Abstract
Loss of Max function in the mouse resulted in generalized developmental arrest of both embryonic and extraembryonic tissues at early postimplantation (∼E5.5–6.5), coincident with loss or dilution of maternal Max stores in the expanding embryo in vivo and in blastocyst outgrowths in vitro. Developmentally arrested embryos were reduced in size and exhibited widespread cytological degeneration and feeble BrdU incorporation. Max and, by extension, the Myc superfamily, serve essential roles in early mammalian development and a maternal reservoir of Max exists in sufficient amount to sustain Myc superfamily function through preimplantation stages of development.
Keywords: Max, mammalian development, Myc function
The Myc superfamily comprises an extended network of related basic region helix–loop–helix/leucine zipper (bHLH/LZ) proteins that interact physically and functionally to regulate diverse biological processes (Henriksson and Lüscher 1996; Facchini and Penn 1998). Current evidence supports the view that in this superfamily, Max serves a central and essential role as obligate dimerization–DNA-binding partner for Myc oncoproteins and Mad transcriptional repressors (Blackwood and Eisenman 1991; Prendergast et al. 1991; for review, see Schreiber-Agus and DePinho 1998). Max appears to be required for all known biological functions of Myc, as evidenced through the use of engineered Myc and Max proteins that do not bind their endogenous partners yet dimerize efficiently with each other (Amati et al. 1993a,b). Such engineered Myc proteins remain biologically inert unless provided with the permissive binding form of Max. Members of the Myc family—c-Myc, N-Myc, and L-Myc—serve important roles in mammalian development as verified by induction of congenital defects brought about through aberrant Myc transgene expression (Morgenbesser and DePinho 1994) or loss of c- or N-Myc function (Charron et al. 1992; Moens et al. 1992; Stanton et al. 1992; Davis et al. 1993; Sawai et al. 1993). In more differentiated tissues, the actions of Myc appear to be countered by those of the Mad family (Mad1, Mxi1, Mad3, and Mad4) via their competitive interaction for Max, occupation of common consensus sites, and recruitment of the mSin3 complex (Ayer et al. 1995; Schreiber-Agus et al. 1995; Foley et al. 1999). These Mad proteins can promote terminal differentiation (Cultrano and Eisenmann 1999; Foley and Eisenmann 1999), maintain organ homeostasis with advancing age (Schreiber-Agus et al. 1998), and suppress the development of cancer in vitro (Lahoz et al. 1994; Koskinen et al. 1995) and in vivo (Schreiber-Agus et al. 1998). The dynamic and contrasting patterns of Myc and Mad family proteins are also consistent with their opposing roles in governing processes of growth and development (Larsson et al. 1994; Chin et al. 1995; Hurlin et al. 1995; Vastrik et al. 1995; for review, see Schreiber-Agus et al. 1998). In contrast, Max has been shown to be present in virtually all cell types and maintained at near constant levels during cellular differentiation, suggesting an important, albeit more passive, role in processes governed by the Myc superfamily (Blackwood and Eisenman 1991; Berberich et al. 1992; Schreiber-Agus et al. 1993).
To assess the role of Max in development, a null Max allele was generated in which the neomycin resistance (Neo) cassette replaced sequences encoding the Max helix II, leucine zipper, and entire carboxyl terminus, including nuclear localization signals (Fig. 1A). Both genomic PCR and Southern blot analyses were used to identify the mutant allele in ES cells, as well as verify its transmission through the germ line via chimera formation (Fig. 1B,C). Primers directed to sequences in the deleted Max region, Neo cassette, and flanking genomic region yielded the expected 1.7- and 1.4-kb amplification products for wild-type and mutant alleles, respectively (Fig. 1B, only tail DNAs shown). Employing a 3′-flanking probe, Southern blot assays of ES- and tail-derived DNA produced the anticipated 9.4- and 5.4-kb BglII restriction fragment for wild-type and mutant alleles, respectively (Fig. 1C, only tail DNAs shown).
Figure 1.
