Despite high conservation of the Notch pathway, its repression appears diverse between organisms. In Drosophila, a high-affinity complex forms between the CSL orthologue Su(H) and Hairless, which is analyzed in great detail in vitro and in vivo. Drosophila Hairless is shown to bind CBF1 and inhibit Notch transcriptional output in mammalian cells.
Abstract
In metazoans, the highly conserved Notch pathway drives cellular specification. On receptor activation, the intracellular domain of Notch assembles a transcriptional activator complex that includes the DNA-binding protein CSL, a composite of human C-promoter binding factor 1, Suppressor of Hairless of Drosophila melanogaster [Su(H)], and lin-12 and Glp-1 phenotype of Caenorhabditis elegans. In the absence of ligand, CSL represses Notch target genes. However, despite the structural similarity of CSL orthologues, repression appears largely diverse between organisms. Here we analyze the Notch repressor complex in Drosophila, consisting of the fly CSL protein, Su(H), and the corepressor Hairless, which recruits general repressor proteins. We show that the C-terminal domain of Su(H) is necessary and sufficient for forming a high-affinity complex with Hairless. Mutations in Su(H) that affect interactions with Notch and Mastermind have no effect on Hairless binding. Nonetheless, we demonstrate that Notch and Hairless compete for CSL in vitro and in cell culture. In addition, we identify a site in Hairless that is crucial for binding Su(H) and subsequently show that this Hairless mutant is strongly impaired, failing to properly assemble the repressor complex in vivo. Finally, we demonstrate Hairless-mediated inhibition of Notch signaling in a cell culture assay, which hints at a potentially similar repression mechanism in mammals that might be exploited for therapeutic purposes.
INTRODUCTION
Notch signaling is an intercellular communication mechanism that is key to cellular differentiation of higher eumetazoa. Through pathway signaling, neighboring cells can adjust for different developmental inputs and differentiate accordingly (reviewed in Artavanis-Tsakonas et al., 1995). A classic example of this process is lateral inhibition, in which one cell is singled out from a group of equipotent cells by forcing its direct neighbors into a secondary cell fate. This cell sends a signal by presenting a membrane-anchored Notch ligand on its surface that activates the Notch receptor in adjacent cells. Consequently, the Notch receptor is cleaved, releasing its biologically active intracellular domain (ICN) from the cell membrane. The ICN travels to the nucleus, where it forms a transcriptional activator complex with Mastermind (Mam) and the DNA-binding protein CSL, a composite of human C-promoter binding factor 1 (CBF1), fly Suppressor of Hairless [Su(H)], and worm lin-12 and Glp-1 phenotype (Lag-1). Subsequently, Notch target genes are activated in the signal-receiving cells, resulting in a down-regulation of the primary fate and allowing for a secondary fate (reviewed in Bray 2006; Kopan and Ilagan, 2009).
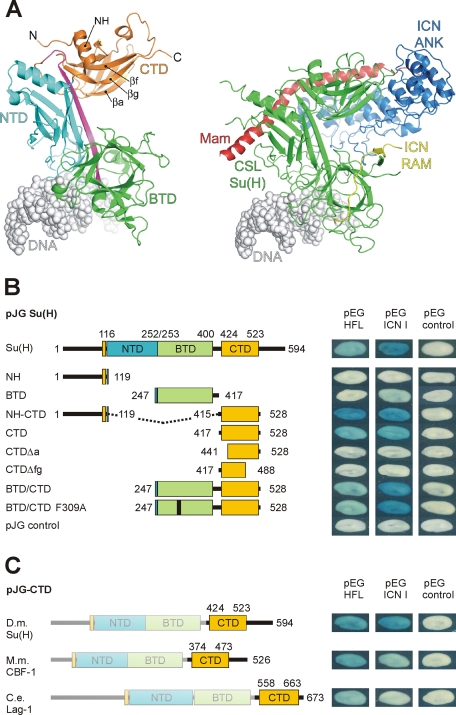
Notch signaling molecules are extremely well conserved in eumetazoa. The DNA-bound activator complex (CSL-ICN-Mam) was crystallized with components derived from the worm Caenorhabditis elegans and from Homo sapiens and turned out to be structurally very similar (Figure 1A; Nam et al., 2006; Wilson and Kovall, 2006). CSL is a rel-type transcription factor, contacting DNA with its N-terminal domain (NTD) and central β-trefoil domain (BTD). ICN contacts the BTD and the C-terminal domain (CTD) of CSL with its RBP-J–associated molecule (RAM) and ankyrin repeats (ANK) domains, respectively. The CTD–ANK domain of Notch interface and the NTD of CSL form a cleft in which Mam fits snugly. Given the high sequence similarity between the involved molecules from Drosophila melanogaster, human, and C. elegans, the respective fly activator complex is presumably structurally very similar (see Supplemental Figure S1).
FIGURE 1:
Defining the Hairless binding domain in Su(H). (A) Left, ribbon diagram of mouse CSL-DNA (Protein Data Bank [PDB] ID: 3IAG) structure, showing overall fold (Friedmann and Kovall, 2010). All CSL proteins, including Drosophila Su(H), have three distinct domains: the N-terminal domain (NTD; cyan), the β-trefoil domain (BTD; green), and the C-terminal domain (CTD; orange). A β-strand that makes hydrogen bonds with all three domains is colored magenta. Both BTD and NTD contact the DNA (gray). Right, ribbon diagram of worm CSL-ICN-Mam-DNA (PDB ID: 2FO1) activation complex (Wilson and Kovall, 2006). CSL is colored green. Notch ICN contacts the BTD with its RAM domain (yellow) and the CTD with its ANK domain (blue). Mam is colored red and binds a groove that is formed by the CTD–ANK interface and the NTD of CSL. (B) Yeast two-hybrid assays demonstrate the interactions between Su(H) and full-length Hairless (HFL) or Notch ICN I, respectively. A set of Su(H) deletion constructs and replacement mutants as indicated were tested to define the domains of Su(H) that interact with Hairless. (C) The CTD domain of CSL proteins derived from Drosophila [D.m., Su(H)], mouse (M.m., CBF1), and C. elegans (C.e., Lag-1) were assayed for interaction with HFL or Notch ICN I. Empty vectors were included as negative controls.
Whereas the activator complex is established in great structural detail, our understanding of Notch target gene inhibition is rather limited and appears surprisingly diverse. Mammalian CSL proteins (human CBF1 and mouse RBP-J) bind to corepressors including SMRT/NCoR, CIR, mSin3A, KyoT2, SHARP (also known as MINT and SPEN), and KDM5a that serve as platforms for chromatin silencers like histone deacetylases (HDACs) and demethylases (HDMs) (reviewed in Borggrefe and Oswald, 2009). A repression domain was identified in CBF1 that roughly corresponds to the BTD and contains several sites relevant for contacting the corepressors SMRT/NCoR and CIR (Hsieh and Hayward, 1995; Hsieh et al., 1997, 1999; Kao et al., 1998; Fuchs et al., 2001), the mouse SHARP homologue MINT (Kuroda et al., 2003; VanderWielen et al., 2011), and presumably mSin3A and KyoT2 (Taniguchi et al., 1998; Zhou et al., 2000). The corepressor binding sites map into or close to the hydrophobic pocket of the BTD that is involved in contacting RAM (Wilson and Kovall, 2006). These data provide an explanation for the observed competition of all identified corepressors and ICN for binding to CBF1/RBP-J (Hsieh and Hayward, 1995; Hsieh et al., 1997; Taniguchi et al., 1998; Fuchs et al., 2001; Kuroda et al., 2003).
