Abstract
The definition of bacterial species is based on genomic similarities, giving rise to the operational concept of genomic species, but the reasons of the occurrence of differentiated genomic species remain largely unknown. We used the Agrobacterium tumefaciens species complex and particularly the genomic species presently called genomovar G8, which includes the sequenced strain C58, to test the hypothesis of genomic species having specific ecological adaptations possibly involved in the speciation process. We analyzed the gene repertoire specific to G8 to identify potential adaptive genes. By hybridizing 25 strains of A. tumefaciens on DNA microarrays spanning the C58 genome, we highlighted the presence and absence of genes homologous to C58 in the taxon. We found 196 genes specific to genomovar G8 that were mostly clustered into seven genomic islands on the C58 genome—one on the circular chromosome and six on the linear chromosome—suggesting higher plasticity and a major adaptive role of the latter. Clusters encoded putative functional units, four of which had been verified experimentally. The combination of G8-specific functions defines a hypothetical species primary niche for G8 related to commensal interaction with a host plant. This supports that the G8 ancestor was able to exploit a new ecological niche, maybe initiating ecological isolation and thus speciation. Searching genomic data for synapomorphic traits is a powerful way to describe bacterial species. This procedure allowed us to find such phenotypic traits specific to genomovar G8 and thus propose a Latin binomial, Agrobacterium fabrum, for this bona fide genomic species.
Keywords: bacterial species, Agrobacterium, ecological niche, bacterial evolution, linear chromosome
Introduction
The species as basic taxonomic unit dates back to Carl Linnaeus and has since been universally used to describe all living organisms, including microbes. In superior Eukaria, the separation of distinct species relies on the occurrence of sexual barriers, as summed up in the famous biological species concept proposed by Mayr (1942). However, in asexually reproducing organisms, species are defined upon similarities of their members contrasted by interspecies genetic discontinuities.
In Bacteria, similarity discontinuities were first revealed through phenotypic traits and used to classify strains in different species by numerical taxonomy (Sneath and Sokal 1973). It was soon discovered that discontinuities also occur at the genomic level, leading to the current genomic species definition. Indeed, empirical studies revealed a gap in the distribution of genomic DNA hybridization ratio for pairwise comparisons of numerous strains around 70% (or around 5 °C for ΔTm) that matched previous phenotype-based distinction of species. Strains displaying genomic similarities above this level are thus considered to belong to the same species (Wayne et al. 1987; Stackebrandt et al. 2002), that are called genomic species. Alternatively, based on sequence data, genomic species can be distinguished through multilocus sequence analysis (Gevers et al. 2005). This is in line with the phylogenetic species concept based on the evolutionary relatedness among organisms that applies to all organisms, including Bacteria and Archaea, as pinpointed by Staley (2004). Although this definition is operational, efficiently leading to the delineation of readily distinguishable genomic species in most taxa, we still need to understand what mechanisms lead to differentiation of such genomic species (Fraser et al. 2009).
In our view, a genomic species is likely descending from a single ancestor that speciated a long time ago consecutively to adaptations to a novel ecological niche. Adaptations of the ancestor to its ecological niche were determined by adaptive mutations that should have been conserved in the progeny as long as they continued to exploit the same primary niche. Traces of adaptation could thus be found in progeny genomes, namely species-specific genes present in genomes of all members of a given species but not in closely related species. Species-specific genes inherited from the ancestor may still be responsible for the adaptation of present species members to a species-specific ecological niche. This hypothesis can be tested using comparative genomics to reveal species-specific genes that likely encode species-specific ecological functions.
Some studies intended to characterize the genomic specificities of bacterial species and understand their evolutionary history (Porwollik et al. 2002; Cai et al. 2009; Touchon et al. 2009; Lefébure et al. 2010; Zhao et al. 2010), other studies aimed to characterize the differences in ecology of ecotypes (or ecovars) among a taxon (Cohan 2002; Sikorski and Nevo 2005; Johnson et al. 2006; Sanjay et al. 2008; Cai et al. 2009; Connor et al. 2010; Zhao et al. 2010). We aimed to combine these approaches to test the hypothesis of genomic species arising from specific ecological adaptations. Good candidate species to test this hypothesis should preferentially display high within-species diversity, so as to capture the most common species characters, with the least possible divergence from their closest neighbors, thus maximizing the chance of detecting specific determinants. The bacterial taxon Agrobacterium tumefaciens fulfils these requirements. According to the current genomic species definition, this taxon displays a too large genomic divergence to be a single species and must be considered as a complex of ten distinct genomic species, currently named genomovar G1 to G9 and G13 (Mougel et al. 2002; Costechareyre et al. 2009). Although, they clearly belong to distinct genomic/genetic lineages, these species have not yet received Latin binomials essentially because they are not easily distinguishable by usual biochemical identification systems. They are, however, bona fide species to test our hypothesis because they are closely related and also have large infraspecies diversity. In addition, agrobacteria are common inhabitants of soils and rhizospheres, with several strains and genomic species commonly found in the same soil samples (Vogel et al. 2003; Costechareyre et al. 2010). Because complete competitor cannot coexist, according to the competitive exclusion principle (Hardin 1960), co-occurring species must be adapted to partly different ecological niches. Hence, often co-occurring and highly diverse Agrobacterium species are choice candidates for testing whether genomic species harbor presumptive determinants of a species-specific ecology. In addition, strain C58 of genomovar G8 is completely sequenced (Goodner et al. 2001; Wood et al. 2001), so a set of reference genes is available for classification according to their level of ubiquity within the entire taxon. The genomic sequence of strain H13-3 from A. tumefaciens genomovar G1 (Wibberg et al. 2011) has been published at the time of submission of this work, providing another reference to validate our results.
In the present work, we looked for genes that could be involved in the ecological specificity of bacterial species. Because we were able to experimentally determine the set of genes specific to genomovar G8, we focused particularly on genomovar G8 as a model species. We then: 1) manually annotated the functions of genes putatively determining ecological specificities, 2) inferred cellular pathways that may be involved in the adaptation of G8 agrobacteria to their ecological niches, and 3) experimentally validated most of the predicted functions and checked that they specifically occurred within all G8 members but not elsewhere. We used this information to precise our representation of the ecological niches of genomic species of the A. tumefaciens complex and develop a scenario for ecology-driven speciation of genomovar G8.
Materials and Methods
Bacterial Strains and Culture Conditions
In the present paper, we used a homogenous nomenclature defined according to the literature as follows: A. tumefaciens for members of the species complex with reference to genomic species, as explained by Costechareyre et al. (2010), A. larrymoorei for strain AF3.10 (Bouzar and Jones 2001), A. vitis for the sequenced strain S4 (Ophel and Kerr 1990; Slater et al. 2009), Rhizobium rhizogenes for the sequenced strain K84 (Slater et al. 2009; Velázquez et al. 2010), and Ensifer meliloti for the sequenced strain 1021 (Galibert et al. 2001; Martens et al. 2007). Strains of the species complex A. tumefaciens and A. larrymoorei tested in the study (table 1) are available at the Collection Française de Bactéries Phytopathogènes (CFBP, INRA, Angers, France). They were routinely grown at 28 °C on YPG medium (yeast extract, 5 g/l; Bacto Peptone, 5 g/l; glucose, 10 g/l, pH 7.2). Genomic DNAs were extracted and purified from 50-ml liquid YPG cultures using the standard phenol–chloroform method (Sambrook and Russell 2001).
Table 1.
Agrobacterium Strains Used in This Study
| Strain Name | CFBP Code | Nb of Detected C58 CDS Homologs |
|||
| CcC58 | LcC58 | pAtC58 | pTiC58 | ||
| Agrobacterium tumefaciens genomovar G1 | |||||
| CFBP 5771 | 2493 | 1392 | 205 | 14 | |
| ICPB TT111 | 5767 | 2461 | 1394 | 101 | 30 |
| A. tumefaciens genomovar G2 | |||||
| CFBP 5495 | 2371 | 1185 | 53 | 0 | |
| CFBP 5494 | 2168 | 944 | 36 | 0 | |
| A. tumefaciens genomovar G3 | |||||
| CFBP 6624 | 2501 | 1104 | 0 | 0 | |
| CFBP 6623 | 2586 | 1388 | 32 | 0 | |
| A. tumefaciens genomovar G4 (bona fide A. radiobacter) | |||||
| B6 | 2413 | 2584 | 1389 | 131 | 32 |
| DC07-012 | 7273 | 2503 | 1283 | 36 | 0 |
| Kerr 14 | 5761 | 2557 | 1376 | 3 | 81 |
| A. tumefaciens genomovar G5 | |||||
| CFBP 6625 | 1751 | 626 | 1 | 0 | |
| CFBP 6626 | 2378 | 1164 | 9 | 1 | |
| A. tumefaciens genomovar G6 | |||||
| NCPPB 925 | 5499 | 2533 | 1507 | 50 | 36 |
| A. tumefaciens genomovar G7 | |||||
| DC07-042 | 7274 | 2370 | 1184 | 5 | 0 |
| RV3 | 5500 | 2516 | 1266 | 1 | 0 |
| Zutra 3/1 | 6999 | 2529 | 1349 | 22 | 169 |
| A. tumefaciens genomovar G8 (A. fabrum nov. sp.) | |||||
| Mushin 6 | 6550 | 2686 | 1693 | 273 | 132 |
| C58T | 1903 | 2765 | 1851 | 542 | 197 |
| DC04-004 | 7272 | 2757 | 1851 | 542 | 197 |
| J-07 | 5773 | 2683 | 1677 | 200 | 0 |
| LMG 46 | 6554 | 2674 | 1669 | 0 | 172 |
| LMG 75 | 6549 | 2681 | 1736 | 198 | 117 |
| T37 | 5503 | 2663 | 1678 | 270 | 134 |
| A. tumefaciens genomovar G9 | |||||
| Hayward 0362 | 5507 | 2565 | 1195 | 7 | 197 |
| Hayward 0363 | 5506 | 2524 | 1213 | 17 | 0 |
| A. tumefaciens genomovar G13 | |||||
| CFBP 6927 | 2517 | 1215 | 10 | 0 | |
| A. larrymoorei | |||||
| AF 3.10T | 5473 | 1032 | 228 | 8 | 0 |
NOTE.—CFBP, Collection Française de Bactéries Phytopathogènes, INRA, Angers, France (http://www.angers.inra.fr/cfbp/). CcC58, circular chromosome of C58; LcC58, linear chromosome of C58; pAtC58, At plasmid of C58; pTiC58, Ti plasmid of C58.
Comparative Genome Hybridization Array Design
Comparative genome hybridizations (CGHs) were performed with DNA microarrays specifically designed for this experiment. Microarrays were made of 389,307 spots of 50-nt probes. All the four replicons of A. tumefaciens str. C58 (GenBank accessions NC_003302, NC_003303, NC_003304, NC_003305) were covered with a probe every 50 nt on each strand and a shift of 25 nt between strands. To obtain a set of control genes known to be absent from the tested strains, the microarrays also included probes designed in the same way to cover some plasmids from diverse Rhizobiaceae members, corresponding to the following GenBank accessions: NC_002377 (pTiA6), DQ058764 (pTiBo542), NC_002575 (pRi1724), NC_006277 (pAgK84), AJ271050 (pRi2659), AF065242 (pTiChry5), and plasmids from R. etli str. CFN42: NC_007 762, NC_007763, NC_007764, NC_004041, NC_007765, NC_007766. In order to model hybridization intensities as a function of levels of DNA base pairing between probe and target DNAs, the microarray contained 50-nt probes designed on the direct strand of all alleles of mutS, recA, and gyrB genes known at that time in A. tumefaciens, A. rubi, A. larrymoorei, A. vitis, and in some remote Rhizobiaceae species including R. rhizogenes and E. meliloti. The microarray also included 39,746 constructor-designed random probes for hybridization control. The C58 whole-genome microarray was constructed by NimbleGen Systems Inc. (Madison, WI), which also performed DNA labeling, hybridization, image capture, and raw data extraction steps according to internal company procedures. Hybridization intensities considered hereafter are log2 transformations of the raw data delivered by the company. Microarray design and experimental raw data are available at http://www.ebi.ac.uk/arrayexpress/ under the accessions A-MEXP-1977 and E-MTAB-558, respectively.