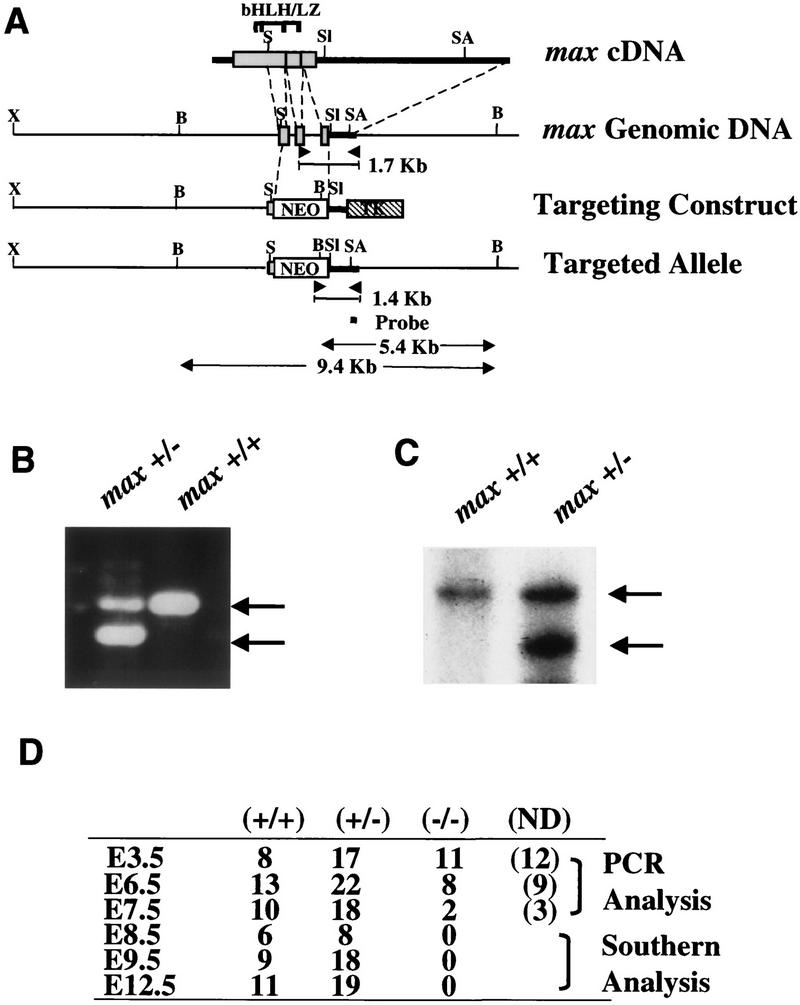
Gene targeting strategy. (A) Alignment of max cDNA and the 3′ genomic region showing exons encoding helix II, the bHLH/LZ region, and the entire carboxyl terminus of the Max protein. Heavy bars indicate 5′- and 3′-untranslated regions. The neo gene replaces a 1.8-kb SmaI–SalI genomic fragment. G418 and Gancyclovir selection produced an enrichment factor of 4 and led to the identification of 1 targeted ES clone from a total of 80 clones screened. The location of the primers or probes used in B and C, and the generated PCR products or restricted fragments corresponding to wild-type or disrupted alleles are indicated. (X) XbaI; (B) BglII; (S) SmaI; (Sl) SalI; (SA) Sau3AI. (B) Three-primer PCR analysis of representative genomic tail DNA from max heterozygous intercrosses showing wild-type (1.7-kb, top arrow) and targeted (1.4-kb, bottom arrow) alleles. (C) Southern blot of BglII-digested tail DNA depicting the wild-type (9.4-kb, top arrow) and mutant (5.4-kb, bottom arrow) banding patterns obtained from live offspring derived from Max+/− intercrosses. The probe was a 250-bp RsaI fragment located in the 3′ UTR outside of the 3′ arm. (D) Genotype distribution of Max+/− intercrosses; (N.D.) A result was not obtained or was inconclusive.
Although a single ES clone was used to generate the mutant Max colony, this clone yielded a high degree of chimerism, arguing against random genetic anomalies. Furthermore, any potential cell culture artifacts were diminished by first backcrossing Max+/− progeny (arising from chimera × C57Bl/6 crosses) to CD1 or C57Bl/6 mice for at least two generations prior to genotype distribution studies and phenotypic characterization. Genotype distribution of mice arising from Max+/− intercrosses failed to show a homozygous null Max mouse in >400 offspring analyzed. Max heterozygotes were indistinguishable from wild-type mice with respect to weight, size, activity, fecundity, and life span—as assessed over a 3- to 4-year period of observation. Histological organ surveys of adult Max+/− mice were also normal (data not shown). These results indicate that loss of Max function is associated with an embryonic lethal condition and that the level of Max in heterozygotes is adequate to permit normal growth and development throughout life.
To determine the basis for embryonic lethality, embryos generated from heterozygous intercrosses were harvested for detailed molecular and morphological studies. Of the 71 embryos analyzed from E8.5 to E12.5, none were Max−/− (Fig. 1D, E8.5–E12.5); and of the 30 E7.5 embryos assayed, only 2 degenerate samples were homozygous null (Fig. 1D, E7.5). In contrast, Max−/− blastocysts were morphologically normal and well represented (Fig. 1D, E3.5), thus pointing to early postimplantation as the period of developmental arrest. Dissection of E6.5 embryos from surrounding maternal tissue followed by PCR genotyping confirmed the presence of Max−/− embryos; however, these null embryos were under-represented (Fig. 1D, E6.5), severely compromised, and 50%–70% smaller than Max+/+ and Max+/− littermates (Fig. 2A). In one series of heterozygous matings, six growth-retarded embryos were determined to be Max−/− either by PCR genotyping (Fig. 2B; n = 3) or by whole-mount immunostaining showing negligible anti-Max signal (Fig. 2D; n = 3). The remaining 21 normal-sized embryos were confirmed to be either Max+/− or Max+/+ on the DNA (n = 7) or protein (n = 14) levels (Fig. 2, B and C, respectively). Furthermore, although wild-type embryos exhibited well-defined extraembryonic ectoderm and endoderm, Max−/− embryos did not possess discernible embryonic features or clear demarcation of embryonic and extraembryonic regions (Fig. 2A). These findings held constant on predominantly C57Bl/6 and CD1 backgrounds. Thus, Max deficiency is associated with generalized developmental arrest soon after implantation.