Of interest, analogous modes of transcriptional repression of Notch target genes have not been reported from C. elegans. In Drosophila, the major antagonist of Notch signaling is a molecule named Hairless that has not yet been identified outside of insects (Maier, 2006); however, it has been suggested that Hairless and MINT may be functional analogues (Kuroda et al., 2003; Oswald et al., 2005). Hairless binds to Su(H) and allows for the assembly of a repression complex by recruiting the two general corepressors Groucho and C-terminal–binding protein (CtBP) (Morel et al., 2001; Barolo et al., 2002; Nagel et al., 2005), which in turn associate with histone deacetylases (reviewed in Ashraf and Yp, 1998). In addition, the Su(H)–Hairless complex recruits histone chaperones, resulting in chromatin inactivation (Goodfellow et al., 2007; Moshkin et al., 2009). However, the Hairless-binding site has been roughly mapped to the C-terminal half of Su(H) (Brou et al., 1994), which is different from the BTD binding described for the mammalian CSL corepressor complexes. In spite of these differences, the current model for Notch target gene regulation is a molecular switch in which CSL either assembles an activator or a repressor complex, depending on the presence or absence of ICN.
In this work we set out to characterize the molecular details of the Notch repressor complex in Drosophila, using the previous structures of the CSL-ICN-Mam activator complexes to guide our mutagenesis and binding studies. To this end, we delineated the relevant regions of interaction in both Su(H) and Hairless. Of interest, Hairless exclusively binds the CTD of Su(H), which is in contrast to the mammalian corepressors, which primarily interact with the BTD of CSL. However, Hairless interacts with different residues on Su(H) than those used by ICN or Mam. Nonetheless, ICN and Hairless compete for Su(H) binding in gel-shift assays and cultured cells. Within the Su(H)-binding domain of Hairless, we mapped a single site that is absolutely essential for stable complex formation with Su(H). We addressed the biological activity of the Hairless mutant in a cellular transcriptional assay and during fly development. As expected from our binding data, the Hairless mutant fails to assemble a stable repressor complex in vivo, strongly indicating the relevance of the mutant site for Hairless activity. Taken together, this study provides for a fuller molecular understanding of the Notch repressor complex in Drosophila, which expands our knowledge of transcriptional regulation in the Notch pathway and potentially identifies new sites of therapeutic intervention for pharmacologically modulating Notch signaling.
RESULTS
The Hairless-binding domain in Suppressor of Hairless
Previously, and prior to the structural determinations of CSL and CSL-ICN-Mam complexes, the Hairless-binding domain had been mapped to the C-terminal half of the Su(H) protein (Brou et al., 1994), which roughly corresponds to the BTD-CTD domains. To further define the interaction domain, we tested the Su(H) constructs BTD-CTD, BTD, and CTD in a yeast two-hybrid assay with Hairless (Figure 1B). Whereas Su(H) CTD-containing constructs bind to Hairless, the BTD domain was neither sufficient for nor enhanced binding in conjunction with CTD. Accordingly, deletion of either β strands a (CTDΔa) or g and f (CTDΔfg) abolished binding (Figure 1, A and B), indicating that proper folding of the CTD was required for the Hairless contact. Moreover, a construct of CTD that contains the first α-helix just N-terminal of NTD (NH-CTD), which, based on the three-dimensional structure of CSL, is in close contact with CTD (Figure 1A), improved the interaction with Hairless. A similarly strong binding of NH-CTD to Notch ICN supports the notion that the N-terminal α-helix may help to stabilize CTD (Figure 1B). On its own the Su(H) NH domain was unable to bind to Hairless or ICN.
Next we engineered a mutation into the BTD of Su(H) (phenylalanine 309 to alanine; F309A) because the corresponding mutant in CBF1 had been shown previously to disrupt its interactions with several corepressors (Hsieh and Hayward, 1995; Kao et al., 1998; Fuchs et al., 2001). This site lies in the hydrophobic pocket of BTD and is important for binding the RAM domain of ICN (Hsieh and Hayward, 1995; Wilson and Kovall, 2006). Notably, the F309A mutation did not interfere with Hairless binding, indicating a different mechanism for repressor complex formation in Drosophila (Figure 1B). It should also be mentioned that our ICN construct does not contain the RAM domain and therefore is not affected by the F309A BTD mutation (Figure 1B).
Binding of Hairless to mouse CBF1 and C. elegans Lag-1
Due to the high sequence conservation among CSL proteins (Supplemental Figure S1), if Hairless binds to conserved regions of Su(H), then one might also expect binding of Hairless to mouse RBP-J/CBF1 and C. elegans Lag-1 proteins. Indeed, this was demonstrated using our yeast two-hybrid assay: the CTD domain of mouse CBF1 showed a strong interaction with Hairless, whereas that of Lag-1 bound markedly less (Figure 1C). Of interest, the Hairless interaction with mouse CTD was qualitatively stronger than the interaction of mouse CTD with fly ICN (Figure 1C).
Hairless- and Notch-binding sites do not overlap
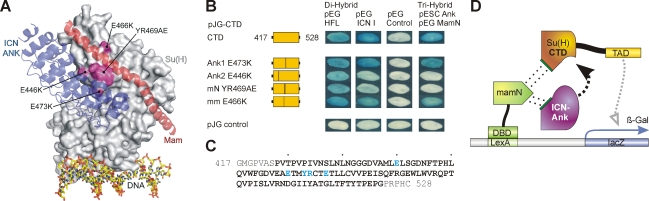
The CTD has been shown to bind to the ankyrin repeats of Notch, forming a groove that binds the N-terminal helix of Mastermind (MamN; Figure 2A; Nam et al., 2006; Wilson and Kovall, 2006). Because Notch and Hairless are genetic competitors in flies (Lindsley and Zimm, 1992; Lyman and Yedvobnick, 1995; Schweisguth et al., 1996), we wondered whether Hairless interacts with the same sites on CTD as ANK. To this end, we mutated four sites in CTD that were predicted to be essential for binding to ANK and/or MamN, based on the structures of the C. elegans and H. sapiens activator complexes (Nam et al., 2006; Wilson and Kovall, 2006); the four CTD mutants are as follows: CTDAnk1, CTDAnk2, CTDmN, and CTDmm (Figure 2, A–C). Of interest, CTDAnk2 and CTDmN were considerably reduced in their binding to Notch ICN in a yeast two-hybrid assay (Figure 2B). In contrast, binding of CTDAnk1 and CTDmm to ICN was nearly identical to wild type (Figure 2B). A yeast three-hybrid assay (CTD, ICN, and Mam; Figure 2, B and D) revealed that CTDmm allowed for reconstitution of the trimeric activator complex, indicating that it bound to MamN as well as to ICN. Both mutant constructs CTDAnk2 and CTDmN completely failed to form the trimeric complex in the yeast three-hybrid assay, again revealing the lack of binding to Notch (Figure 2, B and D). However, Hairless bound to all of these CTD mutants similar to wild-type CTD (Figure 2B).
FIGURE 2:
Fine mapping of the Hairless contact site on Su(H) CTD. (A) Structure of the human CSL-ICN-Mam activator complex (PDB ID: 2F8X) (Nam et al., 2006). CSL is represented as a gray surface, and ICN and Mam are colored blue and red, respectively. Mutations in the CTD that lie at the interface with ICN and Mam are colored magenta. (B) Mutant CTD constructs were tested in a yeast two-hybrid assay for binding to full-length Hairless (HFL) and to intracellular Notch (ICN I). Moreover, the mutant CTD constructs were tested in a yeast three-hybrid assay for their potential to assemble the trimeric activator complex with Notch ANK and Mam. Empty vectors served as negative controls. Position of mutations within CTD is indicated. Note that mutations Ank2 and mN are competent for binding HFL but are considerably reduced for ICN binding and also fail to assemble the trimeric activator complex. Hence, Notch and Hairless contact different sites on Su(H). (C) Primary sequence of the Su(H)–CTD construct; the CTD is shown in bold. Amino acids interacting with ANK/Mam that were mutated are colored blue. (D) Schematic for yeast three-hybrid assay; the N-terminal domain of Mam (mamN) was fused to the LexA DNA-binding domain, and the CTD of Su(H) was fused with the trans-activation domain (TAD). Notch ANK was provided from the plasmid pESC. Su(H) CTD constructs carried mutations as indicated in B. Assembly of the trimeric complex results in activation of the lacZ reporter.
