Abstract
Toll-like receptors (TLRs) are key receptors in innate immunity and trigger responses following interaction with pathogen-associated molecular patterns (PAMPs). TLR3, TLR4 and TLR9 recognize double stranded RNA, lipopolysaccharide (LPS) and CpG DNA, respectively. These receptors differ importantly in downstream adaptor molecules. TLR4 signals through MyD88 and TRIF; in contrast, the TLR3 pathway involves only TRIF while TLR9 signals solely through MyD88. To determine how differences in downstream signaling could influence gene expression in innate immunity, gene expression patterns were determined for the RAW264.7 macrophage cell line stimulated with LPS, poly (I:C), or CpG DNA. Gene expression profiles 6 and 24 hrs post-stimulation were analyzed to determine genes, pathways and transcriptional networks induced. As these experiments showed, the number and extent of genes expressed varied with stimulus. LPS and poly (I:C) induced an abundant array of genes in RAW264.7 cells at 6 hrs and 24 hrs following treatment while CpG DNA induced many fewer. By analyzing data for networks and pathways, we prioritized differentially expressed genes with respect to those common to the three TLR ligands as well as those shared by LPS and poly (I:C) but not CpG DNA. The importance of changes in gene expression was demonstrated by experiments indicating that RNA interference-mediated inhibition of two genes identified in this analysis, PLEC1 and TPST1, reduced IL-6 production by J774A.1 and RAW264.7 macrophages stimulated with LPS. Together, these findings delineate macrophage gene response patterns induced by different PAMPs and identify new genes that have not previously been implicated in innate immunity.
Keywords: innate immunity, gene expression, RNA interference, lipopolysacharide, poly (I:C), CpG DNA, PLEC1, TPST1
1. Introduction
Toll-like receptors are key receptors in the innate immune system and play a critical role in host defense against infection. As pattern recognition receptors (PRRs), TLRs interact with molecular components of bacterial, viral and fungal organisms that collectively are called pathogen-associated molecular patterns (PAMPs). These interactions have high specificity and involve ligands that are biochemically different (Akira and Takeda, 2004; Ishii et al., 2008; Takeuchi and Akira, 2010). Among PRRs, TLR4 is the receptor for bacterial lipopolysaccharide (LPS), TLR3 binds dsRNA including the viral mimic poly (I:C), while TLR9 binds to bacterial CpG DNA (Akira and Takeda, 2004; Ishii et al., 2008; Takeuchi and Akira, 2010). These receptors differ in intracellular location as well as downstream signaling pathways. TLR4 signaling involves the adaptor molecules MyD88 and TRIF. In contrast, the TLR3 pathway involves only the TRIF adaptor while TLR9 signals solely through MyD88. Signaling through the MyD88 adaptor leads to early NF-κB activation and pro-inflammatory cytokine production while signaling through the TRIF adaptor gives rise to late NF-κB activation and production of type I interferons as well as other cytokines.
Genome-wide transcriptional profiling of host cells in response to PAMPs or intact microorganisms using microarrays allows for the identification of novel candidate genes to elucidate the biology of the innate immune system and to develop new targets for therapeutic intervention (Jenner and Young, 2005). Previous genomic studies using microarrays have identified differentially regulated transcripts in murine macrophages in response to stimulation by PAMPs (Gao et al., 2003; Schmitz et al., 2004). Schmitz et al. described genomic signatures attributable to signaling through MyD88 and TRIF adaptors (Schmitz et al., 2004). In a related study, Gao et al. compared transcriptional profiles of macrophages in response to LPS and CpG DNA stimulation and showed that the CpG DNA transcriptional profile is a subset of the LPS profile (Gao et al., 2003). A more recent study combined gene expression profiling of macrophages stimulated with 6 PAMPs (LPS, Pam2CSK4, Pam3CSK4, CpG DNA, poly I:C and R484) with transcription factor binding site analysis to infer a network of associations between transcription factors and clusters of co-expressed target genes; this analysis identified TGIF1 as a novel regulator of macrophage activation (Ramsey et al., 2008). Tross et al. showed that co-stimulation with poly(I:C) and CpG DNA accelerated gene expression and synergistically activated genes primarily associated with immune function (Tross et al., 2009). Finally, a recent study of gene expression following in vivo CpG administration identified two discrete peaks of gene activation (at 3h and 5 days); initial gene up-regulation corresponded to a period in which genes stimulated by TLR9 ligation were functionally associated with the generation of innate and adaptive immune responses; in contrast, the second peak reflected processes associated with cell division (Klaschik et al., 2010).
In our recent studies, we have investigated the response of the RAW264.7 murine macrophage cell line to TLR stimulation by assessing the translocation and extracellular release of HMGB1, a non-nuclear protein that is a prototype alarmin. These studies showed that HMGB1 is released in response to stimulation by LPS and poly (I:C) but not CpG DNA (Jiang et al., 2007; Jiang and Pisetsky, 2006). We have also shown that HMGB1 release in response to LPS and poly (I:C) is correlated with the occurrence of apoptosis (Jiang et al., 2007). Together, these studies indicate complexity in the innate immune response to TLR stimulation, highlighting the differences in response induced by different TLR ligands and their consequences with respect to gene expression, alarmin release, and apoptosis.
Elucidating differences in the macrophage response to TLR stimulation thus requires more detailed genomic analysis in terms of genes, networks and pathways that are involved in the innate immune response. We have therefore conducted a gene expression profiling study of the effects of TLR stimulation using RAW264.7 macrophages as a model in view of the many prior studies with this cell type, including our investigation of alarmin release. For this purpose, RAW264.7 cells were stimulated with LPS, poly (I:C), or CpG DNA for either 6 or 24 hrs, and RNA was profiled on Agilent microarrays containing probes for approximately 20,000 mouse genes. Gene expression data were analyzed to determine genes, pathways and transcriptional networks that are in common and unique to each of the three TLR stimuli. Potentially novel candidates revealed by this analysis were tested for their role in innate immunity using RNA interference. As the results presented herein show, gene expression profiles differ among TLR ligands in patterns that likely depend on downstream signaling adaptors. Furthermore, this analysis has also revealed two novel genes PLEC1 and TPST1 as genes involved in regulation of macrophage response to innate immune stimuli.
2. Materials and Methods
2.1 Cell Culture
RAW 264.7 cells were maintained in RPMI 1640 supplemented with 10% FBS and 200 µg/ml gentamicin (Invitrogen Life Technologies, Carlsbad, CA). For stimulation of the RAW 264.7 cell, cells were plated in 6-well culture plates (3 × 106 cells/well) overnight. The overnight growth was performed to allow the cells to rest prior to. Cells were then washed twice with Opti-MEM (Invitrogen Life Technologies, Carlsbad, CA) and stimulated with either 0.05 µg/ml Escherichia coli 0111:B4 Ultrapure LPS (List Biological Laboratories, Campbell, CA), 0.25 µg/ml poly (I:C) (InvivoGen, San Diego, CA), 1.5 µM CpG ODN 1826 (Midland Certified Reagents, Midland, TX), or no stimulation (media control). All treatments were performed in triplicate. Cells were harvested 6 or 24 hours post-treatment and RNA was extracted using Trizol following the manufacturer’s protocols (Invitrogen Life Technologies, Carlsbad, CA).
Concentrations of the PAMPs used in the array study were chosen based on our prior titration experiments to produce similar levels of stimulation by the three PAMPs as measured by concentrations of TNF-α, NO and HMGB1 release (Jiang et al., 2005). It should be noted, however, that stimulation with CpG DNA, while inducing TNF-α and NO, was shown to induce less HMGB1 release concentration used than did either LPS or poly (I:C). Of note, as shown previously (Jiang et al, 2007), at 20 hours after stimulation, the amount of apoptosis varied among the PAMPs, with the frequency of apoptotic cells much greater with poly (I:C) stimulation than with either LPS or CpG DNA. Thus, while the concentrations of stimuli were chosen on the basis of the production of TNF-α and NO, other differences (i.e., HMGB1, apoptosis) in cellular responses can occur under these conditions.
