Abstract
The blade and sheath of a maize leaf are separated by a linear epidermal fringe, the ligule, and two wedge-like structures, the auricles. In plants homozygous for the null mutation, liguleless2-reference (lg2-R), the ligule and auricles are often absent or positioned incorrectly and the blade–sheath boundary is diffuse. This phenotype is in contrast to that of liguleless1-reference (lg1-R) mutant plants, which have a more defined boundary even in the absence of the ligule and auricles. Additionally, mosaic analysis indicates the lg2-R phenotype is cell-nonautonomous and the lg1-R phenotype is cell-autonomous. Using scanning electron microscopy we show that lg2-R mutant plants are affected before the first visible sign of ligule and auricle formation. We have cloned the Lg2+ gene through a Mutator8 transposon insertion allele, and verified it with five independently derived alleles. The comparison of genomic DNA and cDNA sequences reveals an open reading frame encoding a protein of 531 amino acids with partial homology to a subclass of plant basic leucine zipper (bZIP) transcription factors. Although a large body of molecular and biochemical characterization exists on this subclass of bZIP proteins, our work represents the first report of a mutant phenotype within this group. A specific reverse transcriptase (RT)–PCR assay shows LG2 mRNA expression in meristem/developing ligule regions. RT–PCR also shows that LG2 mRNA accumulation precedes that of LG1 mRNA. The mutant phenotype and expression analysis of lg2 suggest an early role in initiating an exact blade–sheath boundary within the young leaf primordia.
Keywords: Maize, plant, bZIP, liguleless2, leaf development, ligule
The maize (Zea mays) leaf contains three regions. The majority of the mature leaf consists of the basal sheath and the distal blade. Positioned between these two regions is the ligular region, represented by a linear epidermal fringe, the ligule, and two wedge-like structures, the auricles. All three regions contain epidermal, ground, and vascular tissues that are continuous with each other but distinct in cell types and patterns (Sharman 1942; Sylvester et al. 1990). The ligular region therefore represents a boundary within the developing leaf primordium.
The developmental stages of the ligular region have been well characterized. The morphology and division pattern of ligular region epidermal cells were determined histologically (Sharman 1942) and by scanning electron microscopy (SEM) analysis (Becraft et al. 1990; Sylvester et al. 1990). Initially, cells in the position where the ligular region will develop divide anticlinally at a faster rate than surrounding cells, resulting in a preligule band of small cells across the primordium. Preligule band cells on either side of the midrib then divide periclinally. A “wave” of periclinal divisions toward the leaf primordium margins and midrib causes an epidermal ridge (ligular ridge) to arise that spans the width of the leaf primordium. The ridge develops into the ligule and cells distal to the ridge form the auricles. At maturity, the ligule and auricle cells are morphologically distinct from each other and from blade and sheath cells.
The transition from sheath to blade involves alteration not only in the epidermis but also in the pattern of the vasculature (Sharman 1942). Throughout the leaf there is a single midvein with multiple lateral veins on either side. Within the blade, several smaller, intermediate veins exist between the laterals and these anastomose (join) at the ligular region. Consequently, the sheath has only one intermediate vein between each lateral vein.
Ligule development occurs within the larger context of leaf primordium development (for review, see Sylvester et al. 1996). Clonal analysis shows that leaf founder cells initially divide equally to form the leaf primordium (Poethig 1984). These divisions are polar, however, as cells at the future midvein position divide first and divisions then follow laterally around the meristem. Such sequential order of development is also observed for vein development. In the primordium, the first visible vein is the midvein, followed by the sequential appearance of the lateral veins, with the first developing closest to the midvein. Both the midvein and the lateral veins develop acropetally (base to tip). Intermediate veins branch from the lateral veins and develop basipetally (tip to base). They are visible only after their bracketing lateral veins have developed entirely to the leaf margins. In the context of vein formation, ligular divisions are first apparent while intermediate veins are developing basipetally (Sharman 1942; Becraft et al. 1990). At this stage, epidermal differentiation is proceeding from tip to base but has not yet reached the ligular region.
Two unlinked recessive null mutants, liguleless1-reference (lg1-R) and liguleless2-reference (lg2-R), alter the development of the ligular region. lg1-R, located at position 11 on chromosome 2, removes the ligule and auricle in all but the uppermost leaves of the plant, where rudimentary ligule forms. Preligule band divisions can occur on lower leaves, however, they are only in the transverse anticlinal dimension (Sylvester et al. 1990). The boundary between the sheath and blade is at a position similar to that in wild type but is less distinct (Emerson 1912; Becraft et al. 1990). The lg1-R mutant phenotype is cell autonomous (Becraft et al. 1990) and LG1 may be involved in the reception and/or propagation of a “make ligule/auricle” signal that originates on either side of the midrib (Becraft and Freeling 1991). LG1 shows sequence homology to a novel class of DNA binding proteins (squamosa-promoter binding proteins) and expresses in the developing ligular region (Moreno et al. 1997).
lg2-R, located at position 101 on chromosome 3, has a less severe phenotype than lg1-R (Brink 1933; Harper and Freeling 1996). lg2-R mutants lack ligule and auricle only on the lower-most leaves of the plant. On higher leaves, ligule and auricle begin to appear at the leaf margins and with each successive leaf the phenotype gradually becomes less severe. Where the ligule and/or auricle is lacking, the sheath can extend into the blade. Mosaic analysis shows that the lg2-R mutant phenotype is cell nonautonomous (Harper and Freeling 1996), indicating the involvement of cell–cell signaling in LG2 function. Double mutant analysis and reciprocal dosage sensitivity studies between Lg1+ and Lg2+ suggest that these genes act in a common circuit of action (Harper and Freeling 1996).
Here we describe the early morphological events leading to the development of the lg2-R mutant blade–sheath boundary. SEM analyses indicate that lg2-R mutant leaves do not form a normal preligule band and suggest that LG2 is functioning either at or before the time of preligule band divisions. We also present the cloning and initial expression studies of Lg2+. We cloned the Lg2+ gene by transposon tagging and it shows sequence homology to the basic leucine zipper (bZIP) class of transcription factors. It is most similar to a subclass of plant DNA-binding proteins characterized biochemically and hypothesized to function in diverse processes (Tabata et al. 1991; Kim et al. 1994; Liu and Lam 1994; Qin et al. 1994; Ulmasov et al. 1994; Zhang and Singh 1994; Zhang et al. 1995; Chengbin et al. 1996). Lg2+ is the first reported gene within this group with an associated mutant phenotype. Reverse transcriptase (RT)–PCR analyses indicate that LG2 mRNA accumulation precedes that of LG1 mRNA.
Results
SEM of developing and mature lg2-R mutant leaves
A SEM study was undertaken to characterize the development of lg2-R mutant ligular regions as compared with wild-type siblings. In this study, we give leaves two designations—leaf number (L#) and plastochron number (P#). The leaf number was counted from the base of the plant toward the apex. Leaf number therefore indicates leaf position on the main axis of the plant. In contrast, plastochron number indicates the relative age of the leaf. The plastochron number was counted from the apical meristem toward the base of the plant. The most recently initiated leaf primordium was termed P1.