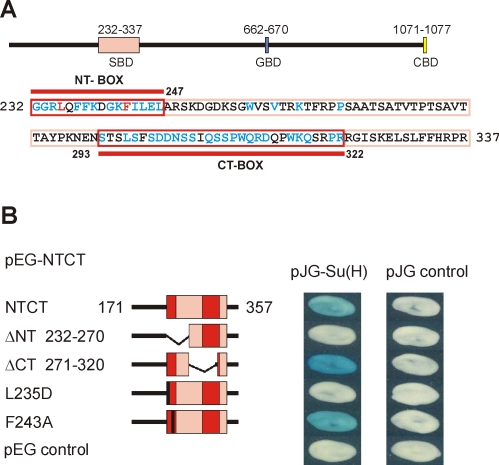
Fine mapping of the Su(H)-binding domain in Hairless
As shown in Figure 3, the Su(H)-binding domain (SBD) in Hairless is contained within a region represented by the NTCT construct. However, our earlier work showed that the N-terminal half is sufficient for binding, whereas the C-terminal half of NTCT does not bind to Su(H) on its own (Maier et al., 2008). Accordingly, a NTCT construct lacking the conserved N-terminal NT box (ΔNT) completely lost Su(H) binding in our yeast two-hybrid assay (Figure 3B). As expected, the corresponding deletion of the C-terminal CT box (ΔCT) had no effect on Su(H) binding (Figure 3B). Circular dichroism (CD) analysis of a purified recombinant protein that corresponds to Hairless NT indicated that in the absence of a binding partner, NT is largely a random coil in solution (Figure 4A). In an attempt to determine residues in Hairless that are important for contacting Su(H), we generated two point mutations and tested for binding to Su(H). Indeed, mutation of leucine 235 to aspartate (L235D) completely abrogated binding to Su(H) (Figure 3B). However, mutation of phenylalanine 243 to alanine (F243A) had no effect on Su(H) binding (Figure 3B). CD analysis of the Hairless mutant L235D protein showed a nearly identical spectrum to wild type (Supplemental Figure S2), indicating that the amino acid substitution did not disrupt the structure or folding of Hairless.
FIGURE 3:
Mapping Su(H)-binding sites in Hairless. (A) Scheme of the HFL protein containing the Su(H)-binding domain (SBD; pink), the Groucho binding domain (GBD; blue) and a binding site for the C-terminal binding protein (CBD; yellow). Bottom, the primary sequence of the SBD is shown; the NT and CT boxes, defined by their conservation in insects, are framed in red. Amino acids that are identical in all known/predicted Hairless proteins are shown in blue and red, with the red residues (L235 and F243) indicating the mutation sites tested for Su(H) binding. (B) Yeast two-hybrid assay on the binding activity of wild-type and mutant NTCT constructs (amino acids 171–357) with full-length Su(H); empty vector served as a negative control. Note that the lack of binding for the ΔNT deletion construct is comparable to the single-site mutant L235D, whereas the F243A mutant binds Su(H) similar to wild-type NTCT.
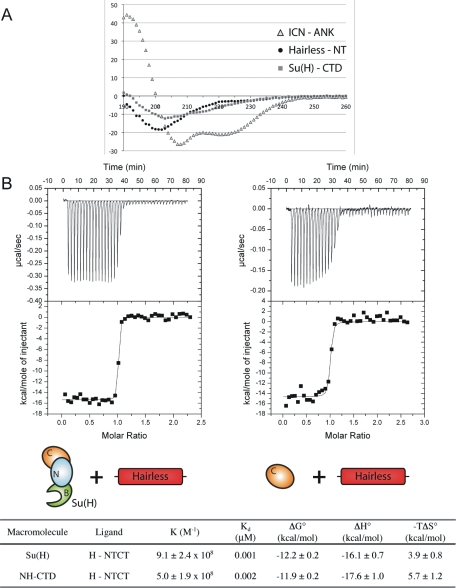
FIGURE 4:
Biophysical analysis of Hairless interactions with Su(H). (A) Far-UV circular dichroism spectroscopy for Hairless (NT), Notch ICN (ANK), and Su(H) NH-CTD. The normalized root-mean-square deviation (NMRSD) parameter values for analysis of the Hairless NT, ANK, and NH-CTD CD data are 0.03, 0.02, and 0.08, respectively. The CD spectrum of Hairless NT has a minimum at ∼200 nm, which is characteristic of random coil. Consistent with its largely helical structure and previously published results (Zweifel et al., 2003), the CD spectrum of ANK shows characteristic minima for α-helix at 207 and 222 nm. The CD spectrum of NH-CTD displays a minimum at 215 nm, consistent with this domain of Su(H) being largely composed of β-sheet. (B) Representative thermograms, raw heat signal, and nonlinear least squares fit to the integrated data for full-length Su(H) and NH-CTD constructs interacting with Hairless (NTCT). Forty titrations were performed per experiment, consisting of 7-μl injections that were spaced 120 s apart. Thermodynamic parameters are shown for each type of experiment; the average N (ligand/macromolecule) values for both experiments were 0.9. Values are the mean of at least three independent experiments and errors represent the SDs of multiple experiments.
Thermodynamics of Su(H)–Hairless interactions
To quantitatively define the affinity and thermodynamic parameters that underlie complexes formed between Su(H) and Hairless, we used isothermal titration calorimetry (ITC) with recombinantly purified Su(H) and Hairless proteins. Our Su(H) construct contains all three conserved domains of CSL (NTD-BTD-CTD; residues 98–523), which is similar to mouse and worm constructs of CSL used previously for ITC binding studies (VanderWielen et al., 2011); our Hairless construct for ITC binding studies corresponds to NTCT (residues 232–358). As shown Figure 4B, Hairless forms a high-affinity 1:1 complex with Su(H) that is enthalpically driven and characterized by a ∼1 nM dissociation constant (Kd). Next we characterized constructs that correspond to the BTD and NH-CTD of Su(H) in order to define the domains of Su(H) that are necessary and sufficient for binding Hairless and validate our yeast two-hybrid studies. As predicted from our yeast two-hybrid results, NH-CTD binds Hairless with nearly identical affinity as full-length Su(H) (Figure 4B). In addition, no binding was observed with BTD or the NTCT mutant L235D.
ICN and Hairless compete for binding to Su(H)
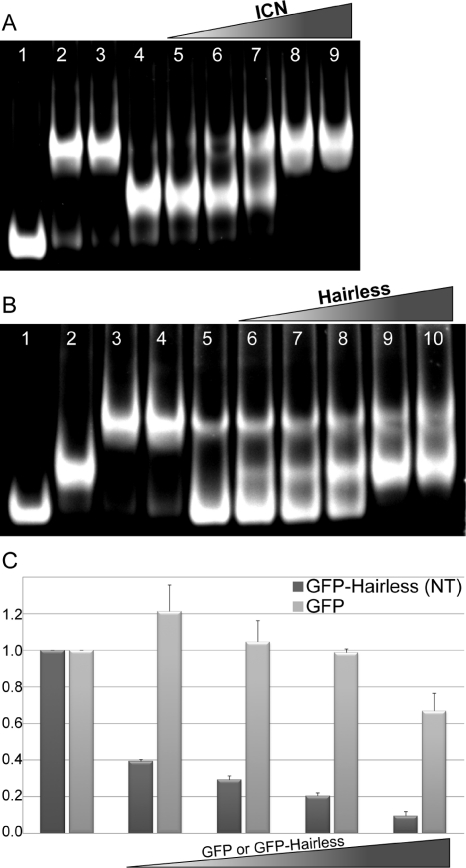
To determine whether Hairless binding and ICN binding to Su(H) are mutually exclusive and competitive, we used electrophoretic mobility shift assays (EMSAs) with purified recombinant Su(H), Hairless, ICN, and Mam proteins. For these experiments we purified a construct of Hairless containing the NT box (residues 232–269). As shown in Figure 5A and Supplemental Figure S3, ICN effectively removed Hairless from Su(H), which was not dependent on the presence of Mam. Conversely, Hairless was not an effective competitor for Su(H) binding, as it was only able to partially supplant ICN from Su(H) (Figure 5B and Supplemental Figure S3). Notably, the EMSAs show no evidence of complexes that contain both ICN and Hairless simultaneously binding Su(H).
