Abstract
Cell migration requires polarization of the cell into the leading edge and the trailing edge. Microtubules (MTs) are indispensable for polarized cell migration in the majority of cell types. To support cell polarity, MT network has to be functionally and structurally asymmetric. How is this asymmetry achieved? In interphase cells, MTs form a dynamic system radiating from a centrosome-based MT-organizing center (MTOC) to the cell edges. Symmetry of this radial array can be broken according to four general principles. Asymmetry occurs due to differential modulation of MT dynamics, relocation of existing MTs within a cell, adding an asymmetric nucleation site, and/or repositioning of a symmetric nucleation site to one side of a cell. Combinations of these asymmetry regulation principles result in a variety of asymmetric MT networks typical for diverse motile cell types. Importantly, an asymmetric MT array is formed at a non-conventional MT nucleation site, the Golgi. Here, we emphasize the contribution of this array to the asymmetry of MT network.
Keywords: microtubules, asymmetry, Golgi, cell polarity, cell motility, CLASP
Importance of MT asymmetry in motile cells
Cell migration in higher organisms is essential for multiple physiological and pathophysiological processes including embryonic development, tissue regeneration, immune responses, and cancer cell metastasis and invasion. The first requirement for directional cell movement is a polarization of the cell into the leading edge and the trailing edge. A highly regulated asymmetric remodeling of the actin cytoskeleton controlled by small GTPases of the Rho family drives polarized motility (for reviews see refs.1, 2). Motility also involves asymmetric formation and turnover of focal adhesions – the cell anchorage sites (for a review see ref.3) and directed post-Golgi trafficking to the protruding lamella (for a review see ref.4).
Microtubules (MTs) are indispensable for polarized cell migration in a variety of cell types, including fibroblasts, astrocytes and neurons5 (for reviews see refs.6, 7) and for major tissue morphogenetic movements such as gastrulation.8 Formation of cell edge protrusions and retractions continues in the absence of MTs but is not distributed in a polarized fashion. MTs are therefore thought to support polarized distribution of regulatory motility signals within a cell.
Some specialized cells, like fish keratocytes or neutrophils, do not need the MT-driven intracellular management for migration, likely due to their small size and simplicity.6, 7 But, although neutrophils are able to move in the absence of MTs, they exhibit impaired directionality of migration.9 The effect of MT disruption on neutrophil motility, though unusual, is likely due to depolarization of signaling events. Thus, in the majority of cell types MTs are needed for the spatial coordination of molecular events leading to efficient cell relocation. In Dictyostelium amoebae that exhibit similar minor MT dependence MTs directed toward the cell rear are responsible for trafficking of adenylyl cyclase-containing vesicles in that direction, which, in turn, coordinates migration of following cells (streaming).10 In this case, asymmetric MT distribution is important for coordination of signals on the multicellular level rather than within each small cell. In many systems, an external signal triggers initial asymmetry of adhesive and actin systems that further need MTs for maintenance.11, 12 In some cases (e.g. in differentiating neurons13), MTs appear to be involved in the initiation of polarity.
Many asymmetric processes essential for cell migration, such as Rac1-dependent actin polymerization14 and focal adhesion turnover,15, 16 are coordinated by MTs. MTs have been shown to deliver and control various functional molecules involved in motility, either as protein complexes or within vesicular membranes. These include β-actin mRNA17 and members of Rho GTPase pathways14, 18 to organize actin and adhesion rearrangements, integrins to initiate adhesion19, membrane to provide building blocks for protrusion (for a review see ref.20) and many others. Some molecules (e.g. integrins) are simply transported to their functional sites by MTs; the functional activity of others, like Rho GEF H1,21 can be modulated by MT binding. Moreover, MT-binding properties of certain active molecules often depend on the dynamic status of MTs. For instance, certain kinesins preferentially bind de-tyrosinated stable MTs22–24 and a number of diverse factors surf growing MT plus ends (for a review see ref.25). Accordingly, the presence of a reliable MT “track” is central for some pathways, while even subtle modulations in MT dynamics can be critical for others. In this regard, it is noteworthy that both MT distribution and dynamics are asymmetric in motile cells.
Variants and principles of MT asymmetry
In order to differentially control diverse domains in a polarized cell, the MT system must be functionally asymmetric. The most logical way to support spatial functional asymmetry is to use a structurally asymmetric regulatory system. How is this asymmetry achieved? In general, MTs form a dynamic system radiating from a perinuclear centrosome-based MT-organizing center (MTOC) to the cell edges. In fibroblasts and other cells with mesenchymal motile phenotype, the distribution of MTs is asymmetric, with a substantial array extended in the direction of movement. This front-oriented array is likely essential for persistent directional motility of these cell types via facilitating actin polymerization,14 adhesion turnover15, 26 and vesicular trafficking27 toward the protrusion. On the contrary, in migrating neutrophils MTs are directed almost exclusively toward the trailing cell edge.28 Upon MT depolymerization, these cells demonstrate defects in finding the shortest way to the chemo-attractant. Interestingly, directional motility of neutrophil disturbed by MT disruption can be restored by inhibiting Rho kinase at the cell rear where MTs are located.9 Similarly, MTs are predominantly directed toward the rear of migrating Dictyostelium amoebae where their major signal distribution function is associated with backward vesicular transport.10 Another example, neurons present an extreme example of MT asymmetry, with principally distinct MT organization in dendrites and axons, which corresponds to distinction in major vesicular transport functions (for reviews see refs.29, 30). In all three scenarios, despite obvious differences, asymmetric MT organization is well suited to provide uneven distribution of signals within a cell and can be achieved by a few variably combined mechanistic principles.
