Abstract
The influenza A M2 protein exhibits inwardly rectifying, pH-activated proton transport that saturates at low pH. A comparison of high-resolution structures of the transmembrane domain at high and low pH suggests that pH-dependent conformational changes may facilitate proton conduction by alternately changing the accessibility of the N-terminal and C-terminal regions of the channel as a proton transits through the transmembrane domain. Here, we show that M2 functionally reconstituted in liposomes populates at least three different conformational states over a physiologically relevant pH range, with transition midpoints that are consistent with previously reported His37 pKas. We then develop and test two similar, quantitative mechanistic models of proton transport, where protonation shifts the equilibrium between structural states having different proton affinities and solvent accessibilities. The models account well for a collection of experimental data sets over a wide range of pHs and voltages and require only a small number of adjustable parameters to accurately describe the data. While the kinetic models do not require any specific conformation for the protein, they nevertheless are consistent with a large body of structural information based on high-resolution NMR and crystallographic structures, optical spectroscopy, and MD calculations.
The influenza virus infects cells by receptor-mediated endocytosis, which is followed by acid-induced fusion of the viral and endosomal membranes. The fusion event is mediated by a conformational change of the influenza hemagglutinin proteins, triggered by the low pH in the endosome lumen (1). It is also necessary, upon fusion, that the viral interior be acidified in order for the viral RNA to be freed in a competent form into the cell cytoplasm (2). Viral acidification within the time of endosomal residence requires a proton transport facilitator denoted “M2” (matrix protein 2), a 97 residue homotetrameric integral membrane protein (3). In addition, it has been shown that influenza strains with higher pH of transition of hemagglutinin from the pre-fusion to the fusion-competent state also require proton transport through M2 when newly synthesized viral proteins are trafficked in the Golgi apparatus (4). At this point in the infection cycle, M2 transports protons out of the slightly acidic Golgi lumen to protect the hemagglutinins from premature transition into the fusion-competent state. The proton transport function of M2 is inhibited by the antiviral compounds amantadine and rimantadine (5), which have been mainstays of influenza treatment for over three decades. However, in recent years, many circulating influenza A virus strains have developed resistance to amantadine and its derivatives (6), prompting renewed interest in the structure and function of the essential M2 protein in the hope of designing alternative inhibitors (see e.g. (7,8)).
The goal of the present work is to understand how M2’s complex proton-dependent conformational transitions contribute to its proton flux activity. M2 is one of the smallest membrane proteins known to mediate ion transport, yet well-defined segments of its extra-membrane sequence have been implicated in other roles in the viral life cycle, indicative of a multi-functional, modular protein (9). Although the current work focuses exclusively on the full-length protein, a very small peptide corresponding to the M2 transmembrane segment was recently shown to reproduce the ion transport, drug sensitivity and tetramer assembly properties of full length M2 (10). This finding paved the way for detailed mechanistic correlation of whole-cell electrophysiology studies of the full-length protein with results of structural and biophysical experiments involving the transmembrane segment.
Extensive electrophysiological characterization (11–20) has established that His37 protonation is part of the conduction process and is responsible for the extreme proton selectivity observed for M2 (21, 22), while Trp41 is required for inward rectification of current (18). Protonation of His37 is important for “activation” of the channel, having a pKa near 6. However, the mechanism of “gating” and conduction through the channel remains to be fully elucidated. There is now general agreement (10, 15, 22–24) that M2 conducts on the order of 1 to 1000 protons per second under conditions of an outer pH (pHout) between 7 and 5, pHin = 7, and an electrical gradient of 0 to 130 mV driving inward proton flux. These rates are far lower than those of potassium or other cation channels that transport ions, under conditions in which their conducted ions are at physiological concentrations, typically 0.15 M. As discussed by Decoursey (25) and Zhou (26), the difference in absolute rate between M2 and traditional typical potassium channels does not imply that protons necessarily need to overcome a large energy barrier while traveling through the M2 channel. In fact, within the physiological pH range of pHout 7 to 5, the overall second order rate constants for proton entry and conduction through M2 are 107 to 109 M−1 sec−1, which are close to the diffusion controlled limit for a proton diffusing through a pore of the dimensions of the M2 channel. Thus, the slow rate of conduction does not necessarily reflect the presence of large energy barriers within the channel, but rather is a simple consequence of the very low proton concentration over the physiological pH range. Therefore, the channel operates within a couple orders of magnitude of the maximal rate possible at the endosomal pH range.
A particularly interesting feature of M2 is that the rate of proton flux through the channel does not scale linearly with the proton concentration when the pH is lowered below the physiologic, but instead levels as the pH is decreased below 5 (11, 27). This behavior contrasts with that of simple cation-selective channels such as gramicidin or model peptides that lack ionizable side chains; with a modest electrical potential these channels have an extrapolated conductance of 100 protons/sec at pH 6 (10−6 M hydronium), and 108 protons/sec at pH 1.0 (0.1 M hydronium ion concentration) (25). A variety of mechanisms can be devised to account for saturation, based on classical channel-like or site-binding models. In a classical channel-gating model (28), the pH-dependent activation of M2 conduction would be associated with protonation of the third His37 residue of the tetrad (pKa approximately 6 (29)), explaining the activation of the channel as the pH is lowered between pH 7 and 5. The saturation of the rate at still lower pHout can occur when the net rate of entry of protons into the channel, which is dependent on proton concentration, is much more rapid than the subsequent transit through the channel, which in the simplest case is independent of the bulk proton concentration. This scenario, which is functionally equivalent to a saturable binding isotherm, leads to accumulation of one or more protons in the pore impeding further flux and accounting for the leveling of the flux at low pHout.
A related model seeks to explain the conductance of the channel in terms of protonation/deprotonation of His37, which serves to shuttle protons through the channel (14, 15). This model has been expanded to include structural and dynamic details as crystallographic, NMR, and spectroscopic studies have become available (26). ssNMR studies of the M2 TM domain in bilayers (29) have shown that the pKa values for successive protonations of His37 in the tetramer (designated +0 through +4) are 8.2, 8.2, 6.3, and ≤ 5; equilibrium studies of full-length M2 in micelles were consistent with these values (10). Moreover, a number of NMR studies of M2 in micelles and bilayers have shown that the backbone conformation of M2 is reasonably well defined in the +0 through +2 states, but becomes more dynamic upon addition of the third proton, with a pKa of 6.3 (30, 31). Conformational dynamics involving both the mainchain amides and the Trp41 side chain occur on the microsecond through millisecond time scale in the +3 state (31). Several important points need to be emphasized from these studies: The fact that the +3 state is seen to be in dynamic equilibrium indicates that there is a relatively small energy difference between the different conformational states (at most a few kcal/mol), and this energetic difference is well within the energetic variation expected for different environments (e.g., crystals, micelles). Thus, high-resolution structures should be viewed as members of a low-energy ensemble, rather than specifically identified with a given protonation state. Secondly, the observed dynamics indicate that the protein undergoes large-scale conformational fluctuations on the same timescale as the rate of proton transport. Finally, various high-resolution structures (31–33) and molecular dynamics calculations (34, 35) have shown that the protein has distinct conformational states that differ with respect to size of the opening of the pore at either ends of the bundle, which dilate and contract in a reciprocal manner.
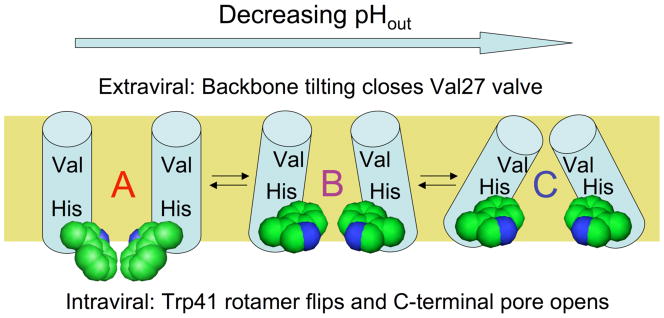
Structure-based models suggest that M2 exists in an equilibrium between conformers that are more open at the N-terminal entryway and C-terminal exit of the channel, encouraging diffusion in and out of the channel (26,34). An analysis of available high-resolution structures (33) and molecular dynamics studies (34) suggests the presence of several conformational variants at equilibrium, described in this work as states “A,” “B,” and “C” (Fig. 1). Here, state “A” resembles the high pH solution NMR structure (31), and states “B” and “C” represent models from intermediate (32, 33) and low pH crystal structures (32). Molecular dynamics simulations suggest that the populations of M2 conformations are sensitive to the state of protonation of His37 (34), and NMR studies show that they are able to change on the same time-scale as proton transit through the channel (31). In the His-relay model (14), the third proton is expected to bind to the His37 tetrad with each turnover of the channel. The saturation of rate occurs at low pH when the site is fully occupied, and is limited by the off-rate for deprotonation. More recent crystallographic studies suggest that this site might instead be considered a cluster of His sidechains and water molecules (33). In either case, the rate of deprotonation of a site with a pKa is given by the ratio of rate constants for deprotonation and protonation, placing an upper limit of 103 sec−1 for a pKa = 6 and a second-order rate constant of 109 M−1sec−1 for diffusion of a proton into a water-filled pore (25). The actual rate will depend on the fractional population of different conformational states and the proton permeabilities of the N-terminal and C-terminal sections of their pores.
