Kif18B is a newly discovered plus-tip-tracking protein that is enriched on astral microtubule (MT) ends during early mitosis. Kif18B binds directly to EB1, and this interaction is required for proper localization of Kif18B and to control astral MT length.
Abstract
Regulation of microtubule (MT) dynamics is essential for proper spindle assembly and organization. Kinesin-8 family members are plus-end-directed motors that modulate plus-end MT dynamics by acting as MT depolymerases or as MT plus-end capping proteins. In this paper, we show that the human kinesin-8 Kif18B functions during mitosis to control astral MT organization. Kif18B is a MT plus-tip-tracking protein that localizes to the nucleus in interphase and is enriched at astral MT plus ends during early mitosis. Knockdown of Kif18B caused spindle defects, resulting in an increased number and length of MTs. A yeast two-hybrid screen identified an interaction of the C-terminal domain of Kif18B with the plus-end MT-binding protein EB1. EB1 knockdown disrupted Kif18B targeting to MT plus ends, indicating that EB1/Kif18B interaction is physiologically important. This interaction is direct, as the far C-terminal end of Kif18B is sufficient for binding to EB1 in vitro. Overexpression of this domain is sufficient for plus-end MT targeting in cells; however, targeting is enhanced by the motor domain, which cooperates with the tail to achieve proper Kif18B localization at MT plus ends. Our results suggest that Kif18B is a new MT dynamics regulatory protein that interacts with EB1 to control astral MT length.
INTRODUCTION
Proper spindle function relies on an underlying organization maintained by proteins that regulate spindle microtubule (MT) dynamics, as well as by motor proteins that move the chromosomes and the MTs. The spindle is composed of several types of MTs that are defined based on their localization and dynamic properties. Within the spindle, there are bundles of more stable MTs linking spindle poles to kinetochores (K-fibers) and more dynamic non–kinetochore MTs emanating from the poles that often overlap midspindle (spindle MTs; Walczak and Heald, 2008). Astral MTs extend from the spindle pole toward the cell cortex, and may be important in spindle positioning (Rosenblatt, 2005; Wuhr et al., 2009; Rankin and Wordeman, 2010). All classes of MTs exhibit dynamic instability, a process in which both growing and shrinking MTs coexist within the population and randomly interconvert between these two states (Mitchison and Kirschner, 1984). However, the mechanisms maintaining these distinct MT populations remain unclear.
MT dynamics are of fundamental importance to the intracellular functions of the MT cytoskeleton and are highly regulated. MTs turn over much more rapidly in cells than do MTs assembled from pure tubulin in vitro (Desai and Mitchison, 1997), highlighting the need for cellular factors to control dynamics. Several families of proteins alter MT turnover in cells, including both MT-stabilizing and MT-destabilizing proteins (Howard and Hyman, 2007; van der Vaart et al., 2009). Of particular interest are members of the end-binding (EB) protein family and the kinesin superfamily that appear to be major MT dynamics regulatory proteins (Akhmanova and Steinmetz, 2008). EB1, the most well-characterized EB family protein member, is a major regulator of MT plus-end polymerization dynamics in cells, acting in part through orchestration of the accumulation of multiple other MT dynamics regulators (Akhmanova and Steinmetz, 2008). Of the kinesin superfamily, the kinesin-13 family members (Kif2A, Kif2B, and MCAK/Kif2C) are strictly MT depolymerases (Desai et al., 1999; Hunter et al., 2003), whereas the kinesin-8 family members (Kif18A, Kif18B, and Kif19) are MT plus-end-directed motors and plus-end-specific destabilizing enzymes (Gupta et al., 2006; Varga et al., 2006; Mayr et al., 2007; Du et al., 2010).
Kinesin-13 family members regulate MT dynamics by binding to the ends of MTs and inducing a catastrophe (Desai et al., 1999; Newton et al., 2004). In mammalian cells, each kinesin-13 plays a distinct role; Kif2a controls spindle bipolarity (Ganem and Compton, 2004; Manning et al., 2007); Kif2b is important for modulating K-fiber MT turnover (Manning et al., 2007; Bakhoum et al., 2009b); and MCAK/Kif2c aids astral MT organization (Rizk et al., 2009), K-fiber dynamics (Kline-Smith et al., 2004; Wordeman et al., 2007; Bakhoum et al., 2009b) and is involved in error correction (Andrews et al., 2004; Kline-Smith et al., 2004; Lan et al., 2004; Ohi et al., 2007). Interestingly, MCAK and Kif2b appear to temporally control K-fiber MT dynamics during prometaphase and metaphase, respectively (Bakhoum et al., 2009a, 2009b), highlighting the intricate nature of the regulation of mitotic MT dynamics.
In contrast to the kinesin-13 proteins, the kinesin-8 family members are much less well understood. Initially, kinesin-8 proteins were studied in Saccharomyces cerevisiae, where the single kinesin-8 protein, Kip3, was shown to be involved in nuclear and spindle positioning (Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Yeh et al., 2000). Kip3 is a MT plus-end motor and plus-end depolymerase (Gupta et al., 2006; Varga et al., 2006) that controls cortical MT dynamics (Gupta et al., 2006). Unlike S. cerevisiae, Schizosaccharomyces pombe has two kinesin-8 genes, Klp5 and Klp6, that function as a heterodimer (Grissom et al., 2009) and are important for accurate chromosome alignment and segregation (Garcia et al., 2002; West et al., 2002). Like S. cerevisiae, Drosophila melanogaster also contains a single kinesin-8 gene, Klp67A, which controls chromosome alignment and spindle length (Goshima and Vale, 2003; Gatt et al., 2005; Goshima et al., 2005; Savoian and Glover, 2010). These studies highlight the diverse roles that kinesin-8 proteins can play.
In human cells, there are three kinesin-8 proteins: Kif18A, Kif18B, and Kif19. Kif18A is the best-characterized family member and functions primarily in chromosome movement. Kif18A is localized to the plus ends of MTs within the spindle, with enrichment on K-fibers (Mayr et al., 2007). Knockdown of Kif18A causes an increase in spindle length (Mayr et al., 2007) and affects the rate of chromosome movement, which may be due to its role in coordinating chromosome oscillations (Stumpff et al., 2008; Jaqaman et al., 2010). The function of Kif18A in mediating chromosome movement may be indirect, as Kif18A mediates the proper targeting of the astrin–CLASP complex to kinetochore MT plus ends (Manning et al., 2010). The biochemical activity of Kif18A remains controversial, as one study suggests it is a plus-end motor and a MT plus-end-depolymerizing enzyme (Mayr et al., 2007), whereas another study suggests it is a plus-end-capping protein (Du et al., 2010). In either case, it is clear that Kif18A is a critical mediator of spindle MT organization and dynamics.
