Abstract
The microsecond folding and unfolding kinetics of ovine prion proteins (ovPrP) were measured under various solution conditions. A fragment comprising residues 94–233 of the full-length ovPrP was studied for four variants with differing susceptibilities to classical scrapie in sheep. The observed biexponential unfolding kinetics of ovPrP provides evidence for an intermediate species. However, in contrast to previous results for human PrP, there is no evidence for an intermediate under refolding conditions. Global analysis of the kinetic data, based on a sequential three-state mechanism, quantitatively accounts for all folding and unfolding data as a function of denaturant concentration. The simulations predict that an intermediate accumulates under both folding and unfolding conditions, but is observable only in unfolding experiments because the intermediate is optically indistinguishable from the native state. The relative population of intermediates in two ovPrP variants, both transiently and under destabilizing equilibrium conditions, correlates with their propensities for classical scrapie. The variant susceptible to classical scrapie has a larger population of the intermediate state than the resistant variant. Thus, the susceptible variant should be favored to undergo the PrPC to PrPSc conversion and oligomerization.
Introduction
Prion diseases are a class of neurodegenerative diseases that includes scrapie in sheep and goats, bovine spongiform encephalopathy in cattle (mad cow disease), as well as fatal familial insomnia, Gerstmann-Straussler-Scheinker syndrome, and Creutzfeldt-Jakob disease in humans. According to the prevailing hypothesis (1), prion diseases result from the misfolding of a naturally occurring prion protein (PrP) from its normal cellular form (PrPC) to a virulent scrapie form (PrPSc). This abnormal isoform is able to catalyze the conversion of PrPC to itself, and has been associated with prion infectivity (1–3). Although the cause of neuronal death in prion diseases remains unclear, all pathogenesis models involve the structural conversion of PrPC to PrPSc (4). PrPC is a copper-binding (5,6), cell-surface glycoprotein (7) composed of an unstructured N-terminal domain and a globular C-terminal domain (residues 126–233) of known structure, comprising three α-helices, a short two-stranded antiparallel β-sheet, and a disulfide bond (Cys182-Cys217, which corresponds to Cys179-Cys214 in human PrP) linking helices 2 and 3 (8–11) (Fig. 1). Relatively little is known about the structure of PrPSc, except that it appears to have significantly more β-sheet and slightly less α-helix (17–30% α-helix; 43–54% β-sheet) compared to PrPC (42% α-helix; 3% β-sheet) (12–14).
Figure 1.
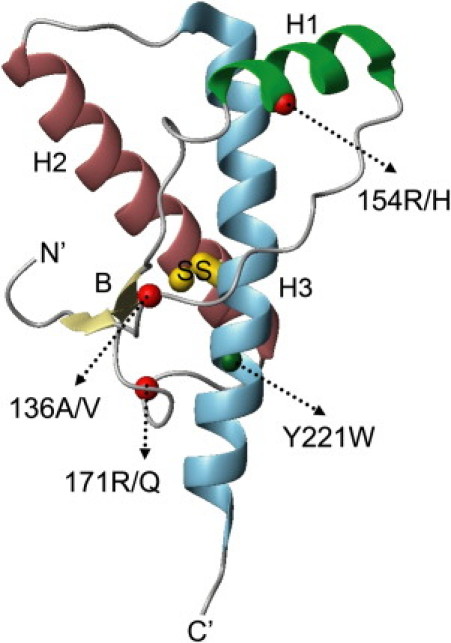
Ribbon representation of the structure of ovPrP (residue 126–233), based on a crystal structure (pdb ID 1UW3 (11)). The structure comprises three α-helices (H1–H3), a short antiparallel β-sheet (B), and a disulfide bridge (SS) linking H2 and H3. The residues involved in genetic modulation in sheep scrapie and the engineered fluorescence probe (Trp-221) are labeled.
Polymorphisms of ovine PrP (ovPrP) have been associated with differing susceptibility to classical scrapie, particularly at positions 136 (A/V), 154 (H/R), and 171 (Q/R). The A136R154R171 (ARR) and A136H154Q171 (AHQ) alleles confer resistance, whereas the V136R154Q171 (VRQ) and A136R154Q171 (ARQ) genotypes are correlated with susceptibility (15–18). Several factors, alone or in combination, might account for this genetic modulation. Variations in conformations and glycosylation patterns between allelic variants have been suggested to modulate scrapie susceptibility by altering solvent accessibility (19,20). PrPC variants susceptible to classical scrapie have been shown to have longer cellular half-lives (21) and higher plasma concentrations (22), when compared to resistant variants, which might give them more opportunities to convert and oligomerize. Finally, the propensity for the PrPC-to-PrPSc conversion, both in vitro and ex vivo, has also been suggested to correlate with susceptibility to classical scrapie (23,24). The PrPC-to-PrPSc conversion efficiencies of ovPrP allelic variants depend on the exogenously introduced PrPSc strain. However, the ARR variant exhibits the lowest conversion efficiency regardless of the strain introduced.
Because this structural conversion requires at least partial unfolding, it would be plausible to hypothesize that more susceptible variants are less stable structurally (25). However, exactly the opposite is observed for classical scrapie: variants with lower structural stability (as measured by the free energy difference ΔGNU between the folded and unfolded states) are less prone to the disease than those with higher structural stability (26,27). Differences in in vivo clearance have been proposed to explain this inverse correlation (21); nevertheless, it cannot explain the higher in vitro conversion propensities displayed by susceptible variants (23). To account for this apparent paradox, we hypothesized that polymorphisms affect the susceptibility by modulating the population and structural properties of the ovPrPSc precursor, which we took to be a folding intermediate. This hypothesis led us to examine two subquestions: First, do the folding and unfolding of ovPrP proceed through an intermediate? Second, if so, is this folding intermediate the precursor of ovPrPSc? A folding intermediate has been linked to human familial prion diseases (28,29). However, it is unclear whether such a PrP intermediate exists in other species, and if so, whether the properties of this intermediate and its role in genetic modulation are consistent across species.
To study these questions and the folding of ovPrP in general, we measured the microsecond folding and unfolding kinetics of these allelic variants under various solution conditions, including two temperatures (5 and 15°C), two pH values (5 and 7), and two denaturants, guanidine hydrochloride (GuHCl) and urea. The unfolding kinetics of ovPrP exhibited two exponentials at pH 7, which indicates the presence of at least one intermediate species besides the native (N) and unfolded (U) species. The most plausible kinetic model seems to be N ↔ I ↔ U in which the native species N unfolds quickly to an intermediate I before unfolding completely to U. Adopting this kinetic model, we found that the population of I in the variants correlate with their differing propensities toward classical scrapie. The variant with higher classical scrapie propensity is characterized by a higher population in I. Our findings support the hypothesis that the formation of this partially unfolded intermediate is an early step in the formation of ovPrPSc.
