Abstract
Lipidic cubic phase (LCP) is a membrane-mimetic matrix suitable for stabilization and crystallization of membrane proteins in lipidic environment. LCP technologies, however, have not yet been fully embraced by the membrane protein structural biology community, primarily because of the difficulties associated with handling viscous materials. Recent developments of pre-crystallization assays and improvements in crystal imaging, successes in obtaining high resolution structures of G protein-coupled receptors, and commercial availability of LCP tools and instruments are beginning to attract many structural biologists to integrate these technologies in their research. This should translate to an increased number of otherwise difficult-to-crystallize membrane protein structures, shedding light on their functional mechanisms and lipid-protein interactions.
Introduction
Crystallization of membrane proteins in lipidic mesophases (also known as lipidic cubic phase (LCP) crystallization, or in meso crystallization) has recently achieved a revived attention among structural biologists due to the formidable success of this method in delivering high-resolution structures of human G protein-coupled receptors (GPCRs) [1–5]. Members of GPCR superfamily transmit signals across the cell membrane in response to a variety of extracellular stimuli, triggering diverse intracellular and physiological responses. These proteins represent some of the most successful drug targets and studying their structure and function is at the forefront of current research.
LCP crystallization was introduced about 15 years ago by Landau and Rosenbusch [6], who demonstrated that an LCP matrix can support membrane protein crystal growth and obtained the first high-resolution structure of bacteriorhodopsin (bR), the protein with a fame of being notoriously difficult to crystallize. After brief excitement, however, came a period of frustration and the method was deemed by some to work only with colored 7TM proteins.
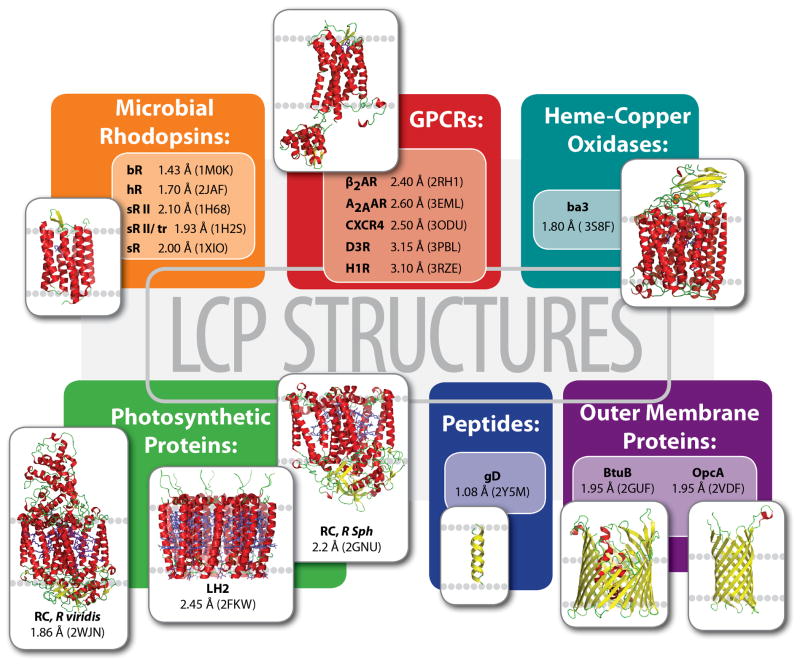
Time has however proven that this technology is not limited to microbial rhodopsins and GPCRs. Membrane proteins from six different classes have been tackled by crystallization in LCP, contributing 79 structures of 17 unique proteins in the Protein Data Bank (Figure 1). The molecular weight of these proteins and peptides ranges from 3.6 to 143 kDa; they represent both α-helical and β-barrel families, including single and multi- subunit proteins with and without chromophores, and covalently bound or diffusible ligands. All of these proteins, however, are bitopic (or transmembrane) and no structure of a monotopic membrane protein has been obtained by this method so far.
Figure 1.
A gallery of protein and peptide structures obtained by LCP crystallization. For each unique protein the highest resolution and corresponding PDB ID are shown.