Much less is known about Kif18B and Kif19. RNA interference (RNAi) analysis of all mitotic kinesins in HeLa cells revealed no function for Kif18B or for Kif19 (Zhu et al., 2005), primarily due to limited expression of these proteins in the tested cells. A more recent study showed that Kif18B is expressed in a cell cycle−specific manner, suggesting a mitotic role (Lee et al., 2010). Kif18B is nuclear during interphase and localizes to astral MTs during early mitosis. There is currently no functional information about the role of Kif18B. In the present study, we show that Kif18B is enriched on astral MT plus ends via a direct interaction with EB1. Loss of Kif18B causes an increase in the number and length of astral MTs, suggesting Kif18B is an important modulator of astral MT dynamics.
RESULTS
Kif18B is a plus-tip-binding protein enriched on astral MTs during early mitosis
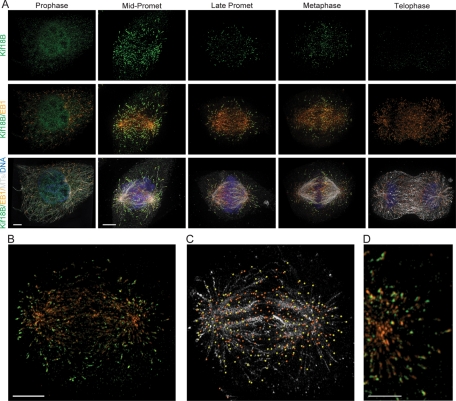
To begin to explore the function of Kif18B, we generated an antibody to the stalk-tail region of Kif18B (aa 378–833) and used it for immunofluorescence localization in HeLa cells. Kif18B was nuclear during interphase (unpublished data) and during prophase (Figure 1A), although some prophase cells also had diffuse cytoplasmic staining. Kif18B was enriched on astral MT plus ends during early prometaphase (Figure 1A); this enrichment decreased as mitosis progressed, consistent with recent studies (Lee et al., 2010). The enrichment on astral MT plus ends versus spindle MT ends was most apparent when imaged in three-dimensional reconstructions, in which we could rotate the spindle as well as the image through the Z-stack (Supplemental Video 1). In addition, treatment of cells with monastrol arrests cells in prometaphase, with astral MT ends extending beyond the chromosomes toward the cell cortex. In monastrol-treated cells, Kif18B was seen on the longer astral MT plus ends in a ring along the outside edge of the aster, unlike EB1, which was found throughout the MT array (Supplemental Video 2).
FIGURE 1:
Kif18B is a plus-tip-tracking protein that is enriched on the plus ends of astral MTs. (A) Kif18B immunostaining of HeLa cells showing Kif18B (green), EB1 (orange), MTs (gray), and DNA (blue) at the indicated mitotic stage. Images were acquired with an Applied Precision OMX Super-Resolution Microscope. All images are taken at equivalent exposure times for a given channel and scaled identically, except for the Kif18B staining in the prophase cell, which is scaled to enhance the nuclear staining. The prophase images are a projection of ∼3-μm z-thickness through the center of the cell, and the rest are full projections of the images. (B) Immunostaining of a prometaphase HeLa cell with Kif18B (green) and EB1 (orange). (C) IMARIS image analysis software was used to identify EB1 and Kif18B staining of MT plus ends. Yellow dots represent EB1 staining that colocalizes with Kif18B, and orange dots represent EB1 staining that is not colocalized with Kif18B. MTs are shown in light gray. (D) An enlarged image showing the distribution of EB1 (orange) and Kif18B (green) at MT ends. Scale bar: (A and B) 4 μm and (D) 2 μm.
An analysis of the distribution of Kif18B relative to EB1 showed that not all MTs that contained EB1 also had Kif18B staining, suggesting Kif18B is only found on a subset of MT ends (Figure 1, B and C). To examine this localization more closely, we used automated image analysis to identify the EB1 comets on MTs in mitotic cells. We then scored whether or not Kif18B was localized within a radius of 0.25 μm of the EB1 staining. We found that EB1 localized on the plus ends of both spindle and astral MTs of prometaphase cells, whereas Kif18B colocalized with ∼40% of the EB1-marked MTs (Figure 1C, n = 12 cells). On MT ends that contained both Kif18B and EB1, there was no clear pattern of arrangement between the two proteins. In some instances Kif18B was localized on the distal side of EB1, whereas in other instances Kif18B was localized on the proximal side of EB1 on other MTs (Figure 1D).
To demonstrate that Kif18B tracks the plus ends of growing MTs, we analyzed the behavior of green fluorescent protein (GFP)-Kif18B expressed in HeLa cells. GFP-Kif18B exhibited classical tip-tracking behavior, in which comets of GFP-Kif18B track the plus ends of both spindle and astral MTs (Supplemental Video 3). Together, our data support the idea that Kif18B is a plus-tip-tracking protein enriched on astral MT plus ends.
Kif18B controls astral MT length
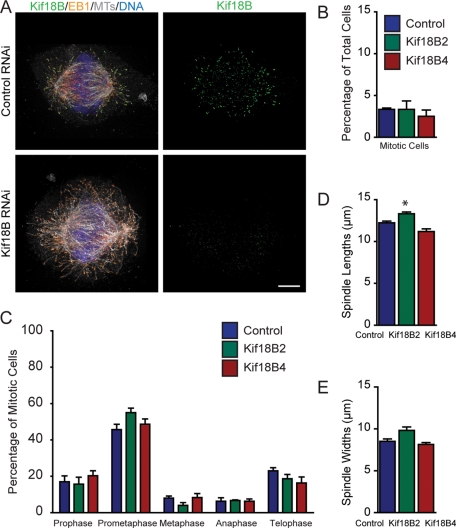
To address how Kif18B affects mitotic progression and spindle organization, we knocked down Kif18B by RNAi and analyzed spindle morphology. Knockdown of Kif18B using two independent small interfering RNAs (siRNAs) eliminated essentially all Kif18B staining (Figure 2A). Because our antibodies were unable to detect a band in control cells by Western blot, we confirmed the knockdown by quantitative real-time PCR analysis. Each of the siRNAs tested knocked down Kif18B by greater than 95% at the mRNA level (96% for Kif18B2 and 99% for Kif18B4), supporting our immunofluorescence analysis. Knockdown of Kif18B did not increase the mitotic index (Figure 2B), nor did it affect the overall timing of mitotic progression (unpublished data). We did find a small increase in the percentage of prometaphase cells with the Kif18B2 siRNA, but this was not reproducible with the Kif18B4 siRNA, suggesting it is not a specific consequence of the Kif18B knockdown (Figure 2C). The overall spindle architecture in the Kif18B RNAi cells also appeared disrupted, resulting in a variety of spindle morphologies, including rounder spindles, and other spindle structures that did not have the typical fusiform shape. However, there was no consistent effect on either the length or the width of the spindles seen with both siRNAs (Figure 2, D and E). These results suggest that Kif18B knockdown does not cause a consistent measurable change in spindle morphology.