Materials and Methods
Protein preparation and purification
Unless stated otherwise, reagents and buffers were purchased from Sigma-Aldrich (St. Louis, MO) in the highest grade available. The plasmid encoding ovPrP94-233, ARQ allele with an N-terminal (His)6 tag was a generous gift from Dr. P. M. Bayley (11). Based on this parental plasmid, ARR, AHQ, and VRQ variants were constructed by site-directed mutagenesis using QuikChange kit (Stratagene, Santa Clara, CA) and appropriate primers (see the Supporting Material, Table S1). To introduce a fluorescence probe, all polymorphic variants contained additional W102F and Y221W mutations. All sequences were confirmed by DNA sequencing. Ovine numbering is used throughout this article unless specified otherwise.
These plasmids were transformed into competent Escherichia coli BL21 (DE3) cells and expressed by isopropyl β-D-1-thiogalactopyranoside induction. Because the expressed ovPrP variants accumulate in inclusion bodies, 8 M urea was used to solubilize the proteins. Purification was carried out by metal-affinity chromatography using nickel-nitrilotriacetic acid resin (Qiagen, Valencia, CA). Protein concentration was determined by the optical density at 280 nm using an extinction coefficient predicted from the amino-acid composition, e.g., 23,505 M−1cm−1 for the wild-type protein (30). The identity of the purified protein was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blotting, and mass spectrometry.
Continuous-flow measurements
For kinetic measurements on the submillisecond timescale, we used a modified version of the continuous-flow capillary mixing instrument described by Shastry et al. (31), including an ultraviolet (UV)-enhanced charge-coupled device camera system (Alta U47-UV, Apogee Instruments, Roseville, CA). The excitation wavelength was 297 nm, and tryptophan fluorescence emission was collected using a 324 nm cut-on filter (Edmund Optics, Barrington, NJ). For most experiments, mixing was carried out at 15°C at a flow rate of 1.8 ml/s, with a final protein concentration of ∼11 μM. The refolding (resp. unfolding) reactions were initiated by diluting the unfolded (resp. native) protein at 1:5 ratio into the refolding (resp. unfolding) buffer containing desired denaturant concentrations that promote refolding (resp. unfolding). The buffer consisted of GuHCl or urea in either 50 mM sodium acetate, pH 5 or 50 mM imidazole, pH 7. The concentration of denaturants was determined from its refractive index.
The instrumental dead time of the continuous-flow mixer was determined by measuring the reaction of N-acetyl-L-tryptophan (NAT) with N-bromosuccinimide (NBS) at several concentrations of NBS. With appropriate amounts of glycerol included to match the viscosity of denaturants in our refolding experiments (Fig. S1), the dead time of the capillary mixer was 39 ± 4 μs. At higher viscosity, matching that of the final GuHCl concentration used for unfolding, the dead time was slightly longer (42 ± 4 μs). The use of glycerol to simulate denaturant solutions was necessary because NBS is unstable in concentrated solutions of GuHCl or urea. We recently found that N-acetyl-L-tryptophan-amide, which has traditionally been used for testing mixer performance (31,32), shows nonexponential kinetics because NBS can react not only with the indole ring, but also with the C-terminal amide protecting group (M. Xu and H. Roder, unpublished data). In contrast, NAT lacks a reactive amide and shows simple pseudo first-order (exponential) kinetics in the presence of a molar excess of NBS.
Kinetic analysis
The fluorescence signal was fit to a sum of exponentials using the least-squares algorithm as implemented in IGOR Pro software (Wavemetrics). In every case, the residuals were examined to confirm the absence of systematic errors. Under strongly denaturing conditions, two exponentials were necessary to fit the kinetic traces. At intermediate denaturant concentrations, one exponential was sufficient to fit unfolding traces. The kinetic traces obtained in refolding experiments were also well represented by a single exponential term.
The observed kinetics of folding and unfolding versus GuHCl concentration was fitted to a kinetic model based on assumptions that are standard in the literature. First, we assume that all reactions are unimolecular. Consistent with this assumption, we detected no systematic dependence of the refolding kinetics on protein concentration (see Results). This implies that the fluorescence signal is a sum of n-1 exponential phases for a kinetic model with n species. Second, the logarithm of each microscopic rate constant kij is assumed to vary linearly with denaturant concentration, c
| (1) |
where kijo is the microscopic rate constant from species i to j in the absence of denaturant. Third, we assume that, under strongly refolding conditions, the folding rate constants kij are significantly larger than their reverse unfolding rate constants kji, whereas the opposite is true under strongly unfolding conditions. This implies that in the unfolding experiments, the protein is initially ∼100% in the native state N; conversely, in the folding experiments, the protein is initially ∼100% in the unfolded state U.
The kinetics of (un)folding as a function of denaturant concentration was modeled using a general matrix approach (33,34). The elementary rate constants at a given denaturant concentration (Eq. 1) define a rate matrix, based on the set of linear differential equations describing a sequential three-state mechanism. The eigenvalues representing the macroscopic rate constants (observable rates λ1 and λ2) and corresponding eigenvectors were calculated by using a program written in Igor Pro. Using a least-squares fitting algorithm in combination with manual constraints, the parameters describing each of the microscopic rate constants (Eq. 1) were varied in an effort to simultaneously fit the observed refolding/unfolding rates at all denaturant concentrations. The set of kinetic parameters thus obtained (four rate constants, kijo, and four kinetic m-values, m‡ij) was used to generate a family of kinetic traces for refolding and unfolding experiments at different denaturant concentrations, c, defined as follows (Eq. 2):
| (2) |
Amplitudes Ai were computed from eigenvectors by solving a set of linear equations for a particular set of initial conditions and fluorescence values for each state, fN/I/U, defined as
| (3) |
where f0N/I/U is the relative fluorescence in the absence of denaturant and sN/I/U is the slope versus denaturant concentration. The intrinsic fluorescence parameters were optimized by using a least-squares algorithm to fit a set of folding or unfolding traces to a three-state model.
