Abstract
Cell division is generally thought to be a process that produces an exact copy of the mother cell by precisely replicating its genomic DNA, doubling organelles, and segregating them into two cells. Yet, many cell types from bacteria to human cells divide asymmetrically to generate daughter cells with distinct characteristics. Such asymmetric divisions are fundamental to a cell’s life span, embryonic development, and stem cell homeostasis. Asymmetric division requires coordination of cellular asymmetry and the cell division machinery. Accumulating evidence suggests that the basic molecular mechanisms that govern this process are conserved from yeast to humans. In this review, we highlight similarities in the mechanisms of asymmetric cell division in yeast and Drosophila male germline stem cells, in the hope of extracting the common themes underlying many systems.
Asymmetric cell division
Asymmetric division is a fundamental process widely observed from bacteria to humans. For example, budding yeast divide asymmetrically into two daughter cells that are different in size, age, and ability to switch mating type. Many embryonic cells undergo asymmetric divisions to create a diverse array of different cell types. Furthermore, many adult stem cells divide asymmetrically to balance self-renewal and commitment to differentiation, contributing to tissue homeostasis. Thus, implications of a failure in asymmetric cell division are huge: yeast cells may age precociously, embryonic development might not succeed, and organisms may develop cancer or suffer degeneration.
Asymmetric cell division is usually achieved by asymmetric segregation of fate determining factors; such factors may be proteins/mRNAs/organelles that are intrinsically expressed in the cell of interest, polarized within the cell, and segregated asymmetrically upon division (Figure 1A, C). Alternatively, the fate determining factors may be the contact between the cell of interest and neighboring cells/extracellular matrix that signal to the cell of interest (Figure 1B).
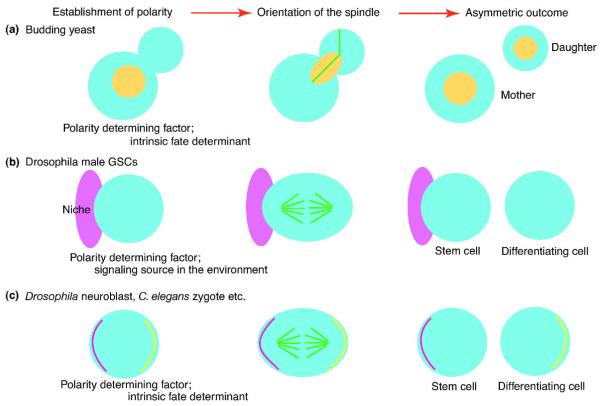
Figure 1.
The mechanism of asymmetric cell division: coordination of cell polarity and spindle orientation.
A) In budding yeast, cells divide asymmetrically by orienting the spindle (green) perpendicularly to the site of cell division (bud neck). Components of the bud neck and cell polarity factors are important for spindle orientation. Importantly, due to the pre-established site of cytokinesis, the mitotic spindle must orient along the mother-daughter polarity axis to allow faithful chromosome segregation. Nuclei are shown in yellow circle/oval.
B) In some cells, such as Drosophila male GSCs, cells divide asymmetrically by orienting the spindle toward the extrinsic fate determinant (signaling source, niche; pink) and segregating contact with the signaling source unequally between two daughter cells.
C) In some cells, such as Drosophila neuroblasts and cells in C. elegans early embryos, the intrinsic fate determinants (yellow and magenta lines) are polarized within the cell and segregated unequally into daughter cells with the aid of the oriented spindle.
Often, the axis of cell asymmetry is predetermined prior to the actual time of division (i.e. at mitosis). For example, in budding yeast, the site of cytokinesis (constriction between mother and daughter cells, named the bud neck) is defined at early stages of the cell cycle (Figure 1A). Also, in asymmetrically dividing cells from multicellular organisms, fate determinants (intrinsic or extrinsic; Figure 1B, C) are often polarized prior to mitosis. This feature — the fact that the establishment of cell asymmetry precedes cell division — has two important implications for the spatial and temporal coordination of spindle orientation with cell polarity cues and the cell cycle control machinery. First, an efficient mechanism must be in place to provide proper orientation of the spindle regarding the mother-daughter polarity axis (spatial coordination). Second, feedback control mechanisms must ensure that mitosis and cytokinesis will not be completed until proper spindle alignment is achieved (temporal coordination).
Although major progress has been made in the understanding of asymmetric cell division in recent years, there still is much to learn about how cells create two daughter cells with distinct fates. Budding yeast has been the leading model organism in which to study the molecular and cellular mechanisms of spindle orientation, and much has been elucidated using these tiny cells. Recent progress in the understanding of asymmetric cell division in multicellular organisms has highlighted striking similarities in molecular and cellular mechanisms utilized in yeast and multicellular organisms. Here, we aim to summarize the parallels between cells from yeast and those from multicellular organisms, using Drosophila male germline stem cells (GSCs) as a major example. Our hope is to facilitate research on asymmetric cell division in higher eukaryotes including humans via side-by-side comparison of yeast and other systems, so that researchers can fully exploit the rich knowledge obtained from studies in yeast.
