Abstract
Transcription factor Histone Nuclear Factor P (HiNF-P; gene symbol Hinfp) mediates cell cycle control of histone H4 gene expression to support the packaging of newly replicated DNA as chromatin. The HiNF-P/p220NPAT complex controls multiple H4 genes in established human cell lines and is critical for cell proliferation. The mouse HinfpLacZ null allele causes early embryonic lethality due to a blastocyst defect. However, neither Hinfp function nor its temporal expression relative to histone H4 genes during fetal development has been explored. Here, we establish that expression of Hinfp is biologically coupled with expression of twelve functional mouse H4 genes during pre- and post-natal tissue-development. Both Hinfp and H4 genes are robustly expressed at multiple embryonic (E) days (from E5.5 to E15.5), coincident with ubiquitous LacZ staining driven by the Hinfp promoter. Five highly expressed mouse H4 genes (Hist1h4d, Histh4f, Hist1h4m and Hist2h4) account for >90% of total histone H4 mRNA throughout development. Post-natal expression of H4 genes in mice is most evident in lung, spleen, thymus and intestine, and with few exceptions (e.g., adult liver) correlates with Hinfp gene expression. Histone H4 gene expression decreases but Hinfp levels remain constitutive upon cell growth inhibition in culture. The in vivo co-expression of Hinfp and histone H4 genes is consistent with the biological function of Hinfp as a principal transcriptional regulator of histone H4 gene expression during mouse development.
Keywords: embryogenesis, blastocyst, cell cycle, histone, HINFP, p220NPAT, CDK2, development, cancer, embryonic stem cells
1. INTRODUCTION
Chromatin packaging of nascent DNA during S phase requires the cell cycle regulated expression of the five histone proteins (H1, H2A, H2B, H3 and H4) (Osley, 1991; Stein et al., 1984; Stein et al., 2006). The timing of human histone gene expression is mechanistically coupled to the formation of nucleosomes during DNA replication in both lineage-committed somatic cells (Holmes et al., 2005) and pristine pluripotent embryonic stem cells (Becker et al., 2007; Ghule et al., 2009). Histone gene expression is regulated by modulating the rate of transcription, pre-mRNA processing and mRNA stability at Histone Locus Bodies (HLBs). HLBs are non-membranous subnuclear organelles associated with multiple clustered human histone genes that bind both transcription factors and 3’ end processing factors to generate functionally integrated macro-molecular structures containing the machinery for production of mature histone mRNAs (Barcaroli et al., 2006; Bongiorno-Borbone et al., 2008; Dominski et al., 2003; Dominski and Marzluff, 2007; Ghule et al., 2007; Ghule et al., 2009; Yang et al., 2009a; Yang et al., 2009b; Yang et al., 2011). The cell cycle controlled transcription of histone H4 genes, as well as other core histone proteins (e.g., H2A, H2B and H3) at HLBs is a key step. This step is initiated in part by the cyclin E/ CDK2 dependent phosphorylation of the p220NPAT/ HINFP complex at the G1/S phase transition (Holmes et al., 2005; Le et al., 2006; Ma et al., 2000; Medina et al., 2006; Medina et al., 2007; Medina et al., 2008; Miele et al., 2005; Mitra et al., 2003; Mitra et al., 2007; Mitra et al., 2009; van Wijnen et al., 1992; Wei et al., 2003; Xie et al., 2007; Ye et al., 2003; Zhao et al., 2000). Activation of DNA damage repair pathways and cell cycle arrest block the transcriptional activation of histone H4 genes by p220NPAT and HINFP (Mitra et al., 2009; Pirngruber and Johnsen, 2010; Su et al., 2004). The cyclin E/ CDK2 / p220NPAT/ HINFP/histone H4 pathway is fundamentally distinct and operates independently of the E2F/pRB pathway (Grana et al., 1998; Stein et al., 2006).
Histone H4 represents the most highly conserved chromatin protein in eukaryotes (Osley, 1991; Stein et al., 1984) and the same protein is encoded by 15 distinct gene copies in human and 12 genes in mouse (Albig and Doenecke, 1997; Braastad et al., 2004; Holmes et al., 2005; Lichtler et al., 1982; Marzluff et al., 2002). Each mammalian histone H4 gene copy is controlled by distinct cis-acting elements (e.g., SP1, ATF1, YY1) (Birnbaum et al., 1995; Guo et al., 1997; Last et al., 1999a; Last et al., 1999b) that result in differences in transcription rates (Holmes et al., 2005; van der Meijden et al., 1998). However, most histone H4 genes contain a single highly conserved HINFP site immediately adjacent to the TATA box that represents the transcriptional endpoint of the cyclin E/CDK2/ p220NPAT/ HINFP axis. Because HINFP can bind other factors involved in cell signaling, transcription or RNA processing (DeRan et al., 2008; Miele et al., 2005; Miele et al., 2007), it may represent a key scaffolding protein that together with p220NPAT integrates cell cycle regulatory cues at HLBs. Other factors including a multi-subunit cyclin A/CDK1/pRB/CUX1 (CDP-cut) complex and IRF proteins (Aziz et al., 1998; Gupta et al., 2003; Last et al., 1999b; Luong et al., 2002; Ramsey-Ewing et al., 1994; van Wijnen et al., 1992; van Wijnen et al., 1994; van Wijnen et al., 1996; Vaughan et al., 1995; Vaughan et al., 1997; Xie et al., 2001; Xie et al., 2002) contribute to histone H4 gene regulation. These DNA binding proteins interact together with HINFP at a multipartite cell cycle regulatory promoter element, Site II, which represents a genomic DNaseI footprint (Hovhannisyan et al., 2003; Pauli et al., 1987). Binding of HINFP to Site II recruits and stabilizes p220NPAT (Medina et al., 2006; Miele et al., 2005) thereby integrating signals emanating from the cyclin E/CDK2 kinase cascade (Ma et al., 2000; Zhao et al., 1998; Zhao et al., 2000) to mediate cell cycle dependent enhancement of histone H4 gene transcription independent of the E2F/pRB switch.
A limited number of studies have examined expression of selected copies of histone genes (i.e., H1, H2A, H2B and H3) during early mouse embryo development (Clarke et al., 1992; Graves et al., 1985). These studies revealed that the bulk maternal histone mRNAs are dramatically reduced after the first one or two divisions. This down regulation occurs presumably by destabilization of histone mRNA when DNA synthesis ceases upon completion of the first S phase (Osley, 1991; Stein et al., 1984). In vivo studies on histone H4 gene transcription have been limited to work with transgenic mice that focused on mid-gestation and later stages of fetal development in both osseous and non-osseous (e.g., liver, spleen) tissues (Gerbaulet et al., 1992; van Wijnen et al., 1991). In addition, a few studies have examined mouse histone H4 gene transcription in cultured cells (Seiler-Tuyns and Paterson, 1987; van der Meijden et al., 1998). Recently, we have shown that a Hinfp null mutation (HinfpLacZ) causes early embryonic lethality at peri-implantation stages in mice (Xie et al., 2009). Cultured blastocysts have a hatching defect and are defective in histone H4 gene expression. Similarly, proviral inactivation of the Npat gene results in early embryonic arrest (Di Fruscio et al., 1997), consistent with a genetic requirement for an intact Hinfp/p220Npat complex to mediate histone gene expression and maintain cell viability. Remarkably, the human HINFP and NPAT genes are located within a chromosomal region (11q23), which encompasses the ATM, MLL and CBL genes, that is frequently rearranged in many cancer cell types (Baysal et al., 2001; Harper and Aplan, 2008; Kalla et al., 2007). The early lethality of HinfpLacZ/LacZ embryos precludes analysis of Hinfp function and its link to histone gene expression at post-implantation stages of mouse development. In this study, we investigated when and where the Hinfp gene is transcribed in relation to histone H4 gene expression throughout pre- and post-natal development.