The kinetic m-values (m‡ij value in Eq. 1) represent the change in the solvation free energy induced by denaturant (35), and thus provides a quantitative measure of the average compactness. For a three-state mechanism, the compactness of the intermediate state relative to the unfolded state can be represented by a α value:
| (4) |
where αUI = 0 represents an intermediate state with the same surface-burial as the unfolded state, whereas αUI = 1 represents an intermediate state that is as compact as the native state.
Results
Tryptophan fluorescence reporter
The kinetics of folding and unfolding of ovPrP variants was monitored by tryptophan fluorescence. The only tryptophan in the 94-233 fragment of the wild-type ovPrP, Trp-102, is located in the unstructured N-terminal segment and is exposed to the solvent under both native and denaturing conditions. As in previous work with human PrP (29,36), we prepared a double mutant of ovPrP, W102F/Y221W, which replaces the endogenous Trp with a conformationally sensitive fluorescence reporter (Fig. 1). Under native conditions the fluorescence emission of Trp-221 is blue-shifted and partially quenched, and shows a 4–6-fold increase in the quantum yield upon unfolding. The secondary structure of native ovPrP and its double mutant W102F/Y221W was assessed by circular dichroism; both spectra exhibit a double minimum at around 208 and 222 nm, which is typical for a α-helical protein (data not shown). Equilibrium unfolding experiments monitored at pH 5 by far-UV circular dichroism and fluorescence indicates that the double mutant has native-like stability. Sedimentation equilibrium experiments were carried out on the ARQ variant of ovPrP using a Beckman XLI analytical ultracentrifuge at 15,000 rpm, 25°C. After 5 days, the sedimentation data (Fig. S2) converged to a classic Boltzmann distribution with an estimated molecular mass of 17 kDa, in close agreement with predicted molecular mass of ovPrP94-233 (16.9 kDa).
Kinetics of (un)folding reactions on the submillisecond timescale
A continuous-flow mixing technique (31) was used to monitor the changes in tryptophan emission (>324 nm) associated with folding and unfolding of ovPrP variants at 15°C as a function of GuHCl concentration (in 50 mM imidazole buffer at pH 7.0). Fig. 2, A and B, show representative refolding traces for the ARQ and ARR, measured after rapid dilution of the unfolded protein (typically in 5 M GuHCl) to final GuHCl concentrations in the range of ∼0.8–2.4 M. Each trace is well described by a single exponential decay (Fig. S3).
Figure 2.
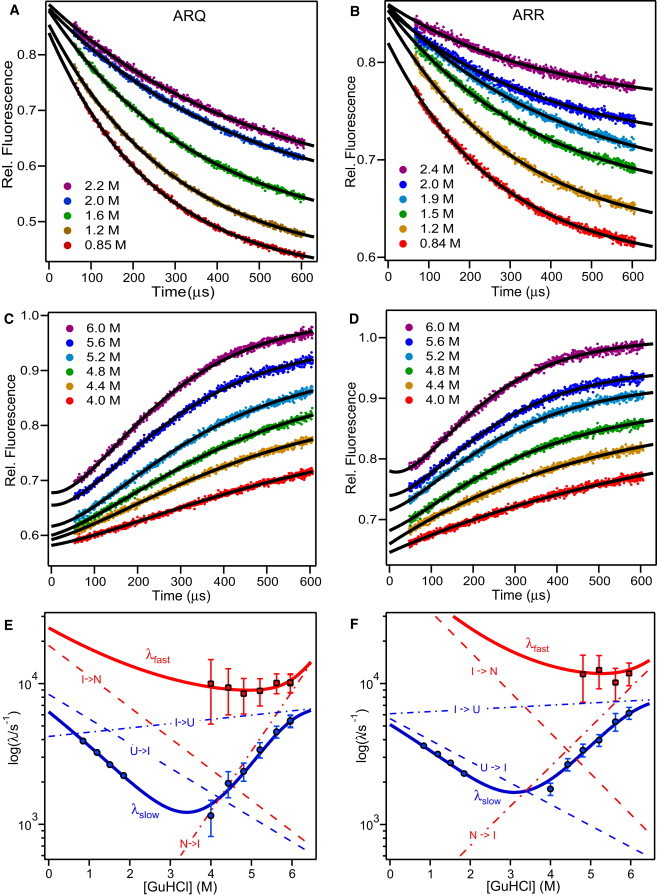
Continuous-flow fluorescence analysis of the kinetic mechanism of folding/unfolding of ovPrP variants ARQ (left panels) and ARR (right panels). The kinetics of folding (A and B) and unfolding (C and D) was measured in the presence of various concentrations of GuHCl at pH 7, 15°C. Refolding (resp. unfolding) traces were initially normalized relative to the fluorescence profiles of the unfolded protein in 5 M GuHCl (resp. native protein in 1.8 M GuHCl). Unfolding traces were then divided by the average signal of the unfolded protein in 5 M GuHCl. Solid lines were obtained by nonlinear least-squares fitting of individual traces, using a single exponential for all experiments up to 4.4 M GuHCl and a sum of two exponentials for unfolding experiments at 4.8 M GuHCl and above. In panels E and F, the logarithm of the observable rate constants are plotted versus GuHCl concentrations. Solid lines in panels E and F represent the two observable rates of folding/unfolding predicted by global analysis of the kinetic data for each variant using a sequential three-state mechanism (U ↔ I ↔ N), and dashed lines indicate the corresponding elementary rate constants.
To determine if the folding behavior of ovPrP changes with solution conditions, we measured the kinetics of refolding of the ARQ, ARR, VRQ, and AHQ variants under several alternative solution conditions, including different temperatures (5 and 15°C), pH values (5 and 7,) and denaturants (GuHCl and urea; see Fig. S4 and Fig. S5). All ovPrP variants exhibit only one refolding phase under these conditions, indicating that there is no detectable intermediate under refolding conditions.
Fig. 2, C and D, show a series of unfolding traces for the ARQ and ARR variants under matching conditions (pH 7, 15°C), measured in continuous-flow fluorescence experiments involving rapid jumps from native conditions (1.8 M GuHCl) to denaturing concentrations of GuHCl (4–6 M). To obtain a satisfactory fit of the unfolding traces for both variants at high denaturant concentration, it was necessary to use a sum of two exponentials. This is illustrated by Fig. 3, which shows that double-exponential fits have substantially smaller, more random, residuals than single-exponential fits. Comparison of the residuals in Table S2 shows that the deviations from single-exponential behavior during unfolding become more pronounced with increasing GuHCl concentration. To rule out the possibility that these deviations reflect mixing artifacts, we performed a series of experiments on a strictly exponential test reaction (oxidation of the indole ring of NAT by NBS) under various solution conditions, using glycerol to match the viscosity changes encountered in GuHCl-induced unfolding experiments (Fig. S1). A representative trace is shown in Fig. 3 C; the quality of the fit using a single exponential is excellent with random residuals down to the ∼40 μs dead time (χ2 = 9·10−4). In contrast, the residuals for single-exponential fits to unfolding traces (Fig. 3, A and B) are much larger and dominated by nonrandom components, confirming that the biphasic nature of the traces reflects complexities in the kinetics of unfolding rather than instrumental artifacts.