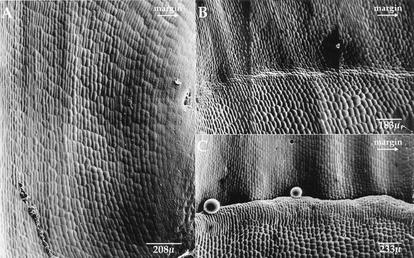
In wild type, no visible sign existed of ligule and auricle formation (Fig. 1A) until an increased region of anticlinal divisions (Fig. 1B) occurred across the adaxial (inner) leaf surface. A subset of these cells then divided periclinally (Fig. 1C) to form an outgrowth of epidermal cells that will eventually form the mature ligule. These two stages of development, the increased anticlinal divisions followed by periclinal divisions, have been termed the preligule and ligule ridge stages (Sylvester et al. 1990).
Figure 1.
Wild-type ligule and auricle development. (A) Leaf 7, plastochron 7: Anticlinal cell divisions giving rise to the ligule and auricle have not been initiated. In the top third of the image, the outer cell files begin to curve toward the margin. The ligule will form below this region. The primordium base can be seen in the lower right-hand corner. (B) Leaf 6, plastochron 8: The preligule band (anticlinal divisions) is formed across the leaf primordium. (C) Leaf 5, plastochron 10: Periclinal divisions are well advanced, causing the ligule ridge to protrude from the adaxial leaf surface. Above the ligule ridge, notice the undulating surface reflecting the underlying venation. The arrow under the word margin indicates the direction of the leaf margin.
In developing lg2-R mutant leaves, the preligule band was often incomplete (Fig. 2A). The number of cells within the preligule band decreased from margin to midrib and the band ended before the midrib. Similarly, during the ligule ridge stage the number of periclinal divisions decreased along the margin to midrib dimension (Fig. 2C). The ligule ridge decreased in height until its appearance resembled that of the preligule band. Immediately distal, a group of small square cells was observed that had undergone more anticlinal divisions than surrounding cells (Fig. 2C, brackets). This pattern, characterized by the ligule diminishing and then displacing into the blade region, was observed frequently in lg2-R mutant leaves.
Figure 2.

lg2-R mutant leaf primordium ligule and auricle development. (A) Leaf 5, plastochron 9: The preligule band (anticlinal divisions) is incompletely formed across the primordium width. (B) The transition area boxed in A is displayed at higher magnification. (C) Leaf 4, plastochron 10: The ligule ridge is not developed equally across the width of the leaf primordium. A small displacement is shown in brackets. Notice the undulating surface reflecting the underlying venation in the blade above the ligule (cf. Fig. 1C) and how this pattern continues above the displacement. The arrow under the word margin indicates the direction of the leaf margin.
The morphology of mature lg2-R leaves was consistent with the morphology of its developing leaves. A mature lg2-R mutant leaf 1 is shown in Figure 3A. At the margin, a small group of cells underwent anticlinal as well as some periclinal divisions, but a normal auricle and ligule did not form. This group of cells is likely part of the ligule region because of the distal blade and proximal sheath cell morphologies. On the remainder of the leaf, the transition from blade-like to sheath-like cells was less distinct. The characteristic blade cells apparent at the top of the figure (Fig. 3A) gradually gave way to the longer cells of the sheath. This is an example of a diffuse blade–sheath boundary.
Figure 3.
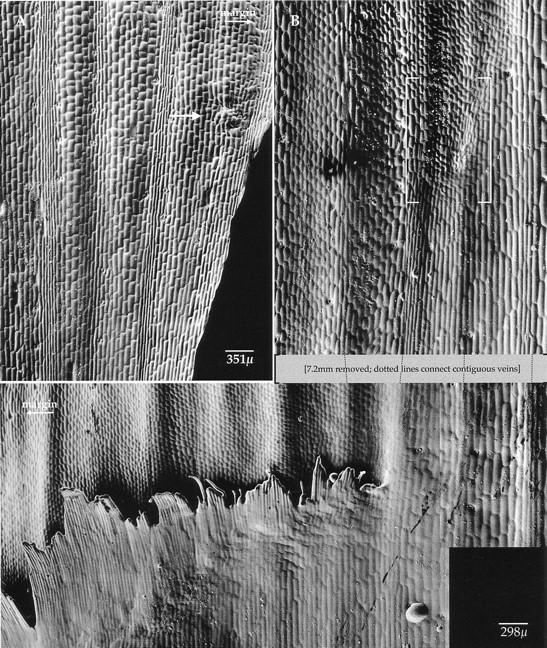
lg2-R mutant plant mature leaves. (A) Leaf 1, plastochron 13: A small ligule/auricle vestige is found at the leaf margin, as indicated by the arrow. Where this structure is missing the transition from blade to sheath is diffuse; long, rectangular sheath cells gradually yield to the short, square blade cells. (B) Leaf 4, plastochron 10: Ligule and auricle are evident at the leaf margin. These mature structures are most developed at the margin and become decreasingly less developed toward the midrib. A region of cells is displaced along a lateral vein, as shown in brackets. The arrow under the word margin indicates the direction of the leaf margin.
Figure 3B shows a mature lg2-R mutant leaf 4. A near-normal auricle and ligule were evident at the margin. The ligule, however, gradually shortened from margin to midrib and finally stopped at the position of a lateral vein. A small group of cells with neither blade nor sheath morphology was found distally and on the midrib side of the vein. This group of cells was composed of small, rounded cells and was pushed up from the surface of the leaf suggesting both anticlinal and periclinal divisions occurred in this region. The vein along which the displacement occurred separated the shorter, more crenulated blade cells (on the margin side) from the longer, noncrenulated sheath cells (on the midrib side).
Identification and genomic cloning of a lg2 transposon-induced allele
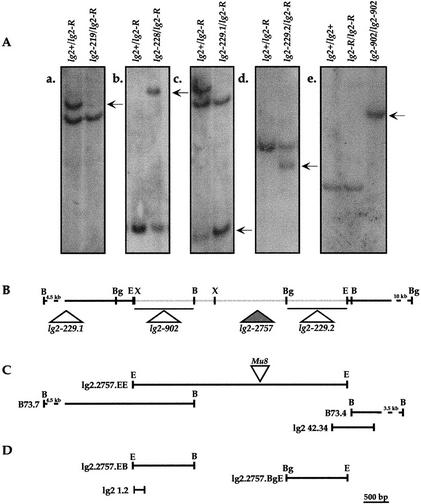
A potential Mutator (Mu) transposon-induced allele of lg2, lg2-2757, was identified in a screen of selfed Mu active families (Harper and Freeling 1996). To identify which Mu element caused the lg2 phenotype, a cosegregation analysis was performed. Genomic DNA from lg2-2757/lg2-R individuals and +/lg2-R siblings was digested with a number of different restriction enzymes. Southern blots were then hybridized sequentially with probes representing the unique interior sequence of the autonomous element MuDR and the nonautonomous elements Mu1, Mu3, Mu4, Mu5, and Mu8. The Mu8 probe hybridized to a 5.5-kb EcoRI restriction fragment that cosegregated with the lg2-2757 allele (data not shown). We cloned this fragment of DNA because EcoRI does not have a recognition site within the Mu8 element, thereby yielding unique flanking sequences.