2.2 Gene Expression Profiling
Total RNA from three separate cell cultures, treatments and isolations (biological replicates) was used for microarray analysis. The eight conditions examined by microarray analysis included the following: media control 6 hrs, LPS 6 hrs, poly (I:C) 6 hrs, CpG DNA 6 hrs, media control 24 hrs, LPS 24 hrs, poly(I:C) 24 hrs, and CpG DNA 24 hrs. Fluorescently labeled cDNA targets were prepared by reverse transcribing 5 µg of total RNA in the presence of aminoallyl-dUTP followed by chemical coupling of NHS-esters of cyanine 3 (Cy3) and cyanine (Cy5) to the aminoallyl linker, according to published protocols (Yang et al., 2002). All samples were co-hybridized with the Mouse Universal Cell Line Reference RNA (Stratagene, La Jolla, CA), and hybridizations were replicated with the two dyes swapped. We used microarrays containing in situ synthesized 60-mer oligoucleotides with sequences that represent over 20,000 unique well-characterized mouse genes (Agilent Technologies mouse oligo array). Arrays were washed using the recommended SSC washes and scanned on the DNA Microarray Scanner (Agilent Technologies, Santa Clara, CA).
2.3 Microarray Analysis
Image analysis was performed using Agilent Feature Extraction software, and lowess-normalized intensities were used in all further analyses performed in TIGR MIDAS and Multiexperiment Viewer (Saeed et al., 2003). All primary data have been deposited to the Gene Expression Omnibus database (accession GSE15066). Dye flip replicas were filtered, merged to produce a single expression ratio measure for each gene, and log-base2-transformed. Differentially expressed genes for TLR ligand-stimulated cells vs. untreated control were identified using significance analysis of microarrays (SAM) with 100 permutations. For each of the six two-group SAM comparison (LPS vs media control at 6 hrs, poly (I:C) vs media control at 6 hours, CpG DNA vs media control at 6 hrs, LPS vs media control at 24 hrs, poly (I:C) vs media control at 24 hours, and CpG DNA vs media control at 24 hrs), we identified differentially expressed genes as those with median false discovery rate (FDR) of 1% and 2-fold change between treatment and control. Lists of differentially expressed genes in response to LPS, poly (I:C) and CpG DNA were compared and analyzed for enriched pathways and protein networks using the Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA).
2.4 RNA Interference
RNA interference was carried out as previously described (Alper et al., 2008). Briefly, pools of 4 siRNA duplexes/gene (Dharmacon, Lafayette, CO) were transfected into J774A.1 or RAW264.7 cells (100,000 cells/well) using the Amaxa Nucleofector 96 well shuttle. 24–36 hours after siRNA transfection, LPS was added to a final concentration of 20 ng/ml LPS (List Biological Labs). Supernatant was collected 5 hours post-LPS treatment and cytokine production was assayed using the Lincoplex kit (Millipore, Billerica, MA) and read on a Luminex system (Bio-Rad, Hercules, CA). All siRNA assays were performed in triplicate, as independent siRNA transfections, followed by PAMP treatments and cytokine assays. The time-point and concentration of LPS used in this study were chosen based on screening experiment for purposes of consistency with our previously optimized siRNA screens (Alper et al., 2008; Fernandez-Botran and Vt'vička, 2001); in this assay, we use to 20 ng/ml LPS for 5 hours, a slightly lower dose than the 50 ng/ml LPS used in array experiments because we can obtain better RNAi responsiveness under these conditions. Cell viability was monitored and cell number normalized using fluorescien diacetate as described (Alper et al., 2008; Fernandez-Botran and Vt'vička, 2001). Cytokine production was normalized relative to a negative control siRNA described in (Sorensen et al., 2003) (first negative control siRNA in Figure 4). The other negative control siRNAs that do not target any mouse gene in Figure 4 in order were non-targeting siRNA pool #1, non-targeting siRNA#1, and non-targeting siRNA #2 (all from Dharmacon). RNA was isolated using the RNAEasy kit (Qiagen, Germantown, MD), and gene knockdown was monitored by real-time RT-PCR with SYBR Green on a 7900HT sequence detection system (Applied Biosystems, Foster City, CA).
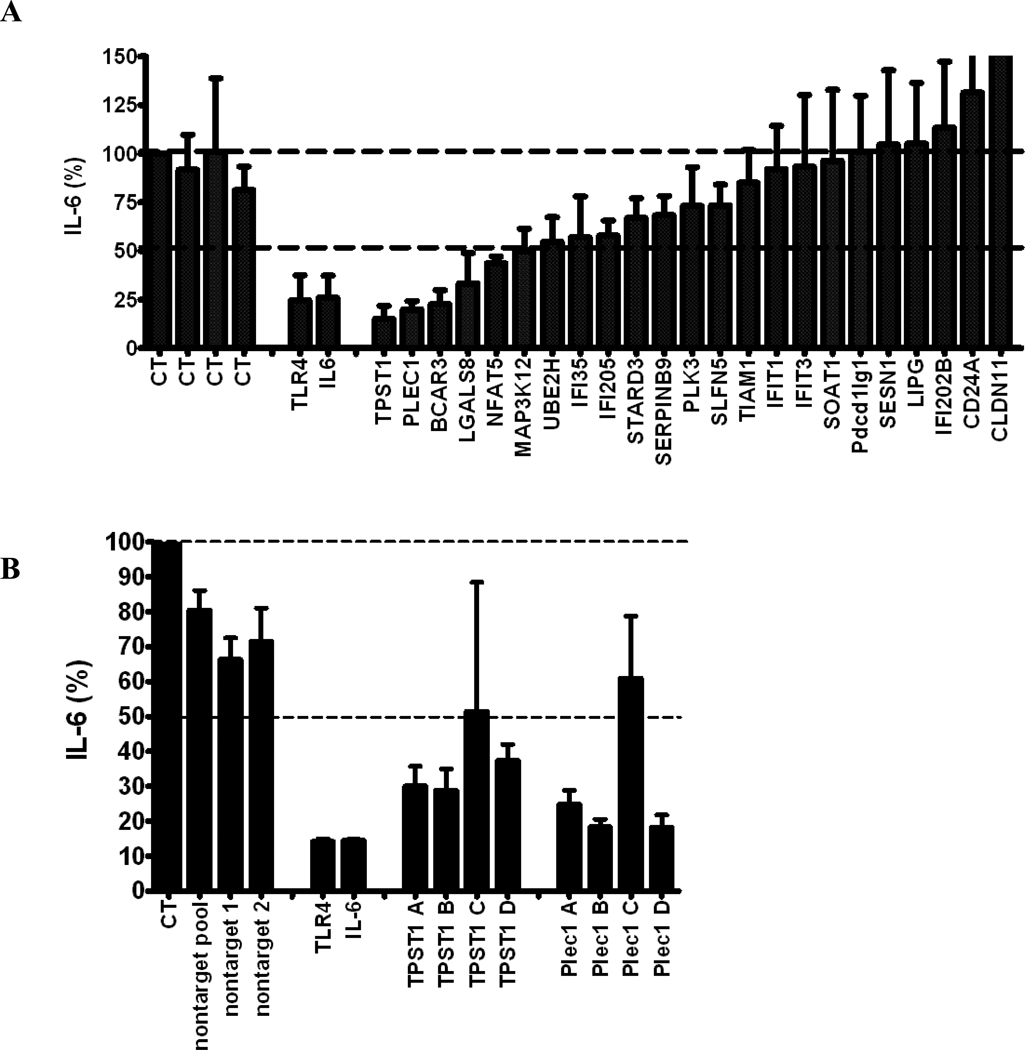
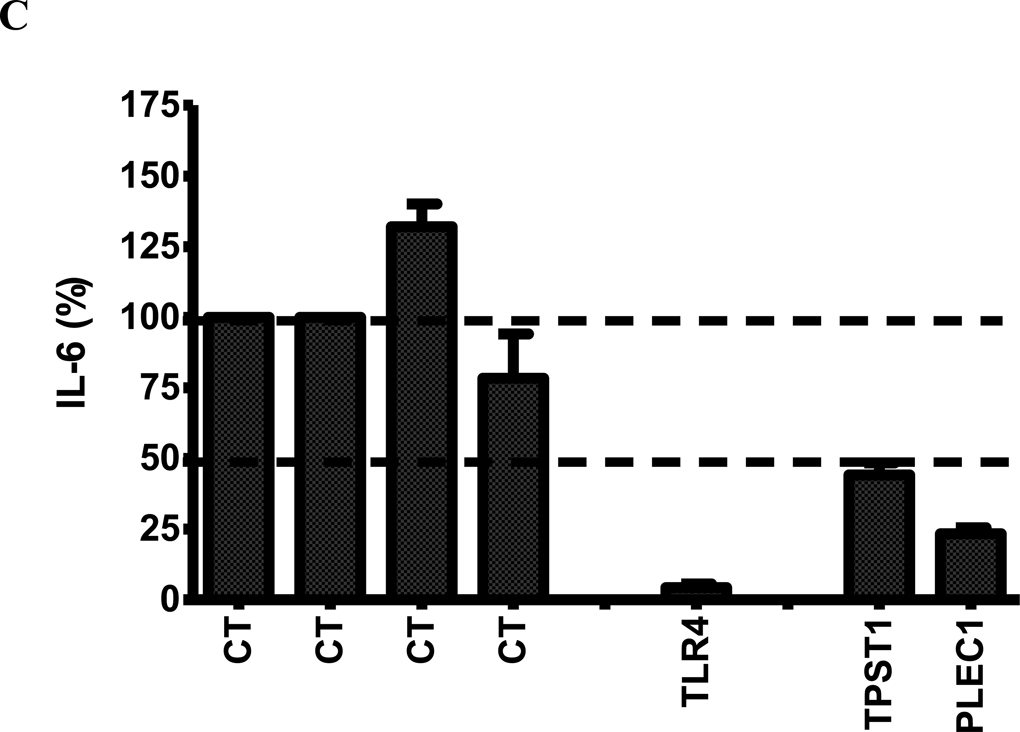
Figure 4.