Figure 3.
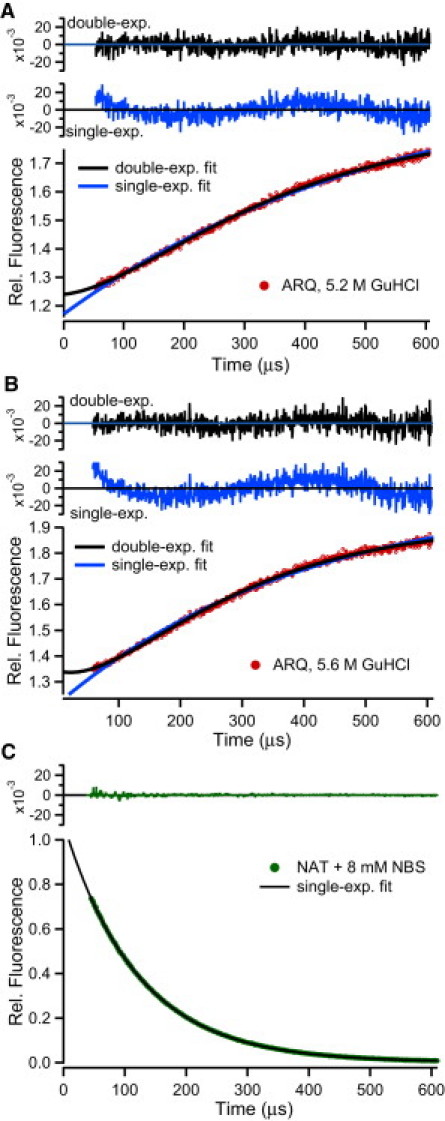
Representative continuous-flow traces illustrating the complex kinetics of unfolding. Solid circles (red) show the time course of unfolding for the ARQ variant at 5.2 M GuHCl (A) and 5.6 M GuHCl (B). Solid lines indicate single- and double-exponential fits and the corresponding residuals (upper: double-exp.; lower: single-exp.). For comparison, panel C shows a representative trace for pseudo first-order (exponential) test reaction in which NAT was mixed with 8 mM NBS under solvent conditions matching those of the protein unfolding experiment in 5.6 M GuHCl (see text and Fig. S2). All residuals are plotted on the same scale (±0.02) to allow direct comparison of the quality of single- versus double-exponential fits.
Fig. 2, E and F, show logarithmic (chevron) plots versus GuHCl concentration for all observable rate constants, including those of the slow and fast unfolding phases (λslow and λfast) as well as the single folding phase, which merges with the slow unfolding phase in the transition region. The solid lines represent a simultaneous fit of both observable rate constants obtained by numerically solving the equations describing a sequential three-state folding mechanism, as detailed in Materials and Methods. The parameters defining the denaturant-dependence of the corresponding elementary rate constants for each transition (dashed lines) are listed in Table 1.
Table 1.
Kinetic parameters for the three-state folding reaction of ovPrP variants∗
| kUI0 | kIU0 | kIN0 | kNI0 | fU | |
|---|---|---|---|---|---|
| ARQ | 8500 | 4100 | 19,000 | 28 | 0.83 |
| ARR | 5700 | 6100 | 65,000 | 190 | 0.76 |
| m‡UI | m‡IU | m‡IN | m‡NI | fI | |
| ARQ | −0.23 | 0.04 | −0.29 | 0.54 | 0.40 |
| ARR | −0.20 | 0.02 | −0.38 | 0.37 | 0.52 |
| ΔGUI | ΔGIN | ΔGUN | αUI | fN | |
| ARQ | −0.42 | −3.72 | −4.1 | 0.25 | 0.40 |
| ARR | 0.05 | −3.33 | −3.4 | 0.23 | 0.52 |
Kinetics of folding/unfolding at pH 7, 15°C, was analyzed using the mechanism U ↔ I ↔ N. Elementary rate constants kij are in s−1, mij is in kcal·mol−1·M−1, and ΔG is in kcal·mol−1 (1 kcal = 4.18 kJ). The last column lists the intrinsic fluorescence values of U (fU), I (fI), and N (fN), normalized relative to the signal of the U-state at 5 M GuHCl, obtained by global fitting of the family of folding and unfolding (a slope of 0.04 M−1 was used to account for the denaturant-dependence of the fluorescence of each state).
To determine whether our observations are affected by protein association, we measured refolding of the ARR variant at different protein concentrations ranging from 3.2 to 13.5 μM (at a final GuHCl concentration of ∼0.8–1.6 M). There was no systematic variation in rates or amplitudes as a function of protein concentration over this range, suggesting that the observed folding kinetics reflects the behavior of monomeric ovPrP, although formally we cannot exclude the possibility that the concentration required for multimeric ovPrP dissociation is lower than 3.2 μM.
Kinetic modeling
Several factors might account for the unusual observation of single-exponential kinetics during refolding despite the presence of an intermediate deduced from the more complex time-course of unfolding, including i), the intermediate is poorly populated under refolding conditions; ii), the second refolding phase occurs too fast to be observed; and iii), the second refolding phase has negligible amplitude. In an effort to discriminate between these possibilities, we used the kinetic parameters derived by fitting a three-state model to the chevron plots (Fig. 2 E and F) to predict the family of kinetic traces as a function of GuHCl concentration (solid lines in Fig. S6). We were able to simultaneously fit all of the observed folding and unfolding traces for a given variant with a fixed set of rate parameters (Table 1) after least-squares optimization of the relative fluorescence amplitudes of U, I, and N states (fU, fI, and fN) and their dependence on denaturant concentration (assumed to be linear). Using the ARQ variant as an example, a preliminary analysis of individual folding/unfolding traces at different GuHCl concentrations gave values of fU = 0.83 ± 0.03 and fI ∼ fN = 0.40 ± 0.015, assuming a constant value of 0.04 for the slope versus GuHCl concentration (the results for ARR were similar; see Table 1). Thus, after allowing for a small amount (<4%) of instrumental variation between individual experiments, we can simultaneously fit the kinetic data using a common set of global kinetic parameters and consistent optical properties for the three states. Our ability to simultaneously fit the raw kinetic traces (Fig. S6) and the empirically determined chevron plots (Fig. 2, E and F) confirms the validity of the kinetic model and provides further confidence in the derived elementary rate parameters (our kinetic modeling procedures involving a combination of least-squares and manual optimization does not lend itself readily to quantitative error estimation).
