table I.
K+ and Na+ contents in wild-type and atmrp5-2 seedlings treated with 0 or 100 mm NaCl
Data represent mean + SD (n = 3).
| K+ Content (% Decrease)
|
Na+ Content (% Increase)
|
Na+:K+ Ratio (% Increase)
|
||||
|---|---|---|---|---|---|---|
| 0 mm NaCl | 100 mm NaCl | 0 mm NaCl | 100 mm NaCl | 0 mm NaCl | 100 mm NaCl | |
| mg g dry wt−1 | ||||||
| WT | 55.2 ± 2.1 | 38.0 ± 1.6 (−31.2%) | 0.4 ± 0.0 | 3.9 ± 0.1 (+975%) | 0.007 | 0.103 (+1471%) |
| atmrp5-2 | 34.7 ± 0.4 | 14.8 ± 0.2 (−57.4%) | 0.3 ± 0.0 | 13.7 ± 0.2 (+4566%) | 0.008 | 0.922 (+11525%) |
The atmrp5-2 Plants Require Higher Concentrations of External K+ to Counteract Salt Stress
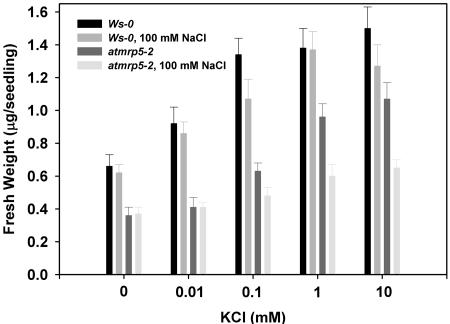
We directly tested the K+ sensitivity with respect to salt stress response by growing plants over a range of K+ levels in the absence or presence of 100 mm NaCl in the medium (Fig. 7). When we compared growth of wild-type and atmrp5-2 plants by measuring fresh weights, which is another valid parameter for seedling growth (Spalding et al., 1999), in the absence of NaCl stress, both plants displayed the typical K+-dependent growth, although atmrp5-2 grew poorly on medium containing less than 100 μm. In the conditions used here, increasing K+ increases growth rate of wild-type seedlings, less so in the mutant. The weaker dependence on K+ of atmrp5-2 growth is consistent with its impaired K+ uptake. NaCl (100 mm) impairs this K+-dependent growth in atmrp5-2 but not in the wild type, possibly accounting for the phenotypic differences in Figure 4.
Figure 7.
Effect of various extracellular concentrations of KCl on growth of wild-type and atmrp5-2 plants. Seedlings were grown in K+-free modified Murashige and Skoog medium supplemented with indicated concentrations of external KCl in the absence or presence of 100 mm NaCl for 9 d. Fresh weights were measured from 25 seedlings and the fresh weight of individual seedling was calculated. Values are the averages of three independent experiments (bars = se [n = 3]).
DISCUSSION
AtMRP5 promoter-GUS studies have revealed that strong AtMRP5 promoter activity is found in the vascular tissues of cotyledons and leaves, including in guard cells (Gaedeke et al., 2001). When we established transgenic plants expressing a 2.4-kb AtMRP5 promoter-GUS fusion construct, we found strong GUS expression in the elongation regions of the roots and in the vascular bundles of the sepals but no expression in other floral organs (data not shown). Interestingly, strong GUS expression was also noted in the pollen (data not shown), which is consistent with the observation that the major determinants of pollen-specific gene expression, namely, cis-acting AGAAA and TCCACCATA elements (Bate and Twell, 1998), are located in the AtMRP5 promoter. However, we did not detect any differences in pollen germination and pollen tube growth between the wild-type plants and the atmrp5-2 mutant using assays previously described by Mouline et al. (2002; data not shown).
