Summary
The Cancer Genome Atlas (TCGA) project has analyzed mRNA expression, miRNA expression, promoter methylation, and DNA copy number in 489 high-grade serous ovarian adenocarcinomas (HGS-OvCa) and the DNA sequences of exons from coding genes in 316 of these tumors. These results show that HGS-OvCa is characterized by TP53 mutations in almost all tumors (96%); low prevalence but statistically recurrent somatic mutations in 9 additional genes including NF1, BRCA1, BRCA2, RB1, and CDK12; 113 significant focal DNA copy number aberrations; and promoter methylation events involving 168 genes. Analyses delineated four ovarian cancer transcriptional subtypes, three miRNA subtypes, four promoter methylation subtypes, a transcriptional signature associated with survival duration and shed new light on the impact on survival of tumors with BRCA1/2 and CCNE1 aberrations. Pathway analyses suggested that homologous recombination is defective in about half of tumors, and that Notch and FOXM1 signaling are involved in serous ovarian cancer pathophysiology.
Background
Ovarian cancer is the fifth leading cause of cancer death among women in the U.S., with 21,880 new cases and 13,850 deaths predicted for 20101. Most deaths are of patients presenting with advanced stage, high grade serous ovarian cancer (HGS-OvCa)2,3 (~70%). The standard of care is aggressive surgery followed by platinum/taxane chemotherapy. After therapy, platinum resistant cancer recurs in approximately 25% of patients within 6 months4 and overall 5-year survival is 31%5. Approximately 13% of HGS-OvCa is attributable to germline mutations in BRCA1 or BRCA26,7, while a smaller percentage can be accounted for by other germline mutations. However, most ovarian cancer can be attributed to a growing number of somatic aberrations8.
The lack of successful treatment strategies led TCGA to comprehensively measure genomic and epigenomic abnormalities on clinically annotated HGS-OvCa samples in order to identify molecular abnormalities that influence pathophysiology, affect outcome, and constitute therapeutic targets. Microarray analyses produced high resolution measurements of mRNA expression, microRNA expression, DNA copy number, and DNA promoter region methylation for 489 HGS-OvCa while massively parallel sequencing coupled with hybrid affinity capture9,10 provided whole exome DNA sequence information for 316 of these samples.
Samples and clinical data
This report covers analysis of 489 clinically annotated stage II-IV HGS-OvCa and corresponding normal DNA (Methods S1, Table S1.1). Patients reflected the age at diagnosis, stage, tumor grade, and surgical outcome of individuals diagnosed with HGS-OvCa. Clinical data were current as of August 25, 2010. HGS-OvCa specimens were surgically resected before systemic treatment but all patients received a platinum agent and 94% received a taxane. The median progression-free and overall survival of the cohort is similar to previously published trials11,12. Twenty five percent of the patients remained free of disease and 45% were alive at the time of last follow-up, while 31% progressed within 6 months after completing platinum-based therapy. Median follow up was 30 months (range 0 to 179). Samples for TCGA analysis were selected to have > 70% tumor cell nuclei and < 20% necrosis.
Coordinated molecular analyses using multiple molecular assays at independent sites were carried out as listed in Table 1. Data are available at http://tcga.cancer.gov/dataportal in two tiers. Tier one datasets are openly available, while tier two datasets include clinical or genomic information that could identify an individual hence require qualification as described at http://tcga.cancer.gov/dataportal/data/access/closed/.
Table 1. Characterization platforms used and data produced.
| Data Type | Platforms | Cases | Data Availability |
|---|---|---|---|
| DNA Sequence of exome | Illumina GAIIxa,b ABI SOLiDc |
236 80 |
Protected Protected |
| Mutations present in exome | 316 | Open | |
| DNA copy number/genotype |
Agilent 244Kd,e Agilent 415Kd Agilent 1Me Illumina 1MDUOf Affymetrix SNP6a |
97 304 539 535 514 |
Open Open Open Protected Protected |
| mRNA expression profiling | Affymetrix U133Aa Affymetrix Exong Agilent 244Kh |
516 517 540 |
Open Protected Open |
| Integrated mRNA expression |
489 | Open | |
| miRNA expression profiling |
Agilenth | 541 | Open |
| CpG DNA methylation | Illumina 27K1 | 519 | Open |
| Integrative analysis | 489 | Open | |
|
Integrative analysis w/ mutations |
309 | Open |
Production Centers: Broad Institute, Washington University School of Medicine, Baylor College of Medicine, Harvard Medical School, Memorial Sloan-Kettering Cancer Center, HudsonAlpha Institute for Biotechnology, Lawrence Berkeley National Laboratory, University of North Carolina, University of Southern California.
Additional data are available for many of these data types at the TCGA DCC.
Mutation analysis
Exome capture and sequencing was performed on DNA isolated from 316 HGS-OvCa samples and matched normal samples for each individual (Methods S2). Capture reagents targeted ~180,000 exons from ~18,500 genes totaling ~33 megabases of non-redundant sequence. Massively parallel sequencing on the Illumina GAIIx platform (236 sample pairs) or ABI SOLiD 3 platform (80 sample pairs) yielded ~14 gigabases per sample (~9×109 bases total). On average, 76% of coding bases were covered in sufficient depth in both the tumor and matched normal samples to allow confident mutation detection (Methods S2, Figure S2.1). 19,356 somatic mutations (~61 per tumor) were annotated and classified in Table S2.1. Mutations that may be important in HGS-OvCa pathophysiology were identified by (a) searching for non-synonymous or splice site mutations present at significantly increased frequencies relative to background, (b) comparing mutations in this study to those in COSMIC and OMIM and (c) predicting impact on protein function.
Two different algorithms (Methods S2) identified 9 genes (Table 2) for which the number of non-synonymous or splice site mutations was significantly above that expected based on mutation distribution models. Consistent with published results13, TP53 was mutated in 303 of 316 samples (283 by automated methods and 20 after manual review), BRCA1 and BRCA2 had germline mutations in 9% and 8% of cases, respectively, and both showed somatic mutations in an additional 3% of cases. Six other statistically recurrently mutated genes were identified; RB1, NF1, FAT3,CSMD3, GABRA6, and CDK12. CDK12 is involved in RNA splicing regulation14 and was previously implicated in lung and large intestine tumors15,16. Five of the nine CDK12 mutations were either nonsense or indel, suggesting potential loss of function, while the four missense mutations (R882L, Y901C, K975E, and L996F) were clustered in its protein kinase domain. GABRA6 and FAT3 both appeared as significantly mutated but did not appear to be expressed in HGS-OvCa (Supplemental Figure S2.1) or fallopian tube tissue so it is less likely that mutation of these genes plays a significant role in HGS-OvCa.