Figure 2.
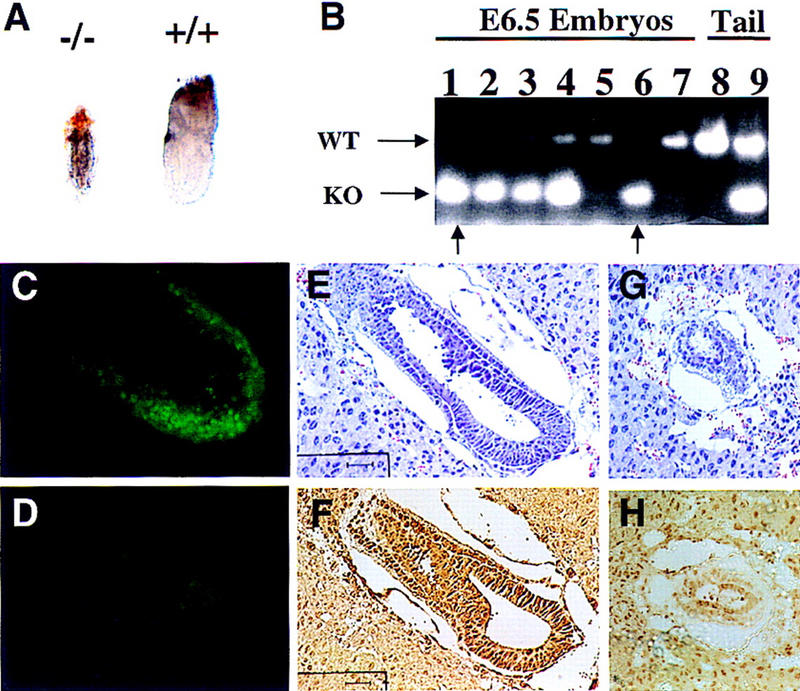
Phenotypic presentation of Max−/− embryos and its correlation with genotype or expression. (A) Gross appearance of PCR-verified E6.5 Max−/− embryo (left) and Max+/+ embryo (right). (B) PCR results of E6.5 embryos derived from Max+/− intercrosses (150-bp mutant allele; 350-bp wild-type allele). Max−/− embryos are indicated with arrows (lanes 1,6); embryos in lanes 1 and 6 are 50% and 70% smaller than Max+/+ and Max+/− littermates, respectively. (C,D) Whole-mount immunofluorescence staining of Max in wild-type (C) or Max−/− (D) E6.5 embryos, showing nuclear staining of normal embryonic cells but dramatically reduced nuclear staining in the mutant. Histological presentation and Max expression analysis of putative wild-type (E,F) and mutant (G,H) E6.5 embryo sections, as performed on adjacent sections. Bars, 40 μm.
Similar to the dissected embryos, the histological analysis of E5.5 and E6.5 decidua generated from heterozygous intercrosses identified two distinct morphological classes. One set of embryos (77%, n = 44) exhibited normal cellularity and cytoarchitecture in all embryonic and extraembryonic structures (Fig. 2E). When tested for anti-Max immunoreactivity, they showed robust anti-Max staining (Fig. 2F). The other embryos (23%, n = 13) were severely growth retarded and displayed signs of cellular degeneration, diminished cellularity, thin germ layers, and overall disorganization of embryonic and extraembryonic structures (Fig. 2G). These abnormal embryos possessed very weak anti-Max signal (Fig. 2H).