Asymmetric division of budding yeast and Drosophila male GSCs
Budding yeast cells divide asymmetrically by producing daughter cells that differ from their mothers in cell size, molecular composition and replicative life span (reviewed in 1, 2). During initial stages of the cell cycle, the mother cell produces a future daughter cell (named the bud), which grows and receives organelles and cellular components (proteins, mRNAs) from its mother. An intrinsic cell polarization mechanism contributes to the targeted secretion and/or asymmetric segregation of sub-cellular components between mother and daughter cells. Shortly after cytokinesis, the cell polarity axis of a newly born yeast cell is defined by the activation of the Rho-like GTPase Cdc42 at the incipient site of bud emergence3. At the bud site, active GTP-bound Cdc42 drives the formation of actin cables that extend from the bud tip into the mother cell. Actin cables are involved in polarized transport of components required for bud growth and microtubule (MT) anchoring 3-5. During this process, a scaffold complex composed of the small GTPases septins accumulate at the boundary between mother and the growing bud (bud neck)6. As the site of cell division is specified by the bud neck, accurate segregation of the genetic material is assured by positioning of the mitotic spindle along the established mother to daughter cell polarity axis (Figure 1A).
Drosophila male GSCs divide asymmetrically by placing the daughter cells into distinct cellular microenvironments (Figure 1B). Asymmetric stem cell division here refers to asymmetry in the fates of two daughter cells: one daughter of the stem cell division remains as a stem cell (stem cell self-renewal) and the other initiates differentiation. Unlike yeast and some other model systems, where cell-intrinsic fate determinants regulate daughters’ fates (Figure 1A, C), no intrinsic fate determinants have been identified in male GSCs so far. Instead, factors that specify GSC identity are provided from the microenvironment. GSCs attach to the somatic support cells called the hub and cyst stem cells (CySCs) that constitute the stem cell niche (Figure 1B, Figure 2B). The term “stem cell niche” refers to a special microenvironment, where stem cells reside and receive factors that are essential for stem cell regulation, such as maintenance of stem cell identity and control of proliferation vs. quiescence. The hub cells secrete a signaling ligand, Unpaired (Upd), to activate the Janus kinase/signal transducer and activators of transcription (JAK-STAT) pathway within GSCs and CySCs and specify their identity7-10. GSCs need to interact with hub cells and CySCs to maintain GSC identity. Adherens junction formed between GSCs and the hub is critical for GSC to stay within the niche and receive the signals from the hub and CySCs. During asymmetric division, one daughter maintains the attachment to the hub and CySCs, retaining stem cell identity, while the other is displaced away from the hub, committing to differentiation. Although Drosophila female GSCs also divide asymmetrically, parallel between them and the yeast system is much less prominent, and thus we do not discuss the details of their asymmetric division (asymmetric female GSC division is reviewed in 7, 8).
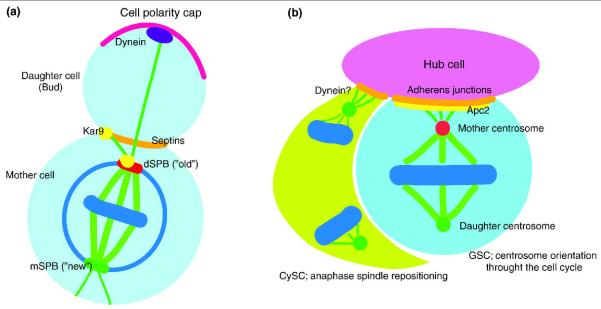
Figure 2.
The mechanism of spindle orientation.
A) In budding yeast, spindle formation starts in the mother cell body with the older centrosome/SPB (red) oriented towards the bud by the action of Kar9 (yellow; Apc2 homolog) and associated proteins (not depicted). The Kar9 pathway positions the nucleus at the bud neck region, which is maintained by the septin scaffold (orange). The dynein-dependent pathway (purple) works at later stages of the cell cycle and provides the forces to move the spindle into the daughter cell, allowing proper spindle orientation. If either the Kar9 or Dyn1 pathways fail, mitotic spindle elongation occurs inside the mother cells and causes the activation of SPOC components (not depicted). Chromosomes, blue; cell polarity cap, magenta; spindle, green.
In Drosophila male GSCs (light blue), the mother centrosome (red circle) is associated with the cell-cell junction between GSCs and hub cells (pink), the major component of the stem cell niche, leading to perpendicular orientation of spindles. As a result, GSCs divide asymmetrically. Apc2 (yellow), a putative Kar9 homolog, concentrates at the cell-cell junction to anchor the mother centrosome, while the daughter centrosome is on the opposite side (green circle). In CySCs (lime) of the Drosophila testis, which associate with GSCs and hub cells, spindle repositioning during anaphase results in asymmetric stem cell division. Spindles are formed with random orientation but rotate during mitosis to acquire the desired orientation of the spindle. Dynein might function to pull the spindle pole (green) to the adherens junction (orange).