Because the I- and N-states are indistinguishable in terms of their fluorescence properties accounts for the absence of a fast transient in refolding experiments and the observation of an apparent lag phase in unfolding (reflecting the delay in the appearance of the U-state due to transient accumulation of I). Although our kinetic evidence for accumulation of an intermediate is based on the unfolding data, the elementary rate constants we obtained by fitting the data to a three-state model (Table 1) predict that the I-state also accumulates under refolding conditions (reaching a maximum at a folding time of ∼200 μs; data not shown) and maintains substantial equilibrium population near the midpoint of the unfolding transition. This is illustrated in Fig. 4 (panels A and B), which shows plots of the equilibrium population (solid lines) and maximal transient population of the I-state (dashed lines) versus GuHCl concentration, calculated on the basis of the kinetic parameters in Table 1. Interestingly, the predicted transient accumulation of the I-state during folding is substantially higher for the ARQ variant (∼21%) than the ARR variant (6.5%). This conclusion is consistent with the statistical analysis of the kinetic traces (Table S2), which reveals more pronounced deviations from single-exponential behavior during unfolding of the ARQ variant compared to ARR. The ARQ variant also shows a somewhat higher population of I at equilibrium (16.7%) compared to the ARR variant (12.9%). The free energy profiles in Fig. 4 (panels C and D) illustrate the effect of denaturant on the relative populations of the three states. Comparison of the profiles for the two variants in the absence of denaturant (Fig. 4 E) explains the difference in I-state population in terms of a lower barrier for the U → I transition (TS1), deeper I-state well, and higher I → N barrier (TS2) for the ARQ variant compared to ARR. TS1 is the dominant barrier during folding of ARR, and λslow approaches kUI at low denaturant concentrations (Fig. 2 F). However, in the case of ARQ the two barriers are more similar in height, and both transitions contribute to the observed folding rate, λslow, which falls below kUI and decreases sharply with increasing GuHCl concentration (Fig. 2 E).
Figure 4.
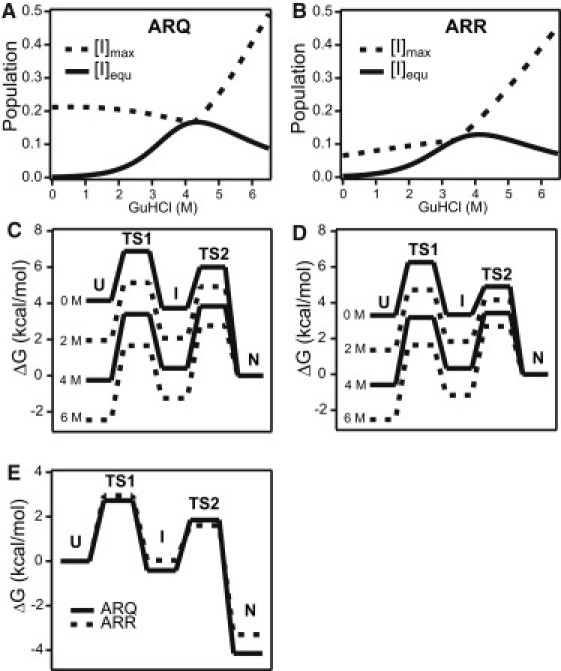
Predicted populations and free energy profiles obtained by three-state analysis of kinetic data for the ARQ (A and C) and ARR (B and D) variants of ovPrP. In panels A and B, the elementary rate constants in Table 1 were used to compute the equilibrium population (solid lines) and maximum level of transient accumulation of the I-state on a 100-μs time (dashed lines). The free energy profiles in panels C and D were calculated using the Arrhenius relationship, assuming a preexponential factor of 1 × 106 s−1. Panel E compares the free energy profiles for three-state folding of the ARQ (solid) and ARR (dashed) variants in the absence of denaturant.
Discussion
The goal of this study was to identify any intermediate conformational state(s) that occur during folding and unfolding of the monomeric ovPrPC to understand their potential role as precursors for the formation of pathogenic isoform, PrPSc. It is generally thought that the subunits of PrPSc are highly ordered, but structurally quite distinct from PrPC (37–39). The (monomeric) precursor to PrPSc is likely to be a transient state with a partially unfolded and/or nonnative structure predisposed to interact with PrPSc particles. Thus, elucidating the structural and energetic properties of partially ordered conformational states occurring during unimolecular folding may be critical for understanding the PrPC-to-PrPSc conversion. Two factors should contribute to the rate of PrPSc formation: the population of the monomeric precursor and its propensity to adopt the PrPSc conformation. On the basis of these considerations, we hypothesized that genetic modulations in classical scrapie determine either the population or structural properties (or both) of the precursor state to modulate the propensity of PrPC to adopt the PrPSc conformation, which has been observed to correlate with disease susceptibility (23,24).
Kinetic intermediate is a plausible PrPSc precursor
In this study we observed that the population of the I-state during folding of the susceptible ARQ variant is about three times higher than that of the resistant ARR variant (Fig. 4). Our observation that the ARQ and ARR variants have similar αUI-values (0.25 and 0.23, respectively; Table 1) suggests that their intermediates are structurally similar; in each case the I-state buries ∼1/4th of the solvent accessible surface area associated with refolding. On the basis of these observations, we argue that I is a likely precursor of PrPSc, and that the genetic modulation in classical scrapie exerts its effect primarily via the population of this kinetic species rather than its structural properties.
We propose the following model of amyloid formation in classical scrapie:
Conformational folding of PrPC: N ↔ I ↔ U
Assembly of PrPSc oligomers: (PrPSc)n + I → (PrPSc)n+1
The rate of adding a subunit to (PrPSc)n is a function of the concentration of the monomeric I-state in solution as well as that of the preexisting PrPSc seed. Thus, variants with well-populated I-states are predicted to be more susceptible to scrapie, and those with poorly populated intermediates are expected to be resistant. Exogenous PrPSc is required for the PrPC-to-PrPSc conversion, since naturally occurring allelic variants of ovPrP, including those that confer high susceptibility to classical scrapie, do not induce the disease spontaneously (40,41).