These phenotypic observations stand in contrast to Myc family knockouts, which survive to much later stages of development, the latter studies fueling speculation that a critical role for the Myc superfamily remains masked due to molecular complementation among the Myc family (Charron et al. 1992; Moens et al. 1992; Stanton et al. 1992; Davis et al. 1993; Hatton et al. 1996). Previous studies have confirmed c-, N-, and L-Myc gene expression in early cleavage and postimplantation embryonic stages (Downs et al. 1989; Domashenko et al. 1997), as well as a coordinate increase in c-Myc transcripts and several Myc-responsive gene targets (Domashenko et al. 1997). If this is the case, the possible existence of maternal Max stores and extreme stability of the protein (Blackwood and Eisenman 1991) could account for progression of Max−/− embryos through early developmental stages. To explore this possibility, Max expression was assayed in staged mouse embryos by immunohistochemical methods utilizing a highly specific anti-Max antibody (see Materials and Methods). As shown in Figure 3A, E0.5 embryos exhibited very intense pronuclear staining, predominantly in the nucleoplasm, as well as modest cytoplasmic signal comparable to that detected in the surrounding granulosa cells and maternal tissues. Because zygotic gene activity initiates at the two-cell stage and paternal protein contributions are thought to be negligible (Schultz 1993), the maternal origin of these abundant Max stores was confirmed by documenting intense Max signal in unfertilized eggs (Fig. 3B). Notably, in contrast to fertilized eggs, Max protein was found to be evenly distributed throughout the entire unfertilized oocyte, indicating that a subcellular shift in Max coincides with fertilization-induced maturation of the pronuclear membrane (Szollosi et al. 1972). These observations are somewhat reminiscent of observations in Xenopus laevis oocytes showing dramatic cytoplasmic to nuclear translocation of c-Myc upon fertilization (Gusse et al. 1989; Lemaitre et al. 1995). However, the Max patterns described here are clearly distinct from those of Xenopus c-Myc in that the latter remains completely sequestered in the cytoplasm prior to fertilization. An assessment of c-Myc distribution in mouse oocytes was not possible with available anti-c-Myc antisera. Together, these observations imply a role for the Myc superfamily during early cleavage stages of development.
Figure 3.
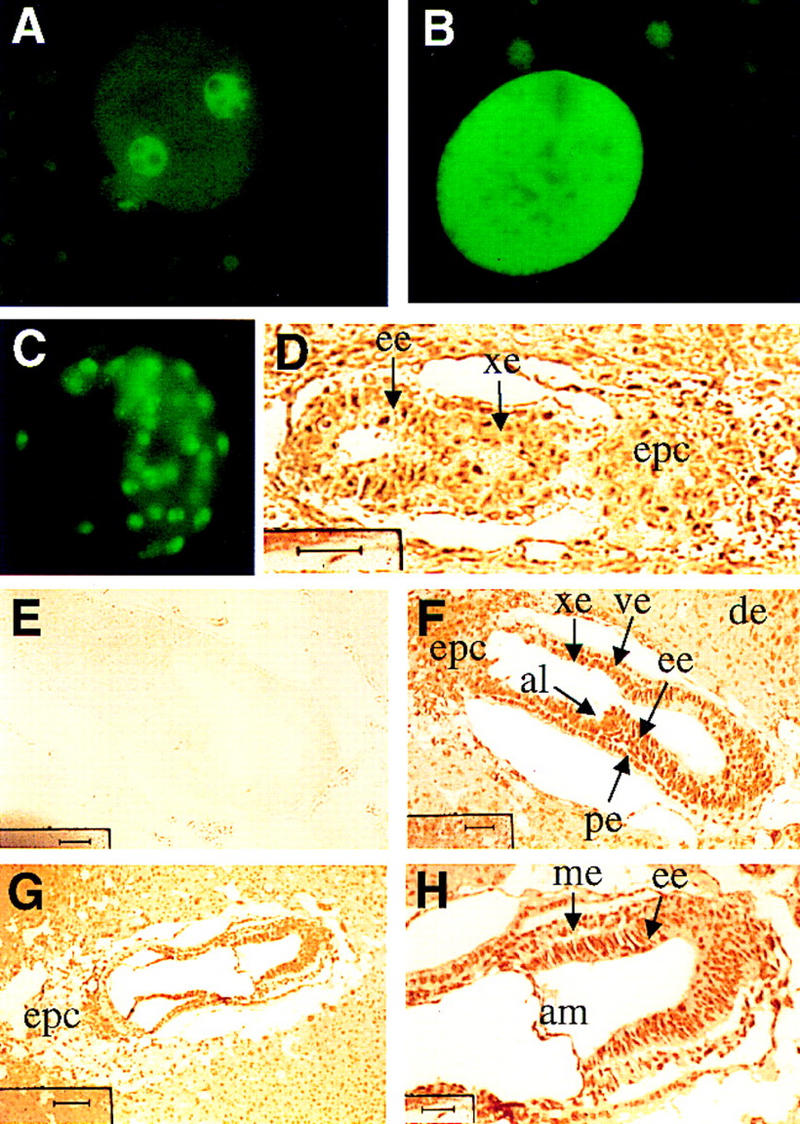
Max expression in early development. (A) Oviduct sections containing one-cell stage embryos; (B) oviduct sections containing unfertilized eggs; (C) whole-mount immunofluorescence of a E3.5 blastocyst; (D) E5.5 embryo section (bar, 40 μm); (E) E6.5 embryo section exposed to excess immunogen peptide (bar, 40 μm); (F) E6.5 embryo section (bar, 40 μm); (G,H) E7.5 embryo sections (G), (bar, 100 μm), (H) (bar, 25 μm). (de) Deciduum; (al) allantois; (am) amnion; (ee) embryonic ectoderm; (xe) extraembryonic ectoderm; (ve) visceral endoderm; (me) mesoderm; (pe) parietal endoderm; (epc) ectoplacental cone.