Figure 1.
Proposed trajectory of structural transitions in the M2 transmembrane domain as a function of extraviral pH. The “A” conformation is based on an NMR structure in micelles at high pH (31), the intermediate “B” conformation is proposed from molecular dynamics experiments on M2 protonation states (34) and intermediate pH crystal forms of the tetramer (33), while the “C” conformation is based on a crystal structure of M2 at low pH (32). The helix bundle is depicted side-on with the front two helices removed. The proposed conformational changes upon lowering N-terminal pH involve closure of the N-terminal Val27 valve by increasing helix tilt at the top of the bundle, while at the C-terminal end of the bundle the Trp41 sidechains (space filling representation) undergo a rotamer shift, then separate as a result of increased helix tilt. These changes control exposure of the proton-binding His37 sidechains near the middle of the bundle to N-terminal and C-terminal pore waters.
Previous attempts to model the highly asymmetric conductance of M2 using a simple single-site model without the possibility of conformational changes resulted in a good fit for most experimental conditions, but either did not consider (26) or failed to explain the low conductance when protons flow in the reverse direction with low pHin and high pHout (36). We therefore wished to examine whether the structural changes, such as those observed in recent NMR and crystallographic structures obtained in detergent micelles (31–33), might better explain the extensive conductance-voltage curves available for the protein (11, 12). It was first important to ascertain that the pKa values previously determined from ssNMR for the TM domain were consistent with the behavior of the full-length protein in a native-like, bilayer environment where proton conduction through M2 could be demonstrated. We therefore conducted measurements of the solvent accessibility of the critical Trp41 residue of full-length A/M2 as a function of pH when the protein was functionally reconstituted in liposomes. The results were fully in accord with the earlier study of the TM peptide that measured His37 pKa values (29) and suggest that the solvent accessibility of Trp41 in the context of functional, full length A/M2 changes with the His37 tetrad protonation state. These pKa values, now validated for full-length, functionally reconstituted protein, provided important experimental input to help constrain fitting of additional parameters necessary to fully define the conductance model. Additionally, the parallel changes of Trp41 solvent accessibility and His37 tetrad protonation suggest that the TM domain of M2 may undergo distinct conformational transitions in bilayers as a function of His37 tetrad protonation state, consistent with the differing conformations seen in high-resolution micelle structures of the TM domain (31–33) obtained at different pH values.
Having constrained the model with experimental parameters, we examined scenarios in which proton dissociation versus conformational changes were modeled to be rate-determining, and found the best agreement to experimentally determined flux values when deprotonation became at least partially rate-determining at low pH. While the results of the fitting do not require any specific structural model, they are consistent with a body of structural data and MD calculations of the M2 proton channel.
Materials and Methods
Monitoring conformational changes of M2 Trp 41 in bilayers
Stern-Volmer fluorescence titrations (37) of M2 Trp41 at varying pH were used to track pH-dependent conformational changes of M2. The WT A/Udorn/72 sequence (accession number CAD22815) has two Trp residues (at positions 15 and 41), which would have complicated spectral analysis. A synthetic gene encoding for a single-Trp, Cys-free full length Udorn A/M2 construct with a 6xHis C-terminal tag was therefore obtained with codon usage optimized for expression in E. coli (Midland Certified Reagent Company, Midland TX.). BL21 cells were transformed with the pET23D(+) plasmid containing the M2 gene using chemical or heat shock methods. The protein was expressed and purified from E. coli as previously described (10). The single-Trp variant used retained the Trp sidechain at position 41 in the transmembrane domain and had a Trp15Phe mutation to simplify data analysis without compromising transport function (10). In addition, this Cys-free construct provided the high culture yields of protein necessary for these experiments.
To enable detection of Trp 41 fluorescence, a target protein monomer:lipid ratio of 1:100 was used. POPC and POPG were purchased as presealed chloroform stocks from Avanti Polar Lipids (Alabaster, AL). Cholesterol powder (Sigma Grade, Sigma-Aldrich, St. Louis, MO) was dissolved in chloroform immediately prior to use. Lipid films (25 μmol total 4:1:2 POPC:POPG:cholesterol), made by evaporating chloroform stocks under a stream of argon, were thoroughly hydrated with ~ 400 μL of MilliQ water and 100 μL of 10x buffer (at 10x, 500 mM KxHxPO4, 100 mM NaCl, pH-adjusted to 7.0 with HCl). The actual volume of water added was adjusted so that following the subsequent addition of protein, the total volume of the sample would be 1 mL. The sample was vigorously vortexed, and the hydrated, protein-free lipid suspensions were subjected to six freeze-thaw cycles (dry ice-ethanol bath and 37°C water bath).
Full length A/M2 (Cys-free, Trp15Phe His-tagged variant as buffer-exchanged eluate from nickel column, in 50 mM HEPES pH 8, 4 mM OG, 20% v/v glycerol, which ran as a single band on a native protein gel) was next added dropwise to the experimental sample. The stock concentration of protein monomer was ~500 μM. For the control liposome sample, an equal volume of protein-free HEPES/OG/glycerol buffer was added. The mixtures were freeze-thawed an additional six times and sized to 100 nm liposome target diameter by 33+ passes through a mini-extruder (Avestin, Ottawa, Canada). To remove residual OG from the samples, the mixtures were dialyzed for 1–2 cycles in 1L of 1x buffer (50 mM KxHxPO4 pH 7.0, 10 mM NaCl) in the presence of the detergent-binding resin Amberlite XAD-4 (Supelco, Bellefonte, PA).
For experiments, 20 μL of liposome suspension was added to 2 mL of 1x buffer set at the desired pH. Measurements were performed in a 1cm × 1cm Hellma (Plainview, NY) QS fluorescence cell. 10 μL of a 4 μM ethanolic stock of the protonophore FCCP (Fluka/Sigma-Aldrich, St. Louis, MO) was added to dissipate the pH gradient. The mixture was stirred for five minutes following FCCP addition prior to the experiment. A reference cuvette similarly configured but with control liposomes added at pH ~7 was used for on-line background signal subtraction from scatter induced by the vesicles.
Experiments were performed while stirring in an Aviv (Lakewood, NJ) ATF-105 spectrofluorometer at 25°C by measuring a baseline Trp fluorescence value at the end of the five minute equilibration period, then adding 50 μL aliquots of a 5 M NaI solution (also containing 1 mM Na2S2O3 to prevent I− photooxidation) and taking measurements following the addition of every aliquot. An automated titrator (Hamilton, Reno, NV) allowed for high precision titrant addition with automated timing of sample equilibration and data acquisition. No titrant was added to the control cuvette, and the subtracted background signal was not corrected for the small dilution effect on reference signal (~10% maximum) that addition of titrant would have caused. To account for any deviation from the pre-set buffer pH as a result of adding other sample components, sample pH was measured at the conclusion of each titration and that value was used as the experimental pH.
To avoid exciting Tyr residues in the protein, fluorescence excitation was performed at 295 nm. Fluorescence emission was measured at 340 nm, near the average emission maximum of indole in both hydrophobic and polar environments.
The measurement of intrinsic Trp fluorescence in bilayers can be complicated by substantial scatter artifacts from the liposome sample (38). Scattering was minimized by installing polarizers in a cross-polarized configuration (excitation polarizer horizontal, emission polarizer vertical) and adding an emission filter that blocked light at wavelengths below 300 nm from reaching the detector. The excitation bandwidth was kept lower (2–3 nm) than the emission bandwidth (6–7 nm). As a control, quenching experiments using this configuration with model Trp-like small molecule compounds were performed (data not shown), and yielded quenching constant values consistent with those available in the literature (39, 40).
