ABSTRACT
Detection of microbial antigens in clinical samples can lead to rapid diagnosis of an infection and administration of appropriate therapeutics. A major barrier in diagnostics development is determining which of the potentially hundreds or thousands of antigens produced by a microbe are actually present in patient samples in detectable amounts against a background of innumerable host proteins. In this report, we describe a strategy, termed in vivo microbial antigen discovery (InMAD), that we used to identify circulating bacterial antigens. This technique starts with “InMAD serum,” which is filtered serum that has been harvested from BALB/c mice infected with a bacterial pathogen. The InMAD serum, which is free of whole bacterial cells, is used to immunize syngeneic BALB/c mice. The resulting “InMAD immune serum” contains antibodies specific for the soluble microbial antigens present in sera from the infected mice. The InMAD immune serum is then used to probe blots of bacterial lysates or bacterial proteome arrays. Bacterial antigens that are reactive with the InMAD immune serum are precisely the antigens to target in an antigen immunoassay. By employing InMAD, we identified multiple circulating antigens that are secreted or shed during infection using Burkholderia pseudomallei and Francisella tularensis as model organisms. Potential diagnostic targets identified by the InMAD approach included bacterial proteins, capsular polysaccharide, and lipopolysaccharide. The InMAD technique makes no assumptions other than immunogenicity and has the potential to be a broad discovery platform to identify diagnostic targets from microbial pathogens.
IMPORTANCE
Effective treatment of microbial infection is critically dependent on early diagnosis and identification of the etiological agent. One means for rapid diagnosis is immunoassay for antigens that are shed into body fluids during infection. Immunoassays can be inexpensive, rapid, and adaptable to a point-of-care format. A major impediment to immunoassay for diagnosis of infectious disease is identification of appropriate antigen targets. This report describes a strategy that can be used for identification of microbial antigens that are shed into serum during infection by the biothreats Burkholderia pseudomallei and Francisella tularensis. Termed InMAD (in vivo microbial antigen discovery), the strategy has the potential for application to a broad spectrum of microbial pathogens.
Introduction
Immunoassay for detection of microbial antigens is a well-established tool for diagnosis of a wide variety of infectious diseases. Rapid diagnosis can lead to selection of an appropriate antimicrobial to treat the infection at an early stage when antibiotics are most effective. Early diagnosis and targeted use of antibiotics may also reduce the development of antibiotic resistance.
A key first step in the development of a diagnostic test that targets antigen is identification of one or more microbial products that are shed into body fluids during infection. In some cases, there are obvious choices, e.g., capsular polysaccharides (CPSs). However, for most microbes, the choice is not obvious. For example, which of the potentially hundreds or thousands of proteins or polysaccharides that are produced by a bacterium or fungus are actually shed into serum or urine in amounts sufficient for detection by immunoassay?
The goal of this study was to design a strategy for identification of potential antigen targets for immunodiagnosis of bacterial infection. There are three requirements for an ideal target discovery strategy. First, the technology should make no assumptions regarding target structure, synthesis, or secretion. Second, the approach must identify only those candidate antigens that are actually present in the body fluid that might be a diagnostic specimen. Third, targets that are identified must be immunogenic to allow for production of the antibodies needed for immunoassay construction.
For this study, we examined the biothreats Burkholderia pseudomallei and Francisella tularensis, the causative agents of melioidosis and tularemia, respectively. B. pseudomallei and F. tularensis are facultative intracellular pathogens that can replicate inside several types of cells (1, 2). Both present diagnostic challenges. Culture of B. pseudomallei is best accomplished by laboratories in areas of disease endemicity with experience in isolating the pathogen. Levels of bacteremia may be very low (~1 CFU/ml blood) (3). Culture and identification of F. tularensis are difficult. F. tularensis is fastidious; isolation is best done using specialized media, and the bacterium grows slowly (4). In both cases, empirical treatment of an infection that often has nonspecific symptoms with antibiotics may yield subsequent clinical samples with negative cultures. Culture of both of these pathogens presents potential hazards to laboratory personnel. Finally, rapid diagnosis of both B. pseudomallei and F. tularensis infection, perhaps in a point-of-care setting, has become an important goal due to the potential use of either bacterium as an agent of bioterrorism (5).
Our results showed that a novel target identification strategy that we have termed in vivo microbial antigen discovery (InMAD) is a means for identification of immunogenic bacterial antigens that are shed into body fluids during infection. The B. pseudomallei capsular polysaccharide (CPS) is one of several antigens identified by InMAD and serves as a proof of principle. Following production of a CPS-specific monoclonal antibody (MAb), this antigen was identified in urine and serum samples from melioidosis patients.
RESULTS
Identification of B. pseudomallei antigens that are secreted during infection.
B. pseudomallei was used as a model pathogen to assess the ability of the InMAD technique to identify bacterial antigens that are circulating in serum during infection (Fig. 1). To begin, BALB/c mice were infected with B. pseudomallei (strain 1026b) by the intranasal route. When the mice became moribund, blood was collected and serum was isolated. The serum was filtered to remove whole bacterial cells, leaving secreted/shed antigens behind. This “InMAD serum,” either undiluted or diluted with phosphate-buffered saline (PBS), was combined with TiterMax adjuvant and used to immunize syngeneic BALB/c mice. “InMAD immune serum” was then collected at various times postimmunization. By varying the immunization dose and time for collection of serum, we were able to determine the optimal amount of InMAD serum for immunization and the time after immunization that yielded the most immunoreactive serum. InMAD immune sera were screened for reactivity by probing Western blots of whole-cell lysates of B. pseudomallei (strain 1026b). InMAD serum diluted 1:2 in adjuvant failed to produce a stable emulsion. However, InMAD serum diluted 1:2 in PBS, followed by the addition of an equal volume of adjuvant, produced a stable emulsion and produced a strong antibody response. We also determined that 8 weeks postimmunization was the optimal time to harvest InMAD immune serum (data not shown).