A size-fractionated genomic bacteriophage library was made from a lg2-2757/lg2-R mutant individual and ∼750,000 clones were screened with the Mu8 probe. Fourteen positive clones were identified and grouped by restriction mapping. For each class of clones, a probe representing non-Mu8 DNA was hybridized to the original segregating family Southern blots. A clone (Fig 4C; lg2.2757.EE) was identified containing a 1.2-kb EcoRI–BamHI fragment (Fig 4D; lg2.2757.EB) that hybridized to the same 5.5-kb fragment as the Mu8 element.
Linkage to other lg2 mutant alleles
To insure that the lg2.2757.EE genomic clone represented part of the lg2 locus and not a linked locus, independently derived lg2 mutants were analyzed. Five lg2 mutations were recovered from Mu-directed tagging experiments. Mutant lg2-902 was identified in a screen of 10,000 plants (L. Harper and M. Freeling, unpubl.) and lg2-219, lg2-228, lg2-229.1, and lg2-229.2 in a separate screen of 18,000 plants. All alleles have a phenotype similar to lg2-R. After two introgressions into the inbred line W23, lg2-229.1 expressed a dramatically milder phenotype. Initial characterization of the lg2-229.1 allele shows a Mu element insertion in the first intron of the gene.
Genomic DNA from the five putative Mu-induced alleles and their respective progenitor alleles was digested with the restriction enzymes EcoRI, BamHI, XhoI, and BglII. Southern blots were hybridized to either the lg2.2757.EB probe or a 1.7-kb BglII–EcoRI probe (Fig. 4D; lg2.2757.BgE). In all cases the new mutant allele is different from the wild-type progenitor allele (Fig. 4A). lg2-902, lg2-228, lg2-229.1, and lg2-229.2 carry some form of rearrangement or insertion and lg2-219 is a deletion. Restriction mapping places the lg2-902 and lg2-229.1 lesions 5′ and the lg2-229.2 lesion 3′ of the lg2-2757 allele Mu8 insertion (Fig. 4B). The end points of the lg2-219 deletion allele have not been mapped. Neither the lg2.2757.EB nor the lg2.2757.BgE probes, however, hybridized to lg2-219 DNA. Both probes hybridize less intensely to other restriction fragments—an example is in Figure 4A(c)—indicating Lg2+ is part of a gene family.
Figure 4.
(A) Five putative Mu transposon-induced liguleless2 alleles differ from their progenitor alleles. (a–d) Restricted DNA (a and c, BamHI; b, XhoI; d, EcoRI) from putative Mu-tagged plants and their respective wild-type siblings hybridized to the lg2.2757.EB probe. (e) EcoRI-restricted DNA from a lg2-902 homozygous plant and plants homozygous for its progenitor alleles hybridized to the lg2.2757.EB probe. The arrows point to the tagged fragments. (B) Multiple independently derived alleles map to the same genomic locus. The positions of lesions are shown on a restriction map of the progenitor genomic locus. Known insertion positions are indicated by triangles and the lg2-2757 Mu8 insertion is shaded. Restriction mapped insertions and/or rearrangements are indicated by a triangle beneath a line spanning the possible region. The minimum extent of the deletion allele (lg2-219) is shown by a shaded line. (C) Cloned fragments from lg2-2757 and B73. The Mu8 position within the lg2-2757 clone is indicated by a triangle (D) DNA fragments used as radiolabeled probes.
Identification of a LG2 cDNA and genomic clones
To identify possible coding sequences, ∼400 bp from the 5′ and 3′ ends of the lg2.2757.EE genomic clone were sequenced. On the basis of a potential 240-bp open reading frame (ORF), two primers (lg2.1 and lg2.2) were synthesized for a PCR assay (Fig. 4D; lg2 1.2). Using a PCR product amplified from the lg2.2757.EE clone as a probe (Fig. 4D; lg2 1.2), ∼1.5 million cDNA clones were screened and six positive hybridizing clones identified. Sequence at the 3′ ends of four of the cDNA clones was identical over 360 bp. Three clones revealed two potential polyadenylation sites 39 bp apart (Fig. 5). The longest of these clones (1825 kb) was sequenced in its entirety. The longest ORF predicts a protein of 531 amino acids (Fig. 5). The nucleotide context of the first methionine, GCCAUGG, is identical to the PuCCAUGG Kozak consensus sequence (Kozak 1989). The cDNA contains a short 5′ untranslated sequence of 5 bp and a 3′ untranslated sequence of 226 bp.
Figure 5.
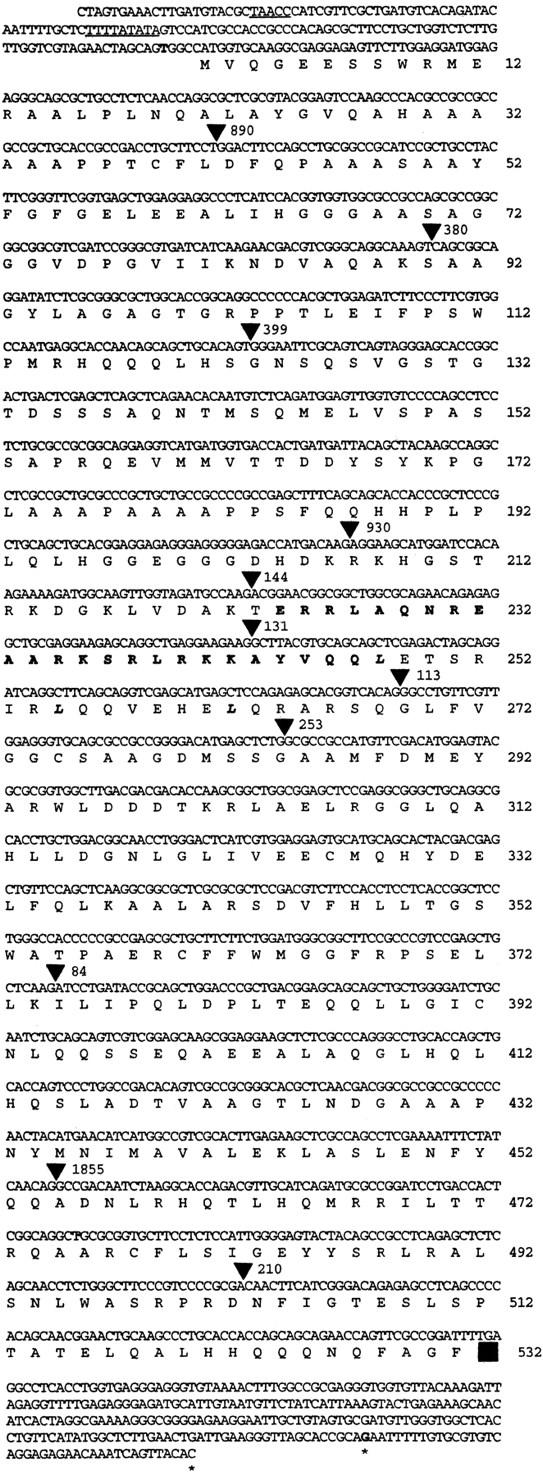
The nucleotide and predicted amino acid sequence of LG2. Numbers indicate amino acid residues. The putative TATA and CAAT boxes are underscored. The basic region (224–248) is indicated in bold and the three leucines within the leucine zipper (248, 255, and 262) are in bold italics. (▪) The stop codon; (▾) intron positions. Intron sizes are given as number of base pairs. The first and last nucleotides of the sequenced cDNA are in boldface type. Polyadenylation sites are indicated by asterisks.