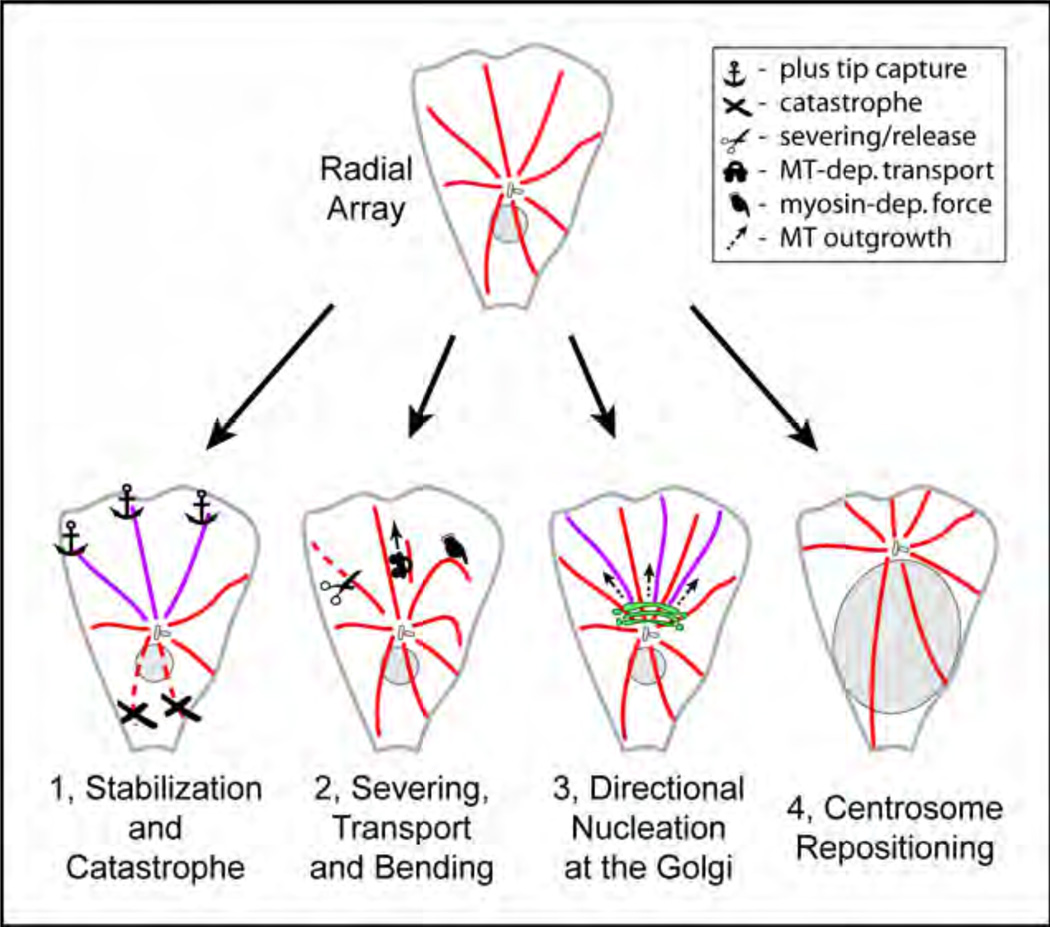
As mentioned above, MTs in interphase cells are nucleated at the centrosome in the cell center. However, MT arrays initially produced by the centrosome are radially symmetric both in vitro31, 32 and in polarized cells.33 Four general principles can be applied to breaking symmetry of the MT network (Fig. 1). First, centrosomal MT array can be modified by differential regulation of MT dynamics at distinct locations. As a result of site-specific modulation of MT dynamics, MTs can be stabilized at one cell side and/or destroyed at the other side. Second, MTs can be moved either within the array or after detachment from the array. Third, an alternative, non-centrosomal source of MTs can produce an additional, asymmetric MT array. Finally, relative positioning of MT nucleation sites within a cell can also modulate the MT asymmetry. For example, locating the centrosome to one cell side would be sufficient to apply a certain degree of asymmetry to the MT network.
Figure 1. Principles of MT asymmetry.
Radial array (top) can be transformed into asymmetric network by 1) modulations of MT dynamics, 2) re-positioning of existing MTs, 3) directional MT formation at alternative MT nucleation sites (bottom, right) and/or 4) repositioning of MTOCs within a cell.
Regulation of MT asymmetry via modulation of plus end dynamics
Dynamic properties of individual MTs in cells are diverse. While many MTs undergo “dynamic instability”34, characterized by particular frequencies of catastrophes (switch to depolymerization) and rescues (switch to polymerization), certain “stable” MTs remain unchanged for hours35. A majority of MTs exhibit persistent growth through the cytoplasm and undergo diverse dynamic changes when they reach cell periphery.36 In a motile cell, MT dynamic parameters are specific for certain cell regions. For example, a particularly long-lived subset of MTs extends from the cell center toward the leading edge in migrating fibroblasts.37 In addition, the time that otherwise dynamic MT ends spend in pauses as well as MT catastrophe frequency varies significantly in distinct PTK1 epithelial cell domains.38 Dynamic diversity of MTs can be regulated through general stabilization of MT lattice by MAP binding or through alteration of plus end MT dynamics as a function of the cell periphery (for a review see ref.39). The latter is highly relevant for migrating cells where properties of peripheral cytoplasm are distributed asymmetrically. Mechanisms of plus end dynamics modulation have been intensively studied over the past decade.
Long MT lifetimes can be achieved by a few complementary mechanisms. A number of phenomena described in variable fibroblastic and epithelial cells, including MT capture, stimulated persistent growth and/or frequent rescues of MTs close to the cell front, lead to longer MT existence. MT capture at the cortex involves association of MT plus-tips with a number of proteins specifically accumulated at the leading edge protrusions, including APC40, 41 and LL5beta42 and is ruled by Rho family of small GTPases through Rac effector IQGAP43 and Rho effector mDia.40 One such local mechanism involves capture of MT plus ends specifically at lipid rafts at the protruding cell edge.41 MT lifetime is also increased by MT-associated proteins that preserve MT from depolymerization by promoting efficient rescues.44–46 Anchoring of MTs at cortical accumulations of LL5β and ELKS proteins, which together with plus tip proteins CLASPs form cortical clusters in close focal adhesion proximity,42 may explain MT capture at focal adhesion phenomenon observed previously.47
Additionally, persistent MT growth can be stimulated by excluding catastrophe activity from a certain cellular area. Small GTPase Rac1 that is locally active at the protruding edges inhibits MT depolymerization factor stathmin through Rac effector PAK1-driven stathmin phosphorylation.48, 49 At the same time, an increasing gradient of stathmin activity extends backwards and may be at least partially responsible for significant increase in MT catastrophe frequency at the trailing edge as compared to the leading edge.50 However, mechanisms that support catastrophes at the cell back are not completely understood. MT plus ends that interact with adhesion sites in the process of targeting in fish fibroblasts are subjected to local catastrophe-inducing activity of unknown origin, which specifically requires presence of a major adhesion protein paxillin.51 Since adhesions are particularly large at the cell rear, and catastrophes at the back are dependent on Rho activity38 that simulates adhesion growth through a mechanosensory mechanism (for a review see ref.52), we speculate that Rho stimulates catastrophes via enlarging adhesions as sites of catastrophe factor accumulation at the cell rear.