We observed that the steady-state Trp fluorescence of M2 in liposomes was somewhat unstable (and declining) as a function of time, especially at certain pH values. A variety of factors were adjusted in an attempt to account for this phenomenon, including the length and frequency of sample irradiation (to rule out photooxidation), length of temperature equilibration, as well as the presence and absence of various ionophores such as FCCP and valinomycin. None of these appeared to be causally involved. The experiment was therefore performed under very stringent and consistent timing by using the autotitrator, and the initial, unquenched fluorescence value in the data analysis was normalized to 1 using a fitted linear error parameter (see below).
Quenching Data Analysis
Fluorophores such as the indole group of tryptophan, when in their excited electronic state (which normally results in fluorescence), can instead non-radiatively transition back to their ground state through a close encounter (collision or static binding) with a quencher (37). The relative potency of the quencher or, conversely, the relative accessibility of the fluorophore being quenched, can be determined through Stern-Volmer analysis (Eq. 1), where the fluorescence lifetimes of the fluorophore in the absence of quencher (τ0) and in the presence (τ) of a certain concentration of quencher [Q] are used to calculate a quenching constant KQ:
| Eq. 1 |
Assuming that the quenching is purely collisional or dynamic (i.e. that no stable, long-lived complexes form between the quencher and the fluorophore), the fluorescence lifetime can be linearly correlated to the fluorescence intensity measured in these experiments (F): t α F, leading to Eq. 2:
| Eq. 2 |
During the titrations, we observed a time-dependent drift of the baseline that was close to linear on the timescale of the experiment. To correct for this, each quenching curve was collected under identical conditions with the addition of quencher at precisely and evenly timed intervals, and analyzed by Eq. 3:
| Eq. 3 |
Eq. 3 represents a rearranged Stern-Volmer equation, with a linear correction with slope b.. In this equation, the b[Q] term represents the time-dependent variation in baseline fluorescence, where time is represented by the quencher concentration, which, in the experimental titration, increases by the same amount at evenly spaced timepoints. In the initial analysis, Eq. 3 was fit to each quenching data set obtained at a given pH. In these fits the experimentally determined values of F0 were used (whose values in volts were close to unity); KQ and b were treated as local variables. A linear function was then fit to a plot of b vs. F0 (for all pH points) resulting from this analysis; the resulting trendline was used to calculate a value of b (−0.491) for F0 = 1. Each individual quenching curve was then re-fit with F0 globally set to 1, and b globally set to nus;0.491, with a sole local variable of KQ.
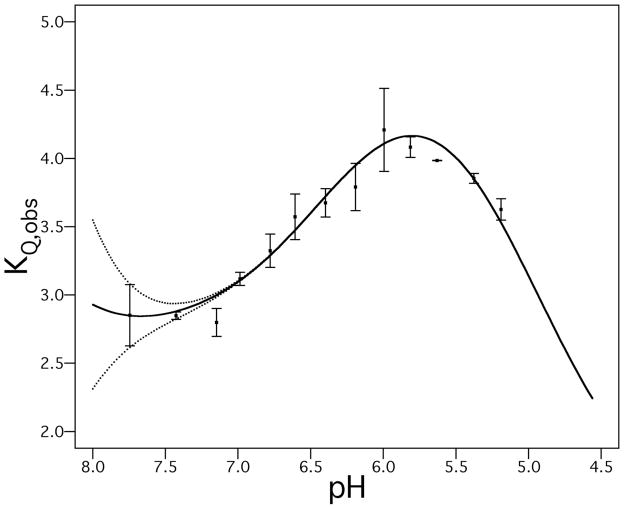
The resulting data points (KQ vs. pH) were arranged into bins spanning 0.2 pH units (representing the estimated error of ± 0.1 pH units associated with changing solvent composition during the titrations); average KQ and pH values for each bin are shown in Fig. 2, with error bars reflecting the standard error for KQ within each bin. A protonation equilibrium function, defined by Eq. 4 and Eq. 5, was fit to the unbinned data (fit function displayed as solid line). The protein was assumed to be fully tetrameric under every condition studied (10).
Figure 2.
Experimentally calculated Stern-Volmer quenching constants of M2 Trp41 as a function of pH over a physiologically encountered pH range. To provide a more robust indication of experimental error, titration data points were binned in groups spanning 0.2 pH units reflecting experimental uncertainty of ~ 0.1 pH units. Square dots represent averages of pH and KQ,obs in each bin; error bars show the standard error associated with each bin (standard deviation of the KQ,obs values within each bin divided by the square root of the number of points in the bin). The data can be deconvoluted into an ensemble of distinct conformations with pH transition midpoints constrained as experimentally observed M2 pKa values (29) (solid line). Because the fully neutral (0+) protonation state is almost completely absent at physiological pH, there is greater uncertainty as to the corresponding conformation’s quencher accessibility (dotted lines represent uncertainty limits of deconvolution fit for the neutral state). The deconvoluted KQ values and fit uncertainities for each protonation state are KQ0 3.4 ± 2.2 M-1, KQ2 2.7 ± 0.1 M-1, KQ3 5.1 ± 0.2 M-1 and KQ4 1.2 ± 0.4 M-1.
| Eq. 4 |
In Eq. 4, Kq0, Kq2, Kq3 and Kq4 represent the quenching constant of the fully deprotonated, doubly protonated, etc. species of the protein. M0, M2, M3 and M4 represent the relative population of each species. When the pKa values governing the protonation events are considered, Eq. 4 is expanded into
| Eq. 5 |
In this model, each protonation state of the tetramer has a preferred equilibrium ensemble of structures, reflected in a specific KQ determined by the intrinsic lifetime of Trp 41 and its accessibility to quencher. Therefore, the observed KQ is deconvolved into population-weighted components made up by KQ0, KQ2, KQ3, and KQ4 (with previously determined experimental pKa values used as constraints (29)). In this previous work, the first two protonation events appeared to occur over a narrow concentration range with a midpoint of 8.2. We therefore treat them as a cooperative two-proton transition. The 4th pKa was estimated to be less than 5.0 in the previous study. Similarly, we find the best fit to the experimental data (for a physically reasonable KQ4 ≥1 M−1) when this parameter is in the range of 4.9 to 5.0. In the fits shown in Fig. 2, the 4th pKa was fixed at 5.0; a very similar best-fit line and fit quality were obtained when it was set to 4.9.
Functional Reconstitution of M2 into liposomes
The Cys-free, Trp15Phe His-tagged M2 construct used for fluorescence studies was expressed as previously described (10). After elution from the nickel column, the protein was desalted, concentrated and exchanged into a final solution of 20 mM HEPES (pH 8), 4 mM OG and 20% (v/v) glycerol using centrifugal concentrators (Millipore, Billerica, MA).
The same lipid mixture of 4:1:2 POPC:POPG:cholesterol was used for functional reconstitution assays, with protein added to a 5 × 10−4 protein (tetramer):lipid mole ratio. Prior to freeze/thaw cycles to form the liposomes, the fluorescent pH indicator hydroxy-pyrene trisulfonic acid (HPTS or pyranine) was added to a concentration of 1 mM. Liposomes were reconstituted in a K+ buffer, consisting of 10 mM K2HPO4, 50 mM K2SO4 and 5 mM MOPS, pH 7.4. Samples were subjected to an additional 10 to 15 freeze/thaw cycles to promote encapsulation of dye molecules. Following extrusion to a target liposome diameter of 100 nm, liposomes were dialyzed for 48 hr against two 2 L vol. of K+ buffer to remove unencapsulated dye molecules and excess OG.
Proton Flux Assay
The measurement was done on an ISS PC1 spectrofluorometer (ISS Inc., Champaign, IL) with 10 mm excitation and 4 mm emission pathlengths and 0.5 mm slit widths for both the excitation and emission monochromators. Internal liposome pH was determined by the ratio of HPTS fluorescence intensities at 460 and 417 nm (460/417) by comparison with an HPTS pH calibration curve. 417 nm is a pH-independent isosbestic point for HPTS. For all measurements, the emission wavelength was set at 515 nm. To assess M2 activity, 10 μl of liposomes reconstituted in K+ buffer were diluted into 990 μl Na+ buffer (10 mM Na2HPO4, 50 mM Na2SO4 and 5 mM MOPS, pH 7.4). Electrochemical gradients across the liposome membranes were generated by addition of 5 μl 5 μM valinomycin. Immediately after addition of valinomycin, time traces were recorded at an excitation wavelength of 460 nm for 200 s at 10 s intervals. Immediately after collection of a time trace at 460 nm, the isosbestic point of HPTS (excitation 417 nm) was also collected for 200 s at 10 s intervals. To probe amantadine inhibition of A/M2 activity, 1 μl 100mM amantadine HCl (Sigma–Aldrich) was added to Na+ buffer and incubated for 5 min with liposomes prior to the addition of valinomycin. Control liposomes (with no A/M2 was added during the reconstitution process) were also included in the assay. Experiments on each condition were performed in duplicate.