To analyze further the structure of the gene, two lg2 genomic BamHI fragments were cloned from the inbred line B73 (Fig. 4C, B73.7 and B73.4). When combined with the lg2.2757.EE genomic clone, the genomic region of the cDNA is almost covered. A 750-bp PCR product (lg2 42.34) was generated to derive contiguous sequence (Fig. 4C). Comparing genomic DNA and cDNA sequences identified 11 introns (Fig. 5), all with 5′ and 3′ consensus splice sites. Intron sizes (Fig. 5) were determined by sequencing or in the case of introns 1, 4, and 10 by comparison of genomic PCR product size with the cDNA.
Alignment of the genomic sequence with the 5′ end of the cDNA sequence allowed the identification of a potential promoter region (Fig. 5). Putative TATA (TTTTATATA) and CAAT boxes (TAACC) are present 49 and 89 bp, respectively, upstream of the start of the cDNA sequence (Fig. 5). Four adenines, representing potential transcription start sites, are present 14, 20, 27, and 29 bp downstream of the putative TATA box. These differ from the consensus start of transcription, Py2CAPy5, by 1, 2, 3, and 3 bp, respectively. The presence of consensus promoter elements, transcription start sites and a translation initiation codon suggests, but does not prove, that the sequence in Figure 5 shows the complete LG2 mRNA.
The LG2 cDNA has homology to the bZIP class of transcription factors
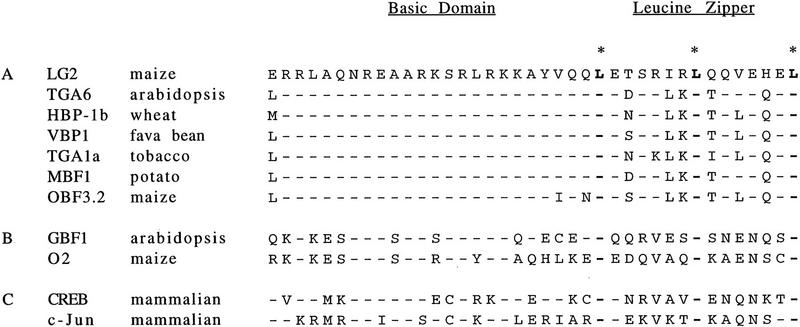
A homology search of the GenBank database using the BLAST algorithm (Altschul et al. 1990) identified the LG2 protein as a member of the bZIP class of transcription factors. The leucine zippers facilitate homo- and/or heterodimerization through hydrophobic interactions, and the adjacent basic domains bind to specific DNA sequences. The LG2 basic domain (residues 224–248) is almost identical to a plant subgroup of bZIP proteins (Fig. 6A), which is similar to a wide range of eukaryotic bZIP proteins (Fig. 6B,C). Within this subgroup, strong homology with LG2 continues through the leucine zipper and ends at the second to last exon of LG2 (residues 252–502). No proteins within the GenBank database show homology to the 224 amino-terminal residues and the 29 carboxy-terminal residues of LG2.
Figure 6.
(A) The LG2 bZIP domain is compared with closely related plant proteins (Katagiri et al. 1989; Tabata et al. 1991; Ehrlich et al. 1992; Foley et al. 1993; Feltkamp et al. 1994; Xiang et al. 1995). (B) The LG2 bZIP domain is compared with other bZIP-containing plant proteins (Hartings et al. 1989; Schmidt et al. 1990; Schindler et al. 1992a). (C) The LG2 bZIP domain is compared with two mammalian bZIP proteins (Hoeffler et al. 1988; Bohman et al. 1987). Identical amino acids are indicated by dashes, and the leucine residues within the leucine zipper by asterisks and boldface type.
RT–PCR analysis of LG2 and LG1 mRNA in wild-type and mutant developing ligular regions
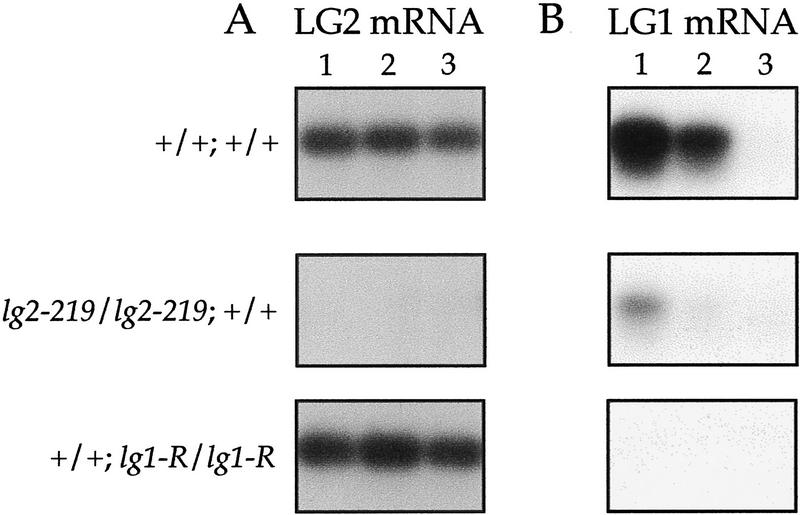
To correlate LG2 mRNA expression with ligule and auricle development, RT–PCR was used to detect LG2 mRNA in developing ligular regions of wild-type, homozygous lg1-R (deletion allele), and homozygous lg2-219 (deletion allele) plants. LG2 mRNA was detected in wild-type and lg1-R fractions (Fig. 7A). The expression is LG2 specific because no message was detected in the lg2-219 samples.
Figure 7.