FIGURE 2:
Kif18B is required for proper astral MT organization. (A) HeLa cells were treated with negative control siRNA (Top) or Kif18B siRNA (Bottom), and then stained for Kif18B (green), EB1 (orange), MTs (gray), and DNA (blue). Images were acquired with an Applied Precision OMX Super-Resolution Microscope. Scale bar: 4 μm. (B) Quantification of the mitotic index of cells using two independent Kif18B siRNAs. Data represent the mean ± SEM from three independent experiments in which 200 cells were counted per experiment. (C) Quantification of mitotic stage distribution using two independent Kif18B siRNAs. Data represent the mean ± SEM from three independent experiments in which 100 mitotic cells were counted per experiment. (D and E) Quantification of spindle lengths (D) and widths (E) of prometaphase cells using two independent Kif18B siRNAs. Data represent the mean ± SEM from three independent experiments in which at least 10 spindles were measured per experiment. *, p < 0.05.
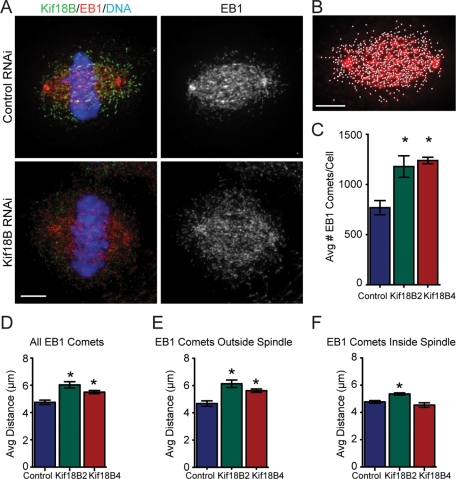
Cells in which Kif18B was knocked down appeared to have an increase in astral MT polymer, suggesting the astral MTs were either longer and/or increased in number (Figure 3A). Because the tracing and measurement of MTs within a spindle is not possible due to the high density of MTs, we developed a method that used EB1 distribution as an indirect measure to score the number of MT ends and to estimate the length of the MTs (Supplemental Figure S1). IMARIS 3D imaging software was utilized to identify the EB1 comets as spots on individual images based on both the intensity and size of the EB1 staining. There was a 1.5- to 1.6-fold increase in the average number of spots/cell with either Kif18B siRNA compared with controls (control, 769 ± 71; Kif18B2, 1179 ± 107; Kif18B4, 1239 ± 33; p < 0.05 between control and experimental; Figure 3C), suggesting that loss of Kif18B caused a stabilizing effect on MTs, thereby increasing the total number of MTs. To estimate the lengths of the MTs, we used the three-dimensional coordinates for the identified EB1 spots and the spindle poles to calculate the distance between each EB1 spot and its closest pole (Figure S1). There was an overall increase in the mean distance of EB1 to the nearest pole in Kif18B knockdown cells relative to controls (control, 4.75 ± 0.16; Kif18B2, 6.03 ± 0.23; Kif18B4, 5.50 ± 0.12; p < 0.05 between control and experimental; Figures 3D and S2). To determine whether this was specific for astral MTs or for spindle MTs, we partitioned the EB1 comets to separate those on the astral MTs from those within the spindle. We first calculated the average spindle width for control cells, and used this value to define the spindle region. We then calculated the angle between the pole axis and one-half of the spindle width, and used this value to partition the EB1 comets as being either outside or inside the control spindle dimensions. The mean distance of the EB1 comets lying outside the spindle on astral MTs was increased relative to control (control, 4.68 ± 0.20; Kif18B2, 6.14 ± 0.27; Kif18B4, 5.63 ± 0.13; p < 0.05 between control and experimental; Figures 3E and S2). There was a slight, albeit significant, increase in the distance within the spindle of the EB1 comets to the closest pole, but only with one of the Kif18B siRNAs (Figures 3F and S2), suggesting any effect on the spindle MTs is either weak or nonspecific. There was no difference in the percentage of spots distributed between the spindle and astral MTs. As a complementary approach, we also scored the effects of GFP-Kif18B overexpression on the number of EB1 comets and the distance from the pole to the EB1 comet. There was a decrease in the number of EB1 comets and an overall decrease in the distance of the EB1 comets to the nearest pole upon GFP-Kif18B overexpression (Figure S3). Together, our data support the idea that Kif18B is a MT-destabilizing enzyme that controls the dynamics of astral MTs.
FIGURE 3:
Kif18B modulates the length of astral MTs. (A) HeLa cells were treated with negative control siRNA (Top) or Kif18B siRNA (Bottom) and then stained for Kif18B (green), EB1 (red), DNA (blue), and MTs (unpublished data). Scale bar: 4 μm. (B) IMARIS software was used to identify EB1 comets (white dots). Scale bar: 4 μm. (C) The average number of identified EB1 comets/cell is plotted for control and Kif18B RNAi. Data represent mean ± SEM from three independent experiments in which at least 10 spindles were analyzed per experiment. (D–F) The average distance between the pole and the EB1 comets was calculated for all EB1 comets (D), those outside the spindle (E), and those within the spindle (F). Data represent mean ± SEM from three independent experiments in which at least 10 spindles were analyzed per experiment. *, p < 0.05. The histograms of the complete data sets are in Figure S2.
Kif18B interacts with EB1 to target to astral MT ends
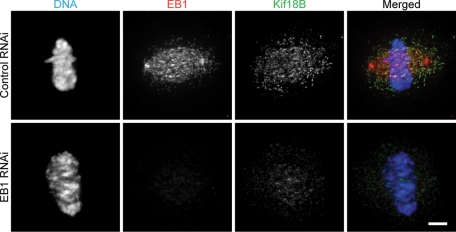
To understand how Kif18B may mediate its effects on astral MTs, we carried out a yeast two-hybrid assay to identify potential Kif18B-interacting proteins. We utilized the stalk-tail domain of Kif18B as bait, screened 10 × 106 colonies, and identified 89 positive clones. Of the 30 sequenced clones, 22 encoded EB1, suggesting that Kif18B interacts with EB1. In support of this idea, we found that knockdown of EB1 caused a significant reduction in the ability of Kif18B to localize to the plus ends of MTs (Figure 4).
FIGURE 4:
Kif18B requires EB1 for targeting to MT plus ends. HeLa cells were treated with negative control siRNA (Top) or EB1 siRNA (Bottom), and then stained for DNA (blue), EB1 (red), and Kif18B (green). Scale bar: 4 μm.