Modeling of Probe Hybridization Behaviors
Hybridization intensity (I) ranged, approximately, from 6 to 16 arbitrary units, including a long range (6–9) for background noise. Even in case of a perfect match, I spanned over a long range (e.g., 8–16 with C58). This complicated the determination of a single presence/absence threshold value indistinctly valid for all probes, especially for strains distantly related to C58. Instead, we used the fact that lacking genes are characterized by long stretches of successive probes mostly delivering a low background signal. Thus, to detect C58 coding DNA sequences (CDSs) homologs in the tested strains, we developed a method to classify segments of C58 replicons according to the homogenous presence or absence of homologous segments in each tested strain by comparison with perfectly matching C58 DNA probes as positive control. For each replicon a, plots of probe hybridization intensities of tested strain i (denoted Ia.i) and reference C58 DNA (denoted Ia.C58) revealed the presence of two populations of points: one, which displayed an almost linear relationship between Ia.i and Ia.C58, corresponding to probed regions that were “present” in the tested strain; and another, that displayed no correlation between Ia.i and Ia.C58, corresponding to regions that were “absent” (supplementary fig. S3, Supplementary Material online). A model (M) fitting these conditions was constructed using a mixture of two linear models, that is, (A) (absent) and (P) (present):
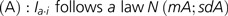 |
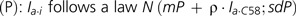 |
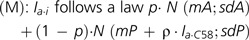 |
where parameters {mA, sdA} and {mP, sdP} are means and standard deviations for normal models (A) and (P), ρ is the slope of the linear relationship between present Ia.i and Ia.C58, and p is the weight of model (A) in (M), which reflects the proportion of probes belonging to the absent population.
For each strain i, given the set of probes representing a C58 replicon a on the microarray, it is straightforward to compute a likelihood function on the basis of this modeling. Then, we looked for the maximum likelihood given this data using the method of Nelder and Mead (1965) as implemented in the “stats” R package (R Development Core Team 2009). This process optimized values for the parameters in three steps:
First, parameters {mA, sdA} were analytically computed from the means and standard deviations of two sets of control probes: a) on probes covering NC_004041 (p42d) which was absent from every tested strain, giving values {mAa, sdAa} or b) on constructor-designed random probes, giving values {mAb, sdAb}.
Secondly, parameters were optimized from both start points {p = 0.5, mA0 = mAx, sdA0 = sdAx, mP = 1, sdP = 1, ρ = 1}, with x = a or x = b. During this first optimization step, {mA, sdA} were fixed in order to find the (P) mode. The posterior likelihood of models was calculated for each set of optimized parameters and the best fit between both sets of optimized parameters a and b was kept.
To adjust the proportions of points recruited by each mode, parameters were again optimized with {mA, mP, ρ} fixed and with a constraint on sdA: sdA ≤ (1.05·sdA0). To maintain the exclusivity of (A) and (P) modes, an additional constraint was set in the case of plasmids NC_003604 and NC_003605: mA + 1.5. sdA + 11. ρ ≤ mP + 2·sdP.
{mA, sdA} parameters were constrained during steps 2) and 3) to avoid a side effect of optimization due to the nonexclusivity of both modes, which may lead to overrecruitment of present points in absent mode (A) in some instances. When mode (A) should recruit very few points, that is, for tested strains very closely related to C58, a greedy optimization algorithm was tented to fill mode (A) by enlarging its boundaries (i.e., increasing sdA) or shifting its mean mA toward present points or conversely to fill mode (P) when plasmids NC_003604 and NC_003605 were completely absent from a strain. Sets of parameters for each microarray are listed in supplementary table S7 (Supplementary Material online).
Segmentation of C58 Replicons into Regions Present/Absent in Tested Strains
Multiple prediction partitioning was performed using Sarment Python modules (Guéguen 2005) to build an incremental partitioning of the sequence of replicon a when hybridized with strain i into segments of consecutive probes of common Absent or Present state given the likelihoods of each probe by models (A) and (P) nested in the optimized model (M). The segmentation process was independent of the sequence annotation; however, it appeared that partitions generally occurred between CDSs. As our interest was to screen for C58 CDSs present or absent in other strains, the incremental process was stopped when the number of CDSs which 100% probes mapped in absent segments was stabilized, typically after a few hundred segmentation iterations. Note that as a result of the segmentation procedure, some probes with high hybridization values surrounded by large number of low hybridization value probes can occur within absent segments. All CDSs located in absent segments are nevertheless considered absent.
Estimate of Genomic DNA-Probe Similarities
Similarity between microarray probes and probed DNA was estimated via probes of alleles of marker genes gyrB, mutS, and recA spotted on the microarray. The actual nucleotidic similarities between probes and known sequences of marker genes of the probed strain were computed using BlastN. The results were imported and parsed using Biopython libraries (Cock et al. 2009). Linear regressions between hybridization intensities and actual nucleotide similarities were done for each microarray while excluding nonhybridized probe noises (empirically determined to be below 80% genomic DNA-probe match) by using the stats R package (R Development Core Team 2009). Linear models (supplementary table S8, Supplementary Material online) were used to estimate the similarity between hybridized DNA and microarray probes (estimated nucleotidic similarity [ES]), thus allowing the calculation of average ESs of all CDSs covered by probes on the microarray (supplementary table S2, Supplementary Material online). For C58 replicons, it was also possible to cope with intensity heterogeneities among CDSs by calculating weighted estimated nucleotidic similarities (WESs). ESs recorded with a given strain were thus divided by the corresponding values obtained with C58, then adjusted according to the actual similarities of sequenced polymerase chain reaction (PCR) products of the tested strain with the C58 genome (supplementary table S3, Supplementary Material online).
Codon Usage Analysis
Effective counts of the 64 codons of the 5,355 CDSs of C58 were calculated using the “seqinR” package from R project (Charif and Lobry 2007). Correspondence analyses were performed and projected using “dudi.coa” and “s.class” functions from the “ade4” package (Dufour and Dray 2007).
Strain Clustering
Agrobacterium tumefaciens strains were clustered on the basis of gene presence/absence characters, as described in Lake (1994), by using logdet distances with C58 as conditioning genome. Logdet/paralinear distances (Lake 1994) were computed using the “binary.dist” function from R project (R Development Core Team 2009). Trees were built using the NEIGHBOR algorithm from PHYLIP package (Felsenstein 1993).
Functional Annotation of Specific Genes
The functional annotation of CDSs included in G8-specific clusters was manually curated using a relational database, that is, AgrobacterScope (in open access at https://www.genoscope.cns.fr/agc/microscope), with the MaGe web interface (Vallenet et al. 2009).
Construction of Deletion Mutants of SpG8-Specific Clusters
Mutants were constructed by mutagenic PCR as described by Choi and Schweizer (2005). Briefly, mutagenic PCR fragments were created by joining three fragments corresponding to the two regions flanking the sequence to be deleted of C58 (ca. 1 kb each) and a fragment encoding the nptII kanamycin resistance gene amplified from plasmid pKD4 (Datsenko and Wanner 2000) by using 70-nt primers consisting of 20 nt priming the kanamycin-resistance gene (3′ segment of the primer) and 50 (±3) nucleotides corresponding to flanking sequence ends of targeted sites (5′ segment of the primer) (supplementary table S9, Supplementary Material online). First and second round PCRs were performed as in Choi and Schweizer (2005), and then PCR fragments were cloned into the pGEM-Teasy vector (Promega, Madison, WI) according to manufacturer’s instructions. After digestion of the resulting plasmids with ApaI and SpeI, fragments were subcloned into pJQ200SK, a plasmid carrying the sacB gene conferring sucrose sensitivity (Quandt and Hynes 1993) digested with the same enzymes. To generate deleted mutants, PCR fragments cloned into pJQ200SK were inserted in C58 by electroporation. Single recombinants were selected on YPG media containing 25 μg/ml kanamycin and 25 μg/ml neomycin. Double crossover events were identified by sucrose resistance on YPG media supplemented with 5% sucrose. Deletion mutants were verified by diagnostic PCR with appropriate primers.
Experimental Validation Assays
Ferulic acid catabolism was tested using the two-step procedure described by Civolani et al. (2000). In a first step, cells were induced for 24 h at 28 °C (optical density [OD]600 nm = 1) in AT minimal medium (Petit et al. 1978) supplemented with 10 mM (NH4)2SO4 and 10 mM succinic acid, and 0.52 mM (0.1 mg·mL−1) ferulic acid (Sigma-Aldrich, St Louis, MO). Cells harvested by centrifugation were then suspended at OD600 nm = 0.1 into AT medium containing 10 mM (NH4)2SO4 and 0.52 mM ferulic acid as sole carbon source. Ferulic acid disappearance was monitored by high-performance liquid chromatography (HPLC) performed on an Agilent 1200 series (Agilent Technologies, Santa Carla, CA) liquid chromatograph associated with a diode array detector. Data acquisition and processing were controlled via Agilent Chemstation software. The separations were carried out on a Kromasil 100-5C18 column (250 × 4.6 mm). Compounds were eluted with a methanol–water gradient (0.4% formic acid) in which the methanol concentration was varied over time as follows: from 0 to 5 min, 20%; 5 to 22 min, increased to 62%; 22 to 25 min, increased to 100%; 25 to 30 min, 100%; 30 to 31 min, decreased to 20%. The flow rate was 1 ml.min−1. Ferulic acid was detected at a wavelength of 320 nm after injection of 5-μl sample. UV spectra and retention time (12.43 min) of ferulic acid were determined by injection of a methanolic suspension of ferulic acid. Identification of ferulic acid in bacterial cultures was confirmed by comparison with this standard.
Curdlan production was assessed by streaking bacteria onto plates containing a modified Congo red medium adapted from Kneen and LaRue (1983) with glucose as sole carbon source, incubated at 28 °C for 48 h and then kept at room temperature for 48 h.
Results
Presence of Homologs of CDSs from C58 Replicons and Other Rhizobiaceae Plasmids
CGH results obtained with an original C58 genome–based microarray were used to detect the presence or absence of genes homologous to C58 in 25 agrobacterial strains. These strains included seven G8 members, one to three for each of the nine other genomic species of the A. tumefaciens complex and one for A. larrymoorei, a sister species of A. tumefaciens (table 1). An original probabilistic method was used to segment C58 replicon sequences into regions that were absent or present in the tested strains, thus allowing us to detect the presence of homologs of C58 CDSs in the tested strains (supplementary table S1, Supplementary Material online).
The absence of detection, however, might have been due to the absence of a real locus or to a weak hybridization signal caused by high sequence divergence between the target genome and C58 DNAs. We thus calculated the ES for CDSs of all replicons probed by the microarray in reference to internal control probes of known mismatch values with tested DNAs (supplementary table S2, Supplementary Material online). In addition, we observed strong intensity heterogeneities among CDSs, even in case of a perfect match with C58. A better similarity estimate was reached by weighting ESs of tested strains by ESs recorded with C58 to provide WESs (supplementary table S3, Supplementary Material online). We used the segmentation method to detect the presence of CDSs displaying WESs as small as 80–86% in G1-ICPB TT111 and G7-DC07-042, respectively, in spite of high hybridization background noise (supplementary table S4, Supplementary Material online). This demonstrated the higher sensitivity of our probabilistic approach over threshold methods.
