Abstract
A bacterial strain, designated BzDS03 was isolated from water sample, collected from Dal Lake Srinagar. The strain was characterized by using 16S ribosomal RNA gene and 16S-23S rRNA internal transcribed spacer region sequences. Phylogenetic analysis showed that 16S rRNA sequence of the isolate formed a monophyletic clade with genera Escherichia. The closest phylogenetic relative was Escherichia coli with 99% 16S rRNA gene sequence similarity. The result of Ribosomal database project's classifier tool revealed that the strain BzDS03 belongs to genera Escherichia.16S rRNA sequence of isolate was deposited in GenBank with accession number FJ961336. Further analysis of 16S-23S rRNA sequence of isolate confirms that the identified strain BzDS03 be assigned as the type strain of Escherichia coli with 98% 16S-23S rRNA sequence similarity. The GenBank accession number allotted for 16S-23S rRNA intergenic spacer sequence of isolate is FJ961337.
Keywords: 16S ribosomal RNA gene, 16S-23S intergenic spacer region, Escherichia coli, Phylogenetic analysis, DNA isolation, DNA sequencing
Background
Access to clean drinking water is key factor underpinning public healths even in areas of the world where significant investments are made to protect water quality [1]. We initiated a systematic screening programme to catalogue the microbial composition of Dal Lake water at the city of heaven, Srinagar India. In the present research communication, we report the genotypic characterization of strain BzDS03 on the basis of 16S rRNA gene sequence and 16S-23S rRNA internal transcribed spacer region analysis. This kind of genotypic technique involves the amplification of any phylogenetically informative targets such as 16S rRNA gene for genera identification [2] and 16S-23S rRNA for species identification [3]. The sequences of the rRNA is essential for the survival of all cells and are highly conserved throughout evolution, because they require complex inter and intramolecular interactions to maintain the protein-synthesizing machinery [4]. A length of 1,184 nt, 16S rRNA sequence of strain BzDS03 was used as query to search for homologous sequence in the GenBank database [5]. Sequence analysis revealed that its closest relative was Escherichia coli DH1 (ME8569) with 99% sequence identity. It is known that most strains of Escherichia coli are harmless and live in the intestines of healthy humans and animals; however some strain produces a powerful toxin and can cause severe illness. One of the hundreds of strains of the bacterium Escherichia coli, O157:H7 is an emerging cause of food-borne and waterborne illness. The presence of Escherichia coli in water sample is implicit evidence for fecal contamination and represents a threat to human and environmental health [6]. Therefore, it is of interest to isolate novel bacterial strain from different environments for potential applications. In this study we describe the isolation and identification of an unknown bacterium from the Dal Lake using 16S rRNA gene sequence and 16S-23S rRNA internal transcribed spacer (ITS) region sequence to characterize the strain BzDS03 as a member of Escherichia coli.
Methodology
Culturing of Bacteria
Water samples were collected in sterile bottles from different location i.e. Nishat Park, Dal Gate, hazrat-Bal of Dal lake, Srinagar. The sample was serially diluted and spread onto peptone/Beef extract/Nacl/Agar-Agar plates followed by incubation at 30°C under anaerobic conditions. Single colonies of bacterial strains were picked and further grown and sub-cultured several times to obtain a pure culture.
DNA isolation of bacteria
Pure culture of the target bacteria was grown overnight in liquid NB medium for the isolation of genomic DNA using a method described by Hiney and colleagues [7].
PCR amplification of 16S rDNA gene
PCR reaction was performed in a gradient thermal cycler (Eppendorf, Germany). The universal primers (Forward primer 5'- AGAGTTTGATCCTGGCTCAG -3' and reverse primer 5'- CTTGTGCGGGCCCCCGTCAATTC-3') were used for the amplification of the 16S rDNA gene fragment. The reaction mixture of 50 μl consisted of 10 ng of genomic DNA, 2.5 U of Taq DNA polymerase, 5 μl of 10X PCR amplification buffer (100 mM Tris-HCl, , 500 mMKCl pH-8.3), 200μM dNTP, 10 p moles each of the two universal primers and 1.5mM MgCl2. Amplification was done by initial denaturation at 94°C for 3 minutes, followed by 30 cycles of denaturation at 94°C for 30 second, annealing temperature of primers was 55°C for 30 second and extension at 72°C for 1 minute. The final extension was conducted at 72°C for 10 minutes.