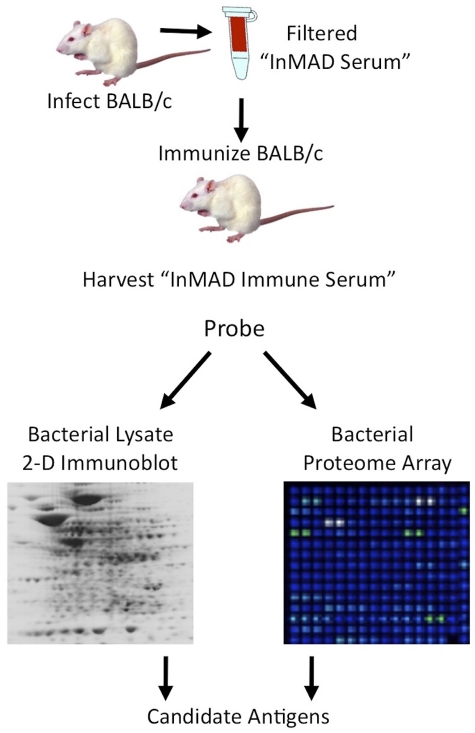
FIG 1 .
InMAD strategy for identifying secreted/shed bacterial antigens. BALB/c mice were infected with a pathogen, and InMAD sera were harvested when mice become moribund. Filtering the InMAD sera removed whole bacteria, and secreted/shed bacterial antigens in the sample passed through the filter. Syngeneic mice were then immunized with the InMAD sera combined with adjuvant. InMAD immune sera were harvested from the immunized mice and used to probe bacterial cell lysates by Western blotting or a bacterial proteome array.
Western blots of whole-cell lysates of B. pseudomallei were probed with InMAD immune serum in an effort to identify circulating B. pseudomallei antigens (Fig. 2). Sera obtained from five representative mice are shown; in every case, the preimmune serum (Fig. 2A, left lane of each mouse sample) was unreactive with the B. pseudomallei lysate. The InMAD immune serum was reactive with multiple bacterial antigens in the immunoblots (Fig. 2A, right lane of each mouse sample). There was considerable mouse-to-mouse variability in production of antibodies reactive with different bacterial antigens.
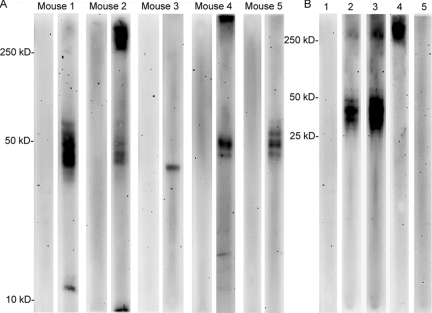
FIG 2 .
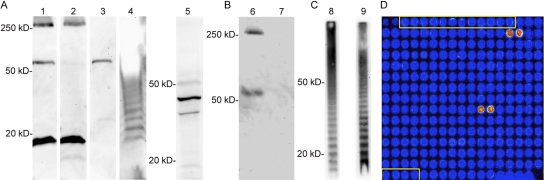
B. pseudomallei proteins and the capsular polysaccharide are reactive with InMAD immune serum. (A) Five representative InMAD immune serum samples (right lane for each mouse) and preimmune serum samples (left lane for each mouse) were used to probe a 1-D immunoblot of a B. pseudomallei whole-cell lysate. (B) Serum from a representative mouse was used to determine if reactive antigens were polysaccharides or proteins. Preimmune serum (lane 1) is not reactive with a 1-D blot of a B. pseudomallei whole-cell lysate. InMAD immune serum from the same mouse is reactive with multiple antigens (lane 2; lane 3 is a longer exposure). Only the high-molecular-weight antigen is reactive when InMAD immune serum is used to probe a blot of a proteinase K-treated lysate (lane 4). The same InMAD immune serum is not reactive with a proteinase K-treated lysate from B. pseudomallei CPS mutant strain SR1015 (lane 5). Antibody detection was done with a goat anti-mouse IgG-HRPO second antibody.
Proteinase K treatment of the cell lysate was used to determine if the InMAD immune serum was reactive with polysaccharides. InMAD immune serum from a representative mouse is shown. This serum was reactive with both high-molecular-weight antigens and multiple antigens between 50 and 25 kDa (Fig. 2B, lanes 2 and 3). Upon treatment of the B. pseudomallei lysate with proteinase K, only the high-molecular-weight antigen remained reactive (Fig. 2B, lane 4). Similar results were found with sera from other immunized mice (data not shown).
The B. pseudomallei capsular polysaccharide (CPS) is a possible candidate for the proteinase K-resistant, high-molecular-weight antigen recognized by the InMAD immune serum. To further characterize this high-molecular-weight antigen, a blot prepared from B. pseudomallei CPS mutant SR1015 (6) was probed with InMAD immune serum. The InMAD immune serum was not reactive with a proteinase K-treated cell lysate prepared from the CPS mutant, suggesting that CPS is the high-molecular-weight antigen recognized by the InMAD serum (Fig. 2B, lane 5). In addition, five other InMAD immune sera that were reactive with the proteinase K-insensitive, high-molecular-weight antigen similarly were not reactive with the proteinase K-treated CPS mutant lysate (data not shown).
Two-dimensional immunoblotting followed by analysis of spots by mass spectrometry was performed to identify the B. pseudomallei proteins that are recognized by InMAD immune sera. Two separate experiments were performed; in each case, a B. pseudomallei lysate was probed with InMAD immune sera pooled from several mice. The preimmune samples were uniformly unreactive by Western blotting (examples shown in Fig. 2A). Table S1 in the supplemental material summarizes the mass spectrometry data; the proteins listed were clearly reactive by two-dimensional (2-D) immunoblotting. Proteins listed in Table S1 include chaparone GroEL (BPSS0477), F0F1 ATP synthase alpha (BPSL3398), and elongation factor Tu (BPSL3228).