Mechanism of spindle orientation: Spatial coordination of cellular asymmetry and spindle
Cellular asymmetries must be coordinated with the division plane to achieve an asymmetric outcome of the division. For this purpose, many cell types use spindle orientation with respect to the cellular asymmetry. Components involved in the spatial control of spindle alignment are conserved from yeast to human cells. Knowledge obtained from detailed studies in yeast has allowed researchers to use candidate approaches to identify genes required for spindle orientation in higher eukaryotes. Collectively, the accumulating evidence illuminates the importance of i) establishment of cortical polarity, ii) association of fate determining factors with the cortical polarity, and iii) linkage of the cell cortex and spindle machinery.
Spindle orientation in yeast
In budding yeast, the mitotic spindle must orient towards the daughter cell already at early stages of the cell cycle. The spatial coordination of spindle alignment with respect to the cell polarity axis is a tightly regulated process that results from the concerted action of two functionally redundant pathways, named the Kar9 and dynein pathways (Figure 2A). The Kar9 pathway functions at an early stage of the cell cycle (pre-anaphase) and is important for the positioning of the nucleus at the bud neck, whereas the dynein pathway functions during anaphase to provide pulling forces to orient the nucleus along the mother to daughter polarity axis11-14,15, 16, 17. Concomitant deletion of genes involved in both pathways is lethal for cell survival, while single deletions of each gene have little effect on survival despite the high rate of spindle mis-alignment.
Spindle orientation in Drosophila male GSC
Drosophila male GSCs always undergo asymmetric division by asymmetrically inheriting the adhesion to the hub cells (Figure 2B) 18. Because of this, the contact site between the hub and GSCs behaves as a “spatial reference point” toward which GSCs are obligated to orient their spindles. Many other stem cells in multicellular organisms are known to have intrinsic/extrinsic polarity toward which their division must be oriented. Therefore, even without a preformed cytokinetic site (e.g., bud neck of the yeast), many cells have spatial restrictions regarding the division plane and thus require a mechanism for spindle orientation.
In Drosophila male GSCs, the centrosome is oriented toward the hub-GSC interface throughout the cell cycle (Figure 2B)18. One centrosome, which is the mother centrosome (see below), is always located close to the hub, while the other (the daughter centrosome) moves to the opposite side: in this manner, the GSC’s spindle orientation is predetermined during interphase, much earlier than the time point of actual commitment to mitosis 19. Correct centrosome orientation toward the hub cells requires adherens junction composed of E-cadherin and β-catenin, Cnn (Centrosomin, a core component of pericentriolar material (PCM) required for recruitment of many PCM proteins and anchorage of astral MTs, to the centrosome 20, 21), and Apc2 [a homolog of the mammalian adenomatous polyposis coli (APC)]18, 22. Apc2 is believed to connect astral MTs to the adherens junction/actin cytoskeleton network formed between hub cells and GSCs to anchor the centrosome.
Similarity between yeast and GSC
Interestingly, yeast Kar9 shows homology to Apc223. Kar9 binds to the MT plus end-associated protein Bim1 (EB1 in other organisms) at the spindle pole body (SPB; the MT organizing center (MTOC) of yeast, Figure 2A), suggesting a striking parallel in the mechanism of spindle orientation in budding yeast and Drosophila male GSCs. Parallel between budding yeast and Drosophila male GSCs extends to the usage of mother vs. daughter centrosome/SPB during asymmetric cell division. In Drosophila male GSCs, the mother centrosome always stays close to the hub cells, while the daughter centrosome migrates toward the opposite side of the cell and is inherited by the differentiating daughter19 (Figure 2B). In budding yeast, the daughter cell inherits the old SPB whereas the mother cell retains the new SPB24. How is a defined pattern of centrosome/SPB inheritance achieved in every cell cycle? Several lines of evidence suggest that the old (mother) centrosome/SPB has higher MT nucleation activity than the new one (daughter). The temporal delay in MT organization allows the old centrosome/SPB to be the first one to be captured by factors that connects them to the defined cortical site (bud cortex in the yeast, or hub-GSC interface in the Drosophila male GSCs) (reviewed in 14,8, 17).