Regulation of MT asymmetry via MT transport
Besides changes in dynamics that influence MT lifetimes, distribution of existing MTs can be changed by moving MTs within a cell. Such movements, driven by molecular motor activity, are often specific for distinct domains a of a motile cell.53,54
In many cases motors move MTs that are still attached with their minus ends to the MTOC. For example, in neutrophils, myosin II-driven MT bending results in complete re-directing of symmetrically nucleated MT array toward the cell rear.28 Similarly, myosin II in neurons and epithelial cells can induce MT bending that results in excluding them from protruding lamella due to actin retrograde flow.54, 55 Myosin contractility also facilitates MT re-organization and bundling behind the leading edge in axonal growth cones.56
Molecular motors can also relocate MTs that are no longer anchored at their nucleation sites. However, MT release from the centrosome in epithelial and fibroblastic cells – a source for both tubulin turnover and free MT fragments36, 57 – has not been shown to exhibit any asymmetry. In neurons, on the contrary, MTs fragments produced by katanin and spastin-dependent severing are transported to distinct cell locations to build up highly asymmetric MT system within axons and dendrites (for reviews see refs.58, 59). Transport of MT fragments can be performed by acto-myosin dependent movements60 or by MT motors (for reviews see refs.30, 59). Strikingly, asymmetric MT transport in neurons can be regulated by kinesin-561 which has been previously well studied as a major force that moves MTs along each other in the mitotic spindle.
Regulation of MT asymmetry via directed non-centrosomal nucleation
The MT pattern depends largely on the process of nucleation at the MTOC. In many motile cells, the centrosome is located in front of the nucleus, and it is thought to be an important stage of polarization of both the MT system and the whole cell.62 Indeed, formation of asymmetric MT arrays in differentiating neurons shows a direct correlation with centrosome positioning.13 It is noteworthy in this regard that MT nucleation can also occur by centrosome-independent means.63 A number of MT-organizing structures have been identified in certain specialized cell types. Among these are the nuclear envelope in myotubes64 and melanosomes in pigment cells.65 A more abundant alternative MT-nucleating site is the Golgi complex as it carries out MT nucleation in retinal pigment epithelial cells66, 67 and in multiple other epithelial cell lines.66
In interphase cells, the Golgi complex is asymmetric. It forms a “ribbon” that consists of membrane cisternae stacks with distinct cis-to-trans polarity.68 MT depolymerization causes disruption of the Golgi ribbon into individual stacks but the polarity within each stack is preserved.69 In the presence of MTs, the Golgi complex accumulates close to the centrosome due to the function of dynein, a minus-end directed MT motor.70, 71 The generally accepted view is that the cis-compartment predominantly faces the centrosome, while the trans-compartment looks toward the cell periphery. Thus, the centrosome, being symmetric, maintains an asymmetric organelle in close proximity. MT array that is formed at the Golgi is also asymmetric: Golgi-derived MTs grow predominantly toward the front of motile cells.66 Thus, the centrosome may influence MT asymmetry indirectly via positioning of the Golgi complex.
Mechanisms and regulation of MT nucleation at the Golgi
Golgi-associated MT nucleation appears to be a significant factor in establishing of MT asymmetry (Fig. 2). It is important to understand molecular machinery that underlies directional mode of MT outgrowth at the Golgi (Fig. 3). MT nucleation at the Golgi continues upon laser ablation of the centrosome indicating that the Golgi acts as a centrosome-independent MTOC.45 However, it requires presence of γ-tubulin, the major component of the MT nucleating γ-tubulin ring complexes (γ-TuRCs).66, 72 Is initial enrichment of γ-TuRC in proximity of CLASP accumulations important for organization of MT arrays? In most cases, levels of γ-tubulin detected at the Golgi membrane do not exceed cytosolic γ-tubulin concentrations. However, γ-tubulin has been found associated with Golgi membranes in vitro72 and in vivo upon overexpression of a potential recruiter GMAP210,73 a cis-Golgi associated protein though it has been a subject of debate.71 Recent evidence suggests that γ-tubulin may be recruited to the Golgi membranes through interaction with AKAP450, a protein involved in MT regulation both at the centrosome and the Golgi.67, 74 Notably, AKAP450 is required for Golgi-derived MT formation and can be found in close association with their minus ends.67. It is possible that AKAP450 stimulates Golgi-derived MT formation by elevating concentration of γ-tubulin at the Golgi membrane.
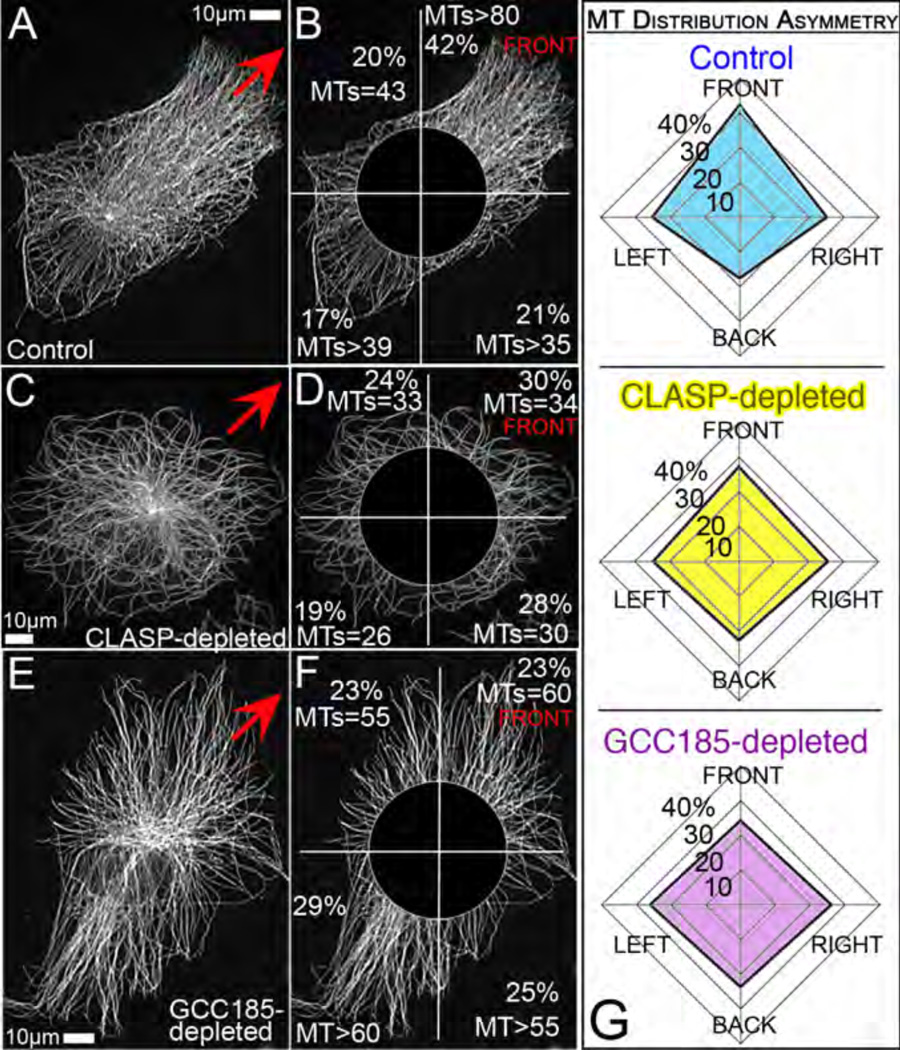
Figure 2. MT asymmetry requires Golgi-derived MTs.