FIGURE 5:
Characterization of ICN and Hairless competition for binding Su(H). For all EMSAs, unless otherwise noted, the concentrations of DNA, Su(H), Hairless (NT), ICN (RAMANK), and Mam are 1, 0.1, 4, 4, and 5 μM, respectively. (A) ICN efficiently displaces Hairless from Su(H); control lanes 1–4: Su(H), Su(H)–ICN, Su(H)–ICN–MAM, and Su(H)–Hairless complexes, respectively; lanes 5–9: preformed Su(H)–Hairless complexes titrated with increasing concentrations of ICN (0.5, 1, 2, 4, and 6 μM). (B) Hairless is less efficient in displacing ICN from Su(H); control lanes 1–5: Su(H), Su(H)–Hairless, Su(H)–ICN–MAM, Su(H)–ICN, and Su(H)–ICN (1 μM ICN) complexes, respectively; lanes 6–10: preformed Su(H)–ICN (1 μM ICN) complexes titrated with increasing concentrations of Hairless (0.5, 1, 2, 5, and 20 μM). (C) Hairless NT competes with ICN for binding to endogenous CSL in mammalian cells. Cultured MEFs were transiently transfected with an activated form of murine Notch1 (ICN1), the 4xCBS luciferase reporter, and increasing concentrations of GFP (light gray bars) or GFP–Hairless (NTCT; dark gray bars) constructs (lanes 2–5). Potent activation of the reporter is observed from the 4xCBS reporter with ICN (lane 1). Increasing concentrations of GFP–Hairless, but not the GFP control, result in strong inhibition from the reporter (lanes 2–5). Lanes 2–5 represent 25, 50, 100, and 250 ng of transfected GFP or GFP–Hairless DNA, respectively. The y-axis represents relative activity derived from normalizing the data to experiments performed in the absence of GFP or GFP–Hairless (lane 1). Data are derived from three independent experiments (n = 3), and the error bars represent the SE of the mean.
To determine whether the competition between Hairless and ICN for CSL occurs in mammalian cells as well, transcriptional reporter assays were performed in mouse embryonic fibroblasts (MEFs) (Figure 5C). MEFs were transiently transfected with the 4xCBS luciferase reporter, which contains four iterative CSL-binding sites, an activated form of mouse Notch1 (ICN1), and a green fluorescent protein (GFP)–Hairless NT construct that was targeted to the nucleus by a NLS sequence. As similarly reported elsewhere (VanderWielen et al., 2011), robust activation of the reporter is observed for cells transfected with ICN1; strikingly, potent inhibition of the reporter is observed upon transfection of increasing amounts of the GFP-Hairless construct but not with the GFP control vector.
Transcriptional activation and repression in S2 Schneider cells
Next we characterized the activity of our Hairless mutant in as S2 cell culture assay using a Notch-dependent transcriptional reporter (Nagel et al., 2005). To this end, the L235D mutation was introduced into a construct that contained the full-length Hairless protein (HLD), and transiently expressed in S2 cells. A luciferase reporter containing Su(H)-binding sites (NRE reporter) (Bray et al., 2005) was used to quantify the activation and repression of transcription by the mutant and wild-type Hairless proteins (Figure 6A). Activation of the NRE reporter by ICN was taken as 100% to normalize the other results. Addition of Hairless repressed transcription from the reporter by ∼70%, similar to what was reported previously (Nagel et al., 2005). In stark contrast, the mutant Hairless protein HLD had lost all repressive activity, presumably due to the lack of a Su(H)–H interaction. The concurrent transient transfection of Su(H) and ICN raised the activation of the reporter about fourfold, in accordance with earlier reports (Matsuno et al., 1997). Wild-type Hairless repressed the activation by Su(H) and ICN nearly to the ICN baseline. In contrast, the Hairless mutant HLD was unable to repress activity from the reporter, despite excessive amounts of Su(H) (Figure 6A). In summary, these data show the inability of the mutant HLD protein to bind to Su(H) and generate a repression complex on Notch target promoters in cultured cells.
FIGURE 6:
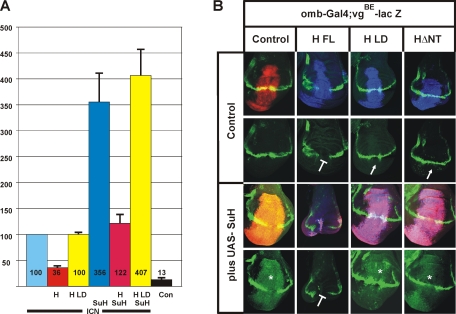
Mutant Hairless fails to assemble functional repressor complexes. (A) Effects of wild-type or mutant Hairless protein, in the absence or presence of Su(H), on Notch ICN-mediated expression from the Notch reporter (NRE) were studied in Drosophila S2 cell culture. S2 cells were transiently transfected with constructs indicated; empty vector was used as a negative control. Luciferase activity is represented on the y-axis, and transfection efficiency was normalized by cotransfection of the Renilla plasmid. Values for NRE expression in the presence of Notch ICN were normalized to 100% (lane 1). (B) Effects of the overexpression of wild-type and mutant Hairless constructs, singly or together with Su(H), on in vivo expression of the Notch target gene vestigial. Wild-type and mutant UAS constructs as indicated were induced in the central region of wing imaginal disks using the omb-Gal4 driver line and visualized by antibody staining. UAS-GFP served as control (red). Expression of vgBE-lacZ reporter is expected along the dorsoventral boundary and shown in green (bottom). In the merged picture (top), overlap appears yellow. Overexpressed Su(H) is shown in red, and Hairless protein is shown in blue; overlap appears magenta, and with vgBE-lacZ (green) it appears white. Overexpression of Su(H) causes an overgrowth of the disk and an activation of vgBE-lacZ (asterisk). Conversely, overexpression of HFL results in a repression of vgBE-lacZ (blunt arrow). However, overexpression of the Hairless mutant constructs HΔNT or HLD has little effect on vgBE-lacZ expression, reflecting the loss of Su(H)-binding activity (arrow). A combination of HFL with Su(H) strongly affects growth of the wing disk and vgBE-lacZ expression. In contrast, a combination of Su(H) with either HΔNT or HLD results in overgrowth and vgBE-lacZ induction that resembles the sole Su(H) overexpression, indicating that these Hairless mutants fail to form repressor complexes in vivo.