The subcellular localization of AtMRP5 in a plant cell has not been determined yet. AtMRP5 seemed to be localized in the vacuolar membrane (Gaedeke et al., 2001). This would fit with the putative function of AtMRP5 as an auxin-conjugate transporter, as has also been postulated by Luschnig (2002). It was also assumed that AtMRP5 was localized in the plasma membrane of guard cells, and it was hypothesized that AtMRP5 may thus act as an ion channel (Klein et al., 2003), as does the mammalian CFTR (Schultz et al., 1996; Akabas, 2000), or, alternatively, it acts as an ion channel regulator, as has been suggested for SUR in animal cells (Schmid-Antomarchi et al., 1987; Babenko et al., 1998; Seino and Miki, 2003). This lack of clarity about the localization of AtMRP5 argues the need for careful and extensive localization studies because this information will help determine the role AtMRP5 plays in plant cells.
In mammalian cells, the response of KATP channel subtypes to sulfonylurea correlates well with the affinity of binding and labeling of receptors. For example, channels reconstituted with SUR1 exhibit IC50 values in the nanomolar range for channel inhibition by glibenclamide, whereas the SUR2A channels typically require a 100-fold higher concentration to reach the same effect (Inagaki et al., 1995, 1996). Biochemical data show that SUR1 binds glibenclamide with a Kd of approximately 1 nm, whereas SUR2A has a Kd value near 1.2 μm (Inagaki et al., 1995, 1996). We found that AtMRP5 expressed in HEK293 cells specifically binds [3H]glibenclamide (Fig. 4). The mean Kd for binding to whole cells was 7.2 ± 1.3 nm (n = 3; Fig. 4B), which is comparable with that observed for native β-cells (range of 0.3-7 nm; Ashcroft and Ashcroft, 1992; Ämmälä et al., 1996). This high affinity of AtMRP5 for glibenclamide suggests that At-MRP5 may represent an SUR1 subtype-like protein in Arabidopsis. At present, it cannot be excluded that other AtMRPs can also bind to glibenclamide with similar affinity when expressed in a heterologous system.
The mrp5-1 mutant was shown to be impaired in the glibenclamide sensitivity of its stomatal movements (Gaedeke et al., 2001). It was also insensitive to several, although not all, modulators such as abscisic acid, Ca2+, and auxin (Klein et al., 2003). As a consequence, Martinoia and colleagues (Gaedeke et al., 2001; Klein et al., 2003) have discussed extensively the possibility that AtMRP5 acts as an ion channel or an ion channel regulator in signaling pathways that involve stomatal movement. Given this debate, additional evidence that supports the notion that AtMRP5 serves as an ion channel or an ion channel regulator would be useful.
In the present study, we found evidence that At-MRP5 is involved in K+ uptake and salt stress tolerance. Particularly revealing was the observation that the root growth of wild-type plants grown in NaCl plus glibenclamide was inhibited to a similar extent as the root growth of the atmrp5-2 mutant grown in the presence of NaCl alone. We speculate that the AtMRP5 protein on the root cell surface acts as a glibenclamide receptor and that the binding of glibenclamide to this receptor results in its conformational change, which affects the downstream signaling pathway. This would explain why the effect of glibenclamide treatment on wild-type plants grown in high-salt concentrations can be mimicked by the atmrp5-2 mutation because this mutation removes the putative target of the drug. Supporting this notion is that when atmrp5-2 plants were grown in high-salt concentrations plus glibenclamide, further inhibition of root growth relative to that of atmrp5-2 plants grown in NaCl alone was not observed. These observations suggest that AtMRP5, of the 15 different AtMRP genes in the Arabidopsis genome (Sánchez-Fernández et al., 2001), may be responsible for most of the glibenclamide-sensitive inhibition of root growth under NaCl stress.
Observations suggest the following model of the involvement of AtMRP5 in K+ uptake: The mutation in the AtMRP5 gene causes a defect in the K+ inwardly rectifying current (which has been shown to follow the K+ equilibrium potential in animal cells), which results in a membrane potential setting that is insufficient for K+ uptake. It will be interesting to test if there is any change in the resting membrane potential in response to low extracellular K+ or to NaCl stress in the root cells of atmrp5-2 plants compared with that of wild-type plants.