Immunostained MTs in control (A,B), CLASP- (C,D) and GCC185-depleted (E,F) cells. CLASP depleted cells lack non-centrosomal MTs, while in GCC185 non-centrosomal MTs are not associated with the Golgi due to CLASP mis-localization66. Red arrows mark the front lamella direction. B,D,F, For analysis, central area is excluded and the rest of the cell divided in 4 sectors. For each sector, detectable MT numbers and average fluorescent intensity as percent of overall intensity are shown. G, Average percentage of edge MTs intensity distributed in 4 cell sectors. 20 cells for each set were analyzed. Note decreased asymmetry of diagrams for CLASP and GCC185-depleted cells. Standard deviations for front to back intensity proportion (not shown) do not overlap between control and knockdown cell populations.
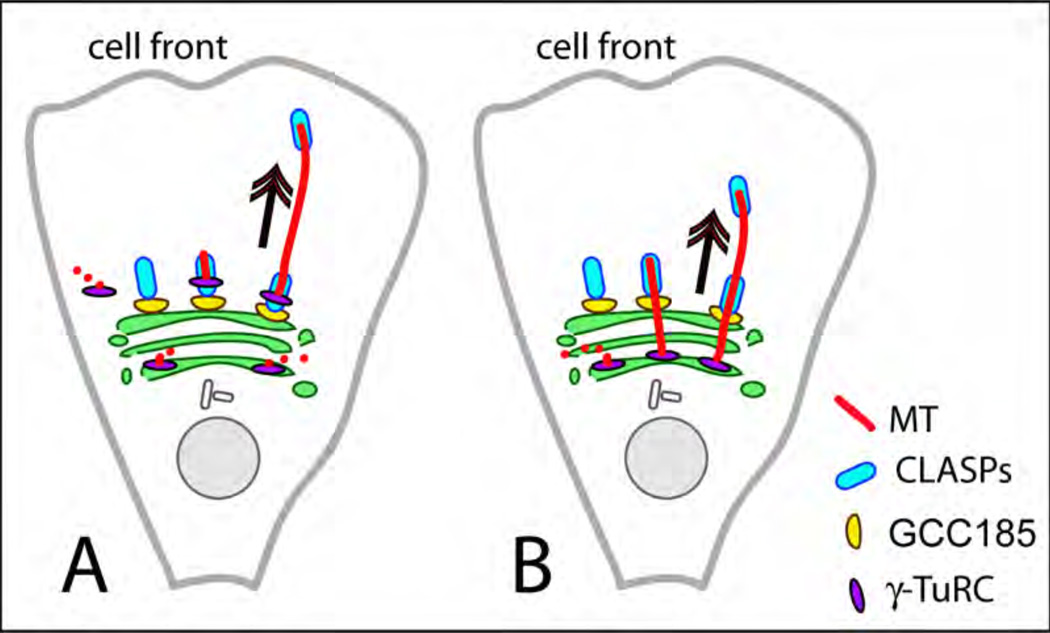
Figure 3. Non-centrosomal asymmetric MT array at the Golgi.
A. After MTs nucleation by γ-TuRC at cis-Golgi or in cytosol, those MT seeds, which redistribute to the TGN continue growing due to CLASP-dependent stabilization and anchoring to the TGN by GCC185. B, After MTs nucleation by γ-TuRC at cis-Golgi, those MTs, which extend to the TGN continue growing due to CLASP-dependent stabilization.
Importantly, nucleation per se appears to be insufficient for MT formation: γ-TuRCs nucleated MT seeds cannot give rise to MTs unless they are associated with Orbit/MAST/CLASP, a well-studied regulator of MT dynamics.45 Depletion or misplacement of this protein from the Golgi membrane leads to elimination of Golgi-derived MT array and impairs MT asymmetry (Fig. 2). In mammalian cells CLASP (Cytoplasmic Linker Associated Protein) is present as two closely related isoforms, CLASP1 and CLASP2. Here, we will refer to both isoforms together as CLASPs. CLASPs are essential regulators of MT dynamics both in mitotic and interphase cells. During mitosis, CLASPs support incorporation of tubulin subunits into kinetochore fibers75, 76 and thus assure correct chromosome segregation. In motile interphase cells, CLASPs laterally anchor MTs at peripheral cortical sites, increasing their stability and growth persistence.42 In both cases, CLASP function is connected with lateral stabilization of MTs that favors polymerization at the plus ends. It is plausible to suggest that CLASP function at the Golgi is accomplished by a similar mechanism. Indeed, CLASPs coat Golgi-associated MTs to trigger their formation.66 Such coating and subsequent stabilization of MT seeds may be regulated by changing CLASP affinity to MTs by phosphorylation.77, 78 Moreover, for MT coating to occur, CLASP molecules undergo fast exchange at the membrane (our unpublished data). Altogether, these data suggest that modulating CLASP association with the Golgi membranes can alter MT-organizing potential of the Golgi. CLASPs are accumulated at the Golgi via TGN (Trans Golgi Network) protein GCC185.66 GCC185, in turn, is recruited to the TGN membranes by cooperative action of two small GTPases, Arl1 and Rab6.79 Thus, being important components of trafficking-organizing signaling (for reviews see refs.80, 81), Arl1 and Rab6 may have an indirect impact on MT organization.