Confirmation of a candidate diagnostic target in melioidosis clinical samples.
The InMAD procedure suggested that CPS is a circulating B. pseudomallei antigen; therefore, experiments were done to produce a CPS MAb. Heat-killed B. pseudomallei strain 1026b was used to immunize BALB/c mice as previously described (7). At least 10 hybridoma clones produced antibodies that were reactive in Western blots with B. pseudomallei whole-cell lysates. Two clones produced antibodies that were reactive with high-molecular-weight, proteinase K-resistant antigens. One clone, 3C5, was chosen for further study on the basis of a strong signal by Western blotting and enzyme-linked immunosorbent assay (ELISA).
MAb 3C5 was reactive with the high-molecular-weight, proteinase K-insensitive antigen identified via InMAD (Fig. 3A, lanes Bp and Bp-PK). This MAb was not reactive with a lysate prepared from the CPS mutant used in Fig. 2; however, it was reactive with the B. pseudomallei parent strain from which the mutant was generated (Fig. 3A, lanes Cap− and Cap+). MAb 3C5 was also reactive with purified CPS (Fig. 3A, lane CPS), confirming that the proteinase K-resistant antigen identified via InMAD was, in fact, CPS. MAb 3C5 was not reactive with lysates of Burkholderia thailandensis but was reactive with Burkholderia mallei lysate, although at a slightly lower molecular weight than the CPS of B. pseudomallei (Fig. 3A, lanes Bt and Bm).
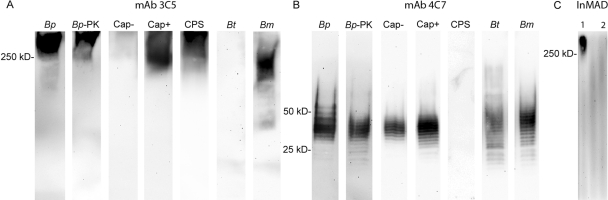
FIG 3 .
Characterization of CPS- and LPS-specific MAbs. Both CPS MAb 3C5 (A) and LPS MAb 4C7 (B) are reactive with B. pseudomallei (lane Bp)- and proteinase K-treated B. pseudomallei (lane Bp-PK)-treated whole-cell lysates. MAb 3C5 is not reactive with a capsule mutant strain SR1015 (lane Cap−) but is reactive with the parental strain from which the mutant was derived (1026b; lane Cap+). MAb 3C5 is also reactive with purified CPS (lane CPS). MAb 4C7 is reactive with the CPS mutant strain, and the parental strain and is not reactive with purified CPS. In addition, MAb 3C5 is not reactive with cell lysates of B. thailandensis (lane Bt), whereas MAb 4C7 is reactive. Both MAbs are reactive with B. mallei (lane Bm). The MAbs were also used to probe filtered InMAD serum (C) harvested from a mouse model of melioidosis; CPS MAb 3C5 was reactive (InMAD lane 1), whereas the LPS MAb 4C7 was unreactive (InMAD lane 2). Lanes 1 and 2 were done with a chamber miniblotter in order to conserve InMAD serum.
During the process of isolating a CPS MAb, we also produced a lipopolysaccharide (LPS)-specific MAb. LPS is an immunogenic antigen that is surface exposed; however, it was not identified by the InMAD process (Fig. 2B). We were therefore interested in determining if the negative LPS InMAD results from the discovery phase would correlate with an inability of an LPS MAb to detect LPS in InMAD sera and filtered samples from melioidosis patients. If so, the InMAD technique may also be useful in ruling out an antigen that one would predict to be shed or secreted. The results (Fig. 3) found the LPS MAb 4C7 to be reactive with proteinase K-insensitive cell lysate antigens that formed a ladder pattern from 20 to 50 kDa (Fig. 3B, lanes Bp and Bp-PK). MAb 4C7 was reactive with a lysate of the CPS mutant (Fig. 3B, lane Cap−) as well as lysates of B. thailandensis and B. mallei (Fig. 3B, lanes Bt and Bm).
Both MAbs 3C5 and 4C7 were used to probe InMAD sera collected from the infected BALB/c mice. As predicted by the InMAD experiments (Fig. 2), the CPS MAb 3C5 was reactive with the InMAD sera, and the LPS MAb 4C7 was not (Fig. 3C, lanes 1 and 2, respectively).
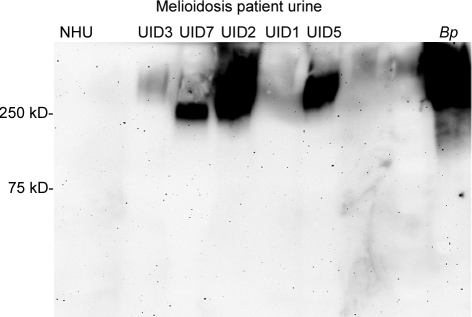
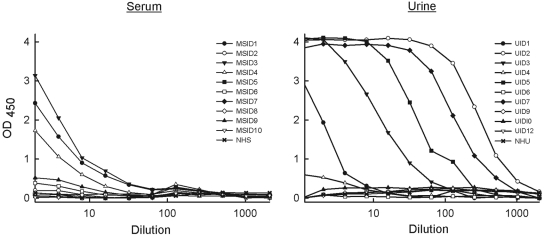
Western blots were prepared from serum (n = 10) or urine (n = 10) samples from 20 melioidosis patients and probed for CPS and LPS with MAbs 3C5 and 4C7, respectively. The serum samples were taken on admission from 10 patients who were blood culture positive for B. pseudomallei; quantitative counts in blood were not determined. The urine samples were taken on admission from 10 patients whose urine cultures were positive for B. pseudomallei. All samples were filter sterilized before analysis. MAb 4C7 was not able to detect LPS in either serum or urine samples by Western blotting (data not shown), a result that is consistent with the absence of reactivity of MAb 4C7 with InMAD sera from infected mice (Fig. 3). The CPS MAb 3C5 was not reactive when used to probe Western blots of patient sera. However, MAb 3C5 was reactive by Western blotting for CPS in urine samples containing greater than 100,000 CFU/ml (prior to filtration) (Fig. 4). Weaker reactivity was seen in a urine sample that contained an estimated 75,000 CFU/ml (Fig. 4, sample UID3). Six additional urine samples that contained less than 75,000 CFU/ml (including sample UID1) (Fig. 4) were negative by Western blotting (data not shown).