Table 2. Significantly mutated genes in HGS-OvCa.
| Gene | Number of Mutations |
Validated | Unvalidated |
|---|---|---|---|
| TP53 | 302 | 294 | 8 |
| BRCA1 | 11 | 10 | 1 |
| CSMD3 | 19 | 19 | 0 |
| NF1 | 13 | 13 | 0 |
| CDK12 | 9 | 9 | 0 |
| FAT3 | 19 | 18 | 1 |
| GABRA6 | 6 | 6 | 0 |
| BRCA2 | 10 | 10 | 0 |
| RB1 | 6 | 6 | 0 |
Validated mutations are those that have been confirmed with an independent assay. Most of them are validated using a second independent WGA sample from the same tumor. Unvalidated mutations have not been independently confirmed but have a high likelihood to be true mutations. An additional 25 mutations in TP53 were observed by hand curation.
Mutations from this study were compared to mutations in the COSMIC17 and OMIM18 databases to identify additional HGS-OvCa genes that are less commonly mutated. This yielded 477 and 211 matches respectively (Table S2.4) including mutations in BRAF (N581S), PIK3CA (E545K and H1047R), KRAS (G12D), and NRAS (Q61R). These mutations have been shown to exhibit transforming activity so we believe that these mutations are rare but important drivers in HGS-OvCa.
We combined evolutionary information from sequence alignments of protein families and whole vertebrate genomes, predicted local protein structure and selected human SwissProt protein features (Methods S3) to identify putative driver mutations using CHASM19,20 after training on mutations in known oncogenes and tumor suppressors. CHASM identified 122 mis-sense mutations predicted to be oncogenic (Table S3.1). Mutation-driven changes in protein function were deduced from evolutionary information for all confirmed somatic missense mutations by comparing protein family sequence alignments and residue placement in known or homology-based three-dimensional protein structures using Mutation Assessor (Methods S4). Twenty-seven percent of missense mutations were predicted to impact protein function (Table S2.1).
Copy number analysis
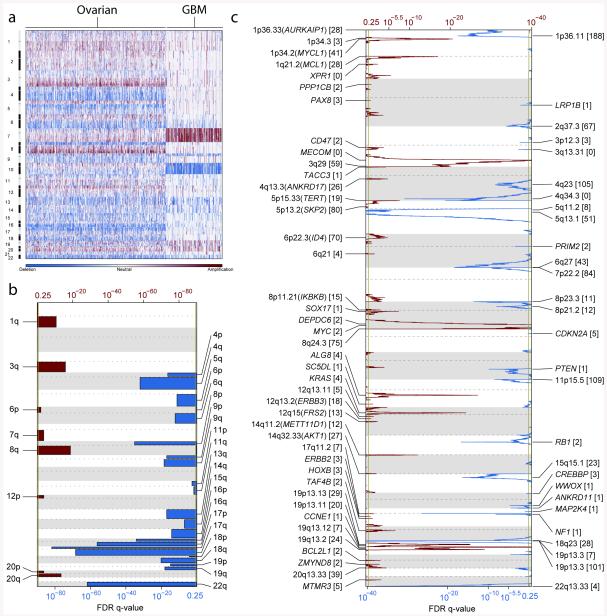
Somatic copy number alterations (SCNAs) present in the 489 HGS-OvCa genomes were identified and compared with glioblastome multiforme data in Figure 1a. SCNAs were divided into regional aberrations that affected extended chromosome regions and smaller focal aberrations (Methods S5). A statistical analysis of regional aberrations (Methods S5)21 identified 8 recurrent gains and 22 losses, all of which have been reported previously22 (Figure 1b and Table S5.1). Five of the gains and 18 of the losses occurred in more than 50% of tumors.
Figure 1. Genome copy number abnormalities.
(a) Copy-number profiles of 489 HGS-OvCa, compared to profiles of 197 glioblastoma multiforme (GBM) tumors46. Copy number increases (red) and decreases (blue) are plotted as a function of distance along the normal genome. (b) Significant, focally amplified (red) and deleted (blue) regions are plotted along the gnome. Annotations include the 20 most significant amplified and deleted regions, well-localized regions with 8 or fewer genes, and regions with known cancer genes or genes identified by genome-wide loss-of-function screens. The number of genes included in each region is given in brackets. (c) Significantly amplified (red) and deleted (blue) chromosome arms.
GISTIC21,23 (Methods S5) was used to identify recurrent focal SCNAs. This yielded 63 regions of focal amplification (Figure 1c, Methods S5, Table S5.2) including 26 that encoded 8 or fewer genes. The most common focal amplifications encoded CCNE1, MYC, and MECOM (Figure 1c, Methods S5, Table S5.2) each highly amplified in greater than 20% of tumors. New tightly-localized amplification peaks in HGS-OvCa encoded the receptor for activated C-kinase, ZMYND8; the p53 target gene, IRF2BP2; the DNA-binding protein inhibitor, ID4; the embryonic development gene, PAX8; and the telomerase catalytic subunit, TERT. Three data sources: http://www.ingenuity.com/, http://clinicaltrials.gov and http://www.drugbank.ca were used to identify possible therapeutic inhibitors of amplified, over-expressed genes. This search identified 22 genes that are therapeutic targets including MECOM, MAPK1, CCNE1 and KRAS amplified in at least 10% of the cases (Table S5.3).
GISTIC also identified 50 focal deletions (Figure 1d). The known tumor suppressor genes PTEN, RB1, and NF1 were in regions of homozygous deletions in at least 2% of tumors. Importantly, RB1 and NF1 also were among the significantly mutated genes. One deletion contained only three genes, including the essential cell cycle control gene, CREBBP, which has 5 non-synonymous and 2 frameshift mutations.
mRNA and miRNA expression and DNA methylation analysis
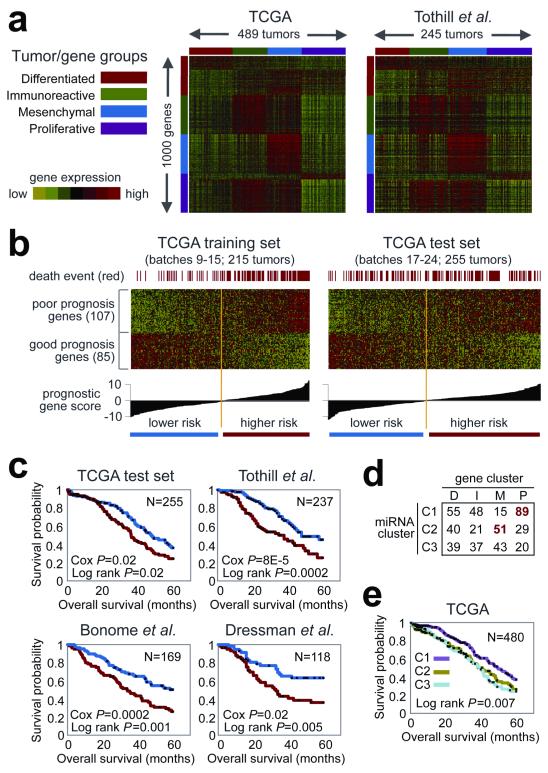
Expression measurements for 11,864 genes from three different platforms (Agilent, Affymetrix HuEx, Affymetrix U133A) were combined for subtype identification and outcome prediction. Individual platform measurements suffered from limited, but statistically significant batch effects, whereas the combined data set did not (Methods S11, Figure S11.1). Analysis of the combined dataset identified ~1,500 intrinsically variable genes24 (Methods S6) that were used for NMF consensus clustering. This analysis yielded four clusters (Methods S6, Figure 2a). Thesame analysis approach applied to a publicly available dataset from Tothill et al. 25, also yielded four clusters. Comparison of the Tothill and TCGA clusters showed a clear correlation (Methods S6, Figure S6.3). We therefore conclude that at least four robust expression subtypes exist in HGS-OvCa.