Sequential versus parallel folding mechanisms
A kinetic model with parallel pathways is often seen in protein refolding due to heterogeneity in the unfolded state, e.g., different isomers of an X-Pro peptide bond (42). However, parallel pathways are less likely to occur in protein unfolding, because the native protein population is generally more homogeneous than the U-state. Indeed, our kinetic simulations show that a three-state mechanism with an obligatory intermediate encountered in both folding and unfolding transitions quantitatively accounts for all experimental data (Fig. 2). The observation of a lag phase further supports our conclusion that the I-state is an obligatory intermediate (43,44). However, a rigorous distinction between these alternative folding models can be made only in rare cases (43,45), and we have not fully explored all possible alternatives (such as triangular schemes) in this study. Nevertheless, the I-state in ovPrP is a strong candidate for an on-pathway intermediate, because the two observable unfolding phases converge at high denaturant concentration, resulting in tight kinetic coupling, which is a key requirement for distinguishing alternative mechanisms (34).
Arguments against a native-state precursor
It has been suggested that native PrPC may act as a precursor to PrPSc under physiological conditions. Zhang et al. (46) proposed that Syrian hamster PrPC possesses a relatively open conformation, which is ready for the PrPC to PrPSc conversion. Welker et al. (47) suggested that oligomerization may proceed through a thiol/disulfide exchange reaction between two native PrP molecules. Although such a dimeric structure has been observed (48), a later report on in vitro conversion argued against this model (49). Direct conversion of the native PrPC structure into PrPSc is also inconsistent with the observation that mutants of human PrP (huPrP) linked to prion diseases do not differ significantly from the wild-type in terms of thermodynamic stability (50–52). In the case of classical scrapie, susceptible alleles even exhibit higher structural stability of their native forms as compared to the resistant ones (26,27). Moreover, pathogenic or protective mutations associated with human or sheep prion diseases do not alter the structure or backbone dynamics of the native state (52,53). Taken together, these studies suggest that genetic modulations associated with prion diseases do not exert their effects on the native form. On the other hand, mutations can strongly modulate the structure, stability, and kinetics of formation/decay of folding intermediates (54,55), which therefore are more plausible PrPSc precursor states.
Species difference in the folding kinetics of ovPrP
At low denaturant concentration the ovPrP variants we studied here refold in a single phase with a time constant of ∼100–300 μs, without resolvable kinetic intermediates. In contrast, in a previous kinetic study of huPrP at 5°C, an early intermediate was found to accumulate with a time constant of ∼50 μs during refolding from the urea-denatured state at pH 4.8 and 7, followed by a rate-determining folding step with a time constant of ∼700 μs (29). Instrumentation plays no role when comparing the present data with the previous huPrP study (29), because the same capillary mixing system was used for both experiments, although with improved detector sensitivity in the current study.
Our quantitative kinetic analysis of the combined folding and unfolding data for the ARQ and ARR variants of ovPrP (Fig. 2) shows that an intermediate accumulates over a wide range of denaturant concentrations, but is not observable in refolding experiments because its fluorescence properties are indistinguishable from those of the N-state. Thus, the apparent differences in kinetic behavior are in part due to species-dependent variation in the properties of the optical probe used to monitor the reaction. In addition, the free energy profiles for ovPrP (Fig. 4) show that, in contrast to huPrP (29), the first barrier encountered (TS1) is rate-limiting during refolding in the absence of denaturant, whereas the second barrier (TS2) becomes rate-limiting at intermediate and high GuHCl concentrations. Thus, relatively small changes in the folding free-energy landscape can account for qualitative differences in kinetic mechanisms. It is likely that the differences in energetic, kinetic, and optical properties of the sheep versus human proteins reflect structural differences in their respective intermediates. Such species differences have been observed for the intermediate in human and Syrian hamster PrP (56).
Structural correlations in PrPSc formation
The PrPSc conformation has been modeled in terms of β-helical and spiral structures where much of the native structure is conserved during the PrPC-to-PrPSc conversion (37,38). Such structural conservation has been observed in an antibody study, in which α-helices 2 and 3 in PrPC remain largely conserved in PrPSc (57). If the interactions in PrPSc mimicked those in PrPC, i.e., if PrPSc has a native-like structure, a kinetic species that retains more native interactions would exhibit higher propensity to convert to PrPSc. Therefore, these structural models suggest that a partially unfolded folding intermediate is a more plausible PrPSc precursor, as compared to the native and unfolded states, since it retains native-like structure (58), but is flexible to undergo conversion. An alternative mechanisms involving refolding of the entire α-helical domain in the transition from PrPC to PrPSc has also been proposed (39,59). In this case, a weakly structured species would be a more plausible PrPSc precursor, which is consistent with the relatively low degree of desolvation of the I-state, based on the αUI we observed (Table 1). However, the precise relation of the structural properties of I to the PrPC-to-PrPSc conversion cannot be determined in the absence of a high-resolution structural model of PrPSc.
Conclusions
We conclude that the structural intermediate, I, observed in our kinetic studies on ovPrP is a likely candidate as a precursor in its conversion from the monomeric cellular form to scrapie particles. The level to which the I-state accumulates transiently and/or at equilibrium is a key factor in determining the efficiency of conversion. This argument is based on the observed correlation between the scrapie susceptibility of the ARQ and ARR variants and I-state population. The higher population of I in the susceptible variant would strongly favor oligomerization. Such a correlation has been observed in human familial prion disease, where pathogenic mutations result in a pronounced increase in the population and hydrophobicity of a kinetic intermediate of human PrP (29). Differences between the folding kinetics of the sheep PrP variants studied here and those reported previously for the human protein (29) can be explained by their distinct optical properties together with (subtle) structural and kinetic differences involving the respective intermediates.
Acknowledgments
We thank the Departments of Physiology and Biochemistry of Michigan State University for their support and Terry Ball for his help with analytical ultracentrifugation.
This work was supported by a grant from the National Science Foundation (MCB 0744607) and the National Institutes of Health (GM056250) to H.R., and a grant from the National Cancer Institute (P30 CA06927) and an Appropriation by the Commonwealth of Pennsylvania to the Fox Chase Cancer Center. K.C.C. was supported by a fellowship from The Lim Pen-Yuan Cultural and Educational Foundation (Taiwan), a thesis-completion fellowship from Michigan State University (MSU), and start-up funds awarded to W.J.W.
Footnotes
Kai-Chun Chen's present address is Department of Neurology, University of Michigan Health System, Ann Arbor, MI.