2. MATERIALS AND METHODS
2.1 Collection of Mouse Embryos and Tissues
Histone H4 and Hinfp mRNA levels during pre- and post-natal development were analyzed in offspring from adult wild type C57BL/6 mice (9-12 weeks). For pre-natal time-points, pregnancies were timed by daily examination for vaginal plugs. Noon of the day that the vaginal plug was observed was considered 0.5 embryonic (E) days of development. Blastocysts (3.5 days post-fertilization) were collected by flushing uteri with medium, while embryos at embryonic days E5.5, E6.5, E10.5, E12.5 and E15.5 were collected by dissection. Tissues of post-natal mice were harvested upon sacrifice by cervical dislocation. All procedures involving the care and use of animals were approved by the Institutional Animal Care and Use Committee at the University of Massachusetts Medical School.
2.2 Cell Culture
Cultures of primary mouse embryonic fibroblasts (MEFs) were established from embryos harvested at E12.5. Embryos were eviscerated, rinsed in fresh PBS and tissues were fragmented. Minced tissues were rinsed twice in PBS and incubated with trypsin/EDTA for 15-20 min at 37°C. Trypsin was neutralized with cell culture medium (complete Dulbecco’s Minimal Esential Medium [DMEM] containing high glucose with 2 mM L-glutamine, 50 U Penicillin/Streptomycin, 15% fetal bovine serum). Cells were gently suspended in 10 ml medium using a pipette and centrifuged for 5 min at 1,000 rpm (IEC centrifuge with swinging bucket rotor). The supernatant was removed and the cell pellet was resuspended in 10 ml fresh complete DMEM. Cells were washed twice and plated in gelatin-coated T175 flasks in 30 ml DMEM. Cells were cultured at 37°C in an incubator with 5% CO2 and 98% humidity. After 48h, confluent MEF cultures were harvested and stored frozen as Passage 1. Continuous cultures of MEFs were split 1:5 when 90% confluent in 100 mm plates. NIH3T3 and MC3T3 cells were obtained from American Type Culture Collection (Manassas VA). Cells were supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin.
2.3 RNA Extraction
RNA was extracted from specified embryos, tissues or cell lines using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Purified total RNA was digested with DNase at 37°C for 30 min, followed by column purification using the DNA Free RNA KitTM (Zymo Research Corp., USA). Eluted total DNA-free RNA was quantitated by spectrophotometry, and 1 μg was added to a reverse transcription reaction using the iScript™ cDNA synthesis kit (Bio-Rad) with random hexamer primers. Because histone H4 genes do not contain introns, we included control reactions without reverse transcriptase. Upon quantitation, cDNA preparations were diluted for qRT-PCR analysis. Total RNA was isolated from the Blastocysts using RNeasy Micro Kit (Qiagen, USA) according to the manufacturer’s instruction. Because blastocysts contain less than 10 μg tissue, synthetic poly-A RNA (20 ng) was added as carrier during RNA extraction.
2.4 Quantitative Reverse Transcription-PCR (qRT-PCR) Analysis of H4 Gene Expression
Expression analysis was performed using qRT-PCR with primer sets against different histone H4 genes, as well as the Hinfp and Hprt1 genes (Figures 1 and 2). We tested seven different internal controls (Pia, β-actin, Hprt, Tbp, Gapdh, β2-microglobulin and 18S rRNA) for normalizing histone H4 gene expression. Hprt was found to be most constant over all tissues in neonatal mice (data not shown). Relative RNA expression was determined using an ABI PRISM 7000 sequence detection system (Applied Biosystems) measuring real-time SYBR Green fluorescence (Biorad) and calculated by the ΔΔCT method. Cycling parameters were: 50°C for 2 min, 95°C for 10 min, then 40 cycles of 95°C for 15 s and 60°C for 1 min. In all experiments, Hprt1 was used as an endogenous reference. The size of the RT-PCR products was confirmed by gel electrophoresis using a standard 1.2% agarose gel.
Figure 1. Histone H4 gene nomenclature and expression in human and mouse.
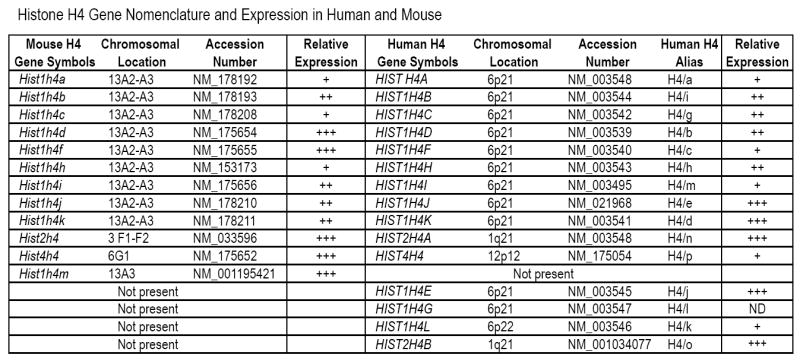
The table summarizes the gene symbols (columns 1 and 5), chromosomal location (columns 2 and 6), accession number (columns 3 and 7), as well as relative expression (columns 4 and 9) of each histone gene present in the mouse (this work) or human genome (Holmes et al., 2005). The table also lists an alternative nomenclature for human histone H4 genes (column 8; ‘Alias’) to permit comparison with previous studies (Albig and Doenecke, 1997; Braastad et al., 2004; Holmes et al., 2005). Five genes are present in only one of the two species (bottom five rows).
Figure 2. Primer sets for qRT-PCR.

The table presents primers pairs used for the analysis of the indicated genes (column 1), including Forward (column 2) and Reverse (column 3) primers.
2.5 In situ Expression of Hinfp by β-Galactosidase Staining
We previously constructed HinfpLacZ/+ mice with a LacZ gene under control of the endogenous Hinfp promoter (Xie et al., 2009) that permits examination of tissues in which the Hinfp gene is expressed. Expression of β-galactosidase driven by the endogenous HiNFP promoter was monitored by 5-bromo-4-chloro-3-indolyl-β- D-galactopyranoside (X-gal) staining. The embryos were fixed in 0.5% glutaraldehyde and stained with a solution containing 1 mg/ml X-gal, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide and 2 mM MgCl2.
2.6 Antibody Generation and Western Blot Analysis
For detection of mouse Hinfp we generated a new rabbit polyclonal antibody against recombinant full length Hinfp. The entire coding sequence of Hinfp was fused in frame with the GST coding sequence in pGEX for expression in E. coli. The recombinant mouse GST-Hinfp fusion protein (predicted molecular mass of ~92 kDa) was purified using Gluthatione Sepharose 4B beads (GE Healthcare Biosciences, Piscataway, NJ). Antisera for GST-Hinfp were generated in rabbit (Cocalico Biologicals, Inc., Reamstown, PA). The immunoglobulin G (IgG) fraction was purified from the resulting antiserum. The specificity of both the antiserum and IgG fraction were validated by Western blot analysis of the GST-Hinfp protein, as well as by immunofluorescence microscopy of mammalian cells in which mouse Hinfp was over-expressed (data not shown). The validated antibody was used to detect the endogenous Hinfp protein (~65 kDa) in mouse cell lines as indicated. Whole-cell extracts were fractionated by SDS-PAGE in a 10% gel and blotted onto polyvinylidene difluoride Immobilon-P membranes (Millipore, Billerica, MA). Immuno-detection was performed with an appropriate dilution of the antibody (1 : 1,000) directed against mouse Hinfp and bands were visualized using the Western Lightning chemiluminescence reagent plus (Perkin Elmer Life Sciences, Waltham, MA).
3. RESULTS
3.1 Expression of Hinfp and multiple histone H4 genes during fetal mouse development
The Zn-finger transcription factor Hinfp is a master regulator of the histone H4 multi-gene family. Histone H4 genes are encoded by a multigene family comprised of 12 distinct gene copies in mouse and 15 copies in human (Fig. 1). Similar to the genomic organization of human histone genes (Albig and Doenecke, 1997; Braastad et al., 2004), the majority of the mouse histone genes are located within two large clusters on mouse chromosome 13, designated Hist1, and mouse chromosome 3, designated Hist2 (Marzluff et al., 2002). The Hist1 and Hist2 clusters are homologous to similar clusters on, respectively, human chromosomes 6 and 1. There is an additional mouse histone H4 gene (Hist4H4) located on mouse chromosome 6, with the homologous human Hist4H4 gene at chromosomal region 12p12 (Marzluff et al., 2002). The multiple histone H4 genes of mouse and human encode identical proteins, and most of these genes have functional Hinfp binding sites. However, the promoter sequences of individual histone H4 genes differ between gene copies and between species (Holmes et al., 2005; van der Meijden et al., 1998). We have previously shown that human histone H4 genes are not equally expressed and that a subset contributes disproportionately to the total pool of H4 mRNA (Holmes et al., 2005). However, the contributions of individual mouse H4 genes to histone H4 mRNA expression and in vivo correlations with Hinfp gene expression have not been described. Therefore, we analyzed the expression of the Hinfp gene and the twelve mouse histone H4 genes during mouse development by real-time quantitative reverse transcriptase PCR (qRT-PCR) assays with H4 gene specific primers (Figs. 2 and 3).