Max expression was detected in the nuclei of virtually all cells of the conceptus at levels comparable to or greater than those detected in the maternally derived cells of the surrounding decidua. In the blastocyst, Max signal was detected in nuclei of the inner cell mass and trophectodermal layer (Fig. 3C). In early postimplantation embryos, strong nuclear-associated and modest cytoplasmic Max signal was maintained in all embryonic and extraembryonic structures including the embryonic ectoderm, visceral and parietal layers of the embryonic endoderm, extraembryonic ectoderm, and ectoplacental cone of the E5.5 and E6.5 conceptus (Fig. 3, D and F, respectively). Additionally, in the face of dynamic differentiation events in E7.5 and E12.5 embryos, strong and constant nuclear-associated Max signal was detected in all embryonic and extraembryonic tissues (Fig. 3G,H; E12.5 not shown). For all stages, nuclear and cytoplasmic anti-Max signals were abolished with preincubation of the antisera with Max peptide (Fig. 3E; data not shown), confirming that the Max signal was specific. These observations are consistent with previous observations that abundant Max is present in virtually all normal and transformed cells and mature tissues (Blackwood and Eisenman 1991; Berberich et al. 1992; Schreiber-Agus et al. 1993), with the singular exception of a cultured neuronal cell line (Hopewell and Ziff 1995). The dynamic and opposing expression patterns of Myc and Mad(Mxi1), in conjunction with the constitutive expression of Max (Larsson et al. 1994; Chin et al. 1995; Hurlin et al. 1995; Vastrik et al. 1995), lend support to the prevailing view of Max as a nodal element through which the opposing actions of myc and mad(Mxi1) are executed.
The capacity of Max-deficient embryos to reach early postimplantation could relate to the abundant maternal Max stores and the stable nature of the protein (half-life > 24 hr; Blackwood and Eisenman 1991). To assess this possibility more rigorously, the relationship between the timing of developmental arrest and depletion of Max stores was determined. All blastocysts (n = 8) derived from heterozygous intercrosses expressed Max, although ∼25% showed slightly reduced staining (data not shown), indicating the presence of Max irrespective of the embryo genotype. Next, blastocysts derived from heterozygous intercrosses were placed in culture, monitored for outgrowth potential, and assayed for Max expression over a 4-day period. Following hatching, all outgrowths assumed the classical appearance of an extended trophectodermal layer and a discernible inner cell mass node (e.g., Fig. 4B). Although ∼75% of the blastocyst outgrowths increased in size (Fig. 4A,B), the growth of the remaining 25% was stunted and ceased by day 3 (Fig. 4C,D). The experiment was terminated on day 4 to permit anti-Max immunohistochemistry and genotyping prior to cellular degeneration. In all cases examined, the stunted inner cell mass phenotype (30%–50% reduction in surface area) correlated with either low or undetectable Max protein expression (n = 2) (Fig. 4, cf. A and C) or PCR-confirmed null genotype (n = 3; data not shown). The arrest of blastocyst outgrowths after 2–3 days in culture parallels the timing of developmental arrest in vivo, ∼5.5–6.5 days (see above). Based upon these results, we conclude that the timing of Max−/− lethality is due to loss and/or dilution of maternal Max at the point when the developing embryo experiences its first burst in growth on E5 and E6.
Figure 4.
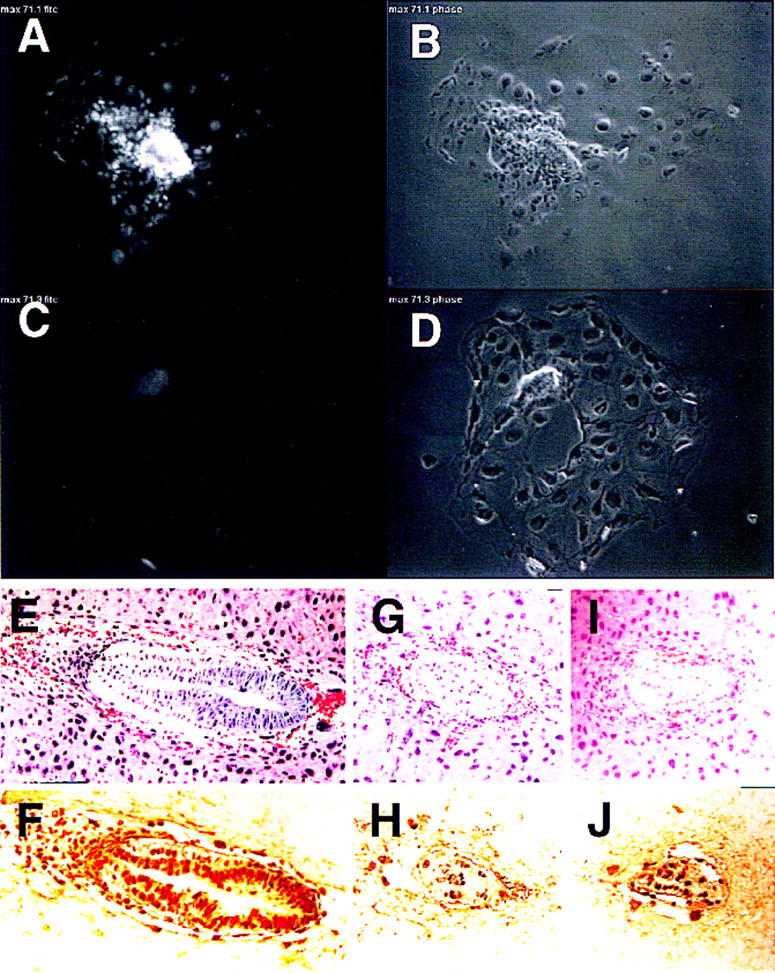