The phenotypic characterization of the atmrp5-2 mutant using various concentrations of K+ and salts in the growth medium showed that this mutant is strikingly similar to sos mutants in many aspects. First, the atmrp5-2 mutant grows poorly on modified Murashige and Skoog medium containing less than 100 μm K+ (Fig. 7). Similar growth phenotypes were observed with sos1, sos2,and sos3 (Liu and Zhu, 1997, 1998). Second, experiments using 86Rb+ demonstrated that atmrp5-2 seedlings have a reduced capacity for K+ uptake (Fig. 6). Similar reduced K+ uptake was observed in sos1, although not in the sos2 and sos3 mutants (Ding and Zhu, 1997). Third, the steady-state levels of the AtMRP5 and SOS1 transcripts are up-regulated by NaCl stress (data not shown; Shi et al., 2000). Fourth, atmrp5-2 seedlings treated with NaCl had decreased K+ and increased Na+ contents (Table I), as sos3-1 seedlings did (Zhu et al., 1998). These similarities suggest that AtMRP5 functions in the same or similar processes that are regulated by SOS1, SOS2, and SOS3 during salinity stress. Thus, it will be interesting to test whether there are any genetic or biochemical interactions between SOS1, 2, or 3 and AtMRP5 in the salt stress response.
MATERIALS AND METHODS
Plant Growth Conditions and the Root-Bending Assay
Wild-type (Wassilewskija ecotype) and atmrp5-2 mutant seeds were surface sterilized and grown on agar medium containing Murashige and Skoog salts (Murashige and Skoog, 1962) with 1% (w/v) Suc and 1% (w/v) agar (pH 5.7). The Murashige and Skoog medium was composed of 1,650 mg L-1 NH4NO3, 1,900 mg L-1 KNO3, 180.54 mg L-1 MgSO4, 170 mg L-1 KH2PO4, 332 mg L-1 CaCl2, 36.7 mg L-1 FeNaEDTA, 0.83 mg L-1 KI, 6.2 mg L-1 H3BO4, 16.9 mg L-1 MnSO4·H2O, 8.6 mg L-1 ZnSO4·7H2O, 0.25 mg L-1 Na2Mo4·2H2O, 0.025 mg L-1 CuSO4·5H2O, and 0.025 mg L-1 CoSO4·6H2O. The modified Murashige and Skoog medium that is free of potassium contains the same ingredients except that KNO3, K2HPO4, and KI were omitted, and 165 mg L-1 (NH4)2HPO4 was added. Different concentrations of potassium were supplemented by adding KCl. For salt and drug treatments, Murashige and Skoog media were supplemented with various concentrations of NaCl, LiCl, mannitol, glibenclamide, and/or diazoxide. Glibenclamide and diazoxide were first dissolved in dimethyl sulfoxide. The final concentration of dimethyl sulfoxide in the incubation solution never exceeded 0.1% (w/v) in these experiments and had no effect on root growth. After 48 h at 4°C to synchronize germination, plates were placed vertically in a growth chamber set to deliver 16 h of light and 8 h of dark at 22°C.
To measure root growth, the root-bending assay was used as described previously by Wu et al. (1996). In brief, 4-d-old seedlings grown in full-strength Murashige and Skoog medium in vertical plates were transferred to Murashige and Skoog agar plates supplemented with various salts or drugs. Treatment plates were placed vertically with seedlings in the upright position. After 7 d, images were captured, and root length was measured using image analysis software (Scion Image 4.02, Scion Corp., Frederick, MD). Three replicates of five seedlings were grown with each treatment.
Expression of Full-Length AtMRP5 cDNA and AtMRP5: GFP Fusion Protein in Cultured HEK293 Cells
HEK293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% (w/v) fetal bovine serum, 100 international units mL-1 penicillin, and 200 international units mL-1 streptomycin at 37°C in humidified 5% (v/v) CO2. Cells were transiently transfected using LipofectAMINE Reagent (Invitrogen, Carlsbad, CA) and mixed in serum-free medium with an expression vector (pcDNA3.1, Invitrogen, Carlsbad, CA) containing AtMRP5 cDNA. Transfection was carried out according to the manufacturer's instructions. Mock-transfected cells received no DNA but were otherwise treated identically. The transfection efficiency was assessed by using a β-galactosidase assay. Cells were allowed to express the transfected DNA for 48 h at 30°C or 37°C and were subsequently used in binding studies and fluorescence image analyses.