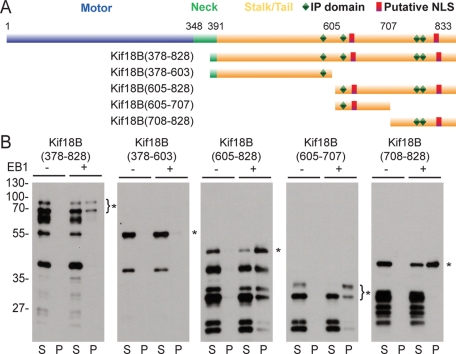
It was shown previously that the consensus sequence SXIP serves as a targeting motif for some EB1-binding proteins (Honnappa et al., 2009). Kif18B does not contain the conical SXIP sequence, but it does contain one SIP sequence and three IP sequences within the stalk-tail domain (Figure 5A), which also can confer EB1 binding (Honnappa et al., 2009). To discover whether the interaction with EB1 was direct and to dissect the domains of interaction, we generated a series of truncation constructs of the Kif18B stalk-tail domain fused to glutathione S-transferase (GST; Figure 5A). The purified GST-tagged Kif18B proteins were incubated with S-tagged EB1 in pulldown experiments. The stalk-tail domain of Kif18B (aa 378–828) bound to EB1, showing that the interaction between these two proteins identified by the yeast two-hybrid screen is direct. Kif18B (aa 378–603) failed to bind to EB1, but Kif18B (aa 605–828) retained binding activity. When we truncated Kif18B (aa 605–828) into two smaller fragments (aa 605–707 and 708–828), both constructs bound to EB1, suggesting that aa 605–828 constitute the region of Kif18B that contains all of the domains that bind EB1.
FIGURE 5:
Kif18B interacts directly with EB1. (A) Schematic diagram of Kif18B indicating the various domains and the locations of the predicted IP and NLS motifs. The deletion constructs used are indicated on the left. (B) Western blots of control (−) and EB1 (+) pulldown experiments with the indicated GST-tagged Kif18B deletion protein. Equivalent amounts of supernatant (S) and pellet (P) fractions from each experiment were separated by SDS–PAGE and analyzed by Western blot with an anti-GST antibody. The asterisk represents the full-length fragment for each truncation protein, and the lower bands represent degradation products present in each purified fusion protein.
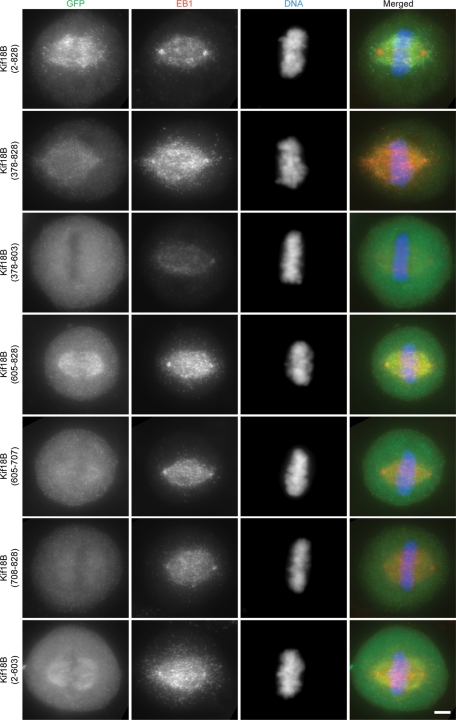
To determine whether the minimal domain of Kif18B that binds to EB1 in vitro is sufficient to target to MT plus ends in vivo, we expressed the same fragments used for the pulldown experiments as GFP fusion proteins in HeLa cells (Figure 6; Figure S4 provides a schematic summary of the data). Full-length GFP-Kif18B targeted robustly to MT plus ends of spindle and astral MTs (Figure 6). We found that the entire stalk-tail domain (aa 378–828) and the minimal EB1-binding domain (aa 605–828) were able to target to MT plus ends, but we never observed binding equivalent to that of the full-length protein. The amount of these proteins found associated with the MTs varied depending upon expression level, and localization was never as consistently robust as seen with the full-length protein. We also saw that the stalk-tail domain sometimes associated with chromatin, especially during anaphase and telophase (unpublished data). These results suggest the minimal EB1-binding domain of Kif18B identified in vitro is sufficient for plus-end MT binding in cells, but robust plus-end accumulation requires both the Kif18B motor domain and the EB1-binding domain within the tail.
FIGURE 6:
Kif18B uses its motor domain and its EB1-binding domain for robust plus-end targeting in cells. HeLa cells were transfected with the indicated GFP-tagged fusion protein construct and then stained with anti-GFP (green), EB1 (red), and DNA (blue). Images are taken at equivalent exposures times and scaled identically for a given channel. Scale bar: 4 μm.
As a complementary experiment, we expressed GFP-Kif18B (aa 2–603), which lacks the minimal Kif18B EB1-binding domain. Surprisingly, GFP-Kif18B (aa 2–603) localized all along MTs of both interphase and mitotic cells (Figures 6 and S5). The amount of binding depended upon the expression level of the protein with increased expression resulting in more binding. These results suggest that Kif18B utilizes both its kinesin-like motor domain as well as its EB1-targeting domain for proper plus-end MT localization. The far C-terminal domain, which is sufficient to bind to EB1 in vitro, is necessary but not sufficient for robust plus-end targeting in cells. In addition, the presence of the far C-terminal domain inhibits localization of Kif18B along the length of MTs.
We also performed an analysis of interphase cells to determine what domains are important for nuclear targeting. We found that both predicted nuclear localization sequences (NLS) can be utilized for nuclear targeting (Figures S4 and S5). Our data show Kif18B needs to be recruited to nuclei prior to nuclear envelope breakdown, so it can target effectively to MT plus ends in early mitosis, where it functions to regulate astral MT organization.
DISCUSSION
We show that Kif18B is a new plus-tip-tracking protein that regulates astral MT length during mitosis. Kif18B can also be added to the growing list of EB1-binding proteins, which highlights the complexities of the control of proper cellular MT dynamics.
Kif18B associates with MT plus ends by a direct interaction with EB1
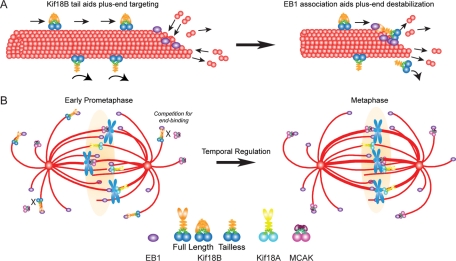
Our data support a model in which Kif18B utilizes its plus-end-directed motor activity to walk toward MT plus ends, where it interacts with EB1 to maintain plus-end association and control MT dynamics (Figure 7A). In support of this idea is the fact that Kif18B requires both the tail domain, which contains the EB1-binding domain, and the motor domain for robust targeting to MT plus ends. In addition, EB1 knockdown also disrupts Kif18B targeting, which shows that Kif18B alone is not sufficient for MT plus-end targeting. It is interesting that Kif18B lacking the EB1-binding domain does not enrich on MT plus ends but rather localizes along the length of MTs in both interphase and mitotic cells. The ability of a kinesin-8 to bind along MTs in the absence of the tail does not appear to be conserved with other members of the kinesin-8 family, as a tailless version of Kif18A is cytoplasmic during interphase and mitosis (Weaver et al., 2011). In addition, Kif18A with a mutated NLS accumulates at plus ends of interphase MTs (Du et al., 2010), suggesting that binding to the lattice versus the ends of MTs is not simply a cell cycle–regulated event. Together, our findings suggest that not only is the Kif18B motor domain not sufficient to accumulate at MT plus ends, but the Kif18B tail is inhibitory to MT lattice binding.