Measurements of pH-dependent currents through M2-expressing Xenopus oocytes
Measurements of current through M2 expressed in the Xenopus laevis oocyte system at varying bathing buffer pH values were performed as previously described (41). Briefly, M2 currents were measured before and after application of 100 μM amantadine for each value of pH. Between each experimental value of pH, the oocyte was bathed in solution of pH 8.5. The amantadine-sensitive net current was calculated by subtracting residual current after amantadine inhibition from maximum currents at each value of pH before amantadine inhibition. Experiments for each condition were performed in triplicate. The resulting amantadine-sensitive current-pHout relationship was used as one of the data sets in mechanistic model fitting (vide infra).
Steady-state kinetic mechanisms of M2 conduction
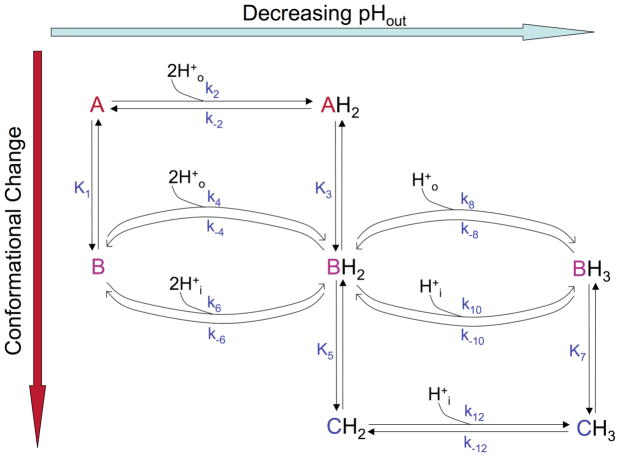
Transporter models were developed to describe flux at steady-state kinetic regimes (i.e. immediately following the imposition of a driving force for proton transport), with the concentrations of H+ inside and outside the membrane clamped at their set values for the analysis. Each model was constructed as a kinetic cycle of interconverting conformational and protonation states, with the simplest arrangement shown in Fig. S1, where each transition is defined by its own rate constant. In Fig. 3, the same scheme is shown but certain rate constant pairs found to be non-rate determining were converted into equilibrium constants for ease of presentation; a similar approach was taken with the more complex mechanism shown in Fig. 5. The derivation of a steady-state proton flux equation from the model schematics is described in detail in the Supporting Information. Briefly, a system of equations was constructed by setting the sum of the rates of formation and depletion of each possible state of the protein to zero so as to reflect steady-state kinetics. The system was solved symbolically for the normalized population of each state as a function of rate constants and proton concentrations, and a flux equation was derived by subtracting the differences of the proton release and binding rates on either side of the membrane.
Figure 3.
Two-state conduction mechanism presented as a kinetic scheme involving activation by low pHout. H+o represent extraviral (N-terminal) protons, while H+i represent intraviral (C-terminal protons). Proton binding and release occur on the horizontal axis, while conformational changes occur on the vertical axis in the scheme. The activating protons can only be bound and released externally. Minor “leak” pathways are described by k5,k−5,k7, and k−7.
Figure 5.
Augmented, multi-state proton transport model, where at least three conformations take part in the conduction cycle. Protonation and deprotonation events are shown on the horizontal axis, while conformational changes are shown on the vertical axis. The first two protons bound can now be conducted by the “A” and “B” conformations, while the 3rd proton is conducted by the “B” and “C” conformations. H+o represent extraviral (N-terminal) protons, while H+i represent intraviral (C-terminal protons). k4, k−4, k10, k−10 represent “leak” pathways.
The resulting flux equation was then globally fitted to M2 functional data from a variety of sources, including data collected by the authors on M2 expressed in Xenopus oocytes, and published experiments from Chizhmakov et al. (11, 12), where M2 was expressed in MEL cells. Global fitting was performed using the Levenberg-Marquardt algorithm implemented in Igor Pro 6.04 (42). Rate constants involving a charged species were modeled as voltage-dependent unless otherwise indicated, using an extension of the single-conformation electrical distance/barrier model developed by Lear (36) with additional conformational states and barriers added to describe the system (see Results and Discussion, and Supporting Information). For kinetic cycles, one of the rate constants was not independently fit but instead calculated using microscopic reversibility from the remaining rate constants in the cycle at zero driving potential and equal pHin and pHout.
Results and Discussion
pH-dependent transitions of Full-length M2 in phospholipid vesicles
In order to correlate structural transitions in M2 with the process of proton conduction, it was important to measure conformational changes under conditions where the protein demonstrates robust proton transport. The Cys-free, single-Trp construct used for fluorescence quenching studies was therefore reconstituted for flux measurements in a similar fashion, into phospholipid vesicles of the same composition and target size. The resulting proteoliposomes demonstrate amantadine-sensitive proton currents (Fig. S2) when flux is triggered by the addition of the ionophore valinomycin (21), unlike protein-free control vesicles that remain essentially proton-impermeable on the timescale of the experiment. This result confirms that M2 is active under the conditions used for spectroscopic studies of its conformation, allowing for correlation of conformational changes with transport phenomena.
Having demonstrated that this M2 variant was active in forming channels, we examined its fluorescence properties as a function of pH. Relatively small changes in M2 Trp fluorescence intensity and emission maximum have been reported previously; the studies of Hay and coworkers (43) were suggestive of a multi-state process, but the degree of spectral perturbation was too small to allow more than a single transition to be observed. We therefore turned to collisional fluorescence quenching to report more fine-grained changes in the Trp41 environment. The great advantage of this method is that in a given condition, the protein is titrated with various concentrations of quencher, allowing a multi-point determination of the quenching constant that is often more sensitive to environmental change as well as more precisely determined than the information derived from taking a single spectral scan. The Stern Volmer quenching constant (KQ) obtained from such experiments reflects the accessibility of a Trp residue along with other properties such as the intrinsic fluorescence lifetime, and environmental factors that can be more difficult to interpret. Here, however, very good structural data already exist for the protein, so we were primarily interested in using KQ as a convenient intrinsic reporter of the Trp41 environment. We therefore examined the quenching of Trp41 by iodide, and automated the titrations to maximize run-to-run reproducibility and to allow a constant correction for instability in the Trp41 signal (see Materials and Methods). It is important to stress, that the pH-dependence of the quenching constant is being measured here as a reporter of conformational change to allow definition of the number of spectroscopically distinct states, rather than in an attempt to provide detailed information about the specific conformational forms.
The relationship of the Stern-Volmer quenching constant for M2 Trp41 (KQobs), calculated by fitting Eq. 4 to quenching titration data sets collected at a pH range of ~8-5, is plotted vs. experimental [H+] in Fig. 2. A triphasic pH dependence of KQobs is observed, with low quencher accessibilities at pH 7–8 that rise to a peak at pH ~6, and decline again at lower pH. These data show that M2 adopts several different protonation states in this physiological pH range, and that in each state the Trp41 residue has a distinct environment.
The position of the peak and inflection points in Fig. 2 are consistent with the pKa values previously reported by Cross and coworkers for His37 in phospholipid bilayers (29). In particular, the low KQ values at high pH would be associated with the neutral to 2+ states (pKa 8.2), the peak near pH 6 is consistent with the formation of the 3+ state (pKa 6.3), and the falloff at low pH is consistent with additional protonation to form the 4+ state. Indeed, a theoretical curve calculated using the pKa values determined by Hu et al. gives a very good fit to the data (Fig. 2, solid line). Because the pKa values were already known from the work of Cross and colleagues, the curve in Fig. 2 was generated by fixing these pKa values and using KQ values for the individual protonation states as the only adjustable parameters. The values of KQ increase from the 2+ to the 3+ states, and then decrease again as the 4+ state becomes populated. It is difficult to place an unambiguous interpretation on the magnitudes of KQ for each species, because the differences in KQ are relatively small (2-fold) over the range studied. Moreover, exhaustive studies using different quenching agents were beyond the scope of this work. There also is a relatively large error associated with the quenching constants for the neutral and 4+ states, because the pKa’s of the system are such that these two states are in low abundance over the physiological pH range studied here. Nevertheless, the data clearly show distinct accessibilities for the different protonation states. It is also interesting to correlate the decrease in quencher accessibility at low pH with the findings of Howard and coworkers, who observed that the C-terminal helix following the M2 transmembrane bundle became more deeply embedded in the bilayer near pH 5 (44). Linked transitions of the C-terminal helix may therefore play some role in the decreased Trp41 accessibility at low pH.