PCR amplification of 16S-23S rDNA intergenic spacer
PCR reaction was performed in a gradient thermal cycler (Eppendorf, Germany). The forward primers 5' GATTAGATACCCTGGTAG 3' and reverse primer 5' AGTCACTTAACCATACAACCC 3' were used to amplify 16S-23S rDNA internal transcribed spacer region sequence. The reaction mixture and reaction steps for PCR were same as for 16S rDNA amplification.
Agarose gel electrophoresis and Purification of PCR product
10 μl of the reaction mixture was then analyzed by submarine gel electrophoresis using 1.0 % agarose with ethidium bromide at 8V/cm and the reaction product was visualized under Gel doc/UV trans-illuminator. The PCR product was purified by Qiagen gel extraction kit.
DNA sequencing of 16S rDNA fragment
The 16S rDNA amplified PCR product (100ng concentration) was used for the sequencing with the single 16S rDNA 27F Forward primer: 5'- AGAGTTTGATCCTGGCTCAG-3' by ABI DNA sequencer (Applied Biosystem Inc).
DNA sequencing of 16S-23 rDNA ITS fragment
PCR amplified product of 16S-23S intergenic pacer (100 ng concentration) was used for the sequencing of 16S-23S ITS region by using 5' GATTAGATACCCTGGTAG 3' as forward primers and 5' AGTCACTTAACCATACAACCC 3' as reverse primer (Applied Biosystem Inc).
Computational analysis
A comparison of the 16s rRNA gene sequence and 16S-23S ITS region sequence of the isolate with the non-redundant collection (Genbank, DDBJ, EMBL & PDB) of sequences was performed using BLAST [8, 9]. A number of sequences of the genus Escherichia coli were aligned with both 16S rRNA gene sequence and 16S-23S ITS region sequence of isolate with 98% sequence similarity. We then developed a multiple sequence alignment (separately for both 16S and 16S-23S ITS sequences) for these homologous sequences using the algorithm described in ClustalW [10]. An evolutionary distance matrix was generated from these nucleotide sequences in the dataset. Further a phylogenetic tree was then drawn using the Neighbour joining method [11] for both 16S rDNA and 16S-23S ITS region. Phylogenetic and molecular evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics analysis) version 4.0 [12]. We compared the 16S rRNA gene sequence of isolate with different set of sequence databases such as small subunit ribosomal RNA (ssu rRNA) and large subunit ribosomal RNA (lsu rRNA) [13] by using Ribosomal RNA BLAST [14]. 16S rRNA gene sequence of isolate was also compared against those sequences in Ribosomal Database Project (RDP) [15] by using the RDP Classifier check Program [16]. The annotated information for the sequence in the database to which 16S rRNA aligns was used for the bacterial identification.
Discussion
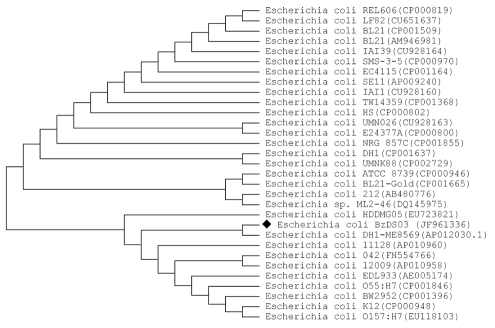
Genomic DNA was extracted from the isolated bacterial strain BzDS03. 16S rDNA and 16S-23S rDNA ITS regions were amplified and sequenced by using their respective primers. A total of 1,183 bp of the 16S rDNA gene and 346 bp of 16S-23S ITS region were sequenced and used for the identification of isolated bacterial strain. Comparison of these test sequences against nonredundant collection of GenBank [05] database was performed with BLAST [8, 9]. The BLAST result showed that the 16S rDNA sequence of isolate BzDS03 has 99% sequence similarity having highest score i.e. 2137 bits with Escherichia coli DH1 (ME8569). The result also showed that isolate BzDS03 has 99% sequence similarity with Escherichia coli DH1, Escherichia coli BW2952, Escherichia coli str. K12 substr. DH10B, Escherichia coli str. K12 substr. W3110 and Escherichia coli str. K-12 substr. MG1655 respectively. The BLAST result of 16S-23S rDNA ITS sequence showed that the isolate has 98% sequence similarity (score=599, Expected=4e-168) with the same strain Escherichia coli DH1 (ME8569). Subsequently, a 16S rDNA gene sequence and 16S-23S rDNA ITS sequence based phylogenetic tree were constructed, which shows the relationship between the isolate BzDS03 and selected representative of genera Escherichia coli (Figure 1& Figure 2). It is evident from phylogenetic analysis of 16S rDNA and 16S-23S rDNA ITS sequences that the isolate BzDS03 represents a strain in genera Escherichia coli. Further the result of similarity rank program classifier [16] available at Ribosomal Database Project [15], for 16S rDNA sequence classified the isolate BzDS03 as a novel genomic species of the genus Escherichia coli with a confidence threshold of 95%. All these results confirm that the isolate BzDS03 belongs to genera Escherichia coli.