The AtMRP5 cDNA fragments used to construct a GFP chimera were subcloned into the pEGFP-N1 (CLONTECH Laboratories, Palo Alto, CA) vector. Enhanced GFP was attached to the carboxyl terminus of AtMRP5 using standard recombinant techniques. In brief, oligonucleotide primers were synthesized that would allow the complete amplification of the coding regions of AtMRP5, and a PCR was performed using the wild-type sequence as the template. The final PCR product was subcloned into the pEGFP-N1 vector so that AtMRP5 was in-frame with GFP, and a three-amino acid linker region was inserted between the C terminus of AtMRP5 and the coding region of GFP. The construct was fully sequenced before it was expressed and used for analysis.
For GFP fluorescence image analyses, cells were grown on glass coverslips and mounted on the imaging chamber. Cells on glass coverslips were washed with phosphate-buffered saline (PBS) and fixed for 20 min at room temperature with 4% (w/v) paraformaldehyde and 5% (w/v) sucrose in PBS (pH 7.2). After a single wash with PBS, coverslips were mounted on microscope slides with 40% (w/v) glycerol in PBS. The distribution of the AtMRP5-GFP chimeric protein was investigated by fluorescence microscopy using an Axioplan microscope (Carl Zeiss, Jena, Germany).
For immunoblot analyses of the AtMRP5:GFP chimeric protein, extracts from cells grown at 30°C or 37°C were prepared in SDS-PAGE Laemmli sample buffer by cell lysis using a 23-gauge syringe. Protein (50 μg) was electrophoresed on a 6% (w/v) SDS-polyacrylamide gel and electroeluted onto PVDF Immobilon filters (Millipore Corporation, Bedford, MA). After staining with 0.05% (w/v) Ponceau S, the filters were blocked for 1 h in phosphate-buffered saline plus Tween 20 (PBST) containing 5% (w/v) nonfat milk, probed with Living color peptide A.v. antibody (1:100 dilution; CLONTECH Laboratories) at room temperature for 1 h, then washed with PBST and incubated with donkey anti-rabbit IgG-horseradish peroxidase (1:2,000 dilution) at room temperature for 1 h. Next, the filters were washed four times with PBST, and antibody binding was visualized with an enhanced chemiluminescence kit (ECL System, Amersham Biosciences, Piscataway, NJ).
Measurement of Sulfonylurea-Binding Activity of AtMRP5 Using an Equilibrium Competition Experiment
The specific binding of the sulfonylurea drug glibenclamide to HEK293 cells expressing AtMRP5 was measured as described by Ämmälä et al. (1996). In brief, transiently transfected HEK293 cells were harvested 48 h after transfection at approximately 80% confluence. The cells were suspended by rinsing with HEPES-Krebs buffer containing 119 mm NaCl, 4.75 mm KCl, 5 mm NaHCO3, 2.54 mm CaCl2, 1.2 mm MgSO4, 1.2 mm KH2PO4, and 20 mm HEPES (pH 7.4 with NaOH) at room temperature. After centrifuging twice at 500g for 5 min, the cells were resuspended in HEPES-Krebs buffer. Next, the cells were incubated for 1 h at room temperature with 10 nm [3H]glibenclamide (New England Nuclear Corporation, Boston, MA) at a density of 2 × 106 cells per assay in 0.5 mL of HEPES-Krebs buffer in the presence of 1 μm unlabeled glibenclamide. In homologous competition assays, various concentrations of unlabeled glibenclamide were used. Bound radioactivity was separated from free radioactive material by rapid filtration under a vacuum over GF/B filters (Whatman, Inc., Clifton, NJ) soaked in HEPES-Krebs buffer. Filters were washed five times with 5 mL of the same buffer at 4°C and counted for 3H in the presence of 3 mL of scintillation fluid (Ready Safe, Beckman Instruments, Inc., Chantilly, VA). Specific binding was determined by subtracting the nonspecific binding measured in the presence of 1 μm unlabeled glibenclamide.