A similar phenomenon is observed in mouse neural glial progenitor cells 25, suggesting that centrosome/SPB asymmetry is an evolutionarily conserved mechanism to orient the mitotic spindle. Interestingly, recent work showed that, in the Drosophila neuroblast, the older centrosome is always inherited by the differentiating cell, while the neuroblast (stem cell) inherits the newly formed centrosome 26, 27. In this case, the daughter centrosome acquires robust MT organizing activity, whereas the mother centrosome, which had active MTOC activity in the previous cell cycle, is inactivated. This elaborate mechanism of switching MTOC activity every cell cycle makes one wonder whether the inheritance of a certain centrosome is critically important for cell behavior, beyond the extent to simply orient the mitotic spindle in a particular way. In summary, these findings, started from study on budding yeast24, illuminate striking similarity in spindle orientation conserved from yeast to mammals.
Other spindle orientation mechanisms
Many other stem cells, including Drosophila neuroblasts, mammalian radial glial progenitor cells, and mammalian skin stem cells, use the evolutionarily conserved polarity complexes Par3/Par6/aPKC and PINS(LGN)/Mud(NuMA)/Gαi for asymmetric division. Many of these polarity complex components were originally identified in a screen of C. elegans mutant embryos28 that were defective in polarization, suggesting that these cell polarity machineries are not specific for stem cells. These systems also rely on the interplay between the MT network and cell cortex-associated components for their spindle orientation. However, homologs of this pathway have not been found in yeast, and, in addition, it is currently unknown whether these systems have a surveillance system for temporal coordination equivalent to the SPOC (spindle position checkpoint) in budding yeast or COC (centrosome orientation checkpoint) in Drosophila male GSCs, as discussed below. Therefore, for the purpose of highlighting parallels in both spatial and temporal coordination, we do not further discuss these mechanisms that depend on Par3/Par6/aPKC, PINS(LGN)/Mud(NuMA)/Gαi and associated proteins. Thus, readers are encouraged to read excellent recent reviews on this topic29,30,31.
Recent work has revealed another type of cellular mechanism for asymmetric stem cell division. Cyst stem cells (CySCs) that encapsulate and regulate GSCs divide asymmetrically by repositioning the mitotic spindle at or around the onset of anaphase (Figure 2B) 32. Like GSCs, CySCs also attach to the hub cells and require the signaling ligand Upd; thus, attachment to the hub cells is the critical determinant of CySC identity 33. CySCs form the mitotic spindle in a random orientation/position within the cell; however, one spindle pole is pulled close to the hub-CySC interface around anaphase, allowing asymmetric segregation of the hub contact site between daughter cells 32. This spindle repositioning requires Cnn (thus, presumably astral MTs), the dynein complex components Lis-1 and Glued, and the actin-cortex anchor moesin, but not Apc2. CySC division might be more analogous to budding yeast spindle orientation in that both involve the pulling of the spindle pole to the cortical site. However, Apc2, a potential Kar9 homolog, is not required in CySC spindle repositioning. It awaits future work to integrate CySC spindle repositioning mechanism in the broader context, allowing the comparison with the mechanisms of spindle orientation in other systems.
Mechanism of temporal coordination: Spindle position checkpoint and centrosome orientation checkpoint
In budding yeast, a failure in spindle orientation along the mother-daughter cell polarity axis activates the spindle position checkpoint (SPOC) (Figure 3A). This checkpoint is particularly important in budding yeast because the site of cytokinesis is specified at a very early stage in the cell cycle, well before SPB duplication and spindle formation (reviewed in 34, 35). Consequently, the mitotic spindle must align perpendicularly to the plane of cell division before the processes of mitotic exit and cytokinesis are initiated. The SPOC provides a cell cycle “wait” signal that allows completion of mitosis only after full elongation of the mitotic spindle between mother and daughter cells. Without this wait signal, the down-regulation of the mitotic cyclins would lead to continued cell cycle progression without proper chromosome segregation between mother and daughter cells, resulting in multi- and anucleated progenies accompanied by cell death.
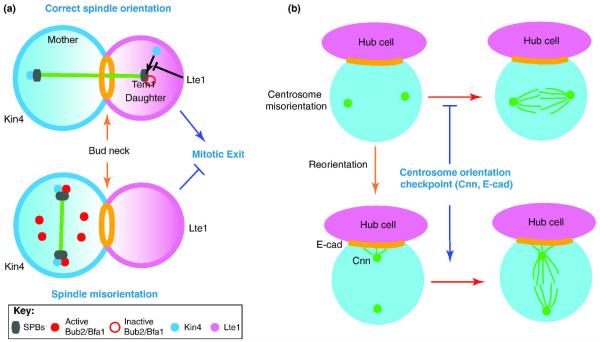
Figure 3.
Mechanisms of SPOC and COC.