Figure 3. Temporal expression of histone H4 and Hinfp expression during murine development.
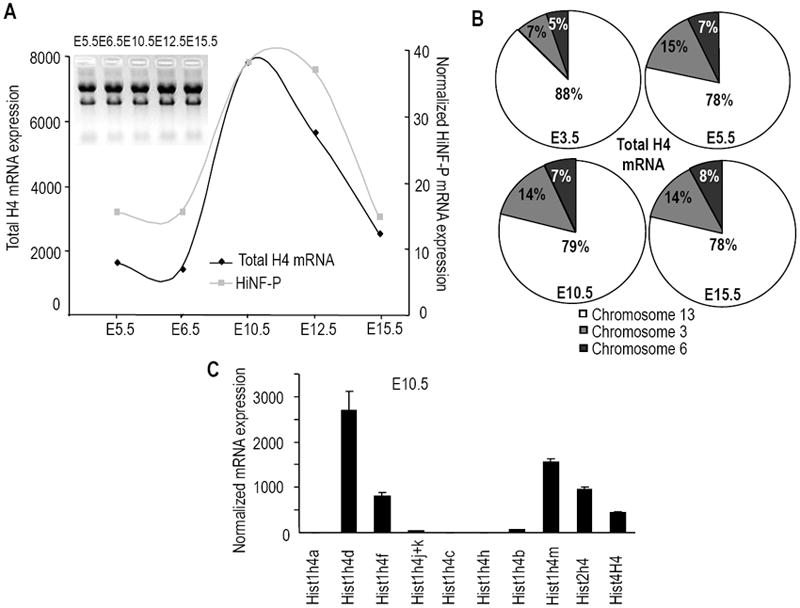
(A) Total RNA of embryos at different stages of pre-natal development was analyzed by qRT-PCR for histone H4 and Hinfp expression (see also Supplementary Fig. S1). The graph provides an overview of the general correlation between total histone H4 mRNA levels (i.e., the sum of all expression values for the individual mouse histone H4 genes) and Hinfp expression relative to Hprt1. The average Ct values for HPRT1 are: 23.46 (E5.5), 23.57 (E6.5), 24.37 (E10.5), 24.12 (E12.5), 24.41 (E15.5). Values for blastocysts (E3.5) are not shown, because RNA samples were supplemented with carrier nucleic acids and cannot be directly compared with other samples. (B) Contribution of distinct mouse histone H4 genes to the total h4 mRNA pool during mouse fetal development as categorized by chromosomal location. The pie charts summarize expression of genes located on chromosome 13 (Hist1h4a, -d, -f, -k, -j, -c, -h, -b, -i, -m; white sector), chromosome 3 (Hist2h4; gray sector), and chromosome 6 (Hist4h4; black sector). Samples at E3.5 were included. Results at E12.5 (data not shown) are very similar to those for E10.5 and E15.5 Note that the actual size of each pie chart is not unit size (as indicated here to permit comparison between days), but differs for each day based on changes in total histone H4 mRNA levels (see Panel A). (C) Representative graph showing expression of the full complement of mouse histone H4 genes at E10.5 when total H4 gene expression is maximal.
Because of the heterogeneity in 5’-flanking sequences of the individual genes, we predicted temporal and/or quantitative differences in the expression of the mouse histone H4 genes at different stages of fetal development. We examined the relative levels of expression of distinct H4 mRNAs in the developing embryo using qRT-PCR at E3.5, E5.5, E6.5, E10.5, E12.5 and E15.5 post-fertilization (Fig. 3). The relative contributions of individual histone H4 genes to the total H4 mRNA pool were determined with forward primers specific to the unique 5’-untranslated region of each H4 mRNA (Fig. 2); the Hist1h4j and Hist1h4k genes are amplified by the same primer pair and their individual expression cannot be resolved. Primer pairs were tested against dilutions of genomic DNA to verify that the amplification efficiencies were similar (data not shown).
We first established that total histone H4 mRNA levels (as defined by the sum of all qRT-PCR signals for each individual histone H4 gene) are low during the early stages of embryonic development (E5.5 and E6.5), but are up-regulated during mid-gestation (E10.5 to 12.5) (Fig. 3A). The relative expression of Hinfp was monitored in parallel to assess the temporal correlation with total histone H4 gene expression. The results show that Hinfp mRNA is expressed at two orders of magnitude less than the total histone H4 mRNA pool. More importantly, Hinfp expression parallels histone H4 gene expression, and Hinfp mRNA levels are maximal at mid-gestation at E10.5 and E12.5 (Fig. 3A). These results demonstrate that the combined expression of the mouse histone H4 gene family is developmentally regulated and temporally associated with the expression of Hinfp as the master regulator of histone H4 genes.
We next addressed whether the two major histone H4 gene clusters are differentially expressed during mouse development. Classification of histone H4 mRNA expression by chromosomal location of the gene copies revealed that the major histone H4 gene cluster on mouse chromosome 13 generates about 78 to 88% of all H4 gene transcripts (Fig. 3B). Interestingly, expression of the histone H4 gene from the chromosome 3 cluster essentially doubled (from 7% to 15%) between E3.5 and E5.5 but remained relatively constant thereafter. The single H4 gene copy at chromosome 6 contributes between 5% and 8% at different stages (Fig. 3B). Thus, while there are modest modulations in the utilization of distinct histone genes at different chromosomal locations, each of the three chromosomal loci remains active throughout mouse development. The results also indicate that there are differences in the extent to which distinct H4 mRNA species are maximally expressed. We find that the Hist1h4d, Hist1h4f, Hist1h4m, Hist2h4 and Hist4h4 genes account for >90% of the detected histone H4 mRNAs at multiple stages of embryonic development. In contrast, Hist1h4a, Hist1h4j+k, Hist1h4c, Hist1h4h and Hist1h4b are minimally expressed, together representing <10% of the detected histone H4 mRNA (Figs. 3C and S1). Thus, five of the histone H4 genes generate the majority of mRNAs that are required to accommodate DNA replication and nascent chromatin assembly.
3.2 Hinfp is ubiquitously expressed during pre- and post-natal development
To understand when and where Hinfp is expressed during mammalian development, we examined β-galactosidase staining in heterozygous HinfpLacZ/+ mice in which the LacZ allele is under control of the native Hinfp promoter (Fig. 4A) (Xie et al., 2009). Embryos at E3.5, E6.5, E9.5 and E14.5 exhibit clear β-galactosidase staining at the gross anatomical level (Xie et al., 2009). LacZ staining is evident in multiple tissues in histological sections from E14.5 embryos, including brain, liver, skin, calvarial bone and brain (Figs. 4B and 1C). Similarly, robust β-galactosidase staining is also evident in representative post-natal tissues (spleen, thymus, heart and liver) (Fig. 4D). The LacZ expression pattern driven by the Hinfp promoter (Fig. 4) is comparable to results obtained with distinct transgenes under control of a representative human histone H4 promoter (Gerbaulet et al., 1992; van Wijnen et al., 1991). The latter studies revealed proliferation-related transgene expression reflecting histone H4 promoter activity in different mouse tissues and cell types, including cells from cranial tissues (e.g., calvaria, brain). Our new results indicate that the Hinfp promoter is actively transcribed in tissues that also express histone H4 genes.
Figure 4. In vivo expression analysis of Hinfp gene during development using HiNFPLacZ/+ mice.