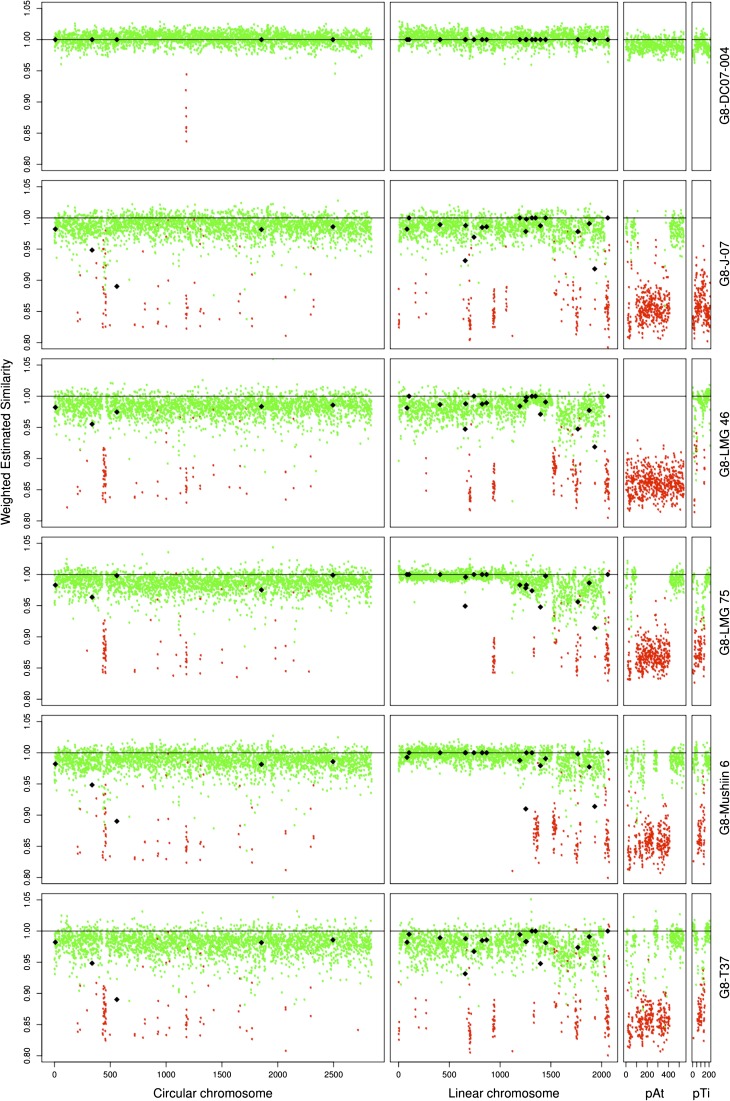
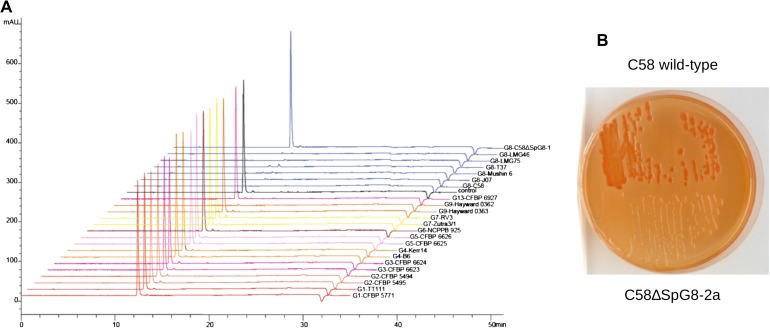
Beyond the analysis in terms of CDS presence, WES allowed us to estimate whether C58 and the tested strains had divergent or identical alleles. WES measures were plotted against CDS positioned along replicons for G8 members (fig. 1). G8-DC07-004 was expected to belong to the same clone or at least the same clonal complex as C58 because both strains have identical alleles for marker genes located on circular and linear chromosomes (recA, mutS, gyrB, chvA, ampC, glgE, gltD, … ) (data not shown). G8-DC07-004 displayed an average WES of 100% with little dispersion (average ΔWES = ±1%) over all four replicons and thus appeared to be identical to C58, except for eight genes that were lacking on the circular chromosome. In contrast, G8-T37, which had different alleles for most marker genes, displayed an average WES clearly below 100% with greater dispersion (average ΔWES = ±4%). In contrast, this allowed us to discover that large regions of the C58 linear chromosome were likely identical in other G8 strains. Remarkably, a region of more than 1 Mb encompassing the left arm of the linear chromosome was identical between C58 and G8-LMG 75 or G8-Mushin 6 (fig. 1). This strongly suggests recent transfer of half of the linear chromosome between members of this species. Although we found such long regions of identity with C58 in several G8 members, such long stretches of genome identity with C58 were not found outside G8 (supplementary fig. S1, Supplementary Material online), suggesting that large transfer events may essentially concern members of the same species.
FIG. 1.—
Presence and estimated similarity of C58 CDS homologs in other genomovar G8 members. Percentage of WES of C58 CDS homologs were plotted against their coordinates on the four C58 replicons. Dot colors indicate the presence (green) or absence (red) of C58 CDS homologs. Diamonds indicate actual similarity values of sequenced PCR products.
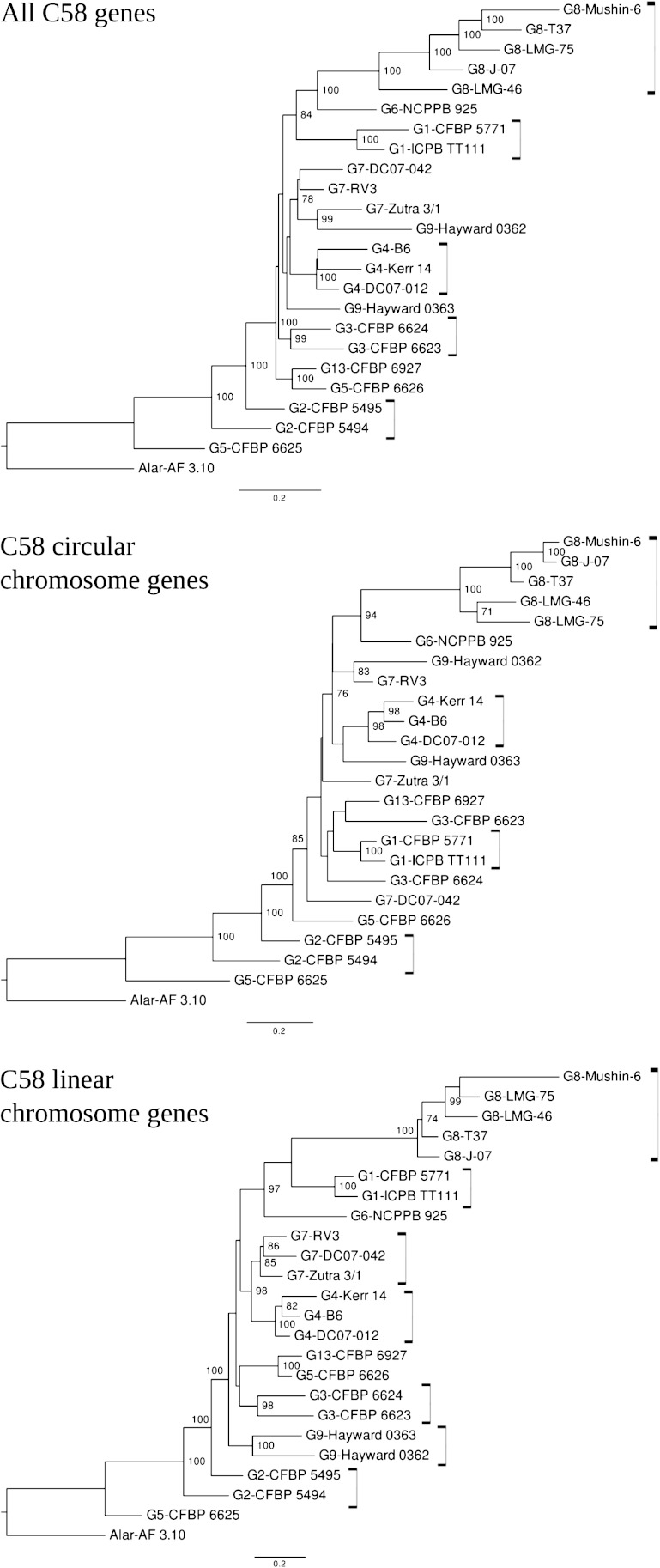
Strains were clustered according to their C58 CDS homolog content. As expected, this clearly allowed significant clustering of all G8 members (fig. 2). Differences in gene content similarities according to chromosomes were observed between genomic species. For six genomic species, the different members significantly grouped when considering CDSs of the C58 linear chromosome, compared with only four grouped genomic species when considering CDSs of the C58 circular chromosome. This suggests that the linear chromosome content better characterizes genomic species than the circular chromosome. Remarkably, G2 members as well as G5-CFBP 6625 were located at a basal position (i.e., far from G8), indicating that they had the lowest number of CDS in common with C58. The remaining species branched together at the same distance from the G8 groups (forking branches), indicating that they had comparable numbers but different sets of C58 CDS homologs.
FIG. 2.—
Clustering of Agrobacterium tumefaciens strains based on absence/presence of C58 CDS homologs. Neighbor-joining trees were constructed using the paralinear distances of Lake (1994) calculated from the presence/absence of C58 CDSs in other strains. With the reference genome being C58, this strain and its nearly identical relative DC07-004 were excluded from the analysis.
The presence of C58 CDS homologs according to their location in C58 replicons confirmed the presence of C58-circular and -linear chromosome CDS homologs in all tested strains (table 1). In contrast, this revealed the complete lack of pTiC58 or pAtC58 homologs in several strains such as for G8-J-07 and G8-LMG 46, which respectively lacked Ti and At plasmids or the absence of large regions of C58 plasmids in numerous strains (supplementary table S1 and fig. S1, Supplementary Material online), which highlights the mosaic nature of these replicons. The segmentation method was not applicable for replicons outside C58, thus hampering detection of barely similar CDS homologs in these cases. Nevertheless, high CDS homologies of around 100% ES were recorded for all CDSs of pTiA6 for both G4-B6 and G1-ICPB TT111, indicating the likely presence of the same Ti plasmid in both strains (supplementary table S2 and fig. S2, Supplementary Material online). The results of the CDS presence/absence analysis were, however, related to the high hybridization stringency conditions used, which may not allow detection of barely similar homologs. For instance, A. larrymoorei AF 3-10 was found to have no detectable CDS homology with pTiC58 (table 1), although this strain is known to be pathogenic with a chrysopine type Ti plasmid (Vaudequin-Dransart et al. 1995). As expected, however, AF3.10 exhibited significant estimated similarity values (ca. 93%) with more similar CDSs of the chrysopine type Ti plasmid pTiChry5 (supplementary table S2 and fig. S2, Supplementary Material online).
Ubiquity Level of C58 CDSs in the A. tumefaciens Complex
Homologs of the 5,355 CDSs of C58 were classed according to their level of ubiquity in Agrobacterium strains and grouped in six classes: only in C58 (“specific to C58,” 166 genes); only in G8 strains but not all (“sporadic in G8,” 151 genes); in all G8 and only G8 strains (“specific to G8,” 196 genes); with no specific presence pattern in A. tumefaciens (“sporadic in At,” 2,846 genes); in all A. tumefaciens strains but not in A. larrymoorei (“specific to At,” 976 genes); or in both A. tumefaciens and A. larrymoorei (“At-Al core genome,” 1,020 genes) (supplementary table S1, Supplementary Material online).