FIG 4 .
Detection of CPS in melioidosis patient urine by Western blotting. MAb 3C5 was used to probe 10 urine samples from melioidosis patients (lanes UID1, -2, -3, -5, and -7) and normal human urine (NHU). Samples not shown were negative by Western blotting. Proteinase K-treated B. pseudomallei cell lysate (lane Bp) was used as a positive control. A summary of the Western blot data is listed in Table 1.
MAbs 3C5 and 4C7 were used to develop antigen capture ELISAs for detection of CPS and LPS antigens in melioidosis patient samples. Unlabeled MAbs were used in the capture (solid) phase of an ELISA plate, and horseradish peroxidase (HRPO)-labeled MAbs were used as indicators. MAb 4C7 was not able to detect LPS in patient serum or urine samples by capture ELISA (data not shown). However, CPS MAb 3C5 was reactive with serum and urine samples obtained from melioidosis patients by ELISA (Fig. 5). Five of the ten serum samples tested were positive (Fig. 5, left). In addition, MAb 3C5 was highly reactive by ELISA with CPS in six of ten urine samples (Fig. 5, right). Titers were recorded as the highest dilutions that produced positive signals (optical density at 450 nm [OD450] ≥ 0.5) and are listed in Table 1.
FIG 5 .
Detection of CPS in melioidosis patient serum and urine samples by antigen capture ELISA. For each ELISA, MAb 3C5 was used in the capture phase, followed by addition of serial diluted patient serum (1:2 starting dilution) and urine (no dilution to start) samples. HRPO-labeled MAb 3C5 was then used in the indicator phase to detect captured CPS. The titer of each of the clinical samples is listed in Table 1.
TABLE 1 .
Analysis of serum and urine samples from melioidosis patients for CPS by Western blotting and antigen capture ELISA
| Patient sample | CFUa | Western blotb | ELISA titerc |
|---|---|---|---|
| Urined | |||
| UID1 | 2.25 × 104 | − | 8 |
| UID2 | >1 × 105 | + | 1,300 |
| UID3 | 7.5 × 104 | + | 59 |
| UID4 | 1.2 × 104 | − | 2.8 |
| UID5 | >1 × 105 | + | 200 |
| UID6 | 3.5 × 103 | − | 0 |
| UID7 | >1 × 105 | + | 550 |
| UID9 | <1 × 103 | − | 0 |
| UID10 | 1 × 103 | − | 0 |
| UID12 | 1 × 103 | − | 0 |
| Serumd | |||
| MSID1 | + | − | 16 |
| MSID2 | + | − | 0 |
| MSID3 | + | − | 20 |
| MSID4 | + | − | 9.8 |
| MSID5 | + | − | 0 |
| MSID6 | + | − | 2.5 |
| MSID7 | + | − | 0 |
| MSID8 | + | − | 0 |
| MSID9 | + | − | 4.6 |
| MSID10 | + | − | 0 |
Quantitative cultures were done with urine samples, and results are reported as the number of CFU/ml. Blood cultures (serum) are reported only as positive or negative. All samples were filtered before analysis for antigen.
Western blotting was prepared from patient urine or serum and was probed with the CPS MAb 3C5.
The ELISA titer is the highest dilution of patient urine or serum that produced an OD450 of ≥0.5 in an antigen capture ELISA prepared from MAb 3C5.
The serum and urine samples were collected from different patients.
Identification of F. tularensis antigens that are secreted during infection.
The ability of the InMAD technique to identify antigens that are shed during infection was confirmed with a mouse model of tularemia. Similar to the B. pseudomallei study, sera harvested from mice infected with F. tularensis were filtered and used to immunize BALB/c mice. The reactivity of InMAD immune sera harvested from five individual mice is shown in Fig. 6A (lanes 1 to 5). A number of F. tularensis antigens were reactive with the immune sera. A high-molecular-weight (≥250-kDa) antigen recognized by serum from mouse 1 was sensitive to treatment with proteinase K (Fig. 6B). The InMAD immune serum from mouse 4 (Fig. 6A, lane 4) was reactive in a “ladder” pattern similar to that produced by F. tularensis LPS (8, 9). The putative LPS reactivity was confirmed by probing a proteinase K-treated lysate with InMAD immune serum from mouse 4 (Fig. 6C, lane 8) and comparing this pattern of reactivity with that produced by probing with MAb FB11, an F. tularensis LPS-specific antibody (Fig. 6C, lane 9). Multiple InMAD immune serum samples from different mice were reactive with LPS (data not shown).
FIG 6 .