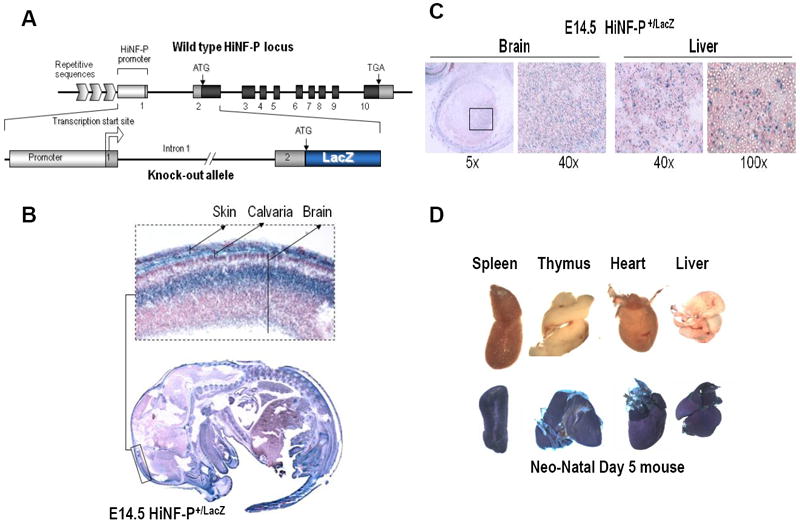
(A) Diagram of the HinfpLacZ allele in which LacZ coding sequences are located immediately downstream of the Hinfp ATG start codon and linked to the native Hinfp promoter (Xie et al., 2009). Mice carrying this allele were examined for expression of β-galactosidase at different embryonic and post-natal stages. (B) Embryos derived from wild type and heterozygous HiNFPLacZ/+ crosses at E14.5 were stained with X-gal in histological sections. One area spanning cranial tissues is enlarged in the inset. (C) Selected tissues as indicated were microscopically examined for β-galactosidase staining in histological sections, (D) Gross anatomical analysis of β-galactosidase staining in tissues from Day 5 neonates derived from wild type and heterozygous HiNFPLacZ/+ mice.
3.3 Expression of multiple mouse histone H4 genes during post-natal development
Expression levels of the mouse histone H4 and Hinfp genes were examined by qRT-PCR with RNA samples from different tissues during post-natal mouse development (days 1, 14 and 60) (Figs. 5, S2 and S3). Figure 5 shows representative results in different tissues for the Hist1h4d and Hist2h4 genes. The Hist1h4d gene is the most abundantly expressed H4 gene at all stages examined, while the Hist2h4 gene was selected because it is the homolog of the human HIST2H4A gene (alias H4/n or FO108) (Braastad et al., 2004; Stein et al., 1984). The latter gene is among the most highly expressed human histone H4 genes and has been extensively investigated as a paradigm for cell cycle control of histone H4 gene transcription (Stein et al., 1984; Stein et al., 2006). In general, the highest mRNA levels for different histone H4 genes were detected in the spleen and thymus of neonates at Day 1 and post-natal mice at Day 14. Maximal histone H4 mRNA levels were detected in intestine and spleen in 2 month-old mice (Day 60) (Figs. 5, S2 and S3), albeit that thymus tissue was not analyzed at Day 60. We note that the overall levels of histone H4 mRNAs are reduced at post-natal Day 60 relative to Days 1 and 14, reflecting reduced cell proliferation in adult tissues. The abundant expression of mouse histone H4 genes in spleen, thymus and intestine is consistent with sustained proliferative activity in these tissues, linked to immune functions and continuous replacement of the intestinal lining in post-natal mice.
Figure 5. Proliferation-related tissue-specific expression of histone genes during post-natal development.
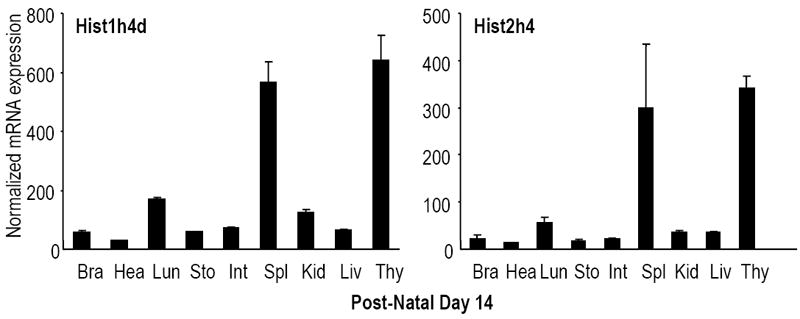
Expression levels of all mouse histone H4 genes were analyzed in selected tissues at multiple days during post-natal development (D1, D14 and D60) by qRT-PCR analysis normalized to Hprt1 (see Supplementary Figs. S2 and S3). The two graphs show representative results for two highly expressed histone H4 genes located in two different histone gene clusters (left, Hist1h4d; right, Hist2h4h) obtained for post-natal Day 14. Histone H4 genes are detectable in all tissues examined, including brain (Bra), heart (Hea), lung (Lun), stomach (Sto), intestine (Int), kidney (Kid), liver (Liv) and thymus (Thy).
3.4 Expression of Hinfp and histone H4 genes is temporally correlated
Cell culture studies clearly indicate that Hinfp is rate-limiting for histone H4 gene transcription and mRNA accumulation. However, this correlation has not yet been validated in vivo. Therefore, we examined the expression of Hinfp in various tissues in relation to the total histone H4 mRNA pool (as defined by the sum of all individual expression values) (Fig. 6). Among the tissues examined in the newborn (Day 1), juvenile (Day 14) and adult (Day 60) mice, Hinfp expression is ubiquitously detected in the thymus, brain, heart, spleen, kidney, lung, stomach, intestine and liver with only moderate variation (Fig. 6 and data not shown). More importantly, Hinfp expression appears to be generally proportional to the total histone H4 mRNA levels in many tissues, except for liver. While histone H4 gene expression is reduced as liver tissue becomes quiescent, Hinfp levels remain elevated. Liver is capable of tissue-regeneration and continued expression of Hinfp may facilitate rapid re-initiation of cell division.
Figure 6. Temporal regulation of histone H4 and Hinfp gene expression during post-natal tissue development.
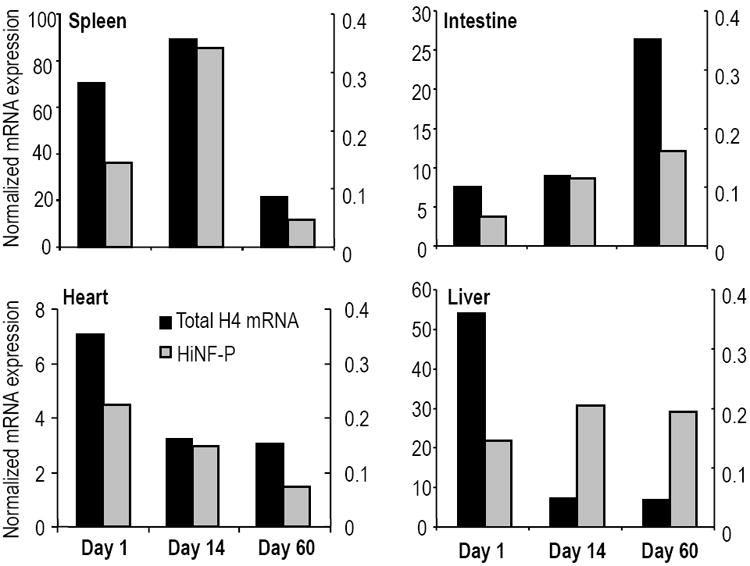
Total H4 mRNA levels (black columns) were obtained by the summation of normalized expression values for individual mouse histone H4 genes and were correlated with expression of the cognate regulatory factor Hinfp in selected tissues as indicated (i.e., spleen, intestine, heart and liver; see also Supplementary Figs. S2 and S3). The left y-axis shows total histone H4 mRNA levels and the secondary y-axis to the right indicates Hinfp gene expression. Experimental error is less than 10% (error bars not indicated).