RT–PCR analysis of LG2 mRNA and LG1 mRNA abundance in developing ligule regions of wild-type and mutant plants. The plant genotypes are indicated to the left. The lg2-219 and lg1-R alleles are molecular deletions. A and B show PCR fragments of LG2 RT–PCR and LG1 RT–PCR, respectively. The numbers 1, 2, and 3 refer to the RNA samples. (1) P6-8 marginal region; (2) P6-8 midrib region; (3) meristem and P1-5; (P) Plastochron.
To determine if LG1 mRNA expression is affected by the absence of LG2, we performed the complementary experiment. LG1 mRNA expression in wild-type and homozygous lg2-219 plants was compared. LG1 expression is specific because no message was detected in the lg1-R samples. LG1 mRNA was detected in wild-type and lg2-219 fractions. In wild type, LG1 mRNA expression is restricted to leaf primordia undergoing preligule band and ligule ridge divisions (Fig. 7B). LG1 wild-type expression was assayed previously by RT–PCR (Moreno et al. 1997), but the youngest developmental fractions included leaf primordia undergoing preligule band divisions. Therefore, the first developmental time at which LG1 mRNA is expressed could not be determined.
Discussion
The blade–sheath boundary incompletely forms in lg2-R mutants
The involvement of LG2 in the normal development of the blade–sheath boundary has been studied previously genetically (Harper and Freeling 1996). To determine when LG2 acts in development we examined the developing ligular region of lg2-R mutant plants. We used SEM analysis to compare the development of lg2-R mutant plants with wild-type siblings. The first visible alteration is the preligule band (anticlinal divisions), which is apparent at the primordium’s margins but does not extend across the full width of the primordium. Consequently, altered ligule ridge (periclinal divisions) and mature ligule develop. Compared to wild type, these truncated structures do not develop equally across the leaf’s width. Additionally, patches of ligule and auricle sometimes displace along lateral veins. All these disturbances may be explained if LG2 functions at or before preligule band divisions. The above lg2-R mutant phenotypes are the result of perturbations in the initial establishment of the preligule band.
An incorrectly established preligule band could explain the longitudinal displacement, as well as the lateral decrease in the extent of ligule/auricle development in lg2-R leaves. The phenotypes could be the result of either incorrect positioning or a slight delay in normal ligule and auricle development, resulting in an uncoupling from the overall development of the leaf primordium. As reviewed in the introduction, several developmental events within the leaf primordium occur temporally along both the lateral and longitudinal dimensions. If a leaf primordium’s ligule and auricle development are not synchronous with other developmental events, then cells may not be equally competent to interpret or carry out determination and differentiation processes. The result is mature structures that do not develop equally and appear at displaced positions. Further investigation is clearly needed to test this hypothesis and to determine the mechanisms.
lg2-R mutant plants also have a tassel phenotype. Preliminary observations show a reduction in lateral tassel branching and the elongation of the upper vegetative internode can be suppressed. We are able to detect LG2 mRNA in immature tassels by RT–PCR. This phenotype suggests that Lg2+ may be involved both in the later events of vegetative development and the early events of inflorescence development.
lg2 encodes a bZIP transcription factor
We cloned the lg2 locus via Mu transposon tagging and confirmed its identity by independently derived mutant alleles. LG2 shows sequence homology to the bZIP transcription factor class of proteins. LG2 is most similar to a subclass that has been found in a wide range of plant species including Arabidopsis (Kawata et al. 1992; Schindler et al. 1992a; Zhang et al. 1993; Miao et al. 1994; Xiang et al. 1995), wheat (Tabata et al. 1991), tobacco (Katagiri et al. 1989), fava bean (Ehrlich et al. 1992), potato (Feltcamp et al. 1994), and maize (Foley et al. 1993). Members of this group were first identified by their ability to bind similar promoter sequences, or by having sequence homology to each other. This bZIP subclass binds promoter sequences that include the octopine synthase (ocs) element within some Agrobacterium promoters, the activation sequence (as-1) within the CaMV 35S promoter and the hexamer (hex) motif of wheat histone gene promoters. A 20-bp DNA-binding consensus sequence has been identified (Bouchez et al. 1989) that is similar to the cAMP response element (CRE) (Lin and Green 1988).
Not only does their binding specificity characterize this subclass but so does their dimerization specificity. This group of bZIP proteins can form homodimers but cannot heterodimerize with another large subclass of plant bZIP proteins called the G-box-binding factors (Tabata et al. 1991; Schindler et al. 1992). Sequences outside of the leucine zipper contribute to dimerization specificity (Katagiri et al. 1992). Carboxy-terminal sequences stabilize dimerization and a full-length protein cannot dimerize with a protein lacking these sequences. LG2 has homology within the bZIP region as well as the carboxy-terminal regions of this group of proteins. Regions of LG2, however, are unique. It is therefore unlikely that an ortholog has been cloned from another species.
Although many plant bZIP proteins have been identified, few have a corresponding mutant phenotype. The first plant bZIP protein to be assigned a biological function by association with a mutant phenotype was the maize protein OPAQUE2 (Hartings et al. 1989; Schmidt et al. 1990). Possible functions for proteins in the subclass of bZIPs that LG2 is a member of have been proposed by studying the cis-binding sequences either transgenically, through fusions with reporter genes, or by studying plant genes with promoters containing the consensus sequence. Hypothesized functions include transcriptional regulation of plant histone (Tabata et al. 1991) and GST genes (Ulmasov et al. 1994; Zhang et al. 1995; Chengbin 1996) as well as auxin, salicylic acid, and methyl jasmonate responses (Kim et al. 1994; Liu and Lam 1994; Qin et al. 1994; Zhang and Singh 1994; Chengbin 1996). The Lg2+ gene has the first published mutant phenotype associated with this group of proteins and adds a developmental function to the above list.
Two family members have been cloned previously from maize, OBF3.1 and OBF3.2 (Foley et al. 1993). These two proteins are closely related and share 95.8% similarity over 338 amino acids. LG2 shares 57% similarity to OBF3.2 over 291 amino acids. We have cloned a fourth member (J. Walsh and M. Freeling, unpubl.) that is more similar to LG2 than to either OBF3.1 or OBF3.2. To obtain mapping information, Foley and coworkers (1993) used the OBF3.1 cDNA as a probe onto recombinant inbred lines. The probe hybridized with four restriction fragments, two of which gave a polymorphism. The two fragments mapped to two separate loci, one on chromosome three at position 105 and another on chromosome eight at position 075. They were unable to find gene specific probes and therefore were unable to determine if either of their clones mapped to these positions. The lg2 locus mapped by a three-point cross to chromosome 3 at position 101 (Brink 1933) and may therefore represent the locus on chromosome 3. The maize chromosomes 3 and 8 show colinearity of homeologous sequences (Helentjaris et al. 1988) that may be the result of an ancient segmental allotetraploidy event (Gaut and Doebley 1997). Given the mapping data, it is possible that lg2 has a closely related family member on chromosome 8 and possibly one on chromosome 3. Whether or not the putative ancient duplication gene is cloned, and whether it is functional, remains to be determined.