(A) The effect of RNAi-mediated inhibition of selected differentially expressed genes in Figure 3 in J77A4.1 macrophages on IL-6 production in response to LPS stimulation. Pools of 4 siRNA duplexes per gene were transfected into J77A4.1 cells, LPS was added, and cytokine production was monitored as described in Materials and Methods. Shown are the results for 4 negative controls (non-targeting siRNAs described in Materials and Methods; all data are normalized to the first negative control), 2 positive controls (TLR4 and IL-6), and 23 genes identified in the network analysis of gene expression data. (B) To verify that the phenotypes observed were caused by specific inhibition of that gene and not an off-target siRNA effect, each of the 4 siRNA duplexes in the pools were transfected individually for the 5 genes that had the strongest effect and IL-6 production was monitored. At least two independent siRNAs induced a phenotype for each gene. (C) The effect of RNAi-mediated inhibition of TPST1 and PLEC1 in RAW264.7 macrophages on IL-6 production in response to LPS stimulation 5 hours post-treatment. Pools of 4 siRNA duplexes per gene were transfected into RAW264.7 cells, LPS was added, and cytokine production was monitored as described in Materials and Methods. Shown are the results for 4 negative controls (non-targeting siRNAs described in Materials and Methods; all data are normalized to the first negative control), a positive controls (TLR4), and TPST1 and PLEC1. In all three panels, results plotted are the means of three independent siRNA experiments with error bars representing standard deviations.
2.5 Taqman Gene Expression Assays
Taqman gene expression assays for PLEC1, TPST1, GAPDH, β-actin and 18S RNA were obtained from Applied Biosystems and performed in a 384-well format according to manufacturer’s protocols and reagents. Briefly, each 10 µL reaction contained 50 ng cDNA template, 1X Taqman probe mix and 1X Taqman Expression PCR mix. Data were collected in a 7900HT sequence detection system and analyzed using the ΔΔCt method (Schmittgen and Livak, 2008). We observed significant changes in the expression of GAPDH and β-actin (raw Ct values) in response to LPS and poly(I:C) treatment while no changes in 18S RNA were observed; we therefore used 18S RNA as the endogenous control.
3. Results
3.1 Gene expression profiles of RAW264.7 macrophages in response to LPS, poly (I:C) and CpG DNA
To study the transcriptional response to TLR stimulation, we treated RAW264.7 macrophages with LPS, poly (I:C) or CpG DNA, and extracted RNA 6 and 24 hours post-treatment for profiling on Agilent oligonucleotide arrays containing probes for approximately 20,000 mouse genes. Differentially expressed genes were identified using the significance analysis of microarrays (SAM) algorithm (Tusher et al., 2001) for each TLR ligand compared to a media control treatment. We used a 1% false discovery rate (FDR) and ≥ 2 fold up- or-down-regulation as criteria for defining differentially expressed genes.
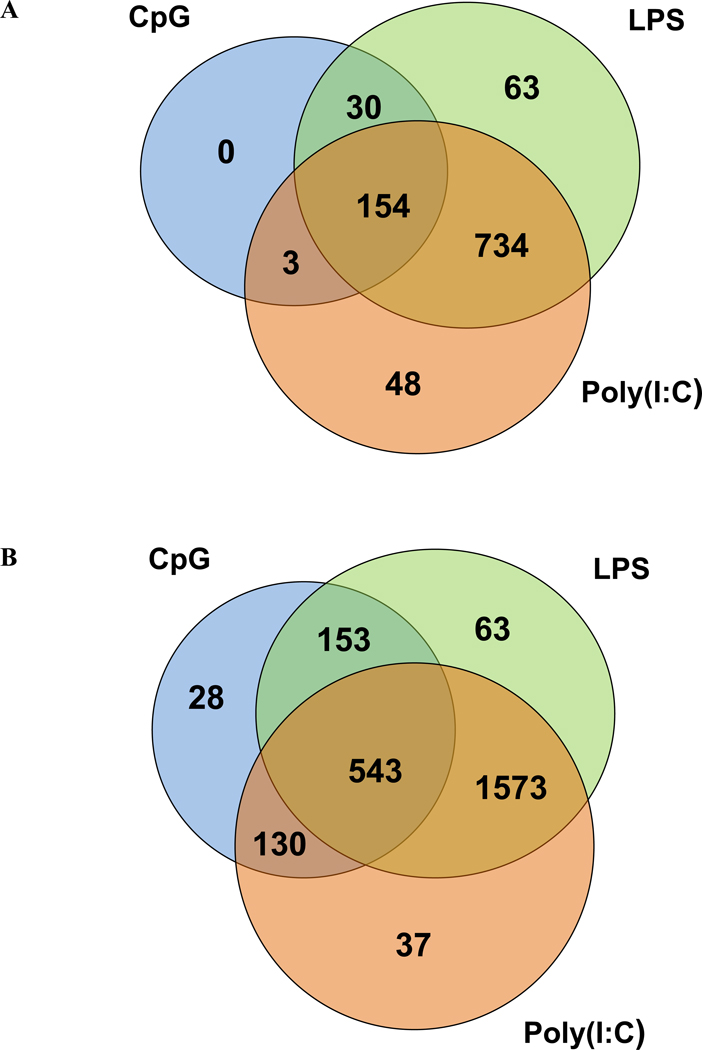
This analysis revealed a large number of differentially expressed genes in response to either LPS or poly (I:C) stimulation after 6 hours, with many genes in common to the two TLR ligands (Figure 1A). CpG DNA, on the other hand, elicited a much less pronounced change in the transcriptional profile with no unique genes induced in response to stimulation with this TLR ligand; this result is consistent with previous findings (Gao et al., 2003). Many more genes were induced or repressed 24 hours post-stimulation for each of the TLR ligands but the pattern of gene expression in response to the three TLR ligands was similar to that at 6 hours (Figure 1B). Complete lists of genes with SAM d scores and TLR ligand/media fold changes are given in Supplemental Tables 2 and 3.