In sum, these findings enable us to correlate physiologically encountered protonation events of the M2 tetramer with conformational transitions of the transmembrane domain observed under conditions where the protein is functional. In particular, our data indicate that binding and release of the 3rd and 4th protons by M2 leads to significant conformational changes in the protein. We next examine mechanistic models for conduction where protonation and deprotonation events drive conformational changes in M2 that affect the rates of permeant proton binding and release.
Conductance models of transport where proton uptake and release rates are mediated by H+-driven changes in structural equilibria yield good fits to multiple electrophysiological data sets
Previous electrophysiological experiments by Chizhmakov et al. and Pinto et al. have provided a wealth of pH-dependent current/voltage curves for the M2 proton channel expressed in mammalian cells and Xenopus laevis oocytes (11, 12, 18, 20). While progress has been made in describing the entire body of electrophysiological results, a single model that provides a complete description of all aspects of the conductance curves has been elusive (11, 15, 26, 35, 36). The current observation of multiple conformational and protonation states over the pH range used in electrophysiology experiments provides a rationale for developing a proton transport model that links His37 protonation events with structural transitions in the M2 transmembrane bundle, which in turn influence proton stability, accessibility to solvent from the N-terminal and C-terminal side of the pore, and maximal rate of proton release. A key conceptual foundation of this analysis is the distinction between conformational equilibria of the M2 tetramer bundle (Fig. 1, where the protein populates some combination of states “A”, “B”, and “C”) and protonation equilibria at the His37 tetrad (states 0+ to 4+), which in the model influence the population of each distinct conformational state. Thus, His37 tetrad protonation states and M2 conformations are distinct, but thermodynamically and functionally interrelated entities.
We initially examined the model shown in Figure 3, which is a refinement of the previously proposed two-state transporter mechanism (34). According to this hypothesis, the first two protons bound from the extraviral compartment activate the protein to enter the conduction cycle; the first two protons themselves are not permeant in this model and cannot be released to the C-terminal side of the membrane. Evidence for protonation-mediated M2 activation comes from experiments (11) where it was shown that with an outward driving transmembrane potential, transport through M2 decreased at more basic pHout in spite of an increasing gradient for outward proton flux and from experiments in which C-terminal pH was lowered to create a pH gradient, but did not result in outward current flow (18). These observations suggested that the protein was populating an inactive state at high external pH. In the model shown in Fig. 3, the low net outward conductance at high pHout can be qualitatively explained by the kinetic trapping of the protein in the nonconducting “A” conformation (Fig. 1), in which the His tetrad has low accessibility to protons from the C-terminal side. Of the known high-resolution structures, this conformation is presumably most similar to the high-pH solution NMR structure (31), which is partially open on the N-terminal side and nearly fully closed at the C-terminus.
At lower pHout we postulated that the first two protons bound convert the “A” conformation, significantly populated only in the 0+ His37 tetrad state, and described by the high pH NMR structure (31), into an ensemble of “B” and “C” conformations. The structures suggest that the “B” and “C” conformers have successively more constricted N-terminal and dilated C-terminal pore regions, but this is not an implicit assumption of the conduction model. Based on structural studies, the “B” conformation is likely similar to an intermediate pH crystal structure (33), a solid-state NMR structure (45,46) and computational models (34, 35). These differ from the “A” conformation by virtue of a more open cytoplasmic end of the pore (Figure 1). Finally, the “C” conformation is likely to resemble the conformations seen in crystal structures, which have splayed C-terminal (inner facing) helices and a closed N-terminal end facing the outside of the virus (32). We therefore schematically show the C-terminal end of the channel to dilate in the figure, although it is important to emphasize that the kinetic conductance model does not presuppose anything other than there are distinct conformational states with differing accessibility and affinities for protons. The stabilities of these conformational states of the M2 bundle depend on pH, in such a way that increasing protonation of the His37 tetrad favors formation of the “C” conformation, thereby allowing hydration and minimizing repulsions between His37 residues on adjacent monomers. However, each distinct protonation state has an equilibrium mixture of conformational states (and vice versa), so it is again important to not equate a given His37 tetrad protonation state with a unique conformational state.
A number of independent experimental restraints constrained the fitting process. The pKa values for binding the first two protons (describing the transition from a fully deprotonated conformation “A” to the doubly protonated conformation “B”) were fixed at 8.2, as measured by Cross and colleagues (29). This double protonation event was treated as a rapid equilibrium, relative to conduction at the pH where the “A” conformation is largely populated, due to the lack of activation of the channel and the low concentrations of permeant protons. Secondly, as supported by our fluorescence quenching results, we expected that binding of the third proton by the doubly protonated conformation “B” shifted the structural equilibrium toward conformation “C”; correspondingly, using the principles of microscopic reversibility and linked equilibria, the model was constrained such that the third proton pKa of “C” was greater than the third proton pKa of “B”. We did not explicitly constrain the actual fitted pKa values for the third protonation step to be anything other than experimentally feasible (in the 2–10 range). However, we used the best-fit results of these values to calculate an averaged third proton pKa weighted according to the relative population of the “B” and “C” conformations predicted by our model (see Supporting Information), and compared it to the value experimentally determined in bilayers (29). In the initial fitting process, on-rates for principal protonation pathways (k3, k9) were allowed to vary within reasonable ranges expected for diffusion-limited processes given the diameter of the pore (106–109 M−1 sec−1) (25). Finally, microscopic reversibility provided another restraint on the system, allowing us to directly calculate one parameter from the values of others. For example k3/k−3 = k5/k−5 and k7/k−7 = k9/k-9, and K4 is known from the values of these constants plus K2. For completeness we included the kinetic pathways, mediated by k5, k−5, k7, k−7, in the fitting, but they did not contribute significantly to the fit, and were eliminated (data not shown). This finding is consistent with the structural model showing that the “B” state would be more accessible to protons on the outside than the “C” state (k3 > k7) and that state “C” is more accessible to protons from the inside (k9 > k5). Thus, these pathways were not described further for this model.
In summary, the following parameters were allowed to float in the fitting process, some with constraints as described above: the third proton pKa for conformations “B” and “C” and the corresponding proton on-rate constants (k3 and k9), and the magnitude of the two rate constants making up the conformational equilibrium constant K4. One of the rate constants making up K2 was initially constrained to result in a rapid equilibrium (the other was calculated by microscopic reversibility). As we will see, even with the very large body of electrophysiological data available, these parameters were not uniquely defined, and multiple minima were observed. However, all but two of these could be ruled out because they predicted pKa values that were far from those measured independently, or the predicted conductance level was too low. Additionally, in order to account for voltage dependence in the target data sets, the rate constants describing the binding, translocation, or release of the third proton were modeled as voltage-dependent using an approach adapted from Lear (36). The relative electrical distances for two conformations and three transition states were constrained to be equal or rise sequentially as the proton made its way from outside to inside the membrane, and to fall between 0 and 1 (36).