The success of using LCP for crystallization can be attributed to several factors. The membrane-like environment of LCP stabilizes sensitive proteins and leads to type I crystal packing, where crystal contacts are formed between both polar and non-polar parts of the protein, contributing to better order and diffraction [7]. Additionally, LCP can act as a size filter, effectively removing large-size impurities and protein aggregates from poisoning crystal growth [8]. Two alternative crystallization techniques, in which protein is crystallized from lipid membrane environment, termed bicelle crystallization [9–10] and crystallization by vesicle fusion [11], have also been used to obtain high-resolution structures of several membrane proteins. While having similar benefits as LCP crystallization, the two latter methods suffer from low mesophase stability, which limits the range of precipitants that can be used to screen for crystallization conditions. A disadvantage of LCP matrices, in turn, is the curved nature of their lipid membranes and their specific microstructure that restricts the size of membrane proteins that can undergo long-range diffusion and thus be able to crystallize. One way to overcome this limitation is to use specific precipitants, such as some non-volatile alcohols, low molecular weight PEGs and some other polymers, that swell LCP and transform it into a sponge phase [12–13].
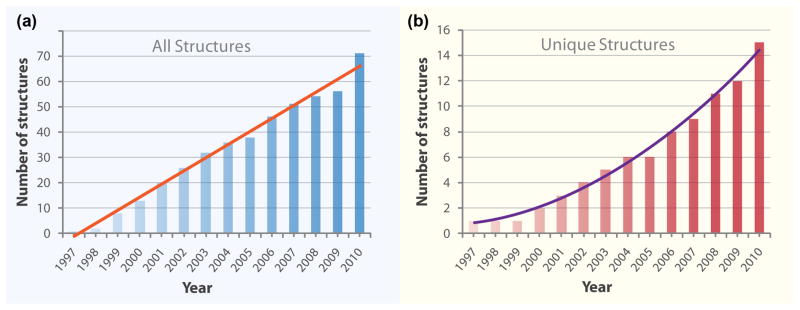
The total number of structures obtained by crystallization in LCP has been increasing steadily since the first bR structure reported in 1997 (Figure 2a). Surprisingly, this growth is linear in time rather than exponential, as was observed for soluble and membrane proteins [14]. A possible reason for the non-exponential growth is a disproportionally large number of bR structures (mutants and photocycle intermediates) that have been obtained during the first 5–10 years. However, even when only unique membrane protein structures are considered, the growth is still non-exponential, and at best could be described by a second-order polynomial (Figure 2b). This trend can likely be explained by the fact that LCP crystallization has never been fully embraced by a broad community of structural biologists. At any given time, no more than a handful of groups have actively pursued this method and incorporated it in their arsenal of crystallization techniques. The obvious reasons for this were the difficulties in preparing and handling the gel-like LCP material and the lack of proper tools and instruments.
Figure 2.
Growth of membrane protein structures obtained by LCP crystallization with time. (a) All structures deposited to PDB. (b) Unique structures (defined as those, in which the protein sequence differs by more than 10%).
This situation however has changed during the last few years. Many tools and instruments for LCP crystallization have become commercially available. Several new assays to monitor properties of membrane proteins directly in LCP were developed, as well as new technologies for setting up crystallization trials and detecting protein crystals have started to emerge.
This review is focused on the recent advancements in LCP-related technologies ranging from pre-crystallization assays to crystallographic data collection. Its purpose is to alert the reader to the progress in the technology and exciting new opportunities associated with using LCP matrices for membrane protein stabilization for biophysical and structural studies.
LCP pre-crystallization assays
LCP crystallization is no different from any other crystallization technique in the sense that it requires extensive screening of precipitant conditions. However, crystallization in LCP is generally more challenging than traditional crystallization in solution because it involves additional variables related to the composition of lipidic phase, which increase the dimensionality of crystallization space. To cope with this problem, more emphasis is placed on the need for pre-crystallization assays capable of reducing the number of crystallization variables and/or limiting their range.
Recently, two such assays for measuring thermal stability (LCP-Tm) [15] and diffusion (LCP-FRAP) [16] of membrane proteins embedded in LCP have been introduced (Figures 3g,h).
Figure 3.