Isolation of a T-DNA-Tagged Plant with a Disrupted AtMRP5 Gene
A T-DNA mutagenized population of Arabidopsis was screened for plants that contain an insertional mutation in the AtMRP5 gene by using the PCR-based reverse genetic method of Krysan et al. (1996). In brief, seeds of T-DNA insertion mutant pools (provided by the Arabidopsis Biological Resource Center; consists of 12,940 independent lines in total, of which 6,500 are in pools of l00 lines [65 pools] and 6,440 are in pools of 20 lines [322 pools]) were grown, and genomic DNA from each pool was extracted. The AtMRP5-specific primers employed were 5′-ctctcgaggcttctagattgttacatcatctccttaa-3′ (forward) and 5′-ctctcgaggataaacatgaaaaccaaagaaactaact-3′ (reverse). Using the AtMRP5-specific forward primer and a T-DNA left border primer (5′-gatgcactcgaaatcagccaattttagac-3′), one positive line was detected. The corresponding plant was selected, and the position of the disrupting T-DNA in the AtMRP5 gene was determined by sequencing PCR-amplified fragments. To ensure that the physiological features observed in the plant carrying the T-DNA are genetically linked to the AtMRP5 disruption, the plants that were homozygous for the disruption were backcrossed with the Wassilewskija ecotype, and further studies were performed on the F2 progeny that are homozygous for the disruption.
Potassium Uptake Assays Using 86Rb+ as a Radioactive Tracer
To measure potassium uptake using 86Rb+ as a tracer, we used the method of Wu et al. (1996), with slight modifications. In brief, 4-d-old seedlings from vertical Murashige and Skoog agar plates were transferred to vertical agar plates containing either normal Murashige and Skoog salt or modified Murashige and Skoog salt with 100 μm potassium. After 3 d, 40 seedlings were collected, rinsed briefly in solution A as described below, and added to 5 mL of solution A supplemented with 100 μm KCl and 0.5 μCi mL-1 86Rb+ (New England Nuclear). The uptake assay was performed for 60 min at room temperature under a white fluorescent light. After the completion of uptake, seedlings were rinsed twice (15 s each) in 15 mL of ice-cold solution A plus 3 mm CaCl2. The seedlings were then blotted dry on filter paper, weighed, and the radioactivity was measured by detection of Cerenkov radiation. Solution A consists of potassium-free 1/20-strength Murashige and Skoog major salts with the normal amounts of minor nutrients. The solution was composed of 82.5 mg L-1 NH4NO3, 22 mg L-1 CaCl2·2H2O, 18.5 mg L-1 MgSO4·7H2O, 7.2 mg L-1 NH4H2PO4, 1.39 mg L-1 FeSO4·7H2O, 1.865 mg L-1 disodium EDTA, 0.7495 mg L-1 NaI, 6.3 mg L-1 H3BO3, 16.9 mg L-1 MnSO4·H2O, 8.6 mg L-1 ZnSO4·7H2O, 0.25 mg L-1 Na2Mo4·2H2O, 0.0016 mg L-1 CuSO4.5H2O, and 0.0267 mg L-1 CoSO4.6H2O (pH 5.7).
Determination of Ion Contents
Plants were cultured in liquid 1× Murashige and Skoog medium (pH 5.6) containing 100 mm NaCl or not, with shaking at 120 rpm for 7 d for whole seedling collection. Plants in hydroponic culture were grown in a growth chamber with a 16-h-light/8-h-dark (22°C) cycle at 75% relative humidity. Plant materials were collected, briefly rinsed three times with deionized water, and dried in a 80°C oven for at least 2 d and weighed. The samples were digested with concentrated HNO3 overnight followed by boiling for approximately 1 h until the solution became completely clear. The K+ and Na+ contents in the solution were determined by atomic absorption spectrophotometer (model 3200A, Analab, Seoul).