Figure 2. Gene and miRNA expression patterns of molecular subtype and outcome prediction in HGS-OvCa.
(a) Tumors from TCGA and Tothill et al. separated into four clusters, based on gene expression. (b) Using a training dataset, a prognostic gene signature was defined and applied to a test dataset. (c) Kaplan-Meier analysis of four independent expression profile datasets, comparing survival for predicted higher risk versus lower risk patients. Univariate Cox p-value for risk index included. (d) Tumors separated into three clusters, based on miRNA expression, overlapping with gene-based clusters as indicated. (e) Differences in patient survival among the three miRNA-based clusters.
We termed the four HGS-OvCa subtypes Immunoreactive, Differentiated, Proliferative and Mesenchymal based on gene content in the clusters (Methods S6) and on previous observations25. T-cell chemokine ligands, CXCL11 and CXCL10, and the receptor, CXCR3, characterized the Immunoreactive subtype. High expression of transcription factors such as HMGA2 and SOX11, low expression of ovarian tumor markers (MUC1, MUC16) and high expression of proliferation markers such as MCM2 and PCNA defined the Proliferative subtype. The Differentiated subtype was associated with high expression of MUC16 and MUC1 and with expression of the secretory fallopian tube maker SLPI, suggesting a more mature stage of development. High expression of HOX genes and markers suggestive of increased stromal components such as for myofibroblasts (FAP) and microvascular pericytes (ANGPTL2, ANGPTL1) characterized the Mesenchymal subtype.
Elevated DNA methylation and reduced tumor expression implicated 168 genes as epigenetically silenced in HGS-OvCa compared to fallopian tube controls26. DNA methylation was correlated with reduced gene expression across all samples (Methods S7). AMT, CCL21 and SPARCL1 were noteworthy because they showed promoter hypermethylation in the vast majority of the tumors. Curiously, RAB25, previously reported to be amplified and over-expressed in ovarian cancer27, also appeared to be epigenetically silenced in a subset of tumors. The BRCA1 promoter was hypermethylated and silenced in 56 of 489 (11.5%) tumors as previously reported (Figure S7.1) 28. Consensus clustering of variable DNA methylation across tumors identified four subtypes (Methods S7, Figure S7.2) that were significantly associated with differences in age, BRCA inactivation events, and survival (Methods S7). However, the clusters demonstrated only modest stability.
Survival duration did not differ significantly for transcriptional subtypes in the TCGA dataset. The Proliferative group showed a decrease in the rate of MYC amplification and RB1 deletion, whereas the Immunoreactive subtype showed an increased frequency of 3q26.2 (MECOM) amplification (Table S6.2, Figure S6.4). A moderate, but significant overlap between the DNA methylation clusters and gene expression subtypes was noted (p<2.2*10−16, Chi-square test, Adjusted Rand Index = 0.07, Methods S7, Table S7.6).
A 193 gene transcriptional signature predictive of overall survival was defined using the integrated expression data set from 215 samples. After univariate Cox regression analysis, 108 genes were correlated with poor survival, and 85 were correlated with good survival (p-value cutoff of 0.01, Methods S6, Table S6.4). The predictive power was validated on an independent set of 255 TCGA samples as well as three independent expression data sets25,29,30. Each of the validation samples was assigned a prognostic gene score, reflecting the similarity between its expression profile and the prognostic gene signature31 (Methods S6, Figure 2c). Kaplan-Meier survival analysis of this signature showed statistically significant association with survival in all validation data sets (Methods S6, Figure 2d).
NMF consensus clustering of miRNA expression data identified three subtypes (Figure S6.5). Interestingly, miRNA subtype 1 overlapped the mRNA Proliferative subtype and miRNA subtype 2 overlaped the mRNAMesenchymal subtype (Figure 2d). Survival duration differed significantly between iRNA subtypes with patients in miRNA subtype 1 tumors surviving significantly longer (Figure 2e).
Pathways influencing disease
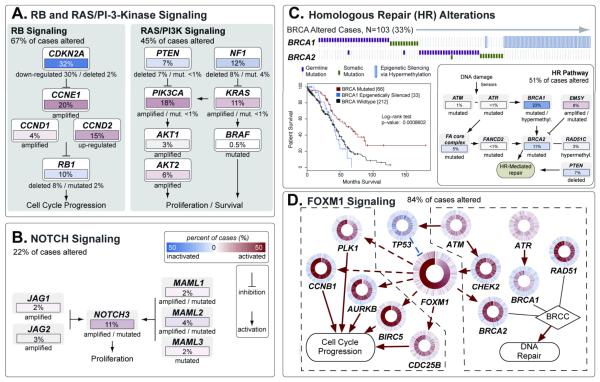
Several analyses integrated data from the 316 fully analyzed cases to identify biology that contributes to HGS-OvCa. Analysis of the frequency with which known cancer-associated pathways harbored one or more mutations, copy number changes, or changes in gene expression showed that the RB1 and PI3K/RAS pathways were deregulated in 67% and 45% of cases, respectively (Figure 3A, Methods S8). A search for altered subnetworks in a large protein-protein interaction network32 using HotNet33 identified several known pathways (Methods S9) including the Notch signaling pathway, which was altered in 23% of HGS-OvCa samples (Figure 3B)34.
Figure 3. Altered Pathways in HGS-OvCa.
(a) The RB and PI3K/RAS pathways, identified by curated analysis and (b) NOTCH pathway, identified by HotNet analysis, are commonly altered. Alterations are defined by somatic mutations, DNA copy-number changes, or in some cases by significant up- or down-regulation compared to expression in diploid tumors. Alteration frequencies are in percentage of all cases; activated genes are red, inactivated genes are blue. (c) Genes in the HR pathway are altered in up to 49% of cases. Survival analysis of BRCA status shows divergent outcome for BRCA mutated cases (exhibiting better overall survival) than BRCA wild-type, and BRCA1 epigenetically silenced cases exhibiting worse survival. (d) The FOXM1 transcription factor network is activated in 87% of cases. Each gene is depicted as a multi-ring circle in which its copy number (outer ring) and gene expression (inner ring) are plotted such that each “spoke” in the ring represents a single patient sample, with samples sorted in increasing order of FOXM1 expression. Excitatory (red arrows) and inhibitory interactions (blue lines) were taken from the NCI Pathway Interaction Database. Dashed lines indicate transcriptional regulation.