FIGURE 7:
Model for Kif18B function. (A) Full-length Kif18B (top of MT) is predicted to translocate along the MT until it reaches the MT end (left), where it interacts with EB1 and destabilizes MT plus ends (right). The presence of the EB1-binding tail inhibits lattice binding, promoting enrichment at the MT plus ends. The tailless motor (bottom of MT) binds along the length of the MT lattice without enrichment to MT plus ends. Without the ability to interact with EB1, tailless Kif18B will dissociate from the MT and be unable to regulate MT plus-end dynamics. (B) During early prometaphase Kif18B accumulates on the plus ends of astral MTs that contain EB1. Perhaps a gradient around chromosomes (light orange) prevents Kif18B association with spindle MT plus ends. Because both Kif18B and MCAK are present, they either bind to distinct MT plus ends, or they compete for binding to the same MT plus ends (left). As the cell transits to metaphase, the overall amount of Kif18B decreases, allowing MCAK to now compete more effectively for binding to EB1-coated MT plus ends.
The concept of autoinhibition of kinesin motors is well documented in the literature (reviewed in Verhey and Hammond, 2009). With conventional kinesin, the tail domain folds over and inhibits the motor domain, insuring kinesin is only active when bound to cargo (Hackney et al., 1992; Coy et al., 1999; Friedman and Vale, 1999). This autoinhibition is mediated by the IAK motif of kinesin, which inhibits ADP release upon interaction of kinesin with the MT (Hackney and Stock, 2000). Analysis of the tail of Kif18B did not reveal an IAK motif, but it is possible that a different sequence of Kif18B plays an analogous role. Exploring whether the tail of Kif18B directly modulates motor activity or affects the ability of Kif18B to modulate MT dynamics will be an important endeavor.
Kif18B regulates MT length
Kif18B appears to regulate MT length and, likely, dynamics. Consistent with this idea, we found that both astral and spindle MT lengths were increased in Kif18B knockdown cells and decreased in GFP-Kif18B–overexpressing cells. While our data reveal that Kif18B inhibition or overexpression affects MT length within the spindle under some conditions, there was no appreciable change in spindle length under these conditions, supporting the idea that Kif18B may primarily exert effects on astral MTs, where it is predominantly localized. The mechanism by which Kif18B regulates dynamics is currently not known. Our data do not support the idea that Kif18B is a robust MT depolymerase, as overexpression/inhibition only caused a modest change in MT length and had no consistent effect on the morphology of the spindle. In addition, mislocalization of Kif18B from the nucleus to the cytoplasm during interphase allowed Kif18B to bind to interphase MTs with no apparent change in the morphology of the MT array.
Overall, the astral phenotype of Kif18B knockdown is similar to that of MCAK inhibition, a kinesin that also interacts with EB1 (Lee et al., 2008; Montenegro Gouveia et al., 2010). This raises the question of how these different kinesins control MT dynamics. One idea is that Kif18B and MCAK bind to spatially distinct MT ends. Alternatively, Kif18B and MCAK may bind to the same MT ends. In this model, both MCAK and Kif18B would compete for binding to MT ends during early prometaphase, such that both proteins cannot bind to EB1 at the same time (Figure 7B). Another level of regulation may come from the possibility that Kif18B and MCAK act temporally to control MT dynamics. Although Kif18B is sequestered in the nucleus in early prophase, Kif18B is most abundantly on astral MTs just after nuclear envelope breakdown (NEB), suggesting that it acts early in mitosis. By the time cells reach metaphase, the overall amount of Kif18B decreases, which would now allow MCAK to be the predominant astral MT dynamics regulator (Figure 7B). Precedence for the temporal regulation comes from the finding that MCAK acts primarily in prometaphase and Kif2B acts predominantly during metaphase to regulate K-fiber dynamics (Bakhoum et al., 2009b), providing an example of how cells temporally control MT dynamics. In the future, it will be important to dissect the individual contributions of Kif18B and MCAK to the regulation of MT dynamics and to explore the mechanisms by which they interact with EB1.
An interesting question is, Why is Kif18B enriched on astral MT ends when EB1 is found on all MT plus ends? (Berrueta et al., 1998). One possibility is that the environment within the spindle disfavors association with EB1. Perhaps this is mediated by the Ran gradient in the spindle (Nachury et al., 2001; Wiese et al., 2001; Ems-McClung et al., 2004; Kalab and Heald, 2008). In support of this idea, Kif18B contains NLS motifs (Lee et al., 2010) and interacts directly with importin α/β (unpublished data), suggesting it could be a target of the Ran pathway. Alternatively, Kif18B targeting to spindle MT ends could be inhibited by the Aurora B gradient in the spindle midzone (Fuller et al., 2008). Kif18B contains multiple Aurora B consensus sites for phosphorylation, and most of these sites have been shown to be phosphorylated in cells in a phosphoproteomic analysis (Dephoure et al., 2008). The finding that Kif18B and EB1 can interact directly in the absence of other proteins, including protein kinases, suggests that phosphorylation is not required for association, but does not rule out a role for kinases providing a mechanism to regulate the binding affinity between Kif18B and EB1. Examining what governs the association of Kif18B with some subsets of EB1-bound MTs is an important avenue for future studies.
Why do cells need so many MT depolymerases?
One fascinating question that remains is, Why do cells need so many different MT depolymerases? It has been clearly established that a single cell expresses all three members of the kinesin-13 family, all of which play distinct roles during mitosis (Rogers et al., 2004; Mennella et al., 2005; Manning et al., 2007), and at least two of the three kinesin-8 proteins, which control distinct aspects of spindle MT dynamics (this study; Mayr et al., 2007; Stumpff et al., 2008; Jaqaman et al., 2010).
It is interesting to speculate about how these dynamics can be regulated. MCAK and other kinesin-13 proteins do not require directed motility to enrich at MT plus ends, and yet they are highly potent depolymerases in vitro (Desai et al., 1999; Hunter et al., 2003; Helenius et al., 2006). In contrast, the kinesin-8 proteins utilize motility as a requisite part of their catalytic cycle (Gupta et al., 2006; Varga et al., 2006; Mayr et al., 2007) and may act as MT-capping proteins (Du et al., 2010). A MT depolymerase is likely to be more potent and may be able to quickly create alterations to the MT cytoskeleton, whereas a capping protein may result in finer-level modulations. In this model, even if MCAK and Kif18B compete for binding to the same MT ends, they would modulate different aspects of MT dynamics. More interesting may be the idea that there are unique classes of astral MTs serving distinct functions in spindle positioning. A key challenge for the future will be the determination of the individual contributions of the multiple MT depolymerases to the intricate regulation of MT dynamics, and the discovery of whether these proteins are functionally redundant or individually critical in the regulation of cytoskeletal dynamics.