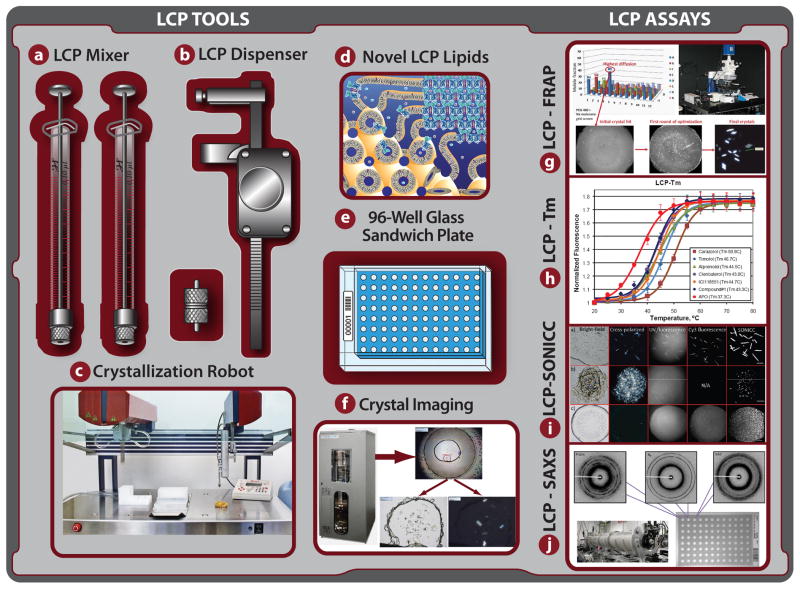
LCP Toolchest: collection of tools and assays for preparation and handling of LCP, and for characterization and crystallization of membrane proteins in LCP. (a) Lipid syringe mixer for LCP preparation [46]; (b) manual LCP dispenser [47]; (c) in meso crystallization robot [21]; (d) Novel LCP forming lipids for host lipid screening [25,27–28]; (e) 96-well glass sandwich crystallization plate [22,33]; (f) Improved LCP crystal imaging; (g) LCP-FRAP assay [16]; (h) LCP-Tm assay [15]; (i) LCP-SONICC [37]; (j) LCP-SAXS [48].
LCP-Tm
Thermal stability of soluble proteins and membrane proteins in detergent solution has been conveniently assessed by using fluorescence and other optical techniques in conjunction with temperature ramping [17]. While LCP is optically transparent when prepared at appropriate conditions, application of these techniques to measure stability of membrane proteins embedded in LCP is not straightforward due to a loss of transparency of LCP upon heating. A new protocol (LCP-Tm) that circumvents this problem has recently appeared [15]. This assay can be used not only to compare the stability of different protein constructs in combinations with different ligands, but more importantly, to select the most promising LCP host lipids and lipid additives that increase protein stability. For example, in the case of β2-adrenergic receptor, monoolein was found to be the most stabilizing LCP host lipid out of a panel of four lipids, and cholesterol was found to be the best additive lipid, in good agreement with crystallization studies [15].
LCP-FRAP
Translational diffusion of membrane proteins embedded in LCP matrix is essential for crystal nucleation and growth. The structural features of LCP, however, can restrict motions of large proteins or oligomeric protein aggregates, thus preventing their crystallization. Fluorescence recovery after photobleaching (FRAP) was used to study the diffusion of integral membrane proteins in LCP [18]. It was found that the diffusion properties within LCP matrix strongly depend on protein stability and screening conditions. Common precipitants often induce fast non-specific aggregation of membrane proteins, limiting their mobility within LCP, and thereby impeding their chances for crystallization. A good correlation between a high protein mobile fraction and successful crystallization conditions was observed. Based on these studies, a high throughput pre-screening assay (HT LCP-FRAP) was developed. This assay can be used to select the most promising protein construct, ligand and LCP host lipid, and rule out conditions that are not conducive to diffusion from subsequent crystallization trials [16]. HT LCP-FRAP was instrumental in obtaining initial crystallization conditions for the CXCR4 chemokine and dopamine D3 GPCRs [3–4].