Published studies have shown that cells with mutated or methylated BRCA1 or mutated BRCA2 have defective homologous recombination (HR) and are highly responsive to PARP inhibitors35-37. Figure 3C shows that 20% of HGS-OvCa have germline or somatic mutations in BRCA1/2, that 11% have lost BRCA1 expression through DNA hypermethylation and that epigenetic silencing of BRCA1 is mutually exclusive of BRCA1/2 mutations (P = 4.4×10−4, Fisher’s exact test). Univariate survival analysis of BRCA status (Figure 3C) showed better overall survival (OS) for BRCA mutated cases than BRCA wild-type cases. Interestingly, epigenetically silenced BRCA1 cases exhibited survival similar to BRCA1/2 WT HGS-OvCa (median OS 41.5 v. 41.9 months, P = 0.69, log-rank test, Methods S8, Figure S8.13B). This suggests that BRCA1 is inactivated by mutually exclusive genomic and epigenomic mechanisms and that patient survival depends on the mechanism of inactivation. Genomic alterations in other HR genes that might render cells sensitive to PARP inhibitors38 (Methods S8, Figure S8.12) discovered in this study include amplification or mutation of EMSY (8%), focal deletion or mutation of PTEN (7%); hypermethylation of RAD51C (3%), mutation of ATM/ATR (2%), and mutation of Fanconi Anemia genes (5%). Overall, HR defects may be present in approximately half of HGS-OvCa, providing a rationale for clinical trials of PARP inhibitors targeting tumors these HR-related aberrations.
Comparison of the complete set of BRCA inactivation events to all recurrently altered copy number peaks revealed an unexpectedly low frequency of CCNE1 amplification in cases with BRCA inactivation (8% of BRCA altered cases had CCNE1 amplification v. 26% of BRCA wild type cases, FDR adjusted P = 0.0048). As previously reported39, overall survival tended to be shorter for patients with CCNE1 amplification compared to all other cases (P = 0.072, log-rank test, Methods, S8 Figure S8.14A). However, no survival disadvantage for CCNE1-amplified cases (P = 0.24, log-rank test, Methods S8, Figure S8.14B) was apparent when looking only at BRCA wild-type cases, suggesting that the previously reported CCNE1 survival difference can be explained by the better survival of BRCA-mutated cases.
Finally, a probabilistic graphical model (PARADIGM40) searched for altered pathways in the NCI Pathway Interaction Database41 identifying the FOXM1 transcription factor network (Figure 3d) as significantly altered in 87% of cases, Methods S10, Figures S10.1-3). FOXM1 and its proliferation-related target genes; AURB, CCNB1, BIRC5, CDC25, and PLK1, were consistently over-expressed but not altered by DNA copy number changes, indicative of transcriptional regulation. TP53 represses FOXM1 following DNA damage42, suggesting that the high rate of TP53 mutation in HGS-OvCa contributes to FOXM1 overexpression. In other datasets, the FOXM1 pathway is significantly activated in tumors relative to adjacent epithelial tissue43-45 (Methods S10, Figure S10.4) and is associated with HGS-OvCa (Methods S10, Figure S10.5)22.
Discussion
This TCGA study provides the first large scale integrative view of the aberrations in HGS-OvCa. Overall, the mutational spectrum was surprisingly simple. Mutations in TP53 predominated, occurring in at least 96% of HGS-OvCa while BRCA1/2 were mutated in 22% of tumors due to a combination of germline and somatic mutations. Seven other significantly mutated genes were identified, but only in 2-6% of HGS-OvCa. In contrast, HGS-OvCa demonstrates a remarkable degree of genomic disarray. The frequent SCNAs are in striking contrast to previous TCGA findings with glioblastoma46 where there were more recurrently mutated genes with far fewer chromosome arm-level or focal SCNAs (Figure 1A). A high prevalence of mutations and promoter methylation in putative DNA repair genes including HR components may explain the high prevalence of SCNAs. The mutation spectrum marks HGS-OvCa as completely distinct from other OvCa histological subtypes. For example, clear-cell OvCa have few TP53 mutations but have recurrent ARID1A and PIK3CA47-49 mutations; endometrioid OvCa have frequent CTTNB1, ARID1A, and PIK3CA mutations and a lower rate of TP5348,49 while mucinous OvCa have prevalent KRAS mutations50. These differences between ovarian cancer subtypes likely reflect a combination of etiologic and lineage effects, and represent an opportunity to improve ovarian cancer outcomes through subtype-stratified care.
Identification of new therapeutic approaches is a central goal of the TCGA. The ~50% of HGS-OvCa with HR defects may benefit from PARP inhibitors. Beyond this, the commonly deregulated pathways, RB, RAS/PI3K, FOXM1, and NOTCH, provide opportunities for therapeutic attack. Finally, inhibitors already exist for 22 genes in regions of recurrent amplification (Methods S5, Table S5.3), warranting assessment in HGS-OvCa where the target genes are amplified. Overall, these discoveries set the stage for approaches to treatment of HGS-OvCa in which aberrant genes or networks are detected and targeted with therapies selected to be effective against these specific aberrations.
Methods Summary
All patient specimens were obtained under appropriate IRB consent. DNA and RNA were collected from samples using the Allprep kit (Qiagen). We used commercial technology for capture and sequencing of exomes from whole genome amplified tumor and normal DNAs. DNA sequences were aligned to Human NCBI build 36; duplicate reads were excluded from mutation calling. Validation of mutations occurred on a separate whole genome amplification of DNA from the same tumor. Data is submitted to dbGaP under accession number PHS000178. Significantly mutated genes were identified by comparing to expectation models based on the exact measured rates of specific sequence lesions. CHASM 20 and MutationAssessor (Methods S4) were used to identify functional mutations. GISTIC analysis of the CBS segmented Agilent 1M feature copy number data was used to identify recurrent peaks comparing to the results from the other platforms to identify likely platform specific artifacts. Consensus clustering approaches were used to analyze mRNA, miRNA, and methylation subtypes as well as predictors of outcome using previous approaches46. HotNet 33 was used to identify portions of the protein-protein interaction network that have more events than expected by chance. Networks that had a significant probability of being valid were evaluated for increased fraction of known annotations. PARADIGM40 was used to estimate integrated pathway activity to identify portions of the network models differentially active in HGS-OvCa.
Supplementary Material
Acknowledgements
We thank Jacqueline Palchik, Anika Mirick, and Julia Zhang for administrative coordination of TCGA activities. This work was supported by the following grants from the United States National Institutes of Health: U54HG003067, U54HG003079, U54HG003273, U24CA126543, U24CA126544, U24CA126546, U24CA126551, U24CA126554, U24CA126561, U24CA126563, U24CA143840, U24CA143882, U24CA143731, U24CA143835, U24CA143845, U24CA143858, U24CA144025, U24CA143882, U24CA143866, U24CA143867, U24CA143848, U24CA143843, and R21CA135877.