3.5 Expression of multiple mouse Histone H4 genes in cell culture
We have previously shown relative differences in human histone H4 gene expression between cells from normal tissue and transformed cells in culture (Holmes et al., 2005). In addition, analysis of gene expression in cell culture permits assessment of direct correlations between gene expression and cessation of cell proliferation. Therefore, we analyzed histone H4 gene expression in two different established mouse cell lines (Fig. 7). We find that mRNAs for about nine of the twelve histone H4 genes are expressed at robustly detectable levels in both actively proliferating NIH3T3 fibroblasts and MC3T3 pre-osteoblasts, while the remaining three H4 genes (Hist1h4a, Hist1h4c and Hist1h4h) are barely detectable or perhaps not expressed at all. The five most highly expressed histone H4 genes in the two cell types are Hist1h4d, Hist1h4f, Hist1h4m, Hist2h4 and Hist4h4. Moderately expressed histone H4 genes are Hist1h4j+k and Hist1h4b, while the Hist1h4a, Hist1h4c and Hist1h4h genes are minimally expressed at levels below that observed for Hinfp (Fig. 7A). The Hist1h4i gene exhibits sporadic modestly high expression in NIH3T3 cells, while this gene is barely expressed in MC3T3 cells (Fig. 7A) or mouse tissues (Figs. 3 and 5). Taken together, the H4 gene expression results for these two immortalized cell lines parallels the profiling data obtained for normal tissues during pre- and post-natal development. The differences in relative mRNA levels of distinct histone H4 gene copies is consistent with the conclusion that differences in H4 gene 5’ regulatory sequences influence the expression levels of mammalian H4 genes in vitro and in vivo (Holmes et al., 2005; van der Meijden et al., 1998).
Figure 7. Cell growth dependent expression of mouse histone H4 genes in cultured cells.
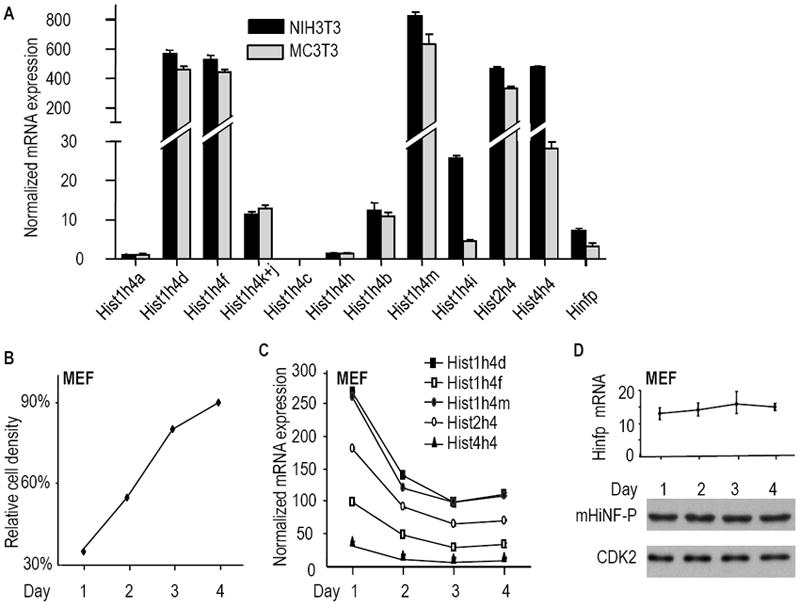
Expression of mouse histone H4 genes and the Hinfp gene was examined in established mesenchymal cell lines (NIH3T3 fibroblasts and MC3T3 osteoblasts; Panel A) and primary mouse embryonic fibroblasts (MEFs; Panels B, C & D). (A) qRT-PCR analysis of individual histone genes. (B) MEFs in cell culture are density inhibited by four days in culture. Near-confluent MEF cultures (90-95% confluent) were split 1:5 and replated (Day 0). The graph shows the approximate cell density (relative cell density as a percentage of a completely confluent culture) as a function of time in culture (Days 1, 2, 3 and 4). (C) Expression analysis of the five most highly expressed histone H4 genes in MEFs during cell density inhibition of growth. (D) Hinfp mRNA (top panel: qRT-PCR assay) and protein levels (bottom panel: western blot analysis) were measured in the same MEF cultures; CDK2 protein was used to account for variation in protein loading.
To assess whether expression levels of the five most prominently expressed mouse histone H4 genes and the cognate regulatory factor Hinfp are coupled to cell proliferation, we analyzed their steady-state mRNA levels in RNA samples prepared from proliferating and confluent mouse embryo fibroblasts (MEFs). Total RNA was isolated from cells harvested on consecutive days (Days 1 to 4) after seeding at a fixed cell density (Figs. 7B and 7C). The results show that MEFs actively proliferate on Days 1 and 2, but begin to reach confluence at Days 3 and 4 cells (Fig. 7B). During the proliferative phase, we find that the five major histone H4 genes (i.e., Hist1h4d, Hist1h4f, Hist1h4m, Hist2h4 and Hist4h4) are highly expressed as expected (Fig. 7C). In addition, all five genes are down regulated with the onset of cell growth inhibition by Days 3 and 4. Thus, expression of H4 genes is functionally linked to cell proliferation in MEFs. Interestingly, while histone H4 mRNAs decline at successive days in culture (Fig. 7C), we find that Hinfp mRNA and protein levels do not immediately decline with decreased cell proliferation (Fig. 7D). This finding corroborates the results obtained with H4 and Hinfp expression in mouse liver tissues (Fig. 5), and indicates that density-inhibited cells may remain poised to transcribe histone H4 genes in a Hinfp dependent manner.
4. DISCUSSION
Histone biosynthesis is coordinated with DNA replication at the onset of S phase through modulations in mRNA availability (Osley, 1991; Stein et al., 1984) that are controlled by the H4 subtype-specific transcription factor HiNF-P (Holmes et al., 2005). The multiple histone H4 genes present in the mouse (n=12) and human (n=15) genes encode the exact same protein (Osley, 1991; Stein et al., 1984). The majority of these genes have validated HiNF-P binding sites immediately upstream from the TATA-box (Holmes et al., 2005; van Wijnen et al., 1992), but upstream auxiliary transcription motifs (e.g., SP1, ATF1 and YY1) (Stein et al., 2006) differ between H4 gene copies (van der Meijden et al., 1998). In this study, we show that five mouse H4 genes (Hist1h4d, Hist1h4f, Hist1h4m, Hist2h4 and Hist4h4) produce the majority of H4 mRNAs. Hinfp is universally expressed in all cells and tissues that express histone H4 genes, providing in vivo support for its role as a master regulator of H4 gene transcription.
Interestingly, we have found that in human there are six H4 genes (HIST1H4D, HIST1H4E, HIST1H4J, HIST1H4K, HIST2H4A and HIST2H4B) that generate almost 80% of all human H4 mRNA (Holmes et al., 2005). Just two of the five major mouse H4 genes (Hist1h4d, homolog of HIST1H4D, and Hist2h4, homolog of HIST2H4A and HIST2H4B) are also highly expressed in human. In contrast, two histone H4 genes that are barely expressed in mouse (Hist1h4j and Hist1hk) are highly expressed in human cells. Finally, two genes that are highly expressed in either mammalian species (mouse Hist1h4m and human HIST1H4E) do not have a homolog in the other species. Comparions of histone H4 gene promoter sequences and the relative expression of individual H4 gene copies reveal that mutations in the Site II cell cycle regulatory element, which comprises both the Hinfp binding motif and the TATA-box, strongly correlate with severely diminished expression (Fig. 8). Specifically, single point mutations in the Hinfp motif and/or the TATA-box may account for the poor expression of the mouse Hist1h4a, Hist1h4c and Hist1h4h genes. Similar observations have been made for human histone H4 genes where decreased expression is linked to natural mutations in the same two elements (Holmes et al., 2005). Taken together, these data are entirely consistent with a transcriptional model in which the interaction of both Hinfp and the TATA-box Binding Protein (TBP) with its associated basal transcription machinery are critical for histone H4 gene transcription.
Figure 8. Comparison of mouse H4 Site II sequences and H4 mRNA levels.