LCP crystallization
LCP crystallization method has evolved over the years from using small glass test tubes and mixing lipid with protein solution by centrifugation to a faster and more efficient way of mixing LCP with a syringe mixer and setting up crystallization trials in 96-well plates. Consumption of protein and lipid per crystallization trial has been reduced by over 100-fold, and the whole process has become about as easy as setting up vapor diffusion trays. A comprehensive guide for using LCP for membrane protein crystallization [19], as well as visual demonstrations [20–21], have recently been published.
The need for screening a vast space in order to identify a crystal lead prompted the work on automation of LCP crystallization trials, which led to the development of an in meso crystallization robot (Figure 3c) [22]. The same automated LCP dispensing technology was recently implemented in several commercial crystallization robots (Table 1). These robots can typically dispense 20–50 nL of LCP per trial and take 5–10 minutes to setup a 96-well plate. Initial crystallization screening usually involves using an array of commercial sparse matrix screens; however, only a few of them have been specifically designed for LCP crystallization (Table 1). Other commercial screens, especially those that target membrane proteins, can also be used, with a caveat that 20–50% of conditions may be incompatible with LCP crystallization [23]. A single LCP host lipid, monoolein, has been used to obtain most structures to date [24]. However, the advantage of performing a host lipid screening with homologous monoacylglycerols (MAGs) has recently been highlighted [25–26], and an improved protocol for MAG synthesis was published [27]. Additionally, synthesis of new isoprenoid-chain lipids capable of forming LCP and useful for host lipid screening have been reported [28]. One of these lipids was used successfully to crystallize bR [29].
Table 1.
Vendors and suppliers of tools and instruments for LCP applications
| Vendor | Website | Supplies |
|---|---|---|
| Avanti Polar Lipids | www.avantilipids.com | Monoacylglycerol lipids |
| Emerald Biosystems | www.emeraldbiosystems.com | Cubic LCP kit, Cubic screen |
| Hampton Research | hamptonresearch.com | LCP Swissci plate, LCP sandwich plate |
| Hamilton Company | www.hamiltoncompany.com | Syringes, needles, syringe dispenser |
| Molecular Dimensions | www.moleculardimensions.com | Laminex plate, LCP Swissci plate, LCP mixing adapter, Sponge phase screen |
| Marienfeld | www.marienfeld-superior.com | LCP sandwich plate |
| MiTeGen | www.mitegen.com | Micromounts |
| Nu-Chek Prep | www.nu-chekprep.com | Monoacylglycerol lipids |
| Qiagen | www.qiagen.com | CubicPhase screens, CubicPhase kit |
| Sigma-Aldrich | www.sigmaaldrich.com | Monoacylglycerol lipids |
| Company | Website | Crystallization Robots |
| Anachem/Gilson | www.gilsonuk.com | Flexus IMP |
| Art Robbins/Rigaku | www.artrobbins.com | Gryphon LCP |
| Formulatrix | www.formulatrix.com | NT8-LCP |
| TTP LabTech | www.ttplabtech.com | Mosquito LCP |
| Zinsser Analytic | www.zinsserna.com | ProCrys Meso |
| Company | Website | Automatic Imagers |
| Formulatrix | www.formulatrix.com | RockImager |
| NEXUS Biosystems | www.irori.com | Crystal Farm |
| TriTek | proteincrystalimaging.com | CrystalPro |
| Thermo Scientific | www.thermo.com | Rhombix |
| Rigaku | www.rigaku.com | Minstrel |
Recently, several approaches to setting up LCP crystallization have emerged. They include novel and unique ways of preparing or dispensing LCP, which take advantage of microfluidics or specific lipid phase behavior in order to simplify or avoid handling of highly viscous materials [30–32]. The validity of these approaches was demonstrated by crystallization of bR and photosynthetic reaction center. It remains to be seen if they will be compatible with a broader selection of protein targets.