MATERIALS AND METHODS
Generation and purification of antibodies
A full-length HsKif18B was made from two nonoverlapping HsKif18B cDNA clones purchased from Open Biosystems (accession #BM470271 and #BC044933). The stalk-tail of Kif18B (aa 378–828) was subcloned into both a 6His-GFP vector and a GST-fusion protein vector for expression in Escherichia coli. Proteins were expressed and purified using standard conditions. Polyclonal antibodies were generated by immunizing a rabbit (Covance Research Products, Denver, PA) with GFP-HsKif18B-ST (aa 378–833) in which one-half of the protein was reacted with 0.5% glutaraldehyde for 45 min at room temperature and then quenched with sodium borohydride. Antibodies were affinity-purified against GST-HsKif18B-3′CT (aa 612–833) as described previously (Walczak et al., 1997). In some experiments, only the rabbit serum was used.
Yeast two-hybrid assay
For the yeast two-hybrid screen, the stalk-tail domain of HsKif18B (aa 378–833) was used as bait by subcloning into the DNA-binding domain pDBLeu vector. The screen was then performed at the IU Yeast-Two-Hybrid Facility (http://sites.bio.indiana.edu/∼michaelslab/yeast_two_hybrid_facility.html) against a human HeLa cell cDNA library in which >10 × 106 clones were screened. A total of 89 out of 95 clones passed the growth test, from which 30 were chosen randomly for sequencing.
Generation of truncation constructs and protein expression
Mammalian expression constructs of Kif18B truncations were generated using Gateway technology, in which Kif18B PCR products were cloned into the pENTR/D-TOPO vector (Invitrogen, Carlsbad, CA). These constructs were then introduced into the Gateway destination vector pEGFP-DEST, which was created by subcloning the EGFP-coding sequence of pEGFPC1 (Clontech, Mountain View, CA) into pcDNA-DEST53 (Invitrogen). To obtain glutathione-S-transferase (GST)-tagged versions of each Kif18B construct, we introduced the corresponding pENTR constructs into the Gateway destination vector, pDEST15. The plasmids were purified using the Plasmid Maxi Kit (Qiagen, Valencia, CA) for transfection. The accuracy of all constructs was checked by sequence analysis.
To express the GST-tagged Kif18B constructs, we transformed pDEST15 plasmids into E. coli-derived competent cells as follows: Kif18B (aa 378–828), Kif18B (aa 378–603), Kif18B (aa 605–828) and Kif18B (aa 605–707) were transformed into Rosetta (EMD Chemicals, Gibbstown, NJ); and Kif18B (708–828) was transformed into Origami-2 (EMD Chemicals). Cultures were grown to 0.4–0.8 OD600 nm in Luria broth with 100 μg/ml ampicillin before induction with 100 μM IPTG for 2–4 h at 37°C. The cells were then pelleted and stored at –80°C until purification. The GST-Kif18B constructs were purified as previously described (Walczak et al., 1997). His-S-EB1 (a gift from Jennifer Tirnauer, University of Connecticut Health Center, Farmington, CT) was transformed into BL21(DE3); grown as for the GST-Kif18B plasmids, but induced at 20°C overnight; and purified on Ni-NTA agarose (Qiagen), as previously described (Walczak et al., 1997). All proteins were dialyzed into XB buffer (10 mM HEPES, pH 7.7, 1 mM MgCl2, 0.1 mM CaCl2, and 100 mM KCl), aliquoted, flash-frozen in liquid nitrogen, and stored at –80°C. Protein was quantified by densitometry of Coomassie Blue–stained SDS–PAGE gels using bovine serum albumin (BSA) as a standard.
Pulldown assays
S-protein pulldown assays were performed in a total volume of 100 μl with 70 pmol of each GST-Kif18B construct, 210 pmol of His-S-EB1, 0.25 mg/ml BSA, and 50 μl of S-protein agarose beads (EMD Chemicals) in XB buffer for 45–60 min at 4°C with rotation. After incubation, the beads were pelleted, and the unbound fraction removed and mixed with an equal volume of 2X sample buffer. The beads were then washed once with 1 ml XB buffer, once with 0.5 ml XB buffer, and once with 0.5 ml Tris-buffered saline (TBS)-TX (20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Triton X-100). The bound protein was eluted with 50 μl 1X sample buffer, and then added to 50 μl XB. Equal volumes of bound and unbound protein fractions were run on 10% SDS–PAGE, transferred to Biotrace nitrocellulose membranes, and probed with 0.52 μg/ml anti-GST primary antibody in AbDil-T (20 mM Tris, pH 7.5, 150 mM NaCl, 2% BSA, 0.1% Tween-20); this was followed by 1:20,000 dilution of horseradish peroxidase–linked donkey anti–rabbit secondary antibody (GE Healthcare Bio-Sciences, Piscataway, NJ) in 5% nonfat dry milk in TBST (TBS, 0.1% Tween-20). Blots were incubated with SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher Scientific, Rockford, IL) and exposed to Amersham Hyperfilm (GE Healthcare Bio-Sciences).
RNAi and plasmid transfections
For RNAi, HeLa cells were plated at 4 × 104 cells/35-mm well over 12-mm poly-l-lysine-coated coverslips and transfected 24 h later with 40 nM siRNA using Lipofectamine RNAiMAX (Invitrogen). siRNAs used: negative control #2 siRNA (UGGUUUACAUGUUGUGUGA; ThermoFisher Scientific); Kif18B2 (GGAAGAAGCUCCAAGUGUAUU; ThermoFisher Scientific); Kif18B4 (CAGUUUCCAUGAAUGCAUUUU; Applied Biosystems/Ambion, Austin, TX); EB1-1 (Louie et al., 2004) or EB1-3 (Wen et al., 2004; ThermoFisher Scientific). After 24 h, the transfection mix was replaced with fresh media, and cells were processed for immunofluorescence 48–52 h posttransfection. For plasmid transfections, HeLa cells were plated at 8 × 104 cells/35-mm well over coverslips or in glass-bottomed dishes (MatTek, Ashland, MA) for live imaging. Cells were blocked with 2 mM thymidine for either one or two 20-h intervals, and then transfected with 250 ng of plasmid DNA 48 h after plating, using Lipofectamine LTX (Invitrogen). The media was replaced 4–6 h posttransfection, and the cells were imaged live or processed for immunofluorescence 28–30 h posttransfection.