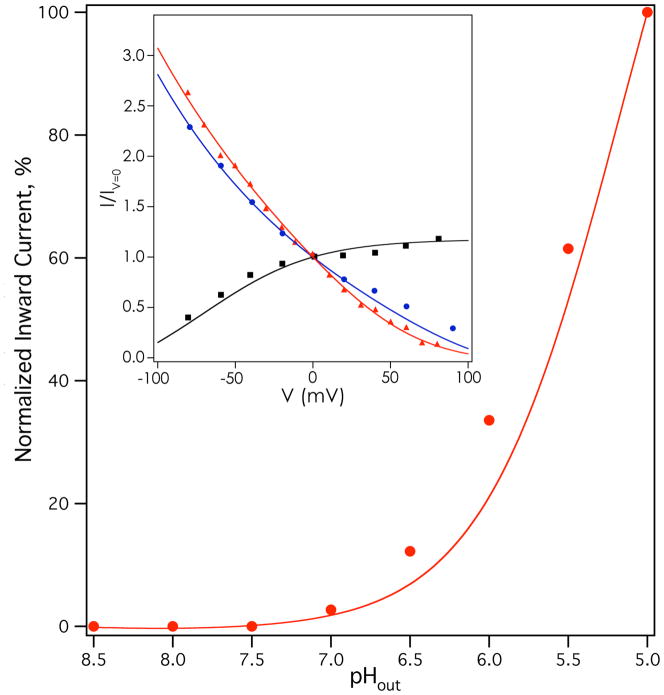
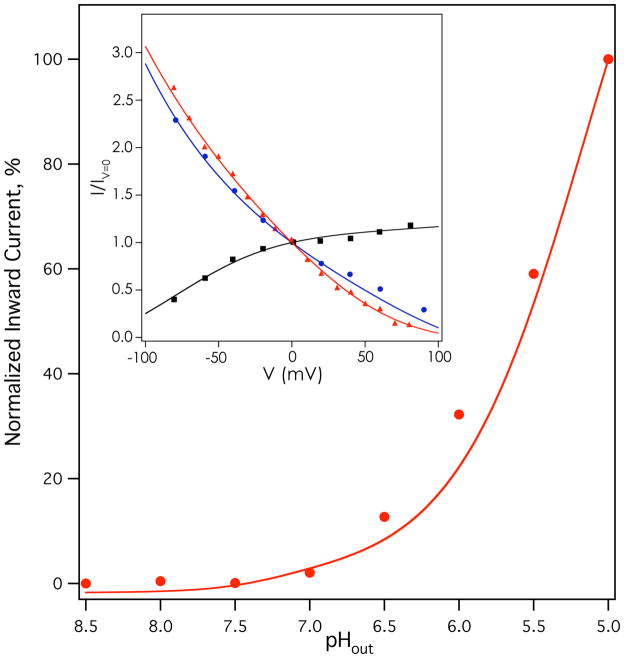
This model was globally fit to current-voltage relationships for Weybridge A/M2 at several physiologically encountered pH conditions (12) (points, Figure 4 inset), pH dependent chord conductance for Weybridge A/M2 (11) (points, Figure S3), and a current-pH relationship for Udorn A/M2 (points, Figure 4); the current-pH relationships of the M2 variants from the Udorn and Weybridge strains are very similar (47). Because the absolute rate of conduction (in flux per tetramer) was not known for most of these experiments, the individually normalized relative rates of conductance were used for each curve. Multiple minima were found that provided adequate fits to the data. Applying the constraints described above, one solution was found that gave an overall, population-averaged third proton pKa (~6.9) near the experimental pKa value (6.3) for the third protonation of the M2 bundle (29) (see Supplementary Information for equation). The conformation-specific third proton pKa value was 6.0 for the “B” state, and was 7.5 for the “C” state (the full list of parameters is shown in Table S1). The results calculated from this solution are shown as solid lines in Figures 4 and S3. Excellent agreement was observed with experimental current-voltage curves (Figure 4 inset), and a very good fit was obtained to pH- and voltage-dependent chord conductances (Fig. S3). A good fit was also seen to the pH-proton flux relationship (Figure 4) shown in the main panel, although the model slightly underestimated flux at intermediate to low pH regimes. These deviations are most likely secondary to the use of global fitting to obtain a single set of parameters. In its global nature, the fit was not optimized to a single buffer condition or changes due to background expression system, and on the whole, the agreement to multiple data sets collected in different cell types and protein strains is remarkable. Encouragingly, the predicted per-tetramer proton flux with the best-fit model parameters (shown in Table S1) was ~0.5 protons per second at pHout=7, ~30 protons/second at pHout=5 (pHin=7.25, V=−20 mV), and ~90 protons/second at pHout=5, pHin=7.25, V=−130 mV, consistent with the range of experimentally determined values in the literature (10, 22, 23).
Figure 4.
Results of fitting activated two-state mechanism to M2 functional data. Inset, rimantadine-sensitive current-voltage relationships of Weybridge A/M2 (data from (12)) at pHin=7 pHout=5 (red triangles), pHin=6 pHout=8 (black squares), and pHin=8 pHout=6 (blue circles). Main panel, amantadine-sensitive current-pHout relationship of Udorn A/M2 at −20 mV. Rimantadine-insensitive leak currents have been subtracted for each measurement in the inset; amantadine-insensitive leak currents have been subtracted for each measurement in the main panel.
We also observed a second good fit with reasonable specific activity and population-normalized pKa when the relationship of the third proton pKa values for the “B” and “C” conformations was reversed (i.e., forcing pKaB > pKaC). The ratio of the best-fit equilibrium constants K2 and K4 was also identically reversed, because of thermodynamic coupling. The fit quality in this case was essentially identical to the solution described above and is not presented graphically. The model parameters for this solution are shown in the right column of Table S1. This result remains consistent with the structural model, but suggests a reverse order of stability for the “C” versus “B” conformations with decreasing pH. Under these conditions, because of the reversal of the ratio of K2 and K4, binding of the third proton now shifts the “B” – “C” conformational equilibrium to favor the “B” state. While this scenario is less consistent with the structural mechanism of progressive N-terminal pore closure and C-terminal pore dilation with decreasing pHout it cannot be ruled out based on electrophysiological data alone.
We also used this model to explore whether the conformational transitions (pathways described by K2 and K4 in Fig. 3), or proton release steps from His37, (described by k−3 and k−9 in Fig. 3) better corresponded to the rate-limiting process in conduction. To this end, when we applied our initial constraints, we treated the conformational transition between BH2 and CH2 (described by K2) as a rapid, non rate determining equilibrium, but allowed the rate constants making up K4, which accounts for the interconversion of BH3 and CH3, to access both the rate determining and non rate determining range. The initial model run converged on a solution where both conformational equilibria were rapid, and thus not rate determining for transport (k−4 > ~ 200 sec−1, k4:k−4 ~ 10:1), leading to the specific activity values described in the paragraph above. In the setting of the constrained, best fit solution, we attempted to force the K4 equilibrium to be rate determining. This was done by gradually lowering the fixed value of k−4 from starting from 100 sec−1 and keeping fixed the best-fit k4:k−4 ratio of approximately 10:1, while simultaneously floating the rate constants for third proton binding (k3 and k9). The solutions under these conditions led to similar fit quality for relative normalized rates, but resulted in significant lowering of predicted specific activity (e.g. ~30 protons/second/tetramer for k−4 = 100, ~12 protons/second/tetramer for k−4 = 50, at pHout=5, pHin=7.25, V=−130 mV). Repeating this simulation, while simultaneously forcing the K2 equilibrium to be slow, resulted in similarly low specific activities or negative best-fit on-rate constants. Similar simulations that primarily constrained the equilibrium described by K2 to be rate determining, and released the rate constants for third proton binding with or without simultaneously slowing the interconversion described by K4, again resulted in negative on-rate constants. We therefore find that under experimental constraints, the model is more consistent with literature specific activity values (10, 15, 22) when the rate of transport is limited by His37 deprotonation.
Some mutants of M2 exhibit biphasic conductance with pH, showing a main transition near pH 6, but also a small transition at higher pH (27). Preliminary models suggested this might arise from a small degree of conduction of the first two protons bound by the tetramer through an equilibrium involving the “A” and “B” conformations, instead of having these protons play a purely activating role. We therefore wondered whether the neglect of conductance through these states by the model shown in Fig. 3 might give rise to a positive deviation of the data from the theoretical curve near pH 6.5 in Fig. 4. To address this issue, we examined an augmented transporter model (Fig. 5), which differs from the simpler model (Fig. 3) in that both “A” and “B” conformations are assumed to be populated in the His-neutral tetrad state. The principal pathways for conduction call for the “A” conformation to bind the first two protons and undergo a conformational change to “B,” which now can mediate their inward release by having some degree of accessibility to C-terminal pore waters. The doubly protonated “B” state can also bind a third proton supplied from the N-terminal side, with inward proton release following conformational change to the “C” state. Once again, minor leak pathways were initially incorporated in the calculations, but did not significantly improve the goodness of fit. The augmented model was constrained essentially as for the simpler model described above, but the on-rates and pKa values for the first two protons bound were now allowed to float with constraints applied similarly to those for the third protonation event in the simpler model. Given the results from the simpler model, conformational transitions were modeled as rapid equilibria. An additional set of electrical distances was fit for the translocation of the first two protons bound by the tetramer. The population-normalized pKas for binding the first two protons and the third proton were again compared for consistency with solid-state NMR measurements (29), and were within 0.7 pKa units for the first two protons, and within 0.6 pKa units for the third. The fitting results are shown in Fig. 6 and Fig. S4, and a full list of fitting parameters is provided in Table S2.
Figure 6.
Results of fitting augmented, multi-conformation model to M2 functional data. Data are shown as in Fig. 4.