LCP imaging and crystal detection
Imaging LCP crystallization trials and reliable detection of crystals growing in lipidic mesophases have represented some of the most difficult tasks since this method was first introduced. Originally, LCP was used only with colored proteins and there was a concern that crystals of colorless proteins could not be easily detected in LCP. Development of a glass sandwich plate (Figure 3e) allowed for high quality imaging of protein crystals growing in LCP using a brightfield transmitted light mode and cross-polarizers (Figure 3f) [22,33]. Nevertheless, even with these plates, detection of crystals in lipidic mesophases is not always straightforward due to formation of various defects in the gel-like LCP, flipping LCP into highly birefringent lamellar or hexagonal phase, or the occurrence of specific crystal space groups and orientations. Intrinsic protein UV fluorescence was shown to be efficient for identification of protein crystals, however, the signal strongly depends on the tryptophan content in the protein molecule and heavy protein precipitation can obscure crystal detection. Additionally, UV illumination may damage protein sample if used extensively. Brightfield, cross-polarizers and UV fluorescence have been implemented in many current automatic crystal imaging systems (Table 1). Most of these imagers also contain other features important for high quality LCP imaging: Köhler illumination, variable zoom lens, temperature control, drop location and extended focus imaging.
To improve detection of initial microcrystals in LCP, trace-labeling of protein with a fluorescent dye has been used successfully [16,34–36]. However, the extra steps required for protein labeling and the possibility that labeling can affect crystallization have made this approach less attractive than UV imaging, despite its superior sensitivity.
A novel method of protein crystal detection, termed SONICC (second order nonlinear imaging of chiral crystals), that is devoid of many drawbacks associated with other imaging techniques, has recently been introduced (Figure 3i) [37]. This method relies on two-photon scattering, which gives virtually no background scattering from randomly oriented molecules, but produces a strong signal from chiral molecules arranged in crystals. SONICC signal is not strongly affected by turbid media, such as certain lipidic mesophases, and it has a high signal-to-noise ratio even in the case of submicron size crystals. These properties make this method suitable for automatic crystal detection using simple and reliable threshold-based algorithms. SONICC, however, does not distinguish between crystals of proteins and other chiral molecules that may be present in the crystallization drop and requires specialized instrumentation, which may be expensive. SONICC imager is available from Formulatrix as a stand-alone instrument or as an attachment to their RockImager line.
Minibeams and X-ray data collection
Crystals that grow in LCP are often small, but due to their superior packing can be highly ordered. In order to collect the maximum crystallographic information possible from small crystals, ideally the size of the X-ray beam should match the size of the crystal. The LCP crystallization method has strongly benefitted from development of microcrystallography, which emerged with the appearance of third generation synchrotron sources about 10–15 years ago. Currently most synchrotron sources have dedicated microfocus beamlines for protein crystallography, with beamsizes ranging between 10 and 20 μm, and going down as low as 5 μm, and even 1 μm [38–39].
Apart from advancements related to beam focusing and collimation, increased accuracy of goniometers and stability of X-ray beams, there was a number of additional developments that were required to efficiently collect data on LCP grown microcrystals. These developments aimed at automation of microcrystal centering using rastering with an attenuated X-ray minibeam, optimization of data collection strategies to obtain better data and minimize radiation damage, and merging together data collected from multiple crystals [34]. The success of these combined approaches can be clearly evidenced by the increasing number of structures of GPCRs and other difficult-to-crystallize membrane proteins [3–5,40–45].
Conclusions
Structural biology of membrane proteins undergoes rapid progress and development, however, the pace of structure determination is still lagging strongly behind that of soluble proteins. Eukaryotic membrane proteins have been particularly difficult to deal with because of their low stability and dependence on specific lipid environment. Development of special tools and membrane-mimetic systems are required to accelerate research work in this area. LCP is one such matrix that can be used to stabilize membrane proteins for biophysical studies and obtain high quality crystals for structure determination. Recent progress in LCP technologies and their commercialization are making them more accessible to researchers. Further developments and improvements that are expected in the next few years include formulations of new fully LCP compatible sparse matrix crystallization screens, design of novel LCP host lipids with different properties, and developments of assays for functional characterization of membrane proteins in LCP.
Microcrystallography will continue to play a strong role in the successfull use of LCP technologies. Several aspects here require further improvements. Centering of frozen crystals embedded in opaque lipid mesophase can be approached with UV fluorescence or SONICC. There is a need for advanced computer algorithms for optimization of data collection and processing from multiple microcrystals, minimizing effects of radiation damage and taking into account anisotropy of diffraction.