Figure 1.
Venn diagrams showing the number of transcripts that are differentially expressed in response to poly (I:C) (orange circles), LPS (green circles), and CpG DNA (blue circles) in RAW 264.7 macrophages at (A) 6 hours and (B) 24 hours post-treatment. Differentially expressed genes are defined as those significant at 1% FDR by SAM and with ≥2 fold up- or down-regulation (compared to media control). Differentially expressed transcripts are listed in Supplemental Tables 2 and 3.
At both time-points, we found that poly (I:C) and LPS elicit a more pronounced transcriptional response than does CpG DNA both 6 and 24 hours post-treatment. To ensure that this observation was not simply a result of the doses of the three PAMPs we chose for this study, we examined gene expression levels of all genes that are differentially expressed in response to all three PAMPs at 6 hours post stimulation (Supplemental Table 1). It is apparent from data in Supplemental Table 1 that fold change in response to CpG DNA stimulation is often comparable and sometimes exceeds that of poly (I:C). This finding strongly suggests that the reduced transcriptional response of macrophages to CpG stimulation is not a result of the dose of CpG DNA used in our study; rather, this is a biological phenomenon that has been described in a previous study (Gao et al., 2003). Moreover, it is important to note that we chose concentrations of LPS, poly (I:C), and CpG DNA based on our previous studies (Jiang et al., 2007; Jiang and Pisetsky, 2006) in which we show comparable cytokine production at the PAMP concentrations but differences in apoptosis and HMGB1 release.
3.2 Different molecular pathways activated/repressed by PAMP stimulation 6 and 24 hours post-treatment
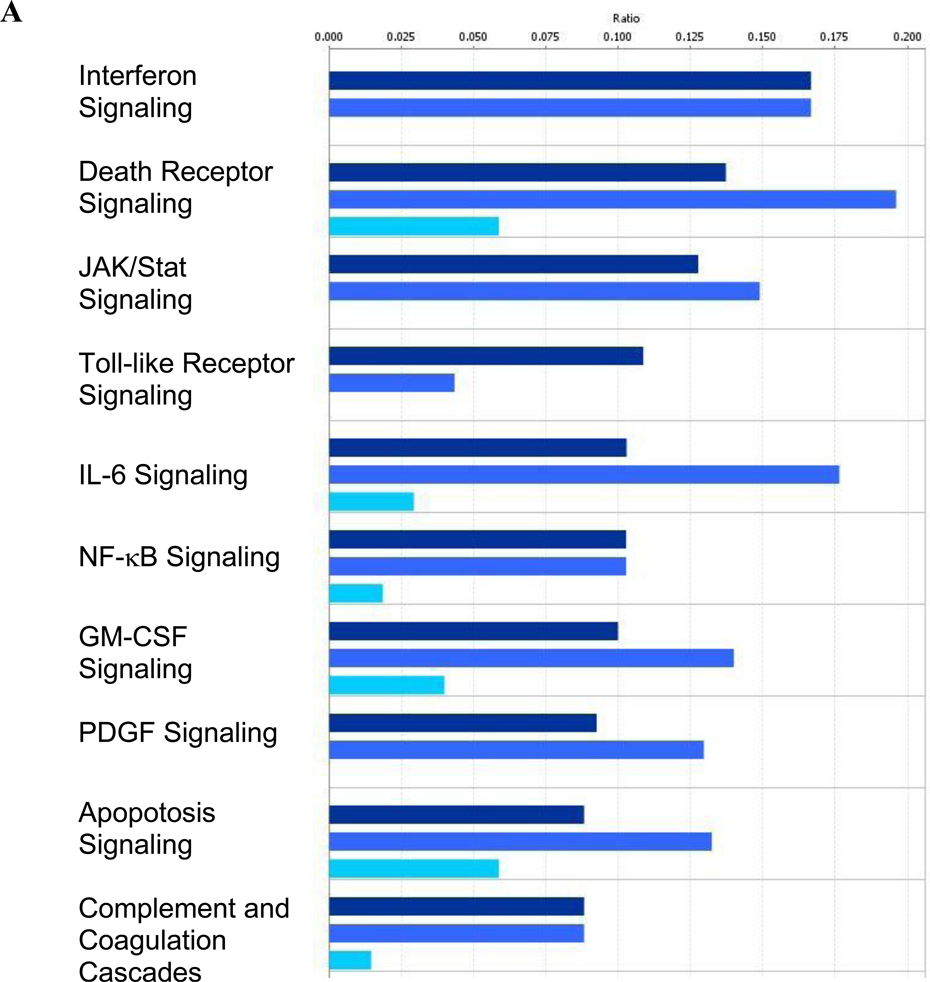
Ingenuity Pathway Analysis (IPA) software and the underlying database (Ganter et al., 2008) were utilized to identify molecular pathways that are activated/repressed in response to stimulation by each TLR ligand. The top 10 significantly altered pathways at the 6 hour time point (Figure 2A) are consistent with the current knowledge of TLR signaling. Interferon signaling is a result of activation of the TRIF-dependent arm of the TLR pathway (Ishii et al., 2008) and is therefore shared by LPS and poly (I:C) but not CpG DNA. The induction of other pathways which are known to be activated in response to TLR stimulation (death receptor, IL-6, NF-κB, apoptosis, and complement/coagulation cascades) (Akira and Takeda, 2004; Raschi et al., 2008; Salaun et al., 2007) were shared by all three TLR ligands but were less pronounced following CpG stimulation.
Figure 2.
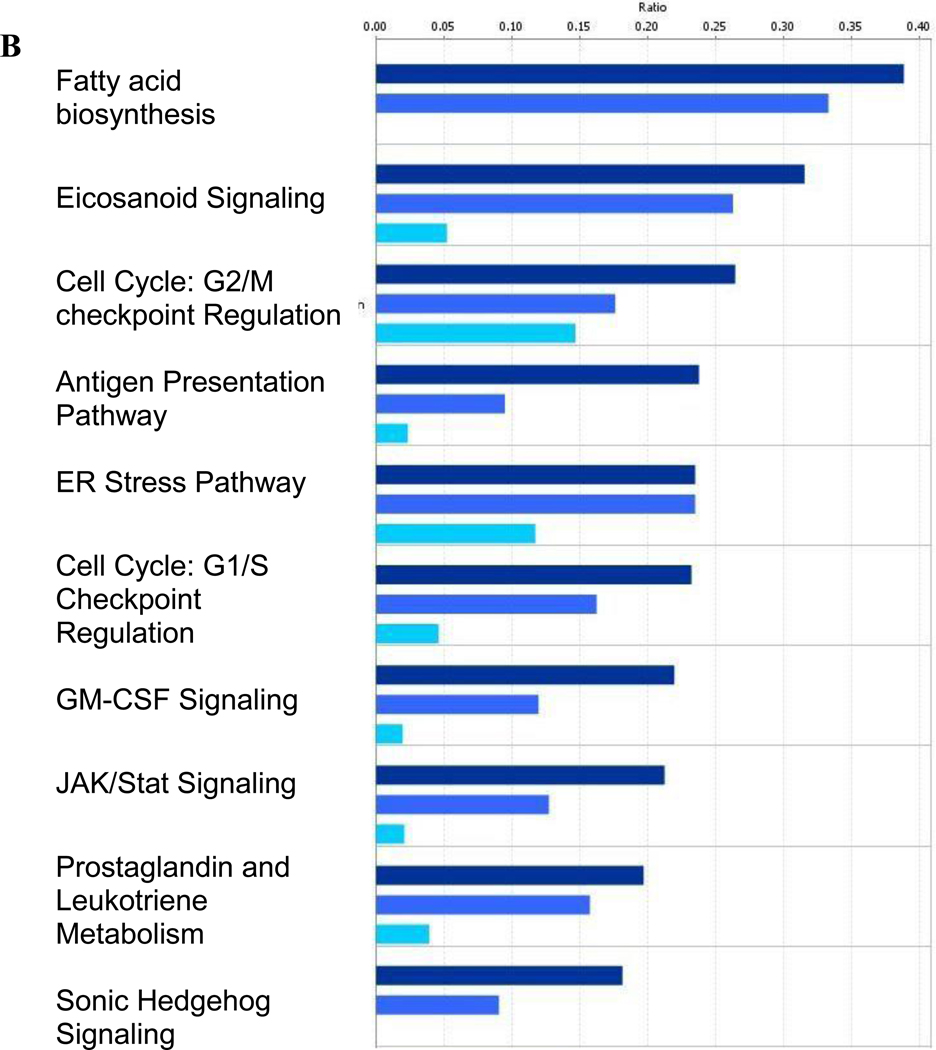
Over-represented canonical pathways identified by Ingenuity Pathway Analysis in RAW264.7 macrophages at (A) 6 hours and (B) 24 hours post-treatment. Differentially expressed genes from the Venn diagrams in Figure 1 were used for the pathway analysis. Poly (I:C) data are shown in navy bars, LPS data are in medium blue, and CpG DNA data are depicted by light blue bars.