Footnotes
Author Contributions: The TCGA research network contributed collectively to this study. Biospecimens were provided by the Tissue Source Sites and processed by the Biospecimen Core Resource. Data generation and analyses were performed by the Genome Sequencing Centers, Cancer Genome Characterization Centers, and Genome Data Analysis Centers. All data were released through the Data Coordinating Center. Project activities were coordinated by the NCI and NHGRI Project Teams. We also acknowledge the following TCGA investigators who contributed substantively to the writing of this manuscript. Leader: Paul T. Spellman(1), Writing Team: Stacey Gabriel(2), Gad Getz(3), Joe W. Gray(1), Elaine R. Mardis(4,5), Databases: Ari Kahn(6), Mutations: Li Ding(4), Kristian Cibulskis(8), David A. Wheeler(7), Michael D. McLellan(4), Michael S. Lawrence(3), Gad Getz(3), Daniel C. Koboldt(4), Andrey Sivachenko(3), Carrie Sougnez(2), Nathan Dees(4), Qunyuan Zhang(4), Cyriac Kandoth(4), Michael C. Wendl(4), Boris Reva(9), Rachel Karchin(10), Hannah Carter(10), Douglas Voet(3), Jinghui Zhang(93), John W. Wallis(4), Dewey Kim(10), Gordon B. Mills(61-62), Copy Number: Gad Getz(3), Craig H. Mermel(3,11), Marcin Imielinski(3,92), Alex H. Ramos(3,11), Wendy Winckler(3), Jingchun Zhu(34), Henrik Bengtsson(13), Jun Li(14), Scott L. Carter(3,11), Michael S. Lawrence(3), Nikolaus Schultz(9), Adam Olshen(15), Barbara A. Weir(3), Joe W. Gray(1), Matthew Meyerson(3,11), Methylation: Peter W. Laird(16), Hui Shen(16), Daniel J Weisenberger(16), Expression: Roel G. W. Verhaak(3,11,12), Chad J. Creighton(7), Hailei Zhang(17), Douglas A. Levine (18), Victoria Wang(13), Katherine A. Hoadley(19-21), John N. Weinstein(22,61), Elizabeth Purdom(13), Andrew Berchuck(23,24), Michael Birrer(25,26), miRNA: D. Neil Hayes(21,27), Shaowu Meng(21), Ying Du(21), Preethi H. Gunaratne(7,29), Douglas A. Levine(18), Nikolaus Schultz(9), Robert Sheridan(9), Anil K. Sood(31,32), Pathways: Chris Sander(9), Ethan Cerami(9), Nikolaus Schultz(9), Giovanni Ciriello(9), Barry S. Taylor(9), Douglas A. Levine(18), Yonghong Xiao(17), David Haussler(34,37), Joshua M. Stuart(34), Charlie J. Vaske(34,36), Steven C. Benz(34), Benjamin J. Raphael(28), Fabio Vandin(28), Peter W. Laird(16), Hui Shen(16)General: Stephen B. Baylin(38), Lynda Chin(17,39), Richard A. Gibbs(7), Raju Kucherlapati(90), Marc Ladanyi(41), Eric S. Lander(3,42,43), Matthew Meyerson(3,11), Richard M. Myers(44), Charles M. Perou(19-21), & Richard K. Wilson(4,5).
The Cancer Genome Atlas (TCGA) Research Network
Disease Working Group and Tissue Source Sites
Debra Bell(45), Andrew Berchuck(23,24), Michael Birrer(25,26), Jeremy Chien(46), Daniel W. Cramer(48), Fanny Dao(49), Rajiv Dhir(50), Philip DiSaia(51), Hani Gabra(52), Pat Glenn(53), Andrew K. Godwin(54), Jenny Gross(55), Lynn Hartmann(56), Min Huang(57), David G. Huntsman(59), Mary Iacocca(58), Steven Kalloger(59), Beth Y. Karlan(55,60), Douglas A. Levine(18), Gordon B. Mills(61,62), Carl Morrison(63,64), David Mutch(65), Narciso Olvera(18), Sandra Orsulic(66), Kay Park(67), Nicholas Petrelli(68), Brenda Rabeno(58), Janet S. Rader(69), Branimir I. Sikic(70), Karen Smith-McCune(53), Anil K. Sood(31,32), David Bowtell (95), Robert Penny(85), & Joseph R. Testa(71).
Genome Sequencing Centers
Baylor College of Medicine – Kyle Chang(7), Huyen H Dinh(7), Jennifer A. Drummond(7), Gerald Fowler(7), Preethi Gunaratne(7), Alica C Hawes(7), Christie L Kovar, Lora R Lewis, Margaret B Morgan(7), Irene F Newsham, Jireh Santibanez, Jeffrey G. Reid(7),Lisa R Trevino(7), Yuan-Qing Wu(7), Min Wang(7), Donna M. Muzny(7), David A. Wheeler(7), Richard A Gibbs(7), Broad Institute of MIT and Harvard: Gad Getz(3), Michael S. Lawrence(3), Kristian Cibulskis(8), Andrey Sivachenko(3), Carrie Sougnez(72), Douglas Voet(3), Jane Wilkinson(73), Toby Bloom(74), Kristin Ardlie(3), Tim Fennell(75), Jennifer Baldwin(73), Stacey Gabriel(2), Eric S. Lander(3,42,43), Washington University in St. Louis: Li Ding(4), Robert S. Fulton(4), Daniel C. Koboldt(4), Michael D. McLellan(4), Todd Wylie(4), Jason Walker(4), Michelle O’Laughlin(4), David J. Dooling(4), Lucinda Fulton(4), Rachel Abbott(4), Nathan D. Dees(4), Qunyan Zhang(4), Cyriac Kandoth(4), Michael Wendl(4), William Schierding(4), Dong Shen(4), Christopher C. Harris(4), Heather Schmidt(4), Joelle Kalicki(4), Kim D. Delehaunty(4), Catrina C. Fronick(4), Ryan Demeter(4), Lisa Cook(4), John W. Wallis(4), Ling Lin(4), Vincent J. Magrini(4), Jennifer S. Hodges(4), James M. Eldred(4), Scott M. Smith(4), Craig S. Pohl(4), George M. Weinstock(4), Elaine R. Mardis(4,5), & Richard K. Wilson(4,5).