Detection of secreted/shed F. tularensis antigens by InMAD. Five naive mice were immunized with InMAD serum from F. tularensis-infected mice. (A) InMAD immune sera was harvested from the five mice, and each sample was used to probe a whole-cell lysate of F. tularensis by Western blotting (lanes 1 to 5). (B) InMAD immune serum from mouse 1 (lane 1) was used to probe F. tularensis cell lysate prepared with proteinase K (lane 6) and without (lane 7) to determine if the reactive high-molecular-weight antigen (>250 kDa) was a polysaccharide. (C) InMAD immune serum from mouse 4 (lane 4) was used to probe proteinase K-treated F. tularensis lysate (lane 8); this reactivity was identical to the ladder pattern induced by anti-F. tularensis LPS MAb FB11 (lane 9). The Western blots shown for mouse 5 (lane 5) were performed with a gradient acrylamide gel, whereas the other blots were performed with a 10% gel. (D) InMAD immune serum was used to probe an F. tularensis proteome array (representative section of the array is shown). The two bright spots on the upper right portion of the array were spotted with FTT0403, and the two spots near the center of the grid were spotted with FTT1043. Spots outlined in yellow did not contain an F. tularensis ORF in the transcription/translation reaction and were used to determine background signal intensity.
An F. tularensis proteome array (10) was probed with InMAD immune serum to identify individual proteins recognized by the immune serum (Fig. 6D). The array was fabricated by (i) PCR amplifying each F. tularensis open reading frame (ORF), (ii) in vivo recombination cloning, and (iii) in vitro transcription/translation and microarray printing. InMAD immune serum samples from individual mice (and pooled samples) were used to probe the proteome array. A transcription/translation mixture that did not contain an F. tularensis ORF (no protein expression) was spotted on the array and was used to determine background signal intensity.
The most reactive antigens are listed in Table S2 in the supplemental material; examples include peptidyl-prolyl cis-trans isomerase (FTT1472), chaperone protein GroEL (FTT1696), and an outer membrane protein (FTT1747). All of the antigens in Table S2 were at least three times higher than the background signal intensity, which ranged from 5,000 to 11,000 (depending on the specific array). The top three proteins were 13 times higher than the background level, and the top 12 proteins were at least 5 times above the background level. Five of the proteins identified by probing of proteome arrays were also identified by 2-D immunoblotting and mass spectroscopy analysis (Table S2). Notably, two proteins identified by probing of immunoblots were among the proteins producing the strongest signals with the proteome arrays (pyruvate dehydrogenase E2 component [FTT1484] and a hypothetical membrane protein [FTT1778]). None of these proteins were reactive by immunoblotting with sera from naive unimmunized mice.
DISCUSSION
In this report, we identified circulating bacterial antigens present in serum following infection with B. pseudomallei or F. tularensis. Sera from infected animals (InMAD sera) were used to immunize syngeneic BALB/c mice. Mice immunized with InMAD sera generated a polyclonal antibody response to soluble bacterial antigens present in the sample.
A B. pseudomallei polysaccharide antigen was identified by InMAD. A polysaccharide composition was inferred from the antigen’s high molecular weight and its resistance to proteinase K treatment. MAb 3C5 produced in response to immunization with B. pseudomallei whole cells was reactive with the same polysaccharide antigen recognized by the InMAD immune sera. The polysaccharide antigen was found to be the CPS, which is an unbranched homopolymer of 1,3-linked 2-O-acetyl-6-deoxy-β-d-manno-heptopyranose residues (11). This conclusion is based on three lines of evidence. First, the InMAD immune sera and MAb 3C5 failed to react with a B. pseudomallei CPS mutant strain. Second, MAb 3C5 was reactive by immunoblotting with purified CPS, whose structure had been confirmed by nuclear magnetic resonance (NMR) analysis. Finally, MAb 3C5 is reactive with lysates of B. mallei but not with lysates of B. thailandensis (Fig. 3). The B. pseudomallei capsular gene cluster is present in both B. pseudomallei and B. mallei; however, this cluster is truncated in B. thailandensis (12).
As a proof of principle for InMAD as a target discovery platform, MAb 3C5 was found to be reactive with antigen in serum from an animal model of inhalational melioidosis and serum and urine samples from melioidosis patients. Detection of capsular polysaccharides in body fluids has been used as an aid to diagnose a variety of bacterial infections. The fact that B. pseudomallei CPS was identified by InMAD as a potential diagnostic target for melioidosis provides further evidence of the power of this approach for target identification.
Unlike results with the murine model of tularemia in which LPS was identified as a likely diagnostic target (Fig. 6), InMAD immune sera derived from the melioidosis model did not produce a Western blot with cell lysates suggestive of LPS. Consistent with this observation, the LPS MAb 4C7 was not reactive (i) with sera from B. pseudomallei-infected mice (Fig. 3) or (ii) in immunoblots or capture ELISA with serum or urine samples from melioidosis patients. This result provides an important negative control for the InMAD process.
Western blotting and antigen capture ELISAs were performed in an attempt to identify CPS in sera from infected mice (InMAD sera) and serum and urine samples from melioidosis patients. CPS was not detectable by Western blotting of patient sera; however, the ELISA proved to be more sensitive, showing that half of the serum samples contained CPS. In addition, ELISA identified CPS in over half of the urine samples. The sensitivity of the assay for diagnosis of melioidosis would likely be enhanced if a more sensitive assay platform was used or if samples, i.e., urine, were concentrated prior to analysis. These results demonstrate that an antigen found via the InMAD process can be found in body fluids beyond serum alone and provides further proof of concept for the discovery strategy. These results are consistent with previous reports of higher levels of bacteria in urine samples than in blood samples of patients with melioidosis (3).
The present study is the first report that a structurally defined CPS is shed into serum and urine in the course of experimental and clinical melioidosis. There have been reports of B. pseudomallei antigen secretion in human disease. In one instance, polyclonal antibodies raised against whole cells recognized antigens in patient urine (13, 14). In a second report, a MAb designated 5F8 recognized B. pseudomallei antigen in culture-positive samples of sputum, pus, pleural fluid, and urine (15). MAb 5F8 was initially reported to be reactive with LPS (16), although subsequent reports have shown it to be reactive with a structurally undefined high-molecular-weight polysaccharide (17, 18), quite likely a previously described exopolysaccharide composed of d-galactose and Kdo (19, 20).