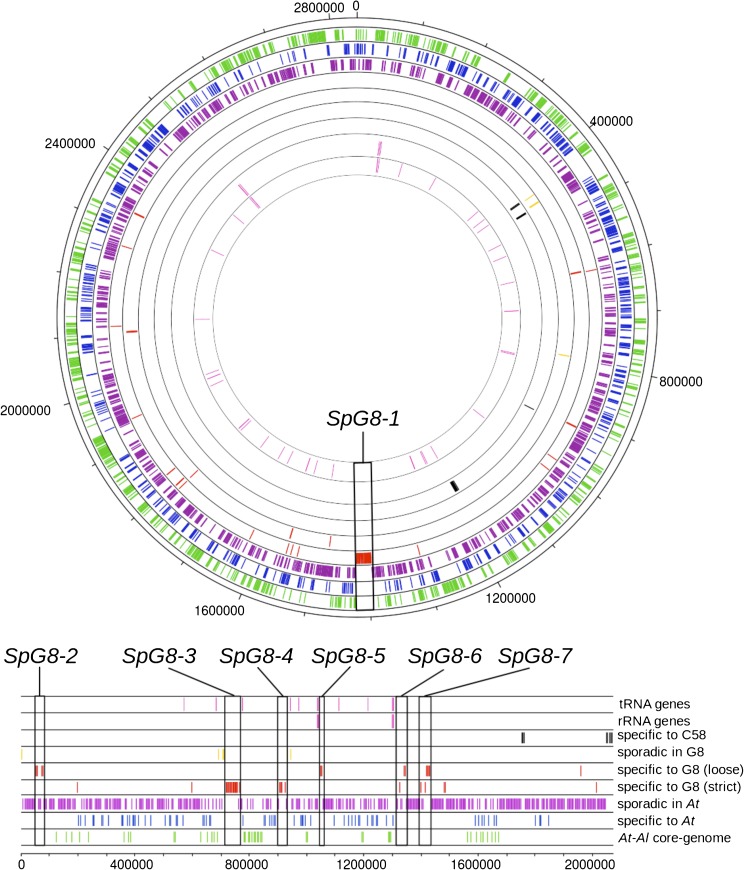
The core genome of A. tumefaciens (“At core genome,” sum of the “specific of At” and At-Al core genome classes) consists of 1,996 genes (37% of the genome). Seventy-five percent and 25% of the At core genome are located on circular and linear chromosomes, respectively (accounting for 56% and 25% of these replicons, respectively), showing clear core genome enrichment on the circular chromosome (fig. 3). As expected, no part of the core genome was found on plasmids because these accessory replicons were lacking in some strains (table 1).
FIG. 3.—
Ubiquity in the Agrobacterium tumefaciens species complex of C58 CDSs according to their localization on C58 chromosomes. Tracks are numbered from inner to outer track (circular chromosome) or top to bottom track (linear chromosome). tRNA and rRNA genes are represented track 1 and 2, respectively (pink). CDSs are represented according to their levels of ubiquity: track 3, specific to C58 (black); track 4, sporadic in G8 (yellow); track 5, strictly specific to G8, and track 6, specific to G8 with a loose criterion (red); track 7, sporadic in At (purple); track 8, specific to At (blue); track 9, “At-Al core-genome” (green). Boxes indicate G8-specific (SpG8) gene clusters.
Genes Specific to C58 and Sporadic in G8
G8-DC07-004 was found to be the same as C58 except for eight CDSs (Atu1183–Atu1190), which were absent in G8-DC07-004 (table 1). These eight CDSs were, strictly speaking, the real C58-specific genes, whereas the remaining 158 genes specific to C58 were specific to both C58 and G8-DC07-004. These genes were mainly grouped in three clusters: Atu1183-Atu1194, Atu4606-Atu4615, and Atu4864-Atu4896. The Atu1183–Atu1194 region, which included the deleted region described above, was located on the circular chromosome. It constituted a prophage (ATPP-2, see below). The last two clusters were located at the right extremity of the linear chromosome close to the telomeric region. They harbored transposase genes, suggesting that they were recombinogenic. The Atu4606–Atu4615 region was annotated as being involved in lipopolysaccharide biosynthesis, a function likely gained by transfer and specific to C58 and G8-DC07-004.
In addition, four regions sporadic in G8 were also identified as probable mobile genomic elements. Three were evidently prophages, which we named A. tumefaciens prophages (ATPP): ATPP-1 (Atu0436–Atu0471), ATPP-2 (Atu1183–Atu1194), and ATPP-3 (Atu3831–Atu3858), which contained genes encoding proteins characteristic of prophages, such as: integrase, excisionase, resolvase, DNA methyl-transferase, phage tail structural and assembly proteins, and DNA-dependent RNA polymerase. By analyzing similarities in their integrase gene sequences with those available in databanks, prophages were assigned to known prophage families: ATPP-1 and ATPP-2 were related to Podoviridae of P22 and T-7 families, respectively, whereas ATPP-3 was related to Myoviridae of the P4 family. The very recent publication of the genomic sequence of strain H13-3 of genomovar G1 (Wibberg et al. 2011) showed that prophages ATPP-1 and ATPP-2 were absent from this strain, but that traces of their past presence could be found at the corresponding loci. The fourth region, ranging from Atu3636 to Atu3665 next to tRNA genes, had a less clear nature. It was apparently undergoing a process of genetic decay and was referred to in this study as a decaying mobile DNA region (DMR). Many CDSs in this region were short CDSs that coded for hypothetical proteins, pseudogenes, and gene remnants, indicating that this region might no longer be under selection pressure.
Genes Specific to G8
In fact, 51 CDSs were strictly specific to the genomovar G8, but 145 CDSs found in all G8 members were also detected in one or two other non-G8 strains. In several instances, those latter genes were contiguous to strictly G8-specific genes (supplementary table S1, Supplementary Material online), suggesting that they cooperate with strict G8-specific genes for their functions. Thus, in order to capture more complete functions, we decided to use a loose definition of species-specific genes by merging the two gene classes for a total of 196 G8-specific CDSs (SpG8) (supplementary table S5, Supplementary Material online). No SpG8 genes were found on plasmids, but they were unevenly dispersed on the two chromosomes: 72% on the linear and 28% on the circular chromosomes, respectively. Remarkably, 61% of SpG8 genes were organized into clusters of five or more contiguous CDSs, whereas others were interspersed within the C58 genome (supplementary table S5, Supplementary Material online). Seven large SpG8 clusters, numbered SpG8-1 to SpG8-7, were located either on the circular chromosome (SpG8-1) or on the linear chromosome for the six others (table 2 and fig. 3). As explained below, some SpG8 clusters were subsequently divided into subclusters encoding homogeneous functions. Sequence data validated the presence of SpG8 clusters in G8 members and the absence of SpG8 genes with a similarity above 70% outside G8 (data not shown).
Table 2.
Characteristics of SpG8 Gene Clusters
| G8-Specific Regions | C58 CDSs | Region Occurrence Outside G8 | Main Predicted Functions | Experimental Validationa |
| SpG8-1a | Atu1398–Atu1408 | G6-NCCPB 925 | Sugar and amino acid transport; sugar metabolism | Not done |
| SpG8-1b | Atu1409–Atu1423 | G9-Hayward 0362 | Ferulic acid uptake and catabolism | Present work |
| SpG8-2a | Atu3054–Atu3059 | r | Curdlan EPS biosynthesis | Present work |
| SpG8-2b | Atu3069–Atu3073 | r | Secondary metabolite biosynthesis | Not done |
| SpG8-3 | Atu3663–Atu3691 | G1-ICPPB TT111 | Siderophore biosynthesis; iron-siderophore uptake | Rondon et al. (2004) |
| G6-NCCPB 925 | ||||
| SpG8-4 | Atu3808–Atu3830 | G6-NCCPB 925 | Ribose transport; monosaccharide catabolism and carbohydrate metabolism | Not done |
| SpG8-5 | Atu3947–Atu3952 | r | Opine-like compounds catabolism | Not done |
| SpG8-6a | Atu4196–Atu4206 | G1-CFBP 5771 | Drug/toxic (tetracycline) resistance | Luo and Farrand (1999) |
| SpG8-6b | Atu4213–Atu4221 | r | Drug/toxic resistance | Not done |
| SpG8-7a | Atu4285–Atu4294 | G6-NCCPB 925 | Environmental signal sensing/transduction | Not done |
| SpG8-7b | Atu4295–Atu4307 | Not present outside G8 | Environmental signal sensing/transduction | Not done |
NOTE.—r, rare occurrence of some CDSs outside G8.
Deleted mutants of C58 were obtained for all regions.
SpG8 regions seem to occur in hotspots of gene insertions. Cluster SpG8-3 adjoins a region containing different types of putative mobile elements referred to here as DMR. SpG8-4 was located next to the putative prophage ATPP-3. Blocks made of SpG8-3 and DMR and SpG8-4 and ATPP-3 are next to tRNA genes. SpG8-5 and SpG8-6 are next to rRNA operons containing tRNA genes (fig. 3).
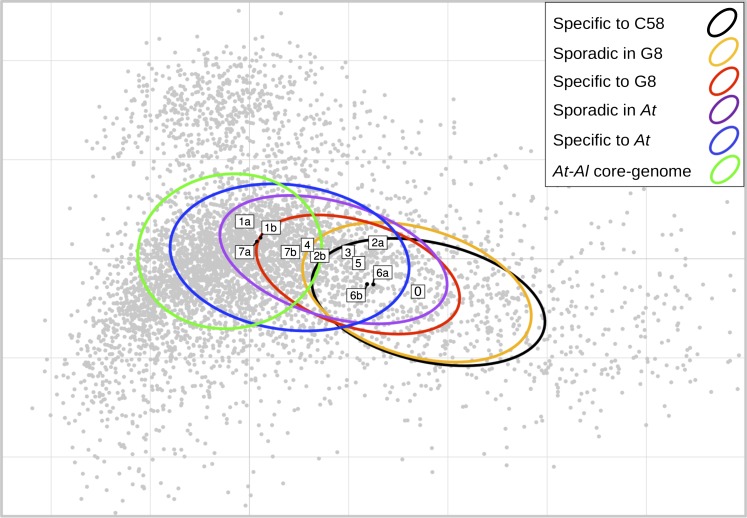
We performed a correspondence analysis on the codon usage of CDSs in C58 to determine whether genes of different ubiquity classes could be differentiated on the basis of their DNA sequence composition. Inertia ellipses of ubiquity classes were found to be dispersed along the first axis of the codon usage space over a gradient reproducing the ubiquity class order, which extends from core-genome genes to sporadic/strain-specific genes (fig. 4). Within the ubiquity class corresponding to SpG8 genes, we could sort SpG8 gene clusters along the “core versus sporadic” axis, from the group of isolated SpG8 genes (i.e., not located in clusters, fig. 4, box #0) at the “sporadic-like” extremity, to loci SpG8-1, SpG8-4, SpG8-7 at the “core gene” extremity. Notably, subclusters of large SpG8 gene clusters with different occurrence patterns in A. tumefaciens displayed different genome signatures. In fact, SpG8-1a and SpG8-7a, which shared genes with the closest relative of G8, that is, G6-NCPPB 925 (Costechareyre et al. 2010), displayed a more marked “core-like” code usage signature than SpG8-1b and SpG8-7b (fig. 4).
FIG. 4.—
Codon usage signatures of SpG8 genes. First factorial plan in the correspondence analysis of C58 CDSs according to their codon usage. First and second axes explain 4.5% and 2.2% of total variance, respectively. Grey dots represent CDSs, ellipses represent the inertia of ubiquity classes, and boxes represent barycenters of SpG8 loci named as detailed in table 2 and supplementary table S6 (Supplementary Material online) (and 0 for interspersed SpG8 CDSs). Codon usage of ubiquity classes were found to gradually vary from core to sporadic genes revealing, in turn, that SpG8 clusters can be distinguished by this criterion from core-like ones to sporadic-like ones. Interestingly, SpG8-1a, SpG8-4, and SpG8-7a—which are shared by the most closely related non-G8 strain G6-NCPPB 925 (table 2)—displayed a core-like codon usage.