Cancer Genome Characterization Centers
Broad Institute/Dana-Farber Cancer Institute – Matthew Meyerson(3,11,12), Wendy Winckler(3), Gad Getz(3), Roel G. W. Verhaak(3,11), Scott L. Carter(3,11), Craig H. Mermel(3,11), Gordon Saksena(3), Huy Nguyen(3), Robert C. Onofrio(3), Michael S. Lawrence(3), Diana Hubbard(3,11,12), Supriya Gupta(3), Andrew Crenshaw(3), Alex H. Ramos(3,11), Kristin Ardlie(3), Harvard Medical School: Lynda Chin(17,39), Alexei Protopopov(17), Juinhua Zhang(17), Tae-Min Kim(40), Ilana Perna(17), Yonghong Xiao(17), Hailei Zhang(17), Georgia Ren(17), Narayan Sathiamoorthy(76), Richard W. Park(40), Eunjung Lee(40), Peter J. Park(40,77), Raju Kucherlapati(90), HudsonAlpha Institute/Stanford University: Devin M. Absher(44), Lindsay Waite(44), Gavin Sherlock(79), James D. Brooks(80), Jun Z. Li(14), Jishu Xu(14), Richard M. Myers(44), University of Southern California / Johns Hopkins: Peter W. Laird(16), Leslie Cope(78), James G. Herman(38), Hui Shen(16), Daniel J. Weisenberger(16), Houtan Noushmehr(16), Fei Pan(16), Timothy Triche, Jr(16), Benjamin P. Berman(16), David J. Van Den Berg(16), Jonathan Buckley(16), Stephen B. Baylin(38), Lawrence Berkeley National Laboratory: Paul T. Spellman(1), Elizabeth Purdom(13), Pierre Neuvial(13), Henrik Bengtsson(13), Lakshmi R. Jakkula(1), Steffen Durinck(1), Ju Han(1), Shannon Dorton(1), Henry Marr(1), Yoon Gi Choi(81), Victoria Wang(29), Nicholas J. Wang(1), John Ngai(81), John G. Conboy(1), Bahram Parvin(1), Heidi S. Feiler(1), Terence P. Speed(1,82), Joe W. Gray(1), Memorial Sloan-Kettering Cancer Center: Douglas A. Levine (18), Nicholas D. Socci(9), Yupu Liang(9), Barry S. Taylor(9), Nikolaus Schultz(9), Laetitia Borsu(41), Alex E. Lash(9), Cameron Brennan(83), Agnes Viale(84), Chris Sander(9), Marc Ladanyi(41). University of North Carolina, Chapel Hill: Katherine A. Hoadley(19-21), Shaowu Meng(21), Ying Du(21), Yan Shi(21), Ling Li(21), Yidi J. Turman(21), Deying Zang(21), Erin B. Helms(21), Saianand Balu(21), Xingliang Zhou(21), Junyuan Wu(21), Michael D. Topal(20,21), D. Neil Hayes(21,27), Charles M. Perou(19-21).
Genome Data Analysis Centers
Broad Institute – Gad Getz(3), Doug Voet(3), Gordon Saksena(3), Juinhua Zhang(17), Hailei Zhang(17), Chang-Jiun Wu(17), Sachet Shukla(17), Kristian Cibulskis(3), Michael S. Lawrence(3), Andrey Sivachenko(3), Rui Jing(3), Richard W. Park(3), Yingchun Liu(17), Peter J. Park(40,77), Michael Noble(3), Lynda Chin(3,17,39)
Lawrence Berkeley National Laboratory: Paul T. Spellman(1), Elizabeth Purdom(13), Pierre Neuvial(13), Henrik Bengtsson(13), Steffen Durinck(1), Ju Han(1), James E. Korkola(1), Laura M. Heiser(1), Raymond J. Cho(1), Zhi Hu(1), Bahram Parvin(1), Terence P. Speed(1,82), Joe W. Gray(1), Memorial Sloan-Kettering Cancer Center: Nikolaus Schultz(9), Ethan Cerami(9), Barry S. Taylor(9), Adam Olshen(15), Boris Reva(9), Yevgeniy Antipin(9), Ronglai Shen(15), Parminder Mankoo(9), Robert Sheridan(9), Giovanni Ciriello(9), William K. Chang(9,35), Joel A. Bernanke(30), Laetitia Borsu(41), Douglas A. Levine (18), Marc Ladanyi(41), Chris Sander(9), University of California, Santa Cruz / Buck Institute: David Haussler(34,37), Christopher C. Benz(91), Joshua M. Stuart(34), Stephen C. Benz(34), J. Zachary Sanborn(34), Charles J. Vaske(34,94), Jingchun Zhu(34), Christopher Szeto(34), Gary K. Scott(91), Christina Yau(91). University of North Carolina: Katherine A. Hoadley(19-21), Ying Du(21), Saianand Balu(21), D. Neil Hayes(21,27), Charles M. Perou(19-21), Matthew Dr. Wilkerson(21).
The University of Texas MD Anderson Cancer Center: Nianxiang Zhang(22), Rehan Akbani(22), Keith A. Baggerly(22), W. K. Yung(89), Gordon B. Mills(61-62), John N. Weinstein(22,61).
Biospecimen Core Resource: Robert Penny(85), Troy Shelton(85), David Grimm(85), Martha Hatfield(85), Scott Morris(85), Peggy Yena(85), Patrick Rhodes(85), Mark Sherman(85), Joseph Paulauskis(85), Sherri Millis(85).
Data Coordination Center: Ari Kahn(6), John M. Greene(6), Robert Sfeir(6), Mark A. Jensen(6), Jessica Chen(6), Jon Whitmore(6), Shelley Alonso(6), Jim Jordan(6), Anna Chu(6).
Project Teams
National Cancer Institute: Anna Barker(86), Carolyn Compton(86), Greg Eley(86), Martin Ferguson(87), Peter Fielding(86), Daniela S. Gerhard(86), Renate Myles(86), Carl Schaefer(86), Kenna R.Mills Shaw(86), Jim Vaught(86), Joseph B. Vockley(86).
National Human Genome Research Institute: Peter J. Good(88), Mark S. Guyer(88), Brad Ozenberger(88), Jane Peterson(88), & Elizabeth Thomson(88).