Acknowledgments
We are grateful to Dr. Julian Schroeder (University of California, San Diego) for insightful discussion. We are grateful to the Arabidopsis Biological Resource Center for providing the Feldmann's T-DNA transformant collection. We also thank Seonghee Ahn (Department of Biology, Yonsei University, Korea) for excellent technical assistance in the construction of the genomic DNA pools from Feldman's T-DNA transformant collection.
This work was supported by the Korean Science and Engineering Foundation (grant no. 981-0510-050-2), by the Yonsei University Research Fund (grant no. 1998-1-0107), by the Plant Diversity Research Center of the 21st Frontier Research Program (grant code no. PF003201-03), and by the Crop Functional Genomics Center funded by the Ministry of Science and Technology of the Korean government (grant code no. CG-134).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.027045.
References
- Akabas MH (2000) Cystic fibrosis transmembrane conductance regulator-structure and function of an epithelia chloride channel. J Biol Chem 275: 3729-3732 [DOI] [PubMed] [Google Scholar]
- Ämmälä C, Moorhouse A, Gribble F, Ashfield R, Proks P, Smith PA, Sakura H, Coles B, Ashcroft SJH, Ashcroft FM (1996) Promiscuous coupling between the sulfhonylurea receptor and inwardly rectifying potassium channels. Nature 379: 545-548 [DOI] [PubMed] [Google Scholar]
- Anderson MP, Gregory RJ, Thompson S, Souza DW, Paul S, Mulligan RC, Smith AE, Welsh MJ (1991) Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science 253: 202-205 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 [DOI] [PubMed] [Google Scholar]
- Ashcroft SJH, Ashcroft FM (1992) The sulfonylurea receptor. Biochim Biophys Acta 1175: 45-59 [DOI] [PubMed] [Google Scholar]
- Babenko AP, Aguilar-Bryan L, Bryan J (1998) A view of SUR/Kir6. X, KATP channels. Annu Rev Physiol 60: 667-686 [DOI] [PubMed] [Google Scholar]
- Bate N, Twell D (1998) Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol Biol 37: 859-869 [DOI] [PubMed] [Google Scholar]
- Bear CE, Li C, Kartner N, Bridges RJ, Jensen TJ, Ramjeesingh M, Riordan JR (1992) Purification and functional reconstitution of the cystic fibrosis transmembrane conductance regulator (CFTR). Cell 68: 809-818 [DOI] [PubMed] [Google Scholar]
- Bohnert HJ, Nelson DE, Jensen RG (1995) Adaptations to environmental stresses. Plant Cell 7: 1099-1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA (1993) Molecular responses to water deficit. Plant Physiol 103: 1035-1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman JC, DeRocher EJ, Bohnert HJ (1990). Gene expression during adaptation to salt stress. In FJ Katerman, ed, Environmental Injury to Plants. Academic Press, New York, pp 173-203
- Davies TGE, Theodoulou FL, Hallahan DL, Forde BG (1997) Cloning and characterisation of a novel P-glycoprotein homologue from barley. Gene 199: 195-202 [DOI] [PubMed] [Google Scholar]
- Demolombe S, Escande D (1996) ATP-binding cassette proteins as targets for drug discovery. Trends Pharmacol Sci 17: 273-275 [DOI] [PubMed] [Google Scholar]
- Ding L, Zhu J-K (1997). Reduced Na+ uptake in the NaCl-hypersensitive sos1 mutant of Arabidopsis thaliana. Plant Physiol 113: 795-799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudler R, Hertig C (1992) Structure of an mdr-like gene from Arabidopsis thaliana. J Biol Chem 267: 5882-5888 [PubMed] [Google Scholar]
- Epstein E (1969) The essential role of calcium in selective cation transport by plant cells. Plant Physiol 36: 437-444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman KA (1991) T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. Plant J 1: 71-82 [Google Scholar]
- Forsthoefel NR, Wu Y, Schultz B, Bennett MJ, Feldmann KA (1992) T-DNA insertion mutagenesis in Arabidopsis: prospects and perspectives. Aust J Plant Physiol 19: 353-366 [Google Scholar]