Immunofluorescence
Cells were rinsed with phosphate-buffered saline (PBS; 12 mM phosphate, 137 mM NaCl, 3 mM KCl, pH 7.4) and fixed for 10 min with cold MeOH. Fixed cells were rinsed with TBS-TX (20 mM Tris, 150 mM NaCl, pH 7.5 plus 0.1% Triton X-100) and blocked in AbDil-TX (2% BSA, 0.1% NaN3 in TBS-TX) for at least 30 min at room temperature. Cells were incubated with 1:1000 dilution rabbit polyclonal anti-Kif18B-CT sera for 30 min, and then incubated 30 min with 0.8–1.0 μg/ml goat anti–rabbit Alexa Fluor 488 (Invitrogen). Cells were then costained with 0.5 μg/ml mouse monoclonal anti-EB1 (BD Transduction Laboratories, San Jose, CA) and 10 μg/ml rat monoclonal antitubulin YL1/2 (Abcam, Cambridge, MA), and then incubated 30 min in 2 μg/ml donkey anti–mouse DyLight 594 (cross-absorbed against rat) and either 7.5 μg/ml donkey anti–rat DyLight 405 or 5 μg/ml donkey anti–rat DyLight 649 (both cross-absorbed against mouse; Jackson ImmunoResearch Laboratories, West Grove, PA). For cells transfected with GFP constructs, MTs and EB1 were visualized using identical methods; however, where indicated, this was preceded with a 30-min incubation in 2 μg/ml rabbit anti-GFP (Hertzer et al., 2006), and then incubated in 0.8 μg/ml donkey anti–rabbit Alexa Fluor 488. DNA was stained for 20 min with either 20 µM DRAQ-5 (Biostatus Limited, Shepshed, Leicester, UK) or for 10 min with 2 μg/ml Hoechst (Sigma-Aldrich, St. Louis, MO) in TBS-TX. All rinses between antibody incubations were performed using TBS-TX, and all antibodies were diluted in AbDil-TX. Coverslips were mounted using ProLong Gold (Invitrogen) and sealed with nail polish.
Imaging
Fixed cells for Kif18B localization were imaged using the OMX 3D-SIM Super-Resolution system (Applied Precision, Issaquah, WA) controlled by DV-OMX software from Applied Precision. Images were captured at 0.125-μm step size with an UNIPLANAPO 100×/1.4 numerical aperture (NA) objective, using 1.514 immersion oil. The 405 λ images were acquired for 100 ms at 1% laser strength, 488 λ images for 50 ms at 10% strength, 561 λ images for 50 ms at 100% strength, and 642 λ images for 50 ms at 100% strength. Images were processed using softWoRx (Applied Precision) and IMARIS 3D imaging software (Bitplane, Saint Paul, MN). All images were processed identically for EB1 and Kif18B visualization. All other fixed cells were imaged using a Nikon Eclipse 90i equipped with a 100× apochromatic PLAN objective (NA 1.4) and a CoolSnap HQ CCD camera (Photometrics, Tucson, AZ) controlled by Metamorph (Molecular Devices, Sunnyvale, CA). Image stacks were collected at 0.5-μm steps through the whole cell volume and then deconvolved using AutoQuant X (Media Cybernetics, Bethesda, MD) for 30 iterations. For live imaging, 30 U/ml oxyrase was added to 2 ml Opti-MEM media (Invitrogen) covering cells in imaging dishes (MatTek), which were then placed in an imaging chamber at 37°C, 5% CO2-controlled by a Live Cell controller (Pathology Devices, Westminster, MD), and imaged with a Yokagawa spinning-disk CSU-10 mounted on a Nikon TE2000 microscope equipped with a 100× objective (apochromatic PLAN, NA 1.4). Images were captured on a Photometrics Cascade II EM-CCD camera set at 3200 gain with 100-ms exposures at 2-s intervals. Contrast was enhanced to visualize the low level of GFP-Kif18B expression.
Image analysis and quantification
To determine mitotic index, 200 cells per condition per experiment were scored, and the mean ± SEM values for at least three independent experiments were graphed. To determine the mitotic distribution, 100 mitotic cells per experiment were scored for mitotic stage. For each condition, the average percentage of mitotic cells at each stage was plotted as mean ± SEM for at least three independent experiments. Significance was considered if p < 0.05, using two-tailed Student's t test performed in Excel (Microsoft, Redmond, WA). Only effects that were seen with both siRNAs were considered to be physiologically important.
To analyze the distribution of EB1 comets within a cell, we imaged and processed control and experimental cells identically, using AutoQuant X 3D deconvolution software. At least 10 cells per condition in each of three independent experiments were analyzed. A basic outline of our experimental protocol is described in Figure S1. Deconvolved image stacks of the EB1 channel were opened in the three-dimensional imaging software, IMARIS. By rotating the three-dimensional image and stepping through the z-series, we defined each pole as the center of the focal point of EB1 staining. The x, y, z coordinates for each pole were used to calculate the spindle length. By stepping through the z-series (slices view in IMARIS), we could measure the spindle widths using the density of spindle EB1 staining to find the plane with the maximal spindle width and draw a line perpendicular to the spindle axis. The EB1 comets and their corresponding x, y, z coordinates were identified using the spots function within IMARIS, setting 0.25 μm (the measured width the EB1 staining) as a minimal diameter, and using the autothreshold values for quality. The coordinates for each EB1 spot and the two poles were then exported to Microsoft Excel and used to calculate the distances of each spot to both poles. The pole that each EB1 spot is actually connected to cannot be determined by this method; therefore, we decided to use the distance to its closest pole. With this method, the calculated data sets would underrepresent those EB1 comets on MTs that crossed the spindle equator and were in reality attached to the farthest pole. This provides a conservative estimate of the MT length. To determine whether an EB1 spot was outside or inside the spindle area, we calculated the spindle angle of each control cell using the following equation: θ = arctan[(average control spindle width/2)/(average control spindle length)/2]. We chose to partition all data to the average angle of the control spindles, because the density of the EB1 fluorescence along the spindle in control cells allowed measurement of the spindle widths within acceptable margins of error, whereas the spindle morphology and subsequent EB1 staining in Kif18B-depleted cells increased the subjectivity of demarcating spindle versus astral EB1 comets. The angle of the EB1 spot defined by the line to its closest pole and the spindle pole was calculated using the cosine law. A spot was considered outside (astral) if this angle was greater than the average control spindle angle, and inside the spindle if the angle was smaller than the average control spindle angle. For each experimental condition, the total number of spots/cell, the spindle lengths, and the spindle widths were averaged for a given day, and then the averages among the three days were compared. The distances to the closest pole were calculated for all the EB1comets, as well as for those outside and inside the spindle; these values were averaged per cell, and then all cells from a given condition were combined to give an average MT length/cell for a given day. The mean ± SEM values for all the days were then calculated for the three independent experiments to account for day-to-day variation. Alternatively, all distances of the EB1 comets to their closest pole for all three independent experiments were plotted in Prism (Irvine, CA) as a relative frequency distribution. The histogram of the frequency distribution was then fit to a Gaussian curve and plotted with the 95% confidence interval. Statistical differences between the means were determined by comparing the Gaussian curves using a F-test in Prism. Two-tailed Student's t tests were performed using Excel as indicated in the text.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant R01-GM059618 to C.E.W., and the Applied Precision OMX microscope was provided by NIH grant S10-RR028697. We thank Jennifer Tirnauer for the EB1 plasmid and Yvonne Brede, Romy Brauer, Jennifer Rawlinson, and Jasmine Newman for early work on the project. Sid Shaw provided many helpful discussions on all aspects of this project. The yeast two-hybrid facility and the Light Microscopy Imaging Center were established by the Indiana METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment. The Light Microscopy Imaging Center is supported in part with funds from Indiana University Office of the Vice Provost for Research.