Affiliations for participants:
1. Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA
2. Genetic Analysis Platform, The Eli and Edythe L. Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, MA 02142 USA
3. The Eli and Edythe L. Broad Institute of Massachusetts Institute of Technology and Harvard University Cambridge, MA 02142 USA
4. The Genome Center at Washington University, Department of Genetics, Washington University School of Medicine, St. Louis, MO 63108 USA
5. Siteman Cancer Center, Washington University School of Medicine, St. Louis, MO 63108 USA
6. SRA International, Fairfax, VA 22033 USA
7. Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX 77030 USA
8. Medical Sequencing Analysis and Informatics, The Eli and Edythe L. Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, MA, 02142 USA
9. Computational Biology Center, Memorial-Sloan Kettering Cancer Center, New York, NY 10065 USA
10. Institute for Computational Medicine, Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 21231 USA
11. Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02115 USA
12. Department of Pathology, Duke University Medical Center, Durham, NC 27710
13. Department of Statistics, University of California at Berkeley, Berkeley, CA 95720 USA
14. Department of Human Genetics, University of Michigan, Ann Arbor, MI 48109 USA
15. Department of Epidemiology and Biostatistics, Memorial-Sloan Kettering Cancer Center, New York, NY 10065 USA
16. University of Southern California Epigenome Center, University of Southern California, Los Angeles, CA 90089 USA
17. Belfer Institute for Applied Cancer Science, Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02115 USA
18. Department of Surgery, Memorial Sloan-Kettering Cancer Center, New York, NY 10065 USA
19. Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA
20. Department of Pathology and Laboratory Medicine, Chapel Hill, NC 27599 USA
21. Lineberger Comprehensive Cancer Center, Chapel Hill, NC 27599 USA
22. Department of Bioinformatics and Computational Biology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030 USA
23. Division of Gynecologic Oncology/Department of Obstetrics and Gynecology, Duke University Medical Center, Durham, NC 27708 USA
24. Duke Institute for Genome Sciences and Policy, Duke University Medical Center, Durham, NC 27708 USA
25. Department of Medicine, Harvard Medical School, Boston, MA 02114 USA
26. Disease Center Leader, Gynecologic Oncology, Massachusetts General Hospital, Boston, MA 02114 USA
27. Department of Internal Medicine, Division of Medical Oncology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA
28. Department of Computer Science and Center for Computational Molecular Biology, Brown University, Providence RI 02912 USA
29. Department of Biology and Biochemistry, University of Houston, Houston, TX 77004 USA
30. Weill Medical College of Cornell University, 1300 York Avenue, New York, NY 10065 USA
31. Department of Gynecologic Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX 77230 USA
32. Center for RNA Interference and Non-Coding RNA, The University of Texas MD Anderson Cancer Center, Houston, TX 77230 USA
33. Department of Molecular Virology and Microbiology, Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX 77030 USA
34. Department of Biomolecular Engineering and Center for Biomolecular Science and Engineering, University of California Santa Cruz, Santa Cruz, CA 95064 USA
35. Department of Physiology and Biophysics, Weill Cornell Graduate School of Medical Sciences, New York, NY 10065 USA
36. Lewis Sigler Institute, Princeton University, Princeton, NJ 08544 USA
37. Howard Hughes Medical Institute, University of California Santa Cruz, Santa Cruz, CA 95064 USA
38. Cancer Biology Division, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University, Baltimore, MD 21231 USA
39. Department of Dermatology, Harvard Medical School, Boston, MA 02115 USA
40. The Center for Biomedical Informatics, Harvard Medical School, Boston, MA 02115 USA
41. Department of Pathology, Human Oncology and Pathogenesis Program, Memorial-Sloan Kettering Cancer Center, New York, NY 10065 USA
42. Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02142 USA
43. Department of Systems Biology, Harvard University, Boston, MA 02115 USA
44. HudsonAlpha Institute for Biotechnology, Huntsville, AL 35806 USA
45. Division of Anatomic Pathology, Mayo Clinic, Rochester, MN 55905 USA
46. Division of Experimental Pathology, Mayo Clinic, Rochester, MN 55905 USA
47. Department of Epidemiology, Harvard School of Public Health, Boston, MA 02115 USA
48. Department of Obstetrics and Gynecology Epidemiology Center, Brigham and Women’s Hospital, Boston, MA 02115 USA
49. Department of Surgery, Memorial Sloan-Kettering Cancer Center, New York, NY 10065 USA
50. Department of Pathology, University of Pittsburgh, Pittsburgh PA 15213 USA
51. Gynecologic Oncology Group, University of California Irvine, Irvine CA 92697 USA
52. Ovarian Cancer Action Research Centre, Department of Surgery and Cancer, Imperial College London Hammersmith Campus, London W12 0NN UK
53. Department of Obstetrics, Gynecology and Reproductive Services, University of California San Francisco, San Francisco CA 94143 USA
54. Women’s Cancer Program, Department of Medical Oncology, Fox Chase Cancer Center, Philadelphia, PA 19111 USA
55. Women’s Cancer Research Institute at the Samuel Oschin Comprehensive Cancer Institute Cedars-Sinai Medical Center, Cedars-Sinai Medical Center, Geffen School of Medicine at UCLA, Los Angeles, CA 90048 USA
56. Division of Medical Oncology, Mayo Clinic, Rochester, MN 55905 USA
57. Department of Pathology, Fox Chase Cancer Center, Philadelphia, PA 19111 USA
58. Department of Pathology, Christiana Care Health Services, Newark, DE 19718 USA
59. Center for Translational and Applied Genomics, British Columbia Cancer Agency, Vancouver, British Columbia, Canada
60. Department of Obstetrics and Gynecology, Cedars-Sinai Medical Center, Geffen School of Medicine at UCLA, Los Angeles, CA 90048 USA
61. Department of Systems Biology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030 USA
62. Kleberg Center for Molecular Markers, The University of Texas MD Anderson Cancer Center, Houston, TX 77030 USA
63. The Department of Pathology and Laboratory Medicine, Roswell Park Cancer Institute, Buffalo, NY 14263 USA
64. Division of Molecular Pathology, Roswell Park Cancer Institute, Buffalo, NY 14263 USA
65. Department of Obstetrics and Gynecology, Division of Gynecologic Oncology, Washington University School of Medicine, St. Louis, MO 3110 USA
66. Women’s Cancer Research Institute, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048 USA
67. Department of Pathology, Memorial Sloan-Kettering Cancer Center, New York, NY 10065 USA
68. Department of Surgery, Helen F Graham Cancer Center at Christina Care, Newark DE 19713 USA
69. Department of Obstetrics and Gynecology, Human and Molecular Genetics Center, Medical College of Wisconsin, Milwaukee, WI 53226 USA
70. Division of Oncology, Department of Medicine, Stanford University School of Medicine, Palo Alto, California 94304 USA
71. Cancer Biology Program, Fox Chase Cancer Center, Philadelphia, PA 19111 USA
72. Cancer Genome & Medical Resequencing Projects, The Eli and Edythe L. Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, MA, 02142 USA
73. Sequencing Platform, The Eli and Edythe L. Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, MA, 02142 USA
74. Sequencing Platform Informatics, The Eli and Edythe L. Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, MA, 02142 USA
75. Directed Sequencing Informatics, The Eli and Edythe L. Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, MA, 02142 USA
76. Partners Center for Personalized Genetic Medicine, Cambridge, MA USA
77. Informatics Program, Children’s Hospital, Boston, MA 02115 USA
78. Biometry and Clinical Trials Division, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University, Baltimore, MD 21231 USA
79. Department of Genetics, Stanford University School of Medicine, Stanford, CA 94305 USA
80. Department of Urology, Stanford University School of Medicine, Stanford, CA 94305 USA