- Gaedeke N, Klein M, Kolukisaoglu U, Forestier C, Müller A, Ansorge M, Becker D, Mammun Y, Kuchler K, Schultz B et al. (2001) The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J 20: 1875-1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Handa AK (1987) Cellular mechanisms of salinity tolerance. Hortic Sci 21: 1317-1324 [Google Scholar]
- Higgins CF (1992) ABC transporters: from microorganisms to man. Annu Rev Cell Biol 8: 67-113 [DOI] [PubMed] [Google Scholar]
- Higgins CF (1995) The ABC of channel regulation. Cell 82: 693-696 [DOI] [PubMed] [Google Scholar]
- Inagaki N, Gonoi T, Clement JP IV, Namba N, Inazawa J, Gonzalez G, Aguilar-Bryan L, Seino S, Bryan J (1995) Reconstitution of IKATP: an inward rectifier subunit plus the sulfonylurea receptor. Science 270: 1166-1170 [DOI] [PubMed] [Google Scholar]
- Inagaki N, Gonoi T, Clement JP IV, Wang CZ, Aguilar-Bryan L, Seino S (1996) A family of sulfonylurea receptors determines the pharmacological properties of ATP sensitive K+ channels. Neuron 16: 1011-1017 [DOI] [PubMed] [Google Scholar]
- Klein M, Perfus-Barbeoch L, Frelet A, Gaedeke N, Reinhart D, Mueller-Roeber B, Martinoia E, Forestier C (2003) The plant multidrug resistance ABC transporter AtMRP5 is involved in guard cell hormonal signaling and water use. Plant J 33: 119-129 [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Tax F, Sussman MR (1996) Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc Natl Acad Sci USA 93: 8145-8150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaHaye PA, Epstein E (1969) Salt toleration by plants: enhancement with calcium. Science 166: 395-396 [DOI] [PubMed] [Google Scholar]
- Leonhardt N, Marin E, Vavasseur A, Forestier C (1997) Evidence for the existence of a sulfonylurea-receptor-like protein in plants: modulation of stomatal movements and guard cell potassium channels by sulfonylureas and potassium channel openers. Proc Natl Acad Sci USA 94: 14156-14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Vavasseur A, Forestier C (1999) ATP binding cassette modulators control abscisic acid-regulated slow anion channels in guard cells. Plant Cell 11: 1141-1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K (1997) An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc Natl Acad Sci USA 94: 14960-14964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K (1998) A calcium sensor homolog required for plant salt tolerance. Science 280: 1943-1945 [DOI] [PubMed] [Google Scholar]
- Lu Y-P, Li Z-S, Drozdowicz YM, Hörtensteiner S, Martinoia E, Rea PA (1998) AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: functional comparisons with AtMRP1. Plant Cell 10: 267-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y-P, Li Z-S, Rea PA (1997) AtMRP1 gene of Arabidopsis encodes a glutathione S-conjugate pump: isolation and functional definition of a plant ATP-binding cassette transport gene. Proc Natl Acad Sci USA 94: 8243-8248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C (2002) Auxin transport: ABC proteins join the club. Trends Plant Sci 7: 329-332 [DOI] [PubMed] [Google Scholar]
- Marin E, Leonhardt N, Vavasseur A, Forestier C (1998) Cloning of AtMRP1, an Arabidopsis thaliana cDNA encoding a homologue of the mammalian multidrug resistance-associated protein. Biochim Biophys Acta 1369: 7-13 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Klein M, Geisler M, Bovet L, Forestier C, Kolukisaoglu Ü, Müller-Röber B, Schulz B (2002) Multifunctionality of plant ABC transporters: more than just detoxifiers. Planta 214: 345-355 [DOI] [PubMed] [Google Scholar]
- Miki T, Nagashima K, Seino S (1999) The structure and function of the ATP-sensitive K+ channel in insulin-secreting pancreatic β-cells. J Mol Endo 22: 113-123 [DOI] [PubMed] [Google Scholar]