SpG8 Functions
We were able to infer global functions for most SpG8 clusters, strengthening the hypothesis that they correspond to coherent functional units. Our expert manual annotations available in the AgrobacterScope database revealed that SpG8 clusters encoded functional units related to environmental sensing (SpG8-7), secreted metabolite production (SpG8-2a, SpG8-3), detoxification (SpG8-6), and metabolite catabolism (SpG8-1, SpG8-4, SpG8-5) (table 2).
SpG8-7: Environmental Signal Sensing
SpG8-7 encoded functions that could be related to environmental signal sensing and transduction: two mechanosensitive channels of the MscS family and a two-component transduction system with a receptor histidine kinase containing a PAS sensory box and two putative response regulators. Genes encoding a two-component system (Atu4300 and Atu4305) were homologous to nwsAB from Bradyrhizobium japonicum USDA 110, whose proteins are involved in plant host recognition during the nodulation process (Lang et al. 2008) and to todST from Pseudomonas putida F1 and styRS from Pseudomonas sp. VLB120, whose proteins recognize toluene and styrene, respectively, and activate related degradation pathways (Lau et al. 1997; Panke et al. 1998). A more comprehensive block was conserved in synteny with genes from Parvibaculum lavamentivorans DS-1 (4 genes, 60.2% amino acid identity on average) and E. meliloti 1021 (7 genes, 64% amino-acid identity on average).
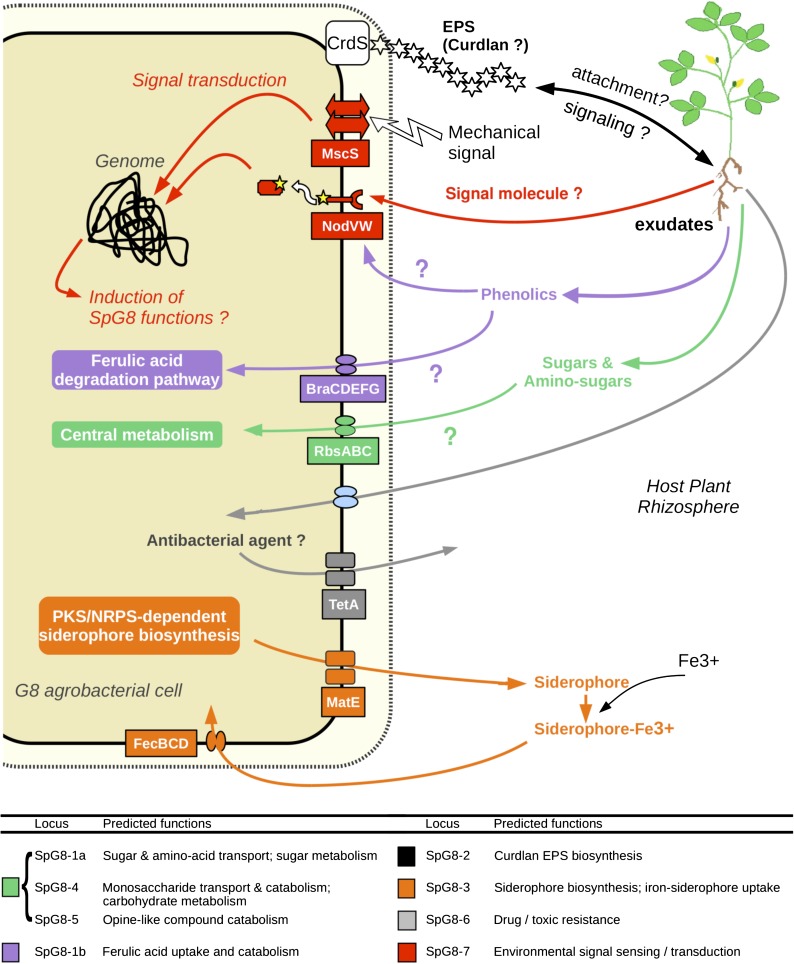
SpG8-2a: Curdlan Biosynthesis
Atu3056 in SpG8-2a codes for a putative beta-1,3-glucan synthase (curdlan synthase, CrdS) that is involved in the synthesis of curdlan, an exopolysaccharide. We experimentally verified this function by deleting the whole locus in C58 (i.e., Atu3054–Atu3059). As a result, colonies formed by the mutant C58ΔSpG8-2a did not bind Congo Red, whereas colonies formed by wild-type C58 were red, indicating that the mutant was affected in polysaccharide production (fig. 5B). In addition, we found that all G8 members similarly accumulated red dye, in contrast with members of other genomic species (data not shown), which demonstrates that this function is specific to G8.
FIG. 5.—
Experimental evidences of G8-specific phenotypes determined by SpG8 loci. (A) Ferulic acid degradation by A. tumefaciens strains determined by HPLC and UV spectrum at 320 nm. mAU, milli absorbance units. All genomovar G8 members were able to catabolize all ferulic acid in 12 h, contrary to non-G8 strains lacking SpG8-1b. (B) Curdlan production revealed by red dye on Congo red medium. C58: Agrobacterium tumefaciens wild-type strain C58 (red colonies), C58ΔSpG8-2a: SpG8-2a-deleted mutant (white colonies).
SpG8-3: Siderophore Biosynthesis, Release, and Reuptake
The largest SpG8 gene cluster, that is, SpG8-3, ranged from Atu3663 to Atu3693. Nearly, all SpG8-3 genes were also shared by G6-NCPPB 925 and G1-ICPB TT111. This region has already been characterized as coding for functions involved in siderophore biosynthesis in C58 (Rondon et al. 2004). It includes eight polypeptides that may form a mega-enzyme complex corresponding to seven nonribosomal peptide synthase (NRPS) modules and three polyketide synthase modules. An isolated NRPS gene (Atu3072) located at the remote locus SpG8-2 may also interact with this mega-enzyme complex. Genes for transporter proteins were also located in SpG8-3: Atu3669 coding for a transporter of the multidrug extrusion transporter family MATE, that includes proteins involved in secondary metabolite transport (Moriyama et al. 2008), and thus is perhaps involved in siderophore release in the medium; and Atu3684–Atu3691 that are homologous to fecABCDE genes involved in TonB-dependent reuptake of the siderophore when it is chelated to iron (Braun et al. 2006). Finally, Atu3684, Atu3692, and Atu3693 (fecAIR) seemed to form a cell surface signaling system, whose homolog in Escherichia coli was proposed to regulate the whole system of biosynthesis, release, and reuptake of the siderophore (Braun et al. 2006). We noted that the whole region was conserved in synteny with A. vitis S4 (30 genes, 51.2% amino-acid identity on average) on its larger plasmid.
SpG8-6: Detoxification
SpG8-6 (Atu4196–4221) contained three putative multidrug transporter systems. Interestingly, one of them (tetR-tetA, Atu4205-Atu4206) was experimentally characterized for tetracycline resistance in G8-C58 and G8-T37 (Luo and Farrand 1999). These authors did not detect this resistance in several other agrobacteria, and some of their genomic species assignments are now known: G4-B6, G4-ATCC15955, and G1-Bo542, thus confirming the G8 specificity of this genomic region. Tetracycline is, however, not the natural inducer of these genes (Luo and Farrand 1999). The TetR-TetA efflux pump system might allow for detoxification of other unknown compounds.
SpG8-4, SpG8-1a, SpG8-5: Carbohydate Catabolism
Among regions involved in carbohydate catabolism, SpG8-4 (Atu3808–3830) seems to constitute a functional unit dedicated to monosaccharide uptake, via the putative ribose-specific ABC transporter encoded by rbsAC1C2B genes, and sugar metabolism involving putative enzymatic functions such as rhamnose mutarotase or D-galactarate dehydrogenase. Four LysR-type transcriptional regulators were found within this locus, which could be involved in substrate-dependant regulation of metabolic pathways (Maddocks and Oyston 2008).
SpG8-1a (Atu1398–1409) also seemed to be involved in sugar metabolism with an ABC transporter operon putatively specific to monosaccharides and genes for glycolate catabolism enzymes. Two other ABC transporter operons were located alongside within SpG8-1, which homologs have been described to specifically import amino acids. These include genes homologous to braBCDEF genes from R. leguminosarum bv. viciae 8401 that are involved in branched-chain amino acid uptake (Hosie et al. 2002).
SpG8-5 (Atu3947–Atu3952) encodes enzymes similar to sarcosine oxidase, ornithine cyclodeaminase, and an alanine racemase with a lectine-like sugar-binding domain likely involved in the catabolism of opine-like compounds. Opines are condensates of an amino acid and a sugar or a cetonic acid that are well known to be involved in the ecology of plant pathogenic Agrobacterium (Vaudequin-Dransart et al. 1995). However, the annotation is not precise enough to ascertain the substrate molecule class. It is thus possible that the concerned substrates belong to another class of condensates of amino acids and sugars called Amadori compounds—a class of molecules produced in decaying plant material and thus common in humic soil.
All functions found in SpG8-1, SpG8-4, SpG8-5 are thus likely to confer G8 agrobacteria a general ability to metabolize sugar and/or opine/Amadori-like compounds. However, many functional annotations are made on the basis of protein similarities with databases. In the case of sugar-binding proteins and ABC transporters, protein families contain many sequences, only a few of which are characterized. Incidentally, although we obtained deletion mutants of these three regions, we could not yet assign precise candidate substrates to improve the annotation or to experimentally verify the predicted functions.
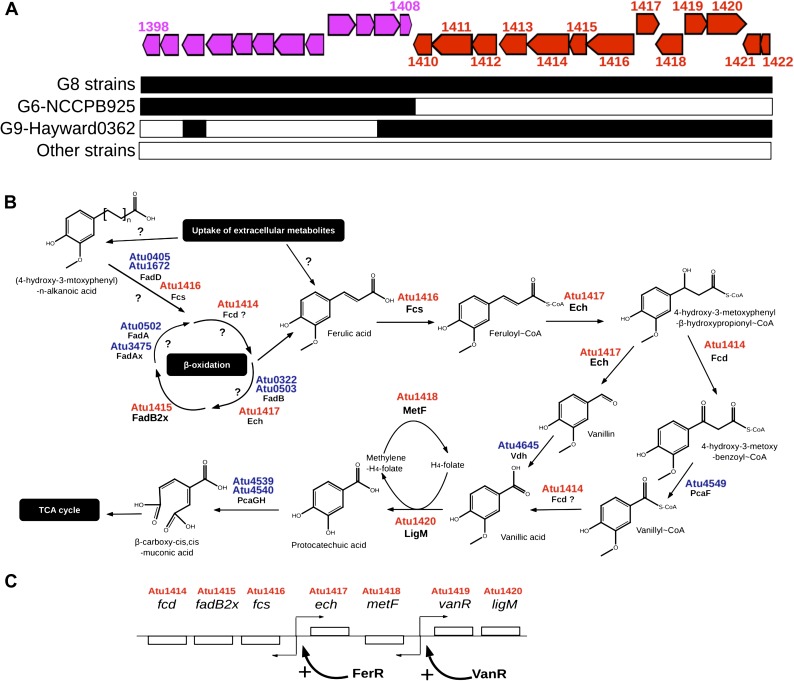
SpG8-1b: Phenolic Catabolism
SpG8-1b (Atu1409–Atu1423) was shared by all G8 strains and also by G9-Hayward 0362. This locus was involved in phenolic catabolism (fig. 6, supplementary table S6, Supplementary Material online for details on homology relationships). Indeed, SpG8-1b includes a gene homologous to fcs (Atu1416), which is involved in a pathway for CoA-dependent, non-beta-oxidative degradation of ferulic acid in Pseudomonas (Overhage et al. 1999; Plaggenborg et al. 2003; Calisti et al. 2008), and other putative enzymatic functions that could be related to the same metabolic pathway, including: an enoyl-CoA hydratase (Ech) (Atu1417), a feruloyl-CoA dehydratase (Fcd) (Atu1414), a tetrahydrofolate-dependent vanillate O-demethylase (LigM) (Atu1420), and a methylenetetrahydrofolate reductase (MetF) (Atu1418), as well as substrate-binding regulators VanR (Atu1419) and FerR (Atu1422). Indeed, we were able to reconstruct a complete ferulic acid degradation pathway (fig. 6B) and to propose a transcriptional regulation scheme (fig. 6C) in C58—and in other G8 strains as well—thanks to the presence of a gene nonspecific to G8 in the linear chromosome, that is, Atu4645 (vdh), encoding vanillin oxidase. The final product of this putative pathway was protocathechuic acid, which can be degraded into metabolites suitable for complete oxidation through the tri-carboxylic acid cycle (Parke 1995). In addition, the SpG8-1 gene Atu1415 encoded a putative n-phenylalkanoyl-CoA dehydratase. This enzyme is involved in a beta-oxidative pathway of long chain substituted phenolic degradation in Pseudomonas (Olivera et al. 2001), yielding short-chain phenylalkanoyl-CoAs such as cinnamoyl-CoA. This suggests alternative entries for this putative G8-specific phenolic degradation pathway: either by uptake of ferulic acid (as one of the cognate transporters could provide this ability) or by transformation of more complex phenolics, for example, by iterative oxidation of long chain–substituted cinnamic acids. G8 strains could thus likely degrade ferulic acid into protocathechuic acid and then assimilate it as a carbon source.