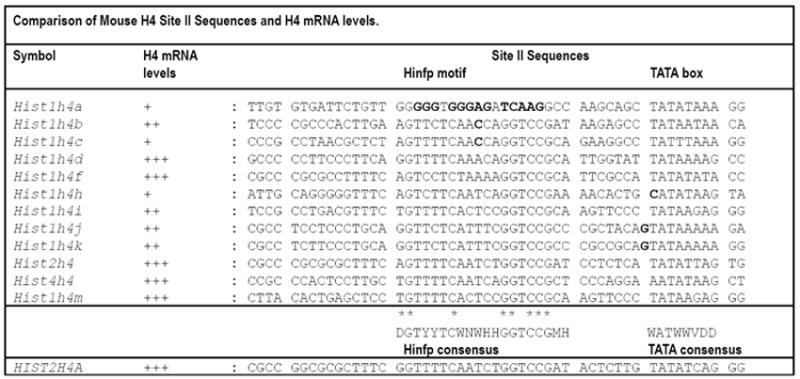
Alignment of the DNA sequences of the Site II cell cycle regulatory domain element (containing Hinfp and TATA elements) in the full complement of mouse histone H4 genes (top) compared to the prototypical human histone H4 gene HIST2H4A (bottom; see (Holmes et al., 2005)). Residues of Hinfp contact within the consensus element are indicated by asterisks (*). Redundant nucleotides (nt) are indicated as follows: D = A, G, or T; H = A, C, or T; M = A or C; W = A or T; Y = C or T; V = A, G or C. Mismatches in mouse Site II sequences (bold), which spans the Hinfp binding site consensus and the TATA box, are indicated in relation to H4 mRNA expression. H4 mRNA expression data in cells, tissues and developmental stages are summarized as follows: +, below average levels; ++, intermediate levels; +++, above average expression. Point-mutations in the Hinfp motif or the TATA-box correlate with decreased H4 gene expression.
The correlation between variation in histone H4 gene expression and Site II sequences in both mouse and human promoter sequences indicates that both the genomic organization and the regulated expression of the H4 multigene family is subjected to considerable evolutionary variation in closely related mammalian species. This conclusion is remarkable in view of the fact that histone H4 is among the most highly conserved proteins in all eukaryotes (Stein et al., 1984). In addition, while in lower eukaryotes and vertebrates (e.g., sea urchin, fly and fish) there are multiple tandemly repeated histones to support histone H4 gene expression (Osley, 1991; Stein et al., 1984), mouse and human have retained only a handful of functionally expressed H4 gene copies ((Holmes et al., 2005); this study). Because mammalian embryos developing in utero are less prone to predation than embryos from lower eukaryotes developing in a less protected external environment, it is tempting to speculate that this degeneration of histone gene organization may relate to reduced evolutionary pressures in mammals for rapid early embryonic growth and maturation.
We have also determined that the genes for histone H4 and Hinfp are most highly expressed in spleen, thymus and intestinal tissues during post-natal development. Because spleen is the largest peripheral immune organ, thymus is a central immune organ in juvenile mice and intestines contain gut-associated lymphoid tissue, it appears that histone H4 gene expression is most prominent in tissues with functions in immunity. Most likely, the expression of Hinfp and H4 genes reflects active lymphocyte proliferation in these tissues. While Hinfp expression positively correlates with histone H4 gene expression and cell proliferation in all tissues including liver tissue, we noted that Hinfp expression persists in adult liver which mostly contains quiescent cells, as well as in confluent mouse embryonic fibroblasts that are cell growth inhibited in confluent cultures. Thus, while H4 mRNA (but not protein) accumulation is restricted to S phase in proliferating cells, Hinfp may be a useful regulatory marker (both mRNA and protein) for proliferating cells, as well as quiescent cells (e.g., adult multi-potent stem cells) that have the potential to enter the cell cycle and transcribe histone H4 genes. Hinfp staining of cells could complement Ki67 staining, which is known to be restricted to actively proliferating cells (reviewed in (Weigel and Dowsett, 2010)) as a marker for quiescent cells that retain proliferative potential. It is conceivable that Hinfp expression in liver is restricted to quiescent adult stem cells that are abundant in this tissue and support rapid tissue-regeneration upon injury (reviewed in (Bergmann and Steller, 2010)).
The Hinfp null mouse suffers from a lethal defect that compromises blastocyst development (Xie et al., 2009). We used β-galactosidase staining of implanted developing embryos and post-natal tissues to how that Hinfp is ubiquitously expressed in multiple tissues at post-blastocyst stages of development. This finding will guide us with the future analysis of Hinfp function in later stages of tissue-development using conditional Hinfp knock-out mice.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01 CA139322. We thank the members of our laboratories including Jennifer Colby, Ricardo Medina, Prachi Ghule, Margaretha van der Deen, Ying Zhang, Shirwin Pockwinse, Klaus Becker and Partha Mitra for stimulating discussions, as well as generously sharing reagents and expertise.
Abbreviations
- HiNF-P
Histone Nuclear Factor P
- CDK2
cyclin-dependent kinase 2
- NPAT
nuclear protein, ataxia-telangiectasia locus
- pRB
retinoblastoma protein
- E2F
adenovirus early gene 2 factor
- CUX
Cut-related homeodomain protein
- CDP
CCAAT Displacement Protein
Footnotes
Disclosure Statement: The authors have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albig W, Doenecke D. The human histone gene cluster at the D6S105 locus. Hum Genet. 1997;101:284–294. doi: 10.1007/s004390050630. [DOI] [PubMed] [Google Scholar]
- Aziz F, van Wijnen AJ, Vaughan PS, Wu S, Shakoori AR, Lian JB, Soprano KJ, Stein JL, Stein GS. The integrated activities of IRF-2 (HiNF-M) CDP/cut (HiNF-D) and H4TF-2 (HiNF-P) regulate transcription of a cell cycle controlled human histone H4 gene: mechanistic differences between distinct H4 genes. Mol Biol Rep. 1998;25:1–12. doi: 10.1023/a:1006888731301. [DOI] [PubMed] [Google Scholar]
- Barcaroli D, Bongiorno-Borbone L, Terrinoni A, Hofmann TG, Rossi M, Knight RA, Matera AG, Melino G, De L V. FLASH is required for histone transcription and S-phase progression. Proc Natl Acad Sci U S A. 2006;103:14808–14812. doi: 10.1073/pnas.0604227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal BE, Willett-Brozick JE, Taschner PE, Dauwerse JG, Devilee P, Devlin B. A high-resolution integrated map spanning the SDHD gene at 11q23: a 1.1-Mb BAC contig, a partial transcript map and 15 new repeat polymorphisms in a tumour-suppressor region. Eur J Hum Genet. 2001;9:121–129. doi: 10.1038/sj.ejhg.5200585. [DOI] [PubMed] [Google Scholar]
- Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. J Cell Physiol. 2007;210:517–526. doi: 10.1002/jcp.20903. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Steller H. Apoptosis, stem cells, and tissue regeneration. Sci Signal. 2010;3:re8. doi: 10.1126/scisignal.3145re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum MJ, Wright KL, van Wijnen AJ, Ramsey-Ewing AL, Bourke MT, Last TJ, Aziz F, Frenkel B, Rao BR, Aronin N, Stein GS, Stein JL. Functional role for Sp1 in the transcriptional amplification of a cell cycle regulated histone H4 gene. Biochemistry. 1995;34:7648–7658. doi: 10.1021/bi00023a011. [DOI] [PubMed] [Google Scholar]
- Bongiorno-Borbone L, De Cola A, Vernole P, Finos L, Barcaroli D, Knight RA, Melino G, De Laurenzi V. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle. 2008;7:2357–2367. doi: 10.4161/cc.6344. [DOI] [PubMed] [Google Scholar]
- Braastad CD, Hovhannisyan H, van Wijnen AJ, Stein JL, Stein GS. Functional characterization of a human histone gene cluster duplication. Gene. 2004;342:35–40. doi: 10.1016/j.gene.2004.07.036. [DOI] [PubMed] [Google Scholar]
- Clarke HJ, Oblin C, Bustin M. Developmental regulation of chromatin composition during mouse embryogenesis: somatic histone H1 is first detectable at the 4-cell stage. Development. 1992;115:791–799. doi: 10.1242/dev.115.3.791. [DOI] [PubMed] [Google Scholar]