In addition to CPS, the InMAD approach identified many proteins that were shed during infection. Each of these is a potential diagnostic target. Diagnostics development that targets proteins will continue by producing polyclonal and monoclonal antibodies reactive with candidate proteins, followed by immunoassay construction. The goal is to target a combination of antigens (possibly CPS and multiple proteins) in order to increase the sensitivity and specificity of the immunoassay.
A study of F. tularensis was done to verify that InMAD can identify circulating antigens from a second bacterium. A full proteome array that was previously used to identify immunodominant F. tularensis antigens in mice (10, 21) was probed to allow for rapid identification of antigens recognized by InMAD immune serum. A number of proteins were identified. Of the ten most immunogenic antigens identified in this earlier report, five were also identified by our InMAD technique as antigens that circulate in serum during an acute infection. LPS from F. tularensis was also identified by InMAD as a circulating antigen. These results confirm results from the melioidosis study that both polysaccharide and protein antigens can be identified via InMAD.
Elements of the InMAD approach were described in two previous reports. First, Lehmann and Reiss found that immunization of rabbits with serum from an immunosuppressed rabbit infected with Aspergillus fumigatus produced antibody that was reactive with a single antigenic moiety (22), later found to be A. fumigatus galactomannan (23). More recently, Homer et al. immunized BALB/c mice with serum from CB-17 SCID mice that were infected with the parasite Babesia microti (24). Sera from the immunized mice was then used to screen a B. microti genomic expression library. Our studies build and expand on these initial reports to describe a potentially broad discovery platform. First, our studies took advantage of the ease of proteomic analysis, the power of protein microarrays, and the inherent specificity of hybridoma technology in target discovery. Second, we demonstrated feasibility using two distinct pathogens. Third, we demonstrate that both polysaccharide and protein antigens can be identified in the same discovery process. Fourth, our studies found that a potential target identified by use of sera from infected animals for target identification identified shed antigen in a second body fluid, i.e., urine. Finally, our studies have moved a potential diagnostic target (CPS) from discovery to MAb production to validation in an animal model to validation with clinical samples.
This report describes a proof of concept for use of InMAD. Further improvements in this strategy might allow for identification of additional or alternative targets. For example, different stages of disease or production of disease in immunosuppressed animals or hosts with different levels of resistance might cause variation in the circulating antigen pool that is presented to the immunized mouse. For example, if one were interested in chronic melioidosis infection, then a C57BL/6 mouse might be a better choice than the BALB/c mouse, in which melioidosis follows a more acute course. In addition, different routes of infection and harvesting serum at different stages of infection may cause variation in the circulating antigen pool. As a consequence, the InMAD process will require modification to suit the pathogenic habit of individual microbes.
Animal-to-animal variation in the polyclonal antibodies present in the InMAD immune serum should also be considered. As shown in Fig. 2 and 6, there are substantial differences in the antigens that are reactive with InMAD immune serum from individual mice. This inherent variability exists even though the mice are syngeneic and are immunized with pooled samples from infected mice. By increasing the number of mice that are immunized with InMAD serum, we were able to generate a consensus set of circulating antigens that were identified from a majority of immunized mice. In addition, the antibody profile generated by the immunized mice will presumably be affected by (i) choice of adjuvant, (ii) boosting, and (iii) use of an outbred strain of mouse to increase antibody diversity.
Finally, the current study used BALB/c mice for both infection and immunization; however, a syngeneic model may not be essential to success. We are currently utilizing InMAD serum generated from outbred mice and guinea pigs to identify circulating antigens from fungal pathogens (25). If the InMAD approach can cross host species lines, it may be possible to use human serum or urine to immunize mice. As a consequence, this method may directly identify circulating antigens present during human disease.
In summary, the InMAD technique is a powerful discovery platform to identify circulating immunogenic proteins and polysaccharides that are present during microbial infections. Identification of such targets may assist in the development of diagnostics that rely on antigen immunoassay for early diagnosis of infection. Transition of this discovery phase to functional immunoassays will require optimization of immunoassays for specific diagnostic targets. In the case of melioidosis, such optimization may include enhancing the affinity of MAb 3C5 by phage display and producing an expanded library of CPS-specific MAbs in order to improve sensitivity. Modification to immunoassay procedures may also further improve sensitivity. Finally, we will be producing MAbs reactive with an expanded set of B. pseudomallei antigens, e.g., the proteins shown in Table S1 in the supplemental material, and utilize diagnostic platforms suitable for rapid point-of-care use, e.g., the lateral flow immunochromatographic assay.
MATERIALS AND METHODS
Bacterial cultures.
B. pseudomallei (1026b) and F. tularensis (SCHU S4) cultures were grown overnight at 37°C in a shaking incubator in brain heart infusion broth or Chamberlain’s broth, respectively. The bacterial cells were collected by sedimentation and resuspended in PBS. The B. pseudomallei cells were heat inactivated at 80°C for 2.5 h, and F. tularensis was heat inactivated at 72°C for 2 h. Both preparations were cultured following inactivation to confirm killing of bacterial cells. Heat-inactivated CPS mutant strain SR1015 was acquired from Donald Woods (University of Calgary).
Animal models.
Murine models of pulmonary melioidosis and tularemia were used to generate sera from infected animals. For the melioidosis model, BALB/c mice were infected by the intranasal route with 5000 CFU of B. pseudomallei strain 1026b. Mice became moribund at 3 to 4 days postinfection, and blood was collected at that time by cardiac puncture. For the tularemia model, BALB/c mice were infected via the intranasal route with 50 CFU of F. tularensis SCHU S4. Four days following infection, blood was collected by cardiac puncture. Serum samples were filtered though a 0.22-µm filter to remove whole bacterial cells.