The fits to current-voltage data and flux-pH data were only very slightly improved as compared to the simpler model (Fig. 6). The fit to chord conductance data was approximately the same (Fig. S5; with these parameters the function exhibits a visible discontinuity at +60 mV because of software rounding error). The fitted pKas for binding the 3rd proton by the “B” and “C” states (6.0 and 7.5 respectively) were identical to those in the simpler model. Allowing transport of the first two protons bound by the “A” and “B” states led to fitted average pKa values of 7.1 and 7.8 respectively (14.3 and 15.6 for the pKa sums for the first two protons). On the whole, when compared to the fit results of the simpler model (Fig. 4, Fig. S3), the added complexity of the augmented multi-state model (Fig. 6, Fig. S4) did not significantly improve the overall quality of the global fit. This finding suggests that for wild-type M2, transport of the first two protons bound contributes fairly little to overall conduction, although this state clearly contributes more to the pH-flux curves exhibited by certain M2 transmembrane domain mutants (27).
Conclusions
In this work, we have shown that M2 functionally reconstituted in bilayers undergoes pH-dependent transitions in its structural equilibrium that are well described by protonation and deprotonation events in the protein tetramer, allowing us to link conducting proton binding and release with structural changes. We find support for at least three conformations or conformational ensembles being involved in activation and/or conduction by wild-type M2 at physiologically encountered pH ranges.
The mechanistic flux models presented here lead to a simple physicochemical explanation for the highly complex electrophysiological behavior of this minimalistic membrane protein, which has been a topic of significant debate since the initial channel-like and shuttle-like mechanistic hypotheses were proposed (14, 28). The fit results shown here suggest that both the saturation of the rate of proton flux through M2 at low pH, as well as its preferred inward proton flux can be explained by protonation-driven shifts in the structural equilibrium that alternately expose the His37 sidechain to external or internal solvent.
The results of fitting binding and rate constants emphasize the need to use multiple starting values to explore different possible solutions, as well as the need to acknowledge the limitations of this type of model. One major difference between the present work, and structure-based models (26) is that while structural models test whether a given set of experimental and MD structures can account quantitatively for the electrophysiological data, we instead search for all possible solutions that are consistent with a set of experimental observations – in this case the finding of three conformational states, and the known pKa values for protonated His37. One of our expectations from the pH dependence of M2 conformations, seen here by fluorescence quenching and suggested in other structural and computational studies (31–35) was that the third protonation of the “C” conformational state would be more favorable than that of the “B” state, resulting in a greater stability of “C” than “B” in the 3+ His37 tetrad state. Although this scenario was found to give a good fit to the data, a second, equally good solution was observed in which the “B” and “C” state pKas were reversed. Thus, while the current study is consistent with results of MD simulations and structural studies that suggest that multi-protonation of His37 results in overall dilation of the C-terminal end of the bundle, it is not possible to distinguish this model from the reverse scenario based on electrophysiological data alone. This finding underscores the need to consider different types of data (i.e. structural and functional) in developing a given model, and indicates that the order of pKas for “B” vs. “C” cannot be determined solely from the functional data sets used to fit the model.
M2 shows some of the characteristics of a transporter, in that its overall rate of conductance is relatively slow at physiological concentrations of its permeant ion (25), it undergoes conformational transitions in response to binding its conducted cargo (protons), and these conformational transitions occur on the same timescale as conductance (31). Moreover, our results suggest that proton dissociation from the channel rather than the modeled “B”-to-“C” conformational change limits the overall rate of conduction. This is consistent with deuterium isotope effects (15), but neither of these results rule out additional conformational transitions within the “C” state that are required for deprotonation to take place. In an environment as crowded as the end of the M2 channel, conformational transitions would almost certainly be linked to protonation/deprotonation at some level and hence are expected to be necessary for the exit of protons from the channel.
The M2 channel also shows many similarities to classical gated ion channels. For sake of simplicity, in the above discussion we suggest that the third proton binds to the His37 tetrad as it transits through the channel. This is clearly an over-simplification, as a new, high-resolution structure of the channel suggests that a conducting proton is statistically and possibly also quantum mechanically delocalized over a cluster of water and His residues (33). In this case, the conduction process through a large segment of the pore would occur over a relatively flat energy surface similar to that envisioned in continuum models for channels (48). Nevertheless, the mathematical modeling used in the present, site-binding approach would be functionally equivalent, but the interpretation of the rates would differ, as the saturation of rate would occur once a given level of occupancy of ions within this region of the channel is reached. Thus, as emphasized by Decoursey (25) and also exemplified in the steady progression of our structural understanding of potassium and cation channels (49, 50), distinguishing site-binding from continuum models of conductance – or even “transporters” from channels – can reduce to a matter of semantics (51).
Finally, these results are also important for efforts to design new M2 inhibitors, since they suggest that the drug may encounter several structural forms of the protein in vivo, thus presenting a spectrum of potential target structures for drug development. Interestingly, the chemical properties of amantadine appear to render it a multi-form inhibitor, with evidence of amantadine binding to both low- (32), intermediate- (45) and high-pH conformations (52). Newer compounds that do not feature this universality would require very tight binding to their target conformation to shift the structural equilibrium to the targetable, drug-bound state.
Supplementary Material
Abbreviations
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- OG
octyl-β-D-glucopyranoside
- TM
transmembrane
- ss
solid state
Footnotes
This research was supported by NIH grants R01AI57363 to L.H.P. and U011074571 to W.F.D. and R.A.L. R.A.L., L.H.P., and W.F.D. are founders and members of the SAB of Influmedix (www.influmedix.com).
SUPPORTING INFORMATION AVAILABLE
Details of transport model derivation, population-averaged pKa calculations, Figures S1–S4, and Tables S1–S2. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Skehel J, Wiley D. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 2.Zhirnov O. Solubilization of matrix protein M1/M from virions occurs at different pH for orthomyxo- and paramyxoviruses. Virology. 1990;176:274–279. doi: 10.1016/0042-6822(90)90253-n. [DOI] [PubMed] [Google Scholar]
- 3.Pinto L, Lamb R. The M2 Proton Channels of Influenza A and B Viruses. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 4.Grambas S, Hay A. Maturation of influenza A virus hemagglutinin--estimates of the pH encountered during transport and its regulation by the M2 protein. Virology. 1992;190:11–18. doi: 10.1016/0042-6822(92)91187-y. [DOI] [PubMed] [Google Scholar]
- 5.Davies W, Grunert R, Haff R, McGahen J, Neumayer E, Paulshock M, Watts J, Wood T, Hermann E, Hoffmann C. Antiviral activity of 1-adamantanamine (amantadine) Science. 1964;144:862–863. doi: 10.1126/science.144.3620.862. [DOI] [PubMed] [Google Scholar]
- 6.Bright R, Shay D, Shu B, Cox N, Klimov A. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. JAMA. 2006;295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Cady S, Balannik V, Pinto L, DeGrado W, Hong M. Discovery of spiro-piperidine inhibitors and their modulation of the dynamics of the M2 proton channel from influenza A virus. J Am Chem Soc. 2009;131:8066–8076. doi: 10.1021/ja900063s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolocouris N, Zoidis G, Foscolos G, Fytas G, Prathalingham S, Kelly J, Naesens L, De Clercq E. Design and synthesis of bioactive adamantane spiro heterocycles. Bioorg Med Chem Lett. 2007;17:4358–4362. doi: 10.1016/j.bmcl.2007.04.108. [DOI] [PubMed] [Google Scholar]