FIG. 6.—
Putative ferulic acid catabolism pathway encoded by SpG8-1b. (A) SpG8-1 CDSs organization in C58 (top): subregions SpG8-1a and SpG8-1b are colored in purple and red, respectively. Presence in other Agrobacterium tumefaciens strains (bottom): presence, black; absence, white. (B) Reconstructed ferulic acid catabolism pathway encoded by SpG8-1b according to similarities to sequences in databases and associated literature: Fcs, feruloyl-CoA synthetase (Overhage et al. 1999; Plaggenborg et al. 2003); Ech, enoyl-CoA hydratase (Pelletier and Harwood 1998); Fcd, feruloyl-CoA dehydratase; LigM, tetrahydrofolate-dependent vanillate O-demethylase (Nishikawa et al. 1998); MetF, methylenetetrahydrofolate reductase (Nishikawa et al. 1998). (C) Putative transcriptional regulation of SpG8-1b genes inferred from sequence similarities in databases: VanR, vanillate catabolism repressor (Morawski et al. 2000); FerR, ferulate catabolism regulator (Breese and Fuchs 1998; Calisti et al. 2008).
We verified this possibility by testing strains for their ability to degrade ferulic acid. After 12 h incubation, strains bearing SpG8-1b (G8 strains and G9-Hayward 0362) degraded all the ferulic acid in a comparable manner, whereas other strains did not (fig. 5A). In addition, a C58 mutant deleted for the whole SpG8-1b (C58ΔSpG8-1b, fig. 5A) was unable to degrade ferulic acid. This locus is therefore clearly involved in ferulic acid degradation.
As a generalization, other genes involved in aromatic compounds catabolism were found in other SpG8 gene clusters: a putative mandelate racemase, in SpG8-1a (Atu1406) and a putative shikimate dehydrogenase in SpG8-7b (Atu4295). These enzymatic reactions were parts of pathways leading to protocathechuic acid production, mandelate degradation, or shikimate degradation, respectively, suggesting that degradation of aromatic compounds into protocathechuic acid may be a crucial synapomorphic trait of genomovar G8.
Occurrence of SpG8 Genes in Other G8 Members
A question could be asked about the overall relevance of the present work. Within the chosen species, that is, the A. tumefaciens genomovar G8, we used the largest set of markedly different strains available when the array was designed. The results were thus valid for this set of strains, but are SpG8 genes also present in other G8 members? Indeed, curdlan biosynthesis genes were already described in the industrial agrobacterial strain ATCC 31749 (Portilho et al. 2006), that we found to be very likely another G8 member based on the high sequence similarities of several genes of ATCC 31749 and C58 as compared with other genomic species (data not shown). Similar observations could also apply to tetA-tetR (Luo and Farrand 1999). In addition, we tracked G8 members in the many agrobacterial strains that can be isolated from various environments. We succeeded in isolating a new one, that is, MKS.01 (CFBP 7336), which differed from all other G8 strains by core gene markers, and we verified that MKS.01 had all the G8-specific genes and phenotypes defined in the present work, including curdlan production and ferulic acid degradation (data not shown). Conversely, the SpG8 genomic islands appear to be absent from the recently published genomic sequence of the genomovar G1 strain H13-3 (Wibberg et al. 2011). All of these a posteriori verifications confirmed the hypothesis of genomic species characterized by common species-specific genes inherited from a common ancestor adapted to a specific primary ecological niche.
Discussion
The aim of the present study was to disclose the speciation mechanism leading to differentiation of genomic species by assessing the presence of species-specific genes in genomes. As this is amenable by comparative genomics, the present study was geared toward detecting C58 gene homologs in other genomes by using a microarray constructed with probes spanning the entire C58 genome. However, although it is easy to detect present genes with CGH arrays when hybridized DNAs are highly similar to C58 DNA (i.e., in genomovar G8), there is a dramatic decrease in the signal over background ratio for more divergent genomes from other genomic species. This difficulty was overcome by taking into account the regional organization of the hybridization signal along the genome because, in spite of their weakness, successive signals of present loci are generally more intense than noises of absent ones. Thanks to this partition procedure, we were able to confidently detect the presence of genes not only in all other species of the A. tumefaciens complex but also in the remote species A. larrymoorei. We, however, failed to obtain accurate results in A. vitis CFBP 5523T, R. rhizogenes K84, or E. meliloti 1021 (data not shown), likely because those bacteria generally diverged beyond the estimated detection threshold (80% similarity at best) of the procedure (supplementary table S4, Supplementary Material online).
G8-Specific Functions Useful for Life in Plant Rhizospheres
Remarkably, SpG8 functions seemed to collectively define an ecological niche of G8 agrobacteria related to commensal interactions with plants (fig. 7). Although agrobacteria are well known to be pathogenic to plants by inducing crown gall disease, this is a secondary ecological trait related to the availability of a dispensable plasmid (i.e., Ti plasmid which harbors the pathogenicity determinants). Agrobacteria are generally Ti plasmid free and are primarily common soilborne organisms able to live commensally in plant rhizospheres (Savka et al. 2002; Hartmann et al. 2009). As we were looking for general adaptive determinants of the species regardless of its pathogenic status, we assumed that species-specific adaptations were more likely related to life in soils or rhizospheres rather than to crown galls.
FIG. 7.—
Hypothetical integrated functioning of SpG8 genes allowing G8 member adaptation to their specific ecological niche. EPS, exopolysaccharide; CrdS, curdlan synthase; McsS, mechano-sensitive channel; NodVW, two-component system sensor kinase and response regulator; BraCDEFG, branched-chain amino acid transporter; RbsABC, ribose transporter; TetA, tetracycline extrusion pump; MatE, multidrug transporter; FecBCD, iron-siderophore transporter.
SpG8 loci code for numerous catabolic pathways of carbohydrates, namely ferulic acid (SpG8-1b), diverse sugars (SpG8-1a, SpG8-4), amino acids (SpG8-1a, SpG8-5), and opine-like/Amadori compounds (SpG8-5). All are typical molecules that can be found in plant rhizospheres, exuded by plants (complex sugars) or derived from plant degradation products (phenolics, opine/Amadori products). Clearly, ferulic acid is a plant compound involved in the lignin biosynthesis present at plant wounds (Humphreys et al. 1999). Degradation products are, however, released in soil and may thus facilitate the survival of agrobacteria in soil as well. The SpG8 loci involvement in life adaptation to plants or soil are therefore not exclusive alternatives. Moreover, sugar, phenolics, and opines are also known to play an important role in the pathogenicity of Ti plasmids harboring agrobacteria. In that sense, the present results support the exaptation hypothesis of Dessaux's team (Vaudequin-Dransart et al. 1995), who proposed that the ability of agrobacteria to use opines selectively arose from a more general ability of this taxon to use opine-like compounds, including Amadori products and other related substrates.
In addition to carbon resources available in the rhizosphere, other factors are important in the bacterial niche definition. For instance, bacterial cells may be able to recognize a favorable environment, reach it (e.g., via positive tropism) and stay inside it (e.g., via physical attachment), or modify it (e.g., by secreting extracellular products or stimulating a plant to modify its exudation spectrum). These functions involve molecular signaling that can be distant (by diffusion of a signal molecule) or by contact between the bacterium and its specific habitat (including a partner plant).
Indeed, production of insoluble β-1,3-glucan exopolysaccharide (curdlan) encoded by SpG8-2a may play a role in attachment (Matthysse and McMahan 1998; Rodríguez-Navarro et al. 2007) and contact signaling. Annotated functions may, however, have pleiotropic effects, and curdlan production may also be important for passive resistance to toxics, especially in plant rhizospheres where antimicrobial agents-like flavonoids are secreted (Palumbo et al. 1998). Other putative defense mechanism of G8 agrobacteria may be provided by SpG8 loci by action of multidrug exporters (SpG8-6), whereas the siderophore biosynthesis locus (SpG8-3) might provide another general fitness gain in competition with other bacteria present in the biotope. Scavenging of limiting resources like iron is known to be a very potent means to outperform competitors, especially in habitats like rhizospheres with dense and diverse populations, as described for plant growth-promoting rhizobacteria like Pseudomonas fluorescens or R. rhizogenes (Penyalver et al. 2001; Siddiqui 2006).
Finally, locus SpG8-7b, which encodes membrane proteins involved in the perception of mechanical and chemical signals, is a candidate to facilitate recognition of favorable environments. Interestingly, those putative environment-sensing genes are homologous to systems of perception of toluene and styrene in Pseudomonas sp. (Lau et al. 1997; Panke et al. 1998) and are conserved in synteny with those from the chromosome of P. lavamentivorans SD-1. This latter species is known to switch from motile to sessile behavior in the presence of phenyl-substituted long-chain fatty acids (Schleheck et al. 2004) that share structural features with ferulic acid. These homology relationships strongly suggest that two-component signaling systems of this family are activated by the presence of some phenolics in the medium. Moreover, these genes code proteins also homologous to NodVW/NwsAB from B. japonicum, which mediate host recognition during the nodulation process. Considering these relationships, we hypothesized that locus SpG8-7b is involved in the perception of signals from the environment that may be responsible for activation of other functions, including, perhaps, SpG8 functions such as phenolic metabolism. The frequent reference to phenolics in annotation of SpG8 genes suggests that these compounds could be of primary importance in the biology of G8 agrobacteria, being both metabolites and signals released by the host plant.
Evolutionary History of SpG8 Genes
The presence of species-specific genes can be understood as due to the conservation of ancestral genes lost in other species or to the acquisition of foreign genes by the most recent common ancestor. Several SpG8 regions (SpG8-1a, SpG8-4, and SpG8-7a) contained genes that were also found in G6-NCCPB925, the closest outgroup of G8. This suggests that these specific regions may have been present in the common ancestor of G8 and G6. Remarkably, these regions tended toward the codon usage signature of core-genome genes (supplementary table S5, Supplementary Material online, fig. 4). Based on these elements, SpG8-1, SpG8-4, and SpG8-7 may be clusters of ancestral genes already present in the genome of an ancient ancestor of agrobacteria, specifically retained in the [G6,G8] clade but lost in other clades. This is especially probable for region SpG8-1. This region was possibly present as an entire cluster in ancestors of G6-NCCPB925 and G9-Hayward0362 and may then have been partially lost, leading to differential retention of subregions SpG8-1a and SpG8-1b in G6-NCCPB925 and G9-Hayward0362, respectively (fig. 6A). In contrast, transfers may be more likely for SpG8 genes shared with more distant genomic species such as G1 (SpG8-3 and SpG8-6a gene clusters) or G4 (Atu4215-4218 in cluster SpG8-6b), which have sporadic-like codon usage signatures. This suggests that lateral gene transfer as well as gene retention contribute to the establishment of a species-specific gene repertoire.