The profile of the top 10 activated/repressed pathways 24 hours post-stimulation is very different from that observed at 6 hours. Most pathways are shared by all three TLR ligands and are involved in fatty acid metabolism (fatty acid biosynthesis, eicosanoid signaling, prostaglandin and leukotriene metabolism) and general stress response pathways (cell cycle regulation, ER stress). The Sonic Hedgehog pathway is activated in response to LPS and poly (I:C) which is consistent with our recent findings on the role that Hedgehog plays in the murine response to systemic LPS (Yang et al., 2010).
3.3 Transcriptional networks
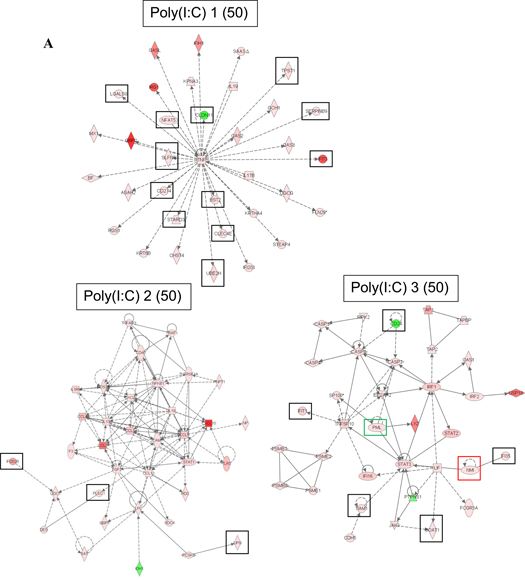
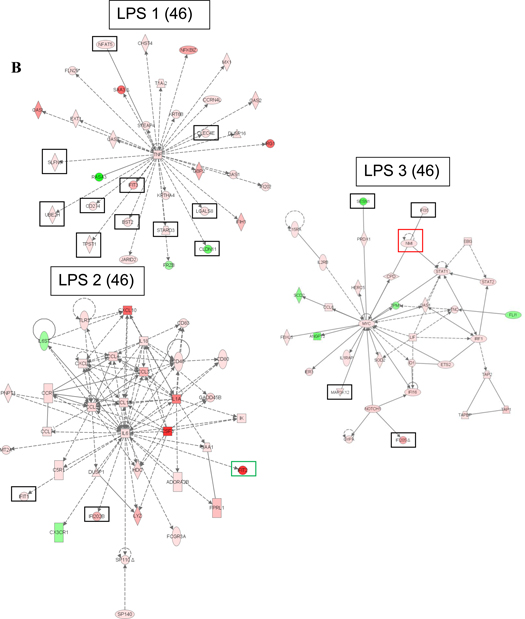
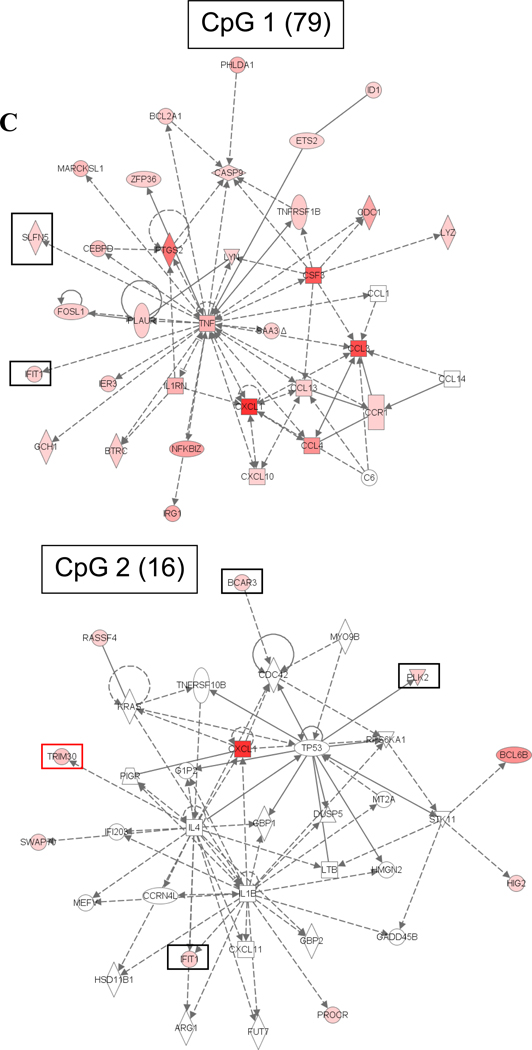
We also used the IPA software to identify the most significant transcriptional networks that were altered in the macrophage response to the TLR ligands at 6 hours post-treatment (Figure 3). The top significantly altered network in response to each of the three stimuli centers around TNF-α and contains many of the genes with known function in TLR signaling. The second altered network in the poly (I:C) gene expression data contains NF-κB and STAT1 transcription factors as well as a number of cytokines and chemokines whose expression is regulated by these two transcription factors. The second altered network in the LPS gene expression data is focused on the IL-6 cytokine but also contains other cytokines and chemokines with known functions in the macrophage response to LPS. The second altered network of genes identified in response to CpG DNA contains very few differentially expressed genes and has a substantially lower score than networks ranked as second in response to poly (I:C) and LPS; the CpG altered network includes the CXCL1 chemokine.
Figure 3.
Top significant transcriptional networks in (A) poly (I:C), (B) LPS, and (C) CpG DNA gene expression data 6 hours post-treatment. Networks are labeled with their IPA ranking and IPA score in parenthesis. The score is the negative exponent of the right-tailed Fisher's exact test p-value that tests the likelihood that the Network Eligible Molecules that are part of a network are found therein by random chance alone. Black boxes denote genes selected for RNA interference assays in macrophages in this study, green boxes denote two genes we have identified in our previous studies whose RNAi-mediated inhibition resulted in significantly lower cytokine production in macrophages (Alper et al., 2008), and red boxes denote two genes we identified in our previous studies whose RNAi-mediated inhibition did not result in significantly lower cytokine production in macrophages (Yang et al., 2010; Yang et al., 2009). IPA network legend is included in the figure.
The third significant network identified in the LPS data involves the STAT1, STAT2, and IRF1 transcription factors, all of which are involved in regulation of interferon-responsive genes. Also identified in this network is myc, an oncogene with an established role in cell cycle progression and apoptosis that has been implicated in host defense. The third significant network in the macrophage response to poly (I:C) contains the STAT3 transcription factor and pro-inflammatory (CASP1, CASP4) and apoptotic (CASP7, CASP8) caspases, among others. Expression profiles of macrophages treated with CpG DNA contain fewer differentially expressed genes than those treated with poly (I:C) or LPS; therefore, only two significant networks were identified in the CpG DNA dataset.