In vivo transcriptional response of Notch target genes
We next addressed the transcriptional activity of the HLD mutant using in vivo assays in the fly. Wild-type Su(H), Hairless, and the mutant versions HLD and HΔNT (ΔNT; deletion of residues G232–S270) were cloned into UAS vectors that can be expressed in a tissue-specific manner during fly development (Brand and Perrimon, 1993). Transgenic fly lines were established using the PhiC31 method, which allows for site-specific integration of the respective mutant and wild-type constructs (Bischof et al., 2007); this circumvents position effects in different transgenic lines. We chose distant integration sites for the Su(H) and Hairless constructs to facilitate subsequent recombination of the transgenes. Su(H) and Hairless variants were induced in the central domain of the wing disk only. Then, expression of a Notch target gene was visualized in the affected tissue and compared with the surrounding wild-type tissue (Figure 6B). We used a reporter vgBE-lacZ, which is specifically activated along the dorsoventral boundary in response to Notch signaling (Kim et al., 1996). Su(H) overexpression [plus UAS-Su(H)] induces this reporter within the entire expression domain, which appears enlarged due to a strong hyperproliferation of the affected tissue (Figure 6B; Nagel et al., 2005). In contrast, HFL overexpression induces a down-regulation of vgBE-lacZ, as expected by its repressor function (Figure 6B; Nagel et al., 2005). However, overexpression of the Hairless mutant variants, HΔNT and HLD, had nearly no effects on vgBE-lacZ expression, in accordance with their lack of binding to Su(H) (Figure 6B). Notably, HLD was indistinguishable from HΔNT, indicating that it had completely lost Su(H)-binding activity. Combined overexpression of Su(H) and HFL severely disturbed growth and development of the wing disk and caused complete repression of vgBE-lacZ (Figure 6B). In this case, a surplus of repression complexes is assembled by the overexpressed proteins, thereby causing an extreme down-regulation of Notch activity (Morel et al., 2001; Nagel et al., 2005). However, the combined overexpression of Su(H) with either HΔNT or HLD was similar to solely overexpressing Su(H), indicating that these Hairless variants failed to assemble a repressor complex in vivo (Figure 6B).
Activity of the Hairless protein variants in the fly
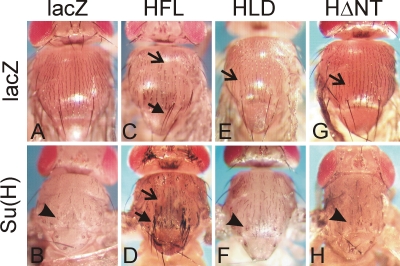
For the in vivo analysis we concentrated on bristle formation on the thorax, a process that involves lateral inhibition and cell type specification that is controlled by Notch signaling (Schweisguth et al., 1996; reviewed in Artavanis-Tsakonas et al., 1995). Overexpression of Su(H) in the thorax causes mild Notch gain-of-function phenotypes, which are recognized by typical shaft-to-socket transformations that affect most of the microchaetae and macrochaetae (Figure 7, A and B). Because Hairless potently antagonizes Notch signaling, overexpression of Hairless induces Notch loss-of-function phenotypes. In the context of bristle development, these include split bristles due to a socket-to-shaft transformation, loss of bristles due to a transformation of external to internal cell fates, and, in extreme cases, ectopic bristles due to a failure of lateral inhibition (Schweisguth et al., 1996; Nagel et al., 2000). The overexpression of wild-type HFL in the thorax caused primarily bristle loss and socket-to-shaft transformations mainly of the macrochaetae (Figure 7C). As expected from their inability to interact with Su(H), the Hairless mutant constructs HΔNT and HLD were nearly inactive in inducing bristle loss or socket-to-shaft transformations, displaying a wild-type phenotype (Figure 7, E and G).
FIGURE 7:
In vivo activity of Hairless mutant constructs. Wild-type and mutant constructs as indicated were expressed in the thorax of the developing fly using the MS1096-Gal4 driver line. Effects of Su(H) and Hairless on bristle development were studied singly and in combination to assay for the formation of a repressor complex. Open arrows (missing bristles due to transformation of external to internal fate) and closed arrows (ectopic bristles or double shafts) point to Notch loss-of-function phenotypes, and arrowheads (loss of bristles, double sockets) point to Notch gain-of-function phenotypes. Compared to the control (A), overexpression of wild-type Su(H) causes typical Notch gain-of-function phenotypes affecting the majority of macrochaetae and microchaetae (B). (C) Overexpression of full-length Hairless HFL results in Notch loss-of-function phenotypes affecting most macrochaetae and many microchaetae. (D) Combined overexpression of HFL and Su(H) enhances this effect strongly, and bushes of bristles appear as a result of reduced lateral inhibition. Flies resulting from overexpression of either Hairless mutant HLD (E) or HΔNT (G) have nearly wild-type phenotype. Moreover, in combination with Su(H) (F, H) the phenotypes are similar to the sole Su(H) overexpression (see B).
A combined overexpression of Hairless with Su(H) results in a strong Notch loss-of-function phenotype since the two assemble a potent repressor complex (Morel et al., 2001; Nagel et al., 2005). Indeed, bristle clusters resulting from incomplete lateral inhibition were observed in the combinations of wild-type Su(H) with HFL (Figure 7D). In contrast, a combination of Su(H) with either HΔNT or HLD caused phenotypes resembling sole overexpression of Su(H) (Figure 7, F and H), indicating that these Hairless mutants are strongly impaired in forming repressor complexes with Su(H).
DISCUSSION
CSL is the nuclear effector of the Notch signaling pathway and is required for both repression and activation of transcription from Notch target genes (Kovall and Blacklow, 2010). In the absence of a signal, CSL functions as a transcriptional repressor by interacting with corepressor proteins, such as SHARP, SMRT/NCoR, KyoT2, and CIR. CSL–corepressor interactions function to localize histone deacetylase and histone demethylase activity at Notch target genes, which converts the local chromatin into a condensed, transcriptionally silent state (reviewed in Borggrefe and Oswald, 2009). On pathway activation, the ICN binds CSL, and together with Mam, forms a transcriptionally active ternary complex that ultimately displaces corepressors from CSL and upregulates transcription from Notch target genes. In mammals, a number of corepressors have been shown to interact with the BTD of CSL, similar to ICN, which provides a model in which ICN displaces or outcompetes corepressors for binding to CSL. Thus, there are potentially two modes of repression mediated by corepressors: 1) at the transcription or chromatin level, in which the recruitment of HDAC/HDM–containing complexes by corepressors silences gene expression—this mode of transcriptional repression is independent of Notch; and 2) at the protein level, by which corepressors and ICN compete for binding to CSL.
Although several of the mammalian corepressors have fly orthologues, these molecules do not seem to be generally involved in repressing transcription from Notch target genes in flies. A complex involving the SMRT homologue SMRTER negatively regulates Notch signaling during the specification of a subset of nonneuronal cell types in the developing Drosophila retina. However, mammalian SMRT is believed to contact CBF1 directly, whereas SMRTER does not bind Su(H) on its own (Tsuda et al., 2002). The Drosophila orthologue of SHARP/MINT, termed Spen, which genetically inhibits Notch signaling in the context of eye development, is presumably not a transcriptional repressor of Notch target genes in this process (Doroquez et al., 2007). Recently it was shown that Spen is required for the activation rather than the repression of Notch target genes during the development of hemocytes (Jin et al., 2009). Moreover, the region of SHARP/MINT that has been defined to interact with CSL is not conserved in Spen (Rebay et al., 2000; Oswald et al., 2002; VanderWielen et al., 2011). Although formally SHARP/MINT might act as a functional Hairless analogue in mammals, the role of the structurally related Spen proteins seems largely diverse in different organisms.
In flies, the major antagonist of Notch signaling is the transcriptional corepressor Hairless, which is ubiquitously expressed in all tissues (Maier et al., 1999). Hairless binds the transcription factor Su(H), as well as the corepressors Groucho and CtBP, which serves to localize the transcriptional repression machinery in the nucleus to Notch target genes, thereby repressing gene expression. Removal of the Groucho and CtBP-binding sites from Hairless does not completely eliminate its activity as a repressor, suggesting that, similar to other corepressors, Hairless might compete with ICN for binding Su(H) (Nagel et al., 2005). However, the molecular mechanism by which Su(H) is converted from a repressor to an activator complex is unclear.