Why is the Golgi-derived MT array asymmetric?
Protein machinery that triggers MT formation at the Golgi can function even in the absence of typical Golgi membrane, for example, upon brefeldin A treatment. Under these conditions MTs form throughout a cell66 and can be often found at ER exit sites.67 Both CLASP coating66 and AKAP450 presence at MT minus ends67 were shown necessary to support non-centrosomal MTs under these conditions. However, mis-placement of MT nucleation from the Golgi (e.g. by GCC185 depletion, Fig. 2) leads to elimination of MT asymmetry in these cells. Thus, specific localization of this MT array to the Golgi is needed for its polarized geometry.
As we discussed above, the Golgi complex is intrinsically asymmetric. Distinct components of MT-initiating machinery are found at distinct compartments within the Golgi. While nucleation has been associated with cis-Golgi markers (e.g. AKAP450), MTs successfully grow from the Golgi membrane only after being stabilized by TGN-associated MT regulators (CLASPs). Thus, plausible scenarios of MT seed location within a polarized Golgi stack include: a) concentration of γ-tubulin at the cis Golgi membrane provides a pool of MT seeds that later redistribute to TGN to serve as MT templates at this location, or b) MT seeds associated with the cis-Golgi give rise to multiple unstable MTs, a certain portion of which reaches out to TGN and is being stabilized by CLASPs (Fig. 3).
In either case, asymmetry of Golgi-derived MTs toward the cell front can be directly connected to the asymmetry of the Golgi ribbon itself because MTs grow from the outer face of the Golgi ribbon66. This model is valid only when the Golgi ribbon is indeed polarized as a whole. However, spatial asymmetry has been strictly shown at a level of a single Golgi stack. On the contrary, the asymmetry of the whole Golgi complex is not a given but requires a certain organization of stacks relative to each other. Our data suggest that both Golgi-derived and centrosomal MTs are necessary for establishment of a polarized Golgi ribbon (Miller et al, in press). A large number of integral Golgi proteins are also required for the proper organization of the Golgi (for a review see ref.81) as they support both cisternal membrane fusion82 and stacking83. Additionally, proteins that regulate centrosome-nucleus attachment84–87 can influence organization and shape of the Golgi complex. In the case of tight centrosome-nucleus attachment, Golgi elements cannot penetrate between the centrosome and nuclear envelope. Such spatial restraint facilitates flat, asymmetric Golgi ribbon shape (our unpublished data).
Alternatively, it is possible that the shape of the Golgi complex is not critical for asymmetry of the MT array. Rather, this asymmetry could depend on differential affinity of CLASP binding to MTs if it is regulated in a polarized fashion across the Golgi complex. CLASP binding to MTS can be diminished by GSK3beta-dependent phosphorylation. This regulation was shown to be spatially specific78 because local inactivation of GSK3beta in protruding lamellae causes extensive CLASP binding to MT lattice. Similar mechanism involving local activation of GSK3beta at the centrosome (evident from GSK3beta localization at the centrosomes88 and phosphorylation of centrosomal proteins89) or at the nuclear envelope90 could restrict CLASP binding to MT seeds at the TGN areas close to the centrosome/nucleus and result in predominantly front-directed MT outgrowth.
Besides a distinct CLASP-dependent formation mechanism, Golgi-derived MTs (and, possibly, also centrosomal MT in the Golgi proximity) possess specific properties that include post-translational modifications and MT-binding proteins. Golgi-associated MTs were described as excessively detyrosinated, acetylated91 and/or polyglutamylated92, properties that can influence MT motors affinity and specificity of these MTs as trafficking routs.22, 92, 93 Certain Golgi-associated proteins may locally modulate MT stability (e.g. Golgi-associated CAP350 that has been implicated in MT stability and anchoring to the centrosome94). If distributed asymmetrically within the Golgi ribbon, such proteins could influence survival of MTs growing in a certain direction.
Role of the organelle positioning and combination of factors
All discussion provided above on the issue of transforming of a radial array into asymmetric one assumed that the centrosome is located in the center of a cell providing a symmetric array. Indeed, in many motile cells the centrosome localized to the cell centroid in dynein-dependent manner.95 However, asymmetry can be introduced by shifting the MTOC (or both MTOCs) to one side of a cell, for example by a large organelle. This can produce a concentrated MT density at the closest cell edge. Such shifting can occur in cells where the nucleus takes a large part of cell volume (e.g. lymphocytes96).
Relative positioning of MTOCs and the nucleus can introduce asymmetry as a result of relocation of the nucleus rearward even while the centrosome stays in the cell centroid.97 In this case, due to the tight attachment of the centrosome to the nucleus, the Golgi ribbon can be shifted in front of the centrosome so the Golgi-derived array becomes front-oriented. According to this model, three factors including 1) maintenance of the centrosome in the cell centroid, 2) rearward nucleus movement and 3) TGN-derived MT array formation are sufficient to provide initial asymmetry to MT network, which can be further strengthened or modified by MT dynamics-driven and MT transport mechanisms.
Assorted combinations of MT network asymmetry principles described here allow highly variable and fine-tunable MT distribution in motile cells. Each of these mechanisms may contribute to microtubule asymmetries in multiple cell types, while some of the mechanisms could be more important in certain cells compared to others. Additionally, some of the mechanisms may be specific to distinct cell types. Here, suggest a generalized systematic approach to MT asymmetry principles which allows to compare their contributions in particular cell types and address how tuning of MT regulations is handled throughout cell differentiation and morphogenesis.
Acknowledgements
Work in the author's laboratory is supported by NIH NIGMS grant 1RO1GM078373-01A2 and pilot grant from NIH NCI SPORE in GI cancer # 5P50 CA095103-07.
Contributor Information
Tatiana Vinogradova, Dept. of Cell and Developmental Biology, Vanderbilt University Medical Center, Nashville, TN.
Paul M. Miller, Dept. of Cell and Developmental Biology, Vanderbilt University Medical Center, Nashville, TN
Irina Kaverina, Dept. of Cell and Developmental Biology, Vanderbilt University Medical Center, Nashville, TN.