We prioritized genes in the most significant networks in the macrophage response to the three TLR ligands based on their known or predicted function. Our goal was to focus our further functional studies on genes that could be candidate innate immune genes based on their function but whose role in innate immunity has not been extensively studied. We generally included kinases, proteases, transcription factors, and genes of unknown function. We excluded genes involved in metabolism. We also focused on genes that are members of gene families which include members with a known role in immune response. Slfn5, for example, is a member of the schlafen family in which schlafen 2 has been identified as a gene with a role in TLR signaling (Sohn et al., 2007). Genes that have black boxes drawn around them in Figure 3 were selected for RNA interference assays based on these criteria. A few genes were excluded from the current investigation based on our published data on the effect of RNAi of these genes on LPS-induced cytokine production by macrophages. PML and IFIT2 (green boxes) are two genes that we identified in our previous studies whose RNAi-mediated inhibition resulted in significantly lower cytokine production in macrophages (Alper et al., 2008); NMI and TRIM30 (red boxes) are two genes that we identified in our previous studies whose RNAi-mediated inhibition did not result in significantly lower cytokine production in macrophages (Yang et al., 2010; Yang et al., 2009). In total, 23 genes were selected for further investigation in this study.
3.4 Novel role of TSPT1 and PLEC1 in the innate immune response
To test novel candidate genes identified by our gene expression analysis for a role in the innate immune response, we inhibited 23 genes using RNA interference in a mouse macrophage cell line and subsequently monitored inflammatory cytokine production following LPS treatment. For this purpose, we initially used the mouse macrophage cell line J774A.1. Our laboratory has conducted extensive RNAi optimization in several macrophage cell lines and found that these cells exhibited a slightly better RNAi response than the RAW264.7 cells [(Alper et al., 2008) and unpublished data]. We transfected J774A.1 cells with a pool of 4 siRNA duplexes for each gene, stimulated cells with LPS, and measured cytokine production 5 hours after LPS treatment. As a control, we inhibited several negative control genes that should not affect cytokine production; this had little effect on cytokine production (Figure 4A). As a positive control for the RNAi, we demonstrated that inhibition of TLR4 and IL-6 strongly decreased the production of IL-6.
Importantly, inhibition of five candidate genes (TPST1, PLEC1, BCAR3, LGALS8, and NFAT5) led to >50% inhibition of IL-6 production. We performed additional siRNA experiments focusing on these five genes to confirm the RNAi results. First, we titrated the siRNA concentration down (1 µM, 0.5 µM, and 0.25 µM siRNA) and monitored IL-6 production and RNA knockdown using qPCR. Inhibition of only two of the candidate genes (TPST1 and PLEC1) consistently inhibited production of IL-6, with the effect lost at lower siRNA concentrations (Supplemental Figure 1A). The ability of the siRNAs to inhibit IL-6 production for these two genes also correlated with the extent of gene expression knockdown (Supplemental Figure 1B). As an additional confirmation of the RNAi results, we transfected each of the four siRNA duplexes in each of these two siRNA pools individually, and demonstrated that at least two out of the four individual siRNAs in each pool significantly inhibited production of IL-6 (Figure 4B). This demonstrates that the effect on IL-6 is likely due to specific inhibition of that target gene and not an off-target siRNA effect.
To validate further findings for PLEC1 and TPST1, we repeated the siRNA assays for these two genes in a second mouse macrophage cell line, RAW 264.7 macrophages, at 5 hours post-LPS treatment. These data confirmed that RNAi-mediated inhibition of PLEC1 and TPST1 inhibited LPS-induced IL-6 production (Figure 4C). To study the effect of RNAi-mediate inhibition of these two genes on cytokine production in response to poly (I:C) and CpG DNA, we first measured the levels of IL-6 and IFN-β produced by RAW 264.7 macrophages in response to these two PAMPs at the same concentrations used in the microarray study at 4, 8, 12, and 24 hours post-treatment. These data showed a peak in IFN-β production at 12 hours post poly (I:C) treatment and very low cytokine production in response to CpG DNA at any of the time-points (data not shown). We therefore chose to examine the effect of inhibition of PLEC1 and TPST1 expression on poly (I:C)-induced IFN-β production 12 hours post treatment; in these experiments, we observed only a modest decrease in poly (I:C)-induced IFN-β production as a result of inhibition of PLEC1 and TPST1 poly (I:C)-induced IFN-β as a result of inhibition of TLR3 (data not shown).
Finally, to validate our microarray data, we examined the expression of PLEC1 and TPST1 at four different time-points (4, 8, 12, and 24 hrs) in response to three different concentrations of each PAMP (concentration used in the microarray study and two additional concentrations) by Taqman gene expression assays. Data in Table 1 demonstrate the induction of TPST1 expression in response to LPS and poly (I:C) at all time-points other than 24 hours; at 24 hrs, we observed a decrease in TPST1 expression compared to no treatment in concordance with microarray data. Similarly, the expression of PLEC1 was induced by poly (I:C) but not LPS at all time-points other than 24 hours, which parallels our microarray findings. We observed downregulation of PLEC1 in response to poly(I:C), a finding that was not measured on the microarray. Finally, in accord with our microarray findings, no significant change in expression of either gene was observed in response to CpG DNA. Together, these studies implicate PLEC1 and TPST1 in the regulation of the innate immune response and show that transcriptional analysis can identify new genes important in the regulation of the macrophage innate immune response.
Table 1.
Expression of PLEC1 and TPST in RAW 264.7 macrophages in response to three concentrations of each PAMP at four timepoints, as assayed by Taqman gene expression assays using 18S RNA as internal reference. All measurements were made in triplicate. Fold changes were calculated by the ΔΔCt method relative to media control (no treatment).
| TPST1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PAMP | Conc, ug/mL |
4 hr Mean |
4 hr SD |
8 hr Mean |
8 hr SD |
12 hr Mean |
12 Hr SD |
24 Hr Mean |
24 Hr SD |
|
| CpG | 1.5 | −1.34 | 0.17 | −1.25 | 0.23 | 1.15 | −0.13 | −2.65 | 1.49 | |
| 10 | −1.25 | 0.03 | −1.21 | 0.17 | 1.36 | −0.09 | −1.86 | 0.06 | ||
| 25 | −1.66 | 0.12 | 1.36 | −0.05 | 1.21 | −0.08 | −1.69 | 0.00 | ||
| LPS | 0.05 | 2.13 | −0.12 | 1.18 | −0.09 | 1.11 | −0.03 | −3.62 | 1.00 | |
| 0.25 | 2.58 | −0.69 | 1.26 | −0.05 | 2.42 | −0.99 | −3.81 | 0.18 | ||
| 0.5 | 3.60 | −0.21 | 1.44 | −0.07 | 1.71 | −0.13 | −3.01 | 0.06 | ||
| PIC | 0.25 | 2.71 | −0.17 | 1.23 | −0.07 | 2.56 | −0.16 | −2.88 | 0.45 | |
| 1 | 2.16 | −0.08 | 1.06 | −0.09 | 3.72 | −0.33 | −2.33 | 0.33 | ||
| 2.5 | 2.09 | −0.08 | 1.89 | −0.13 | 4.28 | −0.38 | −3.04 | 0.86 | ||
| PLEC1 | ||||||||||
| PAMP |
Conc, ug/mL |
4 hr Mean |
4 hr SD |
8 hr Mean |
8 hr SD |
12 hr Mean |
12 Hr SD |
24 Hr Mean |
24 Hr SD |
|
| CpG | 1.5 | 1.19 | 0.06 | −1.14 | 0.02 | −1.38 | 0.14 | −1.31 | 0.25 | |
| 10 | −1.65 | 0.04 | 1.27 | 0.12 | 2.76 | 0.32 | −2.01 | 0.17 | ||
| 25 | −1.65 | 0.03 | 1.02 | 0.04 | 1.71 | 0.33 | −2.39 | 0.26 | ||
| LPS | 0.05 | −1.77 | 0.15 | −1.26 | 0.12 | −2.16 | 0.51 | −8.50 | 1.62 | |
| 0.25 | −2.83 | 0.25 | −2.48 | 0.37 | −1.61 | 0.05 | −10.92 | 2.41 | ||
| 0.5 | −2.99 | 0.20 | −2.66 | 0.23 | −1.73 | 0.10 | −8.66 | 1.19 | ||
| PIC | 0.25 | 1.52 | 0.05 | 2.17 | 0.16 | 2.18 | 0.91 | 1.19 | 0.11 | |
| 1 | 1.56 | 0.48 | 2.40 | 0.10 | 2.57 | 0.28 | 0.98 | 0.12 | ||
| 2.5 | 1.11 | 0.08 | 2.33 | 0.15 | 4.53 | 0.25 | 1.78 | 0.12 | ||
4. Discussion
Our analyses provide new insights into the transcriptional response of macrophages to TLR stimulation and have identified genes, pathways, and transcriptional networks that are differentially expressed in response to stimulation with poly (I:C), LPS, or CpG DNA. Using the well-characterized mouse macrophage cell line RAW264.7 as a model and standard criteria for defining differential gene expression (1% FDR and 2 fold change), we identified a large number of transcriptional changes in macrophages in response to PAMP stimulation. We chose not to restrict our further analysis to a smaller number of genes that are differentially expressed using more stringent criteria (higher fold change, for example) because pathway and network analysis provides a second statistical analysis and associated significance levels, i.e. identifies pathways/networks that are enriched in differentially expressed genes.