Emerging microfluidics approaches capable of handling LCP and other viscous lipidic mesophases will mature and will likely represent the next major step in minimizing the consumption of valuable protein material and may allow for integration of different steps, such as cell-free expression, purification and crystallization, in a single LCP-lab-on-a-chip.
Finally, the expectations are that LCP and other lipid-based crystallization technologies will continue to gain wider community acceptance, resulting in expansion of structural information that will provide further insights into intricate relationship between membrane protein structure and function.
Acknowledgments
The author is indebted to Ray Stevens, Martin Caffrey and Valentin Gordeliy for useful discussions, Enrique Abola, Jerry Joseph and Angela Walker for careful reading the manuscript and Katya Kadyshevskaya for illustrations.
This work was supported in parts by the National Institute of Health grants GM073197 and RR025336.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest published within the period of review have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2. 6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3••.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. High-resolution crystal structures of the human chemokine CXCR4 receptor obtained by LCP crystallization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4••.Chien EY, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. Structure of the human dopamine D3 receptor obtained by LCP crystallization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5••.Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, Abagyan R, Cherezov V, Liu W, Han GW, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011 doi: 10.1038/nature10236. in press. Structure of the human histamine H1 receptor obtained by LCP crystallization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landau EM, Rosenbusch JP. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc Natl Acad Sci U S A. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Caffrey M. Crystallizing Membrane Proteins for Structure Determination: Use of Lipidic Mesophases. Annu Rev Biophys. 2009;38:29–51. doi: 10.1146/annurev.biophys.050708.133655. Recent in-depth review covering lipid phase behavior, LCP crystallogenesis, and practical aspects of using lipidic mesophases for biophysical characterization and crystallization of membrane proteins. [DOI] [PubMed] [Google Scholar]
- 8.Kors CA, Wallace E, Davies DR, Li L, Laible PD, Nollert P. Effects of impurities on membrane-protein crystallization in different systems. Acta Crystallogr D Biol Crystallogr. 2009;65:1062–1073. doi: 10.1107/S0907444909029163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 10.Faham S, Ujwal R, Abramson J, Bowie JU. Practical aspects of membrane proteins crystallization in bicelles. Curr Top Membr. 2009;63:109–125. [Google Scholar]
- 11.Takeda K, Sato H, Hino T, Kono M, Fukuda K, Sakurai I, Okada T, Kouyama T. A novel three-dimensional crystal of bacteriorhodopsin obtained by successive fusion of the vesicular assemblies. J Mol Biol. 1998;283:463–474. doi: 10.1006/jmbi.1998.2103. [DOI] [PubMed] [Google Scholar]
- 12.Cherezov V, Clogston J, Papiz MZ, Caffrey M. Room to move: crystallizing membrane proteins in swollen lipidic mesophases. J Mol Biol. 2006;357:1605–1618. doi: 10.1016/j.jmb.2006.01.049. [DOI] [PubMed] [Google Scholar]
- 13.Wadsten P, Wohri AB, Snijder A, Katona G, Gardiner AT, Cogdell RJ, Neutze R, Engstrom S. Lipidic sponge phase crystallization of membrane proteins. J Mol Biol. 2006;364:44–53. doi: 10.1016/j.jmb.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 14.White SH. Biophysical dissection of membrane proteins. Nature. 2009;459:344–346. doi: 10.1038/nature08142. [DOI] [PubMed] [Google Scholar]