(A) In budding yeast, the spindle position checkpoint (SPOC) prevents exit from mitosis and cytokinesis if the mitotic spindle (green bar) fails to orient along the mother-daughter axis. The function of the SPOC is to keep the small GTPase Tem1 in the GDP-inactive state on the daughter-bound SPB. Tem1 activation is required to trigger the mitotic exit network (MEN) signaling cascade (a pathway that leads to inactivation of mitotic Cdk1). The most downstream SPOC component, Bub2 and Bfa1, form a GAP complex that keeps Tem1 inactive. If the spindle is misoriented, the mother cell enriched kinase Kin4 prevents the inactivation of the Bub2-Bfa1 GAP complex through the phosphorylation of Bfa1 at the SPBs. If the spindle is correctly aligned, Kin4 activity is inhibited, in part by the Lte1 protein (which is confined in the daughter cell body). Exclusion of Kin4 from the daughter cell compartment allows the inactivation of the Bub2-Bfa1 GAP complex by the polo-like kinase Cdc5, thereby facilitating mitotic exit (see text for details).
(B) In Drosophila male GSCs, a surveillance mechanism (centrosome orientation checkpoint; COC) monitors correct centrosome orientation with respect to the hub cells. When the centrosomes (green dots) are misoriented (i.e. neither of the two centrosomes is located close to the hub-GSC junction), GSCs do not enter mitosis until the correct centrosome orientation is re-acquired. Cnn is a component of the centrosome (green dots), while E-cadherin is a component of the adherens junction (orange).
Drosophila male GSCs appear to have a similar checkpoint mechanism to delay the onset of mitosis when centrosomes are not correctly oriented 36, which we named the “centrosome orientation checkpoint (COC)” (Figure 3B). An interesting difference between SPOC and COC is the timing of cell cycle arrest. While SPOC arrests the cell cycle at the point of mitotic exit, COC arrests the cell cycle before commitment to mitosis. Due to the COC, mutations or conditions that increase centrosome misorientation do not necessarily lead to spindle misorientation. Live observation revealed that GSCs with misoriented centrosomes do not divide for a long time, but enter mitosis within 20 minutes upon correction of centrosome orientation 36. This tight temporal coordination between centrosome orientation and mitotic commitment points to the existence of the COC. However, readers should be warned that the COC is not yet molecularly well dissected; thus, it is not established whether phenomena pointing to the presence of the COC fulfills the classical definition of the checkpoint 37 (i.e. whether “fooling” COC can lead to cell cycle progression without correct centrosome orientation). It remains elusive whether CySCs have any checkpoint that monitors spindle repositioning. If such a checkpoint exists, this would link the position of the mitotic spindle to the completion of mitosis (likely cytokinesis), possibly more similar to the SPOC in the budding yeast.
The identification of the SPOC in budding yeast was facilitated by the analysis of mutants that impaired nuclear positioning (e.g. kar9Δ and dyn1Δ cells). In these mutants, the defective capture of astral MTs (also named cytoplasmic MTs in yeast) at the bud neck and/or bud tip causes an accumulation of cells, in which the mitotic spindle fails to align along the mother to daughter cell axis yet it extends inside the mother cell compartment during anaphase. Microscopic analysis revealed that in cells with mis-aligned spindles, chromosome segregation was normal but cells neither exited mitosis until the spindle re-aligned along the mother-bud polarity axis 15. Later studies established that BUB2 was involved in the mitotic delay of cells with misaligned spindles 38-42. BUB2 was originally identified, together with components of the spindle assembly checkpoint (SAC), as a checkpoint gene required for cell cycle arrest upon MT depolymerization by benzimidazole 43. It should be emphasized that the SAC and SPOC are two distinct mitotic checkpoints that monitor distinct defects (reviewed in 44). Briefly, SAC components localize at kinetochores and inhibit the metaphase to anaphase transition until all sister chromatids establish correct bipolar attachment, ensuring accurate chromosome segregation. In contrast, SPOC components localize at the SPB and inhibit the anaphase to G1 transition (mitotic exit) if the spindle fails to align along the mother-daughter cell polarity axis (reviewed in 44). Bub2 belongs to the TBC (Tre-2/Bub2/Cdc16) family of GTPase-activating proteins (GAPs) and it requires the binding partner Bfa1 to function as a GAP45, 46,11, 42. The Bub2-Bfa1 GAP complex is the most downstream SPOC effector, maintaining the GTPase Tem1 in the GDP-bound inactive state (reviewed in 47). Tem1 is the uppermost component of the mitotic exit network (MEN), which is a SPB associated signal transduction pathway required to promote the full activation of the phosphatase Cdc14. Cdc14 in turn triggers the down-regulation of mitotic Cdk1 activity allowing the transition out of mitosis (reviewed in 47).Thus, by keeping Tem1 inactive, the Bub2-Bfa1 GAP efficiently prevents cells from exiting mitosis if the mitotic spindle is misaligned.