We found that poly (I:C) and LPS elicit a more pronounced transcriptional response than does CpG DNA both 6 and 24 hours post-treatment. A large proportion of induced/repressed genes are shared by the two PAMPs; these common genes are presumably induced or repressed through the TRIF-mediated arm of the TLR pathway. Similarly, genes regulated by LPS and CpG DNA are presumably induced/repressed through the MyD88-mediated arm of the TLR pathway. However, the majority of the genes that are differentially expressed in response to CpG DNA are also regulated by LPS and poly (I:C) (middle section of Venn diagrams in Figure 1); these are most likely genes regulated by NF-κB, which acts downstream of both the MyD88 and TRIF adaptors. No genes found are unique to CpG DNA stimulation at 6 hrs; this finding is in agreement with the study by Gao et al. showing that the CpG transcriptional profile is a subset of the LPS profile (Gao et al., 2003).
Similar conclusions about the differences in the consequences of TLR stimulation emerge from the pathway analysis. For example, interferon signaling, which is a result of TLR signaling through TRIF, is shared by LPS and poly (I:C) but not CpG DNA. Other pathways known to be activated in response to TLR stimulation (death receptor, IL-6, NF-κB, apoptosis, and complement/coagulation cascades) are shared by all three PAMPs but are less pronounced in the response to CpG DNA. These patterns are consistent with our studies showing the consequences of TLR stimulation with respect to release of HMGB1 and the induction of apoptosis (Jiang et al., 2007; Pisetsky, 2007). Of note, the top 10 pathways that are over-represented in gene expression profiles 24 hours post-treatment are very different from those at 6 hours post-treatment. This change most likely reflects the secondary response of macrophages at this later time point to cytokines and other molecules produced in response to PAMP stimulation.
Our analysis also identified pathways whose role in innate immunity has been less well documented. One example concerns the role of the Hedgehog signaling pathway. Hedgehog is one of the key regulators of animal development, is conserved from flies to humans, and has also been implicated in the development of some cancers. Our recent studies identified Hedgehog signaling as a component of the innate immune response to systemic LPS in mice (Yang et al., 2010).
Network analysis of gene expression profiles allowed us to identify 23 genes as candidate genes for regulation of the innate immune response. We chose to use network analysis to focus our attention on genes that may or may not be differentially expressed themselves but that interact with genes that are known to be important in the innate immune response; TNFα is for example in the center of the top network for all three PAMPs and we aimed to identify novel genes that are involved in regulation of TNFα production. We selected these genes based on their known or predicted function and on the lack of previous studies on their involvement in innate immunity. By utilizing RNAi-mediated suppression of gene expression in cultured macrophages, two genes with no previously known role in innate immunity were shown to affect cytokine production in macrophages, namely, TPST1 and PLEC1.
TPST1 expression is upregulated in response to poly (I:C) and LPS, and is in the top significantly altered transcriptional networks in both gene expression datasets. IPA shows an indirect action of TNF-α on this gene. TPST1 is one of the two known Golgi tyrosylprotein sulfotransferases that mediate protein tyrosine sulfation, a posttranslational modification that plays a role in strengthening protein-protein interactions. Tyrosine sulfation has been shown to increase activity of C4 in the complement cascade (Hortin et al., 1989) and contribute to the binding of the CCR5 chemokine receptor to MIP-1α, MIP-1β, and HIV-1 gp120/CD4 complexes and to the ability of HIV-1 to enter cells (Seibert et al., 2002). In mice, it has been shown that tyrosine sulfation is required for normal pulmonary function at birth (Westmuckett et al., 2008). Based on these published studies, it is possible that tyrosine sulfation may be involved in the macrophage response to TLR ligand stimulation. Further studies will be necessary to determine which proteins in the TLR or other signaling pathways are sulfated and how this posttranslational modification affects innate immune response.
In response to poly (I:C) stimulation of macrophages, PLEC1 is also upregulated and is in the second significant transcriptional network in this dataset, with the CCL5 chemokine having an indirect action on this gene. PLEC1 is a prominent member of a family of structurally and functionally related proteins, termed plakins or cytolinkers, that are capable of interlinking different elements of the cytoskeleton. Plakins play crucial roles in maintaining cell and tissue integrity and orchestrating dynamic changes in cytoarchitecture and cell shape; they also serve as scaffolding platforms for the assembly, positioning, and regulation of signaling complexes. PLEC1 has been shown to interact with CXCR4 and play a significant role in CXCR4 signaling and trafficking in HEK293 and Jurkat T cells (Ding et al., 2008); it thus may be an important factor in HIV-1 infections. Targeted deletion of plectin isoform 1 in mice results in reduced leukocyte infiltration during wound healing; this study established a role of cytolinker proteins in leukocyte recruitment (Abrahamsberg et al., 2005). These previous studies provide a basis for the potential role of PLEC1 in innate immune signaling in macrophages.
The remaining 21 genes tested in the RNAi assay did not significantly and consistently affect cytokine production by macrophages. However, genes whose inhibition by RNAi did not affect cytokine production in J774A.1 macrophages should not be disregarded as potentially important in innate immunity. There are at least two limitations to the RNAi assay utilized in this study. First, some of our candidate genes may regulate innate immunity through mechanisms that do not affect cytokine production; as such, their function may be revealed in assays that measure other responses such as production of superoxide or nitric oxide, phagocytosis, apoptosis, etc. Second, RNA interference is not always effective in silencing target genes due to a number of factors, including the chosen siRNA sequence and the structure of the siRNA. Finally, it should be noted that genes identified as differentially expressed by microarrays that we did not prioritize for siRNA analysis could be important in innate immunity and are good candidates for further studies.
3 Conclusions
In summary, our study identified genes, pathways and transcriptional networks that characterize the macrophage response to three TLR stimuli, LPS, poly (I:C) and CpG DNA and demonstrated both common and unique patterns. By means of RNAi-mediated gene inhibition, we showed that two genes identified in this way, TPST1 and PLEC1, play a role in LPS-induced pro-inflammatory cytokine production by macrophages. Expression of TPST1 and PLEC1 were both regulated by poly (I:C) and LPS but not CpG DNA, suggesting that these genes may be downstream of the TRIF adaptor molecule in the TLR signaling pathway. Further studies in macrophages and in in vivo mouse models are needed to provide insight into how these genes participate in innate immunity. Moreover, studies in human cohorts will be necessary to determine whether polymorphisms in these two genes play a role in infections caused by microbial or viral pathogens.