- 9.Chen B, Leser G, Jackson D, Lamb R. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol. 2008;82:10059–10070. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma C, Polishchuk A, Ohigashi Y, Stouffer A, Schon A, Magavern E, Jing X, Lear J, Freire E, Lamb R, DeGrado W, Pinto L. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc Natl Acad Sci U S A. 2009;106:12283–12288. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chizhmakov I, Geraghty F, Ogden D, Hayhurst A, Antoniou M, Hay A. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J Physiol. 1996;494:329–336. doi: 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chizhmakov I, Ogden D, Geraghty F, Hayhurst A, Skinner A, Betakova T, Hay A. Differences in conductance of M2 proton channels of two influenza viruses at low and high pH. J Physiol. 2003;546:427–438. doi: 10.1113/jphysiol.2002.028910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holsinger L, Nichani D, Pinto L, Lamb R. Influenza A Virus M2 Ion Channel Protein: a Structure-Function Analysis. J Virol. 1994;68:1551–1563. doi: 10.1128/jvi.68.3.1551-1563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinto L, Dieckmann G, Gandhi C, Papworth C, Braman J, Shaughnessy M, Lear J, Lamb R, DeGrado W. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Nat Acad Sci U S A. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mould J, Li H-C, Dudlack C, Lear J, Pekosz A, Lamb R, Pinto L. Mechanism for Proton Conduction of the M2 Ion Channel of Influenza A Virus. J Biol Chem. 2000;275:8592–8599. doi: 10.1074/jbc.275.12.8592. [DOI] [PubMed] [Google Scholar]
- 16.Pinto L, Holsinger L, Lamb R. Influenza Virus M2 Protein Has Ion Channel Activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 17.Shimbo K, Brassard D, Lamb R, Pinto L. Ion selectivity and activation of the M2 ion channel of influenza virus. Biophys J. 1996;70:1335–1346. doi: 10.1016/S0006-3495(96)79690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Y, Zaitseva F, Lamb R, Pinto L. The Gate of the Influenza Virus M2 Proton Channel Is Formed by a Single Tryptophan Residue. J Biol Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 19.Wang C, Lamb R, Pinto L. Direct measurement of the influenza A virus M2 protein ion channel activity in mammalian cells. Virology. 1994;205:133–140. doi: 10.1006/viro.1994.1628. [DOI] [PubMed] [Google Scholar]
- 20.Wang C, Lamb R, Pinto L. Activation of the M2 Ion Channel of Influenza Virus: A Role for the Transmembrane Domain Histidine Residue. Biophys J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schroeder C, Ford C, Wharton S, Hay A. Functional reconstitution in lipid vesicles of influenza virus M2 protein expressed by baculovirus: evidence for proton transfer activity. J Gen Virol. 1994;75:3477–3484. doi: 10.1099/0022-1317-75-12-3477. [DOI] [PubMed] [Google Scholar]
- 22.Lin T, Schroeder C. Definitive assignment of proton selectivity and attoampere unitary current to the M2 ion channel protein of influenza A virus. J Virol. 2001;75:3647–3656. doi: 10.1128/JVI.75.8.3647-3656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moffat J, Vijayvergiya V, Gao P, Cross T, Woodbury D, Busath D. Proton Transport through Influenza A Virus M2 Protein Reconstituted in Vesicles. Biophys J. 2008;94:434–445. doi: 10.1529/biophysj.107.109082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pielak R, Schnell J, Chou J. Mechanism of drug inhibition and drug resistance of influenza A M2 channel. Proc Natl Acad Sci U S A. 2009;106:7379–7384. doi: 10.1073/pnas.0902548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Decoursey T. Voltage-gated proton channels and other proton transfer pathways. Physiol Rev. 2003;83:475–579. doi: 10.1152/physrev.00028.2002. [DOI] [PubMed] [Google Scholar]
- 26.Zhou HX. Diffusion-Influenced Transport of Ions across a Transmembrane Channel with an Internal Binding Site. J Phys Chem Lett. 2010;1:1973–1976. doi: 10.1021/jz100683t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balannik V, Carnevale V, Fiorin G, Levine B, Lamb R, Klein M, DeGrado W, Pinto L. Functional studies and modeling of pore-lining residue mutants of the influenza A virus M2 ion channel. Biochemistry. 2009;49:696–708. doi: 10.1021/bi901799k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sansom M, Kerr I, Smith G, Son H. The Influenza A Virus M2 Channel: A Molecular Modeling and Simulation Study. Virology. 1997;233:163–173. doi: 10.1006/viro.1997.8578. [DOI] [PubMed] [Google Scholar]
- 29.Hu J, Fu R, Nishimura K, Zhang L, Zhou HX, Busath D, Vijayvergiya V, Cross T. Histidines, heart of the hydrogen ion channel from influenza A virus: Toward an understanding of conductance and proton selectivity. Proc Natl Acad Sci U S A. 2006;103:6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Qin H, Gao P, Cross T. Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Biochim Biophys Acta. 2007;1768:3162–3170. doi: 10.1016/j.bbamem.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnell J, Chou J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stouffer A, Acharya R, Salom D, Levine A, Di Constanzo L, Soto C, Tereshko V, Nanda V, Stayrook S, DeGrado W. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acharya A, Carnevale V, Fiorin G, Levine BG, Polishchuk A, Balannick V, Samish I, Lamb RA, Pinto LH, DeGrado WF, Klein ML. Structural mechanism of proton transport through the influenza A M2 protein. Proc Natl Acad Sci. 2010;107:15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khurana E, Dal Peraro M, DeVane R, Vemparala S, DeGrado W, Klein M. Molecular dynamics calculations suggest a conduction mechanism for the M2 proton channel from influenza A virus. Proc Natl Acad Sci U S A. 2009;106:1069–1074. doi: 10.1073/pnas.0811720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi M, Cross T, Zhou H. Conformational heterogeneity of the M2 proton channel and a structural model for channel activation. Proc Natl Acad Sci U S A. 2009;106:13311–13316. doi: 10.1073/pnas.0906553106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lear J. Proton conduction through the M2 protein of the influenza A virus; a quantitative, mechanistic analysis of experimental data. FEBS Lett. 2003;552:17–22. doi: 10.1016/s0014-5793(03)00778-6. [DOI] [PubMed] [Google Scholar]
- 37.Lakowicz J. Principles of Fluorescence Spectroscopy. 3. Springer; New York, NY: 2006. Quenching of Fluorescence; pp. 278–330. [Google Scholar]
- 38.Ladokhin A, Jayasinghe S, White S. How to measure and analyze tryptophan fluorescence in membranes, and why bother? Anal Biochem. 2000;285:235–245. doi: 10.1006/abio.2000.4773. [DOI] [PubMed] [Google Scholar]
- 39.Dvorin E, Mantulin W, Rohde M, Gotto A, Jr, Pownall H, Sherrill B. Conformational properties of human and rat apolipoprotein A-IV. J Lipid Res. 1985;26:38–46. [PubMed] [Google Scholar]
- 40.Stoeva S, Dolashka P, Bankov B, Voelter W, Salvato B, Genov N. Spectroscopic properties of Callinectes sapidus hemocyanin subunits. Spectrochimica Acta. 1995;A51:1965–1974. [Google Scholar]
- 41.Ohigashi Y, Ma C, Jing X, Balannik V, Pinto L, Lamb R. An amantadine-sensitive chimeric BM2 ion channel of influenza B virus has implications for the mechanism of drug inhibition. Proc Natl Acad Sci U S A. 2009;106:18775–18779. doi: 10.1073/pnas.0910584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.WaveMetrics. Igor Pro 6.04. Lake Oswego, OR: 2008. [Google Scholar]
- 43.Czabotar P, Martin S, Hay A. Studies of structural changes in the M2 proton channel of influenza A virus by tryptophan fluorescence. Virus Res. 2004;99:57–61. doi: 10.1016/j.virusres.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen P, Soto C, Polishchuk A, Caputo G, Tatko C, Ma C, Ohigashi Y, Pinto L, DeGrado W, Howard K. pH-induced conformational change of the influenza M2 protein C-terminal domain. Biochemistry. 2008;47:9934–9936. doi: 10.1021/bi801315m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cady S, Schmidt-Rohr K, Wang J, Soto C, DeGrado W, Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–693. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishimura K, Kim S, Zhang L, Cross TA. The closed state of a H+ Channel Helical Bundle Combining Precise Orientational and Distance Restraints from Solid State NMR. Biochemistry. 2002;41:13170–13177. doi: 10.1021/bi0262799. [DOI] [PubMed] [Google Scholar]
- 47.Wang C, Takeuchi K, Pinto L, Lamb R. The ion channel activity of the influenza A virus M2 protein: chacterization of the amantadine block. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levitt D. Interpretation of biological ion channel flux data -- reaction-rate versus continuum theory. Annu Rev Biophys Biophys Chem. 1986;15:29–57. doi: 10.1146/annurev.bb.15.060186.000333. [DOI] [PubMed] [Google Scholar]
- 49.Gouaux E, MacKinnon R. Principles of selective ion transport in channels and pumps. Science. 2005;310:1461–1465. doi: 10.1126/science.1113666. [DOI] [PubMed] [Google Scholar]
- 50.MacKinnon R. Potassium channels. FEBS Lett. 2003;555:62–65. doi: 10.1016/s0014-5793(03)01104-9. [DOI] [PubMed] [Google Scholar]
- 51.Miller C. ClC chloride channels viewed through a transporter lens. Nature. 2006;440:484–489. doi: 10.1038/nature04713. [DOI] [PubMed] [Google Scholar]
- 52.Hu J, Asbury T, Achuthan S, Li C, Bertram R, Quine J, Fu R, Cross T. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from Influenza A virus. Biophys J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.