Human samples.
Urine and serum samples from patients with culture-positive melioidosis or negative controls were obtained from sample archives (no identifiable private information supplied) at Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand. Culturing of serum and urine samples and quantitative bacterial counts (urine samples only) were performed as previously described (3).
Immunization of mice with sera from infected mice.
Sera from infected mice (6.2 to 50 µl) were brought to a final volume of 100 µl with PBS or used undiluted (100 µl), mixed with 100 µl TiterMax gold adjuvant (TiterMax USA, Inc.), and injected into BALB/c mice via the subcutaneous route. Blood was collected at 4, 6, 8, 10, and 12 weeks postimmunization by retro-orbital bleeding and cardiac puncture. This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the University of Nevada, Reno.
Western blot analysis.
A standard Western blot procedure was done using semidry blotting (MAb 4C7) or tank blotting (InMAD immune serum or MAb 3C5). Briefly, 8 × 106 heat-inactivated bacterial cells were suspended in Laemmli sample buffer (Sigma) and incubated for 10 min at 100°C. If required, 1 volume of a proteinase K solution (3.3 mg in 1 ml Laemmli sample buffer) was added, and the sample was further incubated for 1 h at 60°C. In the case of human serum or urine, the sample (20 µl) was mixed with 6× Laemmli sample buffer and treated as described above. Samples were run on either a 10 to 20% SDS gradient gel or a 10% SDS gel, followed by transfer onto a polyvinylidene difluoride (PVDF) membrane. MAbs 4C7 and 3C5 were labeled with horseradish peroxidase (EZ-Link Plus activated peroxidase kit; Pierce) to yield a product at approximately 0.8 mg/ml and used at a dilution of 1:10,000; signal was detected with a chemiluminescent substrate (Pierce). MAb FB11 (Abcam) specific for F. tularensis LPS was used at a 1:100,000 dilution (2 mg/ml stock). InMAD immune serum was used at a dilution of 1:30. MAb FB11 and mouse antibodies within the InMAD serum were detected with a horseradish peroxidase-labeled anti-mouse antibody (1:10,000 dilution).
Proteomic analysis.
Heat-killed B. pseudomallei cells were lysed and separated by 2-D gel electrophoresis. Two duplicate gels were produced; one gel was blotted onto nitrocellulose. InMAD immune serum was used to probe the membrane, reactive spots were cut from the duplicate gel, and proteins were identified by mass spectrometry. Detailed protocols have been included in Text S1 in the supplemental material.
F. tularensis proteome array.
Protein microarray fabrication and probing methods have been described previously (10). Probing methods were modified slightly in that InMAD immune serum used to probe the proteome array ranged from 1:10 to 1:100 dilutions.
Production of MAbs.
Production of MAbs 4C7 (LPS) and 3C5 (CPS) began with immunization of BALB/c mice with heat-inactivated B. pseudomallei (strain 1026b). An intraperitoneal injection of 2 × 108 bacteria was administered every 2 weeks for an 8-week period (7). An ELISA, with heat-inactivated B. pseudomallei strain 1026b in the solid phase, was used to assess antibody levels (7). Three days prior to harvesting of spleens, one final intraperitoneal immunization with 2 × 108 bacteria was administered. Hybridoma cells were produced as previously described (26). Western blot analysis was done to identify hybridoma cell lines that were producing anti-LPS (ladder pattern) or anti-CPS (high-molecular-weight, proteinase K-insensitive) MAbs. Hybridoma cell lines were grown in Integra CL 1000 culture flasks (Integra Biosciences), and MAbs were purified by affinity chromatography over a protein A column.
Antigen capture immunoassay for CPS and LPS.
Microtiter plates were coated overnight with either MAb 3C5 or 4C7 at 2 µg/ml in PBS, washed with PBS-Tween (PBS containing 0.5% Tween 20), and blocked by incubation for an additional 90 min with PBS-Tween. Serial dilutions of serum, urine, or B. pseudomallei lysate were prepared in PBS-Tween and incubated for 90 min at room temperature with the MAb-coated wells. The wells were washed with PBS-Tween, incubated for 90 min with HRPO-labeled MAb 3C5 or 4C7 (2 µg/ml), washed, and incubated with tetramethylbenzidine substrate (Kirkegaard & Perry Laboratories).
CPS purification.
CPS was purified essentially as previously described (27) using an O antigen-deficient derivative of B. pseudomallei strain DD503. CPS isolated by this method was confirmed by NMR analysis to be a polymer of 1,3-linked 2-O-acetyl-6-deoxy-β-d-manno-heptopyranose residues (P. J. Brett, unpublished data).
SUPPLEMENTAL MATERIAL
Supplemental methods. Download Text S1, DOCX file, 0.14 MB.
B. pseudomallei protein antigens reactive with InMAD immune serum by 2-D Western blotting.
F. tularensis antigens reactive with InMAD immune serum by proteome array.
ACKNOWLEDGMENTS
This project was supported by award U54AI065359 from the National Institute of Allergy and Infectious Diseases. Additional support was provided by award P20RR016464 from the INBRE Program of the National Center for Research Resources. N.C. holds a Wellcome Trust Career Development award in Public Health and Tropical Medicine, United Kingdom. S.J.P. receives support from the Wellcome Trust and the NIHR Cambridge Biomedical Research Centre.
The content is the sole responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Citation Nuti DE, et al. 2011. Identification of circulating bacterial antigens by in vivo microbial antigen discovery. mBio 2(4):e00136-11. doi:10.1128/mBio.00136-11.