- Mouline K, Anne-Aliénor V, Gaymard F, Boucherez J, Pilot G, Devic M, Bouchez D, Thibaud J-B, Sentenac H (2002) Pollen tube development and competitive ability are impaired by disruption of a shaker K+ channel in Arabidopsis. Genes Dev 16: 339-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473-497 [Google Scholar]
- Noh B, Murphy AS, Spalding EP (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441-2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma A (1983) ATP-regulated K+ channels in cardiac muscle. Nature 305: 147-148 [DOI] [PubMed] [Google Scholar]
- Rea PA (1999) MRP subfamily ABC transporters from plants and yeast. J Exp Bot 50: 895-913 [Google Scholar]
- Rea PA, Li Y-P, Drozdowicz YM, Martinoia E (1998) From vacuolar GS-X pumps to multispecific ABC transporters. Annu Rev Plant Physiol Plant Mol Biol 49: 727-760 [DOI] [PubMed] [Google Scholar]
- Sánchez-Fernández R, Ardiles-Díaz W, Van Montague M, Inzé D, May MJ (1998) Cloning and expression analyses of AtMRP4, a novel MRP-like gene from Arabidopsis thaliana. Mol Gen Genet 258: 655-662 [DOI] [PubMed] [Google Scholar]
- Sánchez-Fernández R, Davis TGE, Coleman JOD, Rea PA (2001) The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J Biol Chem 276: 30231-30244 [DOI] [PubMed] [Google Scholar]
- Schmid-Antomarchi H, De Weille J, Fosset M, Lazdunski D (1987) The receptor for antidiabetic sulfonylureas controls the activity of the ATP-modulated K+ channel in insulin-secreting cells. J Biol Chem 262: 15840-15844 [PubMed] [Google Scholar]
- Schultz BD, Deroos ADG, Venglarik CJ, Singh AK, Frizzell RA, Bridges RJ (1996) Glibenclamide blockade of CFTR chloride channels. Am J Physiol 15: L192-L200 [DOI] [PubMed] [Google Scholar]
- Seino S, Miki T (2003) Physiological and pathophysiological roles of ATP-sensitive K+ channels. Prog Biophys Mol Biol 81: 133-176 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu J-K (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896-6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidler M, Hassa P, Hasan S, Ringli C, Dudler R (1998) Involvement of an ABC transporter in a developmental pathway regulating hypocotyls cell elongation in the light. Plant Cell 10: 1623-1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriver K, Mundy J (1990) Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2: 503-512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart CC, Fleming AJ (1996) Hormonal and environmental regulation of a plant PDR5-like ABC transporter. J Biol Chem 271: 19351-19357 [DOI] [PubMed] [Google Scholar]
- Spalding EP, Hirsch RE, Lewis DR, Qi Z, Sussman MR, Lewis BD (1999) Potassium uptake supporting plant growth in the absence of AKT1 channel activity: inhibition by ammonium and stimulation by sodium. J Gen Physiol 113: 909-918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou FL (2000) Plant ABC transporters. Biochim Biophys Acta 1465: 79-103 [DOI] [PubMed] [Google Scholar]
- Tommasini R, Vogt E, Fromenteau M, Hörtensteiner S, Matile P, Amrhein N, Martinoia E (1998) An ABC-transport of Arabidopsis thaliana has both glutathione-conjugate and chlorophyll catabolite transport activity. Plant J 13: 773-780 [DOI] [PubMed] [Google Scholar]
- Tommasini R, Vogt E, Schmid J, Fromenteau M, Amrhein N, Martinoia E (1997) Differential expression of genes coding for ABC transporters after treatment of Arabidopsis thaliana with xenobiotics. FEBS Lett 411: 206-210 [DOI] [PubMed] [Google Scholar]
- Wu S-J, Ding L, Zhu J-K (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8: 617-627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K, Hasegawa PM, Bressan RA (1997) Molecular aspects of osmotic stress in plants. Crit Rev Plant Sci 16: 253-277 [Google Scholar]
- Zhu J-K (2000) Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol 124: 941-948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K, Liu J, Xiong L (1998) Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell 10: 1181-1192 [DOI] [PMC free article] [PubMed] [Google Scholar]