An important regulatory step in SPOC is the regulation of the Bub2-Bfa1 GAP complex. Under unperturbed cell cycle conditions, Bub2 and Bfa1 bind preferentially to the daughter (bud)-directed SPB (dSPB). The GAP activity of Bub2-Bfa1 is in part inhibited in anaphase through phosphorylation of Bfa1 by the polo-like kinase Cdc5, most likely at the dSPB, allowing mitotic exit 48, 49. Interestingly, in cells in which the spindle is misoriented, Cdc5 cannot phosphorylate Bfa1, although Cdc5 is catalytically active at both SPBs50. Under such a circumstance, the kinase Kin4, a critical component of the SPOC 51, 52, phosphorylates Bfa1, preventing the Bub2-Bfa1 GAP from being inhibited by Cdc5 50. Kin4 is a mother cell enriched protein and follows a complex pattern of regulation that requires its phosphorylation and control of localization (reviewed in 34). Recently, the bud neck-associated protein Elm1 and the Protein Phosphatase 2A (PP2A) subunit Rts1 have been shown to activate Kin4 through different mechanisms 53-55. Elm1 is a conserved serine/threonine protein kinase that can activate the yeast homologue of AMP-activated kinase Snf1 (reviewed in 56, 57). Elm1 activates Kin4 through phosphorylation of a conserved residue in Kin4’s activation loop 53, 55. On the other hand, PP2A-Rts1 does not control Kin4 catalytic activity; rather, it supports Kin4 localization at the mother cortex and mother localized SPB and hence SPOC function 53, 54. Whereas cells need to keep active and localized Kin4 in the mother cell compartment to support SPOC function, the artificial targeting of Kin4 in the daughter cell blocks mitotic exit when spindles are correctly aligned 50, 51, 58. Therefore, mechanisms are in place to prevent accumulation of active Kin4 in the daughter cell compartment. Such a mechanism involves the cortical protein Lte1, a positive regulator of mitotic exit that belongs to the Cdc25-family of guanine nucleotide exchange factors (GEF)59, 60. Lte1 is enriched at the daughter cell cortex during most phases of the cell cycle and misplacement of Lte1 in the mother cell causes inappropriate mitotic exit in cells with mis-aligned spindles 38, 61,62. This differential distribution of Kin4 and Lte1 was proposed to be essential to control SPOC function38, 58. However, this scenario might be more complex, as we recently established that Lte1 is a direct inhibitor of Kin4 catalytic activity and required to exclude Kin4 from the dSPB, most likely by regulating Kin4 phosphorylation63. It is thus clear that cells must assure a low level of Lte1 in the mother cell body to avoid inactivation of Kin4 by Lte1. The fact that disrupting the bud neck region of yeast cells causes SPOC deficiency in a Lte1-dependent manner64, argues in favor of a role of the bud neck in checkpoint control via Kin4-Lte1 compartmentalization.
Despite these advances, very little is known about how the spatial information of the mitotic spindle is translated into the biochemical signal that controls Kin4 function at the mother compartment for checkpoint function and/or restricts Kin4 function at the daughter cell body to facilitate mitotic exit. Thus, understanding Kin4 and Lte1 regulation will represent an important step forward in the SPOC field. Intriguingly, our recent study on Drosophila male GSCs revealed that Par-1 kinase, which belongs to AMP-related kinase family, like Kin4, is a critical component of the COC, although molecular details must be worked out to enable comprehensive comparison of the SPOC and COC (Y. Yamashita, unpublished results). As it was suggested to exist only recently, we do not know much about the molecular mechanism of the COC. Apc2 does not appear to play a major role in this checkpoint, although it is an important determinant for centrosome orientation: centrosomes are highly misoriented in GSCs mutant for apc2 but apc2 mutant GSCs appear to be capable of sensing it and delaying mitotic entry, resulting in only a very mild effect on spindle orientation 22. Interestingly, GSCs mutant for cnn or GSCs overexpressing dominant-negative E-cadherin profoundly compromise the COC. As a result of inappropriate entry into mitosis of GSCs with misoriented centrosomes, a high frequency of misoriented spindle was observed. Puzzlingly, cnn mutants, which lack the majority of the pericentriolar material and astral MTs but retains centrioles, are defective in the COC 18, 22, whereas the complete loss of centrosomes/centrioles in the mutant of sas-4, a core component of the centriole, results in normal spindle orientation (65 and unpublished). sas-4 mutant do not have any centrosomes, the very organelle that is monitored by the COC. Our unpublished results indicate that in the complete absence of the centrosome in sas-4 mutant, an alternative mechanism that is normally dormant in the presence of the centrosome functions to orient acentrosomal mitotic spindle. Elucidation of more COC components and their function/localization in future study may reveal how COC molecularly operates, illuminating why cnn mutants show defective COC while sas-4 mutants can correctly orient the spindle. Dominant-negative E-cadherin that leads to COC defects lacks an extracellular domain, which is localized to the GSC cortex evenly rather than concentrated at the hub-GSC interface as in the wild type. It is possible that the centrosome is ectopically anchored via astral MTs to a random cortical site containing dominant-negative E-cadherin, which may create a false signal that the centrosome is anchored correctly to the hub-GSC interface, clearing the COC.