Contributor Information
William J. Wedemeyer, Email: weeds@alum.mit.edu.
Heinrich Roder, Email: roder@fccc.edu.
Supporting Material
References
- 1.Prusiner S.B. Prions. Proc. Natl. Acad. Sci. USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner S.B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 3.Griffith J.S. Self-replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 4.Aguzzi A., Calella A.M. Prions: protein aggregation and infectious diseases. Physiol. Rev. 2009;89:1105–1152. doi: 10.1152/physrev.00006.2009. [DOI] [PubMed] [Google Scholar]
- 5.Brown D.R., Qin K., Kretzschmar H. The cellular prion protein binds copper in vivo. Nature. 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 6.Wadsworth J.D., Hill A.F., Collinge J. Strain-specific prion-protein conformation determined by metal ions. Nat. Cell Biol. 1999;1:55–59. doi: 10.1038/9030. [DOI] [PubMed] [Google Scholar]
- 7.Stahl N., Borchelt D.R., Prusiner S.B. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 8.Riek R., Hornemann S., Wüthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23-231) FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 9.Donne D.G., Viles J.H., Dyson H.J. Structure of the recombinant full-length hamster prion protein PrP(29-231): the N terminus is highly flexible. Proc. Natl. Acad. Sci. USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zahn R., Liu A., Wüthrich K. NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. USA. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haire L.F., Whyte S.M., Bayley P.M. The crystal structure of the globular domain of sheep prion protein. J. Mol. Biol. 2004;336:1175–1183. doi: 10.1016/j.jmb.2003.12.059. [DOI] [PubMed] [Google Scholar]
- 12.Caughey B.W., Dong A., Caughey W.S. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 13.Gasset M., Baldwin M.A., Prusiner S.B. Perturbation of the secondary structure of the scrapie prion protein under conditions that alter infectivity. Proc. Natl. Acad. Sci. USA. 1993;90:1–5. doi: 10.1073/pnas.90.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan K.M., Baldwin M., Cohen F.E. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldmann W., Hunter N., Hope J. Two alleles of a neural protein gene linked to scrapie in sheep. Proc. Natl. Acad. Sci. USA. 1990;87:2476–2480. doi: 10.1073/pnas.87.7.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belt P.B., Muileman I.H., Smits M.A. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J. Gen. Virol. 1995;76:509–517. doi: 10.1099/0022-1317-76-3-509. [DOI] [PubMed] [Google Scholar]
- 17.Tranulis M.A., Osland A., Ulvund M.J. Prion protein gene polymorphisms in sheep with natural scrapie and healthy controls in Norway. J. Gen. Virol. 1999;80:1073–1077. doi: 10.1099/0022-1317-80-4-1073. [DOI] [PubMed] [Google Scholar]
- 18.Elsen J.M., Amigues Y., Laplanche J.L. Genetic susceptibility and transmission factors in scrapie: detailed analysis of an epidemic in a closed flock of Romanov. Arch. Virol. 1999;144:431–445. doi: 10.1007/s007050050516. [DOI] [PubMed] [Google Scholar]
- 19.Moudjou M., Treguer E., Laude H. Glycan-controlled epitopes of prion protein include a major determinant of susceptibility to sheep scrapie. J. Virol. 2004;78:9270–9276. doi: 10.1128/JVI.78.17.9270-9276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thackray A.M., Yang S., Bujdoso R. Conformational variation between allelic variants of cell-surface ovine prion protein. Biochem. J. 2004;381:221–229. doi: 10.1042/BJ20040351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenzer S., Stoltze L., Schild H. Quantitative analysis of prion-protein degradation by constitutive and immuno-20S proteasomes indicates differences correlated with disease susceptibility. J. Immunol. 2004;172:1083–1091. doi: 10.4049/jimmunol.172.2.1083. [DOI] [PubMed] [Google Scholar]
- 22.Thackray A.M., Fitzmaurice T.J., Bujdoso R. Ovine plasma prion protein levels show genotypic variation detected by C-terminal epitopes not exposed in cell-surface PrPC. Biochem. J. 2006;400:349–358. doi: 10.1042/BJ20060746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bossers A., de Vries R., Smits M.A. Susceptibility of sheep for scrapie as assessed by in vitro conversion of nine naturally occurring variants of PrP. J. Virol. 2000;74:1407–1414. doi: 10.1128/jvi.74.3.1407-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabuncu E., Petit S., Vilette D. PrP polymorphisms tightly control sheep prion replication in cultured cells. J. Virol. 2003;77:2696–2700. doi: 10.1128/JVI.77.4.2696-2700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly J.W. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr. Opin. Struct. Biol. 1998;8:101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]
- 26.Rezaei H., Choiset Y., Haertle T. Amyloidogenic unfolding intermediates differentiate sheep prion protein variants. J. Mol. Biol. 2002;322:799–814. doi: 10.1016/s0022-2836(02)00856-2. [DOI] [PubMed] [Google Scholar]
- 27.Fitzmaurice T.J., Burke D.F., Bujdoso R. The stability and aggregation of ovine prion protein associated with classical and atypical scrapie correlates with the ease of unwinding of helix-2. Biochem. J. 2008;409:367–375. doi: 10.1042/BJ20071122. [DOI] [PubMed] [Google Scholar]
- 28.Apetri A.C., Surewicz K., Surewicz W.K. The effect of disease-associated mutations on the folding pathway of human prion protein. J. Biol. Chem. 2004;279:18008–18014. doi: 10.1074/jbc.M313581200. [DOI] [PubMed] [Google Scholar]
- 29.Apetri A.C., Maki K., Surewicz W.K. Early intermediate in human prion protein folding as evidenced by ultrarapid mixing experiments. J. Am. Chem. Soc. 2006;128:11673–11678. doi: 10.1021/ja063880b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasteiger E., Gattiker A., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shastry M.C., Luck S.D., Roder H. A continuous-flow capillary mixing method to monitor reactions on the microsecond time scale. Biophys. J. 1998;74:2714–2721. doi: 10.1016/S0006-3495(98)77977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterman B.F. Measurement of the dead time of a fluorescence stopped-flow instrument. Anal. Biochem. 1979;93:442–444. doi: 10.1016/s0003-2697(79)80176-1. [DOI] [PubMed] [Google Scholar]