References
- 1.Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Small JV, Resch GP. The comings and goings of actin: coupling protrusion and retraction in cell motility. Curr Opin Cell Biol. 2005;17:517–523. doi: 10.1016/j.ceb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Kaverina I, Krylyshkina O, Small JV. Regulation of substrate adhesion dynamics during cell motility. Int J Biochem Cell Biol. 2002;34:746–761. doi: 10.1016/s1357-2725(01)00171-6. [DOI] [PubMed] [Google Scholar]
- 4.Mellor H. Cell motility: Golgi signalling shapes up to ship out. Curr Biol. 2004;14:R434–R435. doi: 10.1016/j.cub.2004.05.038. [DOI] [PubMed] [Google Scholar]
- 5.Vasiliev JM, Gelfand IM, Domnina LV, Ivanova OY, Komm SG, Olshevskaja LV. Effect of colcemid on the locomotory behaviour of fibroblasts. J Embryol Exp Morphol. 1970;24:625–640. [PubMed] [Google Scholar]
- 6.Small JV, Geiger B, Kaverina I, Bershadsky A. How do microtubules guide migrating cells? Nat Rev Mol Cell Biol. 2002;3:957–964. doi: 10.1038/nrm971. [DOI] [PubMed] [Google Scholar]
- 7.Etienne-Manneville S. Actin and microtubules in cell motility: which one is in control? Traffic. 2004;5:470–477. doi: 10.1111/j.1600-0854.2004.00196.x. [DOI] [PubMed] [Google Scholar]
- 8.Kwan KM, Kirschner MW. A microtubule-binding Rho-GEF controls cell morphology during convergent extension of Xenopus laevis. Development. 2005;132:4599–4610. doi: 10.1242/dev.02041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, Wang F, Van Keymeulen A, Rentel M, Bourne HR. Neutrophil microtubules suppress polarity and enhance directional migration. Proc Natl Acad Sci U S A. 2005;102:6884–6889. doi: 10.1073/pnas.0502106102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kriebel PW, Barr VA, Rericha EC, Zhang G, Parent CA. Collective cell migration requires vesicular trafficking for chemoattractant delivery at the trailing edge. J Cell Biol. 2008;183:949–961. doi: 10.1083/jcb.200808105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Condeelis J, Singer RH, Segall JE. The great escape: when cancer cells hijack the genes for chemotaxis and motility. Annu Rev Cell Dev Biol. 2005;21:695–718. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- 12.Oliferenko S, Kaverina I, Small JV, Huber LA. Hyaluronic acid (HA) binding to CD44 activates Rac1 and induces lamellipodia outgrowth. J Cell Biol. 2000;148:1159–1164. doi: 10.1083/jcb.148.6.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Anda FC, Pollarolo G, Da Silva JS, Camoletto PG, Feiguin F, Dotti CG. Centrosome localization determines neuronal polarity. Nature. 2005;436:704–708. doi: 10.1038/nature03811. [DOI] [PubMed] [Google Scholar]
- 14.Waterman-Storer CM, Worthylake RA, Liu BP, Burridge K, Salmon ED. Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nat Cell Biol. 1999;1:45–50. doi: 10.1038/9018. [DOI] [PubMed] [Google Scholar]
- 15.Kaverina I, Krylyshkina O, Small JV. Microtubule targeting of substrate contacts promotes their relaxation and dissociation. J Cell Biol. 1999;146:1033–1044. doi: 10.1083/jcb.146.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol. 2005;7:581–590. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- 17.Oleynikov Y, Singer RH. RNA localization: different zipcodes, same postman? Trends Cell Biol. 1998;8:381–383. doi: 10.1016/s0962-8924(98)01348-8. [DOI] [PubMed] [Google Scholar]
- 18.Krendel M, Zenke FT, Bokoch GM. Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton. Nat Cell Biol. 2002;4:294–301. doi: 10.1038/ncb773. [DOI] [PubMed] [Google Scholar]
- 19.Caswell PT, Norman JC. Integrin trafficking and the control of cell migration. Traffic. 2006;7:14–21. doi: 10.1111/j.1600-0854.2005.00362.x. [DOI] [PubMed] [Google Scholar]
- 20.Nabi IR. The polarization of the motile cell. J Cell Sci. 1999;112(Pt 12):1803–1811. doi: 10.1242/jcs.112.12.1803. [DOI] [PubMed] [Google Scholar]
- 21.Zenke FT, Krendel M, DerMardirossian C, King CC, Bohl BP, Bokoch GM. p21-activated kinase 1 phosphorylates and regulates 14-3-3 binding to GEF-H1, a microtubule-localized Rho exchange factor. J Biol Chem. 2004;279:18392–18400. doi: 10.1074/jbc.M400084200. [DOI] [PubMed] [Google Scholar]
- 22.Dunn S, Morrison EE, Liverpool TB, Molina-Paris C, Cross RA, Alonso MC, et al. Differential trafficking of Kif5c on tyrosinated and detyrosinated microtubules in live cells. J Cell Sci. 2008;121:1085–1095. doi: 10.1242/jcs.026492. [DOI] [PubMed] [Google Scholar]
- 23.Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat Neurosci. 2009;12:559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- 24.Zekert N, Fischer R. The Aspergillus nidulans kinesin-3 UncA motor moves vesicles along a subpopulation of microtubules. Mol Biol Cell. 2009;20:673–684. doi: 10.1091/mbc.E08-07-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akhmanova A, Hoogenraad CC. Microtubule plus-end-tracking proteins: mechanisms and functions. Curr Opin Cell Biol. 2005;17:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Rid R, Schiefermeier N, Grigoriev I, Small JV, Kaverina I. The last but not the least: the origin and significance of trailing adhesions in fibroblastic cells. Cell Motil Cytoskeleton. 2005;61:161–171. doi: 10.1002/cm.20076. [DOI] [PubMed] [Google Scholar]
- 27.Prigozhina NL, Waterman-Storer CM. Protein kinase D-mediated anterograde membrane trafficking is required for fibroblast motility. Curr Biol. 2004;14:88–98. doi: 10.1016/j.cub.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Eddy RJ, Pierini LM, Maxfield FR. Microtubule asymmetry during neutrophil polarization and migration. Mol Biol Cell. 2002;13:4470–4483. doi: 10.1091/mbc.E02-04-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arimura N, Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat Rev Neurosci. 2007;8:194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- 30.Goldstein LS, Yang Z. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu Rev Neurosci. 2000;23:39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- 31.Bergen LG, Kuriyama R, Borisy GG. Polarity of microtubules nucleated by centrosomes and chromosomes of Chinese hamster ovary cells in vitro. J Cell Biol. 1980;84:151–159. doi: 10.1083/jcb.84.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holy TE, Dogterom M, Yurke B, Leibler S. Assembly and positioning of microtubule asters in microfabricated chambers. Proc Natl Acad Sci U S A. 1997;94:6228–6231. doi: 10.1073/pnas.94.12.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salaycik KJ, Fagerstrom CJ, Murthy K, Tulu US, Wadsworth P. Quantification of microtubule nucleation, growth and dynamics in wound-edge cells. J Cell Sci. 2005;118:4113–4122. doi: 10.1242/jcs.02531. [DOI] [PubMed] [Google Scholar]