Blastocyst outgrowth, proliferation, and apoptosis assays. (A,B) Max immunofluorescence and appearance of a normal blastocyst outgrowth with phase-contrast view revealing a growing inner cell mass node and a single layer of trophectoderm cells. (C,D) Max immunofluorescence and appearance of a stunted blastocyst outgrowth showing reduced size of inner cell mass and diminished Max expression. (E–J) In vivo proliferation in E6.5 embryos. (E,F) Adjacent sections of a wild-type E6.5 embryo with hematoxylin and eosin (H&E) staining (E) and BrdU incorporation (F). (G–J) Adjacent sections of two putative Max-deficient E6.5 embryos with H&E staining (G,I) and BrdU incorporation (H,J).
The growth deficit of Max−/− embryos could result from generalized metabolic collapse, excess cell death, decreased cell proliferation, or a combination of these processes. To compare the proliferative and apoptotic profiles in the presence or absence of Max, BrdU incorporation and TUNEL assays were performed on sections of E6.5 decidua. The embryos were classified as normal (Max+/+ or Max+/−) or mutant (Max−/−) according to histological appearance of an adjacent section or staining with anti-Max antibody. As shown in Figure 4, the normal conceptus exhibits both robust anti-Max staining (see, e.g., Fig. 2F) and many BrdU-positive nuclei throughout the embryonic and extraembryonic structures (Fig. 4E,F). The putative mutants showed reduced size (Fig. 4G,I), weak anti-Max staining (see, e.g., Fig. 2H), and fewer BrdU-positive nuclei (Fig. 4H,J). The ratio of proliferating cells (BrdU-positive nuclei) to total cell number on the adjacent section was calculated to be 84% for four normal embryos versus 69% for four mutant embryos, suggesting that the decline in Max leads to a corresponding reduction in cell proliferation in early postimplantation mouse embryos. Because cell division cycles have been estimated to be 10–12 hr during this period (Snow 1981), a 10%–20% decline in the proportion of S-phase cells during early postimplantation could result in a 50% decline in embryo size over a period of 2 days. Severe cellular degeneration is also rampant throughout the mutant embryos and likely contributes to the observed reduction in size. This more generalized process is suggestive of a role for Myc in cellular metabolism (Dang 1999). TUNEL assays showed minimal apoptosis in the histologically normal and compromised E6.5 embryos, suggesting that programmed cell death does not contribute to the null phenotype, although it is possible that an earlier wave of apoptosis was missed.
The finding that Myc family transcripts are abundant from early cleavage stages onward (Domashenko et al. 1997), together with the capacity for c-, N-, or L-Myc null embryos to progress well beyond early development, has stimulated a continuing debate centered on whether survival relates to redundancy among Myc family members or to dispensability of Myc family function during early embryonic development. Here, appreciable Myc and Max expression and early arrest of Max-deficient embryos suggests that the Myc superfamily, Myc/Max in particular, serve vital roles in early development and thereafter.
Materials and methods
Gene targeting
A 15-kb genomic clone containing the 3′ region of Max was isolated from a 129/Sv mouse genomic library and used to generate a positive–negative type targeting construct, pMaxKO, in which the Neo and the HSV thymidine kinase (TK) genes were driven by the PGK promoter (Fig. 1A). pMaxKO was electroporated into WW6 mouse ES cells (Ioffe et al. 1995) and clones selected in 150 μg/ml active G418 and 2 μm gancyclovir. Max+/− ES cell clones were identified by PCR using a common external primer (5′-CAGGTAAGTCGCTCTTGGTA-3′) in combination with a Neo-specific primer (5′-GCCTTCTTGACGAGTTCTTC-3′) and a Max coding primer (5′-CTGCATGTAGGGGTGTCTGT-3′). Germ-line transmission was achieved as depicted in Figure 1. For PCR-based genotyping, E3.5–E10.5 embryos underwent lysis by boiling in 10 μl of lysis buffer containing 0.035 n NaOH and 0.5% SDS.