Highlights.
Our analysis identified pathways and transcriptional networks involved in macrophage response to three TLR ligands: LPS, poly (I:C), and CpG DNA.
RNA interference-mediated inhibition of two genes identified in this analysis, PLEC1 and TPST1, reduced IL-6 production by J774A.1 and RAW264.7 macrophages stimulated with LPS.
These findings delineate macrophage gene response patterns induced by different PAMPs and identify new genes that have not previously been implicated in innate immunity.
Supplementary Material
Acknowledgments
This study was funded by the Intramural Research Program of the NIH, National Institute of the Environmental Health Sciences and National Heart Lung and Blood Institute, a VA Merit Review grant, and American Lung Association grant RG-169529-N.
Abbreviations
- IPA
Ingenuity Pathway Analysis
- LPS
Lipopolysacharide
- PAMP
Pathogen-associated molecular pattern
- PRR
Pattern recognition receptors
- RNAi
RNA interference
- TLR
Toll-like receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahamsberg C, Fuchs P, Osmanagic-Myers S, Fischer I, Propst F, Elbe-Burger A, Wiche G. Targeted ablation of plectin isoform 1 uncovers role of cytolinker proteins in leukocyte recruitment. Proc Natl Acad Sci U S A. 2005;102:18449–18454. doi: 10.1073/pnas.0505380102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Alper S, Laws R, Lackford B, Boyd WA, Dunlap P, Freedman JH, Schwartz DA. Identification of innate immunity genes and pathways using a comparative genomics approach. Proc Natl Acad Sci U S A. 2008;105:7016–7021. doi: 10.1073/pnas.0802405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Zhang L, Goodwin JS, Wang Z, Liu B, Zhang J, Fan GH. Plectin regulates the signaling and trafficking of the HIV-1 co-receptor CXCR4 and plays a role in HIV-1 infection. Exp Cell Res. 2008;314:590–602. doi: 10.1016/j.yexcr.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Botran R, Vt'vička V. Methods in Cell Immunology. Boca Raton, FL: CRC; 2001. [Google Scholar]
- Ganter B, Zidek N, Hewitt PR, Muller D, Vladimirova A. Pathway analysis tools and toxicogenomics reference databases for risk assessment. Pharmacogenomics. 2008;9:35–54. doi: 10.2217/14622416.9.1.35. [DOI] [PubMed] [Google Scholar]
- Gao JJ, Diesl V, Wittmann T, Morrison DC, Ryan JL, Vogel SN, Follettie MT. Bacterial LPS and CpG DNA differentially induce gene expression profiles in mouse macrophages. J Endotoxin Res. 2003;9:237–243. doi: 10.1179/096805103225001431. [DOI] [PubMed] [Google Scholar]
- Hortin GL, Farries TC, Graham JP, Atkinson JP. Sulfation of tyrosine residues increases activity of the fourth component of complement. Proc Natl Acad Sci U S A. 1989;86:1338–1342. doi: 10.1073/pnas.86.4.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe. 2008;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bell CW, Pisetsky DS. The relationship between apoptosis and high-mobility group protein 1 release from murine macrophages stimulated with lipopolysaccharide or polyinosinic-polycytidylic acid. J Immunol. 2007;178:6495–6503. doi: 10.4049/jimmunol.178.10.6495. [DOI] [PubMed] [Google Scholar]
- Jiang W, Li J, Gallowitsch-Puerta M, Tracey KJ, Pisetsky DS. The effects of CpG DNA on HMGB1 release by murine macrophage cell lines. J Leukoc Biol. 2005;78:930–936. doi: 10.1189/jlb.0405208. [DOI] [PubMed] [Google Scholar]
- Jiang W, Pisetsky DS. The role of IFN-alpha and nitric oxide in the release of HMGB1 by RAW 264.7 cells stimulated with polyinosinic-polycytidylic acid or lipopolysaccharide. J Immunol. 2006;177:3337–3343. doi: 10.4049/jimmunol.177.5.3337. [DOI] [PubMed] [Google Scholar]
- Klaschik S, Tross D, Shirota H, Klinman DM. Short- and long-term changes in gene expression mediated by the activation of TLR9. Mol Immunol. 2010;47:1317–1324. doi: 10.1016/j.molimm.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisetsky DS. The role of nuclear macromolecules in innate immunity. Proc Am Thorac Soc. 2007;4:258–262. doi: 10.1513/pats.200701-027AW. [DOI] [PubMed] [Google Scholar]
- Ramsey SA, Klemm SL, Zak DE, Kennedy KA, Thorsson V, Li B, Gilchrist M, Gold ES, Johnson CD, Litvak V, Navarro G, Roach JC, Rosenberger CM, Rust AG, Yudkovsky N, Aderem A, Shmulevich I. Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics. PLoS Comput Biol. 2008;4 doi: 10.1371/journal.pcbi.1000021. e1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschi E, Borghi MO, Grossi C, Broggini V, Pierangeli S, Meroni PL. Toll-like receptors: another player in the pathogenesis of the anti-phospholipid syndrome. Lupus. 2008;17:937–942. doi: 10.1177/0961203308095140. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Salaun B, Romero P, Lebecque S. Toll-like receptors' two-edged sword: when immunity meets apoptosis. Eur J Immunol. 2007;37:3311–3318. doi: 10.1002/eji.200737744. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schmitz F, Mages J, Heit A, Lang R, Wagner H. Transcriptional activation induced in macrophages by Toll-like receptor (TLR) ligands: from expression profiling to a model of TLR signaling. Eur J Immunol. 2004;34:2863–2873. doi: 10.1002/eji.200425228. [DOI] [PubMed] [Google Scholar]
- Seibert C, Cadene M, Sanfiz A, Chait BT, Sakmar TP. Tyrosine sulfation of CCR5 N-terminal peptide by tyrosylprotein sulfotransferases 1 and 2 follows a discrete pattern and temporal sequence. Proc Natl Acad Sci U S A. 2002;99:11031–11036. doi: 10.1073/pnas.172380899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn WJ, Kim D, Lee KW, Kim MS, Kwon S, Lee Y, Kim DS, Kwon HJ. Novel transcriptional regulation of the schlafen-2 gene in macrophages in response to TLR-triggered stimulation. Mol Immunol. 2007;44:3273–3282. doi: 10.1016/j.molimm.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Sorensen DR, Leirdal M, Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J Mol Biol. 2003;327:761–766. doi: 10.1016/s0022-2836(03)00181-5. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tross D, Petrenko L, Klaschik S, Zhu Q, Klinman DM. Global changes in gene expression and synergistic interactions induced by TLR9 and TLR3. Mol mmunol. 2009;46:2557–2564. doi: 10.1016/j.molimm.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmuckett AD, Hoffhines AJ, Borghei A, Moore KL. Early postnatal pulmonary failure and primary hypothyroidism in mice with combined TPST-1 and TPST-2 deficiency. Gen Comp Endocrinol. 2008;156:145–153. doi: 10.1016/j.ygcen.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang IV, Alper S, Lackford B, Rutledge H, Warg LA, Burch LH, Schwartz DA. Novel Regulators of the Systemic Response to Lipopolysaccharide (LPS) Am J Respir Cell Mol Biol. 2010 doi: 10.1165/rcmb.2010-0342OC. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang IV, Chen E, Hasseman JP, Liang W, Frank BC, Wang S, Sharov V, Saeed AI, White J, Li J, Lee NH, Yeatman TJ, Quackenbush J. Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-research0062. research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang IV, Wade CM, Kang HM, Alper S, Rutledge H, Lackford B, Eskin E, Daly MJ, Schwartz DA. Identification of novel genes that mediate innate immunity using inbred mice. Genetics. 2009;183:1535–1544. doi: 10.1534/genetics.109.107540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.