- 34.Kirschner MW, Mitchison T. Microtubule dynamics. Nature. 1986;324:621. doi: 10.1038/324621a0. [DOI] [PubMed] [Google Scholar]
- 35.Gundersen GG, Bulinski JC. Selective stabilization of microtubules oriented toward the direction of cell migration. Proc Natl Acad Sci U S A. 1988;85:5946–5950. doi: 10.1073/pnas.85.16.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komarova YA, Vorobjev IA, Borisy GG. Life cycle of MTs: persistent growth in the cell interior, asymmetric transition frequencies and effects of the cell boundary. J Cell Sci. 2002;115:3527–3539. doi: 10.1242/jcs.115.17.3527. [DOI] [PubMed] [Google Scholar]
- 37.Gundersen GG, Kalnoski MH, Bulinski JC. Distinct populations of microtubules: tyrosinated and nontyrosinated alpha tubulin are distributed differently in vivo. Cell. 1984;38:779–789. doi: 10.1016/0092-8674(84)90273-3. [DOI] [PubMed] [Google Scholar]
- 38.Wadsworth P. Regional regulation of microtubule dynamics in polarized, motile cells. Cell Motil Cytoskeleton. 1999;42:48–59. doi: 10.1002/(SICI)1097-0169(1999)42:1<48::AID-CM5>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 39.Amos LA, Schlieper D. Microtubules and maps. Adv Protein Chem. 2005;71:257–298. doi: 10.1016/S0065-3233(04)71007-4. [DOI] [PubMed] [Google Scholar]
- 40.Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, et al. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe T, Wang S, Noritake J, Sato K, Fukata M, Takefuji M, et al. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell. 2004;7:871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 42.Lansbergen G, Grigoriev I, Mimori-Kiyosue Y, Ohtsuka T, Higa S, Kitajima I, et al. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev Cell. 2006;11:21–32. doi: 10.1016/j.devcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Fukata M, Watanabe T, Noritake J, Nakagawa M, Yamaga M, Kuroda S, et al. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 2002;109:873–885. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- 44.Komarova YA, Akhmanova AS, Kojima S, Galjart N, Borisy GG. Cytoplasmic linker proteins promote microtubule rescue in vivo. J Cell Biol. 2002;159:589–599. doi: 10.1083/jcb.200208058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morrison EE, Wardleworth BN, Askham JM, Markham AF, Meredith DM. EB1, a protein which interacts with the APC tumour suppressor, is associated with the microtubule cytoskeleton throughout the cell cycle. Oncogene. 1998;17:3471–3477. doi: 10.1038/sj.onc.1202247. [DOI] [PubMed] [Google Scholar]
- 47.Kaverina I, Rottner K, Small JV. Targeting, capture, and stabilization of microtubules at early focal adhesions. J Cell Biol. 1998;142:181–190. doi: 10.1083/jcb.142.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of leading edge microtubule and actin dynamics downstream of Rac1. J Cell Biol. 2003;161:845–851. doi: 10.1083/jcb.200303082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of microtubule destabilizing activity of Op18/stathmin downstream of Rac1. J Biol Chem. 2004;279:6196–6203. doi: 10.1074/jbc.M307261200. [DOI] [PubMed] [Google Scholar]
- 50.Niethammer P, Bastiaens P, Karsenti E. Stathmin-tubulin interaction gradients in motile and mitotic cells. Science. 2004;303:1862–1866. doi: 10.1126/science.1094108. [DOI] [PubMed] [Google Scholar]
- 51.Efimov A, Schiefermeier N, Grigoriev I, Ohi R, Brown MC, Turner CE, et al. Paxillin-dependent stimulation of microtubule catastrophes at focal adhesion sites. J Cell Sci. 2008;121:196–204. doi: 10.1242/jcs.012666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geiger B, Bershadsky A. Exploring the neighborhood: adhesion-coupled cell mechanosensors. Cell. 2002;110:139–142. doi: 10.1016/s0092-8674(02)00831-0. [DOI] [PubMed] [Google Scholar]
- 53.Yvon AM, Wadsworth P. Region-specific microtubule transport in motile cells. J Cell Biol. 2000;151:1003–1012. doi: 10.1083/jcb.151.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waterman-Storer CM, Salmon ED. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J Cell Biol. 1997;139:417–434. doi: 10.1083/jcb.139.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaefer AW, Kabir N, Forscher P. Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J Cell Biol. 2002;158:139–152. doi: 10.1083/jcb.200203038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burnette DT, Ji L, Schaefer AW, Medeiros NA, Danuser G, Forscher P. Myosin II activity facilitates microtubule bundling in the neuronal growth cone neck. Dev Cell. 2008;15:163–169. doi: 10.1016/j.devcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keating TJ, Peloquin JG, Rodionov VI, Momcilovic D, Borisy GG. Microtubule release from the centrosome. Proc Natl Acad Sci U S A. 1997;94:5078–5083. doi: 10.1073/pnas.94.10.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baas PW. The role of motor proteins in establishing the microtubule arrays of axons and dendrites. J Chem Neuroanat. 1998;14:175–180. doi: 10.1016/s0891-0618(98)00012-x. [DOI] [PubMed] [Google Scholar]
- 59.Baas PW, Vidya Nadar C, Myers KA. Axonal transport of microtubules: the long and short of it. Traffic. 2006;7:490–498. doi: 10.1111/j.1600-0854.2006.00392.x. [DOI] [PubMed] [Google Scholar]
- 60.Hasaka TP, Myers KA, Baas PW. Role of actin filaments in the axonal transport of microtubules. J Neurosci. 2004;24:11291–11301. doi: 10.1523/JNEUROSCI.3443-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Myers KA, Baas PW. Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J Cell Biol. 2007;178:1081–1091. doi: 10.1083/jcb.200702074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Etienne-Manneville S, Hall A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 2003;421:753–756. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