In this work we set out to investigate the molecular details of the Notch repressor complex in Drosophila. Our analysis was multidisciplinary in nature, using biophysical, biochemical, cellular, and in vivo assays to characterize the protein–protein interface between Hairless and Su(H). We demonstrate that Hairless forms a high-affinity 1:1 complex with Su(H) (∼1 nM Kd) but only interacts with the CTD, which is in stark contrast to mammalian CSL–corepressor interactions, which are largely mediated through BTD contacts. Previous ITC binding studies of the mammalian Notch components Notch1 (ICN) and RBP-J showed that the Kd of the ICN/RBP-J complex is ∼10 nM (Friedmann et al., 2008), suggesting that Su(H)–Hairless and Su(H)–Notch interactions are likely of comparable affinity.
Given the similar affinities of ICN and Hairless for Su(H), the question arises whether ICN and Hairless compete for binding to CSL. On one hand, our gel-shift assays with purified protein components showed that ICN can displace Hairless from Su(H) independent of Mastermind. On the other hand, we showed that residues on Su(H) that are important for Notch ANK and Mam binding to CTD do not affect interactions with Hairless. These data suggest that the ICN- and Hairless-binding sites on Su(H) do not overlap. If the ANK domain of ICN and Hairless are competing for binding to the CTD of Su(H), then there is an additional factor to consider: based on binding studies of the mammalian proteins, the vast majority of the binding energy for the Su(H)–ICN complex comes from the RAM domain interaction with the BTD of Su(H), whereas the isolated CTD–ANK interaction is of very low affinity (Kd > 20 μM) (Delbianco et al., 2008; Friedmann et al., 2008). This represents at least a 10,000-fold difference in the affinities of ANK and Hairless for CTD, which suggests that the ANK domain of ICN would seem to be a very poor competitor for removing Hairless from Su(H).
How then is ICN able to supplant Hairless from Su(H) in order to activate transcription from Notch target genes? Certainly additional experiments will be required to fully address this question; however, at present we favor the following hypothesis: the binding of ICN to Su(H), that is, the RAM–BTD interaction, results in allosteric changes in Su(H) that decreases its overall affinity for Hairless, thereby making ANK a more effective competitor for CTD. Consistent with this notion, our gel-shift experiments showed that Hairless was far less effective at removing ICN from Su(H), even when Hairless was present in vast excess (Figure 5B).
Our studies also analyzed two absolutely conserved residues in Hairless (L235 and F243) for their contribution to binding Su(H). Whereas F243 was dispensable for binding, L235 was absolutely required for binding Su(H) in vitro. Mutation of this site to aspartate abrogated binding but did not change the secondary structure content of Hairless, which suggests that L235 lies at the Su(H)–Hairless interface. Given the conservation of F243 but its dispensability for Su(H) binding, this perhaps suggests that this residue is important for interacting with other nuclear factors. Consistent with our in vitro binding results, the Hairless mutant L235D failed to assemble a repressor complex with Su(H) in cellular and in vivo assays in the fly. In fact, the L235D mutant was as deficient in repression as the Hairless deletion mutant ΔNT, which removes residues 232–270, emphasizing the importance of this contact in forming the Su(H)–Hairless complex.
In conclusion, the fly Notch repressor complex shows similarities and differences compared with the mammalian complex. Despite the high degree of sequence and likely structural conservation, Su(H) in Drosophila differs from mammalian CBF1/RBP-J in that it has no repressor activity on its own; overexpression of Su(H) in cell culture and in vivo results in a Notch gain-of-function phenotype. It is not until the binding of Hairless that Su(H) is transformed into a repressor. Of interest, we showed that Hairless bound the mammalian CSL orthologue CBF1 nearly as avidly as Su(H), which suggests that the Hairless-binding site on the CTD has been conserved in mammals. In accordance, we find a potent repression of Notch transcriptional activity in cultured mammalian cells by the Hairless NT construct. This raises the possibility of identifying Hairless homologues in other organisms or potentially other transcriptional coregulators that use the Hairless-binding site on CTD, which may be indicative of an as-yet-unidentified mode in mammals to repress Notch signaling. Nonetheless, we can now use our detailed knowledge of Su(H)–Hairless interactions to develop molecules that target Notch transcription complexes and either enforce or disrupt their activity, thereby opening new therapeutic avenues.
MATERIALS AND METHODS
Generation of Hairless constructs
Single mutations of individual codons were introduced into NTCT (amino acids 171–357) (Maier et al., 2008) with the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA). The mutant NTCT constructs served to replace the respective wild-type sequences in the full-length Hairless cDNA to generate mutant full-length constructs in pRmHa-3 (Bunch et al., 1988) and pUAST-attB-vectors (Bischof et al., 2007) for further analysis of in vivo activity. The two deletions ΔNT (deleted from amino acids 232–270) and ΔCT (deleted from amino acids 271–320) were made in a ClaI fragment of the Hairless cDNA by using the ExSite PCR Based Site-Directed Mutagenesis Kit (Stratagene). Primer sequences are available upon request. The truncated ClaI fragments were reinserted into the full-length Hairless cDNA. The same two deletions were introduced into NTCT by replacement of the respective wild-type NarI/HindIII fragment. The mutant NTCT constructs were subsequently shuttled as a BamHI/XhoI fragment into a pEG vector (Gyuris et al., 1993) for yeast two-hybrid analysis. All mutants were sequence verified (StarSeq, Mainz, Germany).
Generation of Su(H) constructs
Truncated Su(H) constructs NH (codons 1–119), BTD (codons 247–417), BTD/CTD (codons 247–528), CTD (codons 417–528), CTDΔa (amino acids 441–528), and CTDΔfg (amino acids 417–488) were generated by PCR amplification from Su(H) cDNA and cloned into BT vectors (Stratagene). Primers included EcoRI and XhoI sites to allow for shuttling into a pJG vector (Gyuris et al., 1993) for subsequent yeast two-hybrid analysis. The Su(H) NH-CTD construct (codons 1–119 and 415–528) was created by introducing an EcoRI site at the fusion position. Afterward the respective PCR products were cloned in two steps into the pJG vector. As consequence of this manipulation an additional phenylalanine codon was created at the fusion site. All mutants were sequence verified (StarSeq). Primer sequences are available upon request.
Analysis of protein–protein interactions
Yeast two-hybrid protein interaction assays were performed as previously described using pEG-H (Nagel et al., 2005), pEG-NTCT (Maier et al., 2008), pEG-ICNI, pEG-mam (Matsuno et al., 1995), and pJG-Su(H) (Nagel et al., 2005) in combination with the newly generated constructs. For yeast three-hybrid protein interactions assays, the coding sequence of the Notch ankyrin repeats (N-ANK, codons 1873–2176) was PCR amplified with primers containing BglII and SacI sites, respectively, and cloned in frame with the FLAG epitope of the pESC-Leu vector (Stratagene). Media lacking leucine allowed a selection for pESC presence. The N-terminal region of Mam (MamN, codons 113–178) was cloned into pEG-vector and combined with pJG-Su(H) to complete the yeast three-hybrid assay, which was performed in analogy to the two-hybrid protocol.
Circular dichroism
CD measurements were taken in triplicate using an Aviv Circular Dichroism Spectrometer, model 215 (Aviv Biomedical, Lakewood, NJ), as described before (VanderWielen et al., 2011). Measurements were collected in a 0.02-cm cuvette at 25°C using 1.0-nm-wavelength steps between 190 and 290 nM. Su(H), Hairless, and Notch ICN proteins were characterized in a buffer containing 50 mM sodium phosphate, pH 6.5, and 75 mM NaCl with protein concentrations ranging from 50 to 100 μM.