Abbreviations used:
- BSA
bovine serum albumin
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- Kif18B
kinesin family 18B
- MT
microtubule
- NA
numerical aperture
- NEB
nuclear envelope breakdown
- NLS
nuclear localization sequence
- TBS
Tris-buffered saline
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0363) on July 7, 2011.
REFERENCES
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol. 2009a;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009b;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrueta L, Kraeft SK, Tirnauer JS, Schuyler SC, Chen LB, Hill DE, Pellman D, Bierer BE. The adenomatous polyposis coli-binding protein EB1 is associated with cytoplasmic and spindle microtubules. Proc Natl Acad Sci USA. 1998;95:10596–10601. doi: 10.1073/pnas.95.18.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy DL, Hancock WO, Wagenbach M, Howard J. Kinesin's tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, English CA, Ohi R. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr Biol. 2010;20:374–380. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Ems-McClung SC, Zheng Y, Walczak CE. Importin alpha/beta and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol Biol Cell. 2004;15:46–57. doi: 10.1091/mbc.E03-07-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DS, Vale RD. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol. 1999;1:293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- Fuller BG, Lampson MA, Foley EA, Rosasco-Nitcher S, Le KV, Tobelmann P, Brautigan DL, Stukenberg PT, Kapoor TM. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Compton DA. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J Cell Biol. 2004;166:473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr Biol. 2002;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- Gatt MK, Savoian MS, Riparbelli MG, Massarelli C, Callaini G, Glover DM. Klp67A destabilises pre-anaphase microtubules but subsequently is required to stabilise the central spindle. J Cell Sci. 2005;118:2671–2682. doi: 10.1242/jcs.02410. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length control of the metaphase spindle. Curr Biol. 2005;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Grissom PM, Fiedler T, Grishchuk EL, Nicastro D, West RR, McIntosh JR. Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement. Mol Biol Cell. 2009;20:963–972. doi: 10.1091/mbc.E08-09-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Hackney DD, Levitt JD, Suhan J. Kinesin undergoes a 9 S to 6 S conformational transition. J Biol Chem. 1992;267:8696–8701. [PubMed] [Google Scholar]
- Hackney DD, Stock MF. Kinesin's IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat Cell Biol. 2000;2:257–260. doi: 10.1038/35010525. [DOI] [PubMed] [Google Scholar]
- Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- Hertzer KM, Ems-McClung SC, Kline-Smith SL, Lipkin TG, Gilbert SP, Walczak CE. Full-length dimeric MCAK is a more efficient microtubule depolymerase than minimal domain monomeric MCAK. Mol Biol Cell. 2006;17:700–710. doi: 10.1091/mbc.E05-08-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J. The Kin I kinesin MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K, et al. Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J Cell Biol. 2010;188:665–679. doi: 10.1083/jcb.200909005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Heald R. The RanGTP gradient—a GPS for the mitotic spindle. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell. 2004;15:1146–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, Hunt DF, Walczak CE, Stukenberg PT. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Lee T, Langford KJ, Askham JM, Bruning-Richardson A, Morrison EE. MCAK associates with EB1. Oncogene. 2008;27:2494–2500. doi: 10.1038/sj.onc.1210867. [DOI] [PubMed] [Google Scholar]
- Lee YM, Kim E, Park M, Moon E, Ahn SM, Kim W, Hwang KB, Kim YK, Choi W, Kim W. Cell cycle-regulated expression and subcellular localization of a kinesin-8 member human KIF18B. Gene. 2010;466:16–25. doi: 10.1016/j.gene.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Louie RK, Bahmanyar S, Siemers KA, Votin V, Chang P, Stearns T, Nelson WJ, Barth AI. Adenomatous polyposis coli and EB1 localize in close proximity of the mother centriole and EB1 is a functional component of centrosomes. J Cell Sci. 2004;117:1117–1128. doi: 10.1242/jcs.00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Bakhoum SF, Maffini S, Correia-Melo C, Maiato H, Compton DA. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 2010;29:3531–3543. doi: 10.1038/emboj.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Mennella V, Rogers GC, Rogers SL, Buster DW, Vale RD, Sharp DJ. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat Cell Biol. 2005;7:235–245. doi: 10.1038/ncb1222. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Montenegro Gouveia S, et al. In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr Biol. 2010;20:1717–1722. doi: 10.1016/j.cub.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin B is a mitotic target of the small GTPase ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Newton CN, Wagenbach M, Ovechkina Y, Wordeman L, Wilson L. MCAK, a Kin I kinesin, increases the catastrophe frequency of steady-state HeLa cell microtubules in an ATP-dependent manner in vitro. FEBS Lett. 2004;572:80–84. doi: 10.1016/j.febslet.2004.06.093. [DOI] [PubMed] [Google Scholar]
- Ohi R, Burbank K, Liu Q, Mitchison TJ. Nonredundant functions of Kinesin-13s during meiotic spindle assembly. Curr Biol. 2007;7:953–959. doi: 10.1016/j.cub.2007.04.057. [DOI] [PubMed] [Google Scholar]
- Rankin KE, Wordeman L. Long astral microtubules uncouple mitotic spindles from the cytokinetic furrow. J Cell Biol. 2010;190:35–43. doi: 10.1083/jcb.201004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk RS, Bohannon KP, Wetzel LA, Powers J, Shaw SL, Walczak CE. MCAK and paclitaxel have differential effects on spindle microtubule organization and dynamics. Mol Biol Cell. 2009;20:1639–1651. doi: 10.1091/mbc.E08-09-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- Rosenblatt J. Spindle assembly: asters part their separate ways. Nat Cell Biol. 2005;7:219–222. doi: 10.1038/ncb0305-219. [DOI] [PubMed] [Google Scholar]
- Savoian MS, Glover DM. Drosophila Klp67A binds prophase kinetochores to subsequently regulate congression and spindle length. J Cell Sci. 2010;123:767–776. doi: 10.1242/jcs.055905. [DOI] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart B, Akhmanova A, Straube A. Regulation of microtubule dynamic instability. Biochem Soc Trans. 2009;37:1007–1013. doi: 10.1042/BST0371007. [DOI] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Hammond JW. Traffic control: regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10:765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Verma S, Mitchison TJ. XCTK2: A Kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J Cell Biol. 1997;136:859–870. doi: 10.1083/jcb.136.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LN, Ems-McClung SC, Stout JR, LeBlanc C, Shaw SL, Gardner MK, Walczak CE. Kif18A utilizes a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr Biol. 2011 doi: 10.1016/j.cub.2011.08.005. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- West RR, Malmstrom T, McIntosh JR. Kinesins klp5+ and klp6+ are required for normal chromosome movement in mitosis. J Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin-β in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- Wordeman L, Wagenbach M, von Dassow G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J Cell Biol. 2007;179:869–879. doi: 10.1083/jcb.200707120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhr M, Dumont S, Groen AC, Needleman DJ, Mitchison TJ. How does a millimeter-sized cell find its center? Cell Cycle. 2009;8:1115–1121. doi: 10.4161/cc.8.8.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.