Concluding remarks
As reviewed here, the molecular and cellular mechanisms that govern asymmetric cell division are strikingly conserved through evolution, suggesting that the necessity of asymmetric division arose early during evolution of unicellular organisms. The common themes appear to be i) the establishment of cell polarity, ii) coordination of the division plane with cell polarity often by spindle orientation, and iii) additional mechanisms to ensure cell division takes place only after cell polarity and the division plane are coordinated. Interestingly, while the nature of cell polarity varies greatly depending on the system (e.g. the bud cortex in the yeast, the stem cell niche in fly GSCs, and intrinsic fate determinants in other cells such as Drosophila neuroblasts), the mechanism to orient the spindle with respect to cell polarity is strikingly conserved. Naturally, it is plausible that each system has its own “adapter” molecule that links its own polarity to the spindle orientation machinery. We know too little to generalize about temporal coordination ensuring correct division orientation (such as the SPOC and COC), and it is not known whether other systems also have such surveillance mechanisms.
Acknowledgements
We apologize to the authors whose work we were not able to include in this review owing to space limitations. This work was supported by the Helmholtz Association Grant (HZ-NG-111) and Marie Curie Fellowship (MEXT-CT-2006-042544) to G.P., and the Searle Scholar Program and NIH R01GM086481 to Y. M. Y.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barral Y, Liakopoulos D. Role of spindle asymmetry in cellular dynamics. Int Rev Cell Mol Biol. 2009;278:149–213. doi: 10.1016/S1937-6448(09)78004-9. [DOI] [PubMed] [Google Scholar]
- 2.Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23:2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pruyne D, Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J Cell Sci. 2000;113(Pt 3):365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- 4.Perez P, Rincon SA. Rho GTPases: regulation of cell polarity and growth in yeasts. Biochem J. 2010;426:243–253. doi: 10.1042/BJ20091823. [DOI] [PubMed] [Google Scholar]
- 5.Pruyne D, Bretscher A. Polarization of cell growth in yeast. J Cell Sci. 2000;113(Pt 4):571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- 6.McMurray MA, Thorner J. Septins: molecular partitioning and the generation of cellular asymmetry. Cell Div. 2009;4:18. doi: 10.1186/1747-1028-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: two versions of immortality. Science. 2007;316:402–404. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita YM, et al. Polarity in stem cell division: asymmetric stem cell division in tissue homeostasis. Cold Spring Harb Perspect Biol. 2010;2:a001313. doi: 10.1101/cshperspect.a001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leatherman JL, Dinardo S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell. 2008;3:44–54. doi: 10.1016/j.stem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leatherman JL, Dinardo S. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat Cell Biol. 2010;12:806–811. doi: 10.1038/ncb2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li R. Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc Natl Acad Sci U S A. 1999;96:4989–4994. doi: 10.1073/pnas.96.9.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller RK, Rose MD. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore JK, Cooper JA. Coordinating mitosis with cell polarity: Molecular motors at the cell cortex. Semin Cell Dev Biol. 2010;21:283–289. doi: 10.1016/j.semcdb.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segal M, Bloom K. Control of spindle polarity and orientation in Saccharomyces cerevisiae. Trends Cell Biol. 2001;11:160–166. doi: 10.1016/s0962-8924(01)01954-7. [DOI] [PubMed] [Google Scholar]
- 15.Yeh E, et al. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh E, et al. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson CG, Bloom K. Dynamic microtubules lead the way for spindle positioning. Nat Rev Mol Cell Biol. 2004;5:481–492. doi: 10.1038/nrm1402. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita YM, et al. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita YM, et al. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Megraw TL, et al. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11:116–120. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- 21.Megraw TL, et al. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 1999;126:2829–2839. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- 22.Inaba M, et al. E-cadherin is required for centrosome and spindle orientation in Drosophila male germline stem cells. PLoS ONE. 2010:5. doi: 10.1371/journal.pone.0012473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bienz M. Spindles cotton on to junctions, APC and EB1. Nat Cell Biol. 2001;3:E67–68. doi: 10.1038/35060140. [DOI] [PubMed] [Google Scholar]
- 24.Pereira G, et al. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. Embo J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conduit PT, Raff JW. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in drosophila neuroblasts. Curr Biol. 2010;20:2187–2192. doi: 10.1016/j.cub.2010.11.055. [DOI] [PubMed] [Google Scholar]