Gene Content Flexibility of the Linear Chromosome
As previously observed at higher taxonomic level by comparing Agrobacterium “biovars” (i.e., A. tumefaciens C58 to A. vitis S4 and R. rhizogenes K84; Slater et al. 2009) and other bacteria such as Vibrio (Chen et al. 2003; Vesth et al. 2010), the second chromosome of A. tumefaciens genomic species also appears as the major spot for innovation in the gene repertoire (fig. 3). This high genomic flexibility of the second chromosome was moreover likely facilitated by a linear architecture as illustrated by the transfer of half of the linear chromosome between C58 and other G8 members (fig. 1). Actually, a bacterial linear chromosome could behave as a standard eukaryotic chromosome requiring a single crossover to exchange almost a complete chromosome branch. Linear chromosomes, which are rare genomic features in bacteria, may facilitate the spread of adaptive genomic innovations and likely played a key role in speciation in the A. tumefaciens complex as suspected in Streptomyces or Borrelia spp. (Volff and Altenbuchner 2000; Chen et al. 2010).
Parapatric Speciation Gives Rise to Genomic Species
We found genes coding ecologically relevant functions present in the genomes of members of a given genomic species but not in its closest relatives. They were likely present in the most recent common ancestor of its members likely allowing ecological isolation. We may in turn speculate this isolation initiated the speciation process. We chose to work with a genomic species with high known diversity (Mougel et al. 2002; Portier et al. 2006; Costechareyre et al. 2009, 2010) and also because this species has very closely related sister species often co-inhabiting the same soil (Costechareyre et al. 2010), even at the very microscale (Vogel et al. 2003). Agrobacteria are moreover common rhizospheric bacteria (Krimi et al. 2002, Costechareyre et al. 2010) which genomic species are differentially trapped according to plant host (Lavire C, unpublished data). Agrobacteria in soils thus form ecological guilds where every species likely taps the same resources (e.g., rhizospheres) in a similar way, except for a few specific traits. As agrobacterial species are not geographically isolated and because they have determinants for species-specific ecological niche, we assume that these species have arisen by parapatric speciation. It is likely that speciations in the same habitat occurred as a consequence of local adaptations to host plants, as suggested by annotations of G8-specific functions.
Adaptations to plants might be related to host specificity as already suggested by the known preferential occurrence of A. rubi, A. vitis, and A. larrymoorei in tumors of Rubus sp., Vitis sp., and Ficus benjamina, respectively. Determination of specific adaptations of A. tumefaciens species may also improve our knowledge about crown gall epidemiology, including preferential spread by some hosts. In the case of G8, we suspected preferential trapping by Medicago truncatula (Lavire C, unpublished data), echoing the homology of SpG8-7 with sensors of E. meliloti—the symbiont of M. truncatula. Adaptation to plant is possibly not confined to commensal adaptation to the root biotope but more generally to ecological features encountered in the whole plant, including tumors. Consequently, it is possible that A. tumefaciens species adaptations to plants may also modulate the epidemiology of pathogenic agrobacteria.
Interest of Ecological Species Concept Investigation for Taxonomy
Agrobacterium tumefaciens genomic species are valid species but they are still awaiting a valid Latin binomial because they were lacking well-characterized distinctive phenotypic traits. Novel G8 members could be identified by phylogenetic analysis of core genes such as recA or chvA, as previously described (Costechareyre et al. 2009, 2010) or by looking for G8-specific genes via CGH microarrays or PCR. However, the present work actually emphasizes several traits such as curdlan production, ferulic acid degradation, resistance to tetracyclin for genomovar G8 that, when combined, would be valuable traits for species distinction. This is why, in agreement with the latest recommendations of Stackebrandt et al. (2002), we propose to valid the status of genomovar G8 as a recognized bacterial species by giving it a Latin binomial and a type strain, C58. We thus propose this novel species be named Agrobacterium fabrum, from the Latin plural genitive of smith, in reference to the use of C58 to construct genetically modified plants, while also honoring the pioneer isolator of Agrobacterium (Smith) as well as other scientists with a Faber-related name in different languages, for example, Smith, Schmidt, Smet, Faivre, Farrand, Faure, Herrera, etc., who studied various aspects of Agrobacterium biology.
Generalization of the Concept of Bacterial Genomic Species as Ecological Species
The question of ecological speciation of bacteria is still in debate (Achtman and Wagner 2008) partly because the bacterial species definition is at the center of this debate. Here, we only consider the genomically based species definition still acknowledged by international taxonomic committees (Wayne et al. 1987; Stackebrandt et al. 2002), even though if there is still named bacterial species—especially in anciently described human pathogens—that do not fit the genomic species criterion. We thus chose a taxon level relevant for the current taxonomy and intended to verify that this taxon level could have specific ecological features that scheme a potential primary niche. This was usually achieved by investigating differential ecological properties of species as for instance within the genus Prochlorococcus (Johnson et al. 2006). We showed here that the discovery of the specific ecological niche of a species is amenable by comparative genomic, when it is performed with several strains within this species compared with strains belonging to closely related species. This was done with Salmonella enterica (Porwollik et al. 2002), Lactobacillus casei (Cai et al. 2009), and Campylobacter coli versus C. jejuni (Lefébure et al. 2010). This should be generalized in future taxonomic investigations in order to improve the biological information attached to novel species. Of course, it is also possible to infer primary ecology of other taxonomic levels such as strain clusters within a species as shown in E. coli (Touchon et al. 2009), genera, or still higher taxa (Philippot et al. 2010). Interestingly, these latter authors showed that the broader the clade, the less defined is the associated ecology.
The present study highlighted the relevance of looking for species-specific genes by assessing genome features. We showed that—at least for the present model—species-specific genes were involved in ecological adaptations to the species primary niche. Consequently, it is likely that in this instance the genomic species was of ecovar descent. Our study benefits from the synergy between bioinformatic treatments of high throughput data and bench works. Both approaches are essential for reconstructing—without a priori knowledge—a reliable ecological niche model for further investigations on bacterial speciation and evolution.
Supplementary Material
Supplementary tables S1–S9 and figures S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
F.L. received a doctoral grant from ENS-Lyon, T.C. from Ministère de l'Éducation nationale de l'Enseignement Supérieur et de la Recherche, and M.S. from Ministère des Affaires Etrangères et Européennes. D.M. was supported by an INRA postdoctoral fellowship. The authors would like to thank P. Oger who allowed the inclusion of pTiBo542 in the microarray design, G. Meiffren at CESN for assistance in HPLC analyses, P. Portier at CFBP (http://www-intranet.angers.inra.fr/cfbp/) for strain repository, A. Calteau at Genoscope (https://www.genoscope.cns.fr/agc/microscope/) for AgrobacterScope platform management, M.K. Lhommé at Lyon 2 University for Latin advice, and translator Dr D. Manley for reading the manuscript and providing suggestions. This work was supported by the EcoGenome project of Agence Nationale de la Recherche (grant number BLAN08-1_335186); Lyon 1 University (grant numbers IFR41-2006_Nesme, BQR-2006_Nesme); Bureau des Ressources Génétiques (grant number BRG-2005_Nesme); and the Département Santé des Plantes et Environnement of INRA (grant number DSPE-2005_Nesme).
References
- Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:439. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- Bouzar H, Jones J. Agrobacterium larrymoorei sp. nov., a pathogen isolated from aerial tumours of Ficus benjamina. Int J Syst Evol Microbiol. 2001;51:1023–1026. doi: 10.1099/00207713-51-3-1023. [DOI] [PubMed] [Google Scholar]
- Braun V, Mahren S, Sauter A. Gene regulation by transmembrane signaling. Biometals. 2006;19:103–113. doi: 10.1007/s10534-005-8253-y. [DOI] [PubMed] [Google Scholar]
- Breese K, Fuchs G. 4-Hydroxybenzoyl-CoA reductase (dehydroxylating) from the denitrifying bacterium Thauera aromatica—prosthetic groups, electron donor, and genes of a member of the molybdenum-flavin-iron-sulfur proteins. Eur J Biochem. 1998;251:916–923. doi: 10.1046/j.1432-1327.1998.2510916.x. [DOI] [PubMed] [Google Scholar]
- Cai H, et al. Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol Evol. 2009;1:239–257. doi: 10.1093/gbe/evp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisti C, Ficca AG, Barghini P, Ruzzi M. Regulation of ferulic catabolic genes in Pseudomonas fluorescens BF13: involvement of a MarR family regulator. Appl Microbiol Biotechnol. 2008;80:475–483. doi: 10.1007/s00253-008-1557-4. [DOI] [PubMed] [Google Scholar]
- Charif D, Lobry JR. 2007. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In: Bastolla U, Porto M, Roman E, Vendruscolo M, editors. Structural approaches to sequence evolution: Molecules networks populations. Heidelberg (Germany): Springer-Verlag. p. 207–232. [Google Scholar]
- Chen CY, et al. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 2003;13:2577–2587. doi: 10.1101/gr.1295503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, et al. Chromosomal instability in Streptomyces avermitilis: major deletion in the central region and stable circularized chromosome. BMC Microbiol. 2010;10:198. doi: 10.1186/1471-2180-10-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Schweizer HP. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 2005;5:30. doi: 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civolani C, Barghini P, Roncetti AR, Ruzzi M, Schiesser A. Bioconversion of ferulic acid into vanillic acid by means of a vanillate-negative mutant of Pseudomonas fluorescens strain BF13. Appl Environ Microbiol. 2000;66:2311–2317. doi: 10.1128/aem.66.6.2311-2317.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJA, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan FM. Sexual isolation and speciation in bacteria. Genetica. 2002;116:359–370. [PubMed] [Google Scholar]
- Connor N, et al. Ecology of speciation in the genus Bacillus. Appl Environ Microbiol. 2010;76(5):1349–1358. doi: 10.1128/AEM.01988-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costechareyre D, Bertolla F, Nesme X. Homologous recombination in Agrobacterium: potential implications for the genomic species concept in bacteria. Mol Biol Evol. 2009;26:167–176. doi: 10.1093/molbev/msn236. [DOI] [PubMed] [Google Scholar]
- Costechareyre D, et al. Rapid and efficient identification of Agrobacterium species by recA allele analysis: Agrobacterium recA diversity. Microb Ecol. 2010;60(4):862–872. doi: 10.1007/s00248-010-9685-7. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour A, Dray S. The ade4 package: implementing the duality diagram for ecologists. J Stat Softw. [Internet] 2007;22(i04) [cited 2011 Jul 28] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. Seattle (WA): Department of Genome Sciences, University of Washington. 1993. [Google Scholar]
- Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. The bacterial species challenge: making sense of genetic and ecological diversity. Science. 2009;323:741–746. doi: 10.1126/science.1159388. [DOI] [PubMed] [Google Scholar]