Figure 1.
Neighbor-joining tree of selected 16S rDNA gene sequence of the genus Escherichia coli obtained from BLAST search of the strain BzDS03 sequence for phylogenetic inference.
Figure 2.
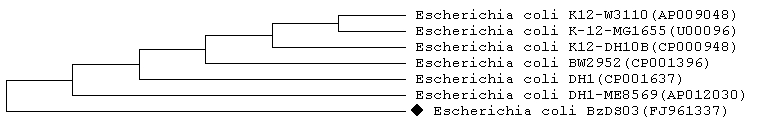
Neighbor-joining tree of selected 16S-23S rDNA ITS region sequence of the genus Escherichia coli obtained from BLAST search of the strain BzDS03 sequence for phylogenetic inference.
Conclusion
Accurate and definitive microorganism identification is essential for a wide variety of application including environmental studies. The rRNA sequence based analysis is a central method to understand not only the microbial diversity within and across the group but also to identify new strains. Bacterial species have at least one copy of the 16S rRNA gene containing highly conserved regions together with hyper variable regions, which is used for identification of new strains. However a considerable variation can occur between species in both the length and the sequence of 16S-23S rDNA ITS region therefore this region is useful in characterization of bacterial species. We used the 16S rRNA gene sequence and 16S-23S rRNA ITS region sequence to characterize the bacterial isolate BzDS03 from Dal Lake, Srinagar India as Escherichia coli strain BzDS03.
Acknowledgments
The Authors are grateful to Mr. Rajeev Srivastava (M.D. Biobrainz, Lucknow, U.P., India) for providing necessary facilities and encouragement.
Footnotes
Citation:Magray et al, Bioinformation 6(10): 370-371 (2011)
References
- 1.L Fewtrell, J Bartram. World Health Organization. London, United Kingdom: IWA Publishing; 2001. [Google Scholar]
- 2.JE Clarridge, et al. 3rd. Clin Microbiol Rev. 2004;17:840. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.V Gürtler, VA Stanisich. Microbiology. 1996;145:2. [Google Scholar]
- 4.CT Sacchi, et al. J Clin Microbiol. 2002;40:4520. doi: 10.1128/JCM.40.12.4520-4527.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. http://www.ncbi.nlm.nih.gov/genbank/
- 6.Z Lu, et al. Appl Environ Microbiol. 2005;71:5992. doi: 10.1128/AEM.71.10.5992-5998.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M Hiney, et al. Appl Environ Microbiol. 1992;58:1039. doi: 10.1128/aem.58.3.1039-1042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. http://blast.ncbi.nlm.nih.gov/Blast.cgi.
- 9.Z Zhang, et al. J Comput Biol. 2000;7:203. [Google Scholar]
- 10.JD Thompson, et al. Nucleic Acids Res. 1994;22:4673. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.N Saitou, M Nei. Mol Biol Evol. 1987;4:406. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 12.K Tamura, et al. Mol Biol Evol. 2007;24:1596. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 13.J Wuyts, et al. Nucleic Acids Res. 2004;32:D101. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. http://bioinformatics.psb.ugent.be/webtools/rRNA/blastrrna.html.
- 15. http://rdp.cme.msu.edu/
- 16.Q Wang, et al. Appl Environ Microbiol. 2007;73:5261. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]