- 27.Januschke J, et al. Drosophila neuroblasts retain the daughter centrosome. Nat Commun. 2011;2:243. doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemphues KJ, et al. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- 29.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 30.Neumuller RA, et al. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008 doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 32.Cheng J, et al. Asymmetric division of cyst stem cells in Drosophila testis is ensured by anaphase spindle repositioning. Development. 2011;138:831–837. doi: 10.1242/dev.057901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voog J, et al. Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature. 2008;454:1132–1136. doi: 10.1038/nature07173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caydasi AK, et al. Monitoring spindle orientation: Spindle position checkpoint in charge. Cell Div. 2010;5:28. doi: 10.1186/1747-1028-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraschini R, et al. The spindle position checkpoint: how to deal with spindle misalignment during asymmetric cell division in budding yeast. Biochem Soc Trans. 2008;36:416–420. doi: 10.1042/BST0360416. [DOI] [PubMed] [Google Scholar]
- 36.Cheng J, et al. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 38.Bardin AJ, et al. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 39.Bloecher A, et al. Anaphase spindle position is monitored by the BUB2 checkpoint. Nat Cell Biol. 2000;2:556–558. doi: 10.1038/35019601. [DOI] [PubMed] [Google Scholar]
- 40.Fraschini R, et al. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J Cell Biol. 1999;145:979–991. doi: 10.1083/jcb.145.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoyt MA. Exit from mitosis: spindle pole power. Cell. 2000;102:267–270. doi: 10.1016/s0092-8674(00)00031-3. [DOI] [PubMed] [Google Scholar]
- 42.Pereira G, et al. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- 43.Hoyt MA, et al. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 44.Lew DJ, Burke DJ. The spindle assembly and spindle position checkpoints. Annu Rev Genet. 2003;37:251–282. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- 45.Geymonat M, et al. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J Biol Chem. 2002;277:28439–28445. doi: 10.1074/jbc.M202540200. [DOI] [PubMed] [Google Scholar]
- 46.Lee SE, et al. The Bub2-dependent mitotic pathway in yeast acts every cell cycle and regulates cytokinesis. J Cell Sci. 2001;114:2345–2354. doi: 10.1242/jcs.114.12.2345. [DOI] [PubMed] [Google Scholar]
- 47.Bardin AJ, Amon A. Men and sin: what’s the difference? Nat Rev Mol Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- 48.Geymonat M, et al. In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J Biol Chem. 2003;278:14591–14594. doi: 10.1074/jbc.C300059200. [DOI] [PubMed] [Google Scholar]
- 49.Hu F, et al. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 2001;107:655–665. doi: 10.1016/s0092-8674(01)00580-3. [DOI] [PubMed] [Google Scholar]
- 50.Maekawa H, et al. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J Cell Biol. 2007;179:423–436. doi: 10.1083/jcb.200705197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D’Aquino KE, et al. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol Cell. 2005;19:223–234. doi: 10.1016/j.molcel.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Pereira G, Schiebel E. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol Cell. 2005;19:209–221. doi: 10.1016/j.molcel.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 53.Caydasi AK, et al. Elm1 kinase activates the spindle position checkpoint kinase Kin4. J Cell Biol. 2010;190:975–989. doi: 10.1083/jcb.201006151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan LY, Amon A. The protein phosphatase 2A functions in the spindle position checkpoint by regulating the checkpoint kinase Kin4. Genes Dev. 2009;23:1639–1649. doi: 10.1101/gad.1804609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moore JK, et al. The spindle position checkpoint is coordinated by the Elm1 kinase. J Cell Biol. 2010;191:493–503. doi: 10.1083/jcb.201006092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 57.Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan LY, Amon A. Spindle position is coordinated with cell-cycle progression through establishment of mitotic exit-activating and -inhibitory zones. Mol Cell. 2010;39:444–454. doi: 10.1016/j.molcel.2010.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shirayama M, et al. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol Cell Biol. 1994;14:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shirayama M, et al. Isolation of a CDC25 family gene, MSI2/LTE1, as a multicopy suppressor of ira1. Yeast (Chichester, England) 1994;10:451–461. doi: 10.1002/yea.320100404. [DOI] [PubMed] [Google Scholar]
- 61.Castillon GA, et al. Septins have a dual role in controlling mitotic exit in budding yeast. Curr Biol. 2003;13:654–658. doi: 10.1016/s0960-9822(03)00247-1. [DOI] [PubMed] [Google Scholar]
- 62.Geymonat M, et al. Lte1 contributes to Bfa1 localization rather than stimulating nucleotide exchange by Tem1. J Cell Biol. 2009;187:497–511. doi: 10.1083/jcb.200905114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bertazzi DT, et al. The cortical protein Lte1 promotes mitotic exit by inhibiting the spindle position checkpoint kinase Kin4. J. Cell Biol. 2011 doi: 10.1083/jcb.201101056. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nelson SA, Cooper JA. A novel pathway that coordinates mitotic exit with spindle position. Mol Biol Cell. 2007;18:3440–3450. doi: 10.1091/mbc.E07-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Riparbelli MG, Callaini G. Male gametogenesis without centrioles. Dev Biol. 2011;349:427–439. doi: 10.1016/j.ydbio.2010.10.021. [DOI] [PubMed] [Google Scholar]