- 15•.Liu W, Hanson MA, Stevens RC, Cherezov V. LCP-Tm: an assay to measure and understand stability of membrane proteins in a membrane environment. Biophys J. 2010;98:1539–1548. doi: 10.1016/j.bpj.2009.12.4296. Development of an assay for measuring thermal stability of membrane proteins embedded in lipid membrane of LCP. This assay allows to select the most stabilizing LCP host lipid or lipid additive for a given membrane protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Xu F, Liu W, Hanson MA, Stevens RC, Cherezov V. Development of an automated high throughput LCP-FRAP assay to guide membrane protein crystallization in lipid mesophases. Cryst Growth Des. 2011;11:1193–1201. doi: 10.1021/cg101385e. High throughput LCP-FRAP assay for measuring mobility of membrane proteins in LCP. This assay can be used to eliminate unfavorable conditions inducing fast non-specific protein aggregation from subsequent crystallization trials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexandrov AI, Mileni M, Chien EY, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Cherezov V, Liu J, Griffith M, Hanson MA, Stevens RC. LCP-FRAP Assay for Pre-Screening Membrane Proteins for in Meso Crystallization. Cryst Growth Des. 2008;8:4307–4315. doi: 10.1021/cg800778j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. Collection of comprehensive step-by-step protocols related to using LCP for membrane proteins characterization and crystallization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Caffrey M, Porter C. Crystallizing membrane proteins for structure determination using lipidic mesophases. J Vis Exp. 2010;45 doi: 10.3791/1712. Visual guide demonstrating reconstitution of a membrane protein into an LCP and setting up crystallization trials in manual mode. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Liu W, Cherezov V. Crystallization of membrane proteins in lipidic mesophases. J Vis Exp. 2011;49 doi: 10.3791/2501. Visual guide demonstrating preparation of an LCP, performing LCP-FRAP pre-crystallization assay, setting up LCP crystallization trials in glass sandwich plates and harvesting crystals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. A robotic system for crystallizing membrane and soluble proteins in lipidic mesophases. Acta Crystallogr D Biol Crystallogr. 2004;60:1795–1807. doi: 10.1107/S0907444904019109. [DOI] [PubMed] [Google Scholar]
- 23.Cherezov V, Fersi H, Caffrey M. Crystallization screens: compatibility with the lipidic cubic phase for in meso crystallization of membrane proteins. Biophys J. 2001;81:225–242. doi: 10.1016/S0006-3495(01)75694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caffrey M, Lyons J, Smyth T, Hart DJ. Monoacylglycerols: the workhorse lipids for crystallizing membrane proteins in mesophases. Curr Top Membr. 2009;63:83–108. [Google Scholar]
- 25.Li D, Lee J, Caffrey M. Crystallizing membrane proteins in lipidic mesophases. A host lipid screen. Cryst Growth Des. 2011;11:530–537. doi: 10.1021/cg101378s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofer N, Aragao D, Lyons J, Caffrey M. Membrane protein crystallization in lipidic mesophases. Hosting lipid effect on the crystallization and structure of a transmembrane peptide. Cryst Growth Des. 2011;11:1182–1192. doi: 10.1021/cg101384p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y, Weng Y, Hong W-X, Zhang Q. Efficient synthesis of unsaturated 1-monoacyl glycerols for in meso crystallization of membrane proteins. Synlett. 2011;6:809–812. doi: 10.1055/s-0030-1259912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hato M, Yamashita J, Shiono M. Aqueous phase behavior of lipids with isoprenoid type hydrophobic chains. J Phys Chem B. 2009;113:10196–10209. doi: 10.1021/jp902883q. [DOI] [PubMed] [Google Scholar]
- 29.Borshchevskiy V, Moiseeva E, Kuklin A, Bueldt G, Hato M, Gordeliy V. Isoprenoid-chained lipid beta-XylOC16+4 - A novel molecule for in meso membrane protein crystallization. J Cryst Growth. 2010;312:3326–3330. [Google Scholar]
- 30.Wohri AB, Johansson LC, Wadsten-Hindrichsen P, Wahlgren WY, Fischer G, Horsefield R, Katona G, Nyblom M, Oberg F, Young G, et al. A lipidic-sponge phase screen for membrane protein crystallization. Structure. 2008;16:1003–1009. doi: 10.1016/j.str.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Perry SL, Roberts GW, Tice JD, Gennis RB, Kenis PJA. Microfluidic Generation of Lipidic Mesophases for Membrane Protein Crystallization. Cryst Growth Des. 2009;9:2566–2569. doi: 10.1021/cg900289d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Fu Q, Kors CA, Stewart L, Nollert P, Laible PD, Ismagilov RF. A Plug-Based Microfluidic System for Dispensing Lipidic Cubic Phase (LCP) Material Validated by Crystallizing Membrane Proteins in Lipidic Mesophases. Microfluid Nanofluidics. 2010;8:789–798. doi: 10.1007/s10404-009-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherezov V, Caffrey M. Nano-volume plates with excellent optical properties for fast, inexpensive crystallization screening of membrane proteins. J Appl Cryst. 2003;36:1372–1377. [Google Scholar]