The identification of LG2 as a member of a bZIP protein family, raises the possibility that the ligule and auricle structures present in lg2 mutants result from related genes either compensating or having a functional redundancy. Functional redundancy can be of particular importance in maize because of its ancient duplication (Gaut and Doebley 1997). In maize, two proteins can jointly perform the functions of a single protein in another plant species (Mena et al. 1996). Consequently, if one gene is mutated then the resulting phenotype can be less severe then if an orthologous gene was mutated in another species. Within animals, compensation has been seen for a number of transcription factor gene families, including CREB/ATF (Hummler et al. 1994; Blendy et al. 1996) and myogenic HLH transcription factors (Rudnicki et al. 1992). If LG2 family members can restore the formation of ligule and auricle structures in lg2 mutants then this ability must be incomplete or we would not observe the recessive mutant phenotype. Another possibility for the ligule and auricle structures in lg2 mutant plants is transposon mosaicism. This possibility is discounted because ligule and auricle are present in plants with a deletion of the lg2 gene.
LG2 and LG1 may act in a common pathway
RT–PCR analysis indicates that LG2 mRNA accumulation precedes that of LG1 mRNA. Harper and Freeling (1996) showed that the lg1-R and the lg2-R mutant phenotypes are sensitive to the wild-type dose of the reciprocal gene, suggesting that they act temporally close to each other. Taken together, these data suggest that LG2 functions before LG1. Our data, however, also show that LG2 mRNA accumulation is neither sufficient nor necessary for LG1 accumulation.
The specific role of LG2 in the definition of the blade–sheath boundary and its associated structures is still under investigation. We can begin to place its function within development. The cell nonautonomous phenotype (Harper and Freeling 1996) of LG2 indicates it is functioning in a cell–cell signaling pathway. The identification of LG2 as a transcription factor suggests a downstream signaling function. Recent reports, however, show that some plant transcription factors can move from cell to cell (Lucas et al. 1995; Perbal et al. 1996). Whether or not LG2 protein is moving or a downstream molecule is involved in the signaling process needs to be determined. SEM analysis implicated LG1 in early stages of ligule and auricle determination and mosaic analysis experiments suggested that it has a role in the propagation or reception of a “make ligule/auricle” signal (Becraft and Freeling 1991). Our expression data is consistent with the idea that LG2 functions before LG1. LG2 may induce the make ligule-auricle signal either by further defining the blade–sheath boundary or by being directly involved in induction of the preligule region.
Materials and methods
Alleles
lg2-R is a spontaneous mutation (Brink 1933) obtained from the Maize Genetics Stock Center. lg2-2757 (also called lg2-rb) was recovered in a screen of families from selfed Mu active plants. lg2-219, lg2-228, lg2-229.1, lg2-229.2, and lg2-902 were obtained from two directed Mu tagging experiments. We crossed lg2-R/lg2-R pollen with +/+ Mu active ears. The progeny was then screened for the rare liguleless plant. A mutation in the wild-type progenitor allele, in combination with the mutant reference allele, results in the mutant phenotype. lg1-R is a spontaneous mutation (Emerson 1912) obtained from the Maize Genetics Stock center.
SEM
Dissections were done on 3-week-old seedlings from a population segregating 1:1 for lg2-R mutant plants and their wild-type siblings in the Mo17 inbred background. Plants were grown in the greenhouses of University of California at Berkeley. Dissections were performed under a dissecting microscope. Leaves were counted in succession, beginning with leaf one and continuing to the apex of the meristem, so that each leaf could be assigned both a leaf and a plastochron number. Each leaf was removed by slicing along the midrib and then gently severing the leaf’s nodal attachment. Leaves were then unrolled onto an adhesive surface with the adaxial surface up. Casts and molds of the leaf surfaces were made as described by Sylvester and coworkers (1990). Casts were mounted on aluminum stubs with epoxy glue, coated with 25 nm of gold-palladium with a Polaron sputter coater, and observed using a Topcon ISI DS130 SEM operating at an accelerating voltage of 10 kV. All images are composite images, assembled from two or more 3 × 4-inch SEM Polaroid negatives. Negatives were scanned using a flatbed scanner at 600–800 ppi, then assembled on Adobe Photoshop 4.0.
DNA isolation and Southern analysis
Maize genomic DNA was isolated from seedling and adult leaf tissues as described (Lisch et al. 1995). Southern analysis was performed with the following modifications according to Schneeberger et al. (1995). The hybridization solution contained 6× SSC, 2 mm EDTA, 10 mm Tris/HCl (pH 7.5), 5× Denhardts, 0.2 mg/ml of salmon sperm DNA, 20 mm sodium phosphate buffer (pH 7), and 1% N-lauryl-sarkosyl. Blots were washed to a final stringency of 0.1× SSC and 0.1% SDS at 65°C. A 530-bp DNA fragment internal to the Mu8 terminal inverted repeats was used for the Mu8 probe. The lg2 probes (lg2.2757.EB and lg2.2757.BgE) are described in Figure 4D.
Genomic cloning
Three genomic libraries were made. For the first library, DNA from a lg2-2757/lg2-R mature leaf was digested with EcoRI and enriched by gel electroelution for 5.5-kb fragments. The library was constructed in EcoRI predigested λ Zap II (Stratagene) vector and packaged using Stratagene’s Gigapack packaging extracts. Nylon or nitrocellulose lifts were hybridized at 65°C with the Mu8 probe and washed with 0.1× SSC and 0.1% SDS. Stratagene’s in vivo excision method was used to generate the lg2.2757.EE insert in the pBluescript SK− vector.
Two separate size-fractionated genomic libraries were made from the maize inbred B73. Immature ear DNA was digested with BamHI and two fractionations of 4 and 7 kb were isolated by gel electroelution. Both libraries were constructed in the BamHI predigested Lambda Zap Express vector (Stratagene). The 7-kb library was screened with a radio-labeled 5′ cDNA probe (Fig. 5, nucleotide 131–880) and the 4-kb library was screened with a 3′ cDNA probe (Fig. 5, nucleotide 1538–1956). In vivo excision resulted in the B73.7 insert and B73.4 insert (Fig. 4D) within the pBK–CMV vector.
PCR was used to amplify a 750-bp product from B73 genomic DNA that contains sequences not included in the above genomic clones. The primers used were 5′-caaagtcgctaggagcgcag and 3′-gttttacaccctccctcaccagg. The product was purified by gel electroelution.
cDNA cloning
As we were not certain of the site of LG2 mRNA expression we chose to make a cDNA library from tissues that included developing ligule and auricle regions. Two-week-old B73 seedlings were dissected by removing the first two leaves and then cutting the seedling below the meristem and above the developing ligule of leaf three. The remaining tissues included the meristem and 9–10 whole or partial leaf primordia with their ligular regions at different stages of development. Total RNA and Poly(A)+ RNA were isolated according to Kloeckener-Gruissem and coworkers. (1992). The cDNA library was constructed using the Lambda Zap II cDNA Synthesis Kit (Stratagene). Primary clones were screened, as described above for the genomic libraries, with the lg2 1.2 probe (Fig. 4D). The lg2 1.2 DNA fragment was obtained by PCR (primer 1, gcagtcagtagggagcaccggca; primer 2, gcgggtggtgctgctgaaagctc) using the lg2.2757.EE clone as template. Clones were in vivo excised into the pBluescript SK− vector.