- DeRan M, Pulvino M, Greene E, Su C, Zhao J. Transcriptional activation of histone genes requires NPAT-dependent recruitment of TRRAP-Tip60 complex to histone promoters during the G1/S phase transition. Mol Cell Biol. 2008;28:435–447. doi: 10.1128/MCB.00607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fruscio M, Weiher H, Vanderhyden BC, Imai T, Shiomi T, Hori TA, Jaenisch R, Gray DA. Proviral inactivation of the Npat gene of Mpv 20 mice results in early embryonic arrest. Mol Cell Biol. 1997;17:4080–4086. doi: 10.1128/mcb.17.7.4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. Formation of the 3’ end of histone mRNA: getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Kaygun H, Dadlez M, Marzluff WF. A 3’ exonuclease that specifically interacts with the 3’ end of histone mRNA. Mol Cell. 2003;12:295–305. doi: 10.1016/s1097-2765(03)00278-8. [DOI] [PubMed] [Google Scholar]
- Gerbaulet SP, van Wijnen AJ, Aronin N, Tassinari MS, Lian JB, Stein JL, Stein GS. Downregulation of histone H4 gene transcription during postnatal development in transgenic mice and at the onset of differentiation in transgenically derived calvarial osteoblast cultures. J Cell Biochem. 1992;49:137–147. doi: 10.1002/jcb.240490206. [DOI] [PubMed] [Google Scholar]
- Ghule PN, Becker KA, Harper JW, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Cell cycle dependent phosphorylation and subnuclear organization of the histone gene regulator p220NPAT in human embryonic stem cells. J Cell Physiol. 2007;213:9–17. doi: 10.1002/jcp.21119. [DOI] [PubMed] [Google Scholar]
- Ghule PN, Dominski Z, Lian JB, Stein JL, van Wijnen AJ, Stein GS. The subnuclear organization of histone gene regulatory proteins and 3’ end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. J Cell Physiol. 2009;220:129–135. doi: 10.1002/jcp.21740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grana X, Garriga J, Mayol X. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene. 1998;17:3365–3383. doi: 10.1038/sj.onc.1202575. [DOI] [PubMed] [Google Scholar]
- Graves RA, Marzluff WF, Giebelhaus DH, Schultz GA. Quantitative and qualitative changes in histone gene expression during early mouse embryo development. Proc Natl Acad Sci U S A. 1985;82:5685–5689. doi: 10.1073/pnas.82.17.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Stein JL, van Wijnen AJ, Stein GS. ATF1 and CREB trans-activate a cell cycle regulated histone H4 gene at a distal nuclear matrix associated promoter element. Biochemistry. 1997;36:14447–14455. doi: 10.1021/bi971781s. [DOI] [PubMed] [Google Scholar]
- Gupta S, Luong MX, Bleuming SA, Miele A, Luong M, Young D, Knudsen ES, van Wijnen AJ, Stein JL, Stein GS. The tumor suppressor pRB functions as a co-repressor of the CCAAT displacement protein (CDP/cut) to regulate cell cycle controlled histone H4 transcription. J Cell Physiol. 2003;196:541–546. doi: 10.1002/jcp.10335. [DOI] [PubMed] [Google Scholar]
- Harper DP, Aplan PD. Chromosomal rearrangements leading to MLL gene fusions: clinical and biological aspects. Cancer Res. 2008;68:10024–10027. doi: 10.1158/0008-5472.CAN-08-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes WF, Braastad CD, Mitra P, Hampe C, Doenecke D, Albig W, Stein JL, van Wijnen AJ, Stein GS. Coordinate control and selective expression of the full complement of replication-dependent histone H4 genes in normal and cancer cells. J Biol Chem. 2005;280:37400–37407. doi: 10.1074/jbc.M506995200. [DOI] [PubMed] [Google Scholar]
- Hovhannisyan H, Cho B, Mitra P, Montecino M, Stein GS, van Wijnen AJ, Stein JL. Maintenance of open chromatin and selective genomic occupancy at the cell-cycle-regulated histone H4 promoter during differentiation of HL-60 promyelocytic leukemia cells. Mol Cell Biol. 2003;23:1460–1469. doi: 10.1128/MCB.23.4.1460-1469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalla C, Scheuermann MO, Kube I, Schlotter M, Mertens D, Dohner H, Stilgenbauer S, Lichter P. Analysis of 11q22-q23 deletion target genes in B-cell chronic lymphocytic leukaemia: evidence for a pathogenic role of NPAT, CUL5, and PPP2R1B. Eur J Cancer. 2007;43:1328–1335. doi: 10.1016/j.ejca.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Last TJ, van Wijnen AJ, Birnbaum MJ, Stein GS, Stein JL. Multiple interactions of the transcription factor YY1 with human histone H4 gene regulatory elements. J Cell Biochem. 1999a;72:507–516. [PubMed] [Google Scholar]
- Last TJ, van Wijnen AJ, de Ridder MC, Stein GS, Stein JL. The homeodomain transcription factor CDP/cut interacts with the cell cycle regulatory element of histone H4 genes packaged into nucleosomes. Mol Biol Rep. 1999b;26:185–194. doi: 10.1023/a:1007058123699. [DOI] [PubMed] [Google Scholar]
- Le XF, Bedrosian I, Mao W, Murray M, Lu Z, Keyomarsi K, Lee MH, Zhao J, Bast RC., Jr Anti-HER2 antibody trastuzumab inhibits CDK2-mediated NPAT and histone H4 expression via the PI3K pathway. Cell Cycle. 2006;5:1654–1661. doi: 10.4161/cc.5.15.3007. [DOI] [PubMed] [Google Scholar]
- Lichtler AC, Sierra F, Clark S, Wells JR, Stein JL, Stein GS. Multiple H4 histone mRNAs of HeLa cells are encoded in different genes. Nature. 1982;298:195–198. doi: 10.1038/298195a0. [DOI] [PubMed] [Google Scholar]
- Luong MX, van der Meijden CM, Xing D, Hesselton R, Monuki ES, Jones SN, Lian JB, Stein JL, Stein GS, Neufeld EJ, van Wijnen AJ. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Mol Cell Biol. 2002;22:1424–1437. doi: 10.1128/mcb.22.5.1424-1437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- Medina R, Buck T, Zaidi SK, Miele-Chamberland A, Lian JB, Stein JL, van Wijnen AJ, Stein GS. The histone gene cell cycle regulator HiNF-P is a unique zinc finger transcription factor with a novel conserved auxiliary DNA-binding motif. Biochemistry. 2008;47:11415–11423. doi: 10.1021/bi800961d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R, van der Deen M, Miele-Chamberland A, Xie RL, van Wijnen AJ, Stein JL, Stein GS. The HiNF-P/p220NPAT cell cycle signaling pathway controls non-histone target genes. Cancer Res. 2007;67:10334–10342. doi: 10.1158/0008-5472.CAN-07-1560. [DOI] [PubMed] [Google Scholar]
- Medina R, van Wijnen AJ, Stein GS, Stein JL. The histone gene transcription factor HiNF-P stabilizes its cell cycle regulatory co-activator p220NPAT. Biochemistry. 2006;45:15915–15920. doi: 10.1021/bi061425m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A, Braastad CD, Holmes WF, Mitra P, Medina R, Xie R, Zaidi SK, Ye X, Wei Y, Harper JW, van Wijnen AJ, Stein JL, Stein GS. HiNF-P directly links the cyclin E/CDK1/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol Cell Biol. 2005;25:6140–6153. doi: 10.1128/MCB.25.14.6140-6153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A, Medina R, van Wijnen AJ, Stein GS, Stein JL. The interactome of the histone gene regulatory factor HiNF-P suggests novel cell cycle related roles in transcriptional control and RNA processing. J Cell Biochem. 2007;102:136–148. doi: 10.1002/jcb.21284. [DOI] [PubMed] [Google Scholar]