- 63.Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- 64.Bugnard E, Zaal KJ, Ralston E. Reorganization of microtubule nucleation during muscle differentiation. Cell Motil Cytoskeleton. 2005;60:1–13. doi: 10.1002/cm.20042. [DOI] [PubMed] [Google Scholar]
- 65.Malikov V, Kashina A, Rodionov V. Cytoplasmic dynein nucleates microtubules to organize them into radial arrays in vivo. Mol Biol Cell. 2004;15:2742–2749. doi: 10.1091/mbc.E03-10-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, et al. Asymmetric CLASP-Dependent Nucleation of Noncentrosomal Microtubules at the trans-Golgi Network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. Embo J. 2009;28:1016–1028. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ladinsky MS, Wu CC, McIntosh S, McIntosh JR, Howell KE. Structure of the Golgi and distribution of reporter molecules at 20 degrees C reveals the complexity of the exit compartments. Mol Biol Cell. 2002;13:2810–2825. doi: 10.1091/mbc.01-12-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vaughan KT. Microtubule plus ends, motors, and traffic of Golgi membranes. Biochim Biophys Acta. 2005;1744:316–324. doi: 10.1016/j.bbamcr.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 71.Barr FA, Egerer J. Golgi positioning: are we looking at the right MAP? J Cell Biol. 2005;168:993–998. doi: 10.1083/jcb.200501088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chabin-Brion K, Marceiller J, Perez F, Settegrana C, Drechou A, Durand G, et al. The Golgi complex is a microtubule-organizing organelle. Mol Biol Cell. 2001;12:2047–2060. doi: 10.1091/mbc.12.7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rios RM, Sanchis A, Tassin AM, Fedriani C, Bornens M. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell. 2004;118:323–335. doi: 10.1016/j.cell.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 74.Larocca MC, Jin M, Goldenring JR. AKAP350 modulates microtubule dynamics. Eur J Cell Biol. 2006;85:611–619. doi: 10.1016/j.ejcb.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 75.Hannak E, Heald R. Xorbit/CLASP links dynamic microtubules to chromosomes in the Xenopus meiotic spindle. J Cell Biol. 2006;172:19–25. doi: 10.1083/jcb.200508180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, et al. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- 78.Wittmann T, Waterman-Storer CM. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J Cell Biol. 2005;169:929–939. doi: 10.1083/jcb.200412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burguete AS, Fenn TD, Brunger AT, Pfeffer SR. Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell. 2008;132:286–298. doi: 10.1016/j.cell.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Munro S. The Arf-like GTPase Arl1 and its role in membrane traffic. Biochem Soc Trans. 2005;33:601–605. doi: 10.1042/BST0330601. [DOI] [PubMed] [Google Scholar]
- 81.Short B, Haas A, Barr FA. Golgins and GTPases, giving identity and structure to the Golgi apparatus. Biochim Biophys Acta. 2005;1744:383–395. doi: 10.1016/j.bbamcr.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 82.Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- 83.Bisel B, Wang Y, Wei JH, Xiang Y, Tang D, Miron-Mendoza M, et al. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J Cell Biol. 2008;182:837–843. doi: 10.1083/jcb.200805045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Collin L, Schlessinger K, Hall A. APC nuclear membrane association and microtubule polarity. Biol Cell. 2008;100:243–252. doi: 10.1042/BC20070123. [DOI] [PubMed] [Google Scholar]
- 85.Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, et al. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 86.Robinson JT, Wojcik EJ, Sanders MA, McGrail M, Hays TS. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J Cell Biol. 1999;146:597–608. doi: 10.1083/jcb.146.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bobinnec Y, Morin X, Debec A. Shaggy/GSK-3beta kinase localizes to the centrosome and to specialized cytoskeletal structures in Drosophila. Cell Motil Cytoskeleton. 2006;63:313–320. doi: 10.1002/cm.20128. [DOI] [PubMed] [Google Scholar]
- 89.Howng SL, Hsu HC, Cheng TS, Lee YL, Chang LK, Lu PJ, et al. A novel ninein-interaction protein, CGI-99, blocks ninein phosphorylation by GSK3beta and is highly expressed in brain tumors. FEBS Lett. 2004;566:162–168. doi: 10.1016/j.febslet.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 90.Miller MW, Caracciolo MR, Berlin WK, Hanover JA. Phosphorylation and glycosylation of nucleoporins. Arch Biochem Biophys. 1999;367:51–60. doi: 10.1006/abbi.1999.1237. [DOI] [PubMed] [Google Scholar]
- 91.Thyberg J, Moskalewski S. Relationship between the Golgi complex and microtubules enriched in detyrosinated or acetylated alpha-tubulin: studies on cells recovering from nocodazole and cells in the terminal phase of cytokinesis. Cell Tissue Res. 1993;273:457–466. doi: 10.1007/BF00333700. [DOI] [PubMed] [Google Scholar]
- 92.Spiliotis ET, Hunt SJ, Hu Q, Kinoshita M, Nelson WJ. Epithelial polarity requires septin coupling of vesicle transport to polyglutamylated microtubules. J Cell Biol. 2008;180:295–303. doi: 10.1083/jcb.200710039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, et al. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 94.Hoppeler-Lebel A, Celati C, Bellett G, Mogensen MM, Klein-Hitpass L, Bornens M, et al. Centrosomal CAP350 protein stabilises microtubules associated with the Golgi complex. J Cell Sci. 2007;120:3299–3308. doi: 10.1242/jcs.013102. [DOI] [PubMed] [Google Scholar]
- 95.Burakov A, Nadezhdina E, Slepchenko B, Rodionov V. Centrosome positioning in interphase cells. J Cell Biol. 2003;162:963–969. doi: 10.1083/jcb.200305082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hulspas R, Houtsmuller AB, Bauman JG, Nanninga N. The centrosome moves out of a nuclear indentation in human lymphocytes upon activation. Exp Cell Res. 1994;215:28–32. doi: 10.1006/excr.1994.1310. [DOI] [PubMed] [Google Scholar]
- 97.Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]