Immunohistochemical analyses
Staged deciduae (E5.5, E6.5, and E7.5) were isolated in ice-cold PBS, fixed in 10% buffered formalin overnight at 4°C, embedded in paraffin, and sectioned at 5 μm. Max expression was detected using a polyclonal antibody (Santa Cruz), a biotinylated secondary antibody, and avidin-conjugated peroxidase (Vector Laboratories). For whole-mount immunofluorescence staining, embryos were fixed in 2% paraformaldehyde and permeabilized with 0.25% Triton X-100. Max was detected using a polyclonal antibody (Santa Cruz) and a FITC-conjugated secondary antibody. The stained embryos were mounted in PBS/glycerol and observed under an Axiophot fluorescence microscope.
Blastocyst outgrowth studies
Blastocysts generated from heterozygous intercrosses were collected by uterine flush and individually cultured in poly-l-lysine-coated chamber dishes in ES media, in 5% CO2 at 37°C. After 4 days in culture, cells were washed twice in HBSS, fixed in 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100 in 2.5% goat serum for 30 min. Max was detected using a polyclonal antibody (Santa Cruz) and a FITC-conjugated secondary antibody. Immunofluorescence was viewed with an Olympus IX70 microscope with Planapochromat 10× N.A. 0.3 or 20× N.A. 0.4 objective. Images were collected with a Photometrics Sensys cooled CCD camera with a grade 1 chip. Intensities were quantitated with I.P. Lab Spectrum software running on a Macintosh G3.
BrdU and TUNEL assays
BrdU incorporation and TUNEL assays were performed as described previously (Morgenbesser et al. 1994).
Acknowledgments
We thank Dr. Ken Chen and Adam Silverman for technical assistance, and Frank Macaluso, Leslie Gunther and Michael Cammer of the Albert Einstein College of Medicine Analytical Imaging Facility for excellent assistance on microscopy and embryo sections. We also thank Dr. Nicole Schreiber-Agus, Dr. Leila Alland, and Dr. Arthur Skoultchi for helpful discussion and critical readings of the manuscript. This work was supported by National Institutes of Health (NIH) grants RO1EY09300 and RO1HD28317 (to R.A.D.), DFCC grant 5P30CA06516 (to R.A.D.), and NIH grant 5P30CA13330 to Albert Einstein Comprehensive Cancer Center. R.A.D. is an American Cancer Society Research Professor.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ron_depinho@dfci.harvard.edu; FAX (617) 632-6069.
References
- Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H, Kretzner L, Blackwood EM. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993a;72:233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- Amati B, Littlewood TD, Evan GI, Land H. The c-Myc protein induces cell cycle progression and apoptosis through dimerization with Max. EMBO J. 1993b;12:5083–5087. doi: 10.1002/j.1460-2075.1993.tb06202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer DE, Lawrence QA, Eisenman RN. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- Berberich S, Hyde-DeRuyscher N, Espenshade P, Cole M. max encodes a sequence-specific DNA-binding protein and is not regulated by serum growth factors. Oncogene. 1992;7:775–779. [PubMed] [Google Scholar]
- Blackwood EM, Eisenman RN. Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, Robertson EJ, Alt FW. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes & Dev. 1992;6:2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- Chin L, Schreiber-Agus N, Pellicer I, Chen K, Lee HW, Dudast M, Cordon-Cardo C, DePinho RA. Contrasting roles for Myc and Mad proteins in cellular growth and differentiation. Proc Natl Acad Sci. 1995;92:8488–8492. doi: 10.1073/pnas.92.18.8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cultraro CM, Bino T, Segal S. Function of the c-Myc antagonist Mad1 during a molecular switch from proliferation to differentiation. Mol Cell Biol. 1997;17:2353–2359. doi: 10.1128/mcb.17.5.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes & Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- Domashenko AD, Latham KE, Hatton KS. Expression of myc-family, myc-interacting, and myc-target genes during preimplantation mouse development. Mol Reprod Dev. 1997;47:57–65. doi: 10.1002/(SICI)1098-2795(199705)47:1<57::AID-MRD8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Downs KM, Martin GR, Bishop JM. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes & Dev. 1989;3:860–869. doi: 10.1101/gad.3.6.860. [DOI] [PubMed] [Google Scholar]
- Facchini LM, Penn LZ. The molecular role of Myc in growth and transformation: Recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- Foley KP, Eisenman RN. Two MAD tails: What the recent knockouts of Mad1 and Mxi1 tell us about the MYC/MAX/MAD network. Biochim Biophys Acta. 1999;1423:M37–M47. doi: 10.1016/s0304-419x(99)00012-8. [DOI] [PubMed] [Google Scholar]