81. Department of Molecular and Cellular Biology, University of California at Berkeley, Berkeley, CA 95720 USA
82. Walter and Eliza Hall Institute, Parkville, Vic 3052 Australia
83. Department of Neurosurgery, Memorial-Sloan Kettering Cancer Center, New York, NY 10065 USA
84. Genomics Core Laboratory, Memorial-Sloan Kettering Cancer Center, New York, NY 10065 USA
85. International Genomics Consortium, Phoenix, AZ 85004 USA
86. National Cancer Institute, National Institutes of Health, Bethesda, MD 20892 USA
87. MLF Consulting, Arlington, MA 02474 USA
88. National Human Genome Research Institute, National Institutes of Health, Bethesda, MD 20892 USA
89. Department of Neuro-Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030 USA
90. Department of Genetics, Harvard Medical School, Boston, MA 02115 USA
91. Buck Institute for Age Research, Novato, CA 94945 USA
92. Disease Center Leader, Gynecologic Oncology, Massachusetts General Hospital, Boston MA 02114 USA
93. Department of Biotechnology, St. Jude Children’s Research Hospital, Memphis TN 38105 USA
94. Lewis-Sigler Institute for Integrative Genomes, Princeton NJ 08544 USA
95. Research Division, Peter MacCallum Cancer Centre, Locked Bag 1 A’Beckett St, Melbourne 8006, VIC. Australia
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Koonings PP, Campbell K, Mishell DR, Jr., Grimes DA. Relative frequency of primary ovarian neoplasms: a 10-year review. Obstet Gynecol. 1989;74:921–926. [PubMed] [Google Scholar]
- 3.Seidman JD, et al. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int J Gynecol Pathol. 2004;23:41–44. doi: 10.1097/01.pgp.0000101080.35393.16. [DOI] [PubMed] [Google Scholar]
- 4.Miller DS, et al. Phase II evaluation of pemetrexed in the treatment of recurrent or persistent platinum-resistant ovarian or primary peritoneal carcinoma: a study of the Gynecologic Oncology Group. J Clin Oncol. 2009;27:2686–2691. doi: 10.1200/JCO.2008.19.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jemal A, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 6.Pal T, et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104:2807–2816. doi: 10.1002/cncr.21536. [DOI] [PubMed] [Google Scholar]
- 7.Risch HA, et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst. 2006;98:1694–1706. doi: 10.1093/jnci/djj465. [DOI] [PubMed] [Google Scholar]
- 8.Bast RC, Jr., Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nature biotechnology. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodges E, et al. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nature protocols. 2009;4:960–974. doi: 10.1038/nprot.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bookman MA, et al. Evaluation of new platinum-based treatment regimens in advanced-stage ovarian cancer: a Phase III Trial of the Gynecologic Cancer Intergroup. J Clin Oncol. 2009;27:1419–1425. doi: 10.1200/JCO.2008.19.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muggia FM, et al. Phase III randomized study of cisplatin versus paclitaxel versus cisplatin and paclitaxel in patients with suboptimal stage III or IV ovarian cancer: a gynecologic oncology group study. J Clin Oncol. 2000;18:106–115. doi: 10.1200/JCO.2000.18.1.106. [DOI] [PubMed] [Google Scholar]
- 13.Ahmed AA, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen HH, Wang YC, Fann MJ. Identification and characterization of the CDK12/cyclin L1 complex involved in alternative splicing regulation. Molecular and cellular biology. 2006;26:2736–2745. doi: 10.1128/MCB.26.7.2736-2745.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aldred MA, Trembath RC. Activating and inactivating mutations in the human GNAS1 gene. Human mutation. 2000;16:183–189. doi: 10.1002/1098-1004(200009)16:3<183::AID-HUMU1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 17.Forbes SA, et al. Current protocols in human genetics / editorial board, Jonathan L. Haines … [et al. 2008. The Catalogue of Somatic Mutations in Cancer (COSMIC) p. 11. Chapter 10, Unit 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. American journal of human genetics. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter H, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter H, Samayoa J, Hruban RH, Karchin R. Prioritization of driver mutations in pancreatic cancer using cancer-specific high-throughput annotation of somatic mutations (CHASM) Cancer biology & therapy. 10 doi: 10.4161/cbt.10.6.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etemadmoghadam D, et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res. 2009;15:1417–1427. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tothill RW, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 26.Dubeau L. The cell of origin of ovarian epithelial tumours. Lancet Oncol. 2008;9:1191–1197. doi: 10.1016/S1470-2045(08)70308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng KW, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- 28.Esteller M, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 29.Bonome T, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dressman HK, et al. An integrated genomic-based approach to individualized treatment of patients with advanced-stage ovarian cancer. J Clin Oncol. 2007;25:517–525. doi: 10.1200/JCO.2006.06.3743. [DOI] [PubMed] [Google Scholar]
- 31.Creighton CJ, et al. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008;26:4078–4085. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keshava Prasad TS, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandin F, Upfal E, Raphael BJ. Algorithms for Detecting Significantly Mutated Pathways in Cancer; 14th International Conference on Research in Computational Molecular Biology; 2010. Springer. pp. 506–521. [DOI] [PubMed] [Google Scholar]
- 34.Choi JH, et al. Jagged-1 and Notch3 juxtacrine loop regulates ovarian tumor growth and adhesion. Cancer Res. 2008;68:5716–5723. doi: 10.1158/0008-5472.CAN-08-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 36.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 37.Veeck J, et al. BRCA1 CpG island hypermethylation predicts sensitivity to poly(adenosine diphosphate)-ribose polymerase inhibitors. J Clin Oncol. 28:e563–564. doi: 10.1200/JCO.2010.30.1010. author reply e565-566. [DOI] [PubMed] [Google Scholar]
- 38.Mendes-Pereira AM, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1:315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakayama N, et al. Gene amplification CCNE1 is related to poor survival and potential therapeutic target in ovarian cancer. Cancer. 116:2621–2634. doi: 10.1002/cncr.24987. [DOI] [PubMed] [Google Scholar]
- 40.Vaske CJ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 26:i237–245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaefer CF, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barsotti AM, Prives C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene. 2009;28:4295–4305. doi: 10.1038/onc.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tone AA, et al. Gene expression profiles of luteal phase fallopian tube epithelium from BRCA mutation carriers resemble high-grade serous carcinoma. Clin Cancer Res. 2008;14:4067–4078. doi: 10.1158/1078-0432.CCR-07-4959. [DOI] [PubMed] [Google Scholar]
- 44.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 45.Wang IC, et al. Deletion of Forkhead Box M1 transcription factor from respiratory epithelial cells inhibits pulmonary tumorigenesis. PLoS One. 2009;4:e6609. doi: 10.1371/journal.pone.0006609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ho ES, et al. p53 mutation is infrequent in clear cell carcinoma of the ovary. Gynecol Oncol. 2001;80:189–193. doi: 10.1006/gyno.2000.6025. [DOI] [PubMed] [Google Scholar]
- 48.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuo KT, et al. Frequent activating mutations of PIK3CA in ovarian clear cell carcinoma. Am J Pathol. 2009;174:1597–1601. doi: 10.2353/ajpath.2009.081000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cuatrecasas M, Villanueva A, Matias-Guiu X, Prat J. K-ras mutations in mucinous ovarian tumors: a clinicopathologic and molecular study of 95 cases. Cancer. 1997;79:1581–1586. doi: 10.1002/(sici)1097-0142(19970415)79:8<1581::aid-cncr21>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.