- Mitra P, Ghule PN, van der Deen M, Medina R, Xie R, Holmes WF, Ye X, Nakayama KI, Harper JW, Stein JL, Stein GS, van Wijnen AJ. CDK inhibitors selectively diminish cell cycle controlled activation of the histone H4 gene promoter by p220(NPAT) and HiNF-P. J Cell Physiol. 2009;219:438–448. doi: 10.1002/jcp.21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P, Xie R, Harper JW, Stein JL, Stein GS, van Wijnen AJ. HiNF-P is a bifunctional regulator of cell cycle controlled histone H4 gene transcription. J Cell Biochem. 2007;101:181–191. doi: 10.1002/jcb.21157. [DOI] [PubMed] [Google Scholar]
- Mitra P, Xie RL, Medina R, Hovhannisyan H, Zaidi SK, Wei Y, Harper JW, Stein JL, van Wijnen AJ, Stein GS. Identification of HiNF-P, a key activator of cell cycle controlled histone H4 genes at the onset of S phase. Mol Cell Biol. 2003;23:8110–8123. doi: 10.1128/MCB.23.22.8110-8123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley MA. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- Pauli U, Chrysogelos S, Stein G, Stein J, Nick H. Protein-DNA interactions in vivo upstream of a cell cycle- regulated human H4 histone gene. Science. 1987;236:1308–1311. doi: 10.1126/science.3035717. [DOI] [PubMed] [Google Scholar]
- Pirngruber J, Johnsen SA. Induced G1 cell-cycle arrest controls replication-dependent histone mRNA 3’ end processing through p21, NPAT and CDK9. Oncogene. 2010;29:2853–2863. doi: 10.1038/onc.2010.42. [DOI] [PubMed] [Google Scholar]
- Ramsey-Ewing A, van Wijnen AJ, Stein GS, Stein JL. Delineation of a human histone H4 cell cycle element in vivo: the master switch for H4 gene transcription. Proc Natl Acad Sci U S A. 1994;91:4475–4479. doi: 10.1073/pnas.91.10.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler-Tuyns A, Paterson BM. Cell cycle regulation of a mouse histone H4 gene requires the H4 promoter. Mol Cell Biol. 1987;7:1048–1054. doi: 10.1128/mcb.7.3.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein GS, Stein JL, Marzluff WF. Histone Genes. John Wiley & Sons, Inc.; New York: 1984. [Google Scholar]
- Stein GS, van Wijnen AJ, Stein JL, Lian JB, Montecino M, Zaidi SK, Braastad C. An architectural perspective of cell-cycle control at the G1/S phase cell-cycle transition. J Cell Physiol. 2006;209:706–710. doi: 10.1002/jcp.20843. [DOI] [PubMed] [Google Scholar]
- Su C, Gao G, Schneider S, Helt C, Weiss C, O’Reilly MA, Bohmann D, Zhao J. DNA damage induces downregulation of histone gene expression through the G1 checkpoint pathway. EMBO J. 2004;23:1133–1143. doi: 10.1038/sj.emboj.7600120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meijden CMJ, Vaughan PS, Staal A, Albig W, Doenecke D, Stein JL, Stein GS, van Wijnen AJ. Selective expression of specific histone H4 genes reflects distinctions in transcription factor interactions with divergent H4 promoter elements. Biochim Biophys Acta. 1998;1442:82–100. doi: 10.1016/s0167-4781(98)00147-x. [DOI] [PubMed] [Google Scholar]
- van Wijnen AJ, Aziz F, Grana X, De Luca A, Desai RK, Jaarsveld K, Last TJ, Soprano K, Giordano A, Lian JB, et al. Transcription of histone H4, H3, and H1 cell cycle genes: promoter factor HiNF-D contains CDC2, cyclin A, and an RB-related protein. Proc Natl Acad Sci USA. 1994;91:12882–12886. doi: 10.1073/pnas.91.26.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen AJ, Choi TK, Owen TA, Wright KL, Lian JB, Jaenisch R, Stein JL, Stein GS. Involvement of the cell cycle-regulated nuclear factor HiNF-D in cell growth control of a human H4 histone gene during hepatic development in transgenic mice. Proc Natl Acad Sci USA. 1991;88:2573–2577. doi: 10.1073/pnas.88.6.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen AJ, van den Ent FM, Lian JB, Stein JL, Stein GS. Overlapping and CpG methylation-sensitive protein-DNA interactions at the histone H4 transcriptional cell cycle domain: distinctions between two human H4 gene promoters. Mol Cell Biol. 1992;12:3273–3287. doi: 10.1128/mcb.12.7.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen AJ, van Gurp MF, de Ridder MC, Tufarelli C, Last TJ, Birnbaum M, Vaughan PS, Giordano A, Krek W, Neufeld EJ, Stein JL, Stein GS. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc Natl Acad Sci USA. 1996;93:11516–11521. doi: 10.1073/pnas.93.21.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan PS, Aziz F, van Wijnen AJ, Wu S, Harada H, Taniguchi T, Soprano KJ, Stein JL, Stein GS. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature. 1995;377:362–365. doi: 10.1038/377362a0. [DOI] [PubMed] [Google Scholar]
- Vaughan PS, van Wijnen AJ, Stein JL, Stein GS. Interferon regulatory factors: growth control and histone gene regulation--it’s not just interferon anymore. J Mol Med. 1997;75:348–359. doi: 10.1007/s001090050120. [DOI] [PubMed] [Google Scholar]
- Wei Y, Jin J, Harper JW. The cyclin E/Cdk2 substrate and Cajal body component p220(NPAT) activates histone transcription through a novel LisH-like domain. Mol Cell Biol. 2003;23:3669–3680. doi: 10.1128/MCB.23.10.3669-3680.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel MT, Dowsett M. Current and emerging biomarkers in breast cancer: prognosis and prediction. Endocr Relat Cancer. 2010;17:R245–R262. doi: 10.1677/ERC-10-0136. [DOI] [PubMed] [Google Scholar]
- Xie R, Medina R, Zhang Y, Hussain S, Colby J, Ghule P, Sundararajan S, Keeler M, Liu L-J, van der Deen M, Mitra P, Lian JB, Rivera-Perez JA, Jones SN, Stein JL, van Wijnen AJ, Stein GS. The histone gene activator HINFP is a non-redundant cyclin E/CDK2 effector during early embryonic cell cycles. Proc Natl Acad Sci USA. 2009;106:12359–12364. doi: 10.1073/pnas.0905651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, van Wijnen AJ, van der Meijden CMJ, Luong MX, Stein JL, Stein GS. The cell cycle control element of histone H4 gene transcription is maximally responsive to interferon regulatory factor pairs IRF-1/IRF-3 and IRF-1/IRF-7. J Biol Chem. 2001;276:18624–18632. doi: 10.1074/jbc.M010391200. [DOI] [PubMed] [Google Scholar]
- Xie RL, Liu L, Mitra P, Stein JL, van Wijnen AJ, Stein GS. Transcriptional activation of the histone nuclear factor P (HiNF-P) gene by HiNF-P and its cyclin E/CDK2 responsive co-factor p220(NPAT) defines a novel autoregulatory loop at the G1/S phase transition. Gene. 2007;402:94–102. doi: 10.1016/j.gene.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie RL, van Wijnen AJ, van der Meijden CM, Stein JL, Stein GS. Forced expression of the interferon regulatory factor 2 oncoprotein causes polyploidy and cell death in FDC-P1 myeloid hematopoietic progenitor cells. Cancer Res. 2002;62:2510–2515. [PubMed] [Google Scholar]
- Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3’ end processing of histone pre-mRNAs. Mol Cell. 2009a;36:267–278. doi: 10.1016/j.molcel.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Sullivan KD, Marzluff WF, Dominski Z. Studies of the 5’ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol Cell Biol. 2009b;29:31–42. doi: 10.1128/MCB.00776-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Xu B, Sabath I, Kunduru L, Burch BD, Marzluff WF, Dominski Z. FLASH is required for the endonucleolytic cleavage of histone pre-mRNAs but is dispensable for the 5’ exonucleolytic degradation of the downstream cleavage product. Mol Cell Biol. 2011 doi: 10.1128/MCB.00979-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Wei Y, Nalepa G, Harper JW. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol Cell Biol. 2003;23:8586–8600. doi: 10.1128/MCB.23.23.8586-8600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Dynlacht B, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S- phase entry. Genes Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