- 33.Berberan-Santos M.N., Martinho J.M.G. The integration of kinetic rate equations by matrix methods. J. Chem. Educ. 1990;67:375–379. [Google Scholar]
- 34.Roder H., Maki K., Cheng H. Early events in protein folding explored by rapid mixing methods. Chem. Rev. 2006;106:1836–1861. doi: 10.1021/cr040430y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen B.L., Baase W.A., Schellman J.A. Folding kinetics of T4 lysozyme and nine mutants at 12 degrees C. Biochemistry. 1992;31:1464–1476. doi: 10.1021/bi00120a025. [DOI] [PubMed] [Google Scholar]
- 36.Apetri A.C., Surewicz W.K. Kinetic intermediate in the folding of human prion protein. J. Biol. Chem. 2002;277:44589–44592. doi: 10.1074/jbc.C200507200. [DOI] [PubMed] [Google Scholar]
- 37.DeMarco M.L., Daggett V. From conversion to aggregation: protofibril formation of the prion protein. Proc. Natl. Acad. Sci. USA. 2004;101:2293–2298. doi: 10.1073/pnas.0307178101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Govaerts C., Wille H., Cohen F.E. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc. Natl. Acad. Sci. USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cobb N.J., Sönnichsen F.D., Surewicz W.K. Molecular architecture of human prion protein amyloid: a parallel, in-register beta-structure. Proc. Natl. Acad. Sci. USA. 2007;104:18946–18951. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bossers A., Harders F.L., Smits M.A. PrP genotype frequencies of the most dominant sheep breed in a country free from scrapie. Arch. Virol. 1999;144:829–834. doi: 10.1007/s007050050548. [DOI] [PubMed] [Google Scholar]
- 41.Hunter N., Cairns D., Donnelly K. Is scrapie solely a genetic disease? Nature. 1997;386:137. doi: 10.1038/386137a0. [DOI] [PubMed] [Google Scholar]
- 42.Wedemeyer W.J., Welker E., Scheraga H.A. Proline cis-trans isomerization and protein folding. Biochemistry. 2002;41:14637–14644. doi: 10.1021/bi020574b. [DOI] [PubMed] [Google Scholar]
- 43.Walkenhorst W.F., Green S.M., Roder H. Kinetic evidence for folding and unfolding intermediates in staphylococcal nuclease. Biochemistry. 1997;36:5795–5805. doi: 10.1021/bi9700476. [DOI] [PubMed] [Google Scholar]
- 44.Sánchez I.E., Kiefhaber T. Evidence for sequential barriers and obligatory intermediates in apparent two-state protein folding. J. Mol. Biol. 2003;325:367–376. doi: 10.1016/s0022-2836(02)01230-5. [DOI] [PubMed] [Google Scholar]
- 45.Capaldi A.P., Shastry M.C., Radford S.E. Ultrarapid mixing experiments reveal that Im7 folds via an on-pathway intermediate. Nat. Struct. Biol. 2001;8:68–72. doi: 10.1038/83074. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H., Stockel J., Cohen F.E. Physical studies of conformational plasticity in a recombinant prion protein. Biochemistry. 1997;36:3543–3553. doi: 10.1021/bi961965r. [DOI] [PubMed] [Google Scholar]
- 47.Welker E., Wedemeyer W.J., Scheraga H.A. A role for intermolecular disulfide bonds in prion diseases? Proc. Natl. Acad. Sci. USA. 2001;98:4334–4336. doi: 10.1073/pnas.071066598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knaus K.J., Morillas M., Yee V.C. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat. Struct. Biol. 2001;8:770–774. doi: 10.1038/nsb0901-770. [DOI] [PubMed] [Google Scholar]
- 49.Welker E., Raymond L.D., Caughey B. Intramolecular versus intermolecular disulfide bonds in prion proteins. J. Biol. Chem. 2002;277:33477–33481. doi: 10.1074/jbc.M204273200. [DOI] [PubMed] [Google Scholar]
- 50.Swietnicki W., Petersen R.B., Surewicz W.K. Familial mutations and the thermodynamic stability of the recombinant human prion protein. J. Biol. Chem. 1998;273:31048–31052. doi: 10.1074/jbc.273.47.31048. [DOI] [PubMed] [Google Scholar]
- 51.Liemann S., Glockshuber R. Influence of amino acid substitutions related to inherited human prion diseases on the thermodynamic stability of the cellular prion protein. Biochemistry. 1999;38:3258–3267. doi: 10.1021/bi982714g. [DOI] [PubMed] [Google Scholar]
- 52.Hosszu L.L., Jackson G.S., Collinge J. The residue 129 polymorphism in human prion protein does not confer susceptibility to Creutzfeldt-Jakob disease by altering the structure or global stability of PrPC. J. Biol. Chem. 2004;279:28515–28521. doi: 10.1074/jbc.M313762200. [DOI] [PubMed] [Google Scholar]
- 53.Bae S.H., Legname G., Dyson H.J. Prion proteins with pathogenic and protective mutations show similar structure and dynamics. Biochemistry. 2009;48:8120–8128. doi: 10.1021/bi900923b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khorasanizadeh S., Peters I.D., Roder H. Evidence for a three-state model of protein folding from kinetic analysis of ubiquitin variants with altered core residues. Nat. Struct. Biol. 1996;3:193–205. doi: 10.1038/nsb0296-193. [DOI] [PubMed] [Google Scholar]
- 55.Capaldi A.P., Kleanthous C., Radford S.E. Im7 folding mechanism: misfolding on a path to the native state. Nat. Struct. Biol. 2002;9:209–216. doi: 10.1038/nsb757. [DOI] [PubMed] [Google Scholar]
- 56.Kremer W., Kachel N., Kalbitzer H.R. Species-specific differences in the intermediate states of human and Syrian hamster prion protein detected by high pressure NMR spectroscopy. J. Biol. Chem. 2007;282:22689–22698. doi: 10.1074/jbc.M701884200. [DOI] [PubMed] [Google Scholar]
- 57.Eghiaian F., Grosclaude J., Knossow M. Insight into the PrPC—>PrPSc conversion from the structures of antibody-bound ovine prion scrapie-susceptibility variants. Proc. Natl. Acad. Sci. USA. 2004;101:10254–10259. doi: 10.1073/pnas.0400014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roder H., Colón W. Kinetic role of early intermediates in protein folding. Curr. Opin. Struct. Biol. 1997;7:15–28. doi: 10.1016/s0959-440x(97)80004-8. [DOI] [PubMed] [Google Scholar]
- 59.Lu X., Wintrode P.L., Surewicz W.K. Beta-sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange. Proc. Natl. Acad. Sci. USA. 2007;104:1510–1515. doi: 10.1073/pnas.0608447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