- Foley KP, McArthur GA, Queva C, Hurlin PJ, Soriano P, Eisenman RN. Targeted disruption of the MYC antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J. 1998;17:774–785. doi: 10.1093/emboj/17.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusse M, Ghysdael J, Evan G, Soussi T, Mechali M. Translocation of a store of maternal cytoplasmic c-myc protein into nuclei during early development. Mol Cell Biol. 1989;9:5395–5403. doi: 10.1128/mcb.9.12.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton KS, Mahon K, Chin L, Chiu FC, Lee HW, Peng D, Morgenbesser SD, Horner J, DePinho RA. Expression and activity of L-Myc in normal mouse development. Mol Cell Biol. 1996;16:1794–1804. doi: 10.1128/mcb.16.4.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson M, Lüscher B. Proteins of the Myc network: Essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- Hopewell R, Ziff EB. The nerve growth factor-responsive PC12 cell line does not express the Myc dimerization partner Max. Mol Cell Biol. 1995;5:3470–3478. doi: 10.1128/mcb.15.7.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlin PJ, Queva C, Koskinen PJ, Steingrimsson E, Ayer DE, Copeland NG, Jenkins NA, Eisenman RN. Mad3 and Mad4: Novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioffe E, Liu Y, Bhaumik M, Poirier F, Factor SM, Stanley P. WW6: An embryonic stem cell line with an inert genetic marker that can be traced in chimeras. Proc Natl Acad Sci. 1995;92:7357–7361. doi: 10.1073/pnas.92.16.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen PJ, Ayer DE, Eisenman RN. Repression of Myc-Ras cotransformation by Mad is mediated by multiple protein-protein interactions. Cell Growth Differ. 1995;6:623–629. [PubMed] [Google Scholar]
- Lahoz EG, Xu L, Schreiber-Agus N, DePinho RA. Suppression of Myc, but not E1a, transformation activity by Max-associated proteins, Mad and Mxi1. Proc Natl Acad Sci. 1994;91:5503–5507. doi: 10.1073/pnas.91.12.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson LG, Pettersson M, Oberg F, Nilsson K, Lüscher B. Expression of mad, mxi1, max and c-myc during induced differentiation of hematopoietic cells: Opposite regulation of mad and c-myc. Oncogene. 1994;9:1247–1252. [PubMed] [Google Scholar]
- Lemaitre JM, Bocquet S, Buckle R, Mechali M. Selective and rapid nuclear translocation of a c-Myc-containing complex after fertilization of Xenopus laevis eggs. Mol Cell Biol. 1995;15:5054–5062. doi: 10.1128/mcb.15.9.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Auerbach AB, Conlon RA, Joyner AL, Rossant J. A targeted mutation reveals a role for N-myc in branching morphogenesis in the embryonic mouse lung. Genes & Dev. 1992;6:691–704. doi: 10.1101/gad.6.5.691. [DOI] [PubMed] [Google Scholar]
- Morgenbesser SD, DePinho RA. Use of transgenic mice to study myc family gene function in normal mammalian development and in cancer. Semin Cancer Biol. 1994;5:21–36. [PubMed] [Google Scholar]
- Morgenbesser SD, Williams BO, Jacks T, DePinho RA. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature. 1994;371:72–74. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- Prendergast GC, Lawe D, Ziff EB. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Sawai S, Shimono A, Wakamatsu Y, Palmes C, Hanaoka K, Kondoh H. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development. 1993;117:1445–1455. doi: 10.1242/dev.117.4.1445. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N, DePinho RA. Repression by the Mad(Mxi1)-Sin3 complex. BioEssays. 1998;20:808–818. doi: 10.1002/(SICI)1521-1878(199810)20:10<808::AID-BIES6>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N, Horner J, Torres R, Chiu FC, DePinho RA. Zebra fish myc family and max genes: differential expression and oncogenic activity throughout vertebrate evolution. Mol Cell Biol. 1993;13:2765–2775. doi: 10.1128/mcb.13.5.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi AI, DePinho RA. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N, Meng Y, Hoang T, Hou H, Jr, Chen K, Greenberg R, Cordon-Cardo C, Lee HW, DePinho RA. Role of Mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature. 1998;393:483–487. doi: 10.1038/31008. [DOI] [PubMed] [Google Scholar]
- Schultz RM. Regulation of zygotic gene activation in the mouse. BioEssays. 1993;15:531–538. doi: 10.1002/bies.950150806. [DOI] [PubMed] [Google Scholar]
- Snow MH. Growth and its control in early mammalian development. Br Med Bull. 1981;37:221–226. doi: 10.1093/oxfordjournals.bmb.a071706. [DOI] [PubMed] [Google Scholar]
- Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes & Dev. 1992;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- Szollosi D, Calarco PG, Donahue RP. The nuclear envelope: Its breakdown and fate in mammalian oogonia and oocytes. Anat Rec. 1972;174:325–339. doi: 10.1002/ar.1091740305. [DOI] [PubMed] [Google Scholar]
- Vastrik I, Kaipainen A, Penttila TL, Lymboussakis A, Alitalo R, Parvinen M, Alitalo K. Expression of the mad gene during cell differentiation in vivo and its inhibition of cell growth in vitro. J Cell Biol. 1995;128:1197–1208. doi: 10.1083/jcb.128.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]