REFERENCES
- 1. Clemens DL, Horwitz MA. 2007. Uptake and intracellular fate of Francisella tularensis in human macrophages. Ann. N. Y. Acad. Sci. 1105:160–186 [DOI] [PubMed] [Google Scholar]
- 2. Woods DE, et al. 1999. Current studies on the pathogenesis of melioidosis. Microbes Infect. 1:157–162 [DOI] [PubMed] [Google Scholar]
- 3. Wuthiekanun V, et al. 2007. Quantitation of B. pseudomallei in clinical samples. Am. J. Trop. Med. Hyg. 77:812–813 [PubMed] [Google Scholar]
- 4. Lindquist D, Chu MC, Probert WS. 2007. Francisella and Brucella. In Murray PR, Barron EJ, Jorgensen JH, Landry ML, Pfaller MA, Manual of clinical microbiology. ASM Press, Washington, DC [Google Scholar]
- 5. NIAID 2007. NIAID strategic plan for biodefense research. National Institute of Allergy and Infectious Diseases, Bethesda, MD. [Google Scholar]
- 6. Reckseidler SL, DeShazer D, Sokol PA, Woods DE. 2001. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect. Immun. 69:34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jones SM, Ellis JF, Russell P, Griffin KF, Oyston PC. 2002. Passive protection against Burkholderia pseudomallei infection in mice by monoclonal antibodies against capsular polysaccharide, lipopolysaccharide or proteins. J. Med. Microbiol. 51:1055–1062 [DOI] [PubMed] [Google Scholar]
- 8. Thirumalapura NR, et al. 2005. Structural analysis of the O-antigen of Francisella tularensis subspecies tularensis strain OSU 10. J. Med. Microbiol. 54:693–695 [DOI] [PubMed] [Google Scholar]
- 9. Prior JL, et al. 2003. Characterization of the O antigen gene cluster and structural analysis of the O antigen of Francisella tularensis subsp. tularensis. J. Med. Microbiol. 52:845–851 [DOI] [PubMed] [Google Scholar]
- 10. Eyles JE, et al. 2007. Immunodominant Francisella tularensis antigens identified using proteome microarray. Proteomics 7:2172–2183 [DOI] [PubMed] [Google Scholar]
- 11. Perry MB, MacLean LL, Schollaardt T, Bryan LE, Ho M. 1995. Structural characterization of the lipopolysaccharide O antigens of Burkholderia pseudomallei. Infect. Immun. 63:3348–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DeShazer D, Waag DM, Fritz DL, Woods DE. 2001. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microb. Pathog. 30:253–269 [DOI] [PubMed] [Google Scholar]
- 13. Desakorn V, et al. 1994. Detection of Pseudomonas pseudomallei antigen in urine for the diagnosis of melioidosis. Am. J. Trop. Med. Hyg. 51:627–633 [DOI] [PubMed] [Google Scholar]
- 14. Haase A, Melder A, Smith-Vaughan H, Kemp D, Currie B. 1995. RAPD analysis of isolates of Burkholderia pseudomallei from patients with recurrent melioidosis. Epidemiol. Infect. 115:115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anuntagool A, Intachote P, Naigowit P, Sirisinha S. 1996. Rapid antigen detection assay for identification of Burkholderia (Pseudomonas) pseudomallei infection. J. Clin. Microbiol. 34:975–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rugdech P, Anuntagool N, Sirisinha S. 1995. Monoclonal antibodies to Pseudomonas pseudomallei and their potential for diagnosis of melioidosis. Am. J. Trop. Med. Hyg. 52:231–235 [DOI] [PubMed] [Google Scholar]
- 17. Anuntagool N, Panichakul T, Aramsri P, Sirisinha S. 2000. Shedding of lipopolysaccharide and 200-kDa surface antigen during the in vitro growth of virulent Ara- and avirulent Ara+ Burkholderia pseudomallei. Acta Trop. 74:221–228 [DOI] [PubMed] [Google Scholar]
- 18. Anuntagool N, Sirisinha S. 2002. Antigenic relatedness between Burkholderia pseudomallei and Burkholderia mallei. Microbiol. Immunol. 46:143–150 [DOI] [PubMed] [Google Scholar]
- 19. Nimtz M, et al. 1997. Structure of an acidic exopolysaccharide of Burkholderia pseudomallei. Eur. J. Biochem. 250:608–616 [DOI] [PubMed] [Google Scholar]
- 20. Steinmetz I, Rohde M, Brenneke B. 1995. Purification and characterization of an exopolysaccharide of Burkholderia (Pseudomonas) pseudomallei. Infect. Immun. 63:3959–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sundaresh S, et al. 2007. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics 23:i508–i518 [DOI] [PubMed] [Google Scholar]
- 22. Lehmann PF, Reiss E. 1978. Invasive aspergillosis: antiserum for circulating antigen produced after immunization with serum from infected rabbits. Infect. Immun. 20:570–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reiss E, Lehmann PF. 1979. Galactomannan antigenemia in invasive aspergillosis. Infect. Immun. 25:357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Homer MJ, et al. 2003. Identification and characterization of putative secreted antigens from Babesia microti. J. Clin. Microbiol. 41:723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chaves S, AuCoin DP, Kozel TK. 2011. Target discovery for immunodiagnosis of invasive aspergillosis, abstr. 11-GM-A-3096-ASM. Abstr. 111th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC. [Google Scholar]
- 26. Kozel TR, et al. 2004. mAbs to Bacillus anthracis capsular antigen for immunoprotection in anthrax and detection of antigenemia. Proc. Natl. Acad. Sci. U. S. A. 101:5042–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brett PJ, et al. 2011. Burkholderia thailandensis oacA mutants facilitate the expression of Burkholderia mallei-like O polysaccharides. Infect. Immun. 79:961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download Text S1, DOCX file, 0.14 MB.
B. pseudomallei protein antigens reactive with InMAD immune serum by 2-D Western blotting.
F. tularensis antigens reactive with InMAD immune serum by proteome array.