- Galibert F, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- Gevers D, et al. Re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- Goodner B, et al. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science. 2001;294:2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- Guéguen L. Sarment: Python modules for HMM analysis and partitioning of sequences. Bioinformatics. 2005;21:3427–3428. doi: 10.1093/bioinformatics/bti533. [DOI] [PubMed] [Google Scholar]
- Hardin G. The competitive exclusion principle. Science. 1960;131:1292–1297. doi: 10.1126/science.131.3409.1292. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Schmid M, Tuinen D, Berg G. Plant-driven selection of microbes. Plant Soil. 2009;321:235–257. [Google Scholar]
- Hosie AHF, Allaway D, Galloway CS, Dunsby HA, Poole PS. Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC family. J Bacteriol. 2002;184:4071–4080. doi: 10.1128/JB.184.15.4071-4080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys JM, Hemm MR, Chapple C. New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci U S A. 1999;96:10045–10050. doi: 10.1073/pnas.96.18.10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZI, et al. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- Kneen BE, LaRue TA. Congo red absorption by Rhizobium leguminosarum. Appl Environ Microbiol. 1983;45:340–342. doi: 10.1128/aem.45.1.340-342.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimi Z, Petit A, Mougel C, Dessaux Y, Nesme X. Seasonal fluctuations and long-term persistence of pathogenic populations of Agrobacterium spp. in soils. Appl Environ Microbiol. 2002;68:3358–33654. doi: 10.1128/AEM.68.7.3358-3365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JA. Reconstructing evolutionary trees from DNA and protein sequences: paralinear distances. Proc Natl Acad Sci U S A. 1994;91:1455–1459. doi: 10.1073/pnas.91.4.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K, Lindemann A, Hauser F, Göttfert M. The genistein stimulon of Bradyrhizobium japonicum. Mol Genet Genomics. 2008;279:203–211. doi: 10.1007/s00438-007-0280-7. [DOI] [PubMed] [Google Scholar]
- Lau PC, et al. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc Natl Acad Sci U S A. 1997;94:1453–1458. doi: 10.1073/pnas.94.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefébure T, Pavinski Bitar PD, Suzuki H, Stanhope MJ. Evolutionary dynamics of complete Campylobacter pan-genomes and the bacterial species concept. Genome Biol Evol. 2010;2:646–655. doi: 10.1093/gbe/evq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZQ, Farrand SK. Cloning and characterization of a tetracycline resistance determinant present in Agrobacterium tumefaciens C58. J Bacteriol. 1999;181:618–626. doi: 10.1128/jb.181.2.618-626.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks SE, Oyston PCF. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- Martens M, et al. Multilocus sequence analysis of Ensifer and related taxa. Int J Syst Evol Microbiol. 2007;57:489–503. doi: 10.1099/ijs.0.64344-0. [DOI] [PubMed] [Google Scholar]
- Matthysse AG, McMahan S. Root colonization by Agrobacterium tumefaciens is reduced in cel, attB, attD, and attR mutants. Appl Environ Microbiol. 1998;64:2341–2345. doi: 10.1128/aem.64.7.2341-2345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Systematics and the origin of species, from the viewpoint of a zoologist. New York (NY): Columbia University Press; 1942. [Google Scholar]
- doi: 10.1111/j.1574-6968.2000.tb09138.x. Morawski B, Segura A, Ornston LN. 2000. Repression of Acinetobacter vanillate demethylase synthesis by VanR, a member of the GntR family of transcriptional regulators. FEMS Microbiol Lett. 187:65–68. [DOI] [PubMed] [Google Scholar]
- Moriyama Y, Hiasa M, Matsumoto T, Omote H. Multidrug and toxic compound extrusion (MATE)-type proteins as anchor transporters for the excretion of metabolic waste products and xenobiotics. Xenobiotica. 2008;38(7–8):1107–1118. doi: 10.1080/00498250701883753. [DOI] [PubMed] [Google Scholar]
- Mougel C, Thioulouse J, Perrière G, Nesme X. A mathematical method for determining genome divergence and species delineation using AFLP. Int J Syst Evol Microbiol. 2002;52:573–586. doi: 10.1099/00207713-52-2-573. [DOI] [PubMed] [Google Scholar]
- Nelder JA, Mead R. A simplex method for function minimization. Comput J. 1965;7:308–313. [Google Scholar]
- Nishikawa S, et al. Cloning and sequencing of the Sphingomonas (Pseudomonas) paucimobilis gene essential for the O demethylation of vanillate and syringate. Appl Environ Microbiol. 1998;64:836–842. doi: 10.1128/aem.64.3.836-842.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera ER, et al. Two different pathways are involved in the beta-oxidation of n-alkanoic and n-phenylalkanoic acids in Pseudomonas putida U: genetic studies and biotechnological applications. Mol Microbiol. 2001;39:863–874. doi: 10.1046/j.1365-2958.2001.02296.x. [DOI] [PubMed] [Google Scholar]
- Ophel K, Kerr A. Agrobacterium vitis sp. nov. for strains of Agrobacterium biovar 3 from grapevines. Int J Syst Bacteriol. 1990;40:236–241. [Google Scholar]
- Overhage J, Priefert H, Steinbuchel A. Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl Environ Microbiol. 1999;65:4837–4847. doi: 10.1128/aem.65.11.4837-4847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo JD, Kado CI, Phillips DA. An isoflavonoid-inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots. J Bacteriol. 1998;180:3107–3113. doi: 10.1128/jb.180.12.3107-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panke S, Witholt B, Schmid A, Wubbolts MG. Towards a biocatalyst for (S)-styrene oxide production: characterization of the styrene degradation pathway of Pseudomonas sp. strain VLB120. Appl Environ Microbiol. 1998;64:2032–2043. doi: 10.1128/aem.64.6.2032-2043.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parke D. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens. J Bacteriol. 1995;177:3808–3817. doi: 10.1128/jb.177.13.3808-3817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier DA, Harwood CS. 2-Ketocyclohexanecarboxyl coenzyme A hydrolase, the ring cleavage enzyme required for anaerobic benzoate degradation by Rhodopseudomonas palustris. J Bacteriol. 1998;180:2330–2336. doi: 10.1128/jb.180.9.2330-2336.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penyalver R, Oger P, López MM, Farrand SK. Iron-binding compounds from Agrobacterium spp.: biological control strain Agrobacterium rhizogenes K84 produces a hydroxamate siderophore. Appl Environ Microbiol. 2001;67:654–664. doi: 10.1128/AEM.67.2.654-664.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit A, et al. Substrate induction of conjugative activity of Agrobacterium tumefaciens Ti plasmids. Nature. 1978;271:570–572. [Google Scholar]
- Philippot L, et al. The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol. 2010;8:523–529. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- Plaggenborg R, Overhage J, Steinbüchel A, Priefert H. Functional analyses of genes involved in the metabolism of ferulic acid in Pseudomonas putida KT2440. Appl Microbiol Biotechnol. 2003;61:528–535. doi: 10.1007/s00253-003-1260-4. [DOI] [PubMed] [Google Scholar]
- Portier P, et al. Identification of genomic species in Agrobacterium biovar 1 by AFLP genomic markers. Appl Environ Microbiol. 2006;72:7123–7131. doi: 10.1128/AEM.00018-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portilho M, Matioli G, Zanin GM, de Moraes FF, Scamparini ARP. Production of insoluble exopolysaccharide of Agrobacterium sp. (ATCC 31749 and IFO 13140) Appl Biochem Biotechnol. 2006;131:864–869. doi: 10.1385/ABAB:131:1:864. [DOI] [PubMed] [Google Scholar]
- Porwollik S, Wong RM, McClelland M. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc Natl Acad Sci U S A. 2002;99:8956–8961. doi: 10.1073/pnas.122153699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt J, Hynes MF. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for statistical computing; 2009. [Google Scholar]
- Rodríguez-Navarro DN, Dardanelli MS, Ruíz-Saínz JE. Attachment of bacteria to the roots of higher plants. FEMS Microbiol Lett. 2007;272:127–136. doi: 10.1111/j.1574-6968.2007.00761.x. [DOI] [PubMed] [Google Scholar]
- Rondon MR, Ballering KS, Thomas MG. Identification and analysis of a siderophore biosynthetic gene cluster from Agrobacterium tumefaciens C58. Microbiology. 2004;150:3857–3866. doi: 10.1099/mic.0.27319-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor (NY): CSHL Press; 2001. [Google Scholar]
- Sanjay AB, Stach JEM, Goodfellow M. Genetic and phenotypic evidence for Streptomyces griseus ecovars isolated from a beach and dune sand system. Antonie Van Leeuwenhoekss. 2008;94:63–74. doi: 10.1007/s10482-008-9246-y. [DOI] [PubMed] [Google Scholar]
- Savka MA, Dessaux Y, Oger P, Rossbach S. Engineering bacterial competitiveness and persistence in the phytosphere. Mol Plant Microbe Interact. 2002;15:866–874. doi: 10.1094/MPMI.2002.15.9.866. [DOI] [PubMed] [Google Scholar]
- Schleheck D, Tindall BJ, Rosselló-Mora R, Cook AM. Parvibaculum lavamentivorans gen. nov., sp. nov., a novel heterotroph that initiates catabolism of linear alkylbenzenesulfonate. Int J Syst Evol Microbiol. 2004;54:1489–1497. doi: 10.1099/ijs.0.03020-0. [DOI] [PubMed] [Google Scholar]
- Siddiqui ZA. PGPR: biocontrol and biofertilization. Dordrecht (The Netherlands): Springer; 2006. [Google Scholar]
- Sikorski J, Nevo E. Adaptation and incipient sympatric speciation of Bacillus simplex under microclimatic contrast at “Evolution Canyons” I and II, Israel. Proc Natl Acad Sci U S A. 2005;102:15924–15929. doi: 10.1073/pnas.0507944102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater SC, et al. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J Bacteriol. 2009;191:2501–2511. doi: 10.1128/JB.01779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath PHA, Sokal RR. Numerical taxonomy. San Fransisco (CA): WH Freeman and Co; 1973. [Google Scholar]
- Stackebrandt E, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- Staley JT. Speciation and bacterial phylospecies. In: Bull AT, editor. Microbial diversity and bioprospecting. Washington (DC): ASM Press; 2004. pp. 40–47. [Google Scholar]
- Touchon M, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D, et al. MicroScope: a platform for microbial genome annotation and comparative genomics. Database. 2009;2009:bap021. doi: 10.1093/database/bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudequin-Dransart V, et al. Novel Ti plasmids in Agrobacterium strains isolated from fig tree and chrysanthemum tumors and their opine-like molecules. Mol Plant Microbe Interact. 1995;8:311–321. doi: 10.1094/mpmi-8-0311. [DOI] [PubMed] [Google Scholar]
- Velázquez E, et al. Analysis of core genes supports the reclassification of strains Agrobacterium radiobacter K84 and Agrobacterium tumefaciens AKE10 into the species Rhizobium rhizogenes. Syst Appl Microbiol. 2010;33:247–251. doi: 10.1016/j.syapm.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Vesth T, et al. On the origins of a Vibrio species. Microb Ecol. 2010;59:1–13. doi: 10.1007/s00248-009-9596-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Normand P, Thioulouse J, Nesme X, Grundmann GL. Relationship between spatial and genetic distance in Agrobacterium spp. in 1 cubic centimeter of soil. Appl Environ Microbiol. 2003;69:1482–1487. doi: 10.1128/AEM.69.3.1482-1487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN, Altenbuchner J. A new beginning with new ends: linearisation of circular chromosomes during bacterial evolution. FEMS Microbiol Lett. 2000;186:143–150. doi: 10.1111/j.1574-6968.2000.tb09095.x. [DOI] [PubMed] [Google Scholar]
- Wayne LG, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- Wibberg D, et al. Complete genome sequencing of Agrobacterium sp. H13–3, the former Rhizobium lupini H13-3, reveals a tripartite genome consisting of a circular and a linear chromosome and an accessory plasmid but lacking a tumor-inducing Ti-plasmid. J Biotechnol. 2011 doi: 10.1016/j.jbiotec.2011.01.010. Forthcoming doi:10.1016/j.jbiotec.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Wood DW, et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- Zhao J, Deng Y, Manno D, Hawari J. Shewanella spp. genomic evolution for a cold marine lifestyle and in-situ explosive biodegradation. PLoS One. 2010;5:e9109. doi: 10.1371/journal.pone.0009109. [DOI] [PMC free article] [PubMed] [Google Scholar]