RT–PCR
RT–PCR analysis was performed on RNA isolated from 16-day-old plants. We used wild-type and mutant plants homozygous for either the lg2-219 deletion allele or the lg1-R deletion allele. Leaf primordia (leaf 4–6; plastochron 6–8) were removed from the shoot and cut 5 mm above their base. The primordia were then cut on either side of the thickened midrib region. The midrib regions of all three plastochrons were pooled, as were the marginal regions. The meristem and remaining five plastochrons were used as a third sample. This dissection resulted in three fractions—the margins of primordia undergoing preligule (anticlinal) and ligule ridge (periclinal) divisions, the midribs of the same primordia, and a third fraction that includes the meristem and leaf primordia at stages before the preligule band divisions. RNA was isolated as described previously (Moreno et al. 1997) for RT–PCR analysis.
RT–PCR was used to detect LG2 and LG1 mRNA. A UBIQUITIN RT–PCR reaction was done on all mRNA samples to confirm their integrity (data not shown). The LG2 and LG1 reverse transcription reactions were performed on 2 μg of total RNA and the UBIQUITIN reactions on 0.5 μg. The LG1 and UBIQUITIN reactions were performed as described previously (Moreno et al. 1997). The same conditions were used for the LG2 reactions. A Lg2 3′ untranslated primer (gttttacaccctccctcaccagg) was used in the RT reaction. The PCR reaction was done with a 5′ primer (tcaacaggccgacaatctaaggc) and a nested 3′ primer (gtgaggcctcaaaatccggc) such that the product spans intron 11. The RNA product is 248 bp and any contaminating genomic DNA is 458 bp. RT–PCR reactions were run on 1.5% agarose gels, Southern blotted, and then hybridized with specific radio-labeled probes.
Sequencing
Plasmid DNA used in sequencing reactions was obtained from overnight liquid cultures using Qiagen’s QIAprep spin miniprep kit. Sequencing was done at the U.C. Berkeley DNA Sequencing Facility with an Applied Biosystems automated sequencer.
Acknowledgments
We are especially grateful to members of the Freeling laboratory for many helpful discussions and suggestions during the course of this work. We thank Barbara Kloeckener-Gruissem, Lisa Harper, Mark Mooney, Lynne Jesaitis, David Braun, Barbara Lane, and Robin MacDiarmid for critical reading and helpful comments on this manuscript. We are grateful to Lisa Harper for providing genetic strains and Barbara Kloeckener-Gruissem for providing the mRNA used for the cDNA library. We thank Scott Poethig for identifying the lg2 tassel phenotype. We thank the Berkeley Electron Microscope Laboratory and the Center For Biological Imaging for providing facilities and technical advice. This work was supported by a grant from the U.S. Department of Energy (DOE grant DE-FG03-91ER20028).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL freeling@nature.berkeley.edu; FAX (510) 642-4995.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Becraft PW, Bongard-Pierce DK, Sylvester AW, Poethig RS, Freeling M. The liguleless-1 gene acts tissue specifically in maize leaf development. Dev Biol. 1990;141:220–232. doi: 10.1016/0012-1606(90)90117-2. [DOI] [PubMed] [Google Scholar]
- Becraft PW, Freeling M. Sectors of liguleless-1 tissue interrupt an inductive signal during maize leaf development. Plant Cell. 1991;3:801–807. doi: 10.1105/tpc.3.8.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blendy JA, Kaestner KH, Schmid W, Gass P, Schutz G. Targeting of the CREB gene leads to up-regulation of a novel CREB mRNA isoform. EMBO J. 1996;15:1098–1106. [PMC free article] [PubMed] [Google Scholar]
- Bohmann D, Bos TJ, Admon A, Nishimura T, Vogt PK, Tjian R. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science. 1987;238:1386–1392. doi: 10.1126/science.2825349. [DOI] [PubMed] [Google Scholar]
- Bouchez D, Tokuhisa JG, Llewellyn DJ, Dennis ES, Ellis JG. The ocs-element is a component of the promoters of several T-DNA and plant viral genes. EMBO J. 1989;8:4197–4204. doi: 10.1002/j.1460-2075.1989.tb08605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink RA. Heritable characters in maize XLVI- Liguleless 2. J Heredity. 1933;24:325–326. [Google Scholar]
- Chengbin X, Miao ZH, Lam E. Coordinated activation of as-1-type elements and a tobacco glutathione S-transferase gene by auxins, salicylic acid, methyl-jasmonate and hydrogen peroxide. Plant Mol Biol. 1996;32:415–426. doi: 10.1007/BF00019093. [DOI] [PubMed] [Google Scholar]
- Cocciolone SM, Cone KC. Pl-Bh an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics. 1993;135:575–588. doi: 10.1093/genetics/135.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich KC, Cary JW, Ehrlich M. A broad bean cDNA clone encoding a DNA-binding protein resembling mammalian CREB in its sequence specificity and DNA methylation sensitivity. Gene. 1992;117:169–178. doi: 10.1016/0378-1119(92)90726-6. [DOI] [PubMed] [Google Scholar]
- Emerson RA. The inheritance of the ligule and auricles of corn leaves. Nebr Agric Exp Stn Annu Rep. 1912;25:81–88. [Google Scholar]
- Feltkamp D, Masterson R, Starke J, Rosahl S. Analysis of the involvement of ocs-like bZip-binding elements in the differential strength of the bidirectional mas1“2” promoter. Plant Physiol. 1994;105:259–268. doi: 10.1104/pp.105.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley RC, Grossman C, Ellis JG, Llewellyn DJ, Dennis ES, Peacock WJ, Singh KB. Isolation of a maize bZIP protein subfamily: Candidates for the ocs-element transcription factor. Plant J. 1993;3:669–679. [PubMed] [Google Scholar]
- Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L, Freeling M. Interactions of liguleless1 and liguleless2 function during ligule induction in maize. Genetics. 1996;144:1871–1882. doi: 10.1093/genetics/144.4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartings H, Maddaloni M, Lazzaroni N, Di Fonzo N, Motto M, Salamini F, Thompson R. The O2 gene which regulates zein deposition in maize endosperm encodes a protein with structural homologies to transcriptional activators. EMBO J. 1989;8:2795–2802. doi: 10.1002/j.1460-2075.1989.tb08425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helentjaris T, Weber D, Wright S. Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics. 1988;118:353–364. doi: 10.1093/genetics/118.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffler JP, Meyer TE, Yun Y, Jameson JL, Habener JF. Cyclic AMP-responsive DNA-binding protein: Structure based on a cloned placental complementary DNA. Science. 1988;242:1430–1433. doi: 10.1126/science.2974179. [DOI] [PubMed] [Google Scholar]