Isothermal titration calorimetry
A MicroCal VP-ITC calorimeter was used for ITC experiments. Experiments were carried out at 25°C in a buffer consisting of 50 mM sodium phosphate, pH 6.5, and 150 mM NaCl. Recombinant core Su(H) (amino acids 98–523) was overexpressed as a glutathione S-transferase fusion protein in bacteria, cleaved, and purified to homogeneity, as described previously (Friedmann and Kovall, 2010); the BTD (amino acids 251–420) and NH-CTD (amino acids 101–119 + 415–523) constructs were purified in a similar manner. A construct of Hairless that corresponds to NTCT (232–358) was generated as an SMT3–Hairless fusion protein and purified in a similar manner as SMT3–MINT fusion proteins described previously (VanderWielen et al., 2011). Hairless NTCT peptides cleaved from SMT3 were not sufficiently soluble under the buffer conditions for ITC, which necessitated the use of the SMT3–NTCT fusion protein to measure binding with Su(H). No binding was detected between SMT3 and Su(H). Proteins were degassed and buffer matched using size exclusion chromatography and/or dialysis. A typical experiment consisted of 10 μM Hairless NTCT in the cell and 100 μM Su(H) in the syringe. Protein concentrations were determined by both UV absorbance at 280 nm and bicinchoninic acid assay (Pierce, Thermo Fisher Scientific, Rockford, IL). The binding data reported are the average of at least three individual experiments (n = 3). The value of c (=Ka[M]N) was ∼5000 and ∼1000 for the core Su(H) and CTD experiments, respectively. The data were analyzed using the ORIGIN software and fit to a one-site binding model.
Reporter assays
Reporter assays were performed as described previously (Nagel et al., 2005). Briefly, Drosophila Schneider S2 cells were transiently transfected with 1 μg of luciferase reporter constructs containing Notch-responsive Su(H) element (NRE) and 0.2 μg of control Renilla expression plasmid (tk-Renilla; Promega, Madison, WI). To stimulate reporter gene expression, the cells were cotransfected with 1 μg of pMT-ICN (Matsuno et al., 1995). Repressor activity of mutant versus wild-type Hairless was tested in the presence or absence of Su(H) by adding 0.5 μg of pMT-H or pMT-HLD and 0.5 μg of pMT-Su(H) alone or in combination. In all cases the total amount of transfected DNA was normalized to 3 μg by use of pMT-A (Invitrogen, Carlsbad, CA). Protein expression was induced by adding 0.5 mM CuSO4 6 h after transfection. Cells were harvested 18 h later. Luciferase activity was measured in duplicate (Lumat LB 9507; EG & G, Salem, MA) normalized to the Renilla control by using the dual-luciferase reporter assay system (Promega).
Luciferase reporter assays in mammalian cells were performed as described previously (VanderWielen et al., 2011). Briefly, mouse embryonic fibroblasts derived from CSL (rbp-j) knockout embryos (MEFs; OT11) (Kato et al., 1997) were transduced with the MigR1 retrovirus containing the gene for rbp-j (mouse CSL) and grown to ∼80% confluence in six-well plates. OT11 MEFs were transiently transfected with an NICD1 construct (murine Notch1, residues 1744–2531) to activate the Notch-responsive luciferase reporter 4xCBS, which contains four iterative CSL-binding sites (Ong et al., 2006). The construct phRL, which expresses Renilla luciferase, was used to normalize for transfection efficiency. Hairless residues 232–269, which correspond to NT, were cloned into the plasmid pEGFP-C1 (Clontech, Mountain View, CA) downstream of GFP. Nuclear localization of the GFP–Hairless fusion protein was ensured with two nuclear localization sequences (DPKKRKV) cloned C-terminal to GFP. Transfections were performed with the SatisFection reagent (Agilent Technologies, Santa Clara, CA) following the manufacturer's protocol, and the amount of transfected DNA was normalized using pBluescript (Stratagene). Luciferase activity was assayed 48 h posttransfection using the Dual Luciferase Kit (Promega). Firefly luciferase activity from the 4xCBS reporter was first normalized to Renilla luciferase expression and reported as relative activity by comparing cells transfected with and without GFP/GFP–Hairless constructs. Average values, errors, and SDs were determined from three independent experiments performed in duplicate.
Electrophoretic mobility shift assays
EMSAs were performed as described previously (Friedmann et al., 2008). Briefly, constructs of core Su(H) (amino acids 98–523), the RAMANK domains of ICN (amino acids 1763–2142) and Mam (amino acids 121–188) from Drosophila, and Hairless NT (amino acids 232–269) were expressed in Escherichia coli and purified in a similar manner as described previously for the mouse Notch components (Friedmann et al., 2008); Notch transcription complexes were bound to an oligomeric 19-mer duplex that contains a single CSL-binding site and separated on a 7% polyacrylamide gel containing 0.5× Tris-borate buffer, pH 7, for several hours at 4°C. Complexes were visualized on the gel using SYBR-GOLD stain (Invitrogen).
In vivo analysis of mutant Hairless and Su(H) constructs
Transgenic flies were established with the PhiC31 integrase-based integration system (Bischof et al., 2007). To this end, the respective wild-type and mutant Su(H) and Hairless constructs were shuttled into a pUAST-attB transformation vector and sequence verified. Hairless constructs were integrated using the ΦX-68E strain and Su(H) construct using the ΦX-96E strain, respectively. These strains carry a landing site at the respective positions (68E, 96E). Correct integration was monitored by PCR. For co-overexpression experiments, the different wild-type and mutant strains were recombined. During the phenotypic analyses, all of the strains were genotyped by PCR and following diagnostic restriction digests.
Tissue-specific expression of respective transgenes was induced with the Gal4/UAS-system (Brand and Perrimon, 1993) using omb-Gal4 and MS1096-Gal4 (Bx-Gal4) (FlyBase, http://flybase.org). Expression of the Notch target gene vestigial (vg) was monitored along the dorsoventral boundary in third-instar imaginal disks using the vgBE-LacZ reporter line (Kim et al., 1996), which was combined with the omb-Gal4 driver line and crossed with the respective UAS lines. Staining of imaginal disks was performed as described before using the following antisera: anti–H-A (Maier et al., 2002) and anti-Su(H) (Santa Cruz Biotechnology, Santa Cruz, CA), as well as anti–β-galactosidase (developed by J. R. Sanes; obtained from Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA). Secondary antibodies coupled to DATF, Cy3, or Cy5 were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Samples were mounted in Vectashield (Vector Labs, Eching, Germany) and analyzed on a Zeiss (Thornwood, NY) Axiophot linked to a Bio-Rad MRC1024 confocal microscope. Flies were photographed with an ES120 camera (Optronics, Goleta, CA), using Pixera (Santa Clara, CA) Viewfinder software, version 2.0. Pictures were assembled with Corel (Mountain View, CA) PhotoPaint and CorelDRAW software, version 9.0.
Supplementary Material
Acknowledgments
We acknowledge S. Artavanis-Tsakonas, S. Bray, and C. Delidakis for sharing DNA constructs and F. Karch for providing the PhiC31 fly lines and vectors. We are grateful to F. Allaudin, S. Bienieck, S. Brentle, and L. Ludwig for generating some yeast constructs used in this study. We thank I. Wech, H. Mastel, and A. Iwanska for invaluable technical help and A. Reyer for protein purification. This work was supported in part by National Institutes of Health Grant CA120199 and a Leukemia and Lymphoma Society Award to R.A.K.
Abbreviations used:
- ANK
ankyrin repeats domain of Notch
- CSL
composite of CBF1 (human C-promoter binding factor 1), Su(H) (Suppressor of Hairless of Drosophila melanogaster), and Lag-1 (lin-12 and Glp-1 phenotype of Caenorhabditis elegans)
- CTD
C-terminal domain of Su(H)
- NTD
N-terminal domain of Su(H)
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-05-0420) on July 7, 2011.