- 34.Cherezov V, Hanson MA, Griffith MT, Hilgart MC, Sanishvili R, Nagarajan V, Stepanov S, Fischetti RF, Kuhn P, Stevens RC. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 micrometer size X-ray synchrotron beam. J R Soc Interface. 2009;6 (Suppl 5):S587–597. doi: 10.1098/rsif.2009.0142.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fry EH, Qin W, Fleck EN, Judge RA, Chiu ML. Improved protein crystal detection in detergent and lipidic meso-phases. Open Struct Biol J. 2009;3:11–15. [Google Scholar]
- 36•.Cherezov V, Abola E, Stevens RC. Recent Progress in the Structure Determination of GPCRs, a Membrane Protein Family with High Potential as Pharmaceutical Targets. Methods Mol Biol. 2010;654:141–168. doi: 10.1007/978-1-60761-762-4_8. Summary of methods used for high-resolution structure determination of G protein-coupled receptors, focused on reconstituting membrane proteins in LCP, conducting LCP-FRAP pre-crystallization assays, setting up LCP crystallization trials, detecting crystals, optimizing crystallization conditions, harvesting crystals and collecting crystallographic data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Kissick DJ, Gualtieri EJ, Simpson GJ, Cherezov V. Nonlinear optical imaging of integral membrane protein crystals in lipidic mesophases. Anal Chem. 2010;82:491–497. doi: 10.1021/ac902139w. Comparison of SONICC imaging with other traditional imaging modes for detecting membrane protein crystals growing in lipidic mesophases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischetti RF, Su SL, Yoder DW, Becker M, Nagarajan V, Sanishvili R, Hilgart MC, Stepanov S, Makarov O, Smith JL. Mini-beam collimator enables microcrystallography experiments on standard beamlines. J Synchrotron Radiat. 2009;16:217–225. doi: 10.1107/S0909049508040612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoder DW, Sanishvili R, Vogt S, Xu S, Makarov O, Benn R, Corcoran S, Fischetti RF. One-Micron Beams for Macromolecular Crystallography at GM/CA-CAT. AIP Conf Proc. 2010;1234:419–422. [Google Scholar]
- 40.Wacker D, Fenalti G, Brown MA, Katritch V, Abagyan R, Cherezov V, Stevens RC. Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J Am Chem Soc. 2010;132:11443–11445. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofer N, Aragao D, Caffrey M. Crystallizing transmembrane peptides in lipidic mesophases. Biophys J. 2010;99:L23–25. doi: 10.1016/j.bpj.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. Structure of a nanobody-stabilized active state of the β2-adrenergic receptor obtained by LCP crystallization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao Z-G, Cherezov V, Stevens RC. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. Structure of an agonist-stabilized active state of the human A2A adenosine receptor obtained by LCP crystallization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiefenbrunn T, Liu W, Chen Y, Katritch V, Stout CD, Fee JA, Cherezov V. High-resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment. PLoS One. 2011 doi: 10.1371/journal.pone.0022348. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng A, Hummel B, Qiu H, Caffrey M. A simple mechanical mixer for small viscous lipid-containing samples. Chem Phys Lipids. 1998;95:11–21. doi: 10.1016/s0009-3084(98)00060-7. [DOI] [PubMed] [Google Scholar]
- 47.Cherezov V, Caffrey M. A simple and inexpensive nanoliter-volume dispenser for highly viscous materials used in membrane protein crystallization. J Appl Crystallogr. 2005;38:398–400. [Google Scholar]
- 48.Joseph JS, Liu W, Kunken J, Weiss T, Tsuruta H, Cherezov V. Characterization of lipid matrices for membrane protein crystallization by high-throughput small angle X-ray scattering. 2011 doi: 10.1016/j.ymeth.2011.08.013. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]