- Hummler E, Cole TJ, Blendy JA, Ganss R, Aguzzi A, Schmid W, Beermann F, Schutz G. Targeted mutation of the CREB gene: Compensation within the CREB-ATF family of transcription factors. Proc Natl Acad Sci. 1994;91:5647–5651. doi: 10.1073/pnas.91.12.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Lam E, Chua NH. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature. 1989;340:727–730. doi: 10.1038/340727a0. [DOI] [PubMed] [Google Scholar]
- Katagiri F, Seipel K, Chua NH. Identification of a novel dimer stabilization region in a plant bZIP transcription activator. Mol Cell Biol. 1992;12:4809–4816. doi: 10.1128/mcb.12.11.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata T, Imada H, Shiraishi K, Okada Y, Shimura Y, Iwabuchi M. A cDNA clone encoding hbp-1b homolog in Arabidopsis thaliana. Nucleic Acids Res. 1992;20:1141. doi: 10.1093/nar/20.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Buckley K, Costa MA, An G. A 20 nucleotide upstream element is essential for the nopaline synthase (nos) promoter activity. Plant Mol Biol. 1994;24:105–117. doi: 10.1007/BF00040578. [DOI] [PubMed] [Google Scholar]
- Kloeckener-Gruissem B, Vogel JM, Freeling M. The TATA box promoter region of maize Adh1 affects organ-specific expression. EMBO J. 1992;11:157–166. doi: 10.1002/j.1460-2075.1992.tb05038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Context effects and inefficient initiation at non-AUG codons in eukaryotic cell-free translation systems. Mol Cell Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YS, Green MR. Interaction of a common cellular transcription factor, ATF, with regulatory elements in both E1a-inducible and cyclic AMP-inducible promoters. Proc Natl Acad Sci. 1988;85:3396–3400. doi: 10.1073/pnas.85.10.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D, Chomet P, Freeling M. Genetic characterization of the mutator system in maize: Behavior and regulation of Mu transposons in a minimal line. Genetics. 1985;139:1777–1796. doi: 10.1093/genetics/139.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lam E. Two binding sites for the plant transcription factor ASF-1 can respond to auxin treatments in transgenic tobacco. J Biol Chem. 1994;269:668–675. [PubMed] [Google Scholar]
- Lucas WJ, Bouche-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, Hake S. Selective trafficking of KNOTTED I homeodomain protein and its mRNA through plasmodesmata. Science. 1995;270:1980–1983. doi: 10.1126/science.270.5244.1980. [DOI] [PubMed] [Google Scholar]
- Mena M, Ambrose BA, Meeley RB, Briggs SP, Yanofsky MF, Schmidt RJ. Diversification of C-function activity in maize flower development. Science. 1996;274:1537–1540. doi: 10.1126/science.274.5292.1537. [DOI] [PubMed] [Google Scholar]
- Miao ZH, Liu X, Lam E. TGA3 is a distinct member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol Biol. 1994;25:1–11. doi: 10.1007/BF00024193. [DOI] [PubMed] [Google Scholar]
- Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M. liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes & Dev. 1997;11:616–628. doi: 10.1101/gad.11.5.616. [DOI] [PubMed] [Google Scholar]
- Perbal M-C, Haughn G, Saedler H, Schwartz-Sommer Z. Non-cell-autonomous function of the Antirrinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development. 1996;122:3433–3441. doi: 10.1242/dev.122.11.3433. [DOI] [PubMed] [Google Scholar]
- Poethig RS. Cellular parameters of leaf morphogenesis in maize and tobacco. In: White RA, Dickinson WC, editors. Contemporary problems of plant anatomy. New York, NY: Academic Press; 1984. pp. 235–238. [Google Scholar]
- Qin XF, Holuigue L, Horvath DM, Chua NH. Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell. 1994;6:863–874. doi: 10.1105/tpc.6.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of Myod in mice leads to up-regulation of the myogenic HLH gene myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Schindler U, Beckmann H, Cashmore AR. TGA1 and G-Box binding factors two distinct classes of Arabidopsis leucine zipper proteins compete for the G-box-like element tgacgtgg. Plant Cell. 1992a;4:1309–1319. doi: 10.1105/tpc.4.10.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR. Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J. 1992b;11:1261–1273. doi: 10.1002/j.1460-2075.1992.tb05170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Burr FA, Auckerman MJ, Burr B. Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA. Proc Natl Acad Sci. 1990;87:46–50. doi: 10.1073/pnas.87.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger RG, Becraft PW, Hake S, Freeling M. Ectopic expression of the knox homeo box gene rough sheath1 alters cell fate in the maize leaf. Genes & Dev. 1995;9:2292–2304. doi: 10.1101/gad.9.18.2292. [DOI] [PubMed] [Google Scholar]
- Sharman BC. Developmental anatomy of the shoot of Zea mays L. Ann Bot. 1942;6:245–284. [Google Scholar]
- Sylvester AW, Cande WZ, Freeling M. Division and differentiation during normal and liguleless-1 maize leaf development. Development. 1990;110:985–1100. doi: 10.1242/dev.110.3.985. [DOI] [PubMed] [Google Scholar]
- Sylvester AW, Smith L, Freeling M. Acquisition of identity in the developing leaf. Annu Rev Cell Dev Biol. 1996;12:257–304. doi: 10.1146/annurev.cellbio.12.1.257. [DOI] [PubMed] [Google Scholar]
- Tabata T, Nakayama T, Mikami K, Iwabuchi M. HBP-1a and HBP-1b: Leucine zipper-type transcription factors of wheat. EMBO J. 1991;10:1459–1468. doi: 10.1002/j.1460-2075.1991.tb07666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle T. The ocs element in the soybean GH2-4 promoter is activated by both active and inactive auxin and salicylic acid analogues. Plant Mol Biol. 1994;26:1055–1064. doi: 10.1007/BF00040688. [DOI] [PubMed] [Google Scholar]
- Xiang C, Miao Z, Lam E. Isolation of TGA6 (accession no. L42327), a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana (PGR 95-063) Plant Physiol. 1995;109:721. [Google Scholar]
- Zhang B, Singh KB. Ocs element promoter sequences are activated by auxin and salicylic acid in Arabidopsis. Proc Natl Acad Sci. 1994;91:2507–2511. doi: 10.1073/pnas.91.7.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Foley RC, Singh KB. Isolation and characterization of two related Arabidopsis ocs-element bZIP binding proteins. Plant J. 1993;4:711–716. doi: 10.1046/j.1365-313x.1993.04040711.x. [DOI] [PubMed] [Google Scholar]
- Zhang B, Chen W, Foley RC, Buttner M, Singh KB. Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell. 1995;7:2241–2252. doi: 10.1105/tpc.7.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]