REFERENCES
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–268. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Ashraf SI, Yp YT. Transcriptional control: repression by local chromatin modification. Curr Biol. 1998;8:R683–R686. doi: 10.1016/s0960-9822(98)70435-x. [DOI] [PubMed] [Google Scholar]
- Barolo S, Stone T, Bang AG, Posakony JW. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 2002;16:1964–1976. doi: 10.1101/gad.987402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Bray SJ, Musisi H, Bienz M. Bre1 is required for Notch signaling and histone modification. Dev Cell. 2005;8:279–286. doi: 10.1016/j.devcel.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Lecourtois M, Vandekerckhove J, Kourilsky P, Schweisguth F, Israel A. Inhibition of the DNA-binding activity of Drosophila Suppressor of Hairless and of its human homolog, KBF2/RBP-J kappa, by direct protein-protein interaction with Drosophila Hairless. Genes Dev. 1994;8:2491–2503. doi: 10.1101/gad.8.20.2491. [DOI] [PubMed] [Google Scholar]
- Bunch TA, Grinblat Y, Goldstein LS. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbianco C, Aster JC, Blacklow SC. Mutational and energetic studies of Notch1 transcription complexes. J Mol Biol. 2008;376:131–140. doi: 10.1016/j.jmb.2007.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroquez DB, Orr-Weaver TL, Rebay I. Split ends antagonizes the Notch and potentiates the EGFR-signaling pathways during Drosophila eye development. Mech Dev. 2007;124:792–806. doi: 10.1016/j.mod.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann DR, Kovall RA. Thermodynamic and structural insights into CSL-DNA complexes. Protein Sci. 2010;19:34–46. doi: 10.1002/pro.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann DR, Wilson JJ, Kovall RA. RAM-induced allostery facilitates assembly of a Notch pathway active transcription complex. J Biol Chem. 2008;283:14781–14791. doi: 10.1074/jbc.M709501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs KP, Bommer G, Dumont E, Christoph B, Vidal M, Kremmer E, Kempkes B. Mutational analysis of the J recombination signal sequence binding protein (RBP-J)/Epstein-Barr virus nuclear antigen 2 (EBNA2) and RBP-J/Notch interaction. Eur J Biochem. 2001;268:4639–4646. doi: 10.1046/j.1432-1327.2001.02387.x. [DOI] [PubMed] [Google Scholar]
- Goodfellow H, Krejci A, Moshkin Y, Verrijzer CP, Karch F, Bray SJ. Gene-specific targeting of the histone chaperone Asf1 to mediate silencing. Dev Cell. 2007;13:593–600. doi: 10.1016/j.devcel.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Hayward SD. Masking of the CBF1/RBPJk transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Nofziger DE, Weinmaster G, Hayward SD. Epstein-Barr virus immortalization: Notch2 interacts with CBF1 and blocks differentiation. J Virol. 1997;71:1938–1945. doi: 10.1128/jvi.71.3.1938-1945.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L-H, Choi JJ, Kim B, Cho HA, Kim J, Kim-Ha J, Kim YJ. Requirement of Split ends for epigenetic regulation of Notch signal-dependent genes during infection-induced hemocyte differentiation. Mol Cell Biol. 2009;29:1515–1525. doi: 10.1128/MCB.01239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MXG. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall R, Blacklow SJ. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr Top Dev Biol. 2010;92:31–71. doi: 10.1016/S0070-2153(10)92002-4. [DOI] [PubMed] [Google Scholar]
- Kuroda K, et al. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity. 2003;18:301–312. doi: 10.1016/s1074-7613(03)00029-3. [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The Genome of Drosophila melanogaster, London: Academic Press; 1992. [Google Scholar]
- Lyman DF, Yedvobnick B. Drosophila Notch receptor activity suppresses Hairless function during adult external sensory organ development. Genetics. 1995;141:1491–1505. doi: 10.1093/genetics/141.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier D. Hairless, the ignored antagonist of the Notch signalling pathway. Hereditas. 2006;143:212–221. doi: 10.1111/j.2007.0018-0661.01971.x. [DOI] [PubMed] [Google Scholar]
- Maier D, Chen AX, Preiss A, Ketelhut M. The tiny Hairless protein from Apis mellifera: a potent antagonist of Notch signalling in Drosophila melanogaster. BMC Evol Biol. 2008;8:175. doi: 10.1186/1471-2148-8-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier D, Nagel AC, Johannes B, Preiss A. Subcellular localization of Hairless protein shows a major focus of activity within the nucleus. Mech Dev. 1999;89:195–199. doi: 10.1016/s0925-4773(99)00208-7. [DOI] [PubMed] [Google Scholar]
- Maier D, Nagel AC, Preiss A. Two isoforms of the Notch antagonist Hairless are produced by differential translation initiation. Proc Natl Acad Sci USA. 2002;99:15480–15485. doi: 10.1073/pnas.242596699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K, Diederich RJ, Go MJ, Blaumüller CM, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Go MJ, Sun XS, Eastman DS, Artavanis-Tsakonas S. Suppressor of Hairless-independent events in Notch signaling imply new pathway elements. Development. 1997;124:4265–4273. doi: 10.1242/dev.124.21.4265. [DOI] [PubMed] [Google Scholar]
- Morel V, Lecourtois M, Massiani O, Maier D, Preiss A, Schweisguth F. Transcriptional repression by Suppressor of Hairless involves the binding of a Hairless-dCtBP complex in Drosophila. Curr Biol. 2001;11:789–792. doi: 10.1016/s0960-9822(01)00224-x. [DOI] [PubMed] [Google Scholar]
- Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JA, Verrijzer CP. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell. 2009;35:782–793. doi: 10.1016/j.molcel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Nagel AC, Krejci A, Tenin G, Bravo-Patiño A, Bray S, Maier D, Preiss A. Hairless mediated repression of Notch target genes requires combined activity of Groucho and CtBP corepressors. Mol Cell Biol. 2005;25:10433–10441. doi: 10.1128/MCB.25.23.10433-10441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel AC, Maier D, Preiss A. Su(H) independent activity of Hairless during mechanosensory organ formation in Drosophila. Mech Dev. 2000;94:3–12. doi: 10.1016/s0925-4773(00)00319-1. [DOI] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R. Target selectivity of vertebrate Notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J Biol Chem. 2006;281:5106–5119. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- Oswald F, et al. SHARP is a novel component of the Notch/RBP-Jκ signalling pathway. EMBO J. 2002;21:5417–5426. doi: 10.1093/emboj/cdf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Winkler M, Cao Y, Astrahantsefs K, Bourteele S, Knöchel W, Borggrefe T. RBP-Jk/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol Cell Biol. 2005;25:10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I, Chen F, Hsiao F, Kolodziej PA, Kuang BH, Laverty T, Suh C, Voas M, Williams A, Rubin GM. A genetic screen for novel components of the Ras/mitogen-activated protein kinase signaling pathway that interacts with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics. 2000;154:695–712. doi: 10.1093/genetics/154.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth F, Gho M, Lecourtois M. Control of cell fate choices by lateral signaling in the adult peripheral nervous system of Drosophila melanogaster. Dev Genet. 1996;18:28–39. doi: 10.1002/(SICI)1520-6408(1996)18:1<28::AID-DVG4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Furukawa T, Tun T, Han H, Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol Cell Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda L, Nagaraj R, Zipurski SL, Banerjee U. An EGFR/Ebi/Sno pathway promotes Delta expression by inactivating Su(H)/SMRTER repression during inductive Notch signalling. Cell. 2002;110:625–637. doi: 10.1016/s0092-8674(02)00875-9. [DOI] [PubMed] [Google Scholar]
- VanderWielen BD, Yuan Z, Friedmann DR, Kovall RA. Transcriptional repression in the Notch pathway: thermodynamic characterization of CSL-MINT complexes. J Biol Chem. 2011;286:14892–14902. doi: 10.1074/jbc.M110.181156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Zhou S, Fujimuro M, Hsieh JJ, Chen L, Hayward SD. A role for SKIP in EBNA2 activation of CBF1-repressed promoters. J Virol. 2000;74:1939–1947. doi: 10.1128/jvi.74.4.1939-1947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifel ME, Leahy DJ, Hughson FM, Barrick D. Structure and stability of the ankyrin domain of the Drosophila Notch receptor. Protein Sci. 2003;12:2622–2632. doi: 